Abstract
Several disease-monitoring techniques are available for the physician treating acute myeloid leukaemia (AML). Besides immunohistochemistry assisted light microscopy, the past 20 years have seen the development and preclinical perfection of a number of techniques, most notably quantitative polymerase chain reaction (PCR) and multicolor flow cytometry. Late additions to the group of applicable assays include next generation sequencing and digital PCR. In this review the principles of use of these modalities at three different time points during the AML disease course are discussed, namely at the time of treatment evaluation, pretransplantation and postconsolidation. The drawbacks and pitfalls of each different technique are delineated. The evidence or lack of evidence for minimal residual disease guided treatment decisions is discussed. Lastly, future strategies in the MRD field are suggested and commented upon.
Keywords: Minimal residual disease, Preemotive treatment, AML, flow cytometry, qPCR, NGS
Introduction
Upon diagnosis, a patient with acute myeloid leukaemia (AML) faces the ordeal of his or her life. Several courses of intensive chemotherapy, complicating infections, and possibly allogeneic bone marrow transplantation awaits, and even then the prognosis is no better than a 5-year survival of 50% even in young adults [Burnett et al. 2013]. For the hematologist aiding the patient through his or her disease course, evidence supports the choice of front-line induction therapy but not much else. In most centers, for the few patients with an especially good prognosis [i.e. core binding factor leukaemias (CBFs), RUNX1–RUNX1T1 and CBFB–MYH11 positive AMLs] consolidation treatment does not include allogeneic bone marrow transplantation. The somewhat more common patients with an especially bad prognosis invariably are considered candidates for transplantation, at least if comorbidities and donor availability permits. Treatment besides this, such as the optimal consolidation treatment for the large majority of intermediate risk patients or the correct treatment upon refractory disease or relapse, still very much depends on the treating hematologist [Cornelissen et al. 2012; Dohner et al. 2010].
Aiding the hematologist in these decisions are a number of prognostic scoring systems, including leukaemia- and patient-specific data, that is, the European Leukaemia Net (ELN) prognostic system [Dohner et al. 2010] the revised Grimwade criteria [Grimwade et al. 2010], and the Wheatley score of the UK Medical Research Council (MRC) program [Wheatley et al. 1999], to name a few. To different degrees, these systems include the rapidly growing amount of data on the different molecular phenotypes of AML. More sophisticated systems will probably emerge in the coming years as prognostic values of the multitudes of different genetic aberration combinations recently discovered can be included.
However, presently, these systems cannot foresee the treatment response at the level of the individual patient with AML. The sheer complexity of both the genetic (and epigenetic) background of the leukaemia as well as of the patient preclude that we anytime soon will be able to predict this.
Thus, during and after treatment, patients are monitored to observe the treatment responses. In this discipline also, a rapidly expanding set of options is available. The choice between different disease-monitoring techniques and the possibilities and caveats of these in the treatment and follow up of AML are the topics of this review. Acute promyelocytic leukaemia presents a special case that will not be covered by this review.
Disease surveillance in the genomic age
Since 1961 [Freireich et al. 1961], the achievement of the state where less than 5% of bone marrow blasts present by morphological examination by light microscopy [morphological complete remission (CR)] has been considered to be of key importance in the long-term prognostication of patients with AML. Since then, technical development has produced a number of advanced techniques to further assess the presence of residual disease. In the 1980s and early 1990s, the term minimal residual disease (MRD) was coined to distinguish residual disease as assessed morphologically from that detected with more sensitive techniques in patients in CR [Campana and Pui, 1995]. Thus, any technique with a sensitivity beyond that of (immunohistochemistry-assisted) light microscopy can in the traditional definition be used as an MRD detection technique. This dichotomy in disease status as evaluable either by standard methods or as MRD is reflected in the current consensus response criteria report from 2003 [Cheson et al. 2003] in which molecular response is included as a provisional response criterion only [Cheson et al. 2003]. With the steadily growing body of evidence that MRD measurements have prognostic value in AML and with trials incorporating MRD measurements in decision making slowly emerging, we advocate the merging of the concepts of morphological residual disease and MRD into the concept of measureable residual disease to reflect the joining of the MRD concept into standard disease surveillance practice [Hokland et al. 2015]. Several studies (e.g. the MRC AML17 and AML19 studies) are currently investigating the beneficial role of MRD in disease surveillance in a large randomized manner to provide definitive evidence for this transition.
The current use of MRD measurements reflects its auxiliary status compared with standard morphology. MRD measurements are most useful at time points when the increased sensitivity compared with light microscopy is exploited to the highest degree. These include when the leukemic clone is disappearing, such as during treatment; when it is imperative that the leukemic burden is low, such as before allogeneic treatment; or when the leukaemia is reappearing after either chemotherapy or transplantation consolidation.
Treatment efficacy evaluation
As the aim of cytoreductive treatment is to eradicate the leukemic clone, it is natural to test the efficacy of this treatment by MRD detection techniques. In the early 2000s a large number of single-center studies confirmed the prognostic value of a low MRD measurement during treatment, either in a selected cohort based on single molecular aberrations [Schnittger et al. 2003, 2007; Stentoft et al. 2006] or using more broadly applicable MRD markers [Ommen et al. 2008; Cilloni et al. 2008, 2009]. In the last 5–8 years the results of a number of pivotal multicenter studies have been published as well [Freeman et al. 2013; Terwijn et al. 2013; Kronke et al. 2011]. While some studies identify a threshold below which relapse risk is low, most commonly 0.1% or 1%, others were powered to show the linear relationship between declining MRD levels and prognosis. Also controversial is the optimal time point for MRD testing. While testing after the first course of chemotherapy is most convenient if intensification of treatment is necessary, data from the HOVON group suggest that some patients, in CR but MRD positive after the first course of chemotherapy, actually achieve a sufficient MRD decrease after the second course of chemotherapy, allowing the inclusion of these patients in the MRD-based low-risk arm [Terwijn et al. 2013]. Further studies will show if this benefit makes up for the obvious risks of administering an additional course of standard chemotherapy to the high-risk patients when prompt treatment intensification could be advocated.
Postchemotherapy early relapse detection
One of the most unsatisfying aspects of disease surveillance based on standard methods is in the follow up of patients after completion of chemotherapy. Patients can be well, with normal hematological counts, and 14 days later they can display signs of relapse with deteriorating bone marrow function. It is unsurprising that longitudinal follow up of patients postchemotherapy with the prospect of early relapse detection was an early venue of the MRD technology. Several points have been made in the last 15 years of testing and perfecting the techniques.
First, evidence suggests that above a certain threshold of MRD positivity, relapse is certain to occur [Ommen et al. 2010; Yin et al. 2012]. This condition is commonly referred to as molecular relapse. Second, surveillance can be done substituting bone marrow aspirations for blood samples, at least in some cases [Cilloni et al. 2008; Yin et al. 2012; Ommen et al. 2010, 2014; Abildgaard et al. 2013]. Third, diligence is required in post-treatment sampling, as many relapses will occur too fast if patients are only occasionally sampled [Ommen et al. 2008, 2010, 2014, 2015; Weisser et al. 2005]. Taken together, early relapse diagnostics is now possible, but evidence is still scarce on how to act upon molecular relapse.
MRD in the allotransplant setting
The use of MRD measurements in the allotransplant setting is a special case. Several studies report the adverse effect of MRD positivity prior to receiving the transplant [Jacobsohn et al. 2009; Walter et al. 2011; Rossi et al. 2015]. These patients are invariably patients who have recently been heavily treated with chemotherapy. Thus, this is a group of patients with suboptimal response to chemotherapy, similar to that noted in ordinary treatment response evaluation. In contrast to treating these patients using chemotherapy alone [Terwijn et al. 2013], allogeneic transplantation has the potential to save 15–20% of these patients [Terwijn et al. 2013]. Based on this, MRD positivity will not preclude transplantation, but could in the future result in the implementation of additional measures reserved for high-risk patients.
In the post-transplant period, the use of MRD measurements is similar to that in patients who have completed chemotherapy consolidation. A couple of important differences are important to note. The cohort of higher-risk patients combined with the delayed onset of the graft versus leukaemia (GvL) effect means that those patients who do relapse, relapse earlier than patients who have not received transplantation [Goswami et al. 2015]. Also, the reconstitution of the donor immune system in the new host and the instigation of the GvL effect fundamentally change the conditions under which the early relapse occurs. At least theoretically, the enhanced immune surveillance exercised by the transplanted immune system could change the relapse pattern, rendering the relapses of two molecularly similar leukaemias different in a transplanted and a nontransplanted host.
Methods
Polymerase chain reaction
The exponential nature of the polymerase chain reaction (PCR) techniques allows for very high sensitivities making these techniques ideal for MRD measurements, especially upon the introduction of the quantitative PCR systems [Kubista et al. 2006]. Recently, development of microfluidics-based systems, such as digital PCR, has allowed for the development of assays with a sensitivity that is even higher (up to 10 fold) than that of the traditional Taqman probe (or similar) based quantitative PCR (qPCR) systems [Hindson et al. 2013]. Additionally, qPCR systems offer relatively easy standardization, and standardized clinically tested assays [Kronke et al. 2011] exist for the commonest fusion transcripts [Gabert et al. 2003], mutated genes and overexpressed genes [Cilloni et al. 2009].
Fusion transcripts
In AMLs harboring chromosomal translocations such as t(8;21), inv(16), t(6;9), t(9;11) and others, the unique fusion transcripts arising from these translocations (RUNX1–RUNX1T1, CBFB–MYH11, DEK–NUP214, MLL–MLLT3) can be used as leukaemia-specific disease markers [Gabert et al. 2003; Ostergaard et al. 2004a; Tobal et al. 2004]. Unfortunately, only about 30% of young adults and older adults with AML harbor translocations [Grimwade and Freeman, 2014] and even the commonest fusion transcripts, RUNX1–RUNX1T1 and CBFB–MYH11 are not seen in more than 5% of cases in population-based surveys [Grimwade and Freeman, 2014]. As such, a large panel of primer-probe sets is necessary to cover only a small fraction of patients with AML, at least in adult AML. For childhood AML, CBF leukaemias, but especially the MLL (HUGO gene) translocations, are commoner, allowing for a coverage of about 51% using fusion transcripts alone [Grimwade and Freeman, 2014]. Fusion-based MRD measurements are among the most sensitive methods available. The sensitivity varies depending on the average number of fusion transcripts per leukemic cell, but often reaches 1 × 10−5 [Gabert et al. 2003].
Mutated genes
Specific qPCR reaction can be constructed to create MRD measurement assays that have a much broader applicability than that of fusion transcript based MRD assays, as all or virtually all AMLs harbor some kind of genetic mutation. However, only in some genes (e.g. NPM1, to some extent DNMT3A) do mutational hotspots exist to allow for the detection of the mutant allele without having to design patient-specific primer and probe sets [Gorello et al. 2006]. In most settings, the additional human capital required to produce patient-specific primer and probe sets is not available. Thus, other commonly mutated genes (e.g. RUNX1, TET2) that require the use of patient-specific primer and probe sets as no mutational hotspots exist are rarely used as MRD markers using qPCR-based techniques.
Overexpressed genes
Fusion transcripts and useful mutated genes do not exist in all AMLs, especially not in childhood AML in which the NPM1 mutation is rare [Grimwade and Freeman, 2014]. Thus, much effort has been put into developing additional assays to cover these remaining patients. This need is further increased by the fact that the commonest fusion transcripts, RUNX1–RUNX1T1, CBFB–MYH11, as well as NPM1 all confer a favorable prognosis to patients, signifying that relapsed or resistant disease actually is rarer in these patients. For patients who have the greatest need for disease surveillance, those at greatest risk of resistant disease or relapse, a far lower fraction of patients can actually be followed by a fusion transcript or mutated gene based MRD marker.
In the transformed homeostasis of the leukemic cell, several mRNA transcripts are expressed to a greater extent than in the healthy bone marrow cell. In diagnosis samples from patients with AML, one or more of these overexpressed genes will in the large majority of cases be several fold higher than in healthy bone marrow (Table 1).
Table 1.
The use of overexpressed genes as MRD markers.
| Gene name | Sensitivity | Applicability (%) |
|---|---|---|
| Wilms tumor gene 1, WT1 | 1:50* | 56 |
| 1:100$ | 23 | |
| 1:200$ | 7 | |
| Preferentially expressed antigen in melanoma, PRAME | 1:50* | 33 |
| 1:100‡ | 26 | |
| Steinbach 7 gene set | 1:30‡ | 74 |
| 1:100‡ | 59 | |
| Goswami 5 gene set | 1:50* | 87 |
Adult AML, compared with median normal expression [Goswami et al. 2015].
Adult AML, compared with 95% upper normal limit [Hokland et al. 2012].
Childhood AML, compared with 90% upper normal limit [Steinbach et al. 2014].
AML, acute myeloid leukaemia; MRD, minimal residual disease.
The downside of using overexpressed genes as MRD markers is that the level of expression of the marker in healthy hematopoiesis has to be taken into account as well. Thus, even for a marker that is upregulated in virtually all cases of AML, the need for 50- or 100-fold upregulation to obtain sufficient sensitivity results in a restriction in the number of patients who can be evaluated. The classical example is the WT1 gene that, even if overexpressed in up to 80% of patients [Ostergaard et al. 2004b], only offers 100-fold upregulation in 23% of adult patients [Hokland et al. 2012] and if this level of overexpression is required, can only be used in that fraction of patients. In the postconsolidation follow-up situation, expression in healthy tissues presents a special problem. One can either establish a safe threshold above which expression is not seen in healthy tissues [Ostergaard et al. 2004; Cilloni and Saglio, 2004] or closely interpret the changes in expression, when a dramatic rise in transcript level will signify an impending relapse [Kristensen et al. 2011]. Most commonly, the two methods are used in combination [Ommen et al. 2008].
With some effort and combination, good gene sets can be constructed to cover almost all leukaemias, allowing overexpressed genes to be used as a universal MRD detection system [Goswami et al. 2015; Steinbach et al. 2006, 2014]. Another possibility is to use the best overexpressed genes (such as WT1) in cases when none of the other PCR-based MRD detection methods (fusion transcripts, mutated genes) are applicable to create an MRD detection system including all three types of MRD markers, possibly supplemented by flow cytometry when especially sensitive flow cytometric markers are present.
The most commonly used genes as well as two published sets of genes and the sensitivities that can be achieved using these are summarized in Table 1.
Flow cytometry
Flow cytometry becomes an obvious MRD measuring tool based on the high-throughput nature of the technique and the possibility for very exact cell characterization offered by multicolor protocols. The principle is to identify leukaemia-associated phenotypes (LAPs) when leukemic cells differ from the large majority of healthy hematopoiesis bone marrow or blood cells. The differing expression of different LAPs can be divided into four different types: cross-lineage expression, when T-cell, B-cell or natural killer cell markers are expressed on the myeloid blasts; overexpression, when normally expressed antigens are expressed to a higher degree on each individual cell; lack of expression, when normally expression antigens are lacking; or finally asynchronous expression, when immature and mature myeloid antigens are expressed together in an aberrant manner [Kern et al. 2010]. Finding a LAP to all AMLs requires an extensive panel of antibodies [Kern et al. 2010], but 80–90% of patients can be followed using a reasonably sized panel [Freeman et al. 2013; Terwijn et al. 2013]. The most common antigens included in LAPs and the frequencies of their possible use are shown in Table 2.
Table 2.
Antigen basis for LAPs in AML. Applicability and detection thresholds in table from Freeman et al. [2013].
| LAP type | Main aberrant antigens | Detection threshold* | Applicability (%) |
|---|---|---|---|
| Cross lineage | CD7 | 0.02–0.1% | 22 |
| CD56 | 0.02–0.1% | 24 | |
| CD19 | 0.02–0.1% | <5$ | |
| Asynchronous | CD13/CD33 | 0.1% | 27 |
| CD38/CD33 | 0.1% | 14 | |
| Overexpression | CD34 | 0.1% | 15 |
| Lack of expression | HLA-DR | 0.02–0.1% | 30 |
LAP only used if present in over 10% of blasts. As at least 20% of blasts are present at diagnosis, a detection threshold of 0.1% compares to a sensitivity of at least 1:20, but often more than 1:100.
Often associated with presence of RUNX1-RUNX1T1 [Gustafson et al. 2009].
AML, acute myeloid leukaemia; LAP, leukaemia-associated phenotype.
LAP selection: immature and stem cell markers
Flow cytometric based MRD detection uses the same principles as overexpressed gene MRD detection and the sensitivity is restricted by the presence of normal cells in the LAP. However, in flow cytometry, the MRD analyst has a lot more control of which cells are included in the comparison to the malignant phenotype than in PCRs when cells are lyzed in the nucleotide (DNA or RNA) purification process. As immature cells are uncommon in healthy bone marrow, the addition of an immature marker to the LAP usually results in normal cell pollution below 0.05%, effectively eliminating the problem [Terwijn et al. 2013].
In many cases surface antigen expression on the leukemic clone varies, probably depending on different states of maturation of the individual clonal cells. Thus, a given LAP will often not comprise all leukemic cells. This raises the problem of whether the cells selected actually have the potential to give rise to a relapse. To amend this, the inclusion of stem cell markers in the LAPs has been proposed [Larsen et al. 2012; Van Rhenen et al. 2007]. This may hamper theoretical sensitivity as stem cells in the leukemic clone are uncommon, but has been shown to perform as well as MRD assays based on standard LAPs or WT1 overexpression [Roug et al. 2013].
Next generation sequencing
Next generation sequencing (NGS) offers the opportunity for detection and follow up of a large number of aberrations. This allows not only classic MRD follow up but also the inclusion of potentially important changes on the subclonal level. As virtually all AMLs harbor genetic mutations and mutational hotspots are not necessary for NGS-based MRD measurements, NGS could potentially be applicable in all AMLs. There is a caveat, however. The current base error rate means that the sensitivity level is about 1% [Kohlmann et al. 2014; Luthra et al. 2014], which cannot compete with any of the above-mentioned MRD measurements techniques, save perhaps the worst overexpressed gene-based assays. Technical development is currently trying to circumvent this [Hokland et al. 2015] and when these endeavors are successful, NGS-based MRD measurements could be a very powerful tool indeed.
Pitfalls in MRD assessment
Phenotype shifts
The dominating malignant clone at diagnosis is often genetically unstable and some genetic evolution will invariantly occur during disease progression. Furthermore, subclone selection by treatment is unavoidable, allowing clones that are only present in small proportions at diagnosis to dominate at relapse. The frequency of phenotype shifts differs substantially between different MRD targets. It is virtually unknown for fusion transcript and NPM1 based MRD detection to display phenotype shifts, whereas other mutations such as FLT3-ITD and WT1 mutation disappears or is added at relapse in up to 20% of cases [Nyvold et al. 2006; Bachas et al. 2010]. The recent paper by Lindsey and colleagues offers an explanation for this [Lindsey et al. 2015]. The authors divide AML mutations into MDS related (e.g. ASXL1, SRSF2), pan-AML (e.g. RUNX1, CEBPA, TET2, WT1, FLT3) and de novo AML related (e.g. NPM1, CBF leukaemia translocations, MLL translocation). De novo AMLs contain relatively few mutations and these mark the leukemogenic events in these patients, whereas pan-AML mutations can occur both as leukomogenic and secondary events, making these markers more prone to loss because of alternative clone outgrowth. This is not to say that a mutation classified by Lindsey and colleagues as pan-AML cannot be a very useful MRD marker, merely to emphasize the power of this model to explain the findings that phenotype shifts are more common for some mutations than for others.
As immunophenotypic markers are not part of the malignant process but rather a product of the disturbed cell homeostasis, these markers are among those most commonly lost [Feller et al. 2004; Voskova et al. 2004]. This can be amended following patients using several markers to lessen the risk of them all disappearing at once. However, this will increase the need for broader antibody panels and the cost of the analysis. Stem cell marker based MRD follow up has been reported to be stable at relapse [Roug et al. 2013] and another possibility is to include some of these markers in the LAPs.
Detection of premalignant clones
The opposite of the risk of using mutations not present in the relapsing subclone as MRD marker is using a marker that is present in the premalignant lesion of the patient.
Thus, Ploen and colleagues found mutated DNMT3A to be present in the leukemic cells of a patient with AML, but also to be continually present even after eradication of the leukemic clone [Ploen et al. 2014]. The DNMT3A mutation in the described case is probably a marker of monoclonal hematopoiesis of unknown significance, as recently described [Jaiswal et al. 2014; Genovese et al. 2014]. DNMT3A is a commonly seen mutation in this context, as is TET2. Diligence is advised if these MRD targets are used as MRD markers to ensure that recurring MRD positivity actually represents recurrence of the leukemic clone.
Post-treatment positivity
Another observation is that when testing patients postconsolidation not all MRD-positive patients relapse [Yin et al. 2012; Ommen et al. 2010, 2014, 2015]. There seems to be a certain threshold below which relapse is not certain to occur, both for fusion transcript based [Yin et al. 2012; Ommen et al. 2010, 2014, 2015], mutation based [Kronke et al. 2011] and flow cytometry based MRD detection [Terwijn et al. 2013]. This phenomenon could have several possible explanations. First, the PCR-based positivity could be simply due to detection of more mature cells not capable of producing a relapse. If so, stem cell based flow cytometric follow up should not display this feature. Further research will show whether this is the case. Second, the low level of positivity could represent a low level of residual leukaemia held in check by the immune system. The observation that the threshold for relapse is higher in transplanted patients [Ommen et al. 2014] could support this notion, but further research is needed before any firm conclusions can be drawn on this point.
Status on the current evidence for the use of MRD assessments in the clinic
How can MRD diagnostics improve the management of patients with AML in morphological CR? Consider a patient at high risk based on pretreatment factors, patient specific or leukaemia specific. If this patient experiences a very good molecular response to induction therapy should consolidation be deescalated and transplantation avoided? Or is it in such patients that transplantation does actually have a chance to provide a cure?
Consider the patient consolidated with chemotherapy experiencing a molecular relapse. Should preemptive treatment be started prior to frank relapse? Or should this life-threatening therapy be reserved for frank relapse?
A number of possible courses of action can be envisioned based on MRD measurements. These are summarized in Table 3. Each of these options will have to be tested in a randomized manner, and few of these studies have been published to date. Some evidence does exist, however, and this is described in the following section.
Table 3.
Possible MRD-directed actions.
| Response evaluation | Trial | Result | p |
|---|---|---|---|
| Treatment intensification, chemotherapy | MRC AML 18 | Pending | |
| Treatment intensification, allogeneic transplant | Zhu et al. [2013] | OS 72% in transplanted high-risk patients OS 27% in nontransplanted high-risk patients |
0.007 |
| Treatment de-escalation | Zhu et al. [2013] | OS 76% in transplanted low-risk patients OS 100% in nontransplanted low-risk patients |
0.013 |
| Molecular relapse | Trial | Result | |
| Wait and watch | AML17+ AML19 |
Pending | |
| Initiation of donor search | AML17+ AML19 |
Pending | |
| Demethylating agent | Sockel et al. [2011] | 7/10 response 3/10 reentered CR |
|
| Platzbecker et al. [2012] | 10/20 reentered CR 4/20 long term survivors |
||
| Leukaemia aberration specific treatment | Boissel et al. [2011] | 1/8 long-term survivors | |
| Discontinuation of immunosuppression | Doubek et al. [2009] | 1/3 long-term survivor | |
| Donor lymphocyte infusion | Doubek et al. [2009] | 3/3 response 1/3 long-term survivor |
|
| Pozzi et al. [2013] | OS 44% 17 DLI-treated patients versus 14% 21 non-DLI-treated patients | 0.004 | |
| Reduced dose standard chemotherapy | Doubek et al. [2009] | 3/15 response No long-term survivors |
|
| Standard chemotherapy | AML17+ AML19 |
Pending | |
| Direct allogeneic transplantation | none |
AML, acute myeloid leukaemia; CR, complete response; MRC, Medical Research Council; MRD, minimal residual disease; OS, overall survival.
Treatment intensification based on MRD detection
Rubnitz and colleagues performed a pivotal study in childhood AML in which risk stratification based on MRD analysis after the first course of chemotherapy determined treatment intensification in the form of an early second course of chemotherapy with the possible addition of gemtuzumab ozogamicin. Furthermore, consolidation was determined based both on MRD measurement and pretreatment risk factors. As is common in childhood AML, the study was not randomized between action upon MRD-directed treatment or not, but the authors concluded that the long-term survival rate is superior to most contemporary protocols, assigning this success to the MRD-directed therapy design [Rubnitz et al. 2010].
In adult AML, Zhu and colleagues performed a study in patients with CBF-positive AML. Whether patients received transplantation or not was based on achievement of MRD at the level of 1 × 10−3. In both the high-risk and low-risk groups, some patients opted not to follow the treatment decision based on the MRD determinations and the authors proved that the patients receiving treatment other than that assigned by the risk status did worse than those who completed their assigned treatment (see Table 3) [Zhu et al. 2013]. Naturally, a study like this is prone to selection bias, especially in the arm where patients did not receive transplantation despite their risk status recommending this treatment. The authors showed that the groups were comparable based on age, sex and comorbidities, but as always, in an unrandomized study, unknown biases are not accounted for.
Currently the UK MRC AML18 trial is recruiting. This trial contains true randomization between two different kinds of intensified treatment in patients who are in the MRD high-risk group and has the potential to further research on MRD-directed treatment intensification significantly.
Relapse kinetics
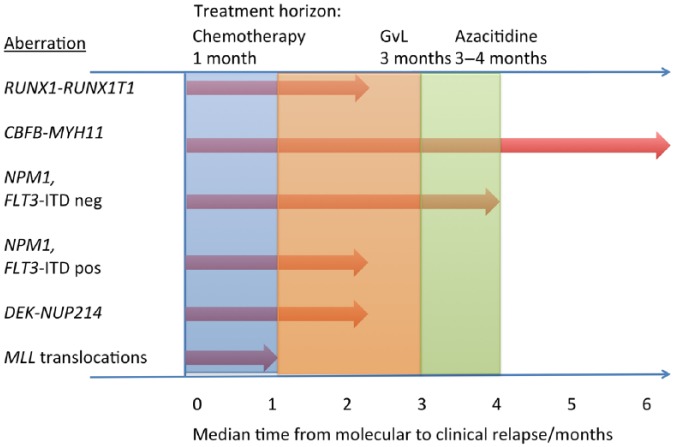
In the evaluation of MRD measurements in patients who no longer receive chemotherapy, the time factor in the possible reappearance of AML becomes a concern. In the case of a negative MRD measurement this indicates the patient is free of relapse in the near future, but since different AML subtypes relapse at very different relapse speeds [Yin et al. 2012; Ommen et al. 2010, 2014, 2015], this time period varies significantly for different AML subtypes. In Figure 1, the median times from molecular to clinical relapse can be seen for those subtypes for which data are published. In the case of a molecular relapse, the relapse kinetics data can aid in the decision on how often to control the relapsing disease if no preemptive treatment is chosen. If sufficient evidence to support preemptive treatment is produced in the future, it is conceivable that relapse kinetics data could be helpful in the choice between different treatment modalities (Figure 1).
Figure 1.
Relapse kinetics of different AML subtypes and relation to treatment onset.
Relapse kinetics from Ommen and colleagues (several papers) [Ommen et al. 2010, 2014, 2015] and Yin et al. [2012]. Time to treatment onset for azacitidine from Silverman et al. [2011]. Time to treatment onset for chemotherapy and GvL effect based on common clinical observations (disappearance of blasts after first course of chemotherapy, full chimerism often 3 months post transplantation). AML, acute myeloid leukaemia; GvL, graft versus leukaemia.
Preemptive treatment
Several smaller studies (see Table 3) have tested the value of preemptive treatment in patients experiencing a molecular relapse. In 2009, Doubek and colleagues published their experiences treating 21 molecular relapses of RUNX1–RUNX1T1, CBFB–MYH11 or MLL gene translocated AML with either gemtuzumab ozogamicin, 5 + 2 like chemotherapy or, in the case of patients who had undergone allogeneic transplantation, prompt discontinuation of immunosuppression or donor lymphocyte infusion (DLI) (see Table 3) [Doubek et al. 2009]. Responses were seen to all treatment modalities, but the treatments were too heterogeneous and the patient cohort too small to draw any firm conclusion. Patients who re-entered molecular CR invariably relapsed, but the duration of response was several hundred days in some cases.
In the venue of post-transplanted patients, Pozzi and colleagues were able to administer DLI in 17/38 patients with molecular relapse [Pozzi et al. 2013]. The survival was superior in the DLI group but the study was not truly randomized; the reason for not administering DLI included random factors such as donor unavailability but also treatment-related factors such as type of transplant (cord blood transplants could not receive DLI).
In another study of preemptive treatment in CBF-positive AML, seven of eight patients with relapsing disease treated with dasatinib upon molecular relapse progressed quickly to clinical relapse at a speed (median time from molecular relapse to clinical relapse, 60 days) indistinguishable from published median times from molecular to clinical relapse [Yin et al. 2012; Ommen et al. 2010; Boissel et al. 2011].
In conclusion, neither reduced dose chemotherapy nor tyrosine kinase inhibitor (TKI) seems an attractive choice for preemptive treatment.
Another possibility is a demethylating agent, even if this option is only attractive in patients in whom sufficient time is available from molecular to clinical relapse for the treatment effect of these agents to commence (see Figure 1). Sockel and colleagues administered azacitidine upon molecular relapse of patients with NPM1 mutation. In 7 of 10 patients, the authors observed a response, either as renewed molecular remission or as a delayed time to clinical relapse [Sockel et al. 2011]. As two-thirds of complete responses were seen in previously transplanted patients, the group initiated the RELAZA trial in which transplanted patients with molecular relapse were treated with azacitidine. Relapse was avoided in 4 of 20 patients but was delayed a median of 230 days in patients with relapsing disease [Platzbecker et al. 2012]. Thus, azacitidine seems promising; especially in the allogeneic transplant setting, but can probably only cure a minority of patients on its own. To follow up on this observation, the NOPHO group of pediatric oncologists are currently starting a study to test whether azacitidine can be useful as a bridging agent between a molecular relapse and an allogeneic transplantation.
Finally, in the UK MRC AML17 and AML19 studies, patients are randomized if they have a useful MRD marker to either MRD follow up or not. The endpoint is overall survival. If successful, this study will probably significantly alter the way we treat molecular relapse as preemptive treatment will have to be seriously considered in these cases.
Future directions in the evaluation of MRD diagnostics
Comparison of flow cytometry versus PCR versus NGS
The different methods of MRD detection offer different advantages and have different disadvantages. In the case of the choice of method in treatment evaluation, two large trials validated the value of flow cytometry [Freeman et al. 2013; Terwijn et al. 2013], but the use of RUNX1–RUNX1T1 and CBFB–MYH11 [Yin et al. 2012] and NPM1 [Kronke et al. 2011] has been validated as well. At treatment evaluation, phenotype shifts are only a lesser concern as selection and outgrowth of a secondary clone has not yet happened. Thus, the expensive procedures of using multiple LAPs requiring both expenditure of multiple antibodies and a significant amount of human capital become less important. The higher sensitivity of fusion transcript or mutated gene based PCR assays is not necessarily useful in the treatment assessment situation as some residual disease is probably acceptable, at least after the first course of chemotherapy [Terwijn et al. 2013]. In the patients without useful fusion transcripts or mutated genes, several works show that one overexpressed gene or a combination of these can be highly prognostically useful in treatment evaluation. Few reports have diligently tested the use of flow cytometry in the post-chemotherapy follow up and problems with phenotype shift are especially prevalent in this setting. However, at least with the current evidence, LAPs including stem cell markers do not seem marred by a great number of LAP losses and the problem with low-level positivity should be reduced using this method, even if evidence does not yet exist.
Flow cytometry and PCR measure two different things, namely malignant cells and mRNA expression in the malignant cells. Using immature or stem cell markers, flow cytometry has the potential to differentiate between cells of the malignant clone with or without relapse potential. However, PCR-based MRD measurement includes information on cell homeostasis as metabolically active cells containing larger amounts of the targeted RNA sequences are more easily detected. It is impossible to theoretically predict which of these two approaches discriminates best between risk groups. Future studies will show if one type of MRD measurement or the other is best at the different MRD measuring situations, but currently flow cytometry and the PCR-based method are probably equally useful at the treatment evaluation time point whereas PCR is probably superior in post-chemotherapy follow up.
Maintenance therapy in AML
In patients not fit for transplantation, the use of MRD measurements could provide the basis for intensification of therapy in the form of addition of maintenance therapy, currently mainly used in the acute lymphoblastic leukaemia (ALL) setting. Also, in frailer patients maintenance therapy could possibly replace intensive chemotherapy consolidation. Evidence for the use of MRD to direct this is currently sparse, but in patients achieving CR but who are not fit for additional intensive treatment, this could definitely be an option.
The drugs to use in this setting remain to be determined, but both demethylating agents and tyrosine kinase inhibitors with activity in AML could be likely candidates. In the case of the use of demethylating agents, follow up by MRD detection methods targeting methylations rather than DNA changes, gene overexpression or flow cytometry could be proposed, but further standardization and testing is awaited before implementation.
The inclusion of relapse kinetics
When evaluating preemptive treatment options, the nature of the relapsing AML should be kept in mind. In the majority of published results for patients treated with preemptive treatment, relapse was not avoided but delayed (Table 3). In some patients not fit for intensive reinduction or transplantation, or without a suitable donor, delaying relapse by several months could be a useful alternative to no treatment.
It is very probable that all molecular relapses should not be treated in the same way. Slower relapsing subtypes could be eligible for azacitidine or direct allogeneic transplantation. Others will have to be reinduced using intensive chemotherapy or experimental treatment by therapies directed at the molecular aberrations in the relapsing leukemic cells since not enough time is available for the initiation of the effect of the slower-acting treatment strategies.
The perfect as the enemy of the good
A large amount of literature exists to support the prognostic value of MRD measurements at several different locations. Current MRD detection techniques are highly sophisticated, allowing detection of minute amounts of residual AML cells. Our current technologies are probably highly sufficient; it is how we act upon a high MRD measurement that is uncertain. Evolving technologies such as NGS, digital PCR or other microfluidics-based assays or even cell-free DNA-based assays will probably refine our MRD measurements further, but in the enthusiasm of the current molecular revolution we should not forget our well tested assays for which the possible time to clinical implementation is shorter. If we use our current understanding of MRD and if we get evidence as to how to act upon MRD measurements, current technologies are fully adequate to help our patients.
Footnotes
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interest statement: The author declares that there is no conflict of interest.
References
- Abildgaard L., Ommen H., Lausen B., Hasle H., Nyvold C. (2013) A novel RT-qPCR assay for quantification of the MLL-MLLT3 fusion transcript in acute myeloid leukaemia. Eur J Haematol 91: 394–398. [DOI] [PubMed] [Google Scholar]
- Bachas C., Schuurhuis G., Hollink I., Kwidama Z., Goemans B., Zwaan C., et al. (2010) High-frequency type I/II mutational shifts between diagnosis and relapse are associated with outcome in pediatric AML: implications for personalized medicine. Blood 116: 2752–2758. [DOI] [PubMed] [Google Scholar]
- Boissel N., Jourdan E., Pigneux A., Blanchet O., Renneville A., Recher C., et al. (2011) Single-agent dasatinib does not prevent hematological relapse in patients with core binding factor (CBF) acute myeloid leukemia (AML) in first complete remission, but persistent or re-appearing molecular minimal residual disease-results of the DASA-CBF trial from the French AML intergroup. Blood 118: 1119. [Google Scholar]
- Burnett A., Russell N., Hills R., Hunter A., Kjeldsen L., Yin J., et al. (2013) Optimization of chemotherapy for younger patients with acute myeloid leukemia: results of the Medical Research Council AML15 trial. J Clin Oncol 31: 3360–3368. [DOI] [PubMed] [Google Scholar]
- Campana D., Pui C. (1995) Detection of minimal residual disease in acute leukemia: methodologic advances and clinical significance. Blood 85: 1416–1434. [PubMed] [Google Scholar]
- Cheson B., Bennett J., Kopecky K., Buchner T., Willman C., Estey E., et al. (2003) Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol 21: 4642–4649. [DOI] [PubMed] [Google Scholar]
- Cilloni D., Messa F., Arruga F., Defilippi I., Gottardi E., Fava M., et al. (2008) Early prediction of treatment outcome in acute myeloid leukemia by measurement of WT1 transcript levels in peripheral blood samples collected after chemotherapy. Haematologica 93: 921–924. [DOI] [PubMed] [Google Scholar]
- Cilloni D., Renneville A., Hermitte F., Hills R., Daly S., Jovanovic J., et al. (2009) Real-time quantitative polymerase chain reaction detection of minimal residual disease by standardized WT1 assay to enhance risk stratification in acute myeloid leukemia: a European LeukemiaNet study. J Clin Oncol 27: 5195–5201. [DOI] [PubMed] [Google Scholar]
- Cilloni D., Saglio G. (2004) WT1 as a universal marker for minimal residual disease detection and quantification in myeloid leukemias and in myelodysplastic syndrome. Acta Haematol 112: 79–84. [DOI] [PubMed] [Google Scholar]
- Cornelissen J., Gratwohl A., Schlenk R., Sierra J., Bornhauser M., Juliusson G., et al. (2012) The European LeukemiaNet AML Working Party consensus statement on allogeneic HSCT for patients with AML in remission: an integrated-risk adapted approach. Nat Rev Clin Oncol 9: 579–590. [DOI] [PubMed] [Google Scholar]
- Dohner H., Estey E., Amadori S., Appelbaum F., Buchner T., Burnett A., et al. (2010) Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 115: 453–474. [DOI] [PubMed] [Google Scholar]
- Doubek M., Palasek I., Pospisil Z., Borsky M., Klabusay M., Brychtova Y., et al. (2009) Detection and treatment of molecular relapse in acute myeloid leukemia with RUNX1 (AML1), CBFB, or MLL gene translocations: frequent quantitative monitoring of molecular markers in different compartments and correlation with WT1 gene expression. Exp Hematol 37: 659–672. [DOI] [PubMed] [Google Scholar]
- Feller N., van der Pol M., van Stijn A., Weijers G., Westra A., Evertse B., et al. (2004) MRD parameters using immunophenotypic detection methods are highly reliable in predicting survival in acute myeloid leukaemia. Leukemia 18: 1380–1390. [DOI] [PubMed] [Google Scholar]
- Freeman S., Virgo P., Couzens S., Grimwade D., Russell N., Hills R., et al. (2013) Prognostic relevance of treatment response measured by flow cytometric residual disease detection in older patients with acute myeloid leukemia. J Clin Oncol 31: 4123–4131. [DOI] [PubMed] [Google Scholar]
- Freireich E., Gehan E., Sulman D., Boggs D., Frei E., 3rd (1961) The effect of chemotherapy on acute leukemia in the human. J Chronic Dis 14: 593–608. [DOI] [PubMed] [Google Scholar]
- Gabert J., Beillard E., van der Velden V., Bi W., Grimwade D., Pallisgaard N., et al. (2003) Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia – a Europe Against Cancer program. Leukemia 17: 2318–2357. [DOI] [PubMed] [Google Scholar]
- Genovese G., Kahler A., Handsaker R., Lindberg J., Rose S., Bakhoum S., et al. (2014) Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371: 2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorello P., Cazzaniga G., Alberti F., Dell’Oro M., Gottardi E., Specchia G., et al. (2006) Quantitative assessment of minimal residual disease in acute myeloid leukemia carrying nucleophosmin (NPM1) gene mutations. Leukemia 20: 1103–1108. [DOI] [PubMed] [Google Scholar]
- Goswami M., McGowan K., Lu K., Jain N., Candia J., Hensel N., et al. (2015) A multigene array for measurable residual disease detection in AML patients undergoing SCT. Bone Marrow Transplant 50: 642–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwade D., Freeman S. (2014) Defining minimal residual disease in acute myeloid leukemia: which platforms are ready for ‘Prime Time’? Blood 124: 3345–3355. [DOI] [PubMed] [Google Scholar]
- Grimwade D., Hills R., Moorman A., Walker H., Chatters S., Goldstone A., et al. (2010) Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 116: 354–365. [DOI] [PubMed] [Google Scholar]
- Gustafson S., Lin P., Chen S., Chen L., Abruzzo L., Luthra R., et al. (2009) Therapy-related acute myeloid leukemia with t(8;21) (q22;q22) shares many features with de novo acute myeloid leukemia with t(8;21)(q22;q22) but does not have a favorable outcome. Am J Clin Pathol 131: 647–655. [DOI] [PubMed] [Google Scholar]
- Hindson C., Chevillet J., Briggs H., Gallichotte E., Ruf I., Hindson B., et al. (2013) Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 10: 1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokland P., Ommen H., Mule M., Hourigan C. (2015) Advancing the minimal residual disease concept in acute myeloid leukemia. Semin Hematol 52: 184–192. doi: 10.1053/j.seminhematol.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokland P., Ommen H., Nyvold C., Roug A. (2012) Sensitivity of minimal residual disease in acute myeloid leukaemia in first remission–methodologies in relation to their clinical situation. Br J Haematol 158: 569–580. [DOI] [PubMed] [Google Scholar]
- Jacobsohn D., Tse W., Chaleff S., Rademaker A., Duerst R., Olszewski M., et al. (2009) High WT1 gene expression before haematopoietic stem cell transplant in children with acute myeloid leukaemia predicts poor event-free survival. Br J Haematol 146: 669–674. [DOI] [PubMed] [Google Scholar]
- Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P., Mar B., et al. (2014) Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371: 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern W., Bacher U., Haferlach C., Schnittger S., Haferlach T. (2010) The role of multiparameter flow cytometry for disease monitoring in AML. Best Pract Res Clin Haematol 23: 379–390. [DOI] [PubMed] [Google Scholar]
- Kohlmann A., Nadarajah N., Alpermann T., Grossmann V., Schindela S., Dicker F., et al. (2014) Monitoring of residual disease by next-generation deep-sequencing of RUNX1 mutations can identify acute myeloid leukemia patients with resistant disease. Leukemia 28: 129–137. [DOI] [PubMed] [Google Scholar]
- Kristensen T., Moller M., Friis L., Bergmann O., Preiss B. (2011) NPM1 mutation is a stable marker for minimal residual disease monitoring in acute myeloid leukaemia patients with increased sensitivity compared to WT1 expression. Eur J Haematol 87: 400–408. [DOI] [PubMed] [Google Scholar]
- Kronke J., Schlenk R., Jensen K., Tschurtz F., Corbacioglu A., Gaidzik V., et al. (2011) Monitoring of minimal residual disease in NPM1-mutated acute myeloid leukemia: a study from the German-Austrian acute myeloid leukemia study group. J Clin Oncol 29: 2709–2716. [DOI] [PubMed] [Google Scholar]
- Kubista M., Andrade J., Bengtsson M., Forootan A., Jonak J., Lind K., et al. (2006) The real-time polymerase chain reaction. Mol Aspects Med 27: 95–125. [DOI] [PubMed] [Google Scholar]
- Larsen H., Roug A., Just T., Brown G., Hokland P. (2012) Expression of the hMICL in acute myeloid leukemia – a highly reliable disease marker at diagnosis and during follow-up. Cytometry B Clin Cytom 82: 3–8. [DOI] [PubMed] [Google Scholar]
- Lindsley R., Mar B., Mazzola E., Grauman P., Shareef S., Allen S., et al. (2015) Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 125: 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra R., Patel K., Reddy N., Haghshenas V., Routbort M., Harmon M., et al. (2014) Next-generation sequencing-based multigene mutational screening for acute myeloid leukemia using MiSeq: applicability for diagnostics and disease monitoring. Haematologica 99: 465–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyvold C., Stentoft J., Braendstrup K., Melsvik D., Moestrup S., Juhl-Christensen C., et al. (2006) Wilms’ tumor 1 mutation accumulated during therapy in acute myeloid leukemia: biological and clinical implications. Leukemia 20: 2051–2054. [DOI] [PubMed] [Google Scholar]
- Ommen H., Hokland P., Haferlach T., Abildgaard L., Alpermann T., Haferlach C., et al. (2014) Relapse kinetics in acute myeloid leukaemias with MLL translocations or partial tandem duplications within the MLL gene. Br J Haematol 165: 618–628. [DOI] [PubMed] [Google Scholar]
- Ommen H., Nyvold C., Braendstrup K., Andersen B., Ommen I., Hasle H., et al. (2008) Relapse prediction in acute myeloid leukaemia patients in complete remission using WT1 as a molecular marker: development of a mathematical model to predict time from molecular to clinical relapse and define optimal sampling intervals. Br J Haematol 141: 782–791. [DOI] [PubMed] [Google Scholar]
- Ommen H., Schnittger S., Jovanovic J., Ommen I., Hasle H., Ostergaard M., et al. (2010) Strikingly different molecular relapse kinetics in NPM1c, PML-RARA, RUNX1-RUNX1T1, and CBFB-MYH11 acute myeloid leukemias. Blood 115: 198–205. [DOI] [PubMed] [Google Scholar]
- Ommen H., Touzart A., MacIntyre E., Kern W., Haferlach T., Haferlach C., et al. (2015) The kinetics of relapse in DEK-NUP214-positive acute myeloid leukemia patients. Eur J Haematol 95: 436–441. [DOI] [PubMed] [Google Scholar]
- Ostergaard M., Stentoft J., Hokland P. (2004a) A real-time quantitative RT-PCR assay for monitoring DEK-CAN fusion transcripts arising from translocation t(6;9) in acute myeloid leukemia. Leuk Res 28: 1213–1215. [DOI] [PubMed] [Google Scholar]
- Ostergaard M., Olesen L., Hasle H., Kjeldsen E., Hokland P. (2004b) WT1 gene expression: an excellent tool for monitoring minimal residual disease in 70% of acute myeloid leukaemia patients – results from a single-centre study. Br J Haematol 125: 590–600. [DOI] [PubMed] [Google Scholar]
- Platzbecker U., Wermke M., Radke J., Oelschlaegel U., Seltmann F., Kiani A., et al. (2012) Azacitidine for treatment of imminent relapse in MDS or AML patients after allogeneic HSCT: results of the RELAZA trial. Leukemia 26: 381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploen G., Nederby L., Guldberg P., Hansen M., Ebbesen L., Jensen U., et al. (2014) Persistence of DNMT3A mutations at long-term remission in adult patients with AML. Br J Haematol 167: 478–486. doi: 10.1111/bjh.13062 [DOI] [PubMed] [Google Scholar]
- Pozzi S., Geroldi S., Tedone E., Luchetti S., Grasso R., Colombo N., et al. (2013) Leukaemia relapse after allogeneic transplants for acute myeloid leukaemia: predictive role of WT1 expression. Br J Haematol 160: 503–509. [DOI] [PubMed] [Google Scholar]
- Rossi G., Carella A., Minervini M., di Nardo F., Waure C., Greco M., et al. (2015) Optimal time-points for minimal residual disease monitoring change on the basis of the method used in patients with acute myeloid leukemia who underwent allogeneic stem cell transplantation: a comparison between multiparameter flow cytometry and Wilms’ tumor 1 expression. Leuk Res 39: 138–143. [DOI] [PubMed] [Google Scholar]
- Roug A., Larsen H., Nederby L., Just T., Brown G., Nyvold C., et al. (2013) hMICL and CD123 in combination with a CD45/CD34/CD117 backbone - a universal marker combination for the detection of minimal residual disease in acute myeloid leukaemia. Br J Haematol 164: 212–222. [DOI] [PubMed] [Google Scholar]
- Rubnitz J., Inaba H., Dahl G., Ribeiro R., Bowman W., Taub J., et al. (2010) Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: results of the AML02 multicentre trial. Lancet Oncol 11: 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger S., Kern W., Schoch C., Haferlach T. (2007) RT-PCR-based MRD detection in NPM1 mutated AML: a prospective follow-up study in 130 patients. 49th Annual Meeting of the American Society of Hematology, 8–11 December 2007 Blood 16: abstract 545. [Google Scholar]
- Schnittger S., Weisser M., Schoch C., Hiddemann W., Haferlach T., Kern W. (2003) New score predicting for prognosis in PML-RARA+, AML1-ETO+, or CBFBMYH11+ acute myeloid leukemia based on quantification of fusion transcripts. Blood 102: 2746–2755. [DOI] [PubMed] [Google Scholar]
- Silverman L., Fenaux P., Mufti G., Santini V., Hellstrom-Lindberg E., Gattermann N., et al. (2011) Continued azacitidine therapy beyond time of first response improves quality of response in patients with higher-risk myelodysplastic syndromes. Cancer 117: 2697–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sockel K., Wermke M., Radke J., Kiani A., Schaich M., Bornhauser M., et al. (2011) Minimal residual disease-directed preemptive treatment with azacitidine in patients with NPM1-mutant acute myeloid leukemia and molecular relapse. Haematologica 96: 1568–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach D., Bader P., Willasch A., Bartholomae S., Debatin K., Zimmermann M., et al. (2014) Prospective validation of a new method of monitoring minimal residual disease in childhood acute myeloid leukemia. Clin Cancer Res 21: 1353–1359. [DOI] [PubMed] [Google Scholar]
- Steinbach D., Schramm A., Eggert A., Onda M., Dawczynski K., Rump A., et al. (2006) Identification of a set of seven genes for the monitoring of minimal residual disease in pediatric acute myeloid leukemia. Clin Cancer Res 12: 2434–2441. [DOI] [PubMed] [Google Scholar]
- Stentoft J., Hokland P., Ostergaard M., Hasle H., Nyvold C. (2006) Minimal residual core binding factor AMLs by real time quantitative PCR–initial response to chemotherapy predicts event free survival and close monitoring of peripheral blood unravels the kinetics of relapse. Leuk Res 30: 389–395. [DOI] [PubMed] [Google Scholar]
- Terwijn M., van Putten W., Kelder A., van der Velden V., Brooimans R., Pabst T., et al. (2013) High prognostic impact of flow cytometric minimal residual disease detection in acute myeloid leukemia: data from the HOVON/SAKK AML 42A study. J Clin Oncol 31: 3889–3897. [DOI] [PubMed] [Google Scholar]
- Tobal K., Frost L., Liu Yin J. (2004) Quantification of DEK-CAN fusion transcript by real-time reverse transcription polymerase reaction in patients with t(6;9) acute myeloid leukemia. Haematologica 89: 1267–1269. [PubMed] [Google Scholar]
- Van Rhenen A., Moshaver B., Kelder A., Feller N., Nieuwint A., Zweegman S., et al. (2007) Aberrant marker expression patterns on the CD34+CD38- stem cell compartment in acute myeloid leukemia allows to distinguish the malignant from the normal stem cell compartment both at diagnosis and in remission. Leukemia 21: 1700–1707. [DOI] [PubMed] [Google Scholar]
- Voskova D., Schoch C., Schnittger S., Hiddemann W., Haferlach T., Kern W. (2004) Stability of leukemia-associated aberrant immunophenotypes in patients with acute myeloid leukemia between diagnosis and relapse: comparison with cytomorphologic, cytogenetic, and molecular genetic findings. Cytometry B Clin Cytom 62: 25–38. [DOI] [PubMed] [Google Scholar]
- Walter R., Gooley T., Wood B., Milano F., Fang M., Sorror M., et al. (2011) Impact of pretransplantation minimal residual disease, as detected by multiparametric flow cytometry, on outcome of myeloablative hematopoietic cell transplantation for acute myeloid leukemia. J Clin Oncol 29: 1190–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser M., Kern W., Rauhut S., Schoch C., Hiddemann W., Haferlach T., et al. (2005) Prognostic impact of RT-PCR-based quantification of WT1 gene expression during MRD monitoring of acute myeloid leukemia. Leukemia 19: 1416–1423. [DOI] [PubMed] [Google Scholar]
- Wheatley K., Burnett A., Goldstone A., Gray R., Hann I., Harrison C., et al. (1999) A simple, robust, validated and highly predictive index for the determination of risk-directed therapy in acute myeloid leukaemia derived from the MRC AML 10 trial. United Kingdom Medical Research Council’s Adult and Childhood Leukaemia Working Parties. Br J Haematol 107: 69–79. [DOI] [PubMed] [Google Scholar]
- Yin J., O’Brien M., Hills R., Daly S., Wheatley K., Burnett A. (2012) Minimal residual disease monitoring by quantitative RT-PCR in core binding factor AML allows risk stratification and predicts relapse: results of the United Kingdom MRC AML-15 trial. Blood 120: 2826–2835. [DOI] [PubMed] [Google Scholar]
- Zhu H., Zhang X., Qin Y., Liu D., Jiang H., Chen H., et al. (2013) MRD-directed risk stratification treatment may improve outcomes of t(8;21) AML in the first complete remission: results from the AML05 multicenter trial. Blood 121: 4056–4062. [DOI] [PubMed] [Google Scholar]