Abstract
The APC/C is the ubiquitin ligase that regulates mitosis by targeting specific proteins for degradation at specific times under the control of the Spindle Assembly Checkpoint (SAC). How the APC/C recognises its different substrates is a key problem in the control of cell division. Here, we have identified the ABBA motif in Cyclin A, BUBR1, BUB1 and Acm1, and show that it binds to the APC/C co-activator CDC20. The ABBA motif in Cyclin A is required for its proper degradation in prometaphase through competing with BUBR1 for the same site on CDC20. Moreover, the ABBA motifs in BUBR1 and BUB1 are necessary for the SAC to work at full strength and to recruit CDC20 to kinetochores. Thus, we have identified a conserved motif integral to the proper control of mitosis that connects APC/C substrate recognition with the SAC.
Introduction
Mitosis in eukaryotic cells is regulated by the activation of the mitotic cyclin-CDKs and their rapid inactivation through ubiquitin-mediated proteolysis (Pines, 2011). The ubiquitin ligase responsible for targeting the mitotic cyclins for degradation is the Anaphase Promoting Complex or Cyclosome (APC/C), which is activated in early mitosis by the CDC20 protein (Chang et al., 2014). CDC20 is regulated by the Spindle Assembly Checkpoint (SAC), which is activated by unattached kinetochores that catalyse the production of an inhibitor called the Mitotic Checkpoint Complex (MCC) (De Antoni et al., 2005; Lara-Gonzalez et al., 2012; Sudakin et al., 2001). CDC20 forms part of the MCC through binding to two SAC components, MAD2 and BUBR1/MAD3. The MCC structure from fission yeast shows that a Lys-Glu-Asn (KEN box) in the N-terminus of Mad3 binds as a pseudo-substrate inhibitor to the top face of the WD40 domain of CDC20 (Chao et al., 2012). Mad2 binds to the Lys-Ile-Leu-Arg (KILR) motif on CDC20 that is required to bind the APC/C (Izawa and Pines, 2012). Because CDC20 is inhibited, most APC/C substrates cannot be degraded while there are any unattached kinetochores; only when all chromosomes have attached to the mitotic spindle does MCC assembly stop, the MCC disassemble (Foster and Morgan, 2012; Mansfeld et al., 2011; Uzunova et al., 2012; Varetti et al., 2011) and CDC20 activate the APC/C. This is crucial to the proper segregation of chromosomes because it controls the time when Securin and Cyclin B1, which prevent sister chromatid separation and mitotic exit, respectively, can be recognised by the APC/C (Clute and Pines, 1999; Hagting et al., 2002).
Some APC/C substrates can be recognized while the SAC is active in prometaphase. These include Cyclin A (Elzen and Pines, 2001; Geley et al., 2001), whose degradation may be required to stabilize microtubule binding to kinetochores before anaphase (Kabeche and Compton, 2013); therefore, elucidating how Cyclin A is degraded while the SAC is active is important to understand proper chromosome segregation.
Some progress has been made in understanding how Cyclin A is degraded while the SAC is active. The N-terminus of Cyclin A competes with the MCC to bind directly to CDC20 (Di Fiore and Pines, 2010); subsequently the CDC20-Cyclin A-CDK-CKS complex binds to the APC/C through the CKS protein, thereby targeting Cyclin A for proteolysis (Wolthuis et al., 2008). But the mechanism by which Cyclin A competes with the MCC for CDC20 is not yet known.
Many cell cycle regulators are intrinsically disordered, some completely - e.g. Securin (Sánchez-Puig et al., 2005) and Acm1 (He et al., 2013), others contain large regions of intrinsic disorder - e.g. the BUB1 family (Bolanos-Garcia and Blundell, 2011) and the Cyclin family (Cox et al., 2002). The function of these disordered regions is frequently conferred by short linear motifs (SLiMs): compact, linear protein interaction sites that typically encode their major binding determinants in three to four key residues within a stretch of less than ten amino acids (Davey et al., 2012b; Diella et al., 2008; Dinkel et al., 2014; Neduva and Russell, 2006; Van Roey et al., 2012). Mitotic regulatory proteins extensively utilize SLiMs: the canonical RxL Cyclin recognition motif binds the hydrophobic patch of Cyclin A, thereby recruiting numerous substrates (Chen et al., 1995; Zhu et al., 1995); and the Destruction box (D-box) and KEN box degrons (Glotzer et al., 1991; Pfleger and Kirschner, 2000) are SLiMs that control protein stability by promoting APC/C dependent poly-ubiquitylation. Despite the importance of the known SLiMs in intrinsically disordered regions of cell cycle proteins, however, the vast majority of these regions remain uncharacterized, and we hypothesize that uncharacterised SLiMs remain to be discovered.
Here we have used bioinformatics to identify a SLiM that mediates binding between mitotic regulators and the APC/C co-activators CDC20 and CDH1. We have named it the ABBA motif because it is found in Cyclin A, BUBR1, BUB1 and Acm1, and we show that it is required for Cyclin A to be degraded correctly, and contributes to the strength of the SAC through BUBR1 and BUB1. Moreover, we show that the ABBA motifs in Cyclin A and BUBR1 bind to the same site on CDC20, which can explain how Cyclin A competes with the MCC to bind CDC20 and be degraded rapidly in mitosis.
Results
An APC/C activator-binding motifs is shared by Cyclin A, BUBR1, BUB1 and Acm1
Three major attributes can be used to identify SLiMs. Firstly, SLiMs are enriched in accessible and natively disordered regions of proteins; this permits the interacting peptide to fold to fit an interacting partner’s binding pocket (Fuxreiter et al., 2007; Sugase et al., 2007). Secondly, residues within SLiMs are generally more conserved than in flanking regions since amino acids in disordered regions are only conserved if they are functional (Davey et al., 2012b). Thirdly, SLiMs can evolve de novo to add a functional interface to a protein; as a result, SLiMs regularly recur in functionally related proteins (Davey et al., 2012b). Thus, strong candidates for previously unidentified SLiM classes can be discovered by searching regions of intrinsic disorder in functionally-associated proteins for comparatively over-conserved groupings of residues (Davey et al., 2012a; Nguyen Ba et al., 2012).
To discover mitotic SLiMs we applied a four stage computational protocol: (i) filter out inaccessible regions; (ii) apply the SLiMPrints tool (Davey et al., 2012a) to discover conserved groupings of residues indicative of a functional motif; (iii) apply the SLiMSearch tool (Davey et al., 2011) to discover repeated instances of the putative motif; and (iv) manually curate the literature for attractive hits. We searched 169 human proteins with the UniProt keyword “mitosis” (Supplementary Table 1), and constructed ortholog alignments for each protein using 69 complete EnsEMBL metazoan proteomes as previously described (Davey et al., 2012a). We analyzed the alignments using the SLiMPrints tool (Davey et al., 2012a), and focused on accessible regions of the proteins by limiting the search to predicted disordered regions and filtering out extracellular and transmembrane regions. This analysis returned 21 significant hits (sig > 0.0001) (Supplementary Table 2A).
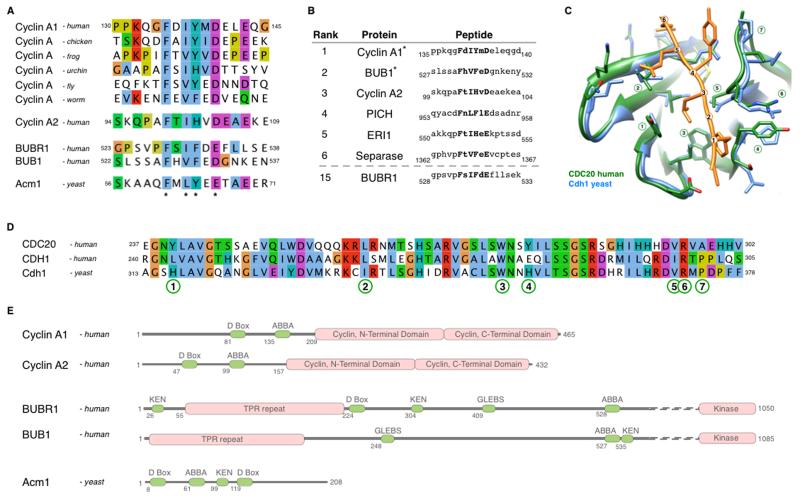
We were particularly drawn to a highly conserved motif in the A-type cyclins: KxxFxxYxDxxE (residues 132-143 in human Cyclin A1, sig=5.3 ×10−5) (Fig. 1A). This motif was conserved in the metazoan A cyclins, representing a time span of 600 million years since the last common ancestor. By contrast, the flanking regions had high rates of insertions, deletions and mutations. Our attention was also drawn to the motif because it was in the region of Cyclin A2 (residues 98-168) important for its CDC20-dependent degradation in the presence of an active SAC (Di Fiore and Pines, 2010). By contrast, the motif was not present in Cyclin B1, which is only destroyed once the SAC has been inactivated. These observations indicated that the KxxFxxYxDxxE motif might be involved in Cyclin A degradation.
Figure 1. Identification of the ABBA motif and its conservation through evolution.
(A) Alignment of the ABBA motif-containing region in Cyclin A1 orthologs, and peptides from human Cyclin A2, BUBR1, BUB1 and yeast Acm1 matching the Fx[ILV][FHY]x[DE] consensus (asterisks). See also supplemental Fig. 1.
(B) Top results, ranked by conservation, of a SLiMSearch analysis of the human proteome using the ABBA motif consensus. Five of the top six instances are found in important mitotic proteins (ERI1 has no known cell cycle function).
(C) Structural alignment of yeast Cdh1 (blue) bound to the ABBA peptide from Acm1 (orange) (He et al., 2013) and human CDC20 (green) (Tian et al., 2012). The ABBA peptide is numbered to define the position of the six core residues in the consensus. CDC20 is numbered to define the key residues in the ABBA motif-binding pocket.
(D) Alignment of the motif-binding pocket in human CDC20, human CDH1, and yeast Cdh1. Numbering refers to the key residues in the ABBA motif-binding pocket defined in (C).
(E) Modular architecture of human Cyclin A1, Cyclin A2, BUBR1, BUB1 and yeast Acm1.
To explore this possibility, we constructed a degenerate consensus of the motif, Fx[ILV][FHY]x[DE], based on the constrained residues in metazoan Cyclins A1 and A2 (Supplementary Figure 1). The Fx[ILV][FHY]x[DE] motif was searched against the human proteome using the SLiMSearch motif discovery tool (Davey et al., 2011). This returned 896 hits in 845 proteins, of which 69 remained after filtering for accessibility, and these were ranked by their relative conservation (Supplementary Table 2B). Several of the most conserved hits in the human proteome were found in important mitotic proteins: Cyclin A1, Cyclin A2, Separase, PICH, and the SAC components BUBR1 and BUB1 (Fig. 1B). A SLiMSearch analysis of the motif pattern against the yeast proteome returned two hits (residues 60-65 and 201-206) in Acm1, which inhibits the APC/C co-activator, Cdh1.
Several reasons indicated that the motif in Cyclin A, BUBR1 and Acm1 might be involved in binding to APC/C activators. Firstly, CDC20 binds to the N-terminus of Cyclin A that contains the motif (Di Fiore and Pines, 2010). Secondly, a peptide overlapping the motif in BUBR1 was required to bind CDC20 (Davenport et al., 2006). Thirdly, the first instance of the motif in Acm1 was required to bind Cdh1, and mutating positions within the motif abolished binding (Burton et al., 2011). Lastly, the structure of Acm1 bound to Cdh1 revealed that the motif peptide (residues 60-65) occupied a pocket between blades 2 and 3 of the WD40 repeats of Cdh1 (He et al., 2013). This structure clarified the conservation of the Fx[ILV][FHY]x[DE] motif as the constrained residues (positions 1, 3, 4 and 6) are in direct contact with binding surfaces, whereas the less constrained residues (positions 2 and 5) point away from the surface (Fig. 1C). The key residues of the binding pocket of Cdh1 contacting the Acm1 peptide are largely conserved between yeast Cdh1 and human CDC20, indicating that the binding mode may be similar (Fig. 1D). The motif seems to have evolved de novo on at least three occasions: in the A-type cyclins, the BUB1-like family and in Acm1 (Fig 1E). It may be significant that on two occasions, in the A-type cyclins and Acm1, the motif appeared at a relatively constant distance away from the D-box (Fig. 1E).
On the basis of its conservation and functional relevance (see below) in A-type cyclins, BUBR1, BUB1 and Acm1 we named the Fx[ILV][FHY]x[DE] motif the ‘ABBA’ motif. (Note that Burton et al., 2011 defined a partially overlapping region containing the ABBA motif in Acm1 as the ‘A-motif’ but this name is not easily searchable, nor does it take into account conservation in other protein families.)
The ABBA motif binds to CDC20
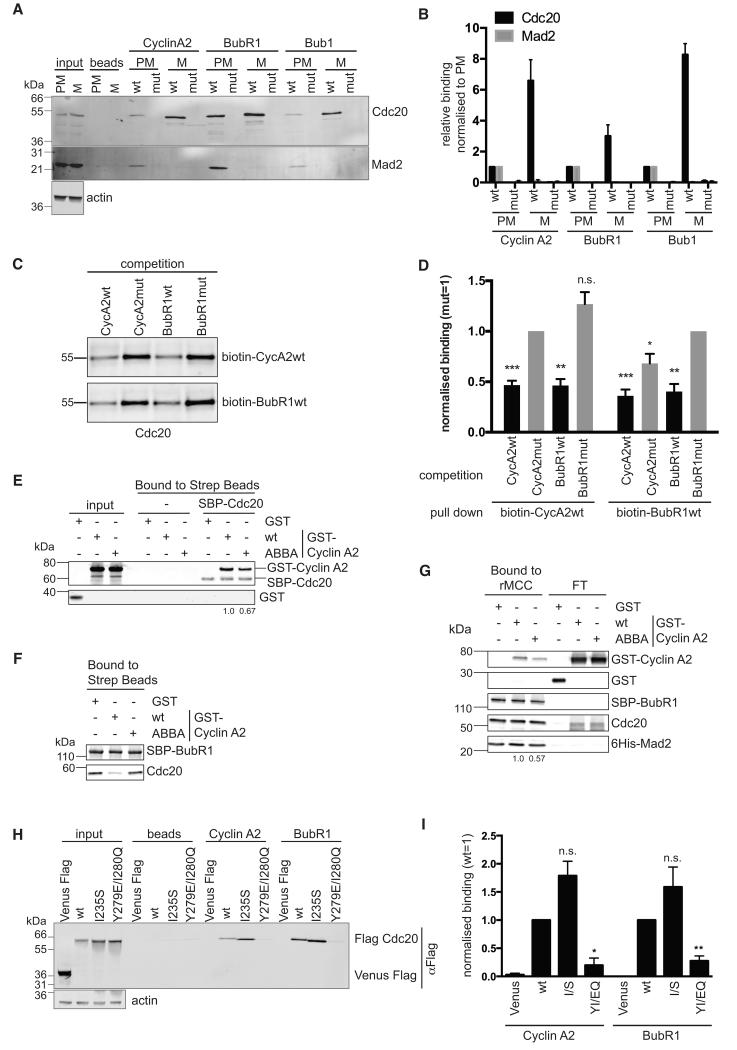
To test whether the ABBA motif does bind to CDC20 we synthesized biotin-conjugated peptides of the motifs in Cyclin A2, BUBR1 and BUB1. As controls we mutated residues 3, 4 and 6 to Ala. We immobilized the peptides on streptavidin beads, incubated the beads in extracts of prometaphase or metaphase cells, and analysed the proteins that bound to the peptides by immunoblotting (Fig 2A). The ABBA motifs from Cyclin A2, BUBR1 and BUB1 could all bind to CDC20 in mitotic cells (Fig. 2A & B). Moreover, they each bound more CDC20 in metaphase extracts than prometaphase (Fig. 2A & B), which would be expected if the peptide had to compete with endogenous Cyclin A2 and BUBR1 to bind to CDC20 in prometaphase. Furthermore, binding was abolished for the ABBA mutant peptides (Fig 2A & B). None of the peptides detectably bound to human CDH1 (data not shown).
Figure 2. The ABBA motif binds to CDC20 between blades 2 and 3 of the WD40 domain.
(A) Immobilized peptides containing wild-type (wt) and mutant (mut) ABBA motif from Cyclin A2, BubR1 and Bub1 were incubated with extracts from prometaphase (PM) and metaphase (M) HeLa cells (see Experimental Procedures). Input signals and background binding to beads are shown. For the input, 1/30 of the extract used for the pulldown was loaded. Actin is the loading control for the input.
(B) Mean and Standard error of the Mean (SEM) from 4 independent experiments as shown in (A). For each peptide the binding is normalised to the binding of the wt peptide in PM.
(C) Sf9 cell extracts expressing Hs CDC20 were incubated with the indicated biotinylated peptides and a 200-fold excess of a competing non-biotinylated peptide followed by purification with streptavidin beads. See also supplemental Fig. 2.
(D) Bar charts show mean and SEM from 3 independent experiments normalized to CDC20 binding after competition with the respective Cyclin A or BubR1 mutant peptide. The competition of wild type Cyclin A and BubR1 peptides to the respective mutant peptide was analysed using a two-tailed paired t-test (*, p<0.05; **, p<0.005; ***, p<0.0005; n.s., not significant).
(E) Streptavidin beads bound to SBP-CDC20 were incubated with GST, GST-Cyclin A2 wt or ABBA mutant. Proteins retained on the beads were analysed by quantitative immunoblotting and normalized values for Cyclin A2 are shown. Results are representative of 2 experiments.
(F) Streptavidin beads were incubated with insect cell lysate expressing SBP-BubR1, unbound proteins washed away, and beads incubated with cell lysate expressing untagged CDC20 with GST, or GST-Cyclin A2 wt or ABBA mutant. Proteins retained on the beads were analysed by quantitative immunoblotting.
(G) Streptavidin beads with rMCC bound via SBP-BubR1 were incubated with GST, GST-Cyclin A2 wt or ABBA mutant. Proteins retained on the beads and in the flow-through (FT) were analysed by quantitative immunoblotting and normalized values for Cyclin A2 are shown. Results representative of 2 experiments.
(H) Peptide pulldown using immobilised peptides and total extract from siRNA CDC20-depleted prometaphase cells expressing Venus-Flag, or siRNA resistant Flag-CDC20 wild-type or Y279E/I280Q mutant. Actin is a loading control.
(I) Mean and SEM from at least 3 independent experiments shown in (H). For each peptide the binding is normalised to the binding of wt CDC20 and values are normalised to input. Paired t-test was used for statistical analysis (*, p<0.05; **, p<0.005; n.s., not significant).
Since the ABBA motifs in Cyclin A2 and BUBR1 both bound to CDC20, it was possible that they bound to the same site, thereby explaining how Cyclin A2 competes with the MCC to bind to CDC20. We tested this with a competition assay. We expressed human CDC20 in insect cells using baculovirus and found that it efficiently bound to biotin-conjugated peptides of Cyclin A2 and BUBR1, but not Acm1, in an ABBA motif dependent manner (Supplementary Fig. S2). We then added competing, unconjugated peptides of the ABBA motif from Cyclin A2 or BUBR1, or mutated control peptides, to the extracts before isolating the biotin-conjugated peptides on streptavidin beads (Fig. 2C). The ABBA motif peptides of Cyclin A and BUBR1 were equally as effective at competing with each other as with their cognate peptides, and competition for CDC20 depended on the ABBA motif (Fig. 2C & D). This supported the idea that the ABBA motifs of Cyclin A and BUBR1 bound to the same, or overlapping, site on CDC20. This site was distinct from the site bound by MAD2 because ABBA motif peptides isolated CDC20 bound to MAD2 in prometaphase extracts (Fig 2A & B).
To strengthen the evidence that Cyclin A and BUBR1 bound to the same site on CDC20 we purified wild-type (wt) and ABBA mutants of SBP-tagged full length BUBR1, and of GST-tagged full length Cyclin A, and tested their ability to bind and compete for purified CDC20 (Fig 2E & F). Wt Cyclin A reproducibly bound better to CDC20 than did the ABBA mutant (30% reduction when the ABBA motif was mutated, Fig 2E), and wt Cyclin A, but not the ABBA mutant, was able to compete with BUBR1 for CDC20 (Fig 2F). We also tested the effect of the ABBA motif on the ability of Cyclin A to bind to CDC20 in the context of the MCC. We generated and purified the MCC using baculovirus expressing MAD2, BUBR1 and CDC20 (Izawa and Pines, 2014) and found that mutating the ABBA motif reproducibly reduced the amount of Cyclin A that could bind to the MCC by about 40% (Fig. 2G).
A stringent test of whether Cyclin A and BubR1 ABBA motifs bound to the same site was to mutate the binding site on CDC20, which should prevent the binding of both proteins. Since the ABBA motif peptides of Cyclin A and BUBR1 did not bind human CDH1, we compared the structure of blades 2 and 3 of the WD40 domain of human CDC20 (Fig. 1C) to human CDH1. This indicated that substituting a Glu for Tyr at position 279 (marked as 4 in Fig. 1C & D) and a Gln at position 280 might alter the ABBA motif receptor specificity of CDC20 to make it resemble CDH1 (Fig. 1C & D). The Y279E/I280Q mutation prevented CDC20 binding to the ABBA motif peptides of both Cyclin A2 and BUBR1 (Fig 2H and I), but CDC20 was properly folded because it could activate the APC/C and substitute for wt CDC20 in mitosis (see Fig. 7). This was strong evidence that the ABBA motif peptides of Cyclin A2 and BUBR1 competed for the same site on CDC20.
The ABBA motif is important for Cyclin A to bind to CDC20 and be rapidly degraded in prometaphase
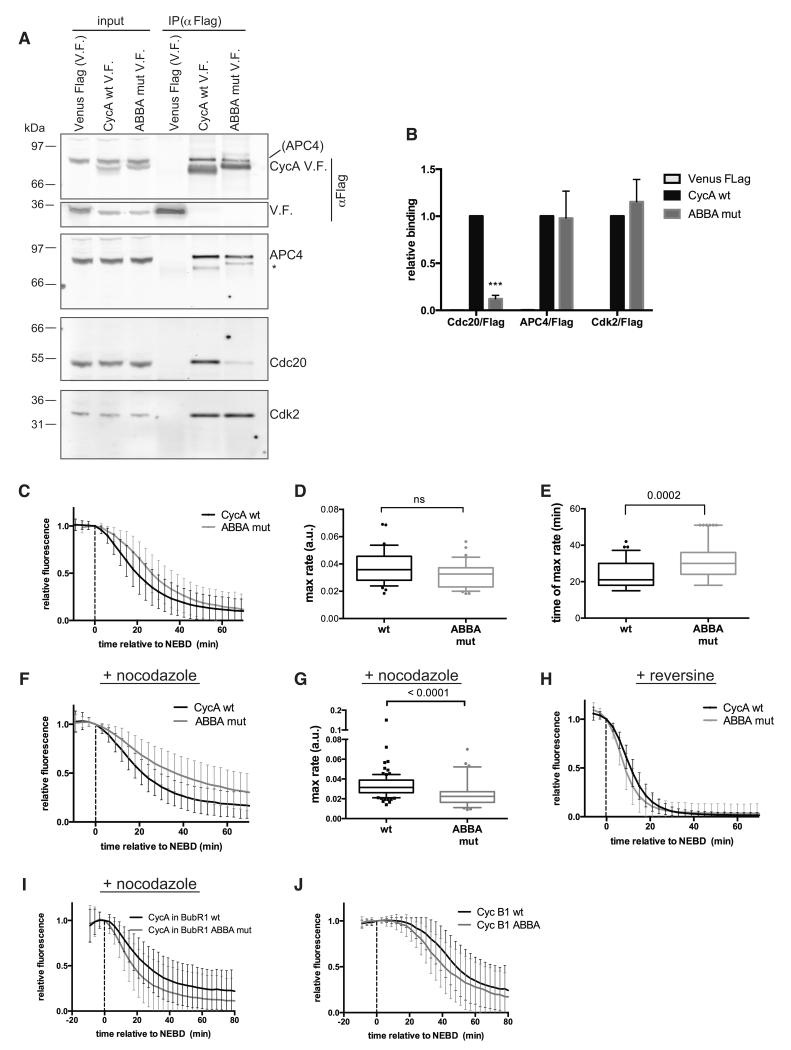
As the ABBA motif was important for Cyclin A to compete with BUBR1 to bind to CDC20 in vitro, we tested whether the ABBA motif was important for Cyclin A to bind to CDC20 in vivo, and to be correctly degraded in mitosis. We mutated positions 1, 3, 4 and 6 of the ABBA motif to Ala in full-length Cyclin A2 (ABBA mut), tagged wt and mutant Cyclin A with Venus fluorescent protein and a triple Flag epitope tag, and generated HeLa cell lines expressing them from an inducible promoter. We found that mutating the ABBA motif in Cyclin A2 dramatically decreased its ability to bind to CDC20 in prometaphase cells (Fig. 3A & B). The effect of mutating the ABBA motif on CDC20 binding in vivo was greater than with purified proteins (Fig 2E), perhaps because in vivo Cyclin A had to compete for CDC20 with BUBR1. (Note that the Cyclin A protein with a mutated ABBA motif was properly folded because binding to Cdk2 and APC4 was unaffected, Fig. 3A & B.)
Figure 3. The ABBA motif is required for Cyclin A to be degraded correctly in prometaphase.
(A) Ant-Flag immunoprecipitations from nocodazole-treated HeLa FRT cell lines expressing inducible Cyclin A Venus-Flag (V.F.) wt and ABBA mutant. Protein in parenthesis indicates signal from a previous blot. Asterisk marks a weak anti-Flag signal due to cross-reaction of the strong Flag signal in the IP with the 2° antibody.
(B) Mean and SEM from 4 independent experiments shown in (A). Paired t-test used for statistical analysis (***, p<0.0005).
(C) Mean degradation curves with SD from Hela cells microinjected with wt or ABBA mutant Cyclin A Venus-Flag. 105 cells (n=6) and 34 cells (n=2) were analysed for Cyclin A wt and ABBA mutant, respectively. Time is relative to nuclear envelope breakdown (NEBD).
(D) & (E) The maximal degradation rate (D) and the time when maximal degradation rate was reached (E) was measured for wt and ABBA mutant Cyclin A from cells in (C) and plotted as a box and whiskers graph. Unpaired t-test used for statistical analysis (ns, not significant).
(F) Cyclin A degradation in cells treated with nocodazole was analysed as in (C). 98 cells (n=5) and 64 cells (n=4) were analysed for Cyclin A wt and ABBA mutant, respectively.
(G) The maximal degradation rate was measured for wt and ABBA mutant Cyclin A, for single cells in (F) and plotted as box and whiskers graphs. Unpaired t-test used for statistical analysis.
(H) Cyclin A degradation in cells treated with reversine was analysed as in (C). 31 cells and 37 cells (n=3) were analysed for Cyclin A wt and the ABBA mutant, respectively.
(I) Cyclin A Venus degradation was analysed as in (C) in Hela FRT cell lines treated with nocodazole expressing wt (n=73) or ABBA mutant mRuby-BUBR1 (n=84) in which the endogenous BUBR1 was depleted by siRNA and Cyclin A Venus-Flag was transiently transfected (n=3).
(J) Cyclin B1 Venus degradation was analysed as in (C) in Hela cells microinjected with wt Cyclin B1 (39 cells) or Cyclin B1 with a N-terminal ABBA motif (48 cells) (n=5).
We predicted that the reduced affinity for CDC20 should cause the ABBA mutant to be degraded less readily than wt Cyclin A in prometaphase, when Cyclin A has to compete for CDC20 with the MCC. This prediction was confirmed: the ABBA mutant was degraded more slowly than wt Cyclin A2 in early mitotic cells (Fig. 3C-E), but at a very similar rate when we prevented the generation of the MCC (Fig. 3H) with reversine (Santaguida et al., 2010). Furthermore, the ABBA mutant was much more stable than wt Cyclin A when we maintained the SAC by treating cells with nocodazole, taking almost twice as long for 50% of the protein to be degraded compared to wild type Cyclin A (Fig. 3F & G). These results supported the idea that the ABBA motif was most important for Cyclin A to be degraded when it had to compete for CDC20 with BUBR1 in the MCC. If so, mutating the ABBA motif in BUBR1 should reduce its ability to compete for CDC20, and Cyclin A should be degraded faster. As predicted, Cyclin A was degraded more quickly in cells where endogenous BUBR1 was depleted and replaced with a BUBR1 with mutations at positions 3, 4 and 6 in the ABBA motif (Fig. 3I)
We concluded that the ABBA motif was important for Cyclin A to be recognized by Cdc20, and considered the possibility that the ABBA motif might act as a portable motif and enhance the degradation of other APC/C-CDC20 substrates. To test this, we introduced the ABBA motif sequence from Cyclin A2 into the N-terminus of Cyclin B1 at approximately the same distance from the D-box as in Cyclin A2. Introducing the ABBA motif did enhance the rate at which Cyclin B1 was degraded, although it did not change the time at which degradation began (Fig. 3J).
The ABBA motif in BUBR1 contributes to binding CDC20 and to the SAC
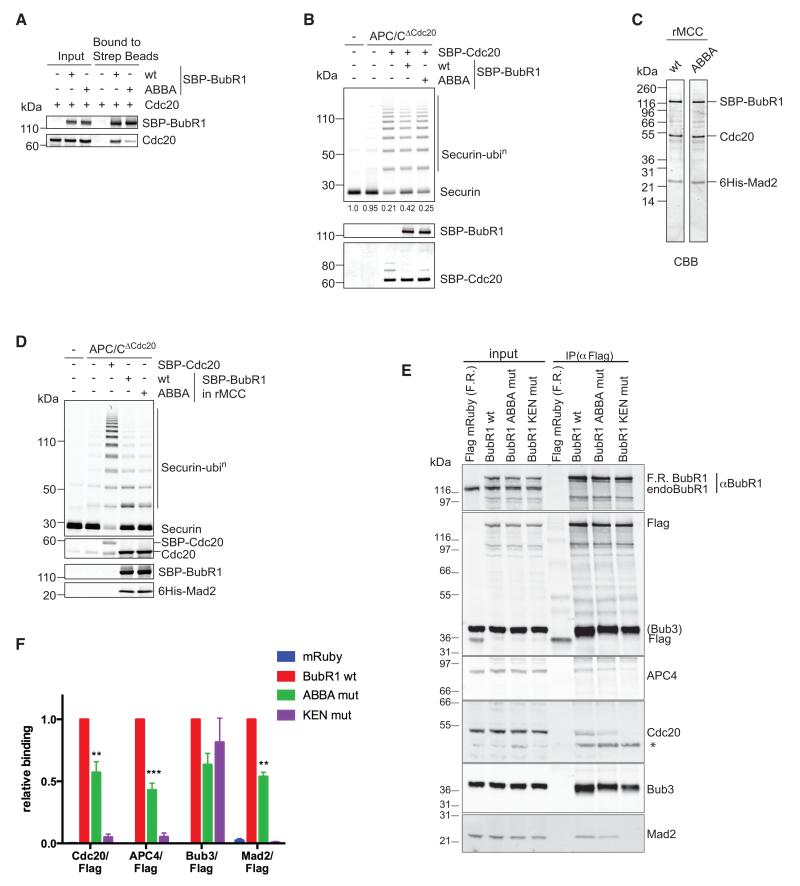
Since the ABBA motif in Cyclin A increased its binding to CDC20, we tested whether the ABBA motif contributed to the ability of BUBR1 to bind CDC20 and form the MCC. We mutated positions 3, 4 and 6 in the BUBR1 ABBA motif to Ala (ABBA mut), expressed the protein using the baculovirus system, and tested its ability to bind to purified CDC20. Mutating the ABBA motif severely compromised the ability of purified BUBR1 to bind to purified CDC20 in vitro (Fig. 4A), and reduced its ability to inhibit APC/C ubiquitylation activity (Fig. 4B), although whether BUBR1 alone is a physiological inhibitor of the APC/C is questionable because the SAC is completely abrogated when MAD2 is depleted (Meraldi et al., 2004). In the context of the MCC, however, BUBR1 requires MAD2 to bind correctly to CDC20 (Chao et al., 2012); therefore, we used the baculovirus system to co-express MAD2, CDC20 and either wt BUBR1 or the ABBA-mutant (Fig. 4C), and assayed the inhibitory activity of the MCC generated. This showed that mutating the ABBA motif in BUBR1 had no effect on the ability of the MCC to inhibit the APC/C in vitro (Fig. 4D).
Figure 4. The ABBA motif of BUBR1 contributes to binding to CDC20.
(A) Cell lysates from Sf9 cells expressing CDC20 and SBP-BubR1 wt or ABBA mutant were incubated with streptavidin beads and proteins retained on the beads analysed by quantitative immunoblotting.
(B) APC/C immunoprecipitated from mitotic siRNA CDC20-depleted HeLa cell extract (APC/CΔCdc20) was incubated with SBP-CDC20, or with SBP-CDC20 preincubated with either BubR1 WT or ABBA mutant. Ubiquitylation reactions were performed with securin as substrate and analysed with a Li-COR Odyssey scanner. Values for unconjugated securin are given below the gel (n=3). SBP-CDC20 and SBP-BubR1 were analysed by quantitative immunoblotting.
(C) His6-Mad2, CDC20, and SBP-BubR1 - either wt or ABBA mutant - were co-expressed in insect cells and recombinant MCC (rMCC) purified using Ni-NTA agarose and streptavidin beads. The purified rMCCs were analysed by SDS-PAGE and Coomassie Blue staining (CBB).
(D) APC/CΔCdc20 purified as in (B) was incubated with SBP-CDC20 and rMCC generated with either wt BubR1 or the ABBA mutant, and securin ubiquitylation assayed as in (B).
(E) Flag-mRuby (F.R.) BubR1 wt and mutants were immunoprecipitated from taxol-arrested HeLa FRT cell lines with an anti-Flag antibody. Asterisk denotes a non-specific band.
(F) Mean and SEM of at least 6 independent experiments shown in (E). A paired t-test was used for statistical analysis (**, p<0.005; ***, p<0.0005). See also supplemental Fig. 3.
Although the ABBA motif was not required for BUBR1 to form a functional MCC in vitro, we tested its effect in living cells. We generated stable cell lines expressing Flag-tagged versions of wt BUBR1, or an ABBA mutant, or a mutant in which we mutated the 1st KEN box (KEN mut) that binds to the top surface of the WD40 domain of CDC20 (Chao et al., 2012) as a positive control. We arrested cells in mitosis with taxol and immunoprecipitated BUBR1 with an anti-Flag antibody to assay its incorporation into the MCC. As previously reported (Burton and Solomon, 2007; Elowe et al., 2010; King et al., 2007; Lara-Gonzalez et al., 2011), the KEN box was essential for BUBR1 to form the MCC (Fig. 4 E & F). In contrast, mutating the ABBA motif allowed the MCC to form, in agreement with our in vitro results, but reduced the amount of MCC by about 50% in taxol-treated cells (Fig. 4 E & F), which have a weakly activated SAC (Collin et al., 2013). Mutating the ABBA motif made no difference to the amount of MCC in doses of nocodazole that strongly activate the SAC (Supplementary Fig. S3). This agrees with our previous finding that differences in MCC stoichiometries are more easily uncovered in taxol-treated cells (Collin et al., 2013).
Although the ABBA motif was not essential for BUBR1 to form the MCC, it did reduce the amount of MCC generated, which could thereby reduce the strength of the SAC (Collin et al., 2013). To test this, we used siRNA to deplete wt BUBR1 and assayed whether the ABBA mutant could restore the SAC and delay cells in prometaphase. We generated siRNA-resistant wt BUBR1, ABBA and KEN box mutants, fused them to a triple Flag epitope tag and mRuby and generated HeLa cell lines expressing the proteins from an inducible promoter. Immunoblot analyses showed that all proteins were expressed at similar levels, and that we could efficiently deplete the endogenous BUBR1 by siRNA (Fig. 5A) without altering the level of MAD2 (Fig. 5A, compare the first two lanes). Using time-lapse DIC microscopy, we assayed the time that cells were able to remain arrested in mitosis in the presence of taxol. As expected, depleting BUBR1 abolished the SAC, and the KEN box was essential for BUBR1 to function in the SAC (Fig. 5B). By contrast, mutating the ABBA motif had an intermediate effect: the ABBA mutant was able to restore a mitotic delay in response to taxol, but cells could not maintain this for as long as cells expressing wt BUBR1 (Fig. 5B). As an alternative means to generate a weak SAC signal we treated cells with DMA to generate monopolar spindles and compromised Mps1 kinase activity with low doses of AZ3146 (Collin et al., 2013). Under these conditions, the ABBA mutant had a similar inability to maintain the checkpoint (Supplementary Fig S4A). Thus, mutating the ABBA motif in BUBR1 appeared to weaken the SAC, and this correlated with a reduction in the amount of MCC.
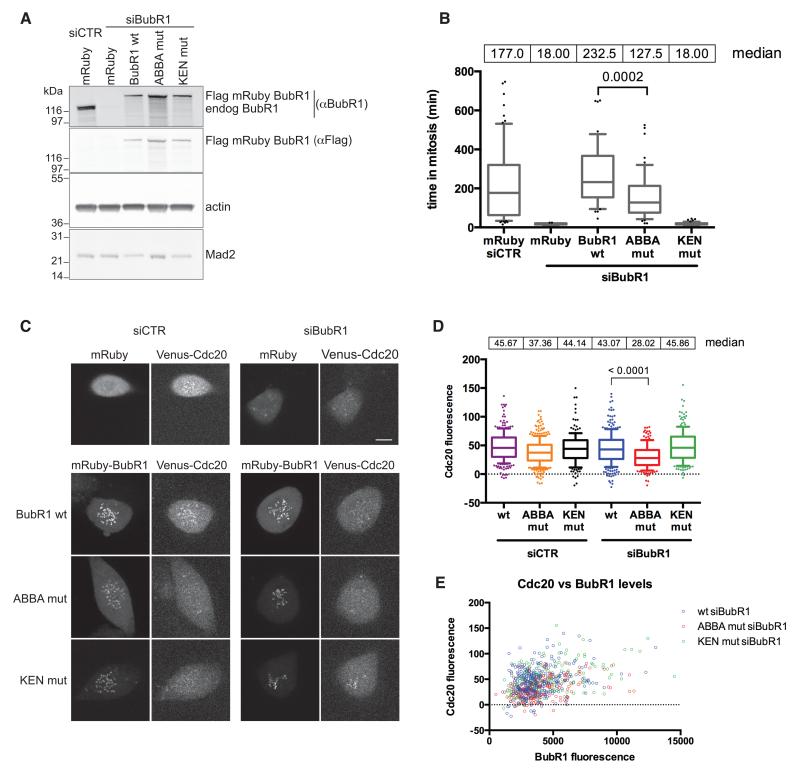
Figure 5. The ABBA motif of BUBR1 is required to recruit CDC20 to kinetochores and contributes to the strength of the SAC.
(A) HeLa FRT cell lines expressing stable inducible siRNA-resistant wt and mutant BubR1 were transfected with control siRNA or siRNA against BubR1. Immunoblotting analysis shows the expression levels compared to endogenous BubR1 and the efficiency of knockdown. Actin is a loading control.
(B) Taxol-treated HeLa FRT cell lines stably expressing inducible siRNA-resistant Flag-mRuby BubR1, were transfected with the indicated siRNA. Cells were imaged by DIC time-lapse microscopy. Median values of the time spent in mitosis are shown and analysed using an unpaired t-test. (n=3.) See also supplemental Fig. 4A.
(C) RPE1 FRT Venus-CDC20 knock-in cell lines stably expressing inducible siRNA resistant Flag-mRuby BubR1 were treated with control siRNA or siRNA against BubR1 and Taxol added. Images of the first frame after NEBD are shown for each BubR1 protein. Scale bars 10 μm. See also supplemental Figs. 4B & 5.
(D) Box and whisker analysis of kinetochore (KT) localised CDC20 in cells transfected with the indicated siRNA and rescued with wt and mutant BubR1, examples shown in (C). KT signal is relative to the background. Median values of the KT intensity are shown. At least 170 KTs were analysed from at least 11 cells. (n=3.) Unpaired t-test used for statistical analysis.
(E) Levels of KT localised CDC20 and BubR1, relative to the background, are shown for the same cells analysed in (D).
The ABBA motif of BUBR1 is important to recruit CDC20 to kinetochores
As the ABBA motif did not appear to contribute directly to the inhibitory activity of the MCC, but reduced the ability of BUBR1 to bind CDC20 in the absence of MAD2, we examined other aspects of SAC protein behavior. When we depleted BUBR1 in cells by siRNA and rescued cells with a BUBR1 ABBA mutant, we were struck by the reduction in CDC20 at unattached kinetochores. We observed this for endogenous CDC20 both by immunofluorescence (Supplemental data Fig. S4B), and by live cell microscopy using a Venus tag knocked into one allele of CDC20 (Fig. 5C & D; see Experimental Procedures and Supplemental data Fig. S5 for the generation and characterization of these cells). By contrast, the BUBR1 KEN box was not required to recruit CDC20 to the kinetochores, although it was essential for the SAC (Fig 5C & D). Moreover, the effects of both mutants on CDC20 recruitment to kinetochores were independent of their expression levels (Fig. 5E). These data indicated both that BUBR1 was required to recruit CDC20 to kinetochores, and that the ABBA motif was important for this recruitment.
The ABBA motif of BUB1 is important for its SAC activity and to localise CDC20 to kinetochores
The effect of the BUBR1 ABBA motif on the SAC and on recruiting CDC20 to kinetochores raised the question of whether the ABBA motif in BUB1 had similar functions. We generated siRNA resistant wt BUB1, and a BUB1 mutant in which we changed positions 1, 3, 4 and 6 of the ABBA motif to Ala. These constructs were tagged with Venus and a triple Flag epitope tag, and expressed in HeLa cell lines from an inducible promoter. Immunoblot analysis showed that we depleted endogenous BUB1 and expressed wt BUB1 and the ABBA mutant to similar levels (Fig. 6A). Unlike wt BUB1, we found the ABBA mutant was completely unable to restore the SAC. There was almost no discernible difference in the amount of time that BUB1-depleted cells delayed in mitosis in the presence of taxol whether we expressed BUB1 lacking its ABBA motif or Venus protein alone (Fig. 6B).
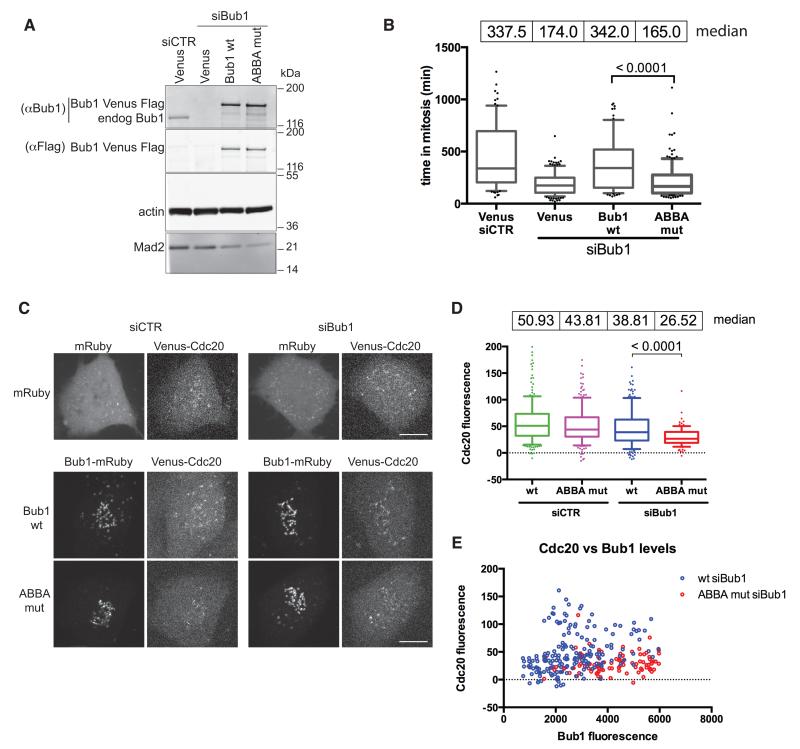
Figure 6. The ABBA motif of BUB1 helps to recruit CDC20 to kinetochores and contributes to the strength of the SAC.
(A) HeLa FRT cell lines expressing inducible siRNA-resistant wt or mutant Bub1 Venus-Flag were transfected with control or Bub1 siRNA. Immunoblot shows expression levels compared to endogenous Bub1. Actin is a loading control.
(B) HeLa FRT cell lines expressing inducible siRNA-resistant Bub1 Venus-Flag, were transfected with the indicated siRNA, then treated and analysed as in Fig 5B. Median values of mitotic duration are shown and analysed by an unpaired t-test (n=3).
(C) RPE1 FRT Venus-CDC20 knock-in cell lines expressing siRNA resistant mRuby-Flag-tagged Bub1, wild-type or ABBA mutant, were treated with control siRNA or siRNA against Bub1 and analysed as in Fig 5C. Scale bars 10 μm. See also supplemental Figs. 5 & 6.
(D) Box and whisker analysis of KT localised CDC20 in cells expressing wt or mutant Bub1 as in (C). Median values of the KT intensity are shown. At least 100 KTs were analysed from at least 7 cell (n=3). Unpaired t-test used for statistical analysis.
(E) Levels of KT localised CDC20 and Bub1 are shown for the cells analysed in (D).
We wondered whether the ABBA motif of BUB1 might also be required to recruit CDC20 to kinetochores, especially since BUB1 is required for BUBR1 to localize to kinetochores (Johnson et al., 2004) (Supplementary Fig. S6A). Just as with BUBR1, and independent of the amount of BUB1 expressed (Fig. 6E), CDC20 levels at kinetochores were reduced in cells expressing BUB1 with a mutant ABBA motif (Fig. 6C & D), although this did not appear to be caused by preventing BUBR1 from localizing to kinetochores (Supplementary Fig. S6A). It was notable that the amount of MAD2 and BUB3 bound to CDC20 did not change markedly, showing that cells were still able to generate the MCC that was also able to bind the APC/C (Supplementary Fig. S6B & C). Thus, we conclude that the ABBA motifs of both BUBR1 and BUB1 are required for a fully functional SAC and for the proper recruitment of CDC20 to unattached kinetochores.
The ABBA motif receptor on CDC20 is required to bind Cyclin A2 and contributes to the response to the SAC
To test whether the ABBA motifs of Cyclin A2, BUBR1 and BUB1 influenced mitosis through their role in binding CDC20, we analysed the effect of replacing CDC20 with a mutant unable to bind the ABBA motif. Introducing Y279E/I280Q mutations into CDC20 compromised the binding of ABBA motif peptides (Fig. 2H & I); therefore we generated siRNA-resistant forms of wt and a Y279E/I280Q mutant of CDC20 tagged with a triple Flag epitope, and generated stable cell lines expressing the proteins from an inducible promoter. In support of our conclusion that the ABBA motif of Cyclin A mediates its binding to CDC20, we found that the Y279E/I280Q mutant was unable to bind to Cyclin A2 (Fig. 7A & B) even though it was expressed at similar levels to wt CDC20 (Fig. 7C). The Y279E/I280Q mutant also bound noticeably less BUBR1 and MAD2 (Fig. 7A & B), indicating that the ABBA motif of BUBR1 contributed to the formation or stability of the MCC. We also observed that less of the Y279E/I280Q mutant was recruited to kinetochores (data not shown), consistent with the idea that CDC20 is recruited to kinetochores through binding to BUBR1.
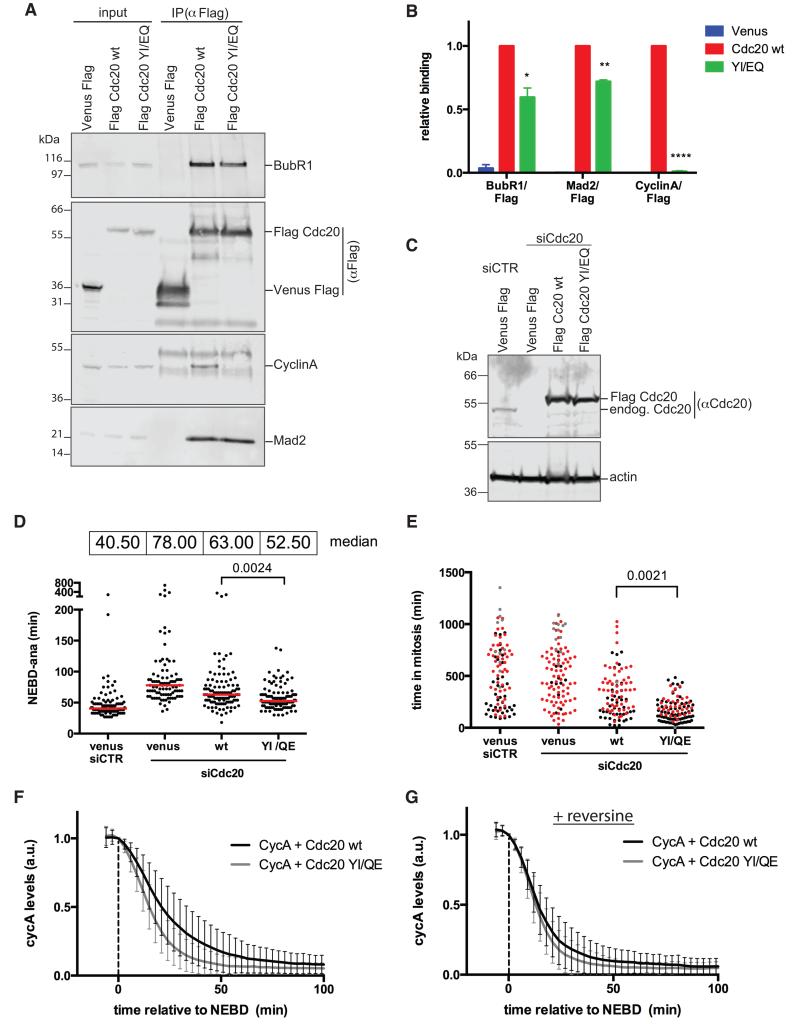
Figure 7. A CDC20 mutant that cannot bind the ABBA motif is unable to bind Cyclin A and is partially refractory to the SAC.
(A) Flag-tagged CDC20 wt and YI/EQ were immunoprecipitated with Flag mAb from taxol-arrested HeLa FRT cell lines and blotted with the indicated antibodies.
(B) Average and SEM of 3 independent experiments shown in (A). Paired t-test used for statistical analysis (*, p<0.05; **, p<0.005; ***, p<0.0005).
(C) HeLa FRT cell lines expressing inducible siRNA-resistant wild-type or YI/EQ mutant CDC20 were transfected with control siRNA or siRNA against CDC20. Immunoblot shows expression levels compared to endogenous CDC20 and the efficiency of knockdown. Actin is a loading control.
(D) Hela FRT cell lines expressing inducible siRNA-resistant Flag-tagged CDC20, were transfected with the indicated siRNA and analysed as in Fig 5B. Red lines indicate the median value. Unpaired t-test used for statistical analysis (n=3).
(E) Rescue experiments as in (D) but in the presence of taxol. Cells that exit mitosis (black), cells that die in mitosis (red), cells arrested in mitosis (grey) are indicated. Unpaired t-test was used for statistical analysis (n=3).
(F) Cyclin A-Venus-Flag degradation in cells transfected with siRNA against CDC20 and microinjected with Venus-Flag- wt Cyclin A together with either wt or YI/EQ mutant CDC20. Average degradation curves with SD were obtained from 48 cells (n=3) for wt CDC20, and 63 cells (n=3) for mutant CDC20. Time is relative to NEBD.
(G) Average curves were obtained as in (F), but in the presence of reversine. 49 cells (n=3) for wt CDC20, and 45 cells (n=3) for mutant CDC20 were analysed.
To determine the consequences on the SAC of the inability of CDC20 to bind the ABBA motif, we depleted CDC20 and replaced it with epitope-tagged wt or Y279E/I280Q mutant CDC20 (Fig. 7C-E). We assayed the SAC by measuring the time from NEBD to anaphase in non-drug treated cells (Fig 7D), and the duration of mitotic arrest in taxol-treated cells (Fig. 7E). The time taken from NEBD to anaphase was slightly faster in cells expressing the Y279E/I280Q mutant, which showed both that the Y279E/I280Q mutation activated the APC/C, and that the SAC appeared to be weaker (Fig. 7D). The compromised SAC was further evident in taxol-treatment where the Y279E/I280Q mutant could not arrest cells for a prolonged period (Fig. 7E). The Y279E/I280Q mutant was fully functional as an APC/C co-activator because it promoted rapid Cyclin A degradation (Fig. 7F) with similar kinetics to wild-type CDC20 in the absence of a functional SAC (Fig. 7G). Thus, we conclude that the ABBA motif promotes physiologically-relevant binding to human CDC20 in mitosis to control both APC/C-dependent degradation and regulation by the SAC.
Discussion
Here we have identified a conserved binding motif for APC/C activators. We have named this the ABBA motif because it is conserved in the human A-type cyclins, BUBR1, BUB1 and in budding yeast Acm1. The ABBA motif in Cyclin A, BUBR1 and BUB1 preferentially binds human CDC20 rather than human CDH1, whereas in Acm1 it preferentially binds to yeast Cdh1 (data not shown). The preference for binding CDC20 or CDH1 is likely to be due to differences in those residues in the cleft between blades 2 and 3 of the WD40 domain that are predicted to contact the ABBA motif (He et al., 2013) (Fig. 1D). We find that mutating this pocket prevents human CDC20 from binding the ABBA motif peptides of Cyclin A and BUBR1, indicating that they bind to the same receptor. We have also identified highly conserved peptides matching the ABBA motif consensus in other mitotic regulators including Separase and PICH, but these peptides bind neither CDC20 nor CDH1 (data not shown). The ABBA motif in these proteins might be a ligand for other WD40-family proteins, although the lack of conservation of the ABBA-motif binding pocket outside the CDC20-like proteins indicates this is unlikely.
Our evidence supports the idea that Cyclin A and BUBR1 compete for CDC20 because they have similar CDC20-binding motifs, and this competition between ABBA motifs is important for Cyclin A to be degraded when the SAC is active. Thus we can now explain our previous observation that in vitro the MCC and Cyclin A compete for CDC20 (Di Fiore and Pines, 2010). The ABBA motif of BUBR1 is, however, in the C-terminal part of the protein that is not strictly required for the SAC, nor to form the MCC (Chao et al., 2012; Davenport et al., 2006; Elowe et al., 2010; Malureanu et al., 2009); therefore, it is not yet clear whether Cyclin A binding prevents CDC20 interacting with MAD2 or with the N-terminal part of BUBR1. One could envisage that, in addition to the ABBA motif of Cyclin A binding to the WD40 domain of CDC20, other low affinity interactions with, for example, the extended D-box, could obscure motifs on CDC20 required to form the MCC, such as the KILR motif (Izawa and Pines, 2012). This could explain our observation that adding the ABBA motif to Cyclin B1 enhances its rate of degradation but does not allow it to be degraded in prometaphase. A series of low affinity interactions confers considerable advantages over a single high affinity binding site in a dynamic system requiring rapid assembly and disassembly of protein complexes, or high turnover of an enzyme-substrate complex.
Although its degradation in prometaphase and in nocodazole-treated cells was compromised, the ABBA mutant of Cyclin A was still degraded; moreover, in the absence of the SAC it was degraded at a similar rate to wt Cyclin A. These results agree with previous observations that the extended D-box and the Cks1-dependent binding of the Cyclin A-Cdk complex to the APC/C are also important determinants in conferring prometaphase degradation (Di Fiore and Pines, 2010; Geley et al., 2001; Wolthuis et al., 2008). It may be important that Cyclin A is degraded relatively quickly in prometaphase because Cyclin A can alter the stability with which kinetochores attach to microtubules, and thus affect the accuracy of chromosome segregation (Kabeche and Compton, 2013). Moreover, the amount of Cyclin A that remains when a cell exits mitosis may influence the subsequent decision whether to proliferate or become quiescent (Spencer et al., 2013).
Our identification of the ABBA motif provides us with a molecular explanation for how Cyclin A can be recognised by the APC/C in a different fashion to Cyclin B (Izawa and Pines, 2011). This is a conserved mechanism because recently Lu et al identified an ABBA motif in the N-terminus of the budding yeast cyclin, Clb5 and found that this contributes to the early recognition of Clb5 by the APC/C in mitosis (Lu et al., 2014). Moreover, we find that incorporating the ABBA motif into the N-terminus of Cyclin B1 caused Cyclin B1 to be degraded faster, but still under the control of the SAC. Thus, the ABBA motif joins the Destruction box and the KEN box as short conserved motifs involved in APC/C substrate recognition. In this respect it is noteworthy that the ABBA motif is placed at fairly uniform spacing from the D-box in the metazoan A-type cyclins, Clb5 and Acm1, possibly allowing these degrons to act cooperatively by binding simultaneously to two separate binding pockets on the CDC20 WD40 domain (as is the case for Acm1).
The ABBA motif is likely to account for previous reports of a Mad2-independent CDC20-binding domain in BUBR1 (Davenport et al., 2006; Elowe et al., 2010; Han et al., 2013; Malureanu et al., 2009; Tang et al., 2001). Although the C-terminal half of BUBR1 is not required for the SAC, the ABBA motif appears to make the SAC more efficient. In complementary experiments, mutating the binding site for the ABBA motif on CDC20 also reduced the strength of the SAC. This supports the conclusion that mutating the ABBA motif affects the SAC through its effect on CDC20 binding, rather than another component of the MCC such as BUB3 (Han et al., 2014). The BUBR1 ABBA mutant is competent to form the MCC in vitro, but in vivo the amount of MCC is reduced in cells with a weak checkpoint. This could indicate that ABBA-motif mediated binding between SAC proteins and CDC20 enhances the rate at which the MCC forms, perhaps through increased recruitment of CDC20 to BUBR1. Alternatively, the SAC may be weaker because the MCC incorporating the ABBA mutant has a reduced ability to bind a second molecule of CDC20, which is important to inhibit the APC/C (Izawa and Pines, 2014).
Mutating the ABBA motif in BUB1 has a very striking effect because it completely abrogates the SAC activity of BUB1. The molecular explanation for the requirement for BUB1 in the SAC is still unclear, but recent evidence points towards a role in recruiting and positioning Mad1 (London and Biggins, 2014; Moyle et al., 2014), which is important to catalyse the formation of the MCC. BUB1 is likely to have a further function, however, because the MCC still forms even though mutating the ABBA motif eliminates SAC activity. One intriguing possibility is that the ABBA motif of BUB1 might help the MCC bind to a second molecule of CDC20 at kinetochores.
Our evidence indicates that the BUBR1 binding to CDC20 mediated by the ABBA-motif is required for CDC20 to accumulate to normal levels at kinetochores, in agreement with recent observations (Han et al., 2014; Lischetti et al., 2014), and that the WD40 repeat of CDC20 (which binds the ABBA-motif) is required for CDC20 to localize to kinetochores (Kallio et al., 2002). We also find that the ABBA motif of BUB1 influences CDC20 recruitment to unattached kinetochores, although we only detect a small amount of CDC20 in BUB1 immunoprecipitates (data not shown). The functional consequences of recruiting CDC20 to kinetochores are unclear, however: CDC20 does not have to bind detectably to kinetochores to form the MCC, nor for the SAC to be operative. Conceivably, kinetochore recruitment could increase the strength of the SAC (Collin et al., 2013), since recruitment would raise the local concentration of the components required to form the MCC and thereby accelerate the speed of the reaction.
In conclusion, we have uncovered a conserved motif that binds to APC/C co-activators that has been adopted by substrates, SAC proteins and APC/C inhibitors. It will be fascinating to discover its role in the other ABBA motif-containing proteins that we have identified, most of which have crucial roles in mitosis.
Experimental Procedures
Cell culture and synchronization
HeLa FRT cells were transfected using the Flp-in system (Invitrogen), to generate stable inducible cell lines. Cells were induced with tetracycline (1μg/ml, Calbiochem) 12 hr before harvesting. HeLa cells were cultured and synchronised by a double thymidine (HeLa FRT) or a thymidin/aphidicolin protocol (HeLa) as previously described (Di Fiore and Pines, 2007). For prometaphase arrest, nocodazole (0.33μM), Taxol (0.1μM, or 10nM when filming HeLa FRT cells), or DMA (10μM) were after a thymidine block and 12hr later cells were either harvested by mitotic shake-off, or harvested 2hr later after adding MG132 (10μM). Prometaphase cells were treated reversine (0.5μM) + MG132 (10μM) for 2 hr to obtain metaphase cells. 3.3μM nocodazole was used for degradation assays in living cells. For siRNA rescue experiments, AZ3146 (0.62 μM) was added to DMA (10μM) treated cells.
RPE-1 FRT/TO cells (Mansfeld et al., 2011) were grown in DMEM/F-12 + 10% fetal bovine serum. To generate the Venus-CDC20 knock in RPE-1 FRT/TO cells were infected with recombinant adeno-associated virus particles harboring mVenus cDNA flanked by ~1500 bp homologous to the CDC20 locus as previously described (Collin et al., 2013). Single positive integrants were selected by flow cytometry and verified by fluorescence microscopy and immunoblot analysis (Supplementary Fig. S5).
Plasmids
Cyclin A, BUBR1, BUB1, CDC20 were cloned into pCDNA5/FRT/TO (Invitrogen), which contained Flag tag alone or linked to a fluorescent protein (Venus or mRuby). Mutants and siRNA-resistant constructs were generated using overlapping PCR. For the ABBA motif mutants positions 1,3,4 & 6 of the motif were replaced by alanine. V94F, E96I, P97H, P99D mutations were made to generate an ABBA motif in the N-terminus of human Cyclin B1. For recombinant GST-Cyclin A2, Cyclin A2 was cloned into pGEX-6P vector. All plasmids were verified by sequencing. Details of cloning are available on request.
Peptide pulldown and competition assays
Peptides were synthesised either with biotin or non-conjugated (Selleck and CovalAb). Wild-type peptides: Cyclin A (ANSKQPAFTIHVDEAEKEAQ), BubR1 (SKGPSVPFSIFDEFLLSEKK), Bub1 (ISSLSSAFHVFEDGNKENYG), Acm1 (QISKAAQFMLYEETAEERNI); in the mutants positions 3,4 & 6 of the ABBA motif were mutated to alanine. Cells were lysed in PBS, 0.5% Triton X- 100 with protease inhibitors cocktail (Roche), extracts cleared by centrifugation and incubated with the biotinylated peptides coupled to streptavidin Ultralink beads (Pierce) for 1 hr at 4°C. Peptide competition experiments were performed using extracts of His-CDC20 expressing SF9 insect cells prepared in PBS, 0.5% Triton X-100. For each condition 30 μg of total extract was incubated with 80 ng of biotinylated peptide (440 nM) and a 200 fold excess of non-biotinylated competing peptide (88 μM), or PBS as a control, for 3 hours at 4°C. Biotinylated peptides were immobilized on 20 μl streptavidin-coupled Dynabeads (Life technologies) for 30 min at room temperature, washed extensively in PBS, 0.5% Triton X-100, eluted at 96°C for 5 min, separated by SDS-PAGE, and analyzed by immunoblotting.
Expression and purification of recombinant proteins
GST and GST-Cyclin A2 were expressed in BL-21 cells for 1 day at 18 °C, and purified via glutathione sepharose 4B (GE Healthcare) according to the manufacturer’s instructions. Recombinant MCC, Cdc20, SBP-BubR1 and 6His-Mad2 was prepared as described (Izawa and Pines, 2014).
All binding assays using recombinant proteins were performed in 50mM TrisHCl, 150mM NaCl buffer supplied with 1% BSA.
In vitro ubiquitylation assay
In vitro ubiquitylation assays were performed as described (Izawa and Pines, 2014) at 37°C for 10 min with purified SBP-Cdc20 (co-activator), and recombinant MCC or SBP-BubR1 (inhibitors). Recombinant securin was labelled with IRDye680 dye (IRDye 680LT Maleimide Infrared Dye: LiCOR) according to the manufacturers instructions and directly scanned with a Li-COR Odyssey scanner after SDS-PAGE.
Microinjection, time-lapse imaging and analysis
Microinjection and time-lapse microscopy were performed as previously described (Di Fiore and Pines, 2010). DIC and fluorescence images were captured every 3 min on a Nikon Ti microscope with an ORCA Flash 4.0 CMOS camera (Hammamatsu, Japan) using Micromanager software. ImageJ software (NIH) was used to quantify the fluorescence after background subtraction. DIC microscopy was used to monitor mitotic phases. Analysis of the rate of degradation was performed as described (Min et al., 2013). To measure intensity of kinetochore localization cells were filmed on a spinning disk (Intelligent Imaging Innovations). Max projection of single z-section at NEBD were obtained and kinetochore intensity quantified using ImageJ.
RNA interference
siRNA duplexes (Dharmacon) against BubR1 (GAUGGUGAAUUGUGGAAUA), Bub1 (CGAAGAGUGAUCACGAUUU), CDC20 (CGGAAGACCUGCCGUUACAUU) and ON-TARGETplus non-targeting siRNA pool (Sigma) were transfected at a final concentration of 50nM with RNAimax (Invitrogen) according to the manufacturer’s protocol. Transfection was carried out 5 hr after release from a thymidine block. For siRNA and plasmid co-transfection Lipofectamine 3000 (Invitrogen) was used.
Immunoprecipitation
Cells were lysed in 50mM Tris-HCl pH 7.8, 150mM NaCl, 0.2% NP40, 1mM EDTA plus protease inhibitor cocktail (Roche) and microcystin (10nM, LGC Promotech) as previously described (Di and Pines, 2010). Complexes were immunoprecipitated for 1 hr at 4°C with anti-FLAG (M2, Sigma) or anti-APC4 (monoclonal antibody (mAb) raised against a C-terminal peptide).
Immunoblotting
Immunoblotting was performed as previously described (Di and Pines, 2010). Primary antibodies were used at the indicated concentration: FLAG (M2, Sigma, 3μg/ml), Mad2 (Bethyl Laboratories, 1μg/ml), actin (AC-15, Sigma, 1:1000), CDC20 (Bethyl Laboratories, 0.5 μg/ml), anti-APC4 (mAb, 1:500), BubR1 (Bethyl Laboratories, 0.5 μg/ml), Bub3 (Bethyl Laboratories, 0.5 μg/ml), Bub1 (Abcam, 1:3000). IRDye680 and IRDye800CW (LI-COR)-conjugated secondary antibodies were used at 1:10000. The signal was detected using an Odyssey scanner (LI-COR) as previously described (Nilsson et al., 2008).
Immunofluorescence
Cells were fixed and stained as previously described except that Tween 20 was used at 0.1% instead of 0.2% (Di Fiore and Pines, 2007). Anti-BubR1 (BD transduction laboratories), CDC20 (E-7, Santa Cruz) antibodies were used at 0.6μg/ml and 1μg/ml, respectively. CREST serum (gift of W. Earnshaw) was used at 1:20000. Secondary antibodies conjugated to Alexa Fluor 568 or Alexa Fluor 647 (Molecular Probes) were diluted 1:400. DNA was stained with Hoechst-33342. Images were collected on a DeltaVision micrososcope (API).
Statistics
Statistical analyses were carried out using GraphPad Prism as detailed in each legend.
Supplementary Material
Highlights.
Identification of the conserved ABBA motif that binds to APC/C co-activators
The ABBA motif of Cyclin A and BUBR1 bind the same site on CDC20
The ABBA motif is required for Cyclin A to be rapidly degraded in prometaphase
The ABBA motifs in BUBR1 and BUB1 are required for a fully functional SAC
Acknowledgements
We are grateful to Philippe Collin for help in generating the CDC20 cell line. We are also grateful to David Morgan for reagents and helpful discussions, and to him and Jakob Nilsson for communicating unpublished results. We thank Mingwei Min for the script to analyse rates of degradation, and members of our laboratories for comments and criticisms. This work was supported by a programme grant to JP from CR UK. JP acknowledges core funding to the Gurdon Institute from the Wellcome Trust and CR UK.
References
- Bolanos-Garcia VM, Blundell TL. BUB1 and BUBR1: multifaceted kinases of the cell cycle. Trends in Biochemical Sciences. 2011;36:141–150. doi: 10.1016/j.tibs.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes & Development. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Xiong Y, Solomon MJ. Mechanisms of pseudosubstrate inhibition of the anaphase promoting complex by Acm1. Embo J. 2011;30:1818–1829. doi: 10.1038/emboj.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014 doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- Chen J, Jackson PK, Kirschner MW, Dutta A. Separate domains of p21 involved in the inhibition of cdk kinase and PCNA. Nature. 1995;374:386–388. doi: 10.1038/374386a0. [DOI] [PubMed] [Google Scholar]
- Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1999;1:82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- Collin P, Nashchekina O, Walker R, Pines J. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat. Cell Biol. 2013;15:1378–1385. doi: 10.1038/ncb2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CJ, Dutta K, Petri ET, Hwang WC, Lin Y, Pascal SM, Basavappa R. The regions of securin and cyclin B proteins recognized by the ubiquitination machinery are natively unfolded. FEBS Letters. 2002;527:303–308. doi: 10.1016/s0014-5793(02)03246-5. [DOI] [PubMed] [Google Scholar]
- Davenport J, Harris LD, Goorha R. Spindle checkpoint function requires Mad2-dependent Cdc20 binding to the Mad3 homology domain of BubR1. Experimental Cell Research. 2006;312:1831–1842. doi: 10.1016/j.yexcr.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Davey NE, Cowan JL, Shields DC, Gibson TJ, Coldwell MJ, Edwards RJ. SLiMPrints: conservation-based discovery of functional motif fingerprints in intrinsically disordered protein regions. Nucleic Acids Research. 2012a;40:10628–10641. doi: 10.1093/nar/gks854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey NE, Haslam NJ, Shields DC, Edwards RJ. SLiMSearch 2.0: biological context for short linear motifs in proteins. Nucleic Acids Research. 2011;39:W56–W60. doi: 10.1093/nar/gkr402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey NE, Van Roey K, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H, Gibson TJ. Attributes of short linear motifs. Mol Biosyst. 2012b;8:268–281. doi: 10.1039/c1mb05231d. [DOI] [PubMed] [Google Scholar]
- De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Di Fiore B, Pines J. Emi1 is needed to couple DNA replication with mitosis but does not regulate activation of the mitotic APC/C. J. Cell Biol. 2007;177:425–437. doi: 10.1083/jcb.200611166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B, Pines J. How cyclin A destruction escapes the spindle assembly checkpoint. J. Cell Biol. 2010;190:501–509. doi: 10.1083/jcb.201001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson TJ. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front. Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- Dinkel H, Van Roey K, Michael S, Davey NE, Weatheritt RJ, Born D, Speck T, Krüger D, Grebnev G, Kuban M, et al. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Research. 2014;42:D259–D266. doi: 10.1093/nar/gkt1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowe S, Dulla K, Uldschmid A, Li X, Dou Z, Nigg EA. Uncoupling of the spindle-checkpoint and chromosome-congression functions of BubR1. J. Cell. Sci. 2010;123:84–94. doi: 10.1242/jcs.056507. [DOI] [PubMed] [Google Scholar]
- Elzen, den N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J. Cell Biol. 2001;153:121–136. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SA, Morgan DO. The APC/C Subunit Mnd2/Apc15 Promotes Cdc20 Autoubiquitination and Spindle Assembly Checkpoint Inactivation. Mol. Cell. 2012;47:921–932. doi: 10.1016/j.molcel.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. APC/C-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 2001;153:137–48. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Hagting A, Elzen, Den N, Vodermaier HC, Waizenegger IC, Peters J-M, Pines J. Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC/C switches from activation by Cdc20 to Cdh1. J. Cell Biol. 2002;157:1125–1137. doi: 10.1083/jcb.200111001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JS, Holland AJ, Fachinetti D, Kulukian A, Cetin B, Cleveland DW. Catalytic Assembly of the Mitotic Checkpoint Inhibitor BubR1-Cdc20 by a Mad2-Induced Functional Switch in Cdc20. Mol. Cell. 2013;51:92–104. doi: 10.1016/j.molcel.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JS, Vitre B, Fachinetti D, Cleveland DW. Bimodal activation of BubR1 by Bub3 sustains mitotic checkpoint signaling. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4185–E4193. doi: 10.1073/pnas.1416277111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Chao WCH, Zhang Z, Yang J, Cronin N, Barford D. Insights into Degron Recognition by APC/C Coactivators from the Structure of an Acm1-Cdh1 Complex. Mol. Cell. 2013;50:649–660. doi: 10.1016/j.molcel.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa D, Pines J. How APC/C-Cdc20 changes its substrate specificity in mitosis. Nat. Cell Biol. 2011;13:223–233. doi: 10.1038/ncb2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa D, Pines J. Mad2 and the APC/C compete for the same site on Cdc20 to ensure proper chromosome segregation. J. Cell Biol. 2012;199:27–37. doi: 10.1083/jcb.201205170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa D, Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature. 2014 doi: 10.1038/nature13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson VL, Scott MI, Holt SV, Hussein D, Taylor SS. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell. Sci. 2004;117:1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- Kabeche L, Compton DA. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature. 2013;502:110–113. doi: 10.1038/nature12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio MJ, Beardmore VA, Weinstein J, Gorbsky GJ. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J. Cell Biol. 2002;158:841–847. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EM, van der Sar SJ, Hardwick KG. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS ONE. 2007;2:e342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Scott MI, Diez M, Sen O, Taylor SS. BubR1 blocks substrate recruitment to the APC/C in a KEN-box-dependent manner. J. Cell. Sci. 2011;124:4332–4345. doi: 10.1242/jcs.094763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr. Biol. 2012;22:R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Lischetti T, Zhang G, Sedgwick GG, Bolanos-Garcia VM, Nilsson J. The internal Cdc20 binding site in BubR1 facilitates both spindle assembly checkpoint signalling and silencing. Nat Commun. 2014;5:5563. doi: 10.1038/ncomms6563. [DOI] [PubMed] [Google Scholar]
- London N, Biggins S. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes & Development. 2014;28:140–152. doi: 10.1101/gad.233700.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Hsiao JY, Davey NE, Van Voorhis VA, Foster SA, Tang C, Morgan DO. Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J. Cell Biol. 2014;207:23–39. doi: 10.1083/jcb.201402041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malureanu LA, Jeganathan KB, Hamada M, Wasilewski L, Davenport J, van Deursen JM. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev. Cell. 2009;16:118–131. doi: 10.1016/j.devcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J, Collin P, Collins MO, Choudhary J, Pines J. APC15 drives the turnover of MCC-Cdc20 to make the Spindle Assembly Checkpoint responsive to kinetochore attachmen. Nat. Cell Biol. 2011;13:1234–1244. doi: 10.1038/ncb2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Min M, Mayor U, Lindon C. Ubiquitination site preferences in anaphase promoting complex/cyclosome (APC/C) substrates. Open Biol. 2013;3:130097. doi: 10.1098/rsob.130097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle MW, Kim T, Hattersley N, Espeut J, Cheerambathur DK, Oegema K, Desai A. A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J. Cell Biol. 2014;204:647–657. doi: 10.1083/jcb.201311015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neduva V, Russell RB. Peptides mediating interaction networks: new leads at last. Curr. Opin. Biotechnol. 2006;17:465–471. doi: 10.1016/j.copbio.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Nguyen Ba AN, Yeh BJ, van Dyk D, Davidson AR, Andrews BJ, Weiss EL, Moses AM. Proteome-wide discovery of evolutionary conserved sequences in disordered regions. Sci Signal. 2012;5:rs1. doi: 10.1126/scisignal.2002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat. Cell Biol. 2008;10:1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes & Development. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Puig N, Veprintsev DB, Fersht AR. Human full-length Securin is a natively unfolded protein. Protein Sci. 2005;14:1410–1418. doi: 10.1110/ps.051368005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SL, Cappell SD, Tsai F-C, Overton KW, Wang CL, Meyer T. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell. 2013;155:369–383. doi: 10.1016/j.cell.2013.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 2001;154:925–36. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell. 2001;1:227–37. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- Tian W, Li B, Warrington R, Tomchick DR, Yu H, Luo X. Structural analysis of human Cdc20 supports multisite degron recognition by APC/C. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18419–18424. doi: 10.1073/pnas.1213438109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzunova K, Dye BT, Schutz H, Ladurner R, Petzold G, Toyoda Y, Jarvis MA, Brown NG, Poser I, Novatchkova M, et al. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nat Struct Mol Biol. 2012;19:1116–1123. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Roey K, Gibson TJ, Davey NE. Motif switches: decision-making in cell regulation. Curr. Opin. Struct. Biol. 2012;22:378–385. doi: 10.1016/j.sbi.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Varetti G, Guida C, Santaguida S, Chiroli E, Musacchio A. Homeostatic control of mitotic arrest. Mol. Cell. 2011;44:710–720. doi: 10.1016/j.molcel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Wolthuis R, Clay-Farrace L, van Zon W, Yekezare M, Koop L, Ogink J, Medema R, Pines J. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol. Cell. 2008;30:290–302. doi: 10.1016/j.molcel.2008.02.027. [DOI] [PubMed] [Google Scholar]
- Zhu L, Harlow E, Dynlacht BD. p107 uses a p21Cip1-related domain to bind cyclin/Cdk2 and regulate interactions with E2F. Genes & Development. 1995;9:1740–1752. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.