Abstract
The zebrafish has a striped skin pattern on its body, and Connexin41.8 (Cx41.8) and Cx39.4 are involved in striped pattern formation. Mutations in these connexins change the striped pattern to a spot or labyrinth pattern. In this study, we characterized Cx41.8 and Cx39.4 after expression in Xenopus oocytes. In addition, we analyzed Cx41.8 mutants Cx41.8I203F and Cx41.8M7, which caused spot or labyrinth skin patterns, respectively, in transgenic zebrafish. In the electrophysiological analysis, the gap junctions formed by Cx41.8 and Cx39.4 showed distinct sensitivity to transjunctional voltage. Analysis of non-junctional (hemichannel) currents revealed a large voltage-dependent current in Cx39.4-expressing oocytes that was absent in cells expressing Cx41.8. Junctional currents induced by both Cx41.8 and Cx39.4 were reduced by co-expression of Cx41.8I203F and abolished by co-expression of Cx41.8M7. In the transgenic experiment, Cx41.8I203F partially rescued the Cx41.8 null mutant phenotype, whereas Cx41.8M7 failed to rescue the null mutant, and it elicited a more severe phenotype than the Cx41.8 null mutant, as evidenced by a smaller spot pattern. Our results provide evidence that gap junctions formed by Cx41.8 play an important role in stripe/spot patterning and suggest that mutations in Cx41.8 can effect patterning by way of reduced function (I203F) and dominant negative effects (M7). Our results suggest that functional differences in Cx41.8 and Cx39.4 relate to spot or labyrinth mutant phenotypes and also provide evidence that these two connexins interact in vivo and in vitro.
Keywords: connexin, connexon (hemichannel), electrophysiology, gap junction, transgenic, zebrafish, skin pattern, stripe
Introduction
Connexin proteins are the major constituents of gap junctions and are four-pass transmembrane proteins. Six connexin proteins make up a hexamer called a connexon, which acts as a hemichannel at the cell membrane. Each gap junction is a large molecular unit formed by the docking of connexons, allowing ∼1,000-Da small molecules to be transferred between neighboring cells (1, 2). Approximately 20 connexins exist in mammalian genomes, and connexins are categorized into five subgroups (α, β, γ, δ, and ζ), depending on their molecular weight and amino acid sequences (3, 4). In zebrafish, ∼36 connexins are predicted to exist (5). This higher gene number might be due to genome duplication events during fish evolution. As in mammals, connexins are expressed in tissue-specific but overlapping patterns. A combination of connexins in the same or adjacent cells can lead to several types of gap junctions. Uniform connexins create homomeric homotypic gap junctions, whereas two or more types of connexins form gap junctions that are heteromeric (different connexins within a connexon), heterotypic (different connexins in two opposing connexons), or a combination of both. The composition of heteromeric and heterotypic gap junctions is expected to create gap junctions with a wide range of properties in vivo, although relatively little is known about their importance or complexity due to technical difficulties associated with their analysis (6–12).
The zebrafish is well known as a model organism for developmental and genetic studies. Because the zebrafish has a striped pattern on its skin (Fig. 1A), this fish is also well known as a useful animal for pattern formation studies (13–22). Zebrafish stripes consist primarily of two types of pigment cells: black pigment cells called melanophores and yellow pigment cells called xanthophores. We proposed that zebrafish stripes are generated by the interaction between melanophores and xanthophores, which satisfies the conditions for the formation of a Turing pattern (18–24). Several molecules involved in cell-cell interactions for skin pattern formation were identified (25–28). The membrane resting potential formed by Kir7.1, the inward rectifier potassium channel 7.1, is important for establishing clear boundaries between melanophores and xanthophores, and this channel functions as a short range factor in the Turing model (22, 29, 30). Notch-Delta signaling from xanthophores (Delta) to melanophores (Notch) is required for melanophore survival and acts as a long range factor in this model (22, 31). The zebrafish has a third type of pigment cell, the iridophore, which has reflecting internal structures and is involved in skin pattern formation by providing the initial conditions of the pattern formation system (32).
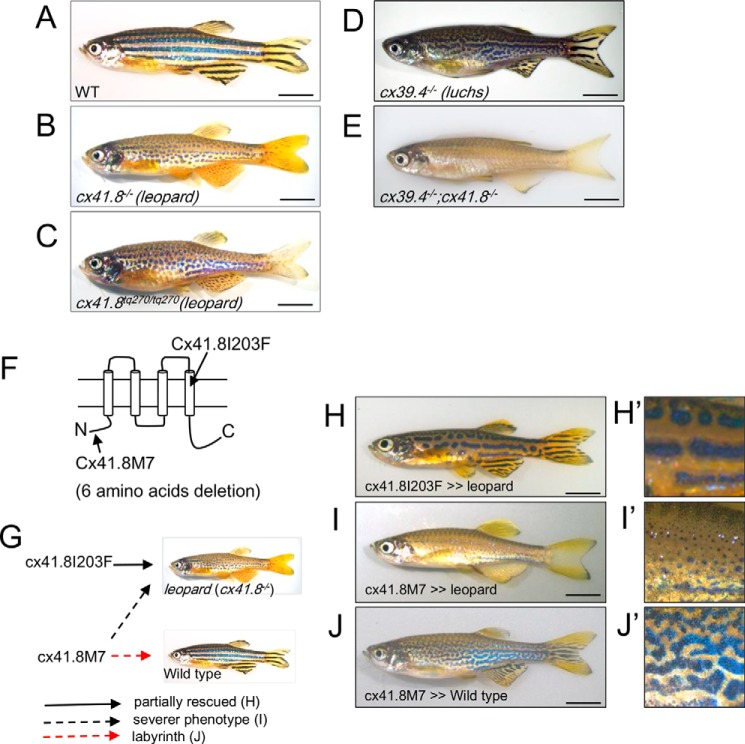
FIGURE 1.
Skin patterns of zebrafish. A, wild type; B, leopard mutant (cx41.8−/−, leot1 allele, null mutant); C, leopard mutant (cx41.8tq270/tq270, leotq270 allele, Cx41.8I203F); D, luchs mutant (cx39.4−/−; knock-out of cx39.4 by TALEN); E, luchs/leopard double homozygous mutant (cx39.4−/−;cx41.8−/−); F, connexin topology sketch highlighting Cx41.8 mutants used in this study. G, summary of the transgenic experiments; H–J, transgenic zebrafish: Tg(mitfa-cx41.8I203F)cx41.8−/− (H), Tg(mitfa-cx41.8M7)cx41.8−/− (I), and Tg(mitfa-cx41.8M7)cx41.8+/+ (J). H′–J′, magnified images of H–J. Scale bar, 5 mm.
Gap junctions also play an important role in pattern formation (22, 33). The gene encoding Cx41.8, an orthologue to the mammalian CX40, was previously identified as a responsible gene for the leopard mutant, which shows a spot pattern phenotype (Fig. 1B) (34). Several alleles in the leopard mutant are known (34, 35). For example, cx41.8t1/t1 is a null mutant allele (Fig. 1B), and cx41.8tq270/tq270 (Fig. 1C) is a dominant mutant allele with an I203F amino acid substitution within the fourth transmembrane domain of Cx41.8 (Fig. 1F). This mutant displays a spot pattern phenotype in both heterozygotic and homozygotic situations. Because the connexin mutant cx41.8tq270/tq270 shows a more severe phenotype than that of the Cx41.8 null mutant, it has been predicted that Cx41.8 makes a heteromeric/heterotypic gap junction with unidentified connexin(s) (34). Recently, we successfully observed changes in Turing-related patterning on fish skin by altering the gene encoding Cx41.8 (33). Cx41.8M7, which has a 6-amino acid deletion from the N terminus of Cx41.8, caused a labyrinth skin pattern when it was introduced into wild-type zebrafish. In summary, Cx41.8 was observed to play a role in tuning the interactions between pigment cells to make Turing patterns in vivo, similar to changing the parameters on the Turing model in silico (33).
Most recently, Cx39.4, a novel connexin specific to zebrafish, was identified as a response gene in the zebrafish skin pattern mutant luchs (Fig. 1D) (36). Cx39.4 is an α-type connexin, as is Cx41.8 (Fig. 2A), and is distributed in the teleost lineage (Fig. 2C). Cx39.4 has an N-terminal sequence 2 amino acids longer, which is predicted to produce unique gap junction characteristics (Fig. 2B). Furthermore, the Cx39.4 null mutant shows a labyrinth pattern (Fig. 1D) (36), similar to that observed in transgenic zebrafish with the Cx41.8M7 mutation (Fig. 1, J and J′) (33). The similar phenotype could suggest that Cx39.4 and Cx41.8 interact in vivo (36).
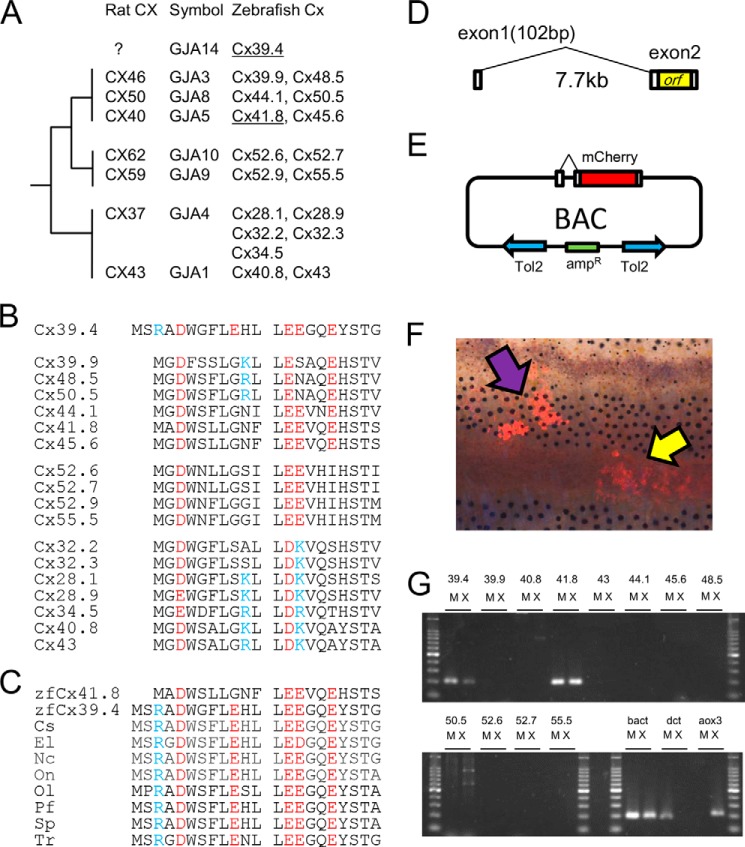
FIGURE 2.
Gap junctions in zebrafish and characterization of Cx39.4. A, phylogenic relationship of α-type connexins in rats and zebrafish. B, alignment of N-terminal sequences of α-type connexins in zebrafish; C, alignment of the N-terminal sequences of GJA14 in several teleosts compared with zebrafish Cx41.8: Cs, Cynoglossus semilaevis (XP_008329622.1); El, Esox lucius (XP_010896375); Nc, Notothenia coriiceps (XP_010777882.1); On, Oreochromis niloticus (XP_005453942.1); Ol, Oryzias latipes (XP_004084568.1); Pf, Poecilia Formosa (XP_007559865.1); Sp, Stegastes partitus (XP_008293308.1); Tr, Takifugu rubripes (XP_011607358.1). B and C, blue letters indicate basic residues, and red letters indicate acidic residues. D, exon-intron structure of cx39.4. E, pTol2-BAC construct for analyzing cx39.4 promoter activity. F, the mCherry signals were detected in melanophores (purple arrow) and xanthophores (yellow arrow), driven by the cx39.4-promoter. G, the mRNA expression of connexin genes in isolated melanophores (M) and xanthophores (X). bact, β-actin, a positive control for RT-PCR. dct is a melanophore marker, and aox3 is a xanthophore marker.
In this study, we performed both transgenic experiments and electrophysiological experiments to assess function and interactions between Cx41.8 and Cx39.4. The oocyte expression system is commonly used to study properties and interactions of mammalian gap junction proteins (37), and zebrafish connexins have previously been expressed and analyzed using dual cell voltage clamp methods after expression in oocytes (38).
Experimental Procedures
Transgenic Fish
All experiments in this study were conducted in accordance with the guidelines and approved protocols for animal care and use (approval number FBS-14-002-1) at Osaka University. Transgenic fish lines were generated using the Tol2 transposon vector system, as described previously (39, 40). The mitfa promoter fragment (41) was used to induce connexin expression in pigment cells (33). The cDNA fragment encoding Cx41.8I203F was amplified with PCR and cloned into the pT2AL200R150G plasmid. Plasmids for wild-type (WT) Cx41.8 and Cx41.8M7 expression were prepared as described previously (33). The plasmid (10 ng/μl) and transposase mRNA (25 ng/μl) synthesized in vitro were co-injected into fertilized eggs of WT or cx41.8 null mutant fish at the single-cell stage (40). The cx39.4 null mutant was generated by the TALEN2 method. TALEN pairs for cx39.4 knock-out were designed by the TALEN Targeter program (available at the Cornell University Web site). The most effective TALEN pair was used for the cx39.4 knock-out. The target sequence was 5′-CCTCTTCCTCTTCCGCatgcttgtgcttggcACGGCTGTGGAATCTGC-3′, in which TALEN binding sequences are shown in capital letters and spacer sequences are in lowercase type. TAL repeats were assembled by Golden Gate TALEN and the TAL Effector Kit 2.0 (42) with some modifications (43) and cloned into the expression vector pCS2TAL3DD or pCS2TAL3RR (44). The TALEN expression plasmids were linearized using the NotI restriction enzyme, and mRNAs were transcribed in vitro using the mMESSAGE mMACHINE SP6 Transcription Kit (Ambion). Synthetic TALEN mRNAs (100 pg) were injected into fertilized eggs at the single cell stage. To confirm the mutagenesis, genomic DNA was extracted from the eggs, and the TALEN target sequence was amplified by PCR using the following primer set: forward primer, 5′-TGTGCAGAGGATGTCCAGAG-3′; reverse primer, 5′-CACTCCCATCTTCGTCTTCC-3′.TALEN-induced mutations were detected by a T7 endonuclease I assay.
5′-RACE Experiment
To disclose the exon-intron structure of the cx39.4 gene, the transcription starting point and 5′-UTR sequence were determined by a 5′-RACE experiment (Fig. 2D) (45). A cDNA library with the cassette fragment was prepared as previously described. Briefly, mRNA extracted from the zebrafish brain with the RNeasy minikit (Qiagen) was subjected to reverse transcription using Superscript III (Invitrogen) and the BamTVTV primer (45). First-strand cDNA was purified with a PCR purification kit (Qiagen) and then reacted with terminal deoxynucleotidyl transferase (TAKARA) and dATP to add a poly(A) fragment to the 3′-end of the cDNA. The poly(A)-tailed single strand cDNA was purified with a PCR purification kit and subjected to PCR using KOD DNA polymerase (TOYOBO) and a 1 mm BamTVTV primer under the following PCR conditions: 95 °C for 30 s, 50 °C for 30 s, and 72 °C for 4 min for 5 cycles. Then the PCR fragment was purified using the PCR purification kit and amplified again with PCR using KOD DNA polymerase and the cassette primer sequence 5′-ACTGACTCTTCGGATCCTTT-3′. The PCR conditions were 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 4 min for 25 cycles. The 5′-UTR fragment was amplified with PCR primers: the cassette primer used above and Cx39.4R (5′-GCAGATTCCACAGCCGTGCCAAG-3′). The DNA sequence of the PCR fragment was determined using an ABI 3100 sequencer (Applied Biosystems).
Construction of pTol2-BAC Construct
To analyze the promoter activity of cx39.4, we modified a bacterial artificial chromosome (BAC) clone harboring a cx39.4 locus with a Tol2 transposon cassette (46). A CH211–261o1 BAC clone was obtained from the BACPAC Resources Center (Children's Hospital Oakland Research Institute, Oakland, CA). The BAC clone was subjected to homologous recombination to integrate the Tol2 cassette and mCherry cassette. Shortly, the Tol2 cassette fragment was amplified with PCR and introduced into Escherichia coli harboring the BAC clone, and the E. coli transformant was selected with chloramphenicol and ampicillin to obtain E. coli harboring pTol2-CH211–261o1. Then the mCherry cassette, which was designed to contain the 5′-UTR of cx39.4-mCherry-poly(A) signal-KmR-3′-UTR of cx39.4, was constructed in a cloning vector and amplified with PCR. The PCR fragment was introduced into E. coli harboring the pTol2-CH211–261o1 clone. The E. coli transformant was selected with chloramphenicol, ampicillin, and kanamycin to obtain the modified BAC clone pTol2-CH211–261o1-mCherry, as shown in Fig. 2E. The BAC clone was purified using the Large-Construct kit (Qiagen).
Collection of Pigment Cells
Melanophores and xanthophores were isolated from fish fins. Caudal and anal fins were collected from zebrafish anesthetized with ethyl-3-aminobenzoate methanesulfonate (Sigma). Black regions and yellow regions on fins were separated manually using a surgical knife under a stereomicroscope. The fin clips were treated with trypsin solution (2.5 mg/ml trypsin (Worthington), 1.2 mg/ml BSA (Sigma), and 1 mm EDTA (Wako) in PBS) in a 2-ml tube for 30 min at 28 °C. The trypsin solution was then removed, and the tissues were rinsed five times with PBS. Then the fin clips were incubated with collagenase solution (1 mg/ml collagenase I (Worthington), 0.1 mg/ml DNase I (Worthington), 0.1 mg/ml soybean trypsin inhibitor (Worthington), 1.2 mg/ml BSA, and 100 nm epinephrine (Sigma) in PBS) for 1 h at 28 °C. Pigment cells were then purified by Percoll gradient centrifugation (80% Percoll for melanophores and 50% Percoll for xanthophores) after filtration with a 25-μm mesh (26, 47).
RT-PCR Analysis of Connexin Expression
The mRNAs were extracted from purified melanophores and xanthophores using the RNeasy minikit, and cDNAs were synthesized using Superscript III (Invitrogen) and oligo(dT)18 primer. Synthesized cDNAs were then subjected to PCR. To rule out the possibility of cross-contamination of melanophores and xanthophores, the expression levels of dct (dopachrome tautomerase; melanophore marker) and aox3 (aldehyde oxidase 3; xanthophore marker) were also examined. The protein β-actin was used as a positive control. PCR amplifications were performed for 35 cycles at 95 °C for 30 s, at 60 °C for 30 s, and at 72 °C for 15 s. Primer sets were designed to span the intron (Table 1).
TABLE 1.
Primer sequences for RT-PCR
Primer pairs were designed to span an exon-exon junction.
| Gene | Forward | Reverse |
|---|---|---|
| cx39.4 | TCTCAGCGGCAGAAGCTCCTCAC | GCAGATTCCACAGCCGTGCCAAG |
| cx39.9 | AAACAGTGAGCTCTGTACC | CACCTCGATAAATGAACCTC |
| cx40.8 | ACAGCTTGTTAATTATTAGG | GCAGTGGAGTACGCCTGGAC |
| cx41.8 | TCATCGGTCAACAGAGATAG | CTAGTACCAAGATCCGGAAG |
| cx43 | TTGGTGACTGAACTTCAGAG | TTGAAAGCTGACTGCTCGTC |
| cx44.1 | CACAAATACTCTGGAGCTCT | GTAGCAAACGTTCTCGCAACC |
| cx45.6 | AGATATCAGAGCTGGGCCAC | CACGGTTAACCACACCTTCC |
| cx48.5 | CATAGACTTGATTTAGAACC | CCGCTCCCAACACCAGAATC |
| cx50.5 | TGACTGGGGGACAGGAGTTC | AACTCAGCAGCCGTGCCCAG |
| cx52.6 | AGTGTGAAAGACAACTGATG | GAATGGTAAGCCAGATTTTG |
| cx52.7 | AACTCCATGGCTGTAAAATG | CAGCCAGATTTTACCCACGATG |
| cx55.5 | TCATTCTGAACTACTATGC | AGAGGATGGGTTAATCAAAG |
| bact | GGAGAAGAGCTATGAGCTGC | ACCTCCAGACAGCACTGTGT |
| dct | ATCAGCCCGCGTTCACGGTT | ACACCGAGGTGTCCAGCTCTCC |
| aox3 | AGGGCATTGGAGAACCCCCAGT | ACACGTTGATGGCCCACGGT |
Preparation of Connexin cRNA and Injection into Xenopus Oocytes
The cDNAs encoding zfCx41.8 and zfCx38.4 were amplified using PCR and cloned into pGEM-HeFx plasmids (48). The plasmids were linearized using restriction enzymes and then used as a template for in vitro synthesis of mRNA (SP6 mMessage mMachine, Ambion) according to the manufacturer's protocol. Xenopus oocytes were collected from Xenopus laevis. An adult Xenopus female was anesthetized with ethyl-3-aminobenzoate methanesulfonate, and the ovarian lobes were collected using a surgical knife and forceps. The eggs were treated with collagenase solution (20 mg/ml collagenase I (Sigma) and 20 mg/ml hyaluronidase (Sigma) in OR2 buffer (82.5 mm NaCl, 2 mm KCl, 1 mm MgCl2, and 5 mm HEPES (pH 7.5, adjusted with NaOH)) at 18 °C for 2 h. Stage V and VI oocytes were collected manually and used for mRNA injection. Then mRNA (5 ng for Cx39.4 or 40 ng for the Cx41.8 series) was co-injected with 10 ng of antisense oligonucleotide DNA for Xenopus Cx38 into Xenopus oocytes. H2O was co-injected with the antisense oligonucleotide as a negative control. Oocytes injected with mRNA were incubated at 18 °C overnight in half-strength L15 medium (Sigma) with 2 mm CaCl2 (adjusted to pH 7.5 with NaOH). Then the vitelline membrane was removed manually using forceps in a hypertonic solution (200 mm aspartic acid, 1 mm MgCl2, 10 mm EGTA, 20 mm KCl, and 10 mm HEPES (pH 7.5)). The oocytes were manually paired with the vegetal poles together in ND96(+) solution (93.5 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 2 mm MgCl2, and 5 mm HEPES (pH 7.5, adjusted with NaOH)).
Voltage Clamp Recording
Transjunctional current was measured using the dual whole-cell voltage clamp technique. Current and voltage electrodes were prepared with a micropipette puller P-1000 (Stuffer) to obtain a resistance of 0.5–1.0 MΩ. The pipette was filled with solution containing 3 m KCl, 10 mm EGTA, and 10 mm HEPES (pH 7.4). Voltage clamp experiments were performed using two iTEV90 multielectrode clamp amplifiers (HEKA). To measure the transjunctional current, both cells were initially clamped at −40 mV, and one cell was then subjected to alternating pulses of 20-mV steps from −140 mV to +60 mV. Currents detected in the second oocyte were recorded, and junctional conductance was calculated using the current value at the end of the steady state. The conductance was obtained by the equation, Gj = Ij/(V1 − V0). Ij was the current value of the second oocyte. V0 was voltage of the first oocyte, and V1 was voltage of the second oocyte whose currents were obtained as Ij at each voltage step, Vj. Vj = V1 − V0. The obtained Gj value was then normalized and plotted against the Vj values. The data were fit to a Boltzmann distribution function. Furthermore, to analyze the time-dependent decay of gap junction current, currents were fit to an exponential curve, y = A0 + A1exp(−kx) using Origin 2015 software (OriginLab) within the first 200 ms.
Non-junctional Currents
Non-junctional (hemichannel) currents were recorded from a single oocyte using an iTEV90 multielectrode clamp amplifier. Modified ND96(+) solution with/without 2 mm CaCl2 was used as a bath solution to assess calcium sensitivity. Cells were initially clamped at −40 mV and then subjected to 10-s voltage steps from −30 to +60 mV in 10-mV increments.
Results
As noted above, Irion et al. (36) recently identified cx39.4 as a gene that is responsible for a zebrafish skin pattern mutant called luchs, which shows an irregular, labyrinth-like skin pattern. They suggested that cx39.4 is a zebrafish-specific gene, and Cx39.4 might partner with Cx41.8 to make a heteromeric gap junction, although no analysis was performed or reported. Here, to examine the relationship between Cx41.8 and Cx39.4 in skin pattern formation, we performed electrophysiological experiments involving Cx41.8 and Cx39.4.
Characterization of cx39.4 Gene
Fig. 2B shows the amino acid alignment of the N-terminal sequences of α-type connexins in zebrafish and indicates that Cx39.4 has an N terminus that is 2 amino acids longer than other connexins. Because Cx39.4 belongs to the GJA subfamily of gap junctions but does not have a mammalian orthologue, we used the designation “GJA14” for Cx39.4 (Fig. 2A). Database analysis revealed that Cx39.4 is not zebrafish-specific but a teleost-specific connexin (Fig. 2C) and that the EXXXE motif, a predicted polyamine binding site, is well conserved among Cx39.4 proteins in teleosts (36, 49). Next, we examined the exon-intron structure of cx39.4 using 5′-RACE methods and found 102 bp of exon 1 and 7.7 kb of intron 1 located upstream of exon 2, as shown in Fig. 2D. Exon 1 includes the 5′-UTR sequence but does not include the ORF sequence, which is encoded only by exon 2. This gene structure is very common for connexin genes (3). Next, we examined the gene expression of cx39.4, not by in situ hybridization or immunostaining, but by transgenic and RT-PCR experiments, because it is very difficult to detect small amounts of mRNA or protein in pigment cells (36). We generated a reporter construct using a BAC clone to detect the promoter activity of cx39.4. The ORF sequence for cx39.4 was replaced with the mCherry gene, and the pTol2 cassette was inserted into the BAC plasmid, as shown in Fig. 2E. The BAC plasmid was co-injected with Tol2 mRNA into fertilized eggs at the one-cell stage, and mCherry signals were detected in pigment cells, melanophores, and xanthophores in adult fish skin at the F0 generation (Fig. 2F). Next, we confirmed the gene expression of cx39.4 by RT-PCR experiments. In this study, we examined the gene expression of α-type connexins and found that cx41.8 and cx39.4 were expressed in both melanophores and xanthophores, and the other α-type connexins tested were not detected (Fig. 2G). To rule out the possibility of cross-contamination of melanophores and xanthophores, the expression levels of dct and aox3 were also examined. Connexins belonging to GJA4 (Fig. 2A) were not analyzed in this study because their exon-intron structures were not elucidated.
Zebrafish Mutants
Mutations in Cx41.8 are known to alter skin patterns in zebrafish (33, 34). In this study, we characterize two Cx41.8 mutations: I203F, a mutant involving an amino acid substitution in the fourth transmembrane domain (Fig. 1F), which induces a spot pattern rather than stripes (34), and Cx41.8M7, an artificially designed mutant with a 6-amino acid deletion from its N terminus (Fig. 1F). The ectopic expression of Cx41.8M7 in wild-type pigment cells caused a labyrinth skin pattern in zebrafish (Fig. 1, J and J′) (33). In this study, we found that the ectopic expression of Cx41.8M7 on the cx41.8−/− background induced an unusual pattern of very small spots (Fig. 1, I and I′). The same pattern was observed in the luchs/leopard double homozygous mutant (cx39.4−/−;cx41.8−/−) (Fig. 1E). Taken together, the similarity of the phenotypes between mutants of luchs or luchs/leopard and Cx41.8M7 transgenic fish lead to the hypothesis that Cx41.8 and Cx39.4 interact in vivo.
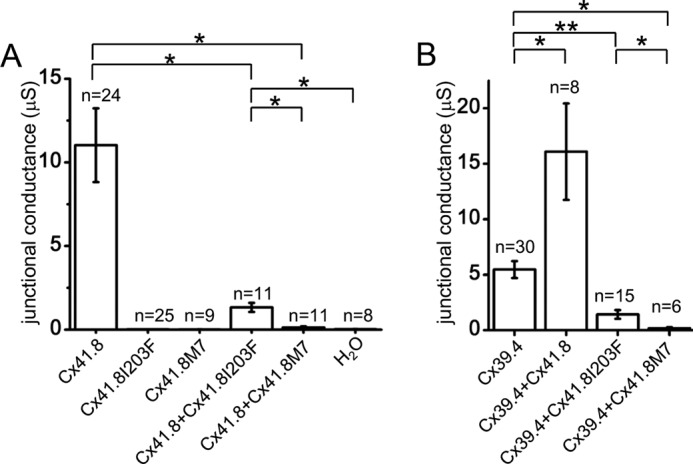
The two mutations Cx41.8I203F and Cx41.8M7 were introduced into the cx41.8 null background fish, and their effects were assessed in the F1 generation (Fig. 1, G–I). When Cx41.8I203F was introduced into the cx41.8 null mutant, the leopard phenotype was partially rescued (Fig. 1, H and H′). However, when Cx41.8M7 was introduced into the cx41.8 null mutant, the mutant phenotype was not rescued; instead, the transgenic fish showed a more severe phenotype (Fig. 1, I and I′) than the Cx41.8 null mutant (Fig. 1B): a smaller spot pattern. These results show that Cx41.8I203F retained the gap junction function to form stripes, but Cx41.8M7 did not, and that Cx41.8M7 had a negative effect on other connexin(s) in vivo.
Expression in Oocytes
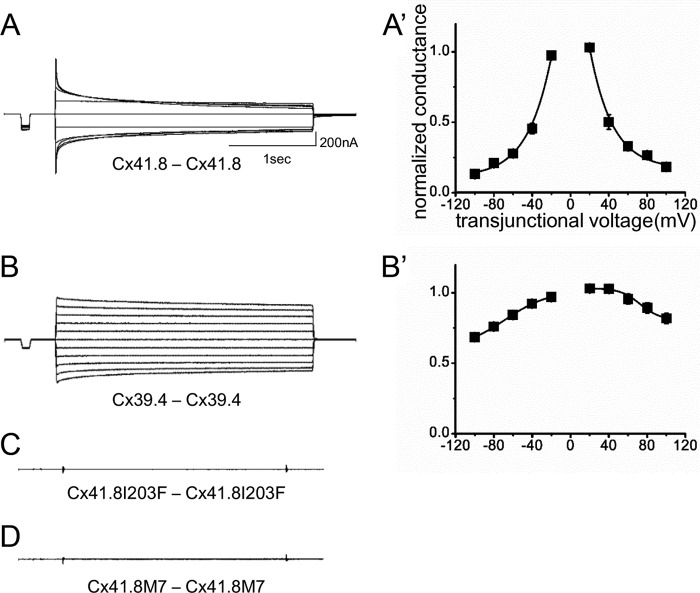
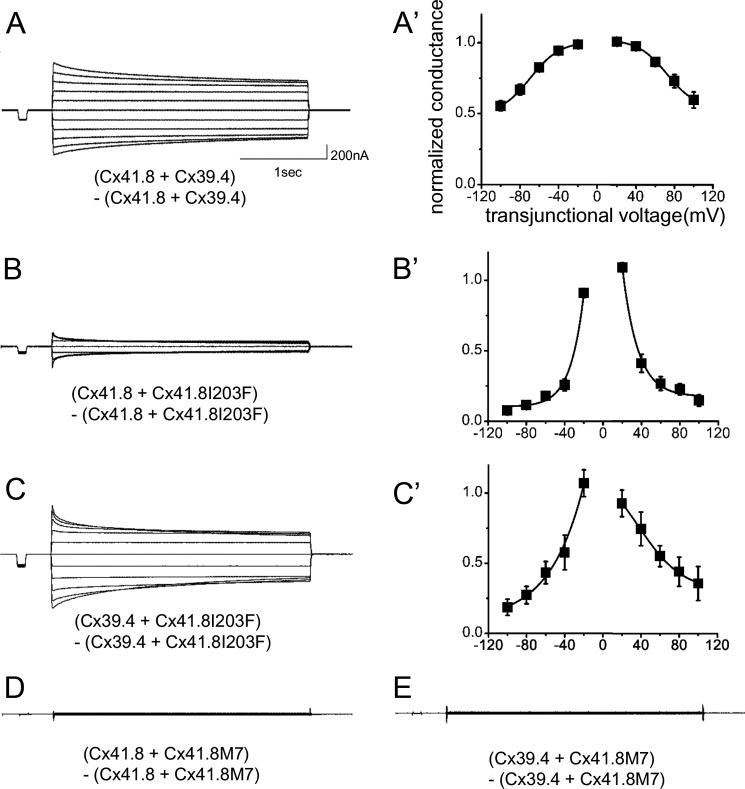
To examine the gap junction properties of Cx41.8 and Cx39.4, the connexins were expressed in Xenopus oocytes. In paired oocytes, junctional currents were induced by both Cx41.8 and Cx39.4. The voltage sensitivity of Cx41.8 was similar to that of zebrafish Cx45.6 (50), a paralogous connexin in zebrafish, and rat CX40 (51), an orthologous connexin in mammals (Fig. 3, A and A′). Cx41.8 displayed relatively strong sensitivity to the transjunctional voltage and relatively quick time-dependent inactivation in response to application of transjunctional voltage (Fig. 3A). As shown in Fig. 3B, Cx39.4 formed a functional gap junction between Xenopus oocytes but showed weaker sensitivity to transjunctional voltage than Cx41.8 (Fig. 3B′); gap junction channels induced by Cx39.4 required higher transjunctional voltages for inactivation and displayed a slower time-dependent decay of the junctional current compared with Cx41.8. For instance, when currents induced by transjunctional voltage of ±100 mV were fitted to the exponential curve, y = A0 + A1exp(−kx), [k] values were [k]Cx41.8 = 2.9 × 10−2 and [k]Cx39.4 = 7.7 × 10−3 (1/s), which were consistent with this observation.
FIGURE 3.
Electrophysiological analysis of homomeric-homotypic gap junctions. Junctional current traces of homotypic gap junctions of Cx41.8 (A), Cx39.4 (B), Cx41.8I203F (C), and Cx41.8M7 (D). A′ and B′, plots of normalized steady state junctional conductance versus transjunctional voltages (A′, n = 24; B′, n = 30). For these experiments, 40 ng of mRNA for Cx41.8 or its mutant or 5 ng of mRNA for Cx39.4 was injected into each oocyte.
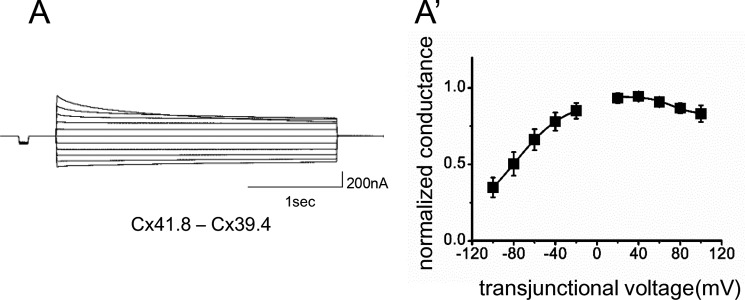
We also examined the formation of heterotypic gap junctions by expressing Cx41.8 and Cx39.4 in apposing cells. When an oocyte injected with mRNA for Cx39.4 was paired with an oocyte injected with mRNA for Cx41.8, transjunctional current with asymmetric sensitivity to voltage was induced (Fig. 4, A and A′).
FIGURE 4.
Cx39.4 and Cx41.8 form heterotypic gap junction channels. Junctional current traces induced by heterotypic gap junctions resulting from the expression of Cx41.8 and Cx39.4 in apposing cells. A′, plot of normalized steady state junctional conductance versus transjunctional voltages (n = 7).
We further assessed the roles and interactions of Cx41.8 and Cx39.4 by expressing Cx41.8 mutants in oocytes. Neither of the Cx41.8 mutants (neither Cx41.8I203F nor Cx41.8M7) induced junctional currents in oocytes (Fig. 3, C and D). To further elucidate differences between Cx41.8I203F and Cx41.8M7, we co-expressed wild-type and mutant connexins in the same oocyte. When 40 ng of mRNA for Cx41.8I203F was co-injected with the same amount of mRNA for WT Cx41.8, a small current was consistently detected (Fig. 5, B and B′). In contrast, when 40 ng of mRNA for Cx41.8M7 was co-injected with mRNA for WT Cx41.8, no current was detected (Fig. 5D). This result indicates that the ability to rescue the leopard phenotype correlates with the formation of gap junction channels in oocytes. The Cx41.8M7 inhibits the function of Cx41.8 junctions in oocytes (Fig. 1, H and I) and induces a small-spot phenotype in the Cx41.8 null mutant (Fig. 1I). This suggests that Cx41.8M7 forms heteromeric channels with Cx41.8.
FIGURE 5.
Electrophysiological analysis of heteromeric gap junctions. Junctional current traces of heteromeric gap junctions of Cx41.8 + Cx39.4 (A), Cx41.8 +Cx41.8I203F (B), Cx39.4 + Cx41.8I203F (C), Cx41.8 + Cx41.8M7 (D), and Cx39.4 + Cx41.8M7 (E). A′–C′, plots of normalized steady state junctional conductance versus transjunctional voltages (A′, n = 8; B′, n = 11; C′, n = 15).
We also used the Xenopus oocyte expression system to examine the possibility that Cx41.8 and Cx39.4 interact, creating heteromeric channels. The junctional currents induced after co-expression of the two connexins displayed intermediate sensitivity to voltage (Fig. 5, A and A′), weaker time- and voltage-dependent inactivation than Cx41.8 (Fig. 3A) but stronger than Cx39.4 (Fig. 3B). This is consistent with the formation of heteromeric channels but not conclusive. The co-expression of mutants such as Cx41.8I203F and Cx41.8M7 with wild type Cx41.8 and Cx39.4 provided stronger evidence for heteromerization (Fig. 5, B–D). Co-expression of the N-terminal mutant abolished currents induced by Cx39.4 (Fig. 5E) and Cx41.8 (Fig. 5D). These results provide strong evidence for interactions between Cx39.4 and Cx41.8 and are consistent with in vivo experiments (Fig. 6).
FIGURE 6.
Electrophysiological analysis of Cx41.8 mutants. Cx41.8 mutants exhibit negative effects on WT Cx41.8 (A) and Cx39.4 (B) in heteromeric gap junctions. Current values of −100 mV at the steady state were used for the comparison. Error bar, S.E. *, p < 0.01; **, p < 0.05, calculated by Student's t test.
Properties of Non-junctional Currents
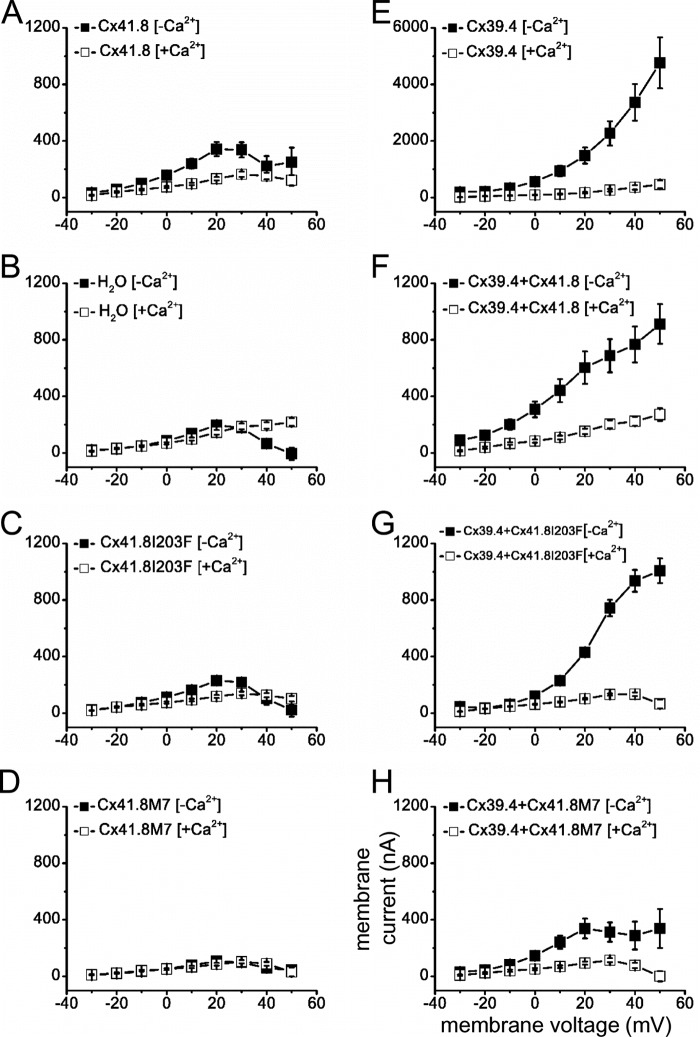
Non-junctional currents carried by connexin channels are often referred to as hemichannel currents. These currents occur across the plasma membrane of single oocytes and were assessed in oocytes injected with RNA encoding either Cx39.4 or Cx41.8 and also oocytes co-injected with RNA encoding both connexins. In addition, both of the Cx41.8 mutants were expressed in single oocytes, and their membrane currents were assessed. Currents induced by Cx41.8, Cx41.8I203F, and Cx41.8M7 were small but differed significantly from each other and from oligonucleotide-injected controls. (Figs. 7 (A–D) and 8A). The relative levels of current induced were well correlated with the junctional current levels induced by Cx41.8, Cx41.8I203F, and Cx41.8M7 (Figs. 6A and 8A and Table 2). In contrast, Cx39.4 RNA induced a large hemichannel current, which increased with membrane depolarization (Fig. 7E). When the Cx41.8 mutants were co-expressed with Cx39.4, membrane currents were reduced significantly compared with those induced by Cx39.4 alone. Specifically, 5 ng of mRNA encoding Cx39.4 was co-injected with 40 ng of RNA for Cx41.8 or the Cx41.8 mutants, and in all cases, current levels were significantly lower than in cells expressing Cx39.4 alone. Fig. 7F shows that the Cx39.4 hemichannel current was reduced by co-expression of Cx41.8, and Fig. 7, G and H, shows that the Cx39.4 hemichannel current was reduced by Cx41.8I203F and Cx41.8M7 (Fig. 8B). These results provide strong evidence that Cx41.8 interacts with Cx39.4 to form heteromeric hemichannels. Furthermore, the results are consistent with the observation that co-expression of Cx41.8M7 significantly reduced gap junction currents carried by Cx39.4 in paired oocytes.
FIGURE 7.
Non-junctional current recordings. For these experiments, 40 ng of mRNA for the Cx41.8 or Cx41.8 mutant and/or 5 ng of mRNA for Cx39.4 was injected into each oocyte. ■, 0 mm [Ca2+]; □, 2 mm [Ca2+]. A, Cx41.8; ■, n = 15; □, n = 14. B, H2O; ■, n = 14; □, n = 13. C, Cx41.8I203F; ■, n = 20; □, n = 28. D, Cx41.8M7; ■, n = 10; □, n = 10. E, Cx39.4; ■, n = 17; □, n = 12. F, Cx39.4 + Cx41.8; ■, n = 14; □, n = 12. G, Cx39.4 + Cx41.8I203F; ■, n = 18; □, n = 15. H, Cx39.4 + Cx41.8M7; ■, n = 15; □, n = 7. Error bars, S.E.
FIGURE 8.
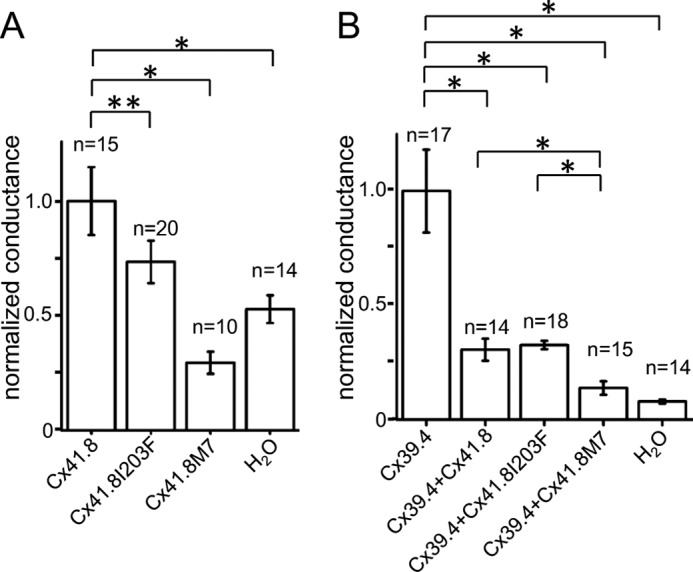
Hemichannel activities of Cx41.8 and Cx39.4. A, Cx41.8 and Cx41.8 mutants. B, effect of Cx41.8 and Cx41.8 mutants on the activity of Cx39.4 in heteromeric hemichannel. Current values of 30 mV at the instantaneous state of recording were used for the comparison. Error bars, S.E.; *, p < 0.01; **, p < 0.05, calculated by Student's t test.
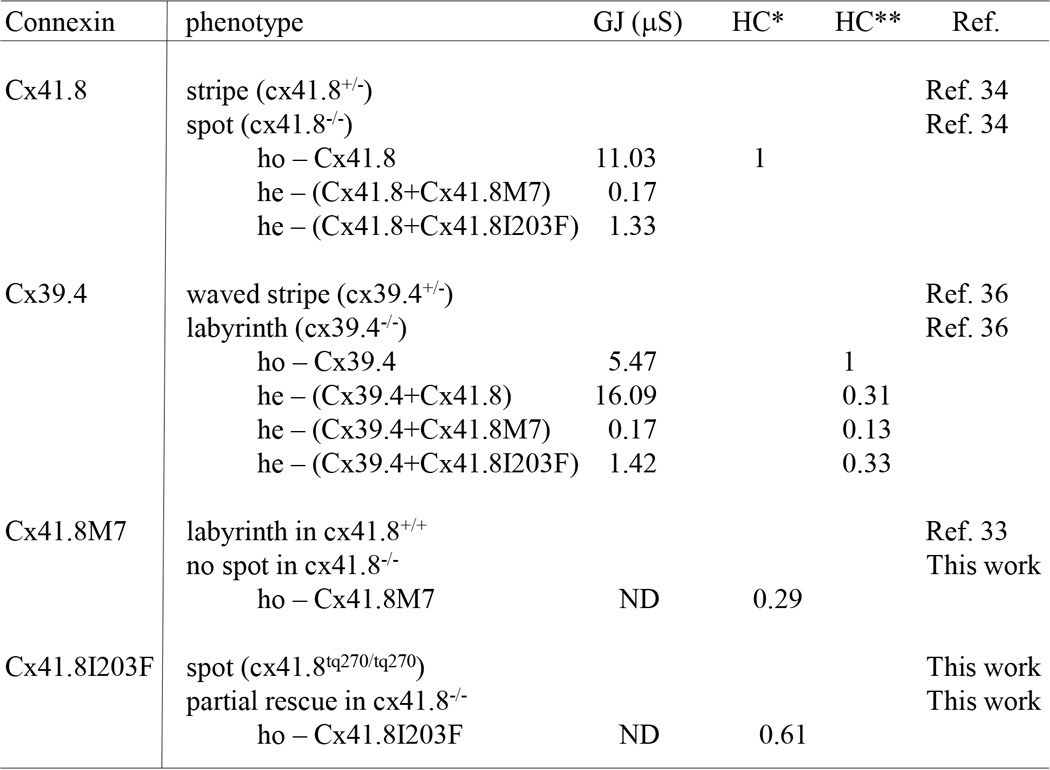
TABLE 2.
Corresponding table of phenotype and properties of gap junction and hemichannel
GJ, gap junction; HC, hemichannel; HC*, normalized conductance to Cx41.8; HC**, normalized conductance to Cx39.4; ho, homomeric gap junction/hemichannel; he, heteromeric gap junction/hemichannel; ND, not detected.
Discussion
When we identified cx41.8 as a gene responsible for the leopard mutant, one or more connexins were predicted to be involved in the zebrafish skin pattern formation (34). In addition, the leotq270 and leotw28 alleles of the leopard mutant, which have the amino acid substitutions I203F and I31F, respectively, showed dominant phenotypes (34). In particular, the leotq270 allele showed smaller spots than did the cx41.8 null mutant (34). Hence, previous results suggested that connexin interactions might be important for skin pattern formation. Specifically, it was hypothesized that the Cx41.8I203F mutant forms a heteromeric and/or heterotypic gap junction with another connexin protein(s), reducing gap junction function. We have shown that the Cx41.8M7 mutant creates a very severe phenotype when introduced into the cx41.8 null mutant, which also suggests the involvement of other connexin(s) in pattern formation. Recently, Irion et al. (36) successfully showed that Cx39.4 is also involved in skin pattern formation and that Cx39.4 functions in melanophores and xanthophores. The expression of cx39.4 in melanophores was independently reported by the Johnson group (52). Cx39.4 expression was detected by the transcriptome analysis of melanophores using NGS (next-generation sequencing) (52). Our RT-PCR results and transgenic experiments also suggested the expression of Cx39.4 in pigment cells.
In this study, we showed that Cx39.4 has different properties from Cx41.8 in both gap junctions and hemichannels. Furthermore, we showed that currents carried by gap junctions and hemichannels correlate to skin pattern phenotypes, although the question of which function is required or whether both are required for pattern formation remains unresolved. By the analysis of hemichannel currents (Fig. 7), we found that Cx41.8 and the mutants Cx41.8I203F and Cx41.8M7 induced very small currents when expressed alone in single oocytes (Fig. 8A). However, both Cx41.8 and Cx41.8I203F mutant reduced Cx39.4 activity when co-expressed in single oocytes (Fig. 8B).
Transplantation experiments suggest that Cx41.8 and Cx39.4 form gap junction(s) between melanophores and xanthophores (35, 36). The mathematical model also suggests interactions between melanophores and xanthophores. By modeling different interactions between melanophores and xanthophores, various skin patterns were easily generated in silico, as can be achieved by changing the function of gap junctions in vivo (33). We conclude that Cx39.4 and Cx41.8 function in a coordinated manner in the interaction between melanophores and xanthophores to create the striped pattern of zebrafish.
The gating mechanisms of gap junctions are quite diverse and include responses to cytoplasmic calcium, protons, and phosphorylation as well as transjunctional (Vj) and transmembrane voltage (53). Two gating models prevail in the literature. One is a ball and chain model, in which the N-terminal domains of six connexins form a plug structure; this plug controls the open/closed states of the gap junction, depending on the resting potential of the cells (54–56). The other is the subunit rotation model, which controls the open/closed states of the gap junction by changing the three-dimensional conformation of the gap junction to alter the channel pore size, depending on the calcium ion concentration (56) and modification of the C-terminal domain of connexin (57, 58). We used the N terminus deletion mutant Cx41.8M7 in this study because it was previously predicted that deletion of the N terminus domain would cause a closed-state gap junction (59, 60). The possibility that the N-terminal deletion might function as a dominant negative connexin form was supported by our electrophysiological analyses showing that Cx41.8M7 inhibits both the gap junction and hemichannel function of Cx41.8 and Cx39.4.
We previously hypothesized that gap junctions involving Cx41.8 might have rectification properties because Cx41.8 has the EXXXE motif in its N terminus, such as rat CX40 (61), which is recognized to be a polyamine binding motif. In the case of zebrafish pigment cells, melanophores express Kir7.1, and this expression is required to make a clear boundary between melanophores and xanthophores in fish skin (30). If Kir7.1 is mutated, melanophores and xanthophores intermingle, and the boundary becomes obscure. Because it is well known that polyamine is required for the rectifier property of Kir channels (62), polyamine is hypothesized to localize around the cell membranes of melanophores and possibly control the channel function of Kir7.1 and gap junctions involving Cx41.8. In Cx39.4, redundant EXXXE motifs exist at the N terminus, and this motif is well conserved among Cx39.4 orthologues in teleosts. This observation suggests that Cx39.4 and Cx41.8 might work together to generate a unidirectional signal flow from xanthophores to melanophores.
This is the first study aimed at correlating gap junction channel function and skin pattern phenotype in zebrafish (Fig. 1). There was a strong correlation between the behavior of connexins expressed in Xenopus oocytes and the phenotypes observed in zebrafish. Both systems provide evidence for interactions between Cx41.8 and Cx39.4. This is also the first study to reveal that Cx39.4 induces calcium-sensitive transmembrane currents that can be regulated (reduced) by co-expression of Cx41.8. This finding may be incorporated into models of pattern formation and suggests that further investigation of connexin function is warranted.
Author Contributions
M. W. and S. K. conceived the study. M. W, R. S., and T. A. performed transgenic experiments. M. W. and I. M. S. performed electrophysiological experiments and analyzed data. M. W. wrote the paper.
This work was supported by Japanese Society for the Promotion of Science, KAKENHI Grants 26291049, 26650078 (to M. W.), by Ministry of Education, Culture, Sports, Science, and Technology in Japan, KAKENHI Grants 25111714 (to M. W.) and 22127003 (to S. K.), and by Core Research for Evolutional Science and Technology (CREST), Japan Science and Technology Agency (to S. K.). The authors declare that they have no conflicts of interest with the contents of this article.
- TALEN
- TAL effector nuclease
- BAC
- bacterial artificial chromosome.
References
- 1.Kumar N. M., and Gilula N. B. (1996) The gap junction communication channel. Cell 84, 381–388 [DOI] [PubMed] [Google Scholar]
- 2.Alexander D. B., and Goldberg G. S. (2003) Transfer of biologically important molecules between cells through gap junction channels. Curr. Med. Chem. 10, 2045–2058 [DOI] [PubMed] [Google Scholar]
- 3.Söhl G., and Willecke K. (2004) Gap junctions and the connexin protein family. Cardiovasc. Res. 62, 228–232 [DOI] [PubMed] [Google Scholar]
- 4.Abascal F., and Zardoya R. (2013) Evolutionary analyses of gap junction protein families. Biochim. Biophys. Acta 1828, 4–14 [DOI] [PubMed] [Google Scholar]
- 5.Eastman S. D., Chen T. H., Falk M. M., Mendelson T. C., and Iovine M. K. (2006) Phylogenetic analysis of three complete gap junction gene families reveals lineage-specific duplications and highly supported gene classes. Genomics 87, 265–274 [DOI] [PubMed] [Google Scholar]
- 6.Ebihara L., Xu X., Oberti C., Beyer E. C., and Berthoud V. M. (1999) Co-expression of lens fiber connexins modifies hemi-gap-junctional channel behavior. Biophys. J. 76, 198–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrio L. C., Suchyna T., Bargiello T., Xu L. X., Roginski R. S., Bennett M. V., and Nicholson B. J. (1991) Gap junctions formed by connexins 26 and 32 alone and in combination are differently affected by applied voltage. Proc. Natl. Acad. Sci. U.S.A. 88, 8410–8414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White T. W., Paul D. L., Goodenough D. A., and Bruzzone R. (1995) Functional analysis of selective interactions among rodent connexins. Mol. Biol. Cell 6, 459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elfgang C., Eckert R., Lichtenberg-Fraté H., Butterweck A., Traub O., Klein R. A., Hülser D. F., and Willecke K. (1995) Specific permeability and selective formation of gap junction channels in connexin-transfected HeLa cells. J. Cell Biol. 129, 805–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werner R., Levine E., Rabadan-Diehl C., and Dahl G. (1989) Formation of hybrid cell-cell channels. Proc. Natl. Acad. Sci. U.S.A. 86, 5380–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao F., Eckert R., Elfgang C., Nitsche J. M., Snyder S. A., Hülser D. F., Willecke K., and Nicholson B. J. (1998) A quantitative analysis of connexin-specific permeability differences of gap junctions expressed in HeLa transfectants and Xenopus oocytes. J. Cell Sci. 111, 31–43 [DOI] [PubMed] [Google Scholar]
- 12.Verselis V. K., Ginter C. S., and Bargiello T. A. (1994) Opposite voltage gating polarities of two closely related connexins. Nature 368, 348–351 [DOI] [PubMed] [Google Scholar]
- 13.Budi E. H., Patterson L. B., and Parichy D. M. (2008) Embryonic requirements for ErbB signaling in neural crest development and adult pigment pattern formation. Development 135, 2603–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelsh R. N., Harris M. L., Colanesi S., and Erickson C. A. (2009) Stripes and belly-spots: a review of pigment cell morphogenesis in vertebrates. Semin. Cell Dev. Biol. 20, 90–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMenamin S. K., Bain E. J., McCann A. E., Patterson L. B., Eom D. S., Waller Z. P., Hamill J. C., Kuhlman J. A., Eisen J. S., and Parichy D. M. (2014) Thyroid hormone-dependent adult pigment cell lineage and pattern in zebrafish. Science 345, 1358–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh A. P., and Nüsslein-Volhard C. (2015) Zebrafish stripes as a model for vertebrate colour pattern formation. Curr. Biol. 25, R81-R92 [DOI] [PubMed] [Google Scholar]
- 17.Kondo S. (2002) The reaction-diffusion system: a mechanism for autonomous pattern formation in the animal skin. Genes Cells 7, 535–541 [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi M., Yoshimoto E., and Kondo S. (2007) Pattern regulation in the stripe of zebrafish suggests an underlying dynamic and autonomous mechanism. Proc. Natl. Acad. Sci. U.S.A. 104, 4790–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamasu A., Takahashi G., Kanbe A., and Kondo S. (2009) Interactions between zebrafish pigment cells responsible for the generation of Turing patterns. Proc. Natl. Acad. Sci. U.S.A. 106, 8429–8434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo S., and Miura T. (2010) Reaction-diffusion model as a framework for understanding biological pattern formation. Science 329, 1616–1620 [DOI] [PubMed] [Google Scholar]
- 21.Yamanaka H., and Kondo S. (2014) In vitro analysis suggests that difference in cell movement during direct interaction can generate various pigment patterns in vivo. Proc. Natl. Acad. Sci. U.S.A. 111, 1867–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe M., and Kondo S. (2015) Is pigment patterning in fish skin determined by the Turing mechanism? Trends Genet. 31, 88–96 [DOI] [PubMed] [Google Scholar]
- 23.Meinhardt H., and Gierer A. (2000) Pattern formation by local self-activation and lateral inhibition. Bioessays 22, 753–760 [DOI] [PubMed] [Google Scholar]
- 24.Turing A. (1952) The chemical basis of morphogenesis. Phil. Trans. R. Soc. Lond. B 237, 37–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fadeev A., Krauss J., Frohnhofer H. G., Irion U., and Nusslein-Volhard C. (2015) Tight junction protein 1a regulates pigment cell organisation during zebrafish colour patterning. eLife 10.7554/eLife.06545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue S., Kondo S., Parichy D. M., and Watanabe M. (2014) Tetraspanin 3c requirement for pigment cell interactions and boundary formation in zebrafish adult pigment stripes. Pigment Cell Melanoma Res 27, 190–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang M. R., Patterson L. B., Gordon T. N., Johnson S. L., and Parichy D. M. (2009) Basonuclin-2 requirements for zebrafish adult pigment pattern development and female fertility. PLoS Genet 5, e1000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parichy D. M., Rawls J. F., Pratt S. J., Whitfield T. T., and Johnson S. L. (1999) Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development 126, 3425–3436 [DOI] [PubMed] [Google Scholar]
- 29.Iwashita M., Watanabe M., Ishii M., Chen T., Johnson S. L., Kurachi Y., Okada N., and Kondo S. (2006) Pigment pattern in jaguar/obelix zebrafish is caused by a Kir7.1 mutation: implications for the regulation of melanosome movement. PLoS Genet. 2, e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inaba M., Yamanaka H., and Kondo S. (2012) Pigment pattern formation by contact-dependent depolarization. Science 335, 677. [DOI] [PubMed] [Google Scholar]
- 31.Hamada H., Watanabe M., Lau H. E., Nishida T., Hasegawa T., Parichy D. M., and Kondo S. (2014) Involvement of Delta/Notch signaling in zebrafish adult pigment stripe patterning. Development 141, 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh A. P., Schach U., and Nüsslein-Volhard C. (2014) Proliferation, dispersal and patterned aggregation of iridophores in the skin prefigure striped colouration of zebrafish. Nat. Cell Biol. 16, 607–614 [DOI] [PubMed] [Google Scholar]
- 33.Watanabe M., and Kondo S. (2012) Changing clothes easily: connexin41.8 regulates skin pattern variation. Pigment Cell Melanoma Res. 25, 326–330 [DOI] [PubMed] [Google Scholar]
- 34.Watanabe M., Iwashita M., Ishii M., Kurachi Y., Kawakami A., Kondo S., and Okada N. (2006) Spot pattern of leopard Danio is caused by mutation in the zebrafish connexin41.8 gene. EMBO Rep. 7, 893–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maderspacher F., and Nüsslein-Volhard C. (2003) Formation of the adult pigment pattern in zebrafish requires leopard and obelix dependent cell interactions. Development 130, 3447–3457 [DOI] [PubMed] [Google Scholar]
- 36.Irion U., Frohnhöfer H. G., Krauss J., Çolak Champollion T., Maischein H. M., Geiger-Rudolph S., Weiler C., and Nüsslein-Volhard C. (2014) Gap junctions composed of connexins 41.8 and 39.4 are essential for colour pattern formation in zebrafish. eLife 3, e05125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swenson K. I., Jordan J. R., Beyer E. C., and Paul D. L. (1989) Formation of gap junctions by expression of connexins in Xenopus oocyte pairs. Cell 57, 145–155 [DOI] [PubMed] [Google Scholar]
- 38.Dermietzel R., Kremer M., Paputsoglu G., Stang A., Skerrett I. M., Gomes D., Srinivas M., Janssen-Bienhold U., Weiler R., Nicholson B. J., Bruzzone R., and Spray D. C. (2000) Molecular and functional diversity of neural connexins in the retina. J. Neurosci. 20, 8331–8343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawakami K., Koga A., Hori H., and Shima A. (1998) Excision of the tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene 225, 17–22 [DOI] [PubMed] [Google Scholar]
- 40.Kawakami K., Shima A., and Kawakami N. (2000) Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. U.S.A. 97, 11403–11408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lister J. A., Robertson C. P., Lepage T., Johnson S. L., and Raible D. W. (1999) nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 126, 3757–3767 [DOI] [PubMed] [Google Scholar]
- 42.Cermak T., Starker C. G., and Voytas D. F. (2015) Efficient design and assembly of custom TALENs using the Golden Gate platform. Methods Mol. Biol. 1239, 133–159 [DOI] [PubMed] [Google Scholar]
- 43.Sakuma T., Ochiai H., Kaneko T., Mashimo T., Tokumasu D., Sakane Y., Suzuki K., Miyamoto T., Sakamoto N., Matsuura S., and Yamamoto T. (2013) Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci. Rep. 3, 3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dahlem T. J., Hoshijima K., Jurynec M. J., Gunther D., Starker C. G., Locke A. S., Weis A. M., Voytas D. F., and Grunwald D. J. (2012) Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8, e1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe M., Kobayashi N., Shin-i T., Horiike T., Tateno Y., Kohara Y., and Okada N. (2004) Extensive analysis of ORF sequences from two different cichlid species in Lake Victoria provides molecular evidence for a recent radiation event of the Victoria species flock: identity of EST sequences between Haplochromis chilotes and Haplochromis sp. “Redtailsheller”. Gene 343, 263–269 [DOI] [PubMed] [Google Scholar]
- 46.Suster M. L., Abe G., Schouw A., and Kawakami K. (2011) Transposon-mediated BAC transgenesis in zebrafish. Nat. Protoc. 6, 1998–2021 [DOI] [PubMed] [Google Scholar]
- 47.Eom D. S., Inoue S., Patterson L. B., Gordon T. N., Slingwine R., Kondo S., Watanabe M., and Parichy D. M. (2012) Melanophore migration and survival during zebrafish adult pigment stripe development require the immunoglobulin superfamily adhesion molecule Igsf11. PLoS Genet. 8, e1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furutani K., Ohno Y., Inanobe A., Hibino H., and Kurachi Y. (2009) Mutational and in silico analyses for antidepressant block of astroglial inward-rectifier Kir4.1 channel. Mol. Pharmacol. 75, 1287–1295 [DOI] [PubMed] [Google Scholar]
- 49.Watanabe M., Watanabe D., and Kondo S. (2012) Polyamine sensitivity of gap junctions is required for skin pattern formation in zebrafish. Sci. Rep. 2, 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Christie T. L., Mui R., White T. W., and Valdimarsson G. (2004) Molecular cloning, functional analysis, and RNA expression analysis of connexin45.6: a zebrafish cardiovascular connexin. Am. J. Physiol. Heart Circ. Physiol. 286, H1623–H1632 [DOI] [PubMed] [Google Scholar]
- 51.Bruzzone R., Haefliger J. A., Gimlich R. L., and Paul D. L. (1993) Connexin40, a component of gap junctions in vascular endothelium, is restricted in its ability to interact with other connexins. Mol. Biol. Cell 4, 7–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higdon C. W., Mitra R. D., and Johnson S. L. (2013) Gene expression analysis of zebrafish melanocytes, iridophores, and retinal pigmented epithelium reveals indicators of biological function and developmental origin. PLoS One 8, e67801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harris A. L. (2001) Emerging issues of connexin channels: biophysics fills the gap. Q. Rev. Biophys. 34, 325–472 [DOI] [PubMed] [Google Scholar]
- 54.Oshima A., Tani K., Hiroaki Y., Fujiyoshi Y., and Sosinsky G. E. (2007) Three-dimensional structure of a human connexin26 gap junction channel reveals a plug in the vestibule. Proc. Natl. Acad. Sci. U.S.A. 104, 10034–10039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maeda S., Nakagawa S., Suga M., Yamashita E., Oshima A., Fujiyoshi Y., and Tsukihara T. (2009) Structure of the connexin 26 gap junction channel at 3.5 Å resolution. Nature 458, 597–602 [DOI] [PubMed] [Google Scholar]
- 56.Oshima A. (2014) Structure and closure of connexin gap junction channels. FEBS Lett. 588, 1230–1237 [DOI] [PubMed] [Google Scholar]
- 57.Grosely R., Kopanic J. L., Nabors S., Kieken F., Spagnol G., Al-Mugotir M., Zach S., and Sorgen P. L. (2013) Effects of phosphorylation on the structure and backbone dynamics of the intrinsically disordered connexin43 C-terminal domain. J. Biol. Chem. 288, 24857–24870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kjenseth A., Fykerud T. A., Sirnes S., Bruun J., Yohannes Z., Kolberg M., Omori Y., Rivedal E., and Leithe E. (2012) The gap junction channel protein connexin 43 is covalently modified and regulated by SUMOylation. J. Biol. Chem. 287, 15851–15861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kyle J. W., Minogue P. J., Thomas B. C., Domowicz D. A., Berthoud V. M., Hanck D. A., and Beyer E. C. (2008) An intact connexin N-terminus is required for function but not gap junction formation. J. Cell Sci. 121, 2744–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oshima A., Tani K., Toloue M. M., Hiroaki Y., Smock A., Inukai S., Cone A., Nicholson B. J., Sosinsky G. E., and Fujiyoshi Y. (2011) Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels. J. Mol. Biol. 405, 724–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Musa H., Fenn E., Crye M., Gemel J., Beyer E. C., and Veenstra R. D. (2004) Amino terminal glutamate residues confer spermine sensitivity and affect voltage gating and channel conductance of rat connexin40 gap junctions. J. Physiol. 557, 863–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hibino H., Inanobe A., Furutani K., Murakami S., Findlay I., and Kurachi Y. (2010) Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol. Rev. 90, 291–366 [DOI] [PubMed] [Google Scholar]