Summary
Significant advances have been made in methods to analyze genomes and transcriptomes of single cells, but to fully define cell states, proteins must also be accessed as central actors defining a cell’s phenotype. Methods currently used to analyze endogenous protein expression in single cells are limited in specificity, throughput, or multiplex capability. Here, we present an approach to simultaneously and specifically interrogate large sets of protein and RNA targets in lysates from individual cells, enabling investigations of cell functions and responses. We applied our method to investigate the effects of BMP4, an experimental therapeutic agent, on early-passage glioblastoma cell cultures. We uncovered significant heterogeneity in responses to treatment at levels of RNA and protein, with a subset of cells reacting in a distinct manner to BMP4. Moreover, we found overall poor correlation between protein and RNA at the level of single cells, with proteins more accurately defining responses to treatment.
Graphical Abstract
Highlights
-
•
Multiplexed protein and RNA measurement in single cells
-
•
Distinct information for defining cell states from RNA and protein data
-
•
Heterogeneity of responses to the putative therapeutic BMP4 in glioblastoma cells
Darmanis et al. present an approach to simultaneously measure levels of up to 96 transcripts and proteins in single cells. They apply this technique to study responses of glioblastoma cells to BMP4, a proposed therapeutic agent, and reveal heterogeneous responses and cell states.
Introduction
The need to understand differences within cellular communities and the nature of heterogeneous cellular responses have prompted development of efficient methods for genomic and transcriptomic analysis at the level of single cells (Macaulay and Voet, 2014, Patel et al., 2014). In order to better understand functional properties of cells, these molecular genetic techniques need to be complemented by high-performance and high-throughput single-cell protein analyses. Current methods to study endogenous protein expression in single cells tend to be limited in throughput or multiplex capability (Bendall et al., 2011, Ståhlberg et al., 2012, Ullal et al., 2014, Yu et al., 2014). Moreover, unlike the state of the art for measuring proteins in e.g., plasma, currently available single cell protein assays rely on target recognition by single antibodies, thus limiting detection specificity.
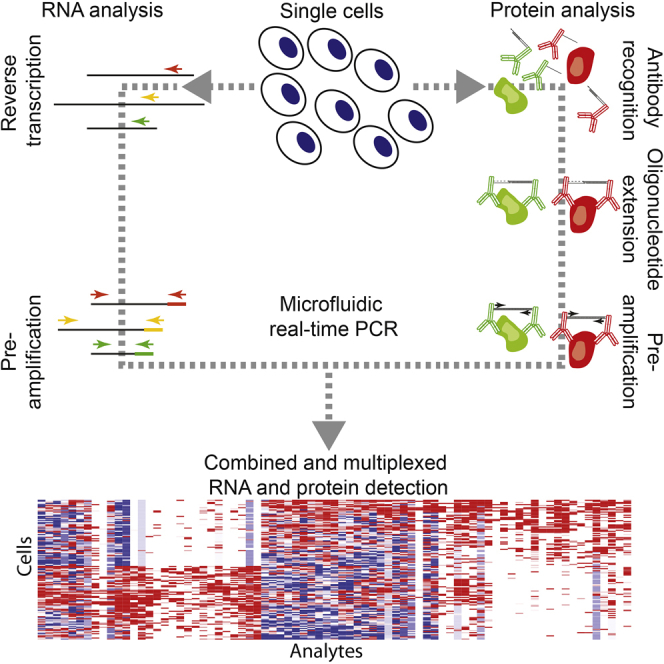
Here, we present a procedure to simultaneously interrogate large sets (∼96) of both RNA and protein targets in single-cell lysates to investigate cell functions and responses. In our approach, single isolated cells are lysed and divided for separate RNA or protein analysis (Figure 1A). Proteins are probed using a homogeneous affinity-based proximity extension assay (PEA) that targets proteins using pairs of antibodies conjugated with oligonucleotides whose free 3′ ends are pairwise complementary (Assarsson et al., 2014). When a cognate antibody pair binds a target protein, the attached oligonucleotides are brought in proximity and can be extended by polymerization to create an amplifiable DNA reporter molecule, which is subsequently quantified by high-throughput real-time PCR. The requirement for pairwise protein detection ensures sandwich immunoassay-quality protein detection. A multiplex readout is achieved by decoding extension-generated DNA reporters by real-time PCR using primer pairs specific for cognate pairs of antibody conjugates. Transcripts are probed using commercial TaqMan Gene Expression Assays using a previously described method (Dalerba et al., 2011).
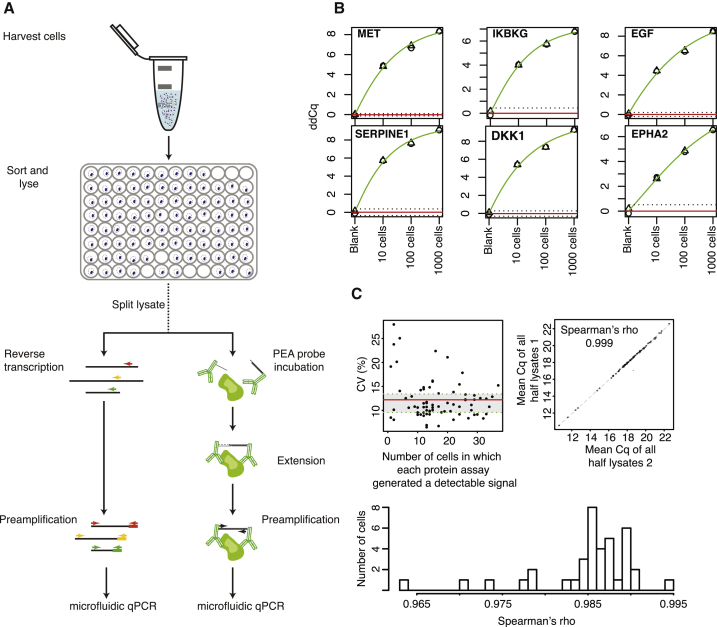
Figure 1.
Experimental Approach and Validation
(A) Illustration of the experimental approach. Single cells are isolated by FACS and lysed immediately. Cell lysates are split for subsequent protein and RNA analysis by PEA and gene-targeted TaqMan assays, respectively.
(B) Standard curves of sorted 1,000, 100, and 10 U3035MG cells plus no cell control (blank) for select PEA assays. The circle and triangle data points represent biological replicates. The red horizontal bar denotes the mean background value, whereas the dashed lines are the mean ± 3 SD. y axis values represent extension control normalized Cq values. See also Figure S1.
(C) Coefficient of variation analysis of n = 40 split U3035MG single cells, where both halves were analyzed with the same PEA single cell protein panel. The coefficient of variation of each assay is plotted as a function of the number cells, out of a total 40 cells, in which the assay generated a detectable signal cells (top left). The top right panel shows the correlation between the mean values for each protein assay, comparing cell half 1 and 2 for each cell. The bottom panel shows the correlation coefficients between each of the two lysate aliquots, calculated for each cell across all assays.
We applied the approach above to characterize the effects of treatment with bone morphogenetic protein 4 (BMP4) on early-passage U3035MG cells, derived from a patient with glioblastoma and grown under neural stem cell conditions. BMP4, a cytokine belonging to the transforming growth factor β (TGF-β) superfamily, is of interest as a potential therapeutic agent in glioblastoma (Duggal et al., 2013). It is believed to reduce numbers of tumor-initiating precursors through induction of astroglial differentiation, thereby potentially limiting tumor propagation and therapeutic resistance (Bao et al., 2006, Piccirillo et al., 2006). During initial screening, U3035MG displayed partial resistance to BMP4 (not shown), motivating a search for factors that contribute to the observed phenotype among all or some of the cells.
Results
Protein and RNA Assay Validation
To construct a multiplex PEA panel for probing cell states, targeting cancer and neuro-oncology pathways of relevance in glioblastoma, we selected polyclonal antibodies raised against all or a significant portion of the proteins of interest. After validating assay sensitivity and specificity, a panel of 75 PEA assays was selected with sensitivity down to ten cells or fewer and no evidence of cross-reactivity in our cell model of interest (Figure 1B; Table S1; Figure S1).
RNA was probed using commercial TaqMan Gene Expression Assays as in a previously described approach (Dalerba et al., 2011). Eighty-two assays were selected to complement the glioblastoma single-cell protein panel in capturing BMP signaling, its interaction with other pathways (e.g., WNT or TGF-β), and cellular processes such as differentiation, proliferation, and apoptosis, expected to be affected by BMP4 (Piccirillo et al., 2006). Twenty-two of the TaqMan assays targeted transcripts for genes whose protein products were included in the protein panel permitting evaluation of RNA-protein correlations (see Table S1 for a list of assays). The final combined RNA and protein panel members had high coverage of gene products involved in major cancer pathways, focal adhesion, cell cycle, apoptosis, and cell development (as identified via KEGG and Reactome Pathways as well as Gene Ontology [GO] terms). The details are listed in Table S1.
To assess the precision of the single-cell protein assays, we sorted 40 individual U3035MG cells using fluorescence-activated cell sorting (FACS). These cells are distinct from the cells sorted and analyzed for BMP4 response. Upon lysis, two halves of each lysate were probed separately with the glioblastoma single-cell protein panel. We then calculated the coefficient of variation (CV) per cell and per assay, excluding samples and targets with no detectable signals. The mean CV across assays was 12.2% (interquartile range [IQR], 9.5%–13.4%). When the percent CV is plotted against the number of single cells for which a signal over background was detected in a given assay, the CV is stable for assays where the molecules were detected in more than five single cells (Figure 1C, top left panel). In addition, the Spearman correlation between the mean Ct values obtained for each assay in all cells revealed a rank correlation coefficient of 0.999, demonstrating excellent reproducibility (Figure 1C, top right). Furthermore, the correlation for the same measurements in the two halves of each of the 40 cells for all assays had an average Spearman rank correlation coefficient of 0.985 (min = 0.9636, max = 0.9941) (Figure 1C, bottom). We similarly determined the reproducibility of RNA detection. Split single-cell lysates from cell lines MCF-7 and Hs578T were probed with 12 different TaqMan assays, revealing a mean CV across assays of 26% (IQR, 14.5%–35.1%). In addition to probing split MCF-7 or Hs578T single cells, we also calculated the CV% for aliquots of lysates prepared from 1,000 (PEA only), 100, or 10 MCF-7 or Hs578T cells. We conclude that the CV% was satisfactory across cell numbers for both the PEA or TaqMan probes. We note, however, that the CV% was stable across cell numbers interrogated with the PEA assays, but the CV% increased with decreasing cell number with the TaqMan assays, although they remained within an acceptable range (mean CV% for 100 cells = 13, for 10 cells = 18, and for single cells = 26). We thus conclude that our assays were reliable and reproducible, allowing accurate measurement of protein and RNA levels in single-cell lysates.
Agreement between Bulk Population and Single Cells
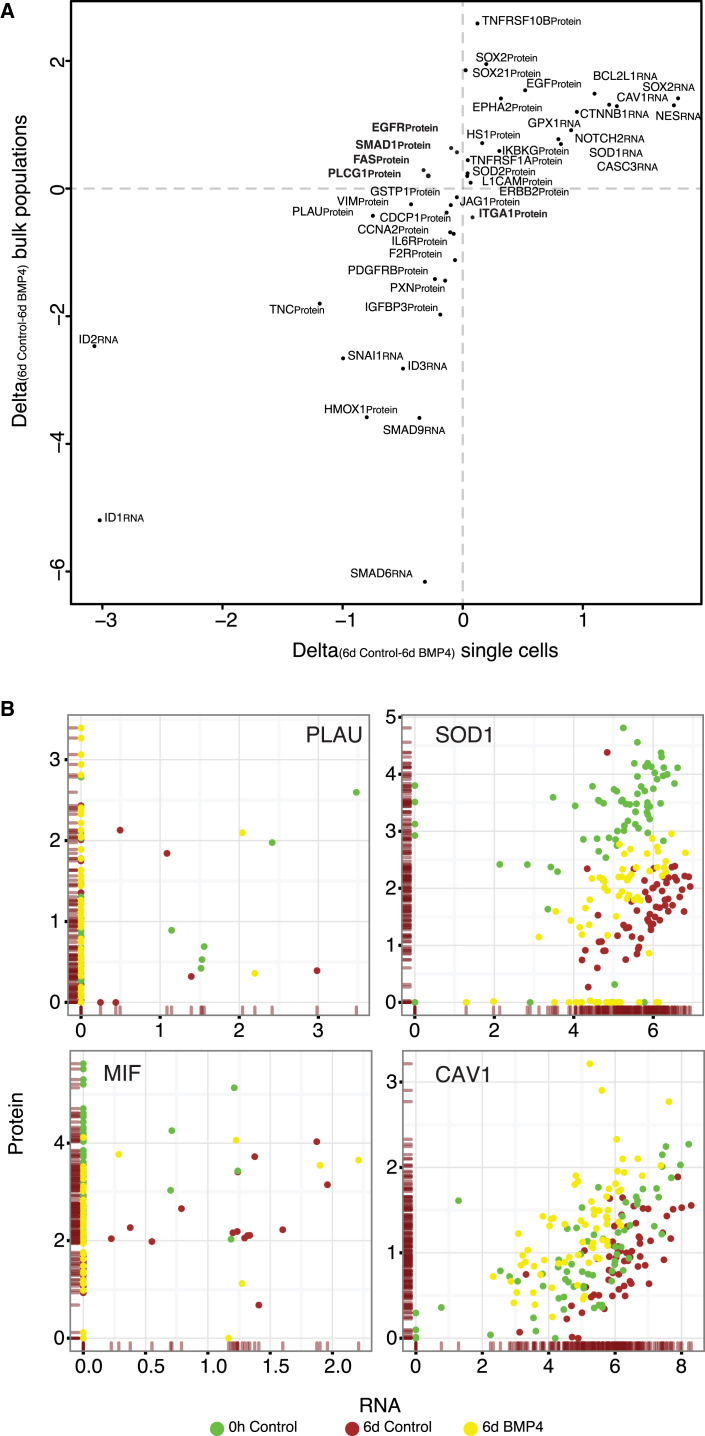
In order to evaluate the agreement of the signals detected in single cells with results from bulk samples, we sorted two sets of 150 cells from the same population sorted for single-cell analysis. There was good agreement between the mean expression across single cells and that observed for pooled cells (Pearson’s r = 0.68; Figure 2A). Five proteins with poor correlation between average single and pooled cells (PLCG1, FAS, SMAD1, EGFR, and ITGA1) were all expressed close to the limits of detection in single cells (∼0.5 ddCq over background), and the differences between treated and untreated cells in the pooled sample were small.
Figure 2.
Analysis of Agreement between Averages of Single Cells and a Bulk Population Sample and Single-Cell RNA-Protein Correlations
(A) Plot of the delta values (6d Control – 6d BMP4) for the means from the single cells (x axis) or the population controls (n = 150 cells) (y axis) sorted at the same time as the single cells (Pearson’s r = 0.68). Gene products that disagree between the average single and bulk controls are in bold.
(B) Correlation plots between RNA and protein levels for selected analytes in all single cells belonging to the 0-hr untreated (green), day-6 untreated (red), and day-6 BMP4-treated (yellow) populations. Distributions of protein and RNA levels are shown as ticks on the y and x axes respectively.
See also Figure S2.
Correlation of RNA and Protein Expression
Across all single cells, the coefficients of determination (R2) between protein and RNA levels were on average 0.04 (min = 0, max = 0.35) for the RNAs and proteins encoded by the same genes (Figure S2). This suggests that for most gene products, only a small portion of the variation of protein levels can be explained by measuring mRNA levels in single cells. The correlation is much lower than what has been reported for experiments performed on populations of cells (Jovanovic et al., 2015, Schwanhäusser et al., 2011). In pools of cells, protein and mRNA levels are averaged over large numbers of cells, ignoring any cell-to-cell variation. Burst-like transcription and generally shorter half-lives for RNA may conspire to render RNA levels less accurate indicators of cell states at the single-cell level, compared to the more stable proteins whose levels may need to be maintained to enact the various functions of cells (Raj and van Oudenaarden, 2009, Schwanhäusser et al., 2011).
Of the protein-RNA pairs that were significantly correlated using the measure Pearson’s r (p < 0.001; CAV1, GFAP, PLAU, PXN, SOD1, and SOX2), all were positively correlated, but they displayed different dynamics of co-expression (Figure 2B). The mesenchymal marker PLAU (Verhaak et al., 2010) (Pearson’s r = 0.16) was detected in most cells at the level of protein, but only a small fraction of cells had detectable RNA expression. That is consistent with a model where RNA levels are transient whereas protein levels are maintained for longer periods of time as required to maintain cellular functions (Schwanhäusser et al., 2011).
SOD1 RNA and protein displayed a strong correlation (Pearson’s r = 0.45–0.52 across time points; Figure 2B), with two notable observations. First, the distribution of RNA expression was similar whether or not the cells had been treated with BMP4, yet protein expression differed between treatment groups. Second, the distribution of protein expression was bimodal in the BMP4-treated cells, with one group of cells (n = 20) displaying no detectable protein but clear RNA expression. Moreover, SOD1PROTEIN was strongly correlated with MIFPROTEIN (Pearson’s r = 0.79), but not with MIFRNA (Pearson’s r = 0.18). The cells that expressed no detectable SOD1PROTEIN were also devoid of MIFPROTEIN, suggesting tight co-regulation of these proteins (Israelson et al., 2015).
BMP4 Treatment and Single-Cell Analysis
BMP4-treated U3035MG cells grown under neural stem cell conditions maintained persistent proliferation in the presence of BMP4, albeit at a reduced rate compared to untreated controls (doubling time 2.5 days rather than 1.8 days) (Figure S3). Other cell lines derived from patients with glioblastoma displayed a heterogeneous response to BMP4, but with significantly reduced cell division in the majority of lines tested (data not shown).
We analyzed single U3035MG cells before treatment (79 cells analyzed) and 6 days after initiation of BMP4 treatment, a time point when control cells (62 cells analyzed) were not confluent, and when BMP4-resistant treated cells (69 cells analyzed) had expanded to constitute a significant fraction of the treated cell population. In total, 210 single cells were monitored for RNA and protein levels (Table S2). The RNA and protein data for U3035MG were processed for quality control (QC) filtering and normalization (see Experimental Procedures). Of the 75 and 82 proteins and transcripts, respectively, that were assayed, 46 PEA assays and 23 TaqMan assays were positive in greater than 50% of the cells interrogated.
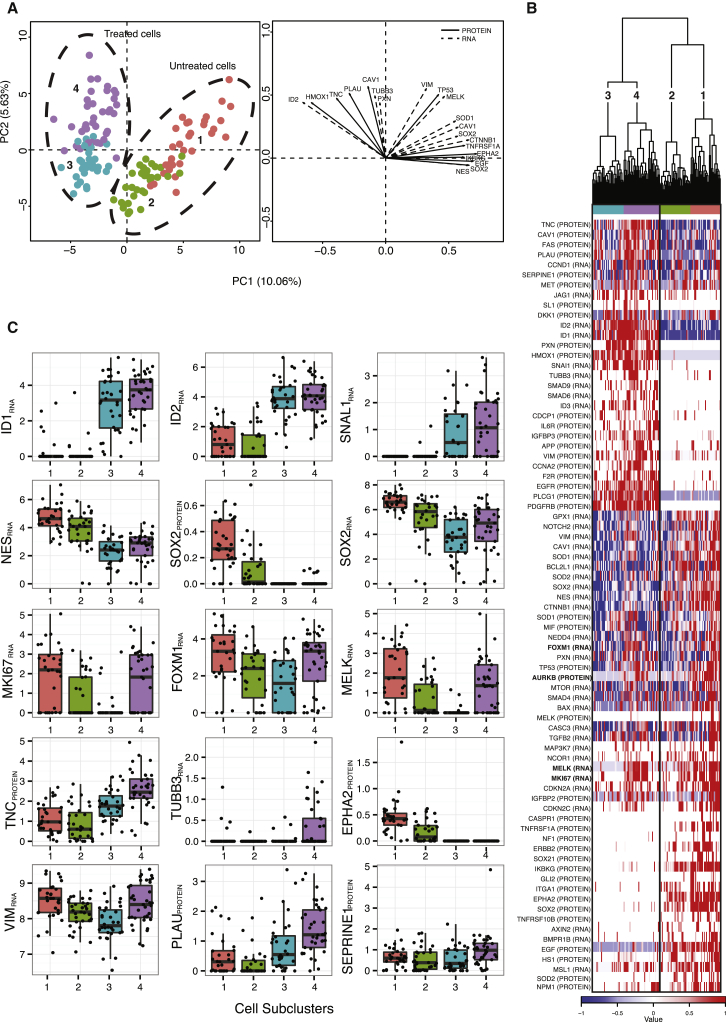
The first dimension (PC1) of a principal component analysis (PCA) of normalized, combined RNA and protein datasets from day 6 clearly separated the control and BMP4-treated cells, whereas the second dimension (PC2) captured variation present within each of the groups (Figure 3A; Table S3). Hierarchical clustering using gene products correlated with PC1 and PC2 (p < 0.001) correctly grouped treated and control cells (Figure 3B), with both RNA and protein contributing to the distinction between the two groups. We also detected clear subclusters among both the control and BMP4-treated cells, highlighting significant heterogeneity within the cell line (Figure 3A and B). Analysis inclusive of the 0-hr untreated cells was comparable to the day-6-only analysis, with a clear distinction between untreated and treated groups, and the 0-hr and day-6 controls clearly overlapping (Figure S3).
Figure 3.
Analysis of BMP4 Response in Glioblastoma Patient Cell Line U3035MG
(A) PCA (left) and gene vector (right) plots for PC1 and PC2 for day-6 untreated and BMP4-treated cells, using combined normalized RNA and protein data. For the PCA plot, values for each marker in individual cells are reflected on a scale from blue to red. The colors and numbers at the top of the heatmap correspond to subclusters identified through the hierarchical clustering analysis shown in (B) with subclusters 1 (orange) and 2 (green) encompassing the day-6 untreated cells and subclusters 3 (turquoise) and 4 (purple) belonging to the day-6 BMP4-treated cells. Subclusters 1 and 4 primarily contain proliferationhigh cells. For the vector plot, vectors illustrate the ten most significant variables, measured at the level of RNA (dashed vector lines) or protein (solid vector lines), correlating with each of the first two principal components. The vector lengths represent the extent of correlation to each of the components.
(B) Hierarchical clustering and heatmap of factors significantly (p < 0.001) associated with PC1 and PC2 from the PCA analysis shown in (A).
(C) Boxplots of select RNA or protein factors divided by subcluster. Subclusters are identified by colors as described in (A). The y axis represent normalized Cq values subtracted from the normalized mean background − 2∗SD of the mean background.
See also Figure S3.
Although the U3035MG cells proved largely refractory to the antiproliferative effect of BMP4 treatment (Figure S3), the cells exhibited clear activation of the known BMP4 response gene products ID1RNA and ID2RNA (Lasorella et al., 2014, Miyazono and Miyazawa, 2002), and a large fraction of cells also had increased expression of ID3RNA (Lasorella et al., 2014, Miyazono and Miyazawa, 2002), SNAI1RNA (Savary et al., 2013), SMAD6RNA (Imamura et al., 1997), and SMAD9RNA (Tsukamoto et al., 2014) (Figures 3B and 3C). The results strongly suggest that the BMP4 pathway was activated in all treated cells. We further observed a decrease of the neural stem cell markers SOX2RNA, SOX2PROTEIN, and NESRNA, as well as EPHA2PROTEIN (Miao et al., 2015). Nonetheless, there were surprisingly few changes in the proportion of cells expressing the astrocyte markers GFAPRNA and GFAPPROTEIN or in their overall levels of expression, consistent with the observation that BMP4 failed to induce terminal differentiation. The only cell marker detectable in an increased number of cells upon treatment was the neuronal marker TUBB3RNA (a change from 8% to 30% detectable cells) (Figure 3C). A marginal increase of TUBB3PROTEIN upon BMP4 exposure was also demonstrated by flow cytometry using day-6 control and BMP4-treated U3035MG cells (Figure S3). These results suggest that a decrease in stemness, as reflected by the decreased expression of SOX2/NES, was accompanied in at least a subset of cells by differentiation toward a neuronal fate, rather than the expected astrocyte-like fate (Piccirillo et al., 2006). This broad differentiation potential is in agreement with recent work showing that BMP4-treated glioma stem cell-enriched cell lines made either an astrocytic or a neuronal lineage choice, depending on specific endogenous or exogenous factors (Videla Richardson et al., 2015).
The combined protein and RNA analysis identified clear differences between treated and untreated cells, but we were also interested in possible heterogeneity within the groups of treated or untreated cells. Hierarchical clustering separated both treated and untreated cells in two subsets each (Figures 3A and 3B). The two subpopulations of treated cells primarily differed in their expression of the proliferation marker MKI67RNA (Whitfield et al., 2006) and the cell-cycle regulators MELKRNA, FOXM1RNA (Joshi et al., 2013), and AURKBPROTEIN (Whitfield et al., 2006) (Figure 3C). Cells expressing high levels of proliferation markers also exhibited significantly higher levels SMAD6RNA, a negative regulator of BMP4 signaling (Imamura et al., 1997). The same cells also had higher expression of a number of genes associated with a mesenchymal signature, specifically VIMRNA, PLAUPROTEIN, SERPINE1PROTEIN, and TNCPROTEIN (Carro et al., 2010, Phillips et al., 2006, Verhaak et al., 2010). Interestingly, the extracellular matrix protein TNC can influence normal neural stem cell development by modulating sensitivity to FGF2 (positively) and BMP4 (negatively) (Garcion et al., 2004). Using flow cytometry, we confirmed that TNCPROTEIN is increased in response to BMP4 and that its expression is enhanced in a subset of proliferation-positive BMP4-treated cells (Figure S3). Together, these results suggest that despite active signaling through the BMP4 pathway, a subset of treated cells partially escape the effects of BMP4 to maintain a proliferative state. An avenue for further investigation using a still larger set of transcripts and proteins is whether the cells also display a shift toward a mesenchymal phenotype (Verhaak et al., 2010).
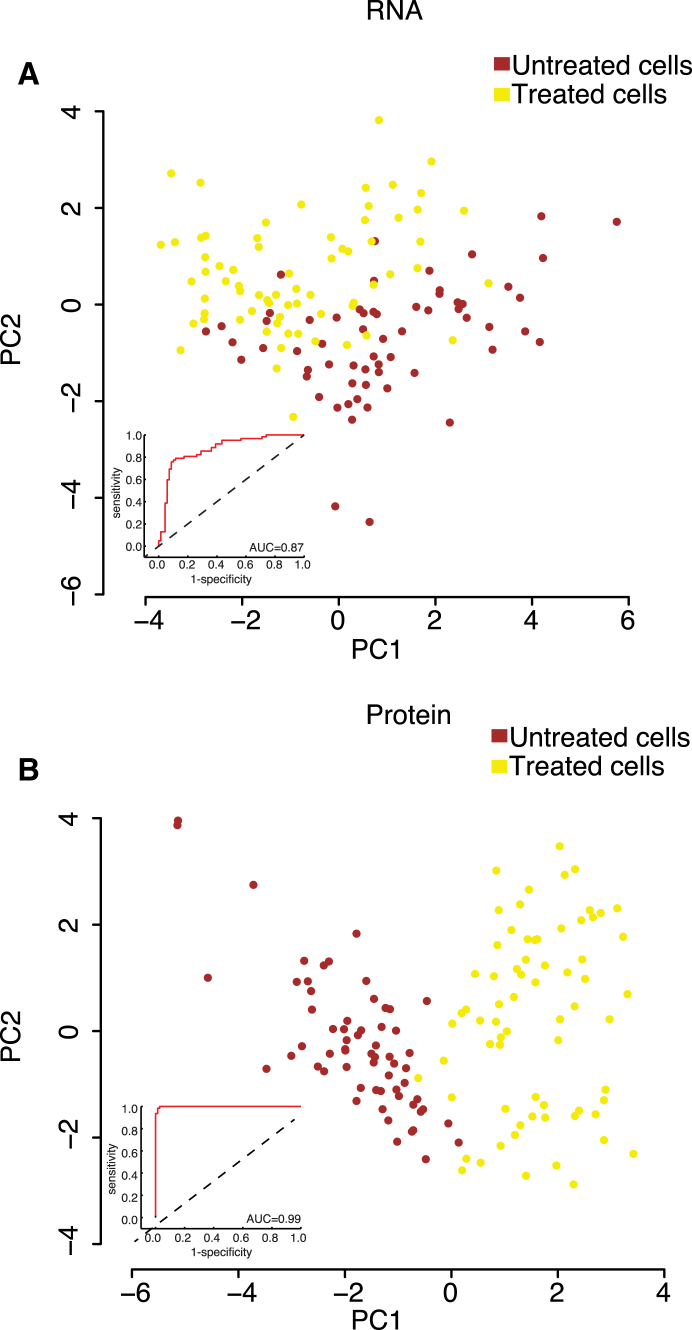
Relative Ability of RNA and Protein Expression Data to Distinguish Treated and Untreated Cells
To examine whether either RNA or protein levels better reflected the effects of BMP4 treatment, we performed PCA separately on the 22 gene products analyzed both at levels of RNA and protein (Figure 4). A combination of the first two principal components for protein and RNA measurements, explaining 25.4% and 22.5% of the variance, respectively, was used as a basis for receiver-operating characteristics (ROC) analysis of treated versus untreated cells (see Experimental Procedures) (Figure 4). The protein measurements accurately distinguished cells according to whether they had been treated with BMP4 or not with an area under the curve (AUC) of 0.99, whereas the corresponding AUC for RNA measurements was lower at 0.87. To ensure that the better distinction between treated and untreated cells for protein rather than RNA measurements was not limited to the first two principal components, we performed the same type of analysis using the principal components from 1 to 30 for both protein and RNA. The results of this analysis are exhibited in Figure S4 and demonstrate that proteins consistently, albeit marginally, outperformed RNA in distinguishing treated from untreated cells. For example, the AUC increased to 1.0 and 0.95 for protein and RNA, respectively, when the first three principal components were included in the analysis. We note that combined protein and RNA gave AUC values higher than those observed when protein and RNA were used separately (Figure S4).
Figure 4.
Distinction of Treated and Untreated Cells Using RNA or Protein Expression Data
PCA plots of BMP4 treated (yellow) and untreated cells (brown) at day 6 using expression of the same 22 genes, measured at the level of RNA (A) or protein (B), illustrating the relative ability of protein and RNA analyses to correctly identify whether cells had been treated with BMP4 or not. Insets show receiver-operating characteristic (ROC) curves derived by performing linear discriminant analysis of the first two principal components. AUC, area under the curve. See also Figure S4 for analysis including more principal components.
Discussion
We report the development of a precise approach to simultaneously quantify large numbers of RNA and proteins molecules in single cells. Our method demonstrates so far the highest degree of multiplexing for protein analysis in single cells. Moreover, the method can be further extended, both on the level of protein and RNA. Protein detection is currently limited only by the selected microfluidic readout (96plex), while it could be possible to record even more proteins and transcripts levels through next-generation sequencing (Darmanis et al., 2010). It has been shown previously that the performance of proximity-based assays does not deteriorate with increased multiplexing, in contrast to other protein assays dependent on recognition by pairs of antibodies (Darmanis et al., 2010). Furthermore, the protein analysis is compatible with established single-cell RNA-sequencing (RNA-seq) protocols (Picelli et al., 2014), thus allowing the analysis of whole transcriptomic signatures along with large numbers of protein molecules.
Using combined RNA and protein data for individual cells, we demonstrated significant heterogeneity within populations of a low-passage cell culture derived from a patient with glioblastoma multiforme and grown under neural stem cell conditions. The expression pattern accurately distinguished control and BMP4-treated cells. Furthermore, we demonstrated evidence that RNA and protein data contribute distinct information in defining cell states. In our model, protein expression was marginally better at defining group membership. This difference could in part reflect that protein levels are more stable over time than RNA expression, as previously reported (Munsky et al., 2012, Schwanhäusser et al., 2011). The greater technical noise in the assays employed here to measure RNA versus protein abundance may also have contributed to this effect. However, considerably more information is available via single-cell RNA-seq than what can be obtained by measuring less than hundred proteins as shown herein, compensating for any greater variability of RNA expression. Nonetheless, as we demonstrate in this study, protein and RNA information can be used in a complementary way, and protein-level measurements may have a special value in analyses of cell states such as the activation status of signaling pathways as reflected in protein modifications and interactions.
Glioblastoma multiforme is a devastating disease, and despite aggressive conventional treatment, including surgical resection followed by radio- and chemotherapy, tumors often relapse. A major obstacle for efficient treatment is the intratumoral heterogeneity of the disease, motivating single-cell studies (Patel et al., 2014, Sottoriva et al., 2013). In this study, we identified distinct BMP4 treatment responses, highlighting a subset of cells that appeared to escape the differentiating effect of BMP4 and failing to become post-mitotic. These results reveal a need for better understanding of the BMP4 effector pathways in cancer, and they temper the prospects of BMP4 as a therapeutic agent in glioblastoma multiforme. The investigation supports a role for single-cell studies at levels of RNA and protein to identify phenotypic variation among subpopulations of cells, as an aide to explore cellular responses, for prognostics, and to design personalized treatment (Patel et al., 2014).
Experimental Procedures
Design of Panels of Proximity Extension Assays
We annotated a list of 1,100 polyclonal antibodies from R&D Systems and Cell Signaling directed against proteins of relevance for known cancer pathways, cell states, and neuro-oncology and available without additives and at a sufficient concentration for conjugation. To design a panel to investigate cell states, we selected 11 GO Slim categories (cumulative versions of the GO categories containing a subset of the whole GO terms) covering a range of cell processes and functions. Among all potential protein targets annotated on the basis of the 11 GO Slim categories, we selected the 110 proteins with the highest number of memberships per category. For the cancer pathways panel, the same 1,100 proteins were annotated using a similar strategy, but this time targeting relevant cancer pathways as described in the KEGG pathway annotation database. For the neuro-oncology panel, we compiled a list of protein targets of interest for glioblastoma tumor biology by curating the literature.
PEA Assays
Each PEA assay is generated by conjugating the 5′ end of two different oligonucleotides to a pair of antibodies forming a PEA A- and B-probe. The A-probe is then hybridized with an oligonucleotide that is partially complementary to the A-oligonucleotide and where the 3′ end is complementary to 3′ end of the conjugated B-oligonucleotide. The antibody pair can either be sourced from two monoclonal antibodies detecting non-overlapping epitopes or from a polyclonal antibody raised against a peptide representing a large-as-possible fraction of the target protein. The list of antibodies used in this study can be found in Table S1. The oligonucleotides contain two different primer pair specific sequences that, when in proximity and extended, can be used for first universal and then target specific PCR amplification.
Protein detection using PEA was performed as follows: 1 μl cell lysate was mixed with 3 μl PEA probe mix. The mix contained 0.3 μl of each the PEA A- and B-probes (final antibody-conjugate concentration 100 pM), 0.2 μl Incubation Stabilizer (Olink Bioscience), and 2.1 μl Incubation Solution (Olink Bioscience). Each 96-well plate included a negative control in triplicate (lysis buffer only), two population controls (two 150-cell lysates sorted at the same time as the single cells), and Olink Bioscience’s inter-plate control in triplicate (Assarsson et al., 2014). The plate was briefly centrifuged, sealed, and incubated overnight at 4°C.
Following overnight incubation, plates were briefly spun down, and 96 μl of a PEA probe extension mix was added to each well. The mix contained 0.2 μl PCR Polymerase, 0.5 μl PEA Enzyme, and 10 μl PEA Solution (all Olink Bioscience) and 85.3 μl molecular grade water. Plates were sealed, gently vortexed, spun down, and then placed in a thermal cycler for the extension reaction (50°C, 20 min) and pre-amplification of extended PEA probes (or DNA reporter molecules) via universal primers (95°C, 5 min; (95°C, 30 s; 54°C, 1 min; and 60°C, 1 min) × 17.
The pre-amplified DNA reporter molecules from the multiplex detection reaction were decoded and quantified using a Fluidigm 96.96 Dynamic Array Integrated Fluidic Circuit on a Biomark HD system. 2.8 μl of each sample was mixed with 5 μl Detection Solution, 0.071 μl Detection Enzyme, and 0.028 μl PCR Polymerase (all Olink Bioscience) and 2.1 μl molecular grade water. 5 μl of each sample plus detection mix was loaded into a primed 96.96 Dynamic Array IFC (right inlets). 5 μl of each of the 96 primer pairs (Olink Bioscience), designed to amplify individual target-specific DNA reporter sequences generated in the PEA reactions, was also loaded in the Dynamic Array (left inlets). The chip was placed in Fluidigm’s IFC HX according to the manufacturer’s instructions and then run in Fluidigm’s Biomark with the following settings (Gene Expression application, ROX passive reference, single-probe assay with FAM-MGB probe) and protocol: thermal mix (50°C, 120 s; 70°C, 1,800 s; 25°C, 600 s), hot start (95°C, 300 s), and PCR cycling for 40 cycles (95°C, 15 s; 60°C, 60 s).
Cell Lysis
Cells lysates were prepared with the following lysis buffer in order to maximize recovery of both proteins and RNA: 1% NP-40, 0.1% Triton X-100, 0.1% sulfobetaine, 150 mM NaCl, 2U SUPERaseIN (Life Technologies), Protease Inhibitor Cocktail cOmplete mini (Roche), and Resuspension Buffer as supplied in the CellsDirect One-Step qRT-PCR Kit (Life Technologies).
Cell Lysates for Assay Validation
Cell lysates were prepared from the cell lines MCF-7 (ATCC HTB-22) and Hs578T (ATCC HTB-126), and from three early-passage glioblastoma cancer stem cell cultures from the Uppsala University Human Glioma Cell Culture biobank (U3013MG, U3017MG, and U3065MG; Xie et al., 2015). Briefly, defined numbers of cells were flow sorted directly into lysis buffer (Figure S1) or lysed in bulk at a concentration of 1,000 cells/μl. The lysates were aliquoted and kept frozen at −80°C until use. Lysates were diluted to contain the equivalent of 1,000, 100, 10, and 1 cell per aliquot. For PEA assays, controls included a mix of recombinant antigens representing a subset of antibody targets (n = 60) and lysis buffer alone to calculate assay-specific background (data not shown).
PEA Assay Validation
The cell lysates were screened with a total of 323 PEA assays against unique targets. Each PEA assay was assessed for sensitivity, cross-reactivity, and appropriate dose-response against cell lysates or recombinant antigens (Figure S1). Assays displaying single- or near-single-cell sensitivity, minimal variation in replicates, and no evidence of cross-reactivity were chosen for subsequent analysis of the U3035MG cells. The final panel included 79 PEA assays plus four spike-in controls. The Olink Bioscience spike-in controls included the two recombinant non-human proteins GFP and phycoerythrin (Protein.SpikeinProteinI_CTRL and Protein.SpikeinProteinII_CTRL, respectively) as incubation controls, an extension control, and a PCR or detection control (Assarsson et al., 2014). The extension control is included to control for intra-plate technical variation and was accordingly used to normalize the real-time PCR measurements. See Figure S1 for pooled cell dilutions from the final panel. The panel was annotated using the Molecular Signatures Database (MSigDB) using curated gene sets KEGG and REACTOME as well as GO gene sets (see Table S1 for details of the annotation).
Gene Expression Assay Validation
TaqMan Gene Expression assays were screened iteratively in multiplex, and assays displaying high signals in negative controls or low sensitivity in dilution series were excluded from subsequent testing. The final panel included 82 assays plus an External RNA Controls Consortium (ERCC) RNA spike-in control (Life Technologies) (Table S1). The ERCC spike-in was read out by a targeted TaqMan Expression assay (Table S1).
Cells for Analysis of BMP4 Treatment
The U3035MG cell line was isolated from a patient diagnosed with grade IV glioblastoma at the University Hospital, Uppsala, Sweden, in accordance with protocols approved by the regional ethical review board and following written consent from the patient. Tumor tissue was dissociated using a scalpel to first mince the tissue, followed by passage through a syringe with a 18G and 22G needle and incubation in a 1:1 mixture of Accutase (eBioscience) and TrypLE (Invitrogen) for 10 min at 37°C. The cells were washed twice in DMEM/F12 medium with centrifugation at 600 rpm for 8 min. The cells were cultured for 5–7 days as spheres on non-coated dishes and then transferred to laminin-coated dishes to establish adherent monolayers. The cells were grown in medium containing a 1:1 mix of Neurobasal (Invitrogen) and DMEM/F12 (Invitrogen) medium supplemented with N2 (Invitrogen), B27 (Invitrogen), 10 ng/ml EGF (Peprotech), and 10 ng/ml FGF-2 (Peprotech).
For BMP4 treatment experiments, U3035MG (passage 8) cells were seeded onto laminin-coated dishes at 1,000 cells/well in 24-well dishes, 3.3 × 104 cells/well in 60-mm dishes, or 1.5 × 104 cells/well in 35-mm dishes. Approximately 48 hr after seeding the cells, the cell media was changed and BMP4 (10 ng/μl) was added to the treatment wells. During the experiment, the cell media was changed and new BMP4 was added on days 2 and 5. The cells in the 35- or 60-mm dishes were used to set the gates for sorting, and those in the 24-well dishes were either sorted and analyzed or counted in triplicate using a hematocytometer (Neubauer improved cell counting chamber). For the flow cytometry confirmatory experiments, U3035MG cells were seeded 3.3 × 104 cells/well in 60-mm plates and treated with BMP4 as described above. Before cell dissociation and fixation on day 6, cells were pulsed with the proliferation marker EdU (10 μM, Life Technologies) for 1 hr.
Flow Cytometry and Cell Sorting
At the analyzed time points, single-cell suspensions of control and BMP4-treated cells were prepared for sorting by FACS. Cells were washed once with PBS and then dissociated from the culture dish using StemPro Accutase (Life Technologies) for maximally 5 min at 37°C. The dissociation agent was diluted using cold PBS; the cells were centrifuged for 5 min at 1,000 rpm and then gently resuspended in 400 μl cold PBS and sorted within 1 hr. Cells were flow sorted into 96-well plates containing 2 μl lysis using the BD FACSAria III. Two population controls of 150 cells/well were included on each plate. Following sorting, plates were centrifuged, snap frozen, and stored at −80°C until processing.
For the flow cytometry confirmation experiments, cells were fixed with 4% paraformaldehyde (PFA) and then blocked and permeabilized with a 5% fetal bovine serum (FBS)/0.1% saponin/PBS solution for 30 min at room temperature (RT). Cells were then probed with either a rabbit anti-TNC (Abcam ab108930) or a rabbit anti-TUBB3 (Sigma, T2200) for 30 min at RT, washed, and then probed with a donkey anti-rabbit Alexa 488 secondary antibody (Invitrogen A21206) for 30 min at RT. As a negative control, cells were probed with only the anti-rabbit Alexa 488 secondary antibody. Incorporation of EdU was resolved following the manufacturer’s protocol. Following a wash step, the cells were resuspended in a 0.5% BSA/4 mM EDTA/PBS solution. Cells were subsequently analyzed on the BD FACSAria III.
Coefficient of Variation Analysis
To assess the precision with which proteins and transcripts could be measured in lysates from half-cells, we sorted 40 single U3035MG, MCF-7, or Hs587T cells for protein analysis and another 40 for RNA analysis. For each single cell, we measured both halves either using TaqMan assays or PEA assays. The CV% for each assay and single cell was calculated using the mean and SD for each assay for each of the two halves of the 40 single cells. In Figure 1C, the CV% was plotted for each PEA assay as a function of the number of cells in which the PEA assay generated a detectable signal. The cells used for this analysis were different from the cells sorted and analyzed for BMP4 response.
cDNA Synthesis and Single-Cell qPCR
Thawed cell lysate plates were gently vortexed and centrifuged. 1 μl cell lysate was transferred to 19 μl RT-PCR mix. The mix contained 1× Cells Direct Reaction Mix and 0.5 μl Superscript III/Platinum Taq from the CellsDirect One-Step qRT-PCR Kit, 2U SUPERaseIN (Life Technologies), 5 μl 0.2× TaqMan Gene Expression assay mix, and TE buffer (pH 8.0) (Life Technologies). cDNA was synthesized at 50°C for 15 min, heated to 70°C for 2 min, and then incubated through 16 cycles of 95°C for 15 s and 60°C for 2 min. The assay mix contained pre-mixed TaqMan Gene Expression Assays (n = 89) plus the ERCC RNA Spike-in Mix (dilution 1:1,000; Ambion). Amplified cDNA was diluted 1:3 with TE buffer before qPCR analysis on the Fluidigm BioMark HD System in a 96.96 Dynamic Array IFC, following the manufacturer’s instructions. After removal of 1 μl cell lysate for RNA expression analysis, 3 μl PEA probe mix was added to the remaining 1 μl half-cell lysate and PEA was performed as described above.
Data QC
The RT-PCR output files from RNA assay determinations in the Fluidigm BioMark microfluidic real-time PCR instrument were processed to eliminate (1) data points labeled as failed by Fluidigm’s Real-Time PCR analysis software, (2) unreliable negative control rows with at least one non-failed value for a non-ERCC assay, and (3) unreliable rows for which the value of the ERCC controls was less than the lower inner fence value (Q1 − 1.5∗IQR) or greater than the upper inner fence value (Q3 + 1.5∗IQR) of the ERCC distribution (0-hr control, n = 2 cells; day-6 control, n = 12 cells; day-6 BMP4, n = 3 cells). The remaining Ct values were normalized as follows: for each sample (plate row), the Ct value of the ERCC was subtracted from the Ct values of the analytes (yielding dCt values), and the resulting values were subtracted from the difference between the LOD (set at 25) and the corresponding Ct value of the ERCC (yielding ddCt values). All ddCt values less than zero were set to zero, and the corresponding signals were deemed undetected.
Similarly, BioMark output files from protein determinations were processed to eliminate (1) data points labeled as Failed in the QC step of the run, (2) unreliable NEGCTRL rows containing a non-failed value in a blank assay well, and (3) unreliable rows for which the value of the extension control (ExtCtl) was smaller than the lower inner fence value (Q1 − 1.5∗IQR) or greater than the upper inner fence value (Q3 + 1.5∗IQR) of the ExtCtl distribution. The remaining Ct values were normalized as follows: for each sample (plate row), the Ct value of the ExtCtr was subtracted from the Ct value of the analyte (yielding dCt values), and for each assay (plate column), the median of the three inter-plate control (Assarsson et al., 2014) samples was subtracted from the resulting value (yielding ddCt values). The inter-plate control is used to minimize inter-plate variation for each individual assay. Then, for each assay, the ddCt values were subtracted from a negative control background value computed as the mean − 2∗SD of at least two NEGCTRL values. This ensures that observed signals for each assay in the presence of a cell are at least 2 SDs away from any signals observed in the absence of any antigen. All resulting values below zero were set to zero, and the signal was deemed undetected.
For both RNA and protein plates, single cells were culled by adding the resulting values across all assays, computing the median and SD of the resulting distribution, and eliminating single-cell samples for which the total values were smaller than the median − 3∗SD or greater than the median + 3∗SD. For the remaining single-cell samples, the normalized values for each column (i.e., analyte) were transformed into Z scores by subtracting the median and dividing by the SD.
Statistical Analysis
The combined Z scores for RNA and protein assays were used as input for a PCA performed using the FactoMineR package in R. Variables contributing to each of the first two components were selected based on the significance (p < 0.001) of their association with each component. Hierarchical clustering was performed in R using Euclidean distances and Ward’s clustering criterion.
For the correlation analysis, data were first filtered to include only matched RNA/protein pairs that fulfilled our QC criteria in a total of 130 cells (69 cells treated with BMP4 [+] and 62 untreated controls [−]). The data were subsequently organized into two matrices (rows = analytes, columns = cells) for RNA and for protein. The data were row-standardized by Z score transformation for each analyte. First, we estimated the first two principal components from the data. To classify treated (+) from untreated (−) cells, we used Linear Discriminant Analysis (LDA; James et al., 2013). In a leave-one-out cross-validation, we systematically excluded each of the 130 cells from the data and fitted an LDA model from the remaining 129 cells (MATLAB fitcdiscr function). From the posterior LDA score assigned to the excluded samples, we calculated an ROC curve in which x = 1 − specificity and y = sensitivity.
Author Contributions
S.D. and C.J.G. conceived of the study and performed the experiments; S.D., C.J.G., and V.D.M. conceived the PEA panel; S.F., E.A., and M.L. prepared the PEA conjugates; S.D., C.J.G., V.D.M., E.A., and M.L. performed the QC analysis; M.N., A.S., and B.W. contributed the glioma cell model; S.D., C.J.G., V.D.M., S.N., M.N., A.S., B.W., and U.L. analyzed the data; and S.D., C.J.G., and U.L. wrote the manuscript. All authors commented on the manuscript.
Conflicts of Interest
U.L. and S.F. are founders of and hold stock in Olink Bioscience.
Acknowledgments
We thank the BioVis Facility at Uppsala University for their help with the flow cytometry experiments. Our research has received support from the European Community’s 7th Framework Program (FP7/2007-2013) under grant agreement 259796 (DiaTools) (U.L.), the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC Grant Agreement 294409 (ProteinSeq) (U.L.), the Swedish Research Council (U.L. and S.N.), Knut and Alice Wallenberg Foundation (B.W.), the Swedish Cancer Society (B.W. and S.N.), the Inga Britt och Arne Lundbergs Forskningsstiftelse (U.L.), and the Swedish Society for Medical Research (S.D. and C.J.G.).
Published: December 31, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes four figures and three tables can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.12.021.
Supplemental Information
Specifically, the normalized protein and RNA expression data for cells measured at 0h and day 6 control or BMP4 treated.
In Figure 3B, we displayed only the top significant genes in the gene vector plot. Here, we provide the correlation coefficients for the first and second dimension of the PCA with the day 6 control and BMP4-treated cells for the full set that showed significance at p < 0.001.
References
- Assarsson E., Lundberg M., Holmquist G., Björkesten J., Thorsen S.B., Ekman D., Eriksson A., Rennel Dickens E., Ohlsson S., Edfeldt G. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE. 2014;9:e95192. doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., Rich J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Bendall S.C., Simonds E.F., Qiu P., Amir A.D., Krutzik P.O., Finck R., Bruggner R.V., Melamed R., Trejo A., Ornatsky O.I. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro M.S., Lim W.K., Alvarez M.J., Bollo R.J., Zhao X., Snyder E.Y., Sulman E.P., Anne S.L., Doetsch F., Colman H. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P., Kalisky T., Sahoo D., Rajendran P.S., Rothenberg M.E., Leyrat A.A., Sim S., Okamoto J., Johnston D.M., Qian D. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat. Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S., Nong R.Y., Hammond M., Gu J., Alderborn A., Vänelid J., Siegbahn A., Gustafsdottir S., Ericsson O., Landegren U., Kamali-Moghaddam M. Sensitive plasma protein analysis by microparticle-based proximity ligation assays. Mol. Cell. Proteomics. 2010;9:327–335. doi: 10.1074/mcp.M900248-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal R., Geissinger U., Zhang Q., Aguilar J., Chen N.G., Binda E., Vescovi A.L., Szalay A.A. Vaccinia virus expressing bone morphogenetic protein-4 in novel glioblastoma orthotopic models facilitates enhanced tumor regression and long-term survival. J. Transl. Med. 2013;11:155. doi: 10.1186/1479-5876-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcion E., Halilagic A., Faissner A., ffrench-Constant C. Generation of an environmental niche for neural stem cell development by the extracellular matrix molecule tenascin C. Development. 2004;131:3423–3432. doi: 10.1242/dev.01202. [DOI] [PubMed] [Google Scholar]
- Imamura T., Takase M., Nishihara A., Oeda E., Hanai J., Kawabata M., Miyazono K. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- Israelson A., Ditsworth D., Sun S., Song S., Liang J., Hruska-Plochan M., McAlonis-Downes M., Abu-Hamad S., Zoltsman G., Shani T. Macrophage migration inhibitory factor as a chaperone inhibiting accumulation of misfolded SOD1. Neuron. 2015;86:218–232. doi: 10.1016/j.neuron.2015.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James G., Witten D., Hastie T., Tibshirani R. Springer-Verlag; 2013. An Introduction to Statistical Learning with Applications in R, First Edition. [Google Scholar]
- Joshi K., Banasavadi-Siddegowda Y., Mo X., Kim S.H., Mao P., Kig C., Nardini D., Sobol R.W., Chow L.M., Kornblum H.I. MELK-dependent FOXM1 phosphorylation is essential for proliferation of glioma stem cells. Stem Cells. 2013;31:1051–1063. doi: 10.1002/stem.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic M., Rooney M.S., Mertins P., Przybylski D., Chevrier N., Satija R., Rodriguez E.H., Fields A.P., Schwartz S., Raychowdhury R. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science. 2015;347:1259038. doi: 10.1126/science.1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasorella A., Benezra R., Iavarone A. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat. Rev. Cancer. 2014;14:77–91. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- Macaulay I.C., Voet T. Single cell genomics: advances and future perspectives. PLoS Genet. 2014;10:e1004126. doi: 10.1371/journal.pgen.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H., Gale N.W., Guo H., Qian J., Petty A., Kaspar J., Murphy A.J., Valenzuela D.M., Yancopoulos G., Hambardzumyan D. EphA2 promotes infiltrative invasion of glioma stem cells in vivo through cross-talk with Akt and regulates stem cell properties. Oncogene. 2015;34:558–567. doi: 10.1038/onc.2013.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K., Miyazawa K. Id: a target of BMP signaling. Sci. STKE. 2002;2002:pe40. doi: 10.1126/stke.2002.151.pe40. [DOI] [PubMed] [Google Scholar]
- Munsky B., Neuert G., van Oudenaarden A. Using gene expression noise to understand gene regulation. Science. 2012;336:183–187. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D., Misra A., Nigro J.M., Colman H., Soroceanu L. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Piccirillo S.G., Reynolds B.A., Zanetti N., Lamorte G., Binda E., Broggi G., Brem H., Olivi A., Dimeco F., Vescovi A.L. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Raj A., van Oudenaarden A. Single-molecule approaches to stochastic gene expression. Annu. Rev. Biophys. 2009;38:255–270. doi: 10.1146/annurev.biophys.37.032807.125928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savary K., Caglayan D., Caja L., Tzavlaki K., Bin Nayeem S., Bergström T., Jiang Y., Uhrbom L., Forsberg-Nilsson K., Westermark B. Snail depletes the tumorigenic potential of glioblastoma. Oncogene. 2013;32:5409–5420. doi: 10.1038/onc.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Sottoriva A., Spiteri I., Piccirillo S.G., Touloumis A., Collins V.P., Marioni J.C., Curtis C., Watts C., Tavaré S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. USA. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhlberg A., Thomsen C., Ruff D., Åman P. Quantitative PCR analysis of DNA, RNAs, and proteins in the same single cell. Clin. Chem. 2012;58:1682–1691. doi: 10.1373/clinchem.2012.191445. [DOI] [PubMed] [Google Scholar]
- Tsukamoto S., Mizuta T., Fujimoto M., Ohte S., Osawa K., Miyamoto A., Yoneyama K., Murata E., Machiya A., Jimi E. Smad9 is a new type of transcriptional regulator in bone morphogenetic protein signaling. Sci. Rep. 2014;4:7596. doi: 10.1038/srep07596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullal A.V., Peterson V., Agasti S.S., Tuang S., Juric D., Castro C.M., Weissleder R. Cancer cell profiling by barcoding allows multiplexed protein analysis in fine-needle aspirates. Sci. Transl. Med. 2014;6:219ra9. doi: 10.1126/scitranslmed.3007361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videla Richardson G.A., Garcia C.P., Roisman A., Slavutsky I., Fernandez Espinosa D.D., Romorini L., Miriuka S.G., Arakaki N., Martinetto H., Scassa M.E., Sevlever G.E. Specific preferences in lineage choice and phenotypic plasticity of glioma stem cells under BMP4 and Noggin influence. Brain Pathol. 2015 doi: 10.1111/bpa.12263. Published online March 23, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield M.L., George L.K., Grant G.D., Perou C.M. Common markers of proliferation. Nat. Rev. Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- Xie Y., Bergström T., Jiang Y., Johansson P., Marinescu V.D., Lindberg N., Segerman A., Wicher G., Niklasson M., Baskaran S. The Human Glioblastoma Cell Culture resource: validated cell models representing all molecular subtypes. EBioMedicine. 2015;2:1351–1363. doi: 10.1016/j.ebiom.2015.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Zhou J., Sutherland A., Wei W., Shin Y.S., Xue M., Heath J.R. Microfluidics-based single-cell functional proteomics for fundamental and applied biomedical applications. Annu. Rev. Anal. Chem. (Palo Alto, Calif.) 2014;7:275–295. doi: 10.1146/annurev-anchem-071213-020323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specifically, the normalized protein and RNA expression data for cells measured at 0h and day 6 control or BMP4 treated.
In Figure 3B, we displayed only the top significant genes in the gene vector plot. Here, we provide the correlation coefficients for the first and second dimension of the PCA with the day 6 control and BMP4-treated cells for the full set that showed significance at p < 0.001.