Abstract
Background
The association between smoking induced chronic obstructive pulmonary disease (COPD) and lung cancer (LC) is well documented. Recent genome-wide association studies (GWAS) have identified 28 susceptibility loci for LC, 10 for COPD, 32 for smoking behavior (SM), and 63 for pulmonary function (PF), totaling 107 non-overlapping loci. Given that common variants have been found to be associated with LC in GWAS, exome sequencing of these high-priority regions has great potential to identify novel rare causal variants.
Patients and Methods
Using a variation of the extreme phenotype approach, we selected 48 sporadic LC patients reporting heavy smoking histories, 37 of whom also exhibited carefully documented severe COPD (in whom smoking is considered the overwhelming determinant), and 54 unique familial LC cases from families with at least three first-degree relatives with LC (who are likely enriched for genomic effects), to search for disease-causing rare germline mutations.
Results
By focusing on exome profiles of the 107 target loci, we identified two key rare mutations. A heterozygous p.Arg696Cys variant in the Coiled-Coil Domain Containing 147 (CCDC147) gene at 10q25.1 was identified in one sporadic and two familial cases. The minor allele frequency (MAF) of this variant in the 1000 Genomes (TG) database is 0.0026. The p.Val26Met variant in Dopamine Beta-Hydroxylase (DBH) gene at 9q34.2 was identified in two sporadic cases; MAF of this mutation is 0.0034 from the TG database. We also observed three suggestive rare mutations on 15q25.1 IREB2/CHRNA5/CHRNB4.
Conclusion
Our results demonstrated highly disruptive risk-conferring CCDC147 and DBH mutations.
Keywords: Exome sequencing, Single nucleotide variants (SNV), Lung cancer (LC), Chronic obstructive pulmonary disease (COPD), Familial and Sporadic
INTRODUCTION
Chronic tobacco-induced airway inflammation provokes a milieu conducive to pulmonary carcinogenesis. We and others have previously shown that tobacco-induced chronic obstructive pulmonary disease (COPD), also characterized by a sustained inflammatory reaction in the airways and lung parenchyma, is a significant contributor to lung cancer (LC) risk in smokers [1]. Likewise, reduced pulmonary function is also reported as an important variable when included in risk prediction models [2]. Recent genome-wide association studies (GWAS) have identified 28 susceptibility loci for LC, 10 loci for COPD, 32 loci for smoking behavior (SM), 63 loci for abnormal pulmonary function (PF) and related phenotypes, totaling 107 unique GWAS susceptibility loci (as of November 2014, Supplemental Table 1). Interestingly, there is considerable overlap among the susceptibility loci for these phenotypes. For example, 6p21.32 MHC class III region (AGER/HLA/BAT/MSH5), 15q24–25.1 CHRNA3/CHRNA5/IREB2, and 19q13.2 CYP2A6 are shared by all four phenotypes; 5p15.33 TERT/CLPTM1L/AHRR, 10q25.1 GSTO2/VTI1A, and 10q23.31 ACTA2/PLCE1 are shared by three of these phenotypes; and over 15 loci are shared by two of the four phenotypes (Supplemental Table 1). Therefore, LC and COPD are not discrete diseases related only through smoking exposure, but genetic predisposition mechanisms may also be shared.
Given the common variants that have been found to be associated with LC in GWAS, exome sequencing with a focused analysis provides a cost-effective approach for further investigation of high-priority regions of the genome and has great potential to identify rare causal variants in GWAS loci, as targeted studies of inflammatory bowel disease [3] and hypertriglyceridemia [4] have demonstrated. Rare variants, with minor allele frequencies (MAFs) of less than 0.01, and with modest to high effect sizes [5–7], might play a crucial role in the etiology of complex traits and could account for missing heritability unexplained by common variants.
Our approach to unveil these hidden rare variants was to sequence selective LC cases adopting a modified extreme phenotype approach. Only about 13 percent of LC cases are reported as familial [8]. However, individuals with a family history of LC are at approximately two- to three fold increased risk of developing this disease [9, 10]. Therefore it could be assumed that LC cases from high risk families would tend to reflect the genetic component of LC etiology more clearly than those not from high risk families. In the present study, we selected: (1) 48 sporadic LC patients reporting heavy smoking histories, 37 of whom exhibited carefully documented severe COPD in whom the environmental factor of smoking is considered overwhelming, and (2) 54 unrelated unique familial LC cases from families with at least three first-degree relatives with LC who are likely enriched for genomic signal, to search for the disease-causing germline rare mutations within the target 107 GWAS loci.
METHODS
Study Population
Familial LC Study Subjects
Phenotype data and biospecimens for 54 LC patients with three or more first-degree relatives affected with histologically-confirmed LC were provided by the Genetic Epidemiology of Lung Cancer Consortium (GELCC) collection. Only one LC patient per family was included in the current study. The selection criteria included availability of adequate amounts and good quality genomic DNA stored at the GELCC biorepository for probands for whom no DNA samples were available on other affected family members. Samples and data were collected by the familial LC recruitment sites of the GELCC, that included the University of Cincinnati, University of Colorado Health Science Center, Karmanos Cancer Institute at Wayne State University, Louisiana State University Health Sciences Center-New Orleans, Mayo Clinic, University of Toledo, Johns Hopkins University, and Saccomanno Research Institute. The GELCC study population and recruitment scheme have been described in detail previously [11]. COPD phenotype was not available on these familial LC patients.
Sporadic LC Study Subjects
Forty-eight sporadic LC patients were selected from enrollees into an on-going study of COPD in current- or former- smokers that was launched in 2002 [12–14]. Ever-smokers aged over 40 were enrolled from three clinics within the Texas Medical Center in Houston, Texas: Ben Taub General Hospital, Houston Methodist Hospital, and Michael E. DeBakey Veterans Affairs Medical Center. The COPD phenotype was carefully defined by irreversible airflow limitation (reduced Forced Expiratory Volume in 1 sec. [FEV1] < 50% predicted and FEV1/FVC < 0.7) assessed by post-bronchodilator spirometry. For this analysis, we selected smokers enrolled into this study who had histologically confirmed LC. Family history of LC information was not available for these sporadic LC patients.
DNA was isolated from peripheral blood from both familial and sporadic LC patients. The study was approved by the institutional review board of all sites accruing participants and by the institutional review board at the Baylor College of Medicine (BCM) for exome sequencing conducted at the BCM Human Genome Sequencing Center (HGSC).
Library Preparation and Capture Enrichment
DNA samples were constructed into Illumina paired-end pre-capture libraries according to the manufacturer’s protocol (Illumina Multiplexing_SamplePrep_Guide_1005361_D). The complete library and capture protocol, as well as oligonucleotide sequences have been described in detail previously [15]. For exome capture, each library pool was hybridized in solution to the BCM-HGSC designed VCRome 2.1 capture reagent according to the manufacturer’s protocol (NimbleGen) with minor revisions.
Exome Sequencing, Alignment, and Variant Calling
The sequencing runs were performed in paired-end mode using the Illumina HiSeq 2000 platform. Sequence analysis was performed using the BCM-HGSC Mercury analysis pipeline [16]. All sequence reads were mapped to the GRCh37 Human reference genome using the Burrows-Wheeler aligner (BWA) [17]. The resulting BAM (binary alignment/map) file underwent quality recalibration using GATK [18]. Putative variants, including single nucleotide variants (SNV), insertions or deletions (Indels), were called using the Atlas2 suite [19]. Read qualities were recalibrated with GATK and a minimum quality score of 30 was required; also, the variant must have been present in > 15% of the reads that cover the position.
Variant Annotation and Filtering
This analysis was restricted to rare mutations mapping to the exons within the 107 selected regions described above (See Supplemental Table 1 for genomic coordinates). Variants were annotated for effect on the protein and predicted function using the SNP & Variation Suite (SVS) software (Golden Helix, Inc). This suite integrates over 378 databases for variant information including: (1) MAF in the European American population in the reference database (dbSNP, 1000 Genomes [TG], Exome Sequencing Project [ESP] 6500) and the UCSC Common SNPs 135/137/141 tracks which include all variants with MAF ≥ 0.01 in the general population; (2) experimental evidence from disease variant databases (such as the COSMIC and ClinVar); and (3) deleterious prediction of variant function determined either by mutation type (truncating, splicing [SP], frame shift [FS], stop gain/loss, or exonic Indels) or mutation effects predicted by dbNSFP Functional Predictions.
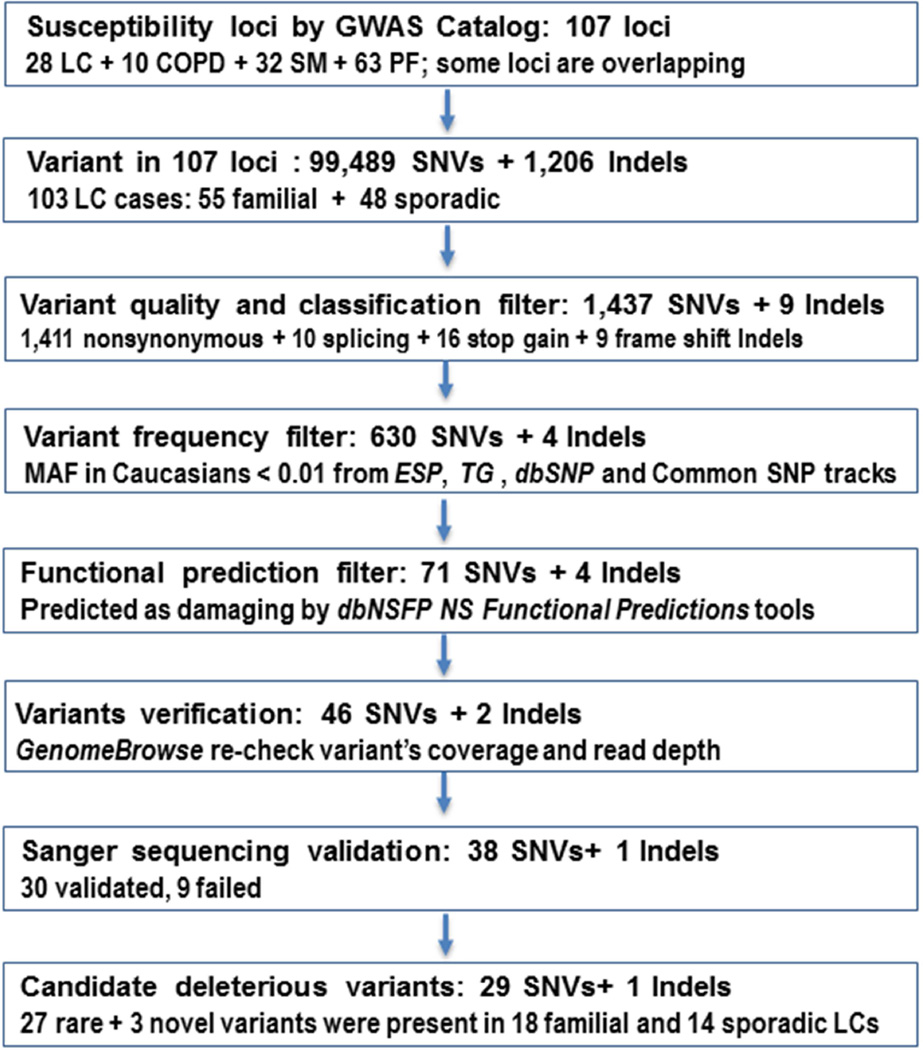
To generate a list of disease causing candidate variants, we focused on identifying genes with rare and novel variants (never having been reported in a publicly available database and UCSC All SNPs 135/137/141 tracks) (Fig. 1). We used scaled C-scores from the Combined Annotation-Dependent Depletion (CADD) method [20] for prioritization of causal variants. A C-score of 10, 20, and 30, indicates variants predicted to be in the top 10%, 1%, and 0.1% of the most deleterious in the human genome, respectively [20]. After implementing the above filtering schema, we used GenomeBrowse (Golden Helix, Inc) to visually confirm the potential candidate variants by re-checking the raw BAM file data. We then tabulated the number of candidate deleterious mutations per gene and within our two study subgroups (familial vs. sporadic), and created a Venn diagram for the list of candidate variants that were significantly associated with the four different phenotypes (LC, COPD, PF, and SM) in previous GWAS.
Figure 1. Workflow and Annotation Pipeline for the Identification of Candidate Variants.
Sanger Validation
The potential candidate variants were verified and segregation examined using Sanger capillary bidirectional sequencing in the selected sample sites. Primers specific to the region containing the variant to be tested were designed, PCR reactions were prepared according to the Qiagen Multiplex PCR Kit protocol (Qiagen), and touchdown PCR was performed (All PCR primers and conditions are available upon request). SNVs were identified using SNP Detector and visually displayed in Sequence Scanner v1.0 (Applied Biosystems).
Candidate Variant Protein Annotation, Structure Modeling and Protein-Protein Interaction
We used online databases Pfam [21] and PRINTS [22] to annotate and classify protein families and domains, the BioGrid and the STRING [23] for predicting protein-protein interaction, and the PHYRE2 server [24] for modeling the 3D structure of the candidate variant gene encoded protein. These resources use sequence-, structure- and systems biology-based features to predict whether the mutation in the protein is likely to have a functional/phenotypic effect.
RESULTS
Demographic information including age, gender, smoking history, and histology is summarized in Table 1. All 54 unrelated familial and 48 sporadic LC cases were adult non-Hispanic whites. The mean age of onset in the familial and sporadic LC cases were 56.0 and 60.9 years, respectively. More than 85% of familial cases and all the sporadic cases (due to study design criteria) reported being ever smokers, with mean pack-years of 52.3, and 60.3, respectively. Overall, 86.0% of the familial and 90.5% sporadic LC cases were diagnosed with non-small cell LC (NSCLC). Adenocarcinoma was diagnosed in 40.5% of the sporadic group, and 30.2% in the familial group for whom histology data were available.
Table 1.
Demographic and Histologic Characteristics of Familial and Sporadic LC Cases
| Characteristics | Familial LC (n = 54) |
Sporadic LC * (n = 48) |
|---|---|---|
| Age of diagnosis | ||
| Mean (SD) | 56.0 (10.1) | 60.9 (4.7) |
| Range | 30 – 70 | 48 – 65 |
| Sex | ||
| Male (%) | 22 (40.0) | 44 (91.7) |
| Female (%) | 32 (60.0) | 4 (8.3) |
| Smoking history | ||
| Ever-smokers (%) | 46 (85.2) | 48 (100) |
| Nonsmoker (%) | 8 (14.8) | 0 (0) |
| Cigarette Pack-years | ||
| Mean (SD) | 52.3 (30.8) | 60.3 (30.9) |
| Range | 0 – 165 | 14 – 150 |
| Histology # | ||
| Non-small cell LC (NSCLC) | 37 (86.0) | 37 (90.5) |
| Adenocarcinoma | 13 (30.2) | 17 (40.5) |
| Squamous cell carcinoma | 17 (39.5) | 16 (38.1) |
| Large cell carcinoma | 7 (16.3) | 5 (11.9) |
| Small cell LC (SCLC) | 6 (14.0) | 4 (9.5) |
Of the 48 sporadic LC, 37 also had severe COPD.
Numbers do not add up because of missing data.
Of 99,489 SNVs and 1,206 Indels located in the exons of the target 107 loci, our stepwise filtering strategy identified 39 potential candidate variants (Fig.1). Of these 39 variants interrogated by Sanger sequencing, 9 mutations failed, and 30 variants (80%) were verified in the original LC samples (Table 2). All the failed mutations were singletons. Of the 30 verified candidate variants, 5 variants were present in two or more patients, three variants were located in highly likely functional sites (CHRNA5 g.78880766 splice donor, MYOZ3 g.150051315 splice acceptor, and C10orf11 p.Ser8 frameshift), and three SNVs were novel (PNPLA8 p.Ile479Ser, PANK1 p.Phe163Ser, and IDE p.Asp9Asn) (Table 2).
Table 2.
List of 30 Candidate Deleterious Germline Mutations in Familial and Sporadic LC Cases
| Region | Disease association |
Gene | Marker * | SNV/Indels | Ref./Alt. | RS ID | MAF in KG/ESP | CADD C-score # |
N mutated Familial vs. Sporadic |
Total N mutated LC cases |
|---|---|---|---|---|---|---|---|---|---|---|
| 10q25.1 | LC+SM+PF | CCDC147 | 10:106163533-SNV | p.Arg696Cys | C/T | rs41291850 | 0.0026/0.0072 | 16.2 | 2 : 1 | 3 |
| 9q34.2 | SM | DBH | 9:136501569-SNV | p.Val26Met | G/A | rs76856960 | 0.0034/0.0045 | 17.3 | 0 : 2 & | 3 |
| 9:136522317-SNV | p.Met563Thr | T/C | rs201973877 | 0.0002/0.0002 | 20.3 | 1 : 0 | ||||
| 15q25.1 | LC+SM+PF+COPD | IREB2 | 15:78783019-SNV | p.Gly747Glu | G/A | rs139092247 | 0.0014/0.0034 | 35 | 1 : 0 | 3 |
| CHRNA5 | 15:78880766-SNV | g. splice donor | G/A | rs200616965 | NA | 22.7 | 0 : 1 & a | |||
| CHRNB4 | 15:78921343-SNV | p.Ala435Val | G/A | rs56317523 | 0.0008/0.0028 | 27.2 | 1 : 0 | |||
| 16q23.1 | PF | KARS | 16:75665388-SNV | p.Arg421Gln | C/T | rs149772470 | 0.0002/0.0018 | 26.1 | 0 : 1 & | 3 |
| 16:75665146-SNV | p.Arg448Cys | G/A | rs77573084 | 0.0006/0.0030 | 19.6 | 0 : 1 & a | ||||
| WWOX | 16:78466521-SNV | p.Arg310Cys | C/T | rs193001955 | 0.0006/0.0006 | 24.1 | 0 : 1 & | |||
| 1q44 | SM | C1orf100 | 1:244541827-SNV | p.Asp71His | G/C | rs41269385 | 0.0022/0.0065 | 14.2 | 2 : 0 | 2 |
| 2q35 | PF | TNS1 | 2:218686643-SNV | p.Glu1027Val | T/A | rs112371945 | 0.0006/0.0013 | 22.7 | 1 : 0 | 2 |
| 2:218669288-SNV | p.Thr1701Met | G/A | rs61740054 | 0.0010/0.0034 | 27.9 | 0 : 1 & | ||||
| 5q32 | LC+PF | FBXO38 | 5:147817940-SNV | p.Pro893Arg | C/G | rs141168806 | NA/0.0001 | 22.9 | 1 : 0 | 2 |
| 5:147821690-SNV | p.Val1108Ile | G/A | rs143682696 | 0.0002/0.0008 | 26.4 | 1 : 0 | ||||
| 7q31.1 | SM | PNPLA8 | 7:108154659-SNV | p.Cys379Gly | A/C | rs141089628 | 0.0002/0.0033 | 15.1 | 0 : 1 & | 2 |
| 7:108137944-SNV | p.Ile479Ser | A/C | Novel | NA | 25.2 | 1 : 0 | ||||
| 10q23.31 | LC+SM+COPD | PANK1 | 10:91359156-SNV | p.Phe163Ser | A/G | Novel | NA | 26.1 | 1 : 0 | 2 |
| IDE | 10:94243061-SNV | p.Asp9Asn | C/T | Novel | NA | 36 | 0 : 1 & b | |||
| 13q12.12 | LC | MIPEP | 13:24448998-SNV | p.Leu197Pro | A/G | rs150167906 | 0.0002/0.0023 | 21.4 | 1 : 1 | 2 |
| 14q22.1 | PF | NID2 | 14:52508948-SNV | p.Thr567Met | G/A | rs150406341 | 0.0006/0.0060 | 19.3 | 1 c : 1 & a | 2 |
| 17q24.2 | LC+PF | BPTF | 17:65889520-SNV | p.Arg823Gln | G/A | rs375975293 | NA/0.0001 | 19.8 | 1 : 0 | 2 |
| 17:65936627-SNV | p.Thr2237Met | C/T | rs372551122 | NA | 17.2 | 0 : 1 & | ||||
| 5q33.1 | LC+PF | MYOZ3 | 5:150051315-SNV | g. splice acceptor | A/G | rs143036945 | 0.0002/0.0005 | 12.3 | 0 : 1 | 1 |
| 10q22.2–3 | PF | C10orf11 | 10:77542754-Deletion | p.Ser8 Frameshift | C/− | rs146123023 | 0.007/0.0013 | NA | 1 : 0 | 1 |
| 12q13.3 | PF | LRP1 | 12:57577915-SNV | p.Arg1993Trp | C/T | rs141826184 | 0.0004/0.0031 | 21.8 | 0 : 1 & | 1 |
| 12q21.2 | SM | NAV3 | 12:78392209-SNV | p.Ser278Ile | G/T | rs755721519 | NA | 31 | 0 : 1 & b | 1 |
| 14q24.2 | PF | SLC8A3 | 14:70515508-SNV | p.Val152Met | C/T | rs144289733 | 0.0004/0.0008 | 27 | 1 : 0 | 1 |
| 15q15.2 | LC | TGM5 | 15:43527092-SNV | p.Tyr502His | A/G | rs146901531 | 0.0002/0.0006 | 18.8 | 0 : 1 & | 1 |
| 18p11.3 | LC | LAMA1 | 18:6965341-SNV | p.Arg2381Cys | G/A | rs142063208 | 0.0028/0.0016 | 25 | 1 c : 0 | 1 |
| 19q13.2 | LC+SM+PF+COPD | EGLN2 | 19:41307024-SNV | p.Val183Met | G/A | rs117916638 | 0.0002/0.0004 | 16.9 | 1 : 0 | 1 |
All are heterozygous mutations.
C-score is the overall measure of deleteriousness. C-score ≥ 20 indicates top 1% deleterious in the human genome.
Sporadic LC patient(s) with severe COPD;
Indicates the same patients.
Overall, the total number and proportion of LC patients (n = 32) carrying these 30 candidate variants was only slightly higher among familial (18 cases, 18/54 = 33.3%) than sporadic cases (14 cases, 14/48 = 29.2%; 11 out these 14 patients also had severe COPD). The mean ages of the familial and sporadic candidate mutation carriers were not different from the overall means. However, in terms of smoking intensity, familial mutation carriers reported lower pack-years than their mean (43 vs. 52) while there was no difference in smoking intensity among sporadic carriers.
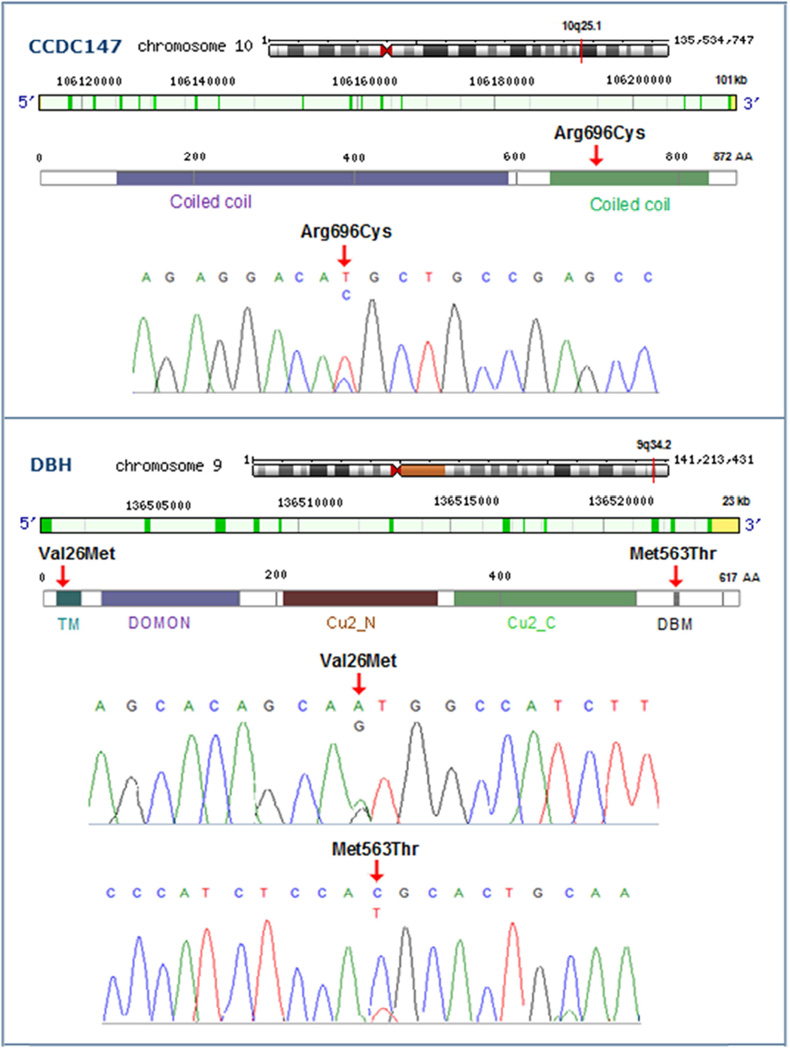
We identified two highly deleterious mutations occurring in more than three LC patients (Table 2 and Supplemental Fig.1). The first was a heterozygous c.2086C>T in the Coiled-coil Domain-containing 147 gene (CCDC147, also named CFAP58), resulting in a p.Arg696Cys substitution. This variant was identified in two familial cases (both female, mean age 54.5, mean pack-years 40, and squamous histology) and one sporadic case (male, age 57, pack-years 88, with adenocarcinoma, without COPD). Notably, the MAF of this variant is 0.0026 from the TG, and 0.0072 from the ESP6500 databases. The mutation is predicted to be protein damaging by PolyPhen-2 (score: 1.0) and highly functional by Mutation taster. This variant has a high scaled CADD C-score of 16.2, which indicates that the Arg696 is predicted to be in the 10% most deleterious of all possible substitutions in the human genome. The CCDC147 spans 101kb, contains 18 exons, and has 872 amino acids (AA) (Fig. 2); the p.Arg696Cys is located in exon 14 and affects a strictly evolutionarily conserved AA residue in the crystal structure of tropomyosin (Protein ID: Q5T655). This p.Arg696Cys mutation is predicted to perturb the tertiary structure (folding of the domain and stability of the three-dimensional shape) of the protein because the Cys696 forms a covalent bond disulfide bridge with 697Cys. It is also very close to the p.Arg698Gln which is a confirmed somatic mutation in cutaneous melanoma patients shown from COSMIC database and the Acetylation modification site Lys692.
Figure 2. Chromosomal Position, Gene Structure, Protein Domain(s), and Sequence of the Candidate Mutations of CCDC147 and DBH Genes.
The mutations were confirmed with Sanger sequencing and indicated with red arrows. CCDC147 includes two coiled coil regions: 106–595 AA, and 642–839 AA. DBH consist of five domains: TM (transmembrane; 20–37 AA), DOMON (dopamine beta-monooxygenase N-terminal; 56–172 AA), Cu2_N (Copper type II, N-terminal; 213–341 AA), Cu2_C (Copper type II, C-terminal; 360–524 AA), and DBM (DB-monooxygenase motif IX; 552–572 AA).
The 2nd candidates were two missense SNVs, p.Val26Met and p.Met563Thr, in the Dopamine beta-hydroxylase (DBH) gene (Table 2 and Supplemental Fig.1). The p.Val26Met presented in two sporadic cases (both male, age 64 and 65, pack-years 40, adenocarcinoma histology, with severe COPD), and p.Met563Thr presenting in one familial case (male, age 30, pack-years 52, histology not specified). MAF of these two variants were 0.0034/0.0045, 0.0002/0.0002 from the TG/ESP6500 databases, respectively. The two mutations were predicted to be protein damaging by PolyPhen-2 (score 0.93 and 0.87), with CADD C-scores (17.3 and 20.3), and exhibited extremely high degrees of sequence conservation (0.96 and 0.99, respectively). The DBH gene contains 12 exons, spans 23 kb, and has 617 AA (Protein ID: P09172; Fig. 2). The p.Val26Met is located within exon 1, lies in the hydrophobic transmembrane (TM) region and possesses helical structure. The p.Met563Thr located in exon 11, lies in a highly conserved region of α helix and the DB monoxgenase (DBM) motif IX that may influence the stability of the enzyme. This somatic mutation is also reported in acute myeloid leukemia patients from the COSMIC database. These observations suggest that the two DBH mutations are likely to have a detrimental effect on the protein.
The other interesting candidates were three SNVs located in the 15q25.1 loci: IREB2 p.Gly747Glu, CHRNA5 g.78880766 splice donor, and CHRNB4 p.Ala435Val. The CHRNA5 splicing variant presenting in a sporadic case (male, 63 years old, 48 pack-years of smoking, NSCLC, with severe COPD), who was also a carrier of another two candidate mutations (NID2 p.Thr567Met and KARS p.Arg448Cys); the IREB2 and CHRNB4 SNVs presented in two familial cases (both female, age 45 and 64, pack-years 45 and 80, small cell and unknown histology).
There were three additional candidate variants, NID2 p.Thr567Met, MIPEP p.Leu197Pro, and C1orf100 p.Asp71His, which were present in multiple LC cases. Other genes that harbored multiple different mutations in different patients included TNS1, FBXO38, PNPLA8, KARS, and BPTF. In addition, a sporadic patient with extremely heavy smoking pack-years (male, 65 years old, 150 pack-years of smoking, adenocarcinoma, with severe COPD) was a carrier of two novel mutations with CADD C-score over 30 (IDE p.Asp9Asn and NAV3 p.Ser278Ile) (Table 2).
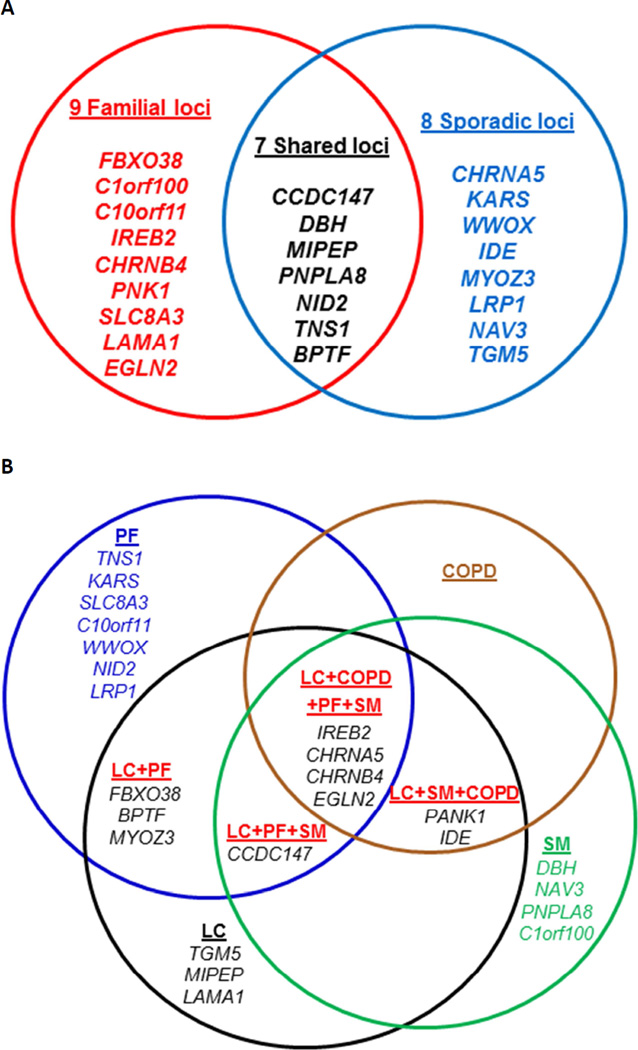
Of the 30 candidate variants belonging to 20 loci and 24 genes (Fig. 3A), seven genes (including CCDC147 and DBH) had candidate variants observed in both the familial and sporadic LC groups. Also, as shown in Fig. 3B, among the candidate genes examined in the current study, the CCDC147, IREB2/CHRNA5/CHRNB4, PANK1/IDE, and EGLN2 genes were shared by three or more phenotypes (LC, COPD, PF, and SM) from the previously published GWAS (Table 2 and Fig. 3B).
Figure 3. Venn Diagrams and Schematic Representations of All Genes with Candidate Mutations in the Familial and Sporadic LC Groups.
A. Shared and specific genes with candidate deleterious variants in the familial, sporadic or both LC groups; B. The list of genes with candidate deleterious variants that were significantly associated with these 4 different phenotypes in previous GWAS studies.
DISCUSSION
Despite previous family-based linkage studies, intensive population-based GWAS analyses and candidate gene screening, a large proportion of the heritability of LC remains unexplained. Using an extreme phenotype design, this report describes the first exome sequencing approach comparing familial and sporadic heavy smoking LC patients evaluating the effects of rare coding variation in the GWAS loci associated with LC, COPD, SM, and PF. Our results showed the familial mutation carriers reported lower pack-years than their group’s mean (43 vs. 52) while there was no difference in smoking intensity among sporadic carriers. Further, we identified two disease-causing rare mutations on 10q25.1 (CCDC147 p.Arg696Cys) and 9q34.2 (DBH p.Val26Met and p.Met563Thr), and three suggestive rare mutations on 15q25.1 (IREB2 p.Gly747Glu, CHRNA5 g.78880766 splice donor, and CHRNB4 p.Ala435Val), although the findings require replication. Familial and sporadic LC cases are indistinguishable upon clinical presentation and our results demonstrated that the two forms of LC may have both shared determinants and distinct components.
Strong evidence for a LC-conferring deleterious mutation was observed at CCDC147 p.Arg696Cys in three LC patients (2 familial and 1 sporadic). Interestingly, the two familial carriers were lighter smokers and had earlier age of onset than the overall means for familial cases. The sporadic carrier was a heavier smoker with 88 pack-years and did not present with documented COPD. Although several genes in 10q25.1 loci have been implicated in susceptibility to LC [25], PF [26], and SM [27] in GWAS, very little is known about the function of CCDC147 gene in humans or mice, although it is expected to produce a functional protein as described in the Proteomics database. CCDC147 protein, also known as cilia- and flagella-associated protein 58 (CFAP58), demonstrates high expression in T cells, nasal epithelium, lungs and alveolar fluids (http://www.genecards.org/cgi-bin/carddisp.pl?gene=CFAP58). It is expected to interact with members of the shelterin complex, the human Telomere Repeat-Binding Factors (TRF1) and Protection of Telomeres 1 (POT1), as reported in the BioGRID database and STRING Interaction Network. Interestingly, recent studies have shown that rare mutations in POT1 are associated with chronic lymphocytic leukemia [28], familial melanoma [29] and familial glioma [30], where it is thought to result in telomere de-protection and length extension associated with cancer. Further, one of the most important functions of shelterin includes modulation of telomerase activity, which has been detected in ~85% of cancers, and is linked to genomic instability and tumorigenesis. Although direct evidence regarding the biological function of CCDC147 is lacking, our finding of CCDC147, as a novel telomere-interacting protein, underscores the need for future work that could elucidate the role of this gene in LC pathogenesis.
Another main finding was the highly disruptive and deleterious rare mutations on 9q34.2 DBH p.Val26Met and p.Met563Thr, in three LC patients (1 familial and 2 sporadic). The familial carrier was very young (age 30). Both sporadic carriers were adenocarcinoma and had severe COPD. Previous GWAS identified DBH rs3025343 as a locus associated with SM [31]. The DBH (OMIM 609312) primarily contributes to conversion of dopamine to noradrenaline. Dopamine is known to be released from neurons in response to nicotine and plays a well-documented role in determining an individual's predisposition to nicotine dependence, through its role in mediating drug reward in the brain [31–34]. The contribution of cigarette smoking to both LC and COPD could invoke a variety of underlying biological processes including inflammation, epithelial-mesenchymal transition, oxidative stress, DNA repair and abnormal cellular proliferation.
NID2 is a known GWAS hit for PF [35] and blood lipid phenotypes [36], and a new biomarker for ovarian cancer [37], hepatocellular carcinoma [38] and oral squamous cell carcinoma [39]. NID2 (OMIM 605399) encodes a member of the highly conserved nidogen family of basement membrane proteins. This protein binds collagens I/IV and laminin, is involved in stabilizing and maintaining the structure of the basement membrane, and plays a key role in cell-extracellular matrix. Unbalanced proteolysis in the extracellular matrix is a potential mechanism to explain inflammatory processes within the emphysematous lung. NID2 mutation in LC patients may favor invasion and metastasis of tumor cells by loosening cell interaction with basal membrane and by weakening the strength of the basement membrane itself, and could be a marker of progression as well.
A main strength of the study is the focus on patients with extreme phenotypes who are most likely to be informative. For quantitative traits, one can select individuals with extreme trait values after adjusting for known covariates. Alternatively, in disease-focused studies, selecting individuals with extreme phenotypes can be conducted on the basis of known risk factors. Smoking, family history of LC and COPD are all well documented risk factors for LC. Because the frequencies of alleles that contribute to the trait/disease are enriched in phenotype extremes (such as familial LC or patients with both LC and COPD), studying extremes has been shown to provide >5 times power (only 20% of the subjects compared to traditional designs) [7]. In the present study, the recurrent rare mutations described herein suggest that it may be possible to identify susceptibility genes in a relatively small sample size, although we cannot rule out the possibility that the results are observed by chance. The small sample size and lack of validation of the identified mutations in a separate large scale cohort limit the relevance of our findings. Another limitation in this analysis is phenotype misclassification between familial and sporadic LCs. For the familial LC patients, we lacked COPD phenotype data, and for the sporadic LC cases, family history of LC was not available. Also, we acknowledge the existence of gender imbalance between familial and sporadic cases that could cause bias and limit applicability of the findings to the general population.
In summary, our results demonstrated highly disruptive germline mutations in CCDC147 and DBH in LC patients that are interesting candidates for LC risk alleles. The overlap in risk loci between familial and sporadic LC, and between COPD and LC may be due to genes and mutations involving telomere maintenance, inflammation, or to a lack of family history in the sporadic cases being due to no smoking exposure in other carriers of the mutation in their families. Therefore, going forward, comprehensive genomic analyses of whole genomes, from point mutations to large structural variants, of a large number of LC samples in diverse race/ethnic groups for validation, and further functional works for the two top candidate genes will be needed to better understand the underlying molecular genetics and to guide screening for mutations in this unique subset of patients to assess their potential LC risk.
Supplementary Material
ACKNOWLEDGEMENT
We would like to thank the patients and their families for participating in this research. We thank Dr. Richard Gibbs, Donna Muzny, Xiaoyun Liao, Van Le, Sandra Lee, and Margi Sheth from the HGSC-BCM for performing the exome sequencing for all the samples in this study.
Funding Support: This work was supported by grants from the National Institutes of Health (R01 CA127219, R01 HL082487, R01 HL110883, K07CA181480, R01 CA060691, R01 CA87895, R01 CA80127, R01 CA84354, R01 CA134682, R01 CA134433, R03 CA77118, P20GM103534, P30CA125123, P30CA023108, P30-ES006096, P30CA022453, N01-HG-65404, U01CA076293, U19CA148127, and HHSN268201 200007C). JEB-W was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. Additional support was provided by the National Library of Medicine T15LM007093 (Davis) and the Population Sciences Biorepository at Baylor College of Medicine (BCM).
Abbreviation
- LC
lung cancer
- COPD
chronic obstructive pulmonary disease
- SM
smoking behavior
- PF
pulmonary function
- GELCC
Genetic Epidemiology of Lung Cancer Consortium
- SNV
Single nucleotide variants
- Indels
Insertions or deletions
- MAF
minor allele frequency
- AA
amino acid
- FS
frameshift
- SP
splicing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Financial Interests: The authors declare no competing financial interests.
Web Resources
GWAS, Genome-wide association studies Catalog, www.genome.gov/gwastudies/
TG, Thousand Genomes, http://www.1000genomes.org
ESP, Exome Sequencing Project, http://evs.gs.washington.edu/EVS/
COSMIC, Catalogue of Somatic Mutations in Cancer, http://cancer.sanger.ac.uk/cosmic
OMIM, Online Mendelian Inheritance in Man, http://www.omim.org
CADD, Combined Annotation Dependent Depletion, cadd.gs.washington.edu/
Pfam, http://pfam.janelia.org/
PRINTS, Protein Motif fingerprint database, http://www.bioinf.manchester.ac.uk/dbbrowser/PRINTS/
BioGRID, Biological General Repository for Interaction Datasets, http://www.thebiogrid.org
STRING, Search Tool for the Retrieval of Interacting Genes, http://string-db.org/
Phyre2, www.sbg.bio.ic.ac.uk/phyre2/
REFERENCE
- 1.Etzel CJ, Kachroo S, Liu M, et al. Development and validation of a lung cancer risk prediction model for African-Americans. Cancer Prev Res (Phila) 2008;1(4):255–265. doi: 10.1158/1940-6207.CAPR-08-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tammemagi CM, Pinsky PF, Caporaso NE, et al. Lung cancer risk prediction: Prostate, Lung, Colorectal And Ovarian Cancer Screening Trial models and validation. J Natl Cancer Inst. 2011;103(13):1058–1068. doi: 10.1093/jnci/djr173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rivas MA, Beaudoin M, Gardet A, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43(11):1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johansen CT, Wang J, Lanktree MB, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42(8):684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang G, Lin D, Hakonarson H, et al. Two-stage extreme phenotype sequencing design for discovering and testing common and rare genetic variants: efficiency and power. Hum Hered. 2012;73(3):139–147. doi: 10.1159/000337300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamina C. Digging into the extremes: a useful approach for the analysis of rare variants with continuous traits? BMC Proc. 2011;5(Suppl 9):S105. doi: 10.1186/1753-6561-5-S9-S105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D, Lewinger JP, Gauderman WJ, et al. Using extreme phenotype sampling to identify the rare causal variants of quantitative traits in association studies. Genet Epidemiol. 2011;35(8):790–799. doi: 10.1002/gepi.20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey-Wilson JE, Amos CI, Pinney SM, et al. A major lung cancer susceptibility locus maps to chromosome 6q23–25. Am J Hum Genet. 2004;75(3):460–474. doi: 10.1086/423857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ooi WL, Elston RC, Chen VW, et al. Increased familial risk for lung cancer. J Natl Cancer Inst. 1986;76(2):217–222. [PubMed] [Google Scholar]
- 10.Jonsson S, Thorsteinsdottir U, Gudbjartsson DF, et al. Familial risk of lung carcinoma in the Icelandic population. JAMA. 2004;292(24):2977–2983. doi: 10.1001/jama.292.24.2977. [DOI] [PubMed] [Google Scholar]
- 11.Liu P, Vikis HG, Wang D, et al. Familial aggregation of common sequence variants on 15q24–25.1 in lung cancer. J Natl Cancer Inst. 2008;100(18):1326–1330. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SH, Goswami S, Grudo A, et al. Antielastin autoimmunity in tobacco smoking-induced emphysema. Nat Med. 2007;13(5):567–569. doi: 10.1038/nm1583. [DOI] [PubMed] [Google Scholar]
- 13.Grumelli S, Corry DB, Song LZ, et al. An immune basis for lung parenchymal destruction in chronic obstructive pulmonary disease and emphysema. PLoS Med. 2004;1(1):e8. doi: 10.1371/journal.pmed.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shan M, Cheng HF, Song LZ, et al. Lung myeloid dendritic cells coordinately induce TH1 and TH17 responses in human emphysema. Sci Transl Med. 2009;1(4):4ra10. doi: 10.1126/scitranlsmed.3000154. [DOI] [PubMed] [Google Scholar]
- 15.Lupski JR, Gonzaga-Jauregui C, Yang Y, et al. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013;5(6):57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reid JG, Carroll A, Veeraraghavan N, et al. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Challis D, Yu J, Evani US, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kircher M, Witten DM, Jain P, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finn RD, Mistry J, Schuster-Bockler B, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Attwood TK, Bradley P, Flower DR, et al. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31(1):400–402. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stark C, Breitkreutz BJ, Reguly T, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 25.Lan Q, Hsiung CA, Matsuo K, et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet. 2012;44(12):1330–1335. doi: 10.1038/ng.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hancock DB, Eijgelsheim M, Wilk JB, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42(1):45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramsay AJ, Quesada V, Foronda M, et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet. 2013;45(5):526–530. doi: 10.1038/ng.2584. [DOI] [PubMed] [Google Scholar]
- 29.Robles-Espinoza CD, Harland M, Ramsay AJ, et al. POT1 loss-of-function variants predispose to familial melanoma. Nat Genet. 2014;46(5):478–481. doi: 10.1038/ng.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bainbridge MN, Armstrong GN, Gramatges MM, et al. Germline mutations in shelterin complex genes are associated with familial glioma. J Natl Cancer Inst. 2015;107(1):384. doi: 10.1093/jnci/dju384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siedlinski M, Cho MH, Bakke P, et al. Genome-wide association study of smoking behaviours in patients with COPD. Thorax. 2011;66(10):894–902. doi: 10.1136/thoraxjnl-2011-200154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiels MS, Huang HY, Hoffman SC, et al. A community-based study of cigarette smoking behavior in relation to variation in three genes involved in dopamine metabolism: Catechol-O-methyltransferase (COMT), dopamine beta-hydroxylase (DBH) and monoamine oxidase-A (MAO-A) Prev Med. 2008;47(1):116–122. doi: 10.1016/j.ypmed.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freire MT, Marques FZ, Hutz MH, et al. Polymorphisms in the DBH and DRD2 gene regions and smoking behavior. Eur Arch Psychiatry Clin Neurosci. 2006;256(2):93–97. doi: 10.1007/s00406-005-0610-x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang XY, Chen da C, Xiu MH, et al. Association of functional dopamine-beta-hydroxylase (DBH) 19 bp insertion/deletion polymorphism with smoking severity in male schizophrenic smokers. Schizophr Res. 2012;141(1):48–53. doi: 10.1016/j.schres.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 35.Wilk JB, Walter RE, Laramie JM, et al. Framingham Heart Study genome-wide association: results for pulmonary function measures. BMC Med Genet. 2007;8(Suppl 1):S8. doi: 10.1186/1471-2350-8-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kathiresan S, Manning AK, Demissie S, et al. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S17. doi: 10.1186/1471-2350-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuk C, Gunawardana CG, Soosaipillai A, et al. Nidogen-2: a new serum biomarker for ovarian cancer. Clin Biochem. 2010;43(4–5):355–361. doi: 10.1016/j.clinbiochem.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng ZX, Huang XH, Wang Q, et al. Clinical significance of decreased nidogen-2 expression in the tumor tissue and serum of patients with hepatocellular carcinoma. J Surg Oncol. 2012;105(1):71–80. doi: 10.1002/jso.22047. [DOI] [PubMed] [Google Scholar]
- 39.Guerrero-Preston R, Soudry E, Acero J, et al. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev Res (Phila) 2011;4(7):1061–1072. doi: 10.1158/1940-6207.CAPR-11-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emond MJ, Louie T, Emerson J, et al. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat Genet. 2012;44(8):886–889. doi: 10.1038/ng.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musunuru K, Pirruccello JP, Do R, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363(23):2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.