Key points
Ribosome biogenesis is the primary determinant of translational capacity, but its regulation in skeletal muscle following acute resistance exercise is poorly understood.
Resistance exercise increases muscle protein synthesis acutely, and muscle mass with training, but the role of translational capacity in these processes is unclear.
Here, we show that acute resistance exercise activated pathways controlling translational activity and capacity through both rapamycin‐sensitive and ‐insensitive mechanisms.
Transcription factor c‐Myc and its downstream targets, which are known to regulate ribosome biogenesis in other cell types, were upregulated after resistance exercise in a rapamycin‐independent manner and may play a role in determining translational capacity in skeletal muscle.
Local inhibition of myostatin was also not affected by rapamycin and may contribute to the rapamycin‐independent effects of resistance exercise.
Abstract
This study aimed to determine (1) the effect of acute resistance exercise on mechanisms of ribosome biogenesis, and (2) the impact of mammalian target of rapamycin on ribosome biogenesis, and muscle protein synthesis (MPS) and degradation. Female F344BN rats underwent unilateral electrical stimulation of the sciatic nerve to mimic resistance exercise in the tibialis anterior (TA) muscle. TA muscles were collected at intervals over the 36 h of exercise recovery (REx); separate groups of animals were administered rapamycin pre‐exercise (REx+Rapamycin). Resistance exercise led to a prolonged (6–36 h) elevation (30–50%) of MPS that was fully blocked by rapamycin at 6 h but only partially at 18 h. REx also altered pathways that regulate protein homeostasis and mRNA translation in a manner that was both rapamycin‐sensitive (proteasome activity; phosphorylation of S6K1 and rpS6) and rapamycin‐insensitive (phosphorylation of eEF2, ERK1/2 and UBF; gene expression of the myostatin target Mighty as well as c‐Myc and its targets involved in ribosome biogenesis). The role of c‐Myc was tested in vitro using the inhibitor 10058‐F4, which, over time, decreased basal RNA and MPS in a dose‐dependent manner (correlation of RNA and MPS, r 2 = 0.98), even though it had no effect on the acute stimulation of protein synthesis. In conclusion, acute resistance exercise stimulated rapamycin‐sensitive and ‐insensitive mechanisms that regulate translation activity and capacity.
Key points
Ribosome biogenesis is the primary determinant of translational capacity, but its regulation in skeletal muscle following acute resistance exercise is poorly understood.
Resistance exercise increases muscle protein synthesis acutely, and muscle mass with training, but the role of translational capacity in these processes is unclear.
Here, we show that acute resistance exercise activated pathways controlling translational activity and capacity through both rapamycin‐sensitive and ‐insensitive mechanisms.
Transcription factor c‐Myc and its downstream targets, which are known to regulate ribosome biogenesis in other cell types, were upregulated after resistance exercise in a rapamycin‐independent manner and may play a role in determining translational capacity in skeletal muscle.
Local inhibition of myostatin was also not affected by rapamycin and may contribute to the rapamycin‐independent effects of resistance exercise.
Abbreviations
- AgNOR
argyrophilic proteins in nucleolar organizing regions
- ERK
extracellular signal‐regulated kinase
- DTT
dithiothreitol
- GAPDH
glyceraldehyde 3‐phosphate dehydrogenase
- GM
growth media
- ITS‐1
internal transcribed spacer 1
- MPB
muscle protein breakdown
- MPS
muscle protein synthesis
- mTORC1
mechanistic/mammalian target of rapamycin
- TA
tibialis anterior
- RRN3
RNA polymerase I transcription factor
- TAF1B
TATA box binding protein‐associated factor RNA polymerase I B
- UBF
upstream binding factor
Introduction
Resistance exercise stimulates muscle protein synthesis (MPS) and the initial increases in protein synthesis are accompanied by enhanced translational signalling. Furthermore, there is evidence that alterations in translational signalling [e.g. through eIF2Bε (Mayhew et al. 2011) and S6K1 (Baar & Esser, 1999)] underpin gains in muscle mass that are acquired through training. However, recently questions have been raised (Atherton & Smith, 2012; Phillips et al. 2013) about the ability of relatively brief changes in individual translation signalling pathways to fully explain (1) the prolonged nature of resistance exercise‐induced enhancement of MPS (Chesley et al. 1992; Phillips et al. 1997), and (2) gains in muscle mass achieved through training.
Hamosch et al. (1967) reported that synergist ablation in the rat hindlimb enhanced ex vivo amino acid incorporation into protein using microsome fractions prepared from hypertrophied soleus muscles. Enhanced amino acid incorporation was accompanied by a robust increase in total RNA content. Given that ribosomal RNA accounts for approximately 85% of total RNA (Young, 1970), increased ribosomal mass, representing increased translational capacity, was hypothesized to be responsible for the increased rate of protein synthesis ex vivo. Collectively, it follows that exercise‐induced increases in MPS, and ultimately muscle mass, may be best explained by the changes in both translational activity (‘ribosome activation’) and translational capacity (‘ribosome abundance’).
Interestingly, the mechanistic/mammalian target of rapamycin complex 1 (mTORC1) is central to enhancing translation activity and also appears to be important in initiating a transcriptional programme that supports ribosome biogenesis. Translationally, in response to anabolic stimuli (e.g. exercise and amino acids), the mTORC1 effectors S6K1 and 4E‐BP1 are phosphorylated, resulting in the formation of a translation preinitiation complex and the initiation of protein synthesis (Holz et al. 2005; Kubica et al. 2005). mTORC1 also regulates ribosome mass through both a transcriptional programme, upregulating ribosomal RNA synthesis and processing, and a translational programme, increasing the translation of ribosomal proteins that contain a 5′ terminal oligopyrimidine tract (Chauvin et al. 2014). Thus, mTORC1 is a potent regulator of muscle hypertrophy (Bodine et al. 2001), because of its dual role in influencing translational activity and capacity. Because S6K1 is a ‘poor quality’ substrate of mTORC1 [i.e. phosphorylation is more dependent on raptor (Kang et al. 2013)], the drug rapamycin completely blocks its phosphorylation at Thr389 by mTORC1; thus, rapamycin can be used as a tool to determine the impact of S6K1 in mediating responses to anabolic stimuli. Rapamycin abolishes the acute increase in MPS that occurs in the first few hours after resistance exercise (Drummond et al. 2009), indicating that this early upregulation of MPS is mTORC1‐dependent. However, the effects of rapamycin on anabolic signalling and MPS at later stages of recovery from acute resistance exercise are unclear. Therefore, an aim of the present study was to determine the impact of mTORC1 signalling on MPS in early and late recovery from acute resistance exercise. Secondly, in light of early (Hamosch et al. 1967) and emerging (Chaillou et al. 2014) evidence that ribosomal content plays an important role in directing anabolic responses, we aimed to determine the mechanisms that regulate ribosome biogenesis after acute resistance exercise. Two targets that directly regulate ribosome biogenesis were brought to the forefront in our analysis: c‐Myc and upstream binding factor (UBF). c‐Myc enhances the expression of numerous genes that regulate ribosome biogenesis (Boon et al. 2001), and UBF is a transcription factor that promotes ribosomal DNA transcription by RNA polymerase I (Kwon & Green, 1994; Voit & Grummt, 2001). Therefore, a secondary aim of the current work was to determine the regulation of UBF and c‐Myc following resistance exercise. Lastly, we measured proteasome activity, the predominant protein degradation pathway, because net protein balance in recovery will be determined by both synthesis and breakdown, and breakdown after resistance exercise is understudied.
Methods
Animal use in acute exercise
Female Fisher 344‐Brown Norway rats (10 months, n = 5 per group) were used according to a protocol approved by the University of California Davis Animal Care and Use Committee. Rats underwent a bout of acute unilateral resistance exercise using a protocol described previously (Baar & Esser, 1999; Hamilton et al. 2010). Briefly, rats were anaesthetized (2.5% isoflurane) and the sciatic nerve was stimulated (100 Hz, 3–6 V, 1 ms pulse, 9 ms delay) using percutaneous wire electrodes to contract the hindlimb muscles for ten sets of six repetitions (repetition length = 2 s). In this model, muscle fibres in the anterior [tibialis anterior (TA), extensor digitorum longus] and posterior (soleus, plantaris, gastrocnemius) compartments are fully recruited and contract simultaneously. As a result, the (smaller) anterior muscles perform high‐force lengthening contractions. Following exercise, animals were given an analgesic (buprenorphine, 0.1 mg kg−1) and returned to their cages for recovery. TA muscles were collected from animals under anaesthesia at 1.5, 3, 6, 12, 18 and 36 h after exercise. For rapamycin treatments, rats were administered rapamycin (1.5 mg kg−1 body weight by i.p. injection) 15 min prior to the exercise protocol, and muscles were collected at 3, 6 and 18 h [note: a 12 h+rapamycin group was added after the completion of the study for the primary purpose of determining the effect of rapamycin on mRNA targets (see Fig. 3 B–D) that were upregulated at this time point]. The non‐exercised contralateral leg served as a within‐animal control for every measure, and a group of animals that did not undergo exercise controlled for any artifacts of muscle collection order between legs (time = 0 in the figures). TA muscles were surgically removed, dissected free of connective tissue, frozen in liquid nitrogen and powdered for further analysis.
Figure 3. Expression of genes associated with ribosome biogenesis after resistance exercise .
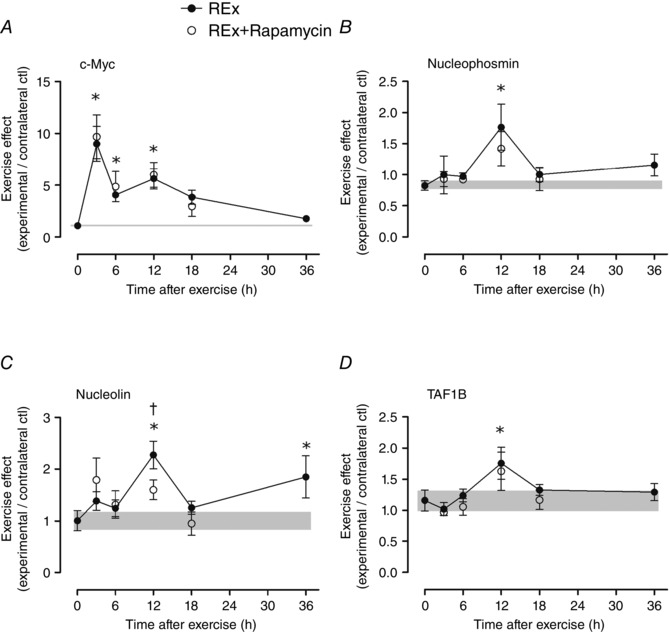
c‐Myc (A), nucleophosmin (B), nucleolin (C) and TATA box binding protein‐associated factor RNA Pol I B (TAF1B; D). The ‘0’ time point is a non‐stimulated control group with its error term shaded to aid visual comparisons. Rapamycin was administered (pre‐exercise, 1.5 mg kg−1 by i.p. injection) to groups shown in open circles. Values (means ± SEM) are expressed relative to GAPDH and the contralateral control using the ΔΔC t method. *Difference between REx and non‐stimulated control group, P < 0.05. †Difference between REx and REx+Rapamycin at the same time point, P < 0.05.
Muscle protein synthesis
Changes in MPS were assessed by measuring the incorporation of exogenous puromycin into nascent peptides as described previously (Goodman et al. 2011). Briefly, puromycin is a tyrosyl tRNA analogue that results in premature release of translation products – puromycin conjugates – that can be subsequently measured by Western blotting using an antibody against puromycin (see Protein expression below). Puromycin (EMD Millipore, Billerica, MA, USA; cat. no. 540222) was dissolved in sterile saline and delivered (0.02 μmol g−1 body weight by i.p. injection) 30 min prior to muscle collection.
Protein expression
Frozen muscle powder was homogenized in a sucrose lysis buffer [50 mm Tris, pH 7.5, 250 mm sucrose, 1 mm EDTA, 1 mm EGTA, 1% Triton X100, 50 mm NaF, 1 mm NaVO4 Na2(PO4)2 and 0.1% dithiothreitol (DTT)]. Samples were centrifuged (10,000 g × 1 min at 4°C) and the supernatant was removed for protein quantification by DC protein assay (Bio‐Rad, Hercules, CA, USA; cat. no. 500‐0116). Protein concentrations were equalized, solubilized in Laemmli sample buffer and incubated at 95°C for 5 min. Samples were loaded on 4–20% gradient polyacrylamide gels with non‐exercised and exercised samples in adjacent wells. Samples were separated by electrophoresis (200 V for 45 min), transferred to nitrocellulose membrane (wet transfer, 100 V for 1 h), blocked in 1% fish skin gelatin, and washed in Tris‐buffered saline 0.1% tween (TBST) before overnight incubation in primary antibody at 4°C. Primary antibodies, diluted 1:1000 in TBST, were from: Cell Signaling Technologies (Danvers, MA, USA) – phospho‐ERK Thr202/Tyr204 (cat. 4370), phospho‐S6K1 Thr389 (cat. 9205), phospho‐rpS6 (cat. 2211), phospho‐eEF2 Thr56 (cat. 2331), total S6K1 (cat. 2708), total S6 (cat. 2217), total eEF2 (cat. 2332), total ERK (cat. 4695); Santa Cruz Biotechnology (Santa Cruz, CA, USA) – phospho‐UBF Ser388 (cat. 21637) and Ser637 (cat. 21639); and EMD Millipore – puromycin (cat. MABE343). Membranes were washed in TBST and incubated in secondary antibody (1:10,000) for 1 h at room temperature before washing in TBST and detection by chemiluminescence (Millipore; cat. WBKLS0500). Images for densitometry analysis were captured with a Bio‐Rad ChemiDoc MP imaging system and quantified using Image Lab Software (Bio‐Rad, v. 5).
RNA isolation and gene expression
Prior to RNA isolation, aliquots of frozen muscle powder were weighed in order to calculate RNA per milligram of wet muscle tissue. RNA was isolated using RNAzol RT reagent (Sigma, cat. R4533) according to the manufacturer's instructions and quantified by absorbance spectrophotometry (Epoch Microplate Spectrophotometer, BioTek Instruments Inc., Wonooska, VT, USA). 1 μg of RNA was converted to cDNA using a reverse transcription kit (Life Technologies, Carlsbad, CA, USA; cat. 4368814) with thermal cycling according to the manufacturer's instructions. cDNA was stored at −80°C and diluted 1:10 in nuclease‐free water prior to quantitative real‐time PCR analysis (SYBR Green Supermix from Bio‐Rad Laboratories, cat. 172‐5121; Eppendorf Light Cycler PCR machine). Primer sequences are available on request. Gene expression was calculated using the delta delta threshold cycle method (Livak & Schmittgen, 2001) using glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) as a housekeeping gene. GAPDH expression did not change significantly at any time point after resistance exercise.
Proteasome and calpain activity
Proteasome and calpain activities were determined as described in detail elsewhere (Gomes et al. 2012; Baehr et al. 2014). Briefly, frozen muscle powder was Dounce‐homogenized in ice‐cold buffer (50 mm Tris, pH 7.5, 1 mm EDTA, 150 mm NaCl, 5 mm MgCl2, 0.5 mm DTT), centrifuged (12,000 g × 3 min at 4°C), protein concentrations were determined by the Bradford method, and samples were loaded onto black 96‐well plates (26S proteasome, 14 μg protein as muscle lysate; 20S proteasome, 20 μg of protein as muscle lysate; calpain, 45 μg of protein as muscle lysate). Activity was determined by measurement of a fluorescent‐tagged substrate (Leu‐Leu‐Val‐Tyr–4‐amino‐7‐methyl coumarin; BACHEM, Dubendorf, Swizterland; cat. I‐1395) in the presence or absence of inhibitors (proteasome: Bortezomib, 2 mm; Calbiochem, Billerica, MA, USA; cat. 504314; calpain: 50 μm calpain inhibitor IV, Calbiochem, cat. 208724). Kinetic reads were taken at 15 min intervals on a Fluoroskan Ascent 2.5 fluorometer for 2 h (excitation wavelength, 390 nm; emission wavelength, 460 nm), and all assays were linear up to 2 h and results were calculated from reads at 60 min.
Cell culture
C2C12 myoblasts were cultured in growth media (GM; high glucose Dulbecco's modified Eagle medium, 10% fetal bovine serum, 0.1% penicillin) until 95% confluence at which point they were differentiated in high glucose media containing 2% horse serum. Experiments were conducted on fully formed myotubes 5 days after the introduction of differentiation media. Long‐term dose–response experiments were conducted using a c‐Myc inhibitor (Sigma‐Aldrich, cat. F3680) whereby myotubes were treated for 8 h with 25, 50 and 100 μm inhibitor, or a solvent control (DMSO). Parallel dose–response experiments were run for analysis of total RNA and gene expression quantification. Three independent experiments were conducted for both MPS and RNA analysis. To determine the role of c‐Myc in the MPS response to an acute anabolic stimulus, a separate cohort of C2C12 myotubes were fasted for 30 min in PBS containing 2% horse serum. After 30 min, the cells were shifted to GM supplemented with 100 nm IGF‐1 in the presence or absence of increasing amounts of the Myc inhibitor for a further 60 min before collection. For MPS analysis, myotubes were incubated with 1 μm puromycin for 5 min before collection in a sucrose lysis buffer (see ‘Protein expression’ above). For total RNA analysis, myotubes were collected in RNAzol RT reagent and RNA was isolated, quantified and analysed for gene expression (see ‘RNA isolation and mRNA abundance’ above).
Staining of argyrophilic proteins in nucleolar organizing regions
Argyrophilic proteins in nucleolar organizing regions (AgNORs) can be stained as a morphological proxy of nucleololar activity and ribosomal DNA transcription (Morton et al. 1983); importantly, nucleolar size is directly related to ribosome production (Hernandez‐Verdun, 2006). C2C12 myotubes were grown on glass coverslips and stained according to a protocol that was adapted from previously described methods (Ploton et al. 1986; Trere, 2000). Briefly, myotubes were fixed (95% ethanol, 5 min at room temperature), incubated in an absolute ethanol/glacial acetic acid mix (3:1, v/v), and hydrated using decreasing concentrations of ethanol, before incubation in a silver nitrate staining solution (33% silver nitrate, 0.33% gelatin, 0.33% formic acid). Next, myotubes were washed in distilled water and counterstained with 1% gold chloride. Myotubes were dehydrated using increasing concentrations of ethanol and coverslips were mounted. Images were obtained at 40× magnification using an Axio Imager M1 light microscope (Zeiss, Oberkochen, Germany), and AgNORs were enumerated in a blinded fashion and according to methods previously described (Trere, 2000).
Statistical analysis
Effects for time after exercise were analysed by one‐way ANOVA, with Dunnett's post‐hoc test for comparisons to the non‐exercised control group. Effects of rapamycin were analysed by two‐way ANOVA (rapamycin × time), with Tukey's post‐hoc test. Western blot data were log‐transformed to correct skewness and unequal variance before statistical analyses; data are given as geometric means ± back‐transformed SEM (Altman et al. 1983; Olivier et al. 2008). The remaining data are arithmetic means ± SEM. Cell culture data were analysed by one‐way ANOVA with Tukey's post‐hoc test. Statistical analyses were performed using SigmaStat software (v.3.1, Systat Software, Point Richmond, CA, USA); statistical significance was set at P < 0.05.
Results
MPS and total RNA
Resistance exercise stimulated a prolonged moderate (∼45%) increase in MPS (P = 0.042 for effect of time after exercise; Fig. 1 A). Rapamycin decreased MPS at 6 h (P = 0.005, REx vs. REx+Rapamycin at 6 h) and at 18 h (P = 0.027, REx vs. REx+Rapamycin at 18 h); however, the inhibitory effect appeared to be complete at 6 h and much less at 18 h (see bar graph inset in Fig. 1 A showing the change of MPS between the initial time point in REx and REx+Rapamycin groups and the 6 and 18 h time points). Total RNA (μg RNA/mg muscle) tended to rise gradually at 18–36 h after exercise (P = 0.085 for main effect of time after exercise; Fig. 1 C). Rapamycin administration reduced the effect of resistance exercise on total RNA (P = 0.016 for REx vs. REx+Rapamycin main effect; Fig. 1 C).
Figure 1. Acute resistance exercise on muscle protein synthesis .
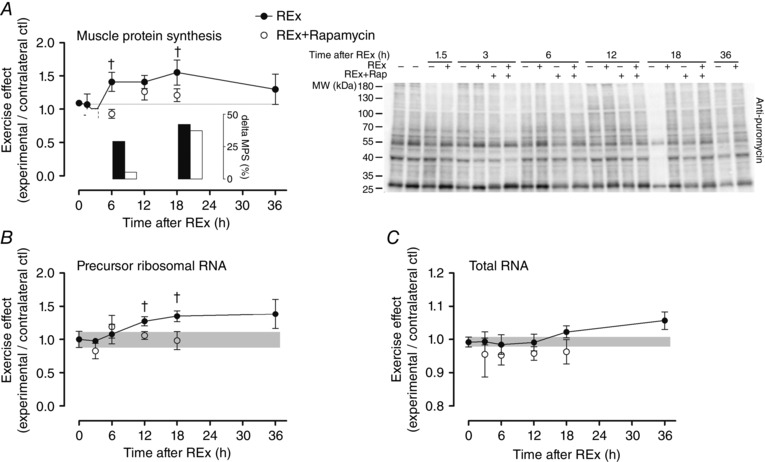
The effect of acute resistance exercise (REx, closed circles) on muscle protein synthesis (MPS; A; P = 0.042, for effect of time; representative blot at right), precursor ribosomal RNA (B; P = 0.054, for effect of time) and total RNA (C; P = 0.085, for effect of time). The ‘0’ time point is a non‐stimulated control group with its error term shaded to aid visual comparisons. Rapamycin was administered (pre‐exercise, 1.5 mg kg−1 by i.p. injection) to groups shown in open circles. MPS was assessed by measuring the incorporation of puromycin into nascent peptides by Western blot; inset shows change in MPS from the initial time point of each group to MPS at 6 and 18 h. Newly synthesized pre‐rRNA was measured using internal transcribed spacer 1 gene expression as a readout. †Difference between REx and REx+Rapamycin at the same time point, P < 0.05. Values are expressed as experimental/contralateral control muscles, means ± SEM.
Translation signalling
Resistance exercise stimulated a rapid and sustained increase in S6K1 phosphorylation (1.5–18 h, all P < 0.05; Fig. 2 A) that was completely inhibited at all time points by rapamycin. Interestingly, S6K1 phosphorylation had returned to basal levels by 18 and 36 h even though protein synthesis was still elevated at these time points. Similarly, S6 phosphorylation was elevated from 1.5–18 h recovery (all P < 0.05 vs. non‐exercised control group; Fig. 2 B). S6 phosphorylation was completely blocked by rapamycin at 6 and 18 h as expected; however, S6 phosphoryation was increased on a relative basis (experimental/contralateral control) at 3 h due to a decrease in the contralateral control, probably reflecting extracellular signal‐regulated kinase (ERK) signalling that was elevated at this time point (Fig. 2 C). Eukaryotic elongation factor 2 (eEF2) phosphorylation was decreased (indicative of activation) following resistance exercise and, interestingly, this occurred to similar levels in both the vehicle and the rapamycin‐treated groups (P = 0.41 for REx vs. REx+Rapamycin; Fig. 2 D). Lastly, UBF phosphorylation at both Ser388 (Fig. 2 E) and Ser637 (Fig. 2 F) increased following resistance exercise and only the 3 h time point (Ser637 only) was sensitive to rapamycin.
Figure 2. Resistance exercise and proteins that regulate translational activity .
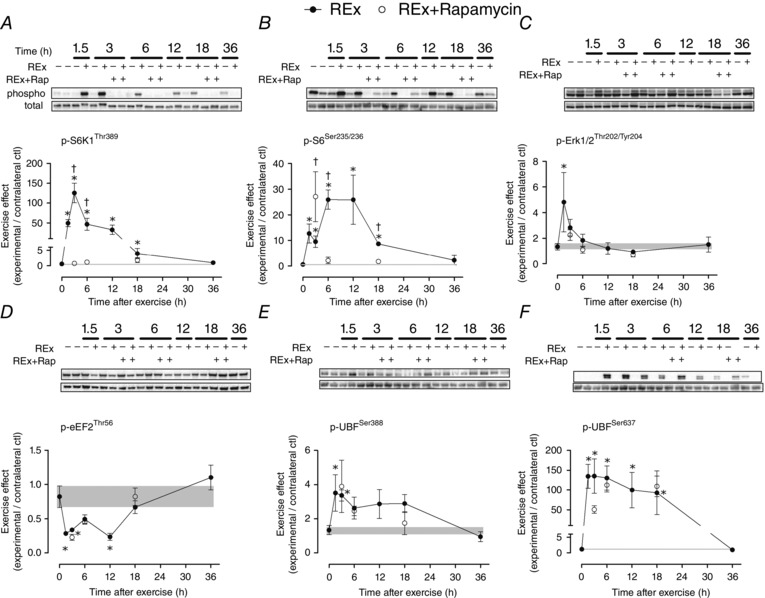
The effect of resistance exercise (REx, closed circles) on the phosphoryation of proteins that regulate translational activity (A–D) and capacity (E and F). The ‘0’ time point is a non‐stimulated control group with its error term shaded to aid visual comparisons. Rapamycin was administered (pre‐exercise, 1.5 mg kg−1 by i.p. injection) to groups shown in open circles. Data are expressed as experimental/contralateral control muscles, with geometric means ± SEM. *Difference between REx and non‐stimulated control group, P < 0.05. †Difference between REx and REx+Rapamycin at the same time point, P < 0.05.
Gene expression
Internal transcribed spacer 1 (ITS‐1) expression, an estimate of ribosomal DNA transcription rate, tended to increase in post‐exercise recovery (P = 0.054; Fig. 1 B) in a manner that preceded changes in total RNA (Fig. 1 C). The increase in ITS‐1 was inhibited by rapamycin at 12 and 18 h (both P < 0.05, REx vs. REx+Rapamycin at the same time point). c‐Myc is a transcription factor for RNA polymerase I, as well as for ribosomal proteins and proteins controlling ribosomal RNA splicing; its expression was elevated at 3, 6 and 12 h (all P < 0.05; Fig. 3 A) and was not affected by rapamycin (P = 0.84). The expression of nucleophosmin and nucleolin, which are targets of c‐Myc that control pre‐ribosomal RNA splicing, were elevated at 12 h (nucleophosmin and nucleolin, P < 0.05; Fig. 3 B and C, respectively) and 36 h of recovery (nucleolin, P < 0.05). The elevation at 12 h of nucleolin, but not nucleophosmin, was rapamycin‐sensitive (nucleolin, P = 0.049; nucleophosmin, P = 0.43; REx vs. REx+Rapamycin at 12 h). Expression of TATA box binding protein‐associated factor RNA polymerase I B (TAF1B), which recruits RNA polymerase I to the ribosomal DNA promoter, was elevated at 12 h (P < 0.05; Fig. 3 D) and was not significantly affected by rapamycin (P = 0.11). Mighty gene expression, a readout of myostatin activity, was increased at 12 and 36 h following exercise (both P < 0.05), suggesting a decrease in myostatin activity; Mighty expression was not affected by rapamycin (P = 0.51 for REx vs. REx+Rapamycin; Fig. 4 B). Furthermore, as we had previously shown, increased Mighty expression corresponded to the cleavage and activation of Notch, which was not different between REx and REx+Rapamycin (P = 0.69; Fig. 4 A).
Figure 4. Notch protein expression and Mighty gene expression after resistance exercise .
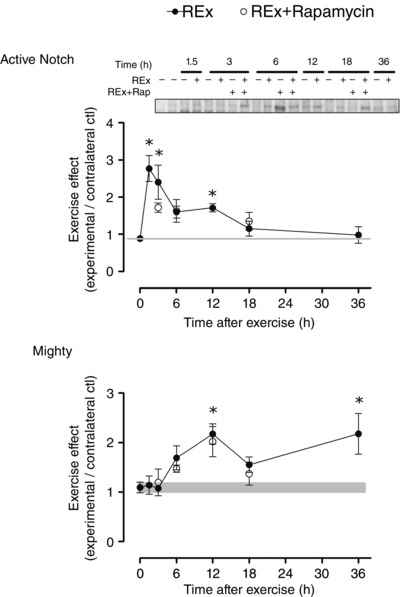
Active Notch (Notch intracellular domain) protein expression (A) and Mighty gene expression (B) after resistance exercise (REx). Notch activation inhibits myostatin signalling, as reflected by an increase in Mighty gene expression. Time points: 0 (non‐stimulated control group), 1.5, 3, 6, 12, 18, 36 h after REx. Rapamycin was administered (pre‐exercise, 1.5 mg kg−1 by i.p. injection) to groups shown in open circles. For active Notch, values are expressed as experimental/contralateral control muscles. For Mighty, values are expressed relative to GAPDH and the contralateral control using the ΔΔC t method. Values are means ± SEM. *Difference between REx and non‐stimulated control group, P < 0.05.
Long‐term c‐Myc inhibition in myotubes
To determine whether c‐Myc could control ribosome mass and protein synthesis in muscle cells, we treated C2C12 myotubes with the inhibitor 10058‐F4. Long‐term (8 h) treatment with the c‐Myc inhibitor had a dose‐dependent effect on both MPS (Fig. 5 B) and total RNA (Fig. 5 C). MPS and total RNA were strongly correlated (r 2 = 0.98, P < 0.001; Fig. 5 D). c‐Myc inhibitor treatment decreased nuclear UBF Ser637 phosphorylation (P = 0.028 at 100 μm), without significantly changing c‐Myc or total UBF abundance in isolated nuclear and cytosolic fractions, respectively (Fig. 5 E and F, respectively). Treatment with increasing concentrations of c‐Myc inhibitor resulted in stepwise decreases in argyrophilic protein staining of nucleolar organizer regions, a morphological proxy of ribosome biogenesis (Fig. 5 G). 10058‐F4 blocks c‐Myc activity by preventing its dimerization with Max and nuclear translocation (Huang et al. 2006). This inhibition/exclusion from the nucleus apparently resulted in a feedback signal that upregulated c‐Myc expression in a stepwise manner at 50 and 100 μm (P < 0.001; Fig. 5 H). Expression of the 5′ external transcribed spacer, part of the pre‐ribosomal RNA transcript, ITS‐1 and RNA polymerase I‐specific transcription initiation factor RRN3 were elevated at the 100 μm c‐Myc inhibitor dose (Fig. 5 I–K, respectively).
Figure 5. Inhibition of c‐Myc in vitro alters translational capacity .
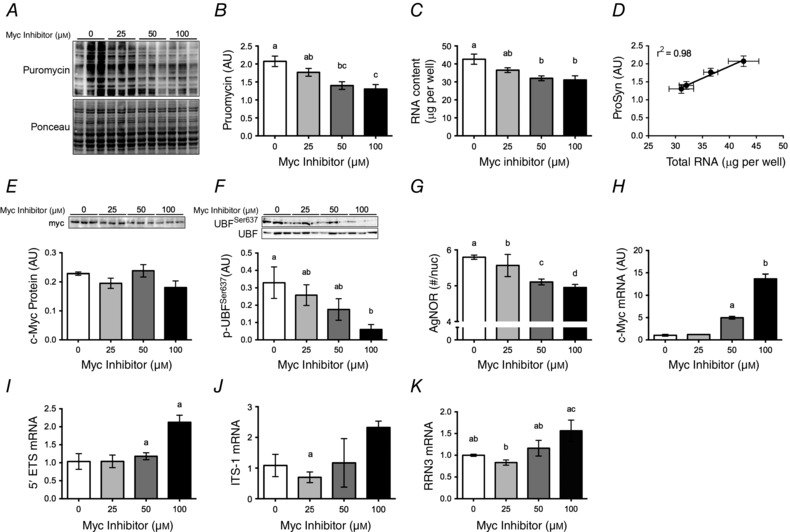
A, differentiated C2C12 myotubes were treated with an Myc inhibitor at the concentrations shown for 8 h (0 μm = solvent control). For MPS analysis, myotubes were incubated with 1 μm puromycin for 5 min before collection and detection of puromycin‐conjugated nascent peptides by Western blot. B–G, muscle protein synthesis (B, MPS) and total RNA in myotubes after c‐Myc inhibitor treatment (C) are directly related (D), c‐Myc (E) and phospho‐UBF Ser637 protein (F), and argyrophylic proteins in the nucleolar organizer region (G) following treatment with the small molecule c‐Myc inhibitor (0 μm = vehicle control). H–K, expression of c‐Myc (H), 5′ external transcribed spacer (5’ETS) (I), internal transcribed spacer 1 (ITS‐1) (J) and RNA polymerase I transcription factor (RRN3) (K) apparently demonstrates feedback upregulation at high inhibitor concentration. Data shown are representative of three independent experiments. Values are means ± SEM. Means with different letters are significantly different from each other, P < 0.01.
c‐Myc and the acute MPS response
To determine whether c‐Myc altered an acute protein synthesis in muscle cells, fasted cells were treated with GM containing 100 nm IGF‐1 for 1 h to stimulate protein synthesis. In this setting, low (25 μm) and moderate (50 μm) concentrations of the c‐Myc inhibitor had no effect on either protein synthesis (Fig. 6 A), or markers of mTORC1 activation (Fig. 6 C–F). At the highest concentration (100 μm) the c‐Myc inhibitor decreased protein synthesis and S6K1 and eEF2 signalling; interestingly, however, 100 μm c‐Myc inhibitor did not block 4E‐BP1 or S6 phosphorylation.
Figure 6. MPS and mTORC1 signalling in myotubes treated with c‐Myc inhibitor .
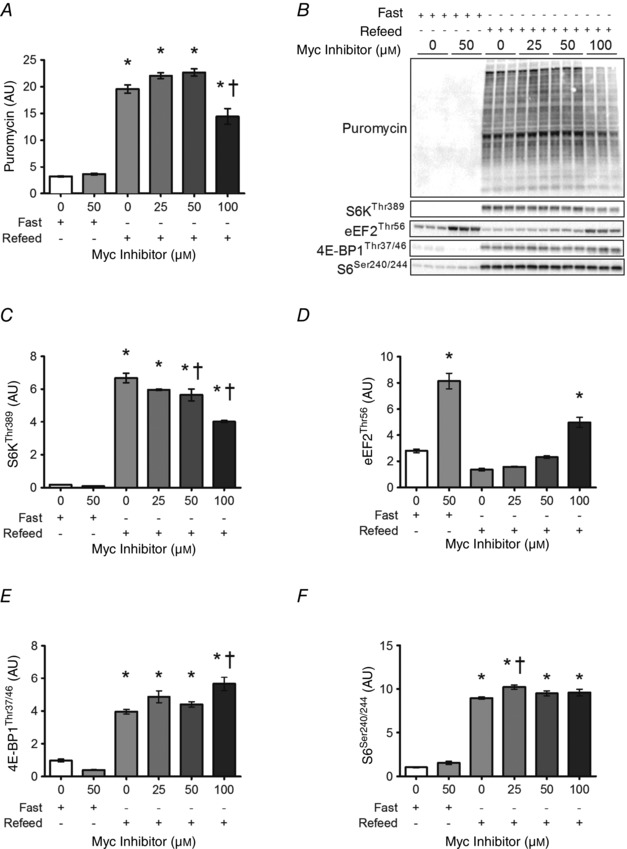
Acute muscle protein synthesis (MPS; A) and mTORC1 signalling (C–F) in myotubes treated with c‐Myc inhibitor. Differentiated C2C12 myotubes were fasted for 30 min in PBS containing 2% horse serum prior to 60 min in GM supplemented with 100 nm IGF‐1 in the presence or absence of increasing doses of Myc inhibitor. For MPS analysis, myotubes were incubated with 1 μm puromycin for 5 min before collection and detection of puromycin‐conjugated nascent peptides by Western blot. Representative data shown are one of two independent experiments. Values are means ± SEM. Means are significantly different from *control, or from †control refeed value, P < 0.01.
Ubiquitin proteasome pathway and calpain activity
ATP‐dependent (26S β5) and ATP‐independent (20S β5) proteasome activities were not increased in early recovery from resistance exercise; 20S proteasome activity was elevated in late post‐exercise recovery period (P < 0.05 at 36 h vs. non‐exercise control group; Fig. 7 B). In the exercised leg, rapamycin increased the activity of the proteasomal subunits 26S β5 at 6 h (P < 0.001 for REx vs. REx+Rapamycin at 6 h; Fig. 7 A) and 20S β5 at 3 and 6 h of recovery (P < 0.001 for REx vs. REx+Rapamycin at the same time point, at 3 and 6 h of recovery; Fig. 7 B). Calpain activity was not significantly altered by resistance exercise or by rapamycin (Fig. 7 C).
Figure 7. Ubiquitin proteasome pathway and calpain activities after resistance exercise .
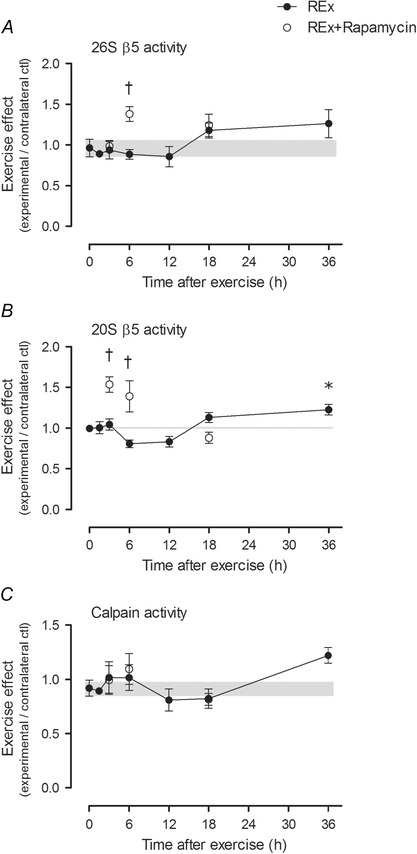
A and B, 26S β5 (A) and 20S β5 (B) reflect ATP‐dependent and ‐independent chymotrypsin‐like activities of the β5 proteasomal subunit (respectively), which is the most important subunit for proteasome function (Jager et al. 1999). C, calpain activity at each time point. The ‘0’ time point is a non‐stimulated control group with its error term shaded to aid visual comparisons. Rapamycin was administered (pre‐exercise, 1.5 mg kg−1 by i.p. injection) to groups shown in open circles. Values are expressed as experimental/contralateral control muscles, means ± SEM. *Difference between REx and non‐stimulated control group, P < 0.05. †Difference between REx and REx+Rapamycin at the same time point, P < 0.001.
Discussion
The effect of acute resistance exercise and the role of mTORC1 on the time course of translational signalling, protein synthesis and the mechanisms of ribosome biogenesis in skeletal muscle were examined in this study. The data provide a mechanistic basis for a hypothesis that skeletal muscle mass is regulated by changes in translational activity and capacity, and that these changes occur through both rapamycin‐sensitive and ‐insensitive mechanisms. Our most important findings are that eEF2, UBF, c‐Myc, TAF1B and nucleophosmin were activated, and that local myostatin activity was inhibited, through rapamycin‐insensitive mechanisms, that rapamycin decreased precursor‐ribosomal RNA, nucleolin and total RNA after resistance exercise, and that rapamycin has a limited effect on protein synthesis late in recovery.
Signalling and protein synthesis after acute resistance exercise
We observed a prolonged elevation of protein synthesis after resistance exercise, as shown previously in humans (Chesley et al. 1992; Phillips et al. 1997). Increases in protein synthesis in early recovery after resistance exercise are underpinned by enhanced translational activity and this translational activity is mTORC1‐dependent (Wong & Booth, 1990 b; Baar & Esser, 1999; Kubica et al. 2005; Glover et al. 2008; Drummond et al. 2009; Kimball & Jefferson, 2010). By comparison, the mechanisms that underpin exercise‐induced elevations in MPS in late recovery are poorly understood. Furthermore, whether specific signalling events can predict the protein synthetic response has recently been challenged (Atherton & Smith, 2012; Phillips et al. 2013). In the present study, mTORC1 signalling returned to near baseline levels by 18 h, a time when protein synthesis was at its peak, suggesting either that the high levels of protein synthesis late in the recovery period were the result of the earlier increase in mTORC1 activity or that mTORC1 was not required for the late rise in protein synthesis. To distinguish between these possibilities, we inhibited mTORC1 using rapamycin and observed that, as in humans (Drummond et al. 2009), protein synthesis was completely blocked 6 h after exercise. However, the late (12–18 h) increase in protein synthesis appeared to be rapamycin‐insensitive, showing an induction from the fully suppressed state at 3–6 h after exercise (see inset to Fig. 1 A); this suggests that prolonged elevation of protein synthesis following resistance exercise involved mTORC1‐independent mechanisms. This differs from the work of Kubica et al. (2005), who showed that rapamycin completely blocked exercise‐induced MPS at 16 h. The contrasting findings could be due to differences in the models that were used. Kubica et al. (2005) operantly trained rats to touch a bar 50 times and the ‘acute’ protocol in fact consisted of four separate sessions with 1 day of rest between sessions and protein synthesis measured 16 h after the fourth session. By contrast, our protocol mimicked a single high intensity resistance exercise training session, involving maximal motor unit recruitment and high‐force lengthening contractions. As rapamycin blocked ribosome biogenesis, as determined by ITS‐1 expression and total RNA content (Fig. 1 B and C), the prolonged elevation of protein synthesis was not the result of an increase in translational capacity. Therefore, our data suggest that a rapamycin‐insensitive process that controls translational activity is activated later in the recovery from acute resistance exercise.
To explore other potential mechanisms that could explain the prolonged increase in protein synthesis, we examined myostatin signalling, a major pathway controlling muscle mass (McPherron & Lee, 1997; Lee, 2004, 2010) that we have previously shown to be downregulated in the exercise model used in the present study (MacKenzie et al. 2013). To our knowledge, the sensitivity of myostatin signalling to rapamycin after resistance exercise is not known. We assessed myostatin activity using one of its transcriptional targets (Mighty) as a readout (Marshall et al. 2008; MacKenzie et al. 2013). In the present study, resistance exercise resulted in a prolonged increase in Mighty expression that peaked in the late recovery period (12–18 h), when protein synthesis was also at its highest. The increase in Mighty expression is thought to occur through a mechanism whereby load across the muscle leads to cleavage and activation of notch (Akiho et al. 2010), which can translocate to the nucleus and inhibit Smad2/3 activity (Carlson et al. 2008). Here, we observed that resistance exercise increased notch activation and Mighty gene expression in a rapamycin‐insensitive manner, thus suggesting local inhibition of myostatin in an mTORC1‐independent manner. Thus, the localized inhibition of myostatin could contribute to the rapamycin‐independent rise in MPS at later (12–18 h) time points of recovery; however, this hypothesis requires further investigation.
Another rapamycin‐insensitive effect that was surprising was the activation of eEF2. This was unexpected as eEF2 is thought to be under the control of mTORC1 through S6K phosphorylation and inhibition of eEF2 kinase (Wang et al. 2001). This finding of rapamycin‐insensitive eEF2 dephosphorylation (activation) adds to data that distinguish the unique ability of mechanical stimuli to stimulate anabolic signalling pathways that are not only independent of phosphoinositide 3‐kinase‐activating hormones/growth factors (Hornberger et al. 2004; Hornberger & Chien, 2006; West et al. 2009; Hamilton et al. 2010), but some of which are also independent of canonical mTORC1 activity (eEF2, UBF, Mighty and c‐Myc in the present study). Interestingly, our data imply that peptide chain elongation may be controlled in an mTORC1‐independent manner after resistance exercise and this might contribute to the prolonged increase in protein synthesis.
Ribosome biogenesis after acute resistance exercise
Another key finding from the current study was that markers of ribosome biogenesis were increased between 12 and 36 h after resistance exercise. We found that pre‐ribosomal RNA, as indicated by ITS‐1 levels, tended to increase gradually and in a sustained manner (plateau at 12–36 h) that preceded a trend toward increased total RNA. This finding is consistent with previous work showing that resistance exercise drives ribosome biogenesis, which is important in the regulation of muscle mass (Adams et al. 2002; Goodman et al. 2011; Chaillou et al. 2014). We have expanded on this previous work to show that the exercise‐induced increases in pre‐ribosomal RNA, in nucleolin (a controller of pre‐rRNA splicing) and in total RNA were prevented by rapamycin. Collectively, this means that the initial rise in protein synthesis was the result of greater translational activity/efficiency (increased peptide synthesis per unit RNA), which is consistent with previous reports (Wong & Booth, 1990 a,b; Fluckey et al. 1996; Bickel et al. 2005), but also suggests that an mTORC1‐dependent increase in translational capacity may be important in the protein accretion that occurs as a result of training (Brook et al. 2015). In general, the 12 and 18 h post‐exercise time points are especially interesting as they appear to be a transition phase between diminishing mTORC1 signalling, sustained MPS and increased ribosome biogenesis. However, it is important to note that even though rapamycin completely blocked ribosome biogenesis, it did not completely block MPS, suggesting that ribosome biogenesis does not explain the prolonged increase in MPS after an acute bout of resistance exercise.
c‐Myc was identified as an important target for investigating ribosome biogenesis given that it regulates RNA polymerase I activity (Arabi et al. 2005; Grandori et al. 2005), ribosomal proteins, and proteins that control ribosomal RNA processing and ribosomal subunit intracellular localization (Boon et al. 2001; Maggi et al. 2008). In the present study, c‐Myc gene expression was highly upregulated after resistance exercise (1.5–12 h of recovery), which is in agreement with earlier work (von Walden et al. 2012), and the upregulation of c‐Myc occurred in a rapamycin‐insensitive manner. Peak c‐Myc expression (3 h of recovery) preceded the upregulation of its targets TAF1B, which recruits RNA polymerase I to the ribosomal DNA promoter (Muth et al. 2001), and nucleophosmin and nucleolin (12 h of recovery), which control c‐Myc nucleolar localization and rDNA transcription (Li & Hann, 2013), splicing of the precursor‐ribosomal RNA transcript (Boon et al. 2001) and nuclear export of ribosomal subunits (Maggi et al. 2008). To help clarify the relationship between c‐Myc, ribosomal content and protein synthesis, we performed experiments in vitro in which we inhibited c‐Myc activity in differentiated myotubes for 8 h. These experiments showed that increasing doses of c‐Myc inhibitor resulted in stepwise decreases in both total RNA and protein synthesis, without causing cellular toxicity (see the ponceau image and the total UBF levels in Fig. 5). The fact that total RNA and protein synthesis rate were strongly correlated suggests that c‐Myc affects the rate of protein synthesis in skeletal muscle by regulating translational capacity. In agreement with this hypothesis, inhibition of c‐Myc had no effect on the acute activation of protein synthesis or mTORC1 in myotubes that had undergone a short fast followed by a strong anabolic stimulus. c‐Myc regulates translational capacity by directly activating ribosomal DNA transcription, as seen by the decreases in argyrophilic protein staining of nucleolar organizer regions, following treatment with the inhibitor (Fig. 5 G). Our in vivo data showed that the increases in c‐Myc, nucleophosmin and TAF1B gene expression are not rapamycin‐sensitive and thus c‐Myc may work together with mTORC1 to alter translational capacity after resistance exercise; however, further mechanistic research in vivo is required to test this hypothesis.
One mTORC1‐regulated mechanism mediating ribosome biogenesis may be activation of UBF, a nucleolar transcription factor that is integral to the formation of a pre‐initiation complex for the transcription of precursor rRNA (Sanij et al. 2008). Phosphorylation of UBF is required for its mediation of ribosomal RNA synthesis (Voit et al. 1995). A previous study (Hannan et al. 2003) in NIH 3T3 cells showed that mTORC1 was required for UBF phosphorylation and subsequent serum‐induced activation of ribosomal DNA transcription. In the present study, however, resistance exercise increased UBF serine 388 and 637 phosphorylation in a largely mTORC1‐independent manner. It was beyond the scope of the present study to determine the upstream kinase responsible for the rapamycin‐insensitive phosphorylation of UBF; previous studies have shown that UBF can be phosphorylated by ERK (Stefanovsky et al. 2001) and cell cycle‐associated kinases (Voit et al. 1992; Voit & Grummt, 2001). A previous resistance exercise study (Figueiredo et al. 2015) highlighted ERK‐mediated activation of ribosome biogenesis via cyclin D1 and TIF‐1A rather than UBF. Here, we report high UBF phosphorylation in the presence of rapamycin, which, combined with rapamycin‐insensitive activation of the transcription of c‐Myc, nucleophosmin and TAF1B, suggests that, even though mTORC1 is required for resistance exercise‐dependent ribosome biogenesis (present data and Hannan et al. 2003; Nader et al. 2005; Jastrzebski et al. 2007; Thoreen et al. 2012), mTORC1 is supported by mTORC1‐independent mechanisms that remain to be fully elucidated.
The exact amount of time required after an acute bout (or number of bouts) to elicit a bona fide increase in translational capacity is unclear. Bickel et al. (2005) reported increased total RNA concentration (μg RNA per mg muscle) after two bouts of exercise, but the plasticity of the ribosomal pool throughout resistance training is understudied (Figueiredo et al. 2015). We hypothesize that the ribosomal pool is responsive to acute resistance exercise and that periodic induction of elements regulating ribosome biogenesis – including UBF phosphorylation as well as expression of c‐Myc, nucleophosmin, nucleolin and TAF1B – during the early stages of training may stimulate increases in translational capacity and facilitate subsequent adaptation (skeletal muscle remodelling and hypertrophy). According to such a model, increased translational capacity, achieved through training, would result in a smaller requirement for mTORC1 signalling to induce an anabolic response after an acute resistance exercise stimulus (Brook et al. 2015). This could explain, at least in part, the paradoxical observation that individuals that exhibit the greatest hypertrophy after training express genes reflecting a downregulation of mTORC1 (Phillips et al. 2013).
Protein degradation after acute resistance exercise
Compared with MPS, muscle protein breakdown (MPB) after resistance exercise is understudied. The ubiquitin proteasome pathway is the major protein degradation pathway in skeletal muscle (Rock et al. 1994), and reduced proteasome activity contributes to pathological (Al‐Khalili et al. 2014) and ageing (Hwee et al. 2014) phenotypes in skeletal muscle. Conversely, proteasome activation accompanies compensatory hypertrophy induced by synergist ablation (Baehr et al. 2014). In contrast to the chronic and supraphysiological stimulus imposed by synergist ablation, acute resistance exercise in the present study did not elevate proteasome and/or calpain activity in early recovery; rather, it was not until the 36 h time point that ATP‐independent chymotrypsin‐like proteasome activity was significantly elevated. A priori, we hypothesized these pathways would be upregulated after exercise in response to contraction‐induced damage, and to facilitate myofibre remodelling; however, this was generally not the case and could be a reflection of the fact that the dynamic range of MPB is small compared to that of MPS after resistance exercise (Phillips et al. 1997; Glynn et al. 2010). Alternatively, a single bout of exercise may not have been sufficient to induce remodelling and/or activation of the ubiquitin proteasome system. While somewhat tangential to the main aims of the study, we noted that resistance exercise in the presence of rapamycin had a stimulatory effect on proteasome activity in early (3–6 h) recovery. The stimulatory effect of exercise plus rapamycin that we observed is in contrast to inhibitory effects reported in vitro (Osmulski & Gaczynska, 2013), but is consistent with more recent experiments in vivo showing that rapamycin activates the proteasome (Rodriguez et al. 2014). Further research is required to explore potential mechanisms and implications of this finding toward therapies aimed at proteostasis and healthy ageing of skeletal muscle. Taken together, the present work showed little evidence of proteasome or calpain activation (e.g. for remodelling or amino acid intracellular recycling) in early recovery from acute resistance exercise.
Summary
Overall, the present findings demonstrate that resistance exercise activates pathways controlling translational activity and capacity through both rapamycin‐sensitive and ‐insensitive mechanisms. Rapamycin completely blocks protein synthesis 6 h after resistance exercise, but has a limited effect at 12 or 18 h. Rapamycin‐insensitive pathways, including eEF2 and localized myostatin inhibition, may contribute to the prolonged rise in protein synthesis. c‐Myc expression is upregulated after resistance exercise and may play a role in controlling translational capacity by stimulating ribosome biogenesis; however, further mechanistic work in vivo is needed to clarify the role of Myc in load‐induced muscle hypertrophy. In a broader context, given that muscle mass is an important clinical indicator of health, understanding how prolonged protein synthesis and ribosome biogenesis are controlled following exercise has the ability to identify potential targets to combat the loss of muscle with ageing and disease.
Additional information
Competing interests
The authors declare no conflicts of interest.
Funding
This work was funded by Veteran Affairs RR&D Merit Grant 1I01RX000673‐01A1 (SCB), US National Institutes of Health 1R01AG045375‐01 (KB), and a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship (DWDW).
Author contributions
Conception and design of the experiments: DW, SB, KB. Collection, analysis and interpretation of the data: all authors. Drafting the article or revising it critically for important intellectual content: DW, SB, KB. All authors read and approved the final version of the manuscript and all authors listed qualify for authorship.
Acknowledgements
Thanks are expressed to Kurt Watson for assistance with myotube cultures.
References
- Adams GR, Caiozzo VJ, Haddad F & Baldwin KM (2002). Cellular and molecular responses to increased skeletal muscle loading after irradiation. Am J Physiol Cell Physiol 283, C1182–1195. [DOI] [PubMed] [Google Scholar]
- Akiho M, Nakashima H, Sakata M, Yamasa Y, Yamaguchi A & Sakuma K (2010). Expression profile of Notch‐1 in mechanically overloaded plantaris muscle of mice. Life Sci 86, 59–65. [DOI] [PubMed] [Google Scholar]
- Al‐Khalili L, de Castro Barbosa T, Ostling J, Massart J, Cuesta PG, Osler ME, Katayama M, Nystrom AC, Oscarsson J & Zierath JR (2014). Proteasome inhibition in skeletal muscle cells unmasks metabolic derangements in type 2 diabetes. Am J Physiol Cell Physiol 307, C774–787. [DOI] [PubMed] [Google Scholar]
- Altman DG, Gore SM, Gardner MJ & Pocock SJ (1983). Statistical guidelines for contributors to medical journals. Br Med J 286, 1489–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, Larsson LG & Wright AP (2005). c‐Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 7, 303–310. [DOI] [PubMed] [Google Scholar]
- Atherton PJ & Smith K (2012). Muscle protein synthesis in response to nutrition and exercise. J Physiol 590, 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baar K & Esser K (1999). Phosphorylation of p70(S6k) correlates with increased skeletal muscle mass following resistance exercise. Am J Physiol 276, C120–127. [DOI] [PubMed] [Google Scholar]
- Baehr LM, Tunzi M & Bodine SC (2014). Muscle hypertrophy is associated with increases in proteasome activity that is independent of MuRF1 and MAFbx expression. Front Physiol 5, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel CS, Slade J, Mahoney E, Haddad F, Dudley GA & Adams GR (2005). Time course of molecular responses of human skeletal muscle to acute bouts of resistance exercise. J Appl Physiol 98, 482–488. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ & Yancopoulos GD (2001). Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo . Nat Cell Biol 3, 1014–1019. [DOI] [PubMed] [Google Scholar]
- Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, Roobeek I, Weis I, Voute PA, Schwab M & Versteeg R (2001). N‐myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J 20, 1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook MS, Wilkinson DJ, Mitchell WK, Lund JN, Szewczyk NJ, Greenhaff PL, Smith K & Atherton PJ (2015). Skeletal muscle hypertrophy adaptations predominate in the early stages of resistance exercise training, matching deuterium oxide‐derived measures of muscle protein synthesis and mechanistic target of rapamycin complex 1 signaling. FASEB J 29, 4485–4496. [DOI] [PubMed] [Google Scholar]
- Carlson ME, Hsu M & Conboy IM (2008). Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature 454, 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillou T, Kirby TJ & McCarthy JJ (2014). Ribosome biogenesis: emerging evidence for a central role in the regulation of skeletal muscle mass. J Cell Physiol 33, 474–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau‐Aveilla C, Dreazen A, Cagnard N, Carpentier W, Kiss T, Meyuhas O & Pende M (2014). Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 229, 1584–1594. [DOI] [PubMed] [Google Scholar]
- Chesley A, MacDougall JD, Tarnopolsky MA, Atkinson SA & Smith K (1992). Changes in human muscle protein synthesis after resistance exercise. J Appl Physiol 73, 1383–1388. [DOI] [PubMed] [Google Scholar]
- Drummond MJ, Fry CS, Glynn EL, Dreyer HC, Dhanani S, Timmerman KL, Volpi E & Rasmussen BB (2009). Rapamycin administration in humans blocks the contraction‐induced increase in skeletal muscle protein synthesis. J Physiol 587, 1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo VC, Caldow MK, Massie V, Markworth JF, Cameron‐Smith D & Blazevich AJ (2015). Ribosome biogenesis adaptation in resistance training‐induced human skeletal muscle hypertrophy. Am J. Physiol Endocrinol Metab 309, E72–82. [DOI] [PubMed] [Google Scholar]
- Fluckey JD, Vary TC, Jefferson LS & Farrell PA (1996). Augmented insulin action on rates of protein synthesis after resistance exercise in rats. Am J Physiol 270, E313–319. [DOI] [PubMed] [Google Scholar]
- Glover EI, Oates BR, Tang JE, Moore DR, Tarnopolsky MA & Phillips SM (2008). Resistance exercise decreases eIF2Bepsilon phosphorylation and potentiates the feeding‐induced stimulation of p70S6K1 and rpS6 in young men. Am J Physiol Regul Integr Comp Physiol 295, R604–610. [DOI] [PubMed] [Google Scholar]
- Glynn EL, Fry CS, Drummond MJ, Dreyer HC, Dhanani S, Volpi E & Rasmussen BB (2010). Muscle protein breakdown has a minor role in the protein anabolic response to essential amino acid and carbohydrate intake following resistance exercise. Am J Physiol Regul Integr Comp Physiol 299, R533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AV, Waddell DS, Siu R, Stein M, Dewey S, Furlow JD & Bodine SC (2012). Upregulation of proteasome activity in muscle RING finger 1‐null mice following denervation. FASEB J 26, 2986–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P & Hornberger TA (2011). Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J 25, 1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Gomez‐Roman N, Felton‐Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN & White RJ (2005). c‐Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7, 311–318. [DOI] [PubMed] [Google Scholar]
- Hamilton DL, Philp A, MacKenzie MG & Baar K (2010). A limited role for PI(3,4,5)P3 regulation in controlling skeletal muscle mass in response to resistance exercise. PloS one 5, e11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosch M, Lesch M, Baron J & Kaufman S (1967). Enhanced protein synthesis in a cell‐free system from hypertrophied skeletal muscle. Science 157, 935–937. [DOI] [PubMed] [Google Scholar]
- Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB & Hannan RD (2003). mTOR‐dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy‐terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol 23, 8862–8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez‐Verdun D (2006). The nucleolus: a model for the organization of nuclear functions. Histochem Cell Biol 126, 135–148. [DOI] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP & Blenis J (2005). mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123, 569–580. [DOI] [PubMed] [Google Scholar]
- Hornberger TA & Chien S (2006). Mechanical stimuli and nutrients regulate rapamycin‐sensitive signaling through distinct mechanisms in skeletal muscle. J Cell Biochem 97, 1207–1216. [DOI] [PubMed] [Google Scholar]
- Hornberger TA, Stuppard R, Conley KE, Fedele MJ, Fiorotto ML, Chin ER & Esser KA (2004). Mechanical stimuli regulate rapamycin‐sensitive signalling by a phosphoinositide 3‐kinase‐, protein kinase B‐ and growth factor‐independent mechanism. Biochem J 380, 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MJ, Cheng YC, Liu CR, Lin S & Liu HE (2006). A small‐molecule c‐Myc inhibitor, 10058‐F4, induces cell‐cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp Hematol 34, 1480–1489. [DOI] [PubMed] [Google Scholar]
- Hwee DT, Baehr LM, Philp A, Baar K & Bodine SC (2014). Maintenance of muscle mass and load‐induced growth in Muscle RING Finger 1 null mice with age. Aging Cell 13, 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Groll M, Huber R, Wolf DH & Heinemeyer W (1999). Proteasome beta‐type subunits: unequal roles of propeptides in core particle maturation and a hierarchy of active site function. J Mol Biol 291, 997–1013. [DOI] [PubMed] [Google Scholar]
- Jastrzebski K, Hannan KM, Tchoubrieva EB, Hannan RD & Pearson RB (2007). Coordinate regulation of ribosome biogenesis and function by the ribosomal protein S6 kinase, a key mediator of mTOR function. Growth Factors 25, 209–226. [DOI] [PubMed] [Google Scholar]
- Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB & Sabatini DM (2013). mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science 341, 1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR & Jefferson LS (2010). Control of translation initiation through integration of signals generated by hormones, nutrients, and exercise. J Biol Chem 285, 29027–29032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubica N, Bolster DR, Farrell PA, Kimball SR & Jefferson LS (2005). Resistance exercise increases muscle protein synthesis and translation of eukaryotic initiation factor 2Bε mRNA in a mammalian target of rapamycin‐dependent manner. J Biol Chem 280, 7570–7580. [DOI] [PubMed] [Google Scholar]
- Kwon H & Green MR (1994). The RNA polymerase I transcription factor, upstream binding factor, interacts directly with the TATA box‐binding protein. J Biol Chem 269, 30140–30146. [PubMed] [Google Scholar]
- Lee SJ (2004). Regulation of muscle mass by myostatin. Ann Rev Cell Devel Biol 20, 61–86. [DOI] [PubMed] [Google Scholar]
- Lee SJ (2010). Extracellular regulation of myostatin: a molecular rheostat for muscle mass. Immunol Endocr Metab Agents Med Chem 10, 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z & Hann SR (2013). Nucleophosmin is essential for c‐Myc nucleolar localization and c‐Myc‐mediated rDNA transcription. Oncogene 32, 1988–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ & Schmittgen TD (2001). Analysis of relative gene expression data using real‐time quantitative PCR and the 2–ΔΔ C T Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- MacKenzie MG, Hamilton DL, Pepin M, Patton A & Baar K (2013). Inhibition of myostatin signaling through Notch activation following acute resistance exercise. PloS one 8, e68743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi LB, Jr , Kuchenruether M, Dadey DY, Schwope RM, Grisendi S, Townsend RR, Pandolfi PP & Weber JD (2008). Nucleophosmin serves as a rate‐limiting nuclear export chaperone for the mammalian ribosome. Mol Cell Biol 28, 7050–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall A, Salerno MS, Thomas M, Davies T, Berry C, Dyer K, Bracegirdle J, Watson T, Dziadek M, Kambadur R, Bower R & Sharma M (2008). Mighty is a novel promyogenic factor in skeletal myogenesis. Exp Cell Res 314, 1013–1029. [DOI] [PubMed] [Google Scholar]
- Mayhew DL, Hornberger TA, Lincoln HC & Bamman MM (2011). Eukaryotic initiation factor 2B epsilon induces cap‐dependent translation and skeletal muscle hypertrophy. J Physiol 589, 3023–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron AC & Lee SJ (1997). Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA 94, 12457–12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton CC, Brown JA, Holmes WM, Nance WE & Wolf B (1983). Stain intensity of human nucleolus organizer region reflects incorporation of uridine into mature ribosomal RNA. Exp Cell Res 145, 405–413. [DOI] [PubMed] [Google Scholar]
- Muth V, Nadaud S, Grummt I & Voit R (2001). Acetylation of TAFI68, a subunit of TIF‐IB/SL1, activates RNA polymerase I transcription. EMBO J 20, 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader GA, McLoughlin TJ & Esser KA (2005). mTOR function in skeletal muscle hypertrophy: increased ribosomal RNA via cell cycle regulators. Am J Physiol Cell Physiol 289, C1457–1465. [DOI] [PubMed] [Google Scholar]
- Olivier J, Johnson WD & Marshall GD (2008). The logarithmic transformation and the geometric mean in reporting experimental IgE results: what are they and when and why to use them? Ann Allergy Asthma Immunol 100, 333–337. [DOI] [PubMed] [Google Scholar]
- Osmulski PA & Gaczynska M (2013). Rapamycin allosterically inhibits the proteasome. Mol Pharmacol 84, 104–113. [DOI] [PubMed] [Google Scholar]
- Phillips BE, Williams JP, Gustafsson T, Bouchard C, Rankinen T, Knudsen S, Smith K, Timmons JA & Atherton PJ (2013). Molecular networks of human muscle adaptation to exercise and age. PLoS genetics 9, e1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips SM, Tipton KD, Aarsland A, Wolf SE & Wolfe RR (1997). Mixed muscle protein synthesis and breakdown after resistance exercise in humans. Am J Physiol 273, E99–107. [DOI] [PubMed] [Google Scholar]
- Ploton D, Menager M, Jeannesson P, Himber G, Pigeon F & Adnet JJ (1986). Improvement in the staining and in the visualization of the argyrophilic proteins of the nucleolar organizer region at the optical level. Histochem J 18, 5–14. [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D & Goldberg AL (1994). Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771. [DOI] [PubMed] [Google Scholar]
- Rodriguez KA, Dodds SG, Strong R, Galvan V, Sharp ZD & Buffenstein R (2014). Divergent tissue and sex effects of rapamycin on the proteasome‐chaperone network of old mice. Front Mol Neurosci 7, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanij E, Poortinga G, Sharkey K, Hung S, Holloway TP, Quin J, Robb E, Wong LH, Thomas WG, Stefanovsky V, Moss T, Rothblum L, Hannan KM, McArthur GA, Pearson RB & Hannan RD (2008). UBF levels determine the number of active ribosomal RNA genes in mammals. J Cell Biol 183, 1259–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky VY, Pelletier G, Hannan R, Gagnon‐Kugler T, Rothblum LI & Moss T (2001). An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol Cell 8, 1063–1073. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS & Sabatini DM (2012). A unifying model for mTORC1‐mediated regulation of mRNA translation. Nature 485, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trere D (2000). AgNOR staining and quantification. Micron 31, 127–131. [DOI] [PubMed] [Google Scholar]
- Voit R & Grummt I (2001). Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc Natl Acad Sci USA 98, 13631–13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit R, Kuhn A, Sander EE & Grummt I (1995). Activation of mammalian ribosomal gene transcription requires phosphorylation of the nucleolar transcription factor UBF. Nucleic Acids Res 23, 2593–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit R, Schnapp A, Kuhn A, Rosenbauer H, Hirschmann P, Stunnenberg HG & Grummt I (1992). The nucleolar transcription factor mUBF is phosphorylated by casein kinase II in the C‐terminal hyperacidic tail which is essential for transactivation. EMBO J 11, 2211–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Walden F, Casagrande V, Ostlund Farrants AK & Nader GA (2012). Mechanical loading induces the expression of a Pol I regulon at the onset of skeletal muscle hypertrophy. Am J Physiol Cell Physiol 302, C1523–1530. [DOI] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR & Proud CG (2001). Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J 20, 4370–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West DW, Kujbida GW, Moore DR, Atherton P, Burd NA, Padzik JP, De Lisio M, Tang JE, Parise G, Rennie MJ, Baker SK & Phillips SM (2009). Resistance exercise‐induced increases in putative anabolic hormones do not enhance muscle protein synthesis or intracellular signalling in young men. J Physiol 587, 5239–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TS & Booth FW (1990. a). Protein metabolism in rat gastrocnemius muscle after stimulated chronic concentric exercise. J Appl Physiol 69, 1709–1717. [DOI] [PubMed] [Google Scholar]
- Wong TS & Booth FW (1990. b). Protein metabolism in rat tibialis anterior muscle after stimulated chronic eccentric exercise. J Appl Physiol 69, 1718–1724. [DOI] [PubMed] [Google Scholar]
- Young VR (1970). The role of skeletal and cardiac muscle in the regulation of protein metabolism In Mammalian Protein Metabolism, ed. Munro HN, pp. 586–657. Academic Press, New York. [Google Scholar]


