Abstract
There are several malignancies of the digestive system (including gastric, pancreatic and colorectal cancers, and hepatocellular carcinoma), which are the most common types of cancer and a major cause of death worldwide. MicroRNA (miR)-7 is abundant in the pancreas, playing an important role in pancreatic development and endocrine function. Expression of miR-7 is downregulated in digestive system malignancies compared with normal tissue. Although there are contrasting results for miR-7 expression, almost all research reveals that miR-7 is a tumor suppressor, by targeting various genes in specific pathways. Moreover, miR-7 can target different genes simultaneously in different malignancies of the digestive system. By acting on many cytokines, miR-7 is also involved in many gastrointestinal inflammatory diseases as a significant carcinogenic factor. Consequently, miR-7 might be a biomarker or therapeutic target gene in digestive system malignancies.
Keywords: MicroRNA-7, Digestive system malignancy, Tumor biomarker, Target gene, Inflammation
Core tip: MicroRNA (miR)-7 targets different genes in various complicated pathways and plays diagnostic, prognostic, anti-metastatic, and therapeutic roles in digestive system malignancies. MiR-7 might be a biomarker or therapeutic target gene in digestive system malignancies, even in the precancerous lesions (inflammatory disease).
INTRODUCTION
MicroRNAs are small noncoding RNAs consisting of 18-25 nucleotides that post-transcriptionally regulate expression of target genes, and are involved in cell proliferation, epithelial-mesenchymal transition (EMT), apoptosis, migration, invasion and metastasis[1-3]. miRNAs have emerged as potential critical regulators of carcinogenesis and tumor progression[4,5].
The digestive system is composed of many ducts and glands, and because of its complicated physiology and anatomy, numerous diseases may occur, especially malignancies including the third, fourth and eighth most common cancers worldwide: Colorectal cancer (CRC), gastric cancer (GC) and esophageal cancer, respectively[6,7], as wells as the leading cause of cancer-related death: Pancreatic cancer (PC)[8]. According to the 2014 cancer statistics, the combined cancer mortality rates have been continuously declining for the past two decades. However, the incidence of some digestive system malignancies, including cancers of the esophagus, liver, anus and pancreas, is increasing. Moreover, with rising death rates for cancers of the liver, anus and pancreas, and other non-digestive cancers, cancer is still the second leading cause of death following heart disease[8]. Therefore, it is necessary for us to explore novel molecular mechanisms, and screen for the most effective therapeutic methods to avoid the majority of patients succumbing to these digestive malignancies.
MicroRNA (miR)-7 is an evolutionarily conserved miRNA that is involved in the development of the eye and pancreas in Drosophila. Li et al[9] reported that miR-7 is repressed by the transcription factor Yan, which is degraded while mediating epidermal growth factor receptor (EGFR) signaling. Also, miR-7 is expressed abundantly in human pancreas and endocrine cells and has a specific role in endocrine cell differentiation and function[10]. It has been demonstrated that miR-7 is a tumor suppressor in breast, lung and ovarian cancers, and glioblastoma, mainly focusing on its relationship with EGFR[11-15]. Accumulating evidence shows that miR-7 can simultaneously target a variety of mRNAs involved in diverse signaling pathways in different tumors. However, no specific review has described the role of miR-7 in digestive tract malignancies. In this review, we focus on current research on miR-7 in order to elucidate its role in digestive system malignancies or their precancerous lesions, with reference to its expression, signaling pathways, and role as a circulatory biomarker.
EXPRESSION OF MIR-7
By comparing the differential expression of miRNAs in pancreatic islets (endocrine) and acinar (exocrine) tissue in rats, using microarray and quantitative polymerase chain reaction (qPCR), Bravo-Egana et al[16] revealed that miR-7 was ranked highest among the 17 miRNAs preferentially expressed in islets, suggesting that it acts as an endocrine miRNA. Another two studies reported that miR-7 was expressed at a high level during human pancreatic islet development[10,17]. For malignancy, it has been demonstrated that miR-7 is downregulated in cancer tissue of digestive malignancies such as GC[18-20], CRC[21,22] and hepatocellular carcinoma (HCC)[23] by comparison with normal tissues, suggesting that it acts as a suppressor. A similar conclusion was drawn in a study of hydroxycamptothecin-resistant GC cells[24]. In some inflammatory diseases, such as gastritis and Crohn’s disease[25], the level of miR-7 is also lower than that in normal tissue, which suggests that it is an inflammation-related miRNA participating in the process of digestive cancer.
In contrast, using the same method, Suto et al[26] discovered that miR-7 level was higher in CRC tissue than in adjacent normal tissue, induced by EGFR mutations. However, it was found that the aforementioned results would be opposite when the EGFR protein expression was positive in CRC. Finally, they concluded that low miR-7 expression resulted in poorer prognosis than high expression. Ahmed et al[27] identified the expression of miR-7 in stool samples from 40 cases of colon cancer (TNM stages 1-4), and found that miR-7 was one of the 12 increased miRNAs, which they then recognized as a diagnostic gene. In HCC, Fang et al[28] speculated that owing to inactivation of the transcriptional regulators and/or failure to promote miR-7 expression, there is no alteration of its expression between tumor and adjacent normal tissues. However, miR-7 and miR-21 are overexpressed in esophageal squamous cell carcinoma (ESCC) and related to its differentiation[29].
From Table 1, we can speculate the reasons for the divergent views about the expression of miR-7 under different conditions, including[30]: (1) heterogeneity of different malignancies/diseases; (2) different study sample sizes; and (3) the standards were not the same (e.g., whether or not to include patients with prior cytotoxic therapy). Based on the published studies, we conclude that miR-7 could be an oncogene or tumor suppressor in the digestive system depending on the specific gene targeted (Table 2).
Table 1.
Expression of microRNA-7 in the digestive system
| Cancer | Sample | Sap No. | Method | Exp | Role | Ref. |
| Colon | Stool | 60 | qPCR | ↑ | Diagnostic | [27] |
| CRC | Tissue | 80 | qPCR | ↓ | Diagnostic/therapeutic | [21] |
| CRC | Tissue | 8 | RT-PCR | ↓ | Therapeutic | [22] |
| CRC | Tissue | 105 | qRT-PCR | ↑ | Prognostic | [26] |
| ESCC | Tissue | 34 | Microarray/qRT-PCR | ↑ | Differentiation | [29] |
| GC | Tissue | 40 | ISH/IHC | ↓ | Inhibits metastasis/EMT | [18] |
| GC | Tissue | 23 | Microarray | ↓ | Inhibits invasion/metastasis | [19] |
| qRT-PCR | ||||||
| GC | Tissue | 28 | RT-PCR | ↓ | Represses inflammation | [20] |
| HCC | Tissue | 10 | Microarray | - | Therapeutic/diagnostic/prognostic | [28] |
| qRT-PCR | ||||||
| HCC | Tissue | 12 | qRT-PCR | ↓ | Tumor suppressor | [23] |
| HCC | Tissue | 429 | Chip assay | ↓ | Prognostic | [43] |
| CD | Tissue | - | RT-PCR | ↓ | Therapeutic | [25] |
↓: MiR-7 is downregulated; ↑: MiR-7 is upregulated; −: There is no alteration for expression of miR-7, or no mention. Sap No.: Sample number; Exp: Expression; ISH/IHC: In situ hybridization/immunohistochemistry; CRC: Colorectal cancer; GC: Gastric cancer; HCC: Hepatocellular carcinoma; ESCC: Esophageal squamous cell carcinoma; EMT: Epithelial–mesenchymal transition; qPCR: Quantitative polymerase chain reaction.
Table 2.
Function of microRNA-7 by targeting diverse genes
| Cells | Function of miR-7 | Target | Ref. |
| CCA | Reduces migration, invasion and metastasis | LAT1 | [41] |
| CRC | Inhibits proliferation, invasion and metastasis, and induces G1 arrest | PAX6 | [21] |
| CRC | Inhibits proliferation and induces apoptosis | XRCC2 | [22] |
| CRC | Suppresses proliferation, induces G1 arrest, and induces apoptosis | YY1 | [37] |
| GC | Suppresses invasion and metastasis | IGF1R | [18] |
| GC | Inhibits proliferation, invasion and metastasis | EGFR | [19] |
| HCC | Decreases invasion and migration | PIK3CD/mTOR/p70S6K | [28] |
| HCC | Suppresses colony formation and induces cell cycle arrest | CUL5 | [23] |
CCA: Cholangiocarcinoma; CRC: Colorectal cancer; GC: Gastric cancer; HCC: Hepatocellular carcinoma; miR-7: MicroRNA; XRCC2: X-ray repair complementing defective repair in Chinese hamster cells 2; IGF1R: Insulin-like growth factor 1 receptor; EGFR: Epidermal growth factor receptor; mTOR: Mammalian target of rapamycin; CUL5: Cullin 5.
GC
The pathogenesis of GC has been extensively studied, and there is a consensus that intestinal gastric carcinogenesis is a multistep process starting with chronic gastritis triggered by Helicobacter pylori, progressing through atrophy, intestinal metaplasia and dysplasia to carcinoma (Correa model)[31]. Thus, inflammation is a significant event in gastric carcinogenesis, whereas miR-7 is an inflammation-mediated miRNA inversely correlated with many proinflammatory cytokines and inflammatory factors such as interleukin-1β and tumor necrosis factor-α. Three genes, LPHN2, BASP1 and MAFG, targeted by miR-7 are induced in the cyclo-oxygenase (COX)-2/prostaglandin (PG) E2 pathways, which shows that miR-7 plays a significant part in gastric tumorigenesis with an inflammatory response[20]. MiR-7 suppresses GC cell invasion and metastasis both in vitro and in vivo by targeting the miR-7/insulin-like growth factor 1 receptor/Snail axis, which shows its EMT function and suggests that it can act as a therapeutic biomarker to prevent GC metastasis[18]. Xie et al[19] demonstrated that restoration of miR-7 significantly inhibited tumor cell viability, invasion and migration by suppressing EGFR expression. These results suggest that targeting miR-7 is a potential therapeutic option for GC (Figure 1).
Figure 1.
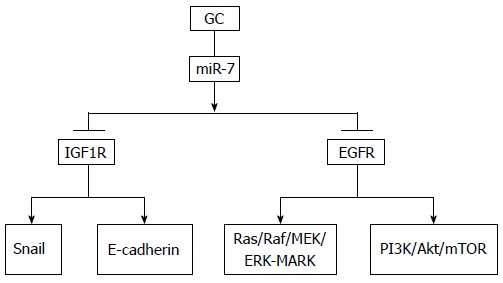
Pathway of microRNA-7 in gastric cancer. It has been revealed that miR-7 targets mainly IGF1R and EGFR. IGF1R: Insulin-like growth factor 1 receptor; EGFR: Epidermal growth factor receptor; GC: Gastric cancer; miR-7: MicroRNA-7; PI3K: Phosphoinositide 3-kinase; mTOR: Mammalian target of rapamycin.
PC
PC is one of the major leading causes of cancer mortality; the 5-year survival rate for pancreatic adenocarcinoma is < 5%, and most patients die within the first 2 years[8]. Therefore, there is an urgent need to explore novel therapeutic methods. In accordance with the expression of miR-7 in the pancreas, miR-7-3, which is one of the three endogenous genes potentially transcribed in the human genome, is upregulated by targeting mitogen-activated protein kinase (MAPK), suggesting that miR-7 is negatively modulated by an EGFR-MAPK feedback loop[32]. In an in vitro study, in which miR-7 targeted SET domain containing 8 leading to increased p53 expression and decreased Bcl-2 level, curcumin suppressed cell growth, migration and invasion, and induced apoptosis in PC cells, indicating that targeting miR-7 is a useful therapeutic option for PC[33]. Although knockdown of LIV-1 (a zinc transporter) can upregulate expression of miR-7 in PC cells, the exact role of miR-7 in the maintenance of cancer-stem-cell-related phenotypes in PC remains unclear[34]. Future research will focus on identifying the exact pathway of miR-7 in PC, and only in this way, can research proceed from bench to bedside (Figure 2).
Figure 2.
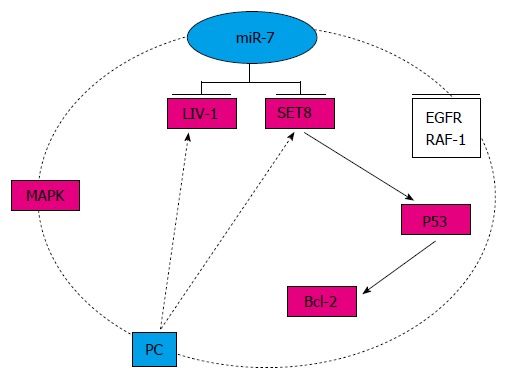
Pathway of microRNA-7 in pancreatic cancer. In PC, there is an EGFR-MAPK-miR-7 negative feedback loop, and LIV-1 and SET8 are two other targets. EGFR: Epidermal growth factor receptor; MAPK: Mitogen-activated protein kinase; PC: Pancreatic cancer; miR-7: MicroRNA-7; SET8: SET domain containing 8.
CRC
CRC is related to the mutation of genes such as P53, APC, SMAD4, PIK3CA, KRAS, ARID1A, SOX9 and FAM123B. Some minimal tailoring of therapy (selecting a chemotherapeutic agent based on toxicity, or not using anti-EGFR in those with KRAS-mutated tumors) can be offered to patients, however, the dream of truly individualized therapy remains elusive[35,36]. Based on the present studies, miR-7 can target specific genes to modulate the correlated pathways, and its decreased expression continuously participates in the process of CRC. MiR-7 is a tumor suppressor, which is mediated through the YY1-P53-Wnt signaling pathway, and plays pivotal roles in many cellular processes, such as development, differentiation, proliferation and apoptosis[37]. By targeting EGFR and v-raf-1 murine leukemia viral oncogene homolog 1 (RAF-1), a low level of miR-7 suggests poor prognosis for CRC, and miR-7 precursor, alone or in combination with a monoclonal antibody, could be a novel therapy against CRC[26]. XRCC2 (X-ray repair complementing defective repair in Chinese hamster cells 2) participates in homologous recombination, and its relationship with miR-7 has been studied. In vitro, overexpression of miR-7 suppressed proliferation and induced apoptosis of CRC cells by directly targeting XRCC2 through decreasing cyclin D1 and increasing p21, caspase-3 and BAX expression[22]. In addition, the expression of paired box (PAX) 6 is inversely correlated with that of miR-7, and simultaneous activation of the extracellular signal-regulated kinase and phosphoinositide 3-kinase (PI3K) signaling pathways and regulation of the levels of matrix metalloproteinase (MMP) 2 and MMP9 could modulate the expression of PAX6 and miR-7 in opposing ways, which suggests that miR-7 is a promising therapeutic target for CRC[21]. Thus, further mechanisms mediated by miR-7 should be explored, which could be a promising approach for individually tailored therapy of CRC (Figure 3).
Figure 3.
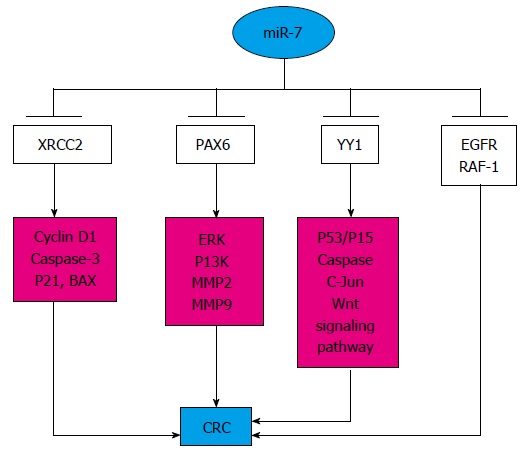
Pathway of microRNA-7 in colorectal cancer. MiR-7 can target various genes involving different pathways and act as a suppressor. CRC: Colorectal cancer; EGFR: Epidermal growth factor receptor; miR-7: MicroRNA-7. XRCC2: X-ray repair complementing defective repair in Chinese hamster cells 2; ERK: Extracellular signal-regulated kinase.
HCC
HCC, the third most common cause of cancer mortality worldwide, which develops from activation of cellular oncogenic pathways and abrogation of tumor suppressor pathways including the p53/p21WAF1 pathway, the p16INK4a/CDK4/RB1/E2F pathway, the Wnt/β-catenin signaling pathway, transforming growth factor-α, c-myc, transcription factor NF-κB, insulin/IGF-I, and receptor tyrosine kinases and their downstream activators[38,39]. Several studies have shown that miR-7 participates in several pathways by targeting different genes in HCC. MiR-7 regulates the PI3K/Akt/mammalian target of rapamycin in vitro and in vivo, which functions downstream of EGFR, suggesting that miR-7 is a potential target for treating or diagnosing/prognosing HCC[28]. Likewise, ectopic expression of cullin 5, a novel target gene of miR-7, inhibits HCC cell proliferation, arrests cell cycle progression, and suppresses colony formation, although the exact pathway remains unclear[23]. Moreover, a member of the highly conserved cyclin family, CCNE1 (cyclin E1), is inversely correlated with miR-7 expression in HCC cell lines and clinical samples, indicating that it is a downstream mediator for miR-7, and miR-7 might be a candidate for the treatment of HCC[40]. Most studies in this field have been in vitro experiments, except one[28]. The detailed mechanisms remain to be elucidated, thus, the exact role of miR-7 in HCC needs further research (Figure 4).
Figure 4.
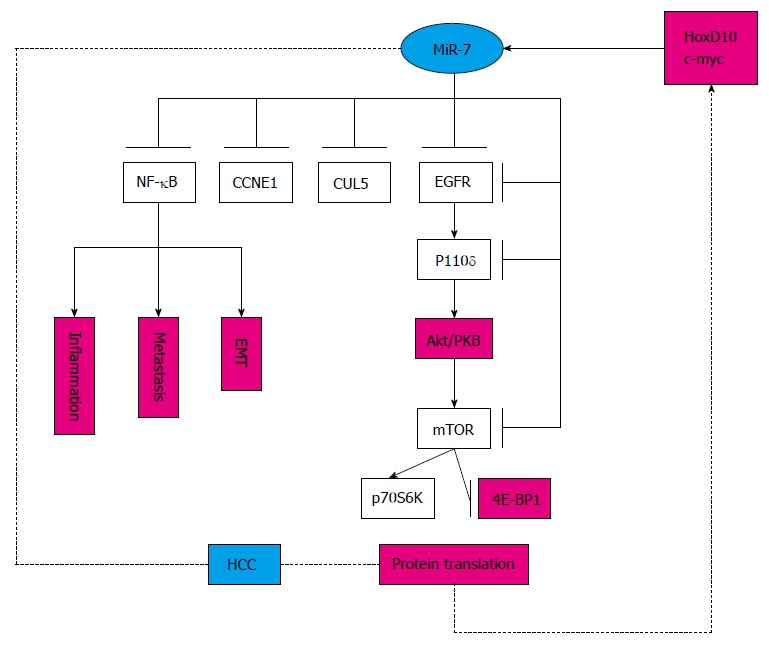
Pathway of miR-7 in hepatocellular carcinoma. MiR-7 can target various genes in the specific signaling pathway. HCC: Hepatocellular carcinoma; miR-7: MicroRNA-7; NF-κB: Nuclear factor κB; CCNE1: Cyclin E1; CUL5: Cullin 5; EGFR: Epidermal growth factor receptor; mTOR: Mammalian target of rapamycin; EMT: Epithelial–mesenchymal transition.
OTHER DIGESTIVE MALIGNANCIES
MiR-7 also plays a role in other digestive tract malignancies such as cholangiocarcinoma and ESCC[29,41]. However, the expression and exact role of miR-7 in these two malignancies need to be verified.
INFLAMMATORY DISEASE
Inflammation makes a significant contribution to carcinogenesis and progression of malignancies[42]. In some conditions, inflammation such as chronic atrophic gastritis is defined as a precancerous lesion of GC. MiR-7 also participates in some inflammatory diseases, in addition to malignancies. The role of miR-7 in the progression from chronic inflammation to GC has been studied more thoroughly compared with other inflammatory diseases. Using established mouse models, Kong et al[20] have demonstrated that downregulation of miR-7 induced by PGE2 associated with inflammation, and activation of EGFR are critical steps in gastric carcinogenesis. Although the COX-2/mPGES-1/PGE2/EP2 pathway has been identified in gastric tumorigenesis, whether there is a similar mechanism mediated by miR-7 has not been established in other malignancies. It has been revealed that the expression of miR-7 is decreased in actively inflamed colonic tissues form patients with Crohn’s disease, which is regulated by hCD98[25]. Similarly, chronic hepatitis has an important influence on HCC development, and hepatocyte nuclear factor 4α and NF-κB form a feedback circuit, for which miR-7 and miR-124 could be the targets[43]. These findings suggest that miR-7 is involved in many inflammatory diseases by activating many inflammatory/proinflammatory cytokines, and it will be intriguing to demonstrate the role of miR-7 in the regulation of alimentary inflammatory responses and carcinogenesis.
CIRCULATORY BIOMARKER
The diagnostic and prognostic biomarkers for some digestive malignancies including GC and CRC are still limited. Common circulatory markers such as carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 have inadequate sensitivity, therefore, exploring specific biomarkers is a significant breakthrough.
MiRNAs are stable in serum, plasma and body fluids (e.g., stools and gastric juice), and their expression differs between tumor and non-tumor tissue. Some miRNAs, like miR-21, have been subjected to meta-analysis and concluded to be diagnostic biomarkers for GC[44]. Wang et al[45] designed their study with three phases. In the discovery phase, they detected 723 miRNAs in 80 serum samples using microarrays; in the training phase they experimented on another 112 plasma samples using qPCR; and finally, they confirmed the results with 49 samples using a logistic model, and screened miR-7 as one of a panel that yielded high diagnostic accuracy to diagnose CRC. Compared with CEA, miR-7 has a higher receiver operating characteristic curve, sensitivity and specificity (0.897, 82% and 89%, respectively). Similarly, by analyzing the serum from 12 acute pancreatitis patients and three healthy controls, Liu et al[46] identified miR-7 as one of the three diagnostic and prognostic biomarkers. Although several systematic reviews[44,47-49] have investigated biomarkers for GC, none has shown that miR-7 could be a biomarker of GC.
CONCLUSION
Several studies have identified possible mechanisms mediated by miR-7 in specific malignancies of the digestive system, including some inflammatory diseases. No study has investigated miR-7 comprehensively, which may explain why different studies have discovered different targets for miR-7, or it may be because miRNA can form one-to-one, one-to-multiple or multiple-to-one relationships with its target genes[50]. Disruption of homeostasis in the digestive system is due to many pathways acting together in a complicated manner, which contributes to the progression from inflammatory diseases to malignancy. Furthermore, the genetic abnormalities in tumors are highly heterogeneous, and no two tumors are exactly alike, which raises a serious challenge. Consequently, more research should be conducted to verify whether miR-7 could be a biomarker or therapeutic target gene for digestive system malignancies.
Footnotes
Supported by National Natural Science Foundation of China, No. 81273735; Science and Technology Planning Project of Guangdong Province, China, No. 2013B021800169; Traditional Chinese Medicine Science and Technology Research Projects of Guangdong Provincial Hospital of Chinese Medicine, China, No. YN2014ZH05.
Conflict-of-interest statement: The authors declared that they have no conflicts of interest in this work.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: April 16, 2015
First decision: June 19, 2015
Article in press: November 19, 2015
P- Reviewer: Kalpaxis DL, Xu YQ S- Editor: Yu J L- Editor: Wang TQ E- Editor: Jiao XK
References
- 1.Calin GA, Croce CM, MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 2.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 4.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285:116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet. 2010;375:1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 7.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 8.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 10.Correa-Medina M, Bravo-Egana V, Rosero S, Ricordi C, Edlund H, Diez J, Pastori RL. MicroRNA miR-7 is preferentially expressed in endocrine cells of the developing and adult human pancreas. Gene Expr Patterns. 2009;9:193–199. doi: 10.1016/j.gep.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Masuda M, Miki Y, Hata S, Takagi K, Sakurai M, Ono K, Suzuki K, Yang Y, Abe E, Hirakawa H, et al. An induction of microRNA, miR-7 through estrogen treatment in breast carcinoma. J Transl Med. 2012;10 Suppl 1:S2. doi: 10.1186/1479-5876-10-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Zheng Y, Sun G, Xiong S. Restoration of miR-7 expression suppresses the growth of Lewis lung cancer cells by modulating epidermal growth factor receptor signaling. Oncol Rep. 2014;32:2511–2516. doi: 10.3892/or.2014.3519. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X, Hu Y, Dai L, Wang Y, Zhou J, Wang W, Di W, Qiu L. MicroRNA-7 inhibits tumor metastasis and reverses epithelial-mesenchymal transition through AKT/ERK1/2 inactivation by targeting EGFR in epithelial ovarian cancer. PLoS One. 2014;9:e96718. doi: 10.1371/journal.pone.0096718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Jiang Z, Huang J, Huang S, Li Y, Yu S, Yu S, Liu X. miR-7 inhibits glioblastoma growth by simultaneously interfering with the PI3K/ATK and Raf/MEK/ERK pathways. Int J Oncol. 2014;44:1571–1580. doi: 10.3892/ijo.2014.2322. [DOI] [PubMed] [Google Scholar]
- 15.Wang B, Sun F, Dong N, Sun Z, Diao Y, Zheng C, Sun J, Yang Y, Jiang D. MicroRNA-7 directly targets insulin-like growth factor 1 receptor to inhibit cellular growth and glucose metabolism in gliomas. Diagn Pathol. 2014;9:211. doi: 10.1186/s13000-014-0211-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bravo-Egana V, Rosero S, Molano RD, Pileggi A, Ricordi C, Domínguez-Bendala J, Pastori RL. Quantitative differential expression analysis reveals miR-7 as major islet microRNA. Biochem Biophys Res Commun. 2008;366:922–926. doi: 10.1016/j.bbrc.2007.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joglekar MV, Joglekar VM, Hardikar AA. Expression of islet-specific microRNAs during human pancreatic development. Gene Expr Patterns. 2009;9:109–113. doi: 10.1016/j.gep.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Zhao X, Dou W, He L, Liang S, Tie J, Liu C, Li T, Lu Y, Mo P, Shi Y, et al. MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene. 2013;32:1363–1372. doi: 10.1038/onc.2012.156. [DOI] [PubMed] [Google Scholar]
- 19.Xie J, Chen M, Zhou J, Mo MS, Zhu LH, Liu YP, Gui QJ, Zhang L, Li GQ. miR-7 inhibits the invasion and metastasis of gastric cancer cells by suppressing epidermal growth factor receptor expression. Oncol Rep. 2014;31:1715–1722. doi: 10.3892/or.2014.3052. [DOI] [PubMed] [Google Scholar]
- 20.Kong D, Piao YS, Yamashita S, Oshima H, Oguma K, Fushida S, Fujimura T, Minamoto T, Seno H, Yamada Y, et al. Inflammation-induced repression of tumor suppressor miR-7 in gastric tumor cells. Oncogene. 2012;31:3949–3960. doi: 10.1038/onc.2011.558. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Li Y, Liu Y, Xie P, Li F, Li G. PAX6, a novel target of microRNA-7, promotes cellular proliferation and invasion in human colorectal cancer cells. Dig Dis Sci. 2014;59:598–606. doi: 10.1007/s10620-013-2929-x. [DOI] [PubMed] [Google Scholar]
- 22.Xu K, Chen Z, Qin C, Song X. miR-7 inhibits colorectal cancer cell proliferation and induces apoptosis by targeting XRCC2. Onco Targets Ther. 2014;7:325–332. doi: 10.2147/OTT.S59364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma C, Qi Y, Shao L, Liu M, Li X, Tang H. Downregulation of miR-7 upregulates Cullin 5 (CUL5) to facilitate G1/S transition in human hepatocellular carcinoma cells. IUBMB Life. 2013;65:1026–1034. doi: 10.1002/iub.1231. [DOI] [PubMed] [Google Scholar]
- 24.Wu XM, Shao XQ, Meng XX, Zhang XN, Zhu L, Liu SX, Lin J, Xiao HS. Genome-wide analysis of microRNA and mRNA expression signatures in hydroxycamptothecin-resistant gastric cancer cells. Acta Pharmacol Sin. 2011;32:259–269. doi: 10.1038/aps.2010.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen HT, Dalmasso G, Yan Y, Laroui H, Dahan S, Mayer L, Sitaraman SV, Merlin D. MicroRNA-7 modulates CD98 expression during intestinal epithelial cell differentiation. J Biol Chem. 2010;285:1479–1489. doi: 10.1074/jbc.M109.057141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suto T, Yokobori T, Yajima R, Morita H, Fujii T, Yamaguchi S, Altan B, Tsutsumi S, Asao T, Kuwano H. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis. 2015;36:338–345. doi: 10.1093/carcin/bgu242. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed FE, Ahmed NC, Vos PW, Bonnerup C, Atkins JN, Casey M, Nuovo GJ, Naziri W, Wiley JE, Mota H, et al. Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genomics Proteomics. 2013;10:93–113. [PubMed] [Google Scholar]
- 28.Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55:1852–1862. doi: 10.1002/hep.25576. [DOI] [PubMed] [Google Scholar]
- 29.Fu HL, Wu de P, Wang XF, Wang JG, Jiao F, Song LL, Xie H, Wen XY, Shan HS, Du YX, et al. Altered miRNA expression is associated with differentiation, invasion, and metastasis of esophageal squamous cell carcinoma (ESCC) in patients from Huaian, China. Cell Biochem Biophys. 2013;67:657–668. doi: 10.1007/s12013-013-9554-3. [DOI] [PubMed] [Google Scholar]
- 30.Gao M, Yin H, Fei ZW. Clinical application of microRNA in gastric cancer in Eastern Asian area. World J Gastroenterol. 2013;19:2019–2027. doi: 10.3748/wjg.v19.i13.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yakirevich E, Resnick MB. Pathology of gastric cancer and its precursor lesions. Gastroenterol Clin North Am. 2013;42:261–284. doi: 10.1016/j.gtc.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Ikeda Y, Tanji E, Makino N, Kawata S, Furukawa T. MicroRNAs associated with mitogen-activated protein kinase in human pancreatic cancer. Mol Cancer Res. 2012;10:259–269. doi: 10.1158/1541-7786.MCR-11-0035. [DOI] [PubMed] [Google Scholar]
- 33.Ma J, Fang B, Zeng F, Pang H, Zhang J, Shi Y, Wu X, Cheng L, Ma C, Xia J, et al. Curcumin inhibits cell growth and invasion through up-regulation of miR-7 in pancreatic cancer cells. Toxicol Lett. 2014;231:82–91. doi: 10.1016/j.toxlet.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Unno J, Masamune A, Hamada S, Shimosegawa T. The zinc transporter LIV-1 is a novel regulator of stemness in pancreatic cancer cells. Scand J Gastroenterol. 2014;49:215–221. doi: 10.3109/00365521.2013.865075. [DOI] [PubMed] [Google Scholar]
- 35.Kaemmerer E, Klaus C, Jeon MK, Gassler N. Molecular classification of colorectal carcinomas: the genotype-to-phenotype relation. World J Gastroenterol. 2013;19:8163–8167. doi: 10.3748/wjg.v19.i45.8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang N, Li X, Wu CW, Dong Y, Cai M, Mok MT, Wang H, Chen J, Ng SS, Chen M, et al. microRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene. 2013;32:5078–5088. doi: 10.1038/onc.2012.526. [DOI] [PubMed] [Google Scholar]
- 38.Kudo M. Signaling pathway/molecular targets and new targeted agents under development in hepatocellular carcinoma. World J Gastroenterol. 2012;18:6005–6017. doi: 10.3748/wjg.v18.i42.6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alves RC, Alves D, Guz B, Matos C, Viana M, Harriz M, Terrabuio D, Kondo M, Gampel O, Polletti P. Advanced hepatocellular carcinoma. Review of targeted molecular drugs. Ann Hepatol. 2011;10:21–27. [PubMed] [Google Scholar]
- 40.Zhang X, Hu S, Zhang X, Wang L, Zhang X, Yan B, Zhao J, Yang A, Zhang R. MicroRNA-7 arrests cell cycle in G1 phase by directly targeting CCNE1 in human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2014;443:1078–1084. doi: 10.1016/j.bbrc.2013.12.095. [DOI] [PubMed] [Google Scholar]
- 41.Janpipatkul K, Suksen K, Borwornpinyo S, Jearawiriyapaisarn N, Hongeng S, Piyachaturawat P, Chairoungdua A. Downregulation of LAT1 expression suppresses cholangiocarcinoma cell invasion and migration. Cell Signal. 2014;26:1668–1679. doi: 10.1016/j.cellsig.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 42.Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014;15:e493–e503. doi: 10.1016/S1470-2045(14)70263-3. [DOI] [PubMed] [Google Scholar]
- 43.Ning BF, Ding J, Liu J, Yin C, Xu WP, Cong WM, Zhang Q, Chen F, Han T, Deng X, et al. Hepatocyte nuclear factor 4α-nuclear factor-κB feedback circuit modulates liver cancer progression. Hepatology. 2014;60:1607–1619. doi: 10.1002/hep.27177. [DOI] [PubMed] [Google Scholar]
- 44.Zeng Z, Wang J, Zhao L, Hu P, Zhang H, Tang X, He D, Tang S, Zeng Z. Potential role of microRNA-21 in the diagnosis of gastric cancer: a meta-analysis. PLoS One. 2013;8:e73278. doi: 10.1371/journal.pone.0073278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S, Xiang J, Li Z, Lu S, Hu J, Gao X, Yu L, Wang L, Wang J, Wu Y, et al. A plasma microRNA panel for early detection of colorectal cancer. Int J Cancer. 2015;136:152–161. doi: 10.1002/ijc.28136. [DOI] [PubMed] [Google Scholar]
- 46.Liu P, Xia L, Zhang WL, Ke HJ, Su T, Deng LB, Chen YX, Lv NH. Identification of serum microRNAs as diagnostic and prognostic biomarkers for acute pancreatitis. Pancreatology. 2014;14:159–166. doi: 10.1016/j.pan.2014.03.019. [DOI] [PubMed] [Google Scholar]
- 47.Shrestha S, Hsu SD, Huang WY, Huang HY, Chen W, Weng SL, Huang HD. A systematic review of microRNA expression profiling studies in human gastric cancer. Cancer Med. 2014;3:878–888. doi: 10.1002/cam4.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang JL, Hu Y, Kong X, Wang ZH, Chen HY, Xu J, Fang JY. Candidate microRNA biomarkers in human gastric cancer: a systematic review and validation study. PLoS One. 2013;8:e73683. doi: 10.1371/journal.pone.0073683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu X, Han Y, Wu Y, Zhu X, Lu X, Mao F, Wang X, He X, Zhao Y, Zhao Y. Prognostic role of microRNA-21 in various carcinomas: a systematic review and meta-analysis. Eur J Clin Invest. 2011;41:1245–1253. doi: 10.1111/j.1365-2362.2011.02535.x. [DOI] [PubMed] [Google Scholar]
- 50.Hashimoto Y, Akiyama Y, Yuasa Y. Multiple-to-multiple relationships between microRNAs and target genes in gastric cancer. PLoS One. 2013;8:e62589. doi: 10.1371/journal.pone.0062589. [DOI] [PMC free article] [PubMed] [Google Scholar]


