Abstract
Introduction: Long non-coding RNA MEG3 (lncRNA MEG3) has been showed to involve in a variety of cancers. However, the association between lncRNA MEG3 expression level and the prognosis of osteosarcoma is still unclear. Methods: The expression levels of lncRNA MEG3 in osteosarcoma tissues and adjacent non-tumor tissues were detected using quantitative real-time PCR (qRT-PCR). Differences in patient survival were determined using the Kaplan-Meier method and a log-rank test. A Cox proportional hazards regression analysis was used for univariate and multivariate analyses of prognostic values. Results: Our findings showed that expression of lncRNA MEG3 was clearly lower in osteosarcoma tissues compared with adjacent non-tumor tissues. The expression of lncRNA MEG3 was associated with clinical stage and distant metastasis (P<0.05). Kaplan-Meier analysis showed that patients with low lncRNA MEG3 expression had a shorter overall survival (log-rank test, P<0.05). Furthermore, multivariate analysis revealed that decreased expression of lncRNA MEG3, advanced clinical stage and distant metastasis were all independent predictors to overall survival of osteosarcoma patients. Conclusions: Downregulation of lncRNA MEG3 was associated with poor overall survival of osteosarcoma. LncRNA MEG3 could be a useful biomarker for progression and prognosis of osteosarcoma.
Keywords: lncRNA MEG3, osteosarcoma, progression, prognosis
Introduction
Osteosarcoma is the most common primary malignancy in children and adolescents, accounting for 20 to 35% of all malignant primary bone tumors [1,2]. Though advances of modern treatments such as surgery, chemotherapy, and the combination of surgery and chemotherapy are improved, long-term survival rate of patients diagnosed with advanced osteosarcoma remains very low [3,4]. Therefore, it is of great significance to investigate the molecular mechanisms involved in osteosarcoma, and to identify novel and effective targets to improve therapeutic efficacy and clinical outcome for osteosarcoma.
Long non coding RNA (lncRNA) is a type of RNA molecule with length of more than 200 nucleotides and lacks an open reading frame of significant length and the capability of coding protein [5,6]. A large number of studies showed that lncRNA act as key molecules regulating gene expression, chromatin remodeling, transcription, and post-transcriptional processing [7,8]. In addition, increasing numbers of reports showed that the disorders of lncRNA are closely related to human diseases, especially cancer [9]. For example, Gupta et al showed that lncRNA HOTAIR was increased in breast tumors and could promote metastasis by the interaction with the Polycomb Repressive Complex 2 [10]. Zhang et al showed that clear cell renal cell carcinoma patients with higher SPRY4-IT1 expression had an advanced clinical stage and poorer prognosis, knockdown of SPRY4-IT1 reduced renal cancer cell proliferation, migration, and invasion [11]. Shi et al reported that GAS5 was downregulated in non-small-cell lung carcinoma and GAS5 overexpression could induce apoptosis and growth arrest, In addition, they found that p53 and E2F1 were key downstream mediators of GAS5 [12]. However, the role of lnRNAs in the development of osteosarcoma remains ambiguous and further studies is needed.
In this study, we investigated the expression of lncRNA MEG3 in osteosarcoma, and analyzed the association of its expression with clinicopathological features and clinical prognosis.
Materials and methods
Patient samples
This study was approved by the Research Ethics Committee of Xinxiang Central Hospital. Written informed consent was obtained from all of the patients. In total, we recruited 64 patients with osteosarcomas from Department of Orthopedics, Xinxiang Central Hospital between May 2005 and April 2007. None of the patients enrolled in this study had received radiotherapy or chemotherapy before surgery. The clinical stage of these osteosarcoma patients was classified according to the sixth edition of the TNM classification of the Union for International Cancer Control (UICC). The clinicopathological information of the patients is summarized in Table 1.
Table 1.
Correlation between lncRNA MEG3 expression and clinicopathological features of osteosarcoma
| Clinicopathological features | Group | Total | MEG3 expression | P value | |
|---|---|---|---|---|---|
|
| |||||
| Low | High | ||||
| Gender | Male | 36 | 19 | 17 | 0.614 |
| Female | 28 | 13 | 15 | ||
| Age (years) | <25 | 40 | 18 | 22 | 0.302 |
| ≥25 | 24 | 14 | 10 | ||
| Tumor size (cm) | <8 cm | 37 | 22 | 15 | 0.076 |
| ≥8 cm | 27 | 10 | 17 | ||
| Anatomic location | Tibia/femur | 44 | 24 | 20 | 0.281 |
| Elsewhere | 20 | 8 | 12 | ||
| Clinical stage | I/II | 31 | 10 | 21 | 0.006 |
| III | 33 | 22 | 11 | ||
| Distant metastasis | Absence | 47 | 19 | 28 | 0.011 |
| Presence | 17 | 13 | 4 | ||
Quantitative real-time PCR
The total RNA isolated from frozen samples using TRIzol reagent (Invitrogen) based on the constructor’s instructions. For quantitative real-time PCR (qRT-PCR), RNA was reverse-transcribed to cDNA by using a reverse transcription kit (Takara). qRT-PCR was performed with Power SYBR Green (Takara). Results were normalized to the expression of GAPDH. The PCR primers for MEG3 or GAPDH were as folows: MEG3 sense, 5’-CTGCCCATCTACACCTCACG-3’ and reverse, 5’-CTCTCCGCCGTCTGCGCTAGGGGCT-3’; GAPDH sense, 5’-GTCAACGGATTTGGTCTGTATT-3’ and reverse, 5’-AGTCTTCTGGGTGGCAGTGAT-3’. qRT-PCR and data collection were performed on ABI 7500. The relative expression of MEG3 was calculaed and normalized using the 2-ΔΔCt method relative to GA PDH.
Statistical analysis
We used SPSS 18.0 software for statistical analysis. Differences between groups were evaluated using Student’s t-test and χ2 test. Survival analysis was done by using the log-rank test and Kaplan-Meier method. Moreover, a Cox proportional hazards model was performed to evaluate prognostic values of clinicopathological features. P<0.05 was statistically significant.
Results
Expression of lncRNA MEG3 is down-regulated in osteosarcoma
To test the effect of lncRNA MEG3 on tumor progression, the expression levels of lncRNA MEG3 in tumor tissues and paired non-tumor tissues from 64 osteosarcoma patients were measured by qRT-PCR. As shown in Figure 1A, lncRNA MEG3 expression was significantly decreased in osteosarcoma tissues compared to adjacent non-tumor tissues (P<0.05). These data suggested that lncRNA MEG3 could act as a tumor suppressor to prevent progression of osteosarcoma.
Figure 1.
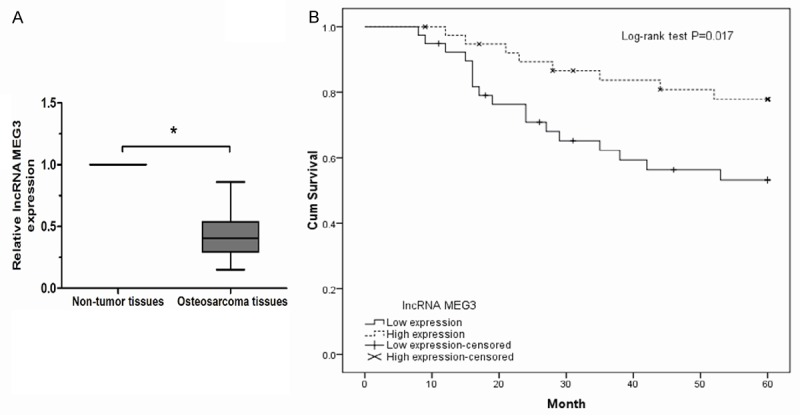
Relative lncRNA MEG3 expression in osteosarcoma tissues and its clinical significance. A. Relative lncRNA MEG3 expression level in osteosarcoma tissues and adjacent non-tumor tissues; the expression of level of lncRNA MEG3 was detected in osteosarcoma patients by qRT-PCR. B. Kaplan-Meier overall survival curves according to lncRNA MEG3 expression level. *P<0.05.
Association between lncRNA MEG3 expression and clinicopathological features
According to the median expression level of lncRNA MEG3, we categorized the patients into low and high expression groups. The correlation between clinicopathological features and lncRNA MEG3 expression in high and low expression groups were summarized in Table 1. Low lncRNA MEG3 expression was observed to be closely correlated with advanced clinical stage and distant metastasis (P<0.05). However, there was no association between lncRNA MEG3 expression and other clinical features, such as gender, age, tumor size, and anatomic location (P>0.05).
Association between lncRNA MEG3 expression and prognosis in osteosarcoma patients
As shown in Figure 1B, Kaplan-Meier analysis showed that the overall survival rate of the osteosarcoma patients with low lncRNA MEG3 expression was significantly lower than that of patients with high lncRNA MEG3 expression (log-rank test, P<0.05). Next, univariate and multivariate analyses were utilized to evaluate whether the lncRNA MEG3 expression level and various clinicopathological features were independent prognostic features of osteosarcoma patient outcomes. Univariate analysis showed that lncRNA MEG3 expression, clinical stage, and distant metastasis were significantly associated with overall survival of osteosarcoma patients (Table 2, P<0.05). Furthermore, multivariate Cox regression analyses revealed that lncRNA MEG3 expression, clinical stage, and distant metastasis were independent prognostic factors for osteosarcoma patients (Table 3, P<0.05).
Table 2.
Univariate analysis of clinicopathological features for overall survival of osteosarcoma patients
| Clinicopathological features | Univariate analysis | ||
|---|---|---|---|
|
| |||
| Hazard ratio | 95% CI | P | |
| Gender | 1.316 | 0.518-3.182 | 0.373 |
| Male vs. Female | |||
| Age (years) | 0.975 | 0.593-2.861 | 0.406 |
| ≥25 vs. <25 | |||
| Tumor size | 2.137 | 0.759-6.068 | 0.098 |
| ≥8 cm vs. <8 cm | |||
| Anatomic location | 1.134 | 0.665-3.672 | 0.184 |
| Elsewhere vs. Tibia/femur | |||
| Clinical stage | 2.945 | 1.815-6.963 | 0.004 |
| III vs. I/II | |||
| Distant metastasis | 3.716 | 2.183-8.476 | 0.002 |
| Presence vs. Absence | |||
| MEG3 | 2.735 | 1.573-7.718 | <0.001 |
| Low vs. High | |||
Table 3.
Multivariate analysis of clinicopathological features for overall survival of osteosarcoma patients
| Clinicopathological features | Multivariate analysis | ||
|---|---|---|---|
|
| |||
| Hazard ratio | 95% CI | P | |
| Clinical stage | 2.713 | 1.592-6.311 | 0.015 |
| III vs. I/II | |||
| Distant metastasis | 3.358 | 1.885-7.392 | 0.011 |
| Presence vs. Absence | |||
| MEG3 | 2.415 | 1.318-6.806 | 0.006 |
| Low vs. High | |||
Discussion
The role of lnRNAs in development of osteosarcoma remains ambiguous and discovery of new specific therapeutic targets may provide effective management of disease. Dysreulation of different lncRNAs have been previously suggested in osteosarcoma, For example, Sun et al showed that increased expression of lncRNA HULC was associated with poor prognosis and promoted cell metastasis in osteosarcoma [13]. Zhang et al revealed that downregulation of lncRNA TUG1 inhibited osteosarcoma cell proliferation and promoted apoptosis [14]. Dong et al suggested that lncRNA MALAT1 could promote the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway [15]. In the present study, we evaluate the expression of lncRNA MEG3 in term of osteosarcoma.
Maternally expressed gene 3 (MEG3), which encoded a non-coding RNA, is an imprinted gene belonging to the DLK1-MEG3 locus located on chromosome 14q32.3 in humans with maternal expression [16]. MEG3 RNA was expressed in many normal tissues, while it was lost in an expanding list of primary human tumors [17]. For example, Lu et al showed that lncRNA MEG3 was decreased in non-small cell lung cancer and associated with advanced clinical features. Furthermore, patients with lower expression of lncRNA MEG3 had a significantly poor prognosis [18]. Yin et al reported that decreased expression of lncRNA MEG3 could promote cell proliferation and predicted a poor prognosis in colorectal cancer patients [19]. Sun et al found that downregulated lncRNA MEG3 was associated with poor prognosis and promoted cell proliferation in gastric cancer [20].
In the present study, our results showed that expression of lncRNA MEG3 was significantly decreased in osteosarcoma, suggesting that lncRNA MEG3 may function as tumor suppressor. Furthermore, we found that lncRNA MEG3 expression was associated with clinical stage and distant metastasis, indicating that lncRNA MEG3 might be involved in the carcinogenesis and metastasis of osteosarcoma. More important, we found patient with low expression of lncRNA MEG3 was significantly associated with a shorter overall survival time, suggesting that low lncRNA MEG3 level is a biomarker of poor prognosis for osteosarcoma patients. However, the precise molecular mechanisms behind the altered expression of lncRNA MEG3 in osteosarcoma and its function are not very clear. Therefore, additional studies are needed to more clearly and comprehensively articulate the molecular mechanisms of both the cause and the effects of altered expression of lncRNA MEG3 in the progression of osteosarcoma.
In conclusion, our result showed that lncRNA MEG3 may function in suppression of tumor whose downregulation of lncRNA MEG3 can be associated with progression and metastasis of osteosarcoma and would applied as a therapeutic agent.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 3.Ta HT, Dass CR, Choong PF, Dunstan DE. Osteosarcoma treatment: state of the art. Cancer Metastasis Rev. 2009;28:247–263. doi: 10.1007/s10555-009-9186-7. [DOI] [PubMed] [Google Scholar]
- 4.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang HM, Yang FQ, Yan Y, Che JP, Zheng JH. High expression of long non-coding RNA SPRY4-IT1 predicts poor prognosis of clear cell renal cell carcinoma. Int J Clin Exp Pathol. 2014;7:5801–5809. [PMC free article] [PubMed] [Google Scholar]
- 12.Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F, Song Y. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2015;54(Suppl 1):E1–E12. doi: 10.1002/mc.22120. [DOI] [PubMed] [Google Scholar]
- 13.Sun XH, Yang LB, Geng XL, Wang R, Zhang ZC. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp Pathol. 2015;8:2994–3000. [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q, Geng PL, Yin P, Wang XL, Jia JP, Yao J. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J Cancer Prev. 2013;14:2311–2315. doi: 10.7314/apjcp.2013.14.4.2311. [DOI] [PubMed] [Google Scholar]
- 15.Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K Akt pathway. Tumour Biol. 2015;36:1477–1486. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48:R45–53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, Zhou Y, Klibanski A. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010;151:939–947. doi: 10.1210/en.2009-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH, Guo RH. Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour Biol. 2015;36:4851–4859. doi: 10.1007/s13277-015-3139-2. [DOI] [PubMed] [Google Scholar]
- 20.Sun XH, Yang LB, Geng XL, Wang R, Zhang ZC. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp Pathol. 2015;8:2994–3000. [PMC free article] [PubMed] [Google Scholar]


