Abstract
Most bacterial RNA polymerases (RNAP) contain five conserved subunits, viz. 2α, β, β′, and ω. However, in many Gram-positive bacteria, especially in fermicutes, RNAP is associated with an additional factor, called δ. For over three decades since its identification, it had been thought that δ functioned as a subunit of RNAP to enhance the level of transcripts by recycling RNAP. In support of the previous observations, we also find that δ is involved in recycling of RNAP by releasing the RNA from the ternary complex. We further show that δ binds to RNA and is able to recycle RNAP when the length of the nascent RNA reaches a critical length. However, in this work we decipher a new function of δ. Performing biochemical and mutational analysis, we show that Bacillus subtilis δ binds to DNA immediately upstream of the promoter element at A-rich sequences on the abrB and rrnB1 promoters and facilitates open complex formation. As a result, δ facilitates RNAP to initiate transcription in the second scale, compared with minute scale in the absence of δ. Using transcription assay, we show that δ-mediated recycling of RNAP cannot be the sole reason for the enhancement of transcript yield. Our observation that δ does not bind to RNAP holo enzyme but is required to bind to DNA upstream of the −35 promoter element for transcription activation suggests that δ functions as a transcriptional regulator.
Keywords: DNA binding protein, gram-positive bacteria, RNA polymerase, transcription, transcription regulation, Bacillus subtilis, delta factor, transcriptional regulator
Introduction
Transcription is the first step in gene regulation in bacteria in which RNA polymerase (RNAP)3 together with different σ factors and transcriptional regulators control the gene expression. Bacterial RNAP core enzyme contains five conserved subunits: 2 α, β, β′, and ω. A specificity factor σ associates with RNAP core enzyme to form RNAP holo enzyme that is able to recognize and initiate transcription at promoters.
In certain Gram-positive bacteria, including Bacillus subtilis and Staphylococcus aureus, an additional factor, called δ, is associated with RNAP. The δ factor was first identified in 1975 during the purification of RNAP from phage (SP01)-infected B. subtilis (1). The protein copurified with RNAP, and therefore it was thought that δ functions as a subunit of RNAP. Attempts were made to characterize the functional role of the protein in transcription. Several reports suggested that δ was involved in promoter selection (2–5) and functioned together with σA as an initiation subunit of RNAP (6, 7) or as an allosteric modulator of RNAP conformation in both initiation and the RNAP core recycling phase (5). Other reports showed that δ and σA bind to RNAP core with negative cooperativity (8, 9), and δ has no effect on transcription initiation, the rate of elongation, or termination (5). Using in vitro transcription assays, several groups showed that δ enhances the production of transcripts from certain promoters. This increase in transcript yield in the presence of δ is attributed to the recycling of RNAP possibly by δ-mediated release of RNAP from the elongation complex following transcription termination or by inhibiting the formation of stable RNAP core-DNA/RNA complex (5, 10).
To characterize the role of δ in vivo, strains lacking δ were studied. It was found that these mutants did not exhibit any distinctive phenotype, except an altered cell morphology and a delayed exit from stationary phase (9) or a block in sporulation (11). Recently, Rabatinová et al. (12) demonstrated that mutant strains lacking δ displayed a decreased ability to survive under a condition where iNTP concentration changes rapidly. This was explained by δ-mediated changes in the requirement of iNTP by RNAP to stabilize the open complex formation at the promoters where the stability of the open complex formation is rate-limiting for transcription. Transcriptome analysis of S. aureus strains with and without δ show that the protein is not only involved in up-regulation of certain genes but is also involved in down-regulation of the genes that encode virulence factors (13).
Despite many attempts to understand the role of this factor in transcription in vitro and in vivo, the function of δ remains ambiguous because of contradictory observations and conclusions. The facts that (i) δ is present in molar excess relative to RNAP and (ii) binding of δ and σ to RNAP are mutually exclusive, whereas both δ and σ are required for δ-mediated transcriptional regulation in B. subtilis raises the possibility that δ may bind to RNAP nonspecifically and remain associated with RNAP but may not function as a subunit of RNAP.
We therefore aimed to delineate the mechanism by which δ regulates transcription and in this report, we show that δ exhibits a moderate affinity for RNAP core but has little or no effect on RNAP core-mediated transcription. On the other hand, δ does not bind to RNAP holo but enhances the transcript yield by RNAP holo-mediated transcription. We further demonstrate that δ functions by binding to the DNA at A-rich sequence immediately upstream of the −35 element of the promoter. We observed that the presence of δ drastically increases the rate of open complex formation allowing RNAP to initiate transcription faster, on the second scale. We further show that removal of the δ binding site from the promoter DNA results in the loss of the protein-mediated enhancement of transcript yield.
Materials and Methods
Cloning of rpoE and rpoD Gene
B. subtilis rpoE (encoding δ) and rpoD (encoding σA) genes were amplified by PCR from genomic DNA (isolated from strain Bs168) using oligonucleotide primers (Table 1) and cloned into pET28a(+) vector using NcoI-EcoRI and NcoI-HindIII (NEB) respectively. The start codon TTG in the rpoE gene was replaced by ATG. rpoE and rpoD genes were further individually or together cloned into pAcYcDuet-1 vector using oligonucleotide primers (Table 1) with EcoRV-KpnI and NcoI-HindIII (NEB), respectively.
TABLE 1.
DNA sequences
Bold letters denote transcription start sites. −10 and −35 elements of the promoters are underlined. Italicized letters represent the DNA region protected by δ in DNaseI footprinting assay.
| Sequence | |
|---|---|
| DNA template | |
| abrB (−95/+144) | GTTTCCAAGACATTACTGACTATAAGAACTAATTCTTACAATCAATAGTAAACAAAATGATTGACGATTATTGGAAACCTTGTTATGCTATGAAGGTAAGGATTTTGTCGAATAATGACGAAGAAAAATATAATTTAAACAAATAAGTATCTCTTGGGAGGAGAATGTTTATGAAATCTACTGGTATTGTACGTAAAGTTGATGAATTAGGACGTGTAGTTATTCCTATCG |
| rrnBP1 (−92/+47) | GAAATCATGGCGAGGATTATAGTTTATTTGTTTTATAGATTTTTTTTAAAAAACTA[underln]TTGCAATAAATAAATACAGGTGTTATATTATTAAACGTCGCTGATGCACAGCGGACACAAACTAGATGCTTCAAAACAACTTG |
| Oligonucleotide primers | |
| rpoE forward | GAAAGGGAGTGTCCGACCATGGGTATCAAAC |
| rpoE reverse | GTCTAAAGTAGAATTCTTGCTAGATACTATTTAATT |
| rpoE forward (EcoRV) | AAGATATCATGGGGAGTGTCCGACCATGGGTATCAAAC |
| rpoE reverse (KpnI) | AAGGTACCCTATATTTAATTTCCTCTTCTTCATCATC |
| sigA forward | GGATCCATGGCTGATAAACCCACG |
| sigA reverse | GAATTCAAGCTTTTATTCAAGGAAATCTTTCAAACGTTTACTTC |
| rpoEL51C forward | GTGAAAAAAGAAGAGTGTGGAGACCGC |
| rpoE L51C reverse | GCGGTCTCCACACTCTTCTTTTTTCAC |
| abrB −95 forward | AAGAATTCGTTTCCAAGACATTACTGACTATAAG |
| abrB −95 forward (KpnI) | AAGGTACCGTTTCCAAGACATTACTGACTATAAG |
| abrB +136 reverse | AAGGATCCCGATAGGAATAACTACACGTCC |
| abrB +30 reverse | TTCTTCGTCATTATTCGACAAAATCC |
| abrB −15 reverse | AAGGATCCCGACAAAATCCTTACCTTCATAGC |
| abrB −66 forward | TAATTCTTACAATCAATAGTAAAC |
| abrB −56 forward | AATCAATAGTAAACAAAATGATTG |
| abrB −47 forward | TAAACAAAATGATTGACGATTATTG |
| abrB −41 forward | AAATGATTGACGATTATTGGAAAC |
| GCabrB forward | GGCCCGGGCCCGGGCCCTTGACGATTATTGGAAAC |
| GCabrB forward (KpnI) | GGTACCGGCCCGGGCCCGGGCCCTTGACGATTATTGG |
| abrB mut −40 forward | CAATCAATAGTAAACACCATGATTGACG |
| abrB mut −40 reverse | CGTCAATCATGGTGTTTACTATTGATTG |
| abrB mut −44 forward | CAATCAATAGTACCCAAAATGATTGACG |
| abrB mut −44 reverse | CGTCAATCATTTTGGGTACTATTGATTG |
| abrB mut −40/−44 forward | CAATCAATAGTACCCACCATGATTGACG |
| abrB mut −40/−44 reverse | CGTCAATCATGGTGGGTACTATTGATTG |
| abrB C14T forward | GGATTTTGTTGAATAATGACGAAG |
| abrB C14T reverse | CTTCGTCATTATTCAACAAAATCC |
| abrB C24T forward | GGATTTTGTTGAATAATGATGAAG |
| abrB C24T reverse | CTTCATCATTATTCAACAAAATCC |
| rrnBP1(−92) forward | AAGGATCCGAAATCATGGCGAGGATTATAG |
| rrnBP1(+47) reverse | CAAGTTGTTTTGAAGCATCTAG |
| rrnBP1 −63 forward | GTTTTATAGATTTTTTTTAAAAAAC |
| rrnBP1 −55 forward | GATTTTTTTTAAAAAACTATTGC |
| rrnBP1 −41 forward | AACTATTGCAATAAATAAATACAGGTG |
| rrnBP1P2(+113) reverse | AAAGCTTGATGATACGCACATCGCAGTGC |
DNA region encompassing abrB promoter (−95/+15, sequence shown in Table 1) and GC-rich upstream derivatives of the abrB promoter (−52/+15) were amplified by PCR from genomic DNA (isolated from Bs168) using oligonucleotide primers (Table 1) and were cloned in pFPVmCherry vector using KpnI-BamHI (NEB). Mutations at positions −40 and −41, positions −44 and −45; and positions −40, −41, −44, and −45 of the abrB promoter into pFPVmCherry plasmid were performed by site-directed mutagenesis (Stratagene Inc) using oligonucleotide primers (Table 1).
Bacterial Strains and Culture Conditions
For purification of B. subtilis (Bs) RNAP core, Escherichia coli B834 (DE3) cells were transformed with respective plasmids and were grown in LB medium supplemented with 0.1% dextrose and antibiotics (100 μg/ml ampicillin and 35 μg/ml chloramphenicol). For purification of δ, pET28-rpoE was transformed into E. coli BL21 (DE3) cells and grown in 2× YT medium (16 g of tryptone, 10 g of yeast extract, and 5 g of NaCl per liter) supplemented with 50 μg/ml kanamycin monosulfate. For the expression of σA, E. coli C43 (DE3) and 2× YT medium was used.
Purification of δ
δ was purified using a protocol essentially as in López de Saro et al. (10) except that instead of pI precipitation, the pH of the protein samples were adjusted to 8.0 by adding a required quantity of 0.1 n NaOH, before fractionation of the samples in FPLC using Mono Q HR 10/10 column with a gradient of 0–1.0 m NaCl in the buffer (20 mm Tris-Cl, pH 8.0). The eluted fractions were collected and concentrated, and glycerol was added to a final concentration of 30% and stored at −20 °C.
Purification of Bs RNAP Core
E. coli B834 (DE3) cells were transformed with plasmids pNG545 (encoding β and α) and pNG540 (encoding ω and β′) (kind gift from Peter J. Lewis, University of Newcastle, Callaghan, Australia (14)) and were grown in 3 liters of LB at 37 °C up to 0.4 OD. Temperature of the growth media was lowered to 16 °C before addition of 0.5 mm IPTG. The cells were further grown at 16 °C for 12 h. Cells were harvested, and RNAP was purified as described by Mukhopadhyay et al. (15).
For purification of Bs RNAP from a δ knock-out strain of B. subtilis HB6010 (CU1065 ΔrpoE::cm; a kind gift from Dr. Helmann (9)), cells were grown in 2 liters of LB at 37 °C up to 1.2 OD. The cells were harvested, and the protein was purified as recombinant Bs RNAP excluding nickel affinity chromatography.
Purification of σA
E. coli C43 (DE3) cells containing pET28-sigA were grown in 1 liter of LB at 37 °C until OD reached 0.5. Protein production was induced by adding 0.5 mm IPTG, followed by growth at 25 °C for 5 h. Cells were harvested by centrifugation (6,000 × g, 10 min, 4 °C), were resuspended in 20 ml of TG buffer (50 mm Tris-Cl, 5% glycerol) containing 200 mm NaCl, 5 mm β-mercaptoethanol, 1 mm PMSF and were disrupted by sonication. The lysates were spun at 18,000 × g for 30 min at 4 °C. The supernatant was diluted to 100 ml with TG buffer and was loaded onto Q-Sepharose column (GE Healthcare) pre-equilibrated with TG buffer. Protein was eluted using a step gradient of NaCl in TG buffer. The eluted fractions enriched with σA were further purified on a Mono Q HR10/10 column in an AKTA purifier (GE Healthcare) using a 0.1–1.0 m NaCl gradient in TG buffer. The purified σA sample was concentrated and kept at −80 °C after adding glycerol to a final concentration of 30%.
Preparation of DNA and RNA Fragments
abrB promoter sequence −95 to +136 and rrnBP1P2 promoter sequence −92 (with respect to +1 of P1) and +112 (with respect to +1 of P2) (sequence listed in Table 1) were amplified by PCR from genomic DNA (isolated from Bs168) using oligonucleotide primers (Table 1) and were cloned in pUC19 vector using EcoRI-BamHI.
Promoter DNA fragments having different lengths of upstream regions were also amplified by PCR using oligonucleotide primers (Table 1) and purified by PAGE. abrB promoter DNA fragment containing mutation at positions −40 and −41; positions −44 and −45; and positions −40, −41, −44, and −45 or GC-rich upstream sequence was prepared by PCR using oligonucleotide primers (Table 1). 144-nucleotides RNA was prepared by in vitro transcription assay as below and was purified after treating the reaction mixture with DNaseI.
In Vitro Transcription Assays
Multiround Transcription
100 nm RNAP core was mixed with 400 nm σA in 10 μl of transcription buffer (18 mm Tris-Cl, pH 8.0, 10 mm NaCl, 8 mm β-mercaptoethanol, 10 mm MgCl2) and were incubated on ice for 30 min followed by 10 min at 25 °C to form the holoenzyme. 50 nm promoter DNA fragments (unless stated otherwise) was added to RNAP holo and incubated at 37 °C for 20 min to form the open complex. Transcription was initiated with NTP (final concentrations: 250 μm of ATP, GTP, and UTP and 25 μm of [α-32P]CTP (0.2 μCi)) at 37 °C for 30 min. The reactions were terminated by the addition of 2.5 μl of FLB dye (80% formamide, 10 mm EDTA, 0.01% bromphenol blue, 0.01% xylene cyanol), resolved in 8% or 12% urea-PAGE (38) and was scanned by storage phosphor scanner (Typhoon trio+; GE Healthcare). When transcription assays were performed in the presence of δ, the protein was added to the open complex mixtures, following incubation at 37 °C for 5 min before the addition of NTP (unless stated otherwise).
Single Round Transcription
Single round transcription was carried out as the multiround assays described above except that heparin (0.25 μg/μl) was added to the reaction mixtures along with NTP. In a separate assay, the open complexes were allowed to form for 1, 5, and 30 min by incubating RNAP and promoter DNA as above followed by the addition of heparin (0.25 μg/μl) and NTP.
Stalled Elongation Complex Assays
performed as above except that transcription initiation reactions were carried out with the abrB promoter derivatives that contains first C residues at +14, +24, and+45, respectively, and with the addition of 250 μm GTP and UTP and 25 μm [α-32P]ATP.
In Vitro Transcription Assay Using Kool NC-45TM Template
1.75 pmol of Kool NC-45TM (Epicenter) template was incubated at 37 °C with 200 nm RNAP core and increasing concentrations of δ in 10 μl of transcription buffer, along with 0.5 mm NTP at 37 °C for 30 min, and fluorescence was monitored as per manufacturer's protocol (Epicenter Biotechnologies).
Fluorescence Anisotropy Assays
Labeling of δ with TMR
δ protein does not contain any cysteine residue. Single cysteine derivative of δ was prepared by introducing a cysteine residue at the amino acid residue 51 of rpoE in pET28a by point mutation, using a site-directed mutagenesis kit (Stratagene Inc). The single-cysteine derivative of δ was purified following the protocol used for δ. 50 μl of 11 μm protein sample was reduced with 10 mm DTT as described by Kim et al. (16) before the labeling reaction. The sample was dissolved in 100 μl of buffer A (100 mm sodium phosphate, pH 7.3, 1 mm EDTA) and reacted with tetramethyl rhodamine (TMR)-6-maleimide for 1 h at 4 °C. The protein sample was centrifuged for 10 min at 18,000 × g and loaded onto a 10-ml BioGel P6 column (Bio-Rad) pre-equilibrated with buffer A to remove the free dye. The labeled δ was eluted in the void volume, mixed with an equal volume of 100% glycerol, and stored at −80 °C. The labeling efficiency is 98%, and activity of the labeled protein was confirmed by in vitro transcription assay.
Fluorescence Anisotropy Measurements
20 nm of TMR-labeled δ in 60 μl of transcription buffer was titrated with increasing concentrations of RNAP or DNA or RNA at 37 °C, and the fluorescence intensity and anisotropy values were measured (λex = 540 nm, λem = 580 nm) using a PTI fluorescence master QM400 system fitted with automatic polarizers. Normalized fluorescence anisotropy increments (ΔA/A0, where A and A0 are the anisotropy value of δ at a particular concentration of RNAP [or DNA] or zero RNAP [or DNA], respectively, and ΔA = A − A0) were plotted against titer concentration using the Sigmaplot software (Systat Software Inc.). The dissociation constants (Kd) of the bindings for RNAP or DNA were determined by fitting the data to single parameter hyperbolic or sigmoidal functions.
EMSA
The primer for abrB DNA fragment was labeled using [γ-32P]ATP and T4 polynucleotide kinase (NEB) following the manufacturer's protocol. The promoter DNA fragment was amplified by PCR and was purified using agarose gel elution. In the first set, 200 nm RNAP holo samples were incubated with 25 nm 32P-labeled DNA in 10 μl of transcription buffer at 37 °C for given time intervals, challenged by 400 nm unlabeled DNA before resolving at 5% PAGE in 0.5× TBE buffer. In the second set, 200 nm of δ was mixed with the RNAP samples before addition of DNA. The gels were scanned by phosphorimaging (Typhoon trio+; GE Healthcare). EMSAs with promoter DNA fragments of different upstream length were performed as above after labeling the DNA with [γ-32P]ATP.
DNaseI Footprinting Assay
0.2 μm abrB promoter DNA fragment (−95/+30) labeled with 32P at the 5′ end of the template strand was mixed with 0, 0.5, 1.0, and 2.0 μm δ in 50 μl of transcription buffer and incubated at 37 °C for 30 min. 1 μl of 100 mm CaCl2 and 0.01 unit of DNase I was added to the reaction mixtures at room temperature. The reactions were stopped after 90 s by the addition of 10 μl of 0.5 m EDTA. The DNA samples were extracted following Sambrook and Russel (2001) (38). Samples containing equal counts were resolved on an 8% urea PAGE gel. The bands were visualized by phosphorimaging.
FeBABE-mediated Protein-DNA Footprinting Assay
The single-cysteine (at position 51) derivative of δ was reacted with FeBABE (Dojindo Molecular Technologies Inc., Japan) in 1:5 molar ratio as described by Rudra et al. (17). The unused probes were removed by gel filtration P-6 column (Bio-Rad), pre-equilibrated with buffer (20 mm Tris-Cl, pH 8.0, 0.2 m NaCl). Labeled proteins were distributed in 20-μl aliquots and stored at −80 °C.
0.2 μm of abrB promoter DNA fragment (−95/+30) labeled with 32P at the 5′ end of the template strand was mixed with 0, 0.1, and 0.2 μm FeBABE-labeled δ in 100 μl of transcription buffer and incubated at 37 °C for 15 min. Note that 0.1–0.2 μm labeled δ was used in FeBABE footprinting assay, 10-fold less than the amount used in DNaseI footprinting to reduce the nonspecific binding of δ to DNA. The cleavage reaction and purification of the products were performed as in Rudra et al. (17). The products were run on 6% urea-PAGE gel.
Recombinant in Vivo Reporter Assay
A recombinant in vivo reporter assay using three-plasmid expression system in E. coli was employed essentially as described by Banerjee et al. (18). Plasmid pNG 219 (a kind gift from Dr. Lewis (14)) containing the genes rpoA, rpoB, and rpoC, respectively, of B. subtilis was used for Bs RNAP core expression. The plasmids pAcYcDuet-rpoD and pAcYcDuet-rpoD-rpoE were used for expression of σA and both σA and δ, respectively. abrB promoter fragment (−95/+15) and its four mutant derivatives were inserted at the upstream of the mCherry gene in the pFPVmCherry vector. Cells (E. coli B834 (DE3)) were transformed with the following plasmids (i) pFPVmCherry-abrB alone (for background); (ii) pFPVmCherry-abrB + pAcYcDuet-rpoE (for control; δ only, no Bs RNAP); (iii) pFPVmCherry-abrB + pNG219 for Bs RNAP core; (iv) pFPVmCherry-abrB + pNG219 + pAcYcDuet-rpoD; and (v) pFPVmCherry-abrB + pNG219 + pAcYcDuet-rpoD-rpoE. Similar set of assays were performed with pFPVmCherry-abrB derivatives carrying mutations at (i) positions −40 and −41; (ii) positions −44 and −45; and (iii) positions −40, −41, −44, and −45; and with (iv) GC-rich upstream sequence. The cells (in 50 ml of LB medium supplemented with 100 μg/ml ampicillin, 35 μg/ml chloramphenicol) were grown at 37 °C up to 0.5 OD, added with 0.5 mm IPTG and were grown further for 16 h at 16 °C. Cells from each set were diluted to obtain equal OD, and their fluorescence intensities were measured at 610 nm with excitation at 592 nm.
Results
δ Binds to RNAP Core but Not to RNAP Holo
Using electrophoretic mobility shift assay, it was previously shown that the binding of δ and σA to RNAP are mutually exclusive, either because of overlapping binding sites of these factors on RNAP or because of negative cooperativity between these factors for their binding to RNAP (8, 9). Using EMSA, López de Saro et al. (9) determined the binding affinity of δ to RNAP (Kd = 400 nm). Because EMSA involves a separation step, the observed affinity may not reflect the actual affinity in the cases of low affinity binding or nonspecific binding. Here, we employed a fluorescence anisotropy assay with TMR-labeled δ to determine the binding constants of the protein to RNAP core and to RNAP holo (Fig. 1, A and B). Because δ does not have any cysteine residue, we first introduced a cysteine residue in δ by site-directed mutagenesis at amino acid residue 51 and subsequently labeled the protein derivative by cysteine specific reaction with TMR maleimide.
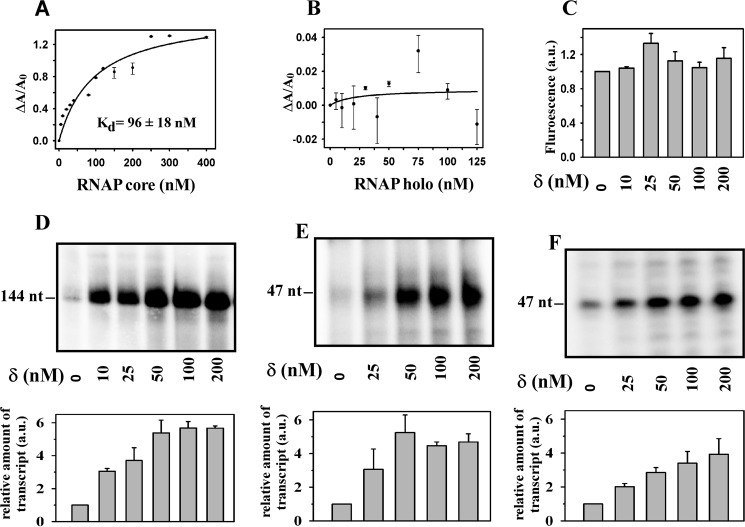
FIGURE 1.
δ binds to RNAP core but has no effect on core-mediated transcription. A, binding of δ to RNAP. For fluorescence anisotropy assay, 20 nm TMR-labeled δ was added with core RNAP. Fluorescence anisotropy of the labeled δ was monitored with excitation at 540 nm and emission at 580 nm. Each data set represents means of three replicates. The dissociation constant (Kd) of δ to RNAP core was estimated by fitting the data using a single parameter hyperbolic function. B, same as A, but for RNAP holo. C, effect of δ on RNAP core-mediated transcription. Fluorescence-based transcription assays were performed using 100 nm RNAP core and 1.75 pmol of Kool NC-45TM template in the presence of δ. Each data set represents mean of three replicates. D, effect of δ on RNAP holo-mediated transcription. Radioactive based transcription assay with 100 nm RNAP holo and 50 nm abrB promoter DNA fragment was used. Run-off transcripts were 144 nucleotides. Each experiment was repeated thrice, and the mean of fold increase in the amount of transcript at each concentration of δ with respect to the amount in the absence of δ was plotted as a bar graph (shown in lower panel). E, same as D, but 50 nm rrnBP1 promoter DNA fragment was used. Run-off transcripts were 47 nucleotides in length. F, same as E, but using Bs RNAP purified from δ knock-out strain of B. subtilis. Run-off transcripts were 47 nucleotides in length.
The labeled δ retained its activity as judged by in vitro transcription assay (data not shown). The dissociation constant (Kd) for binding of δ to RNAP core by the anisotropy assay was estimated to be 96 ± 18 nm which is 4-fold lower than the previously reported value (9). However, we did not observe any affinity of δ to RNAP holo, consistent with the previous observation (8, 9).
δ Has No Effect on RNAP Core-mediated Transcription but Enhances the Yield of RNAP Holo-mediated Transcription
Because δ binds to RNAP core, we tested whether the protein has any effect on RNAP core-mediated transcription. Because Bs RNAP core does not produce any transcripts from double-stranded linear DNA fragments or tailed template DNA fragments, we monitored the yield of transcripts in a fluorescence based in vitro transcription assay in which RNAP core generates transcripts from a Kool template. It is reported that RNAP core enzyme produces transcripts from the Kool template by an unknown mechanism (19). We observed that δ has little or no effect on the level of transcripts produced (Fig. 1C). On the other hand, when a radioactive based in vitro transcription assay with RNAP holo and abrB (20) or rrnBP1 (21) promoter DNA fragments were performed, we observed a significant increase in the level of transcripts in the presence of δ (Fig. 1, D and E). In these transcription assays, 100 nm RNAP and 50 nm promoter DNA were used in the presence of increasing concentrations of δ from 10 to 200 nm. The result showed that substoichiometric amount of δ (50 nm versus 100 nm) relative to RNAP had the same effect on the yield of transcription as compared with the yield in the presence of equimolar or higher level of δ (Fig. 1, D and E). To test whether the results were due to any artifact arising from the use of recombinant Bs RNAP purified from E. coli, we isolated Bs RNAP from a δ knock-out strain of B. subtilis (HB6010 (CU1065 ΔrpoE :: cm (9)) and performed an identical assay. The result showed that δ activated transcription from the rrnBP1 promoter by the native Bs RNAP with similar efficiency as the recombinant version (Fig. 1F). We note that this result contradicts the previous observation by Rabatinová et al. (12). The above observations that (i) δ binds to RNAP core but fails to induce core-mediated transcription and (ii) δ does not show any affinity toward RNAP holo but requires RNAP holo for its function suggested that δ may function as a transcriptional regulator.
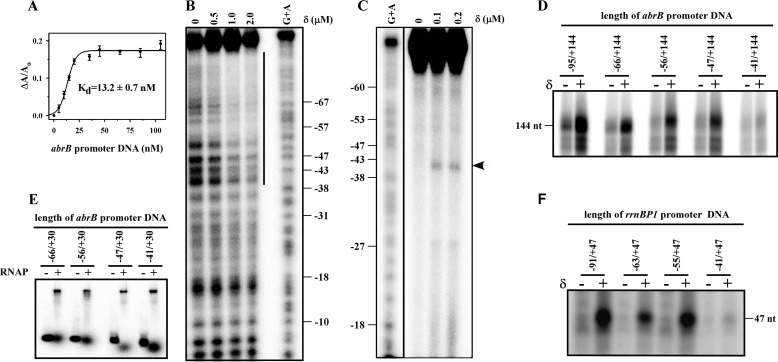
δ Binds to DNA Upstream of the Promoter Element
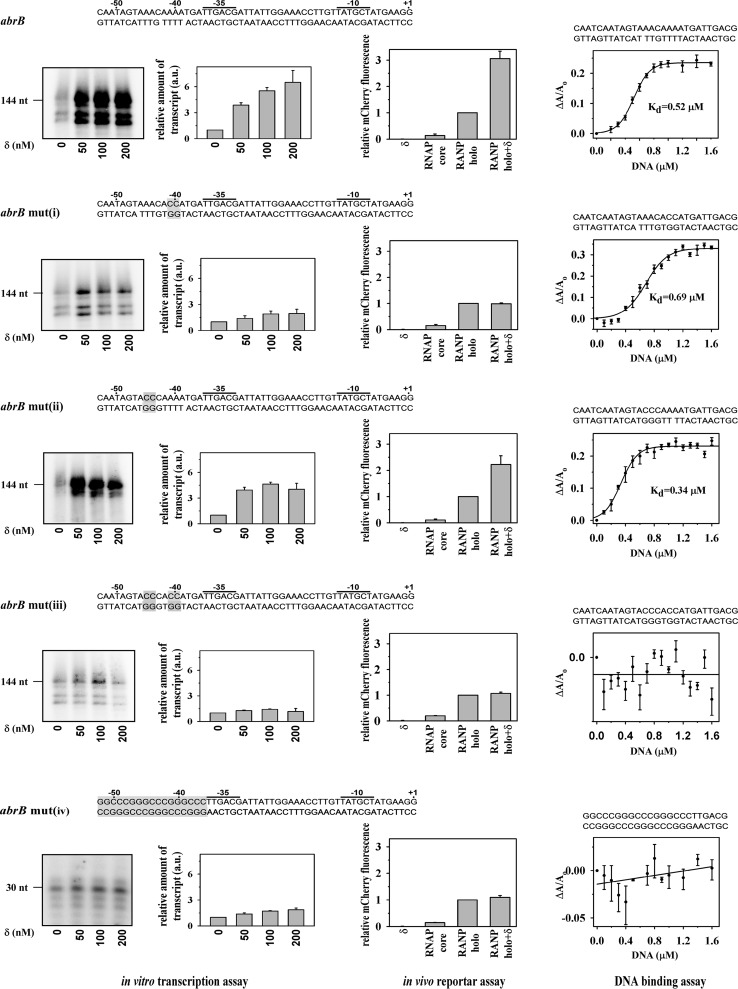
Because most transcriptional regulators bind to DNA at specific sites, we therefore examined the ability of δ to bind DNA. We used a fluorescence anisotropy assay with TMR-labeled δ to monitor its ability to bind to any promoter DNA. The data showed that δ exhibits strong affinity for the abrB promoter DNA fragment (apparent Kd = 13 ± 0.4 nm; Fig. 2A) that spans from an upstream position of −95 to a downstream position of +144. Because we used 20 nm labeled δ in this assay, and the Kd value was estimated using the sigmoidal function, the actual dissociation constant of binding of δ to DNA should be less than the estimated value. The sigmoidal nature of binding data also suggests cooperative binding of δ to the promoter DNA, possibly because of binding of the protein at multiple sites of DNA. To map the binding site of δ on DNA, DNase I footprinting was performed with radiolabeled promoter DNA fragment (abrB promoter, −95/+30). In the presence of δ, a broad region upstream of the −35 region is protected (Fig. 2B). As the exact location of the binding site of δ on DNA could not be mapped from this assay, we carried out protein-DNA footprinting assay using FeBABE-labeled δ. The results showed a δ-induced cleavage at −41 of abrB promoter (Fig. 2C) DNA, suggesting that the possible binding site of δ on the promoter DNA lies around −41 bp of the promoter. To verify whether this upstream element has any effect on the function of δ, we used several derivatives of the same promoter DNA fragment in which the upstream region was deleted stepwise from the 5′ end and monitored the yield of transcripts from these DNA fragments in the presence and absence of δ. Our results showed that removal of DNA beyond −41 completely abolished the δ-mediated increase in the transcript yield (Fig. 2D). To test whether removal of the upstream DNA region impaired the ability of RNAP to form the open complex on these DNA fragments, we performed EMSA. It was found that the removal of upstream DNA did not affect the open complex formation by RNAP (Fig. 2E). The effect of the upstream region on δ-mediated transcription was also tested with rrnBP1 promoter DNA fragments with a similar result: a complete loss of transcriptional activity by δ was observed when the upstream region beyond −41 was removed from the promoter DNA fragment (Fig. 2F). As the stretch of DNA immediately upstream of the −35 element of both the promoters contain AT-rich regions, we tested whether these AT-rich sequences are required by δ for transcriptional activation. We prepared four derivatives of the abrB promoter DNA fragment where (i) AA sequence at positions −40 and −41 was replaced by CC (abrB mut(i)), (ii) AA at positions −44 and −45 were replaced by CC (abrB mut(ii)), (iii) AA at positions −41 and −42 and AA at positions −44 and −45 both were replaced by CC (abrB mut(iii)), and (iv) a 17-bp GC-rich sequence was inserted immediately upstream of −38 bp (abrB mut(iv)) (Fig. 3). We observed that the replacement of AA by CC around −40 abrogated δ-mediated increase in transcript yield but did not have any effect on the initial transcript level without δ (Fig. 3, left panel). On the other hand, the replacement of AA by CC around −44 had no adverse effect on δ function (Fig. 3, left panel). Similarly for the same promoter, when both the AA bases around both −40 and −44 were replaced by CC or the 17 bp AT-rich sequence upstream of the −35 element was replaced by GC-rich sequence, the effect of δ on transcription at this promoter derivative was completely lost (Fig. 3, left panel). Therefore, we infer that the A-rich sequence around −40 is required for δ-mediated increase in transcript yield on the abrB promoter. This also explains why δ was unable to enhance the transcription yield at the rrnBP1 promoter DNA derivative in which the 6 A bases at upstream of the −35 element (starting from −40) were removed (Fig. 2E). The data clearly indicate that binding of δ at the upstream DNA site is essential for its function.
FIGURE 2.
δ binds to DNA upstream of promoter element. A, binding of δ to promoter DNA fragment. For fluorescence anisotropy assay, 20 nm TMR-labeled δ was added to the abrB promoter DNA (−95/+144) (Table 1). Each data set represents mean of three replicates. The Kd values of δ to abrB promoter was estimated by fitting the data using the sigmoidal function. B, protection of upstream promoter DNA sequence in the presence of δ. For DNase I footprinting assay, 0.2 μm 32P-labeled (at the 5′ end of the template strand) abrB promoter DNA (−95/+30) was incubated with δ in transcription buffer as indicated. The products were separated on 8% urea-PAGE gel. The line indicates the protected region on DNA by δ. C, location of binding region of δ on abrB promoter DNA. For FeBABE-induced protein-DNA footprinting assay, 0.2 μm of abrB promoter DNA fragment (as above) was incubated with FeBABE labeled δ in transcription buffer. The products were separated on 6% urea-PAGE gel. G+A DNA ladder and samples were run on same gel but visualized with different contrast. The arrow indicates the cleavage product. D, effect of upstream DNA on δ-mediated transcription. For in vitro transcription assay, 200 nm RNAP holo, 100 nm abrB promoter DNA fragments having different length of upstream region were used, in the absence and presence of δ. Run-off transcripts sizes were 144 nucleotides. E, removal of upstream DNA has no effect on open complex formation. For EMSA, 200 nm of RNAP holo samples were incubated at 37 °C for 20 min with 20 nm 32P-labeled promoter DNA fragments having different length of upstream region. The products were challenged with 400 nm of the same unlabeled DNA and run on 5% PAGE. The bands were visualized by PhosphorImager scanning. F, same as D, but for rrnBP1 promoter DNA. Run-off transcripts were 47 nucleotides in length.
FIGURE 3.
Effect of upstream A-rich sequences on δ function. Left panel, in vitro transcription assay. 200 nm RNAP holo, 50 nm promoter DNA both (wt and mutants) and in the presence and absence of δ. Sequences of the promoter DNA templates were shown at each panel; the mutated bases are highlighted. Run-off transcripts were 144 nucleotides. Each experiment was repeated thrice, and the mean of fold increase in the amount of transcript at each concentration of δ with respect to the amount in the absence of δ was plotted as a bar graph. Middle panel, in vivo recombinant reporter assay; three-plasmid expression system in E. coli. The bars represent relative mCherry fluorescence of E. coli cells containing the pFPVmCherry-abrB/abrB mutants and plasmids encoding (i) δ (pAcYcDuet-rpoE), (ii) Bs RNAP core (pNG219), (iii) Bs RNAP holo (pNG219 + pAcYcDuet-rpoD), and (iv) Bs RNAP holo + δ (pNG219 + pAcYcDuet-rpoD-rpoE). DNA fragments (−95/+15) of abrB or abrB mut(i)–(iii) and fragment (−52/+15) of abrB mut(iv) were inserted upstream of mCherry gene. Each set of assay was repeated thrice, and the mean values of relative mCherry fluorescence of the cells were plotted. Fluorescence of the cells containing Bs RNAP holo were normalized to 1. Right panel, binding of δ to A-rich DNA fragments. For fluorescence anisotropy assay, 20 nm TMR-labeled δ was added with double-stranded DNA (sequence shown above the graph) containing putative δ binding sites and its mutants. Each data set represents mean of three replicates. The Kd values of δ to A-rich DNA template was estimated using the sigmoidal function.
To test the effect of these mutations at abrB promoter on the activity of δ in vivo, we employed a recombinant reporter assay in E. coli using three-plasmid expression system as in Banerjee et al. (18). The abrB promoter fragment (−95/+15) and its four mutant derivatives were inserted upstream of the mCherry gene in pFPVmCherry vector. The plasmid pNG219 was used for expression of Bs RNAP core along with either pAcYcDuet-rpoD or pAcYcDuet-rpoE or pAcYcDuet-rpoD-rpoE for expression of σA, δ, or both. All three plasmids were transformed in E. coli B834 (DE3), and the cells were grown at 16 °C for 16 h after IPTG induction. To rule out the possible interference by E. coli (Ec) RNAP on the mCherry expression, we performed control assays by omitting Bs RNAP expressing plasmid, pNG219. The assays were carried out with the wild type abrB promoter and its all four derivatives (Fig. 3, middle panels). The levels of mCherry expression from all the promoter derivatives by Bs RNAP holo were comparable and normalized to 1. Our results showed that mCherry fluorescence from the control assays (without Bs RNAP) were comparable to the background fluorescence obtained with E. coli harboring pFPVmCherry-abrB only, thus establishing the fact that δ does not function with Ec RNAP as observed in our in vitro transcription assay (data not shown). However, expression of Bs RNAP core increased the level of mCherry expression compared with the background. This increase probably occurred because of leaky expression. In contrast, expression of Bs RNAP holo resulted in a ∼5-fold increase in the levels of mCherry expression from the abrB promoter and its derivatives. The presence of δ increased the mCherry expression from the wild type abrB promoter by 3-fold compared with Bs RNAP holo. However, the presence of δ did not change the level of mCherry fluorescence for all three abrB mutant derivatives: abrB mut(i), abrB mut(iii), and abrB mut(iv). On the other hand, expression level on abrB mut(ii) showed a wild type-like (2.5-fold) increase upon coexpression of δ. We note that the levels of mCherry expression from the promoters in this assay could be different if being performed in B. subtilis. However, we previously (18) showed that the recombinant reporter assay in E. coli is sufficient to test the in vivo interactions of promoters with RNAP and transcriptional regulators if there is no interference from E. coli RNAP. These experiments therefore provide further evidence that the mutation on abrB promoter around −40 abrogates the δ-mediated transcriptional activation in vivo as well, corroborating with our in vitro findings that A-rich sequence around −40, immediately upstream of −35 element is required for δ function.
To test whether δ is able to bind the small DNA fragment containing the proposed DNA binding site, we prepared double-stranded DNA fragment spanning from −57 to −30 of the abrB promoter and its mutant derivatives (Fig. 3, sequences in right panels). The binding of δ to these DNA fragments was monitored using fluorescence anisotropy assay. For GC-rich abrB promoter derivative, we used the DNA fragment spanning from −52 to −30. (Fig. 3, right panels). The data (Fig. 3) showed that δ was unable to bind to abrB mut(iii) containing the double mutation (around −40 and −44) and abrB mut(iv) that contains GC-rich sequence but was able to bind to the wild type abrB promoter (approximate Kd value 0.52 μm), abrB mut(i) (approximate Kd value 0.69 μm), and abrB mut(ii) (approximate Kd value 0.34 μm). The Kd value for δ to the small DNA fragments are much higher than the apparent Kd (13 nm) observed with longer abrB DNA fragment spanning from −95 to +144. We presume that this is possibly due to the binding of δ at multiple sites within the DNA. Interestingly, δ binds to the abrB mut(i) that contains mutation around −40 but is unable to activate transcription from this mutant promoter. The result further confirms that binding of δ to A-rich sequence at −40, not at −44, is critical for its function.
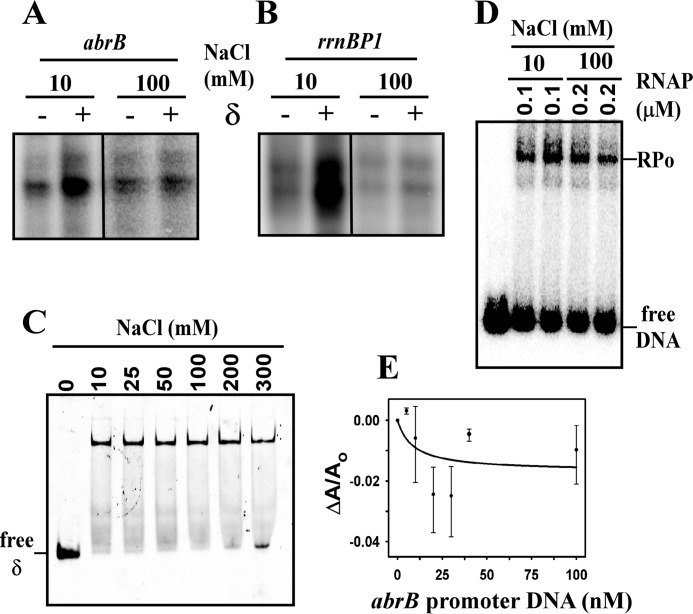
The requirement of δ to bind DNA for transcriptional activity was further demonstrated by the in vitro transcription assay carried out at higher salt concentration (100 mm as opposed to 10 mm of NaCl; Fig. 4, A and B). At 100 mm salt, δ retained its affinity toward RNAP core as observed by EMSA (Fig. 4C) but lost its ability to bind DNA as observed by fluorescence anisotropy assay (Fig. 4E). At high salt concentration, RNAP was able to bind promoter DNA as confirmed by EMSA (Fig. 4D) and was able to produce transcripts from both the promoters used in the assay (Fig. 4, A and B), although there was lower yield of transcripts compared with that at 10 mm NaCl concentration. In the presence of δ, there was no change in the yield of transcripts from both the promoter DNA fragments (Fig. 4, A and B). Therefore, at high salt concentration, δ loses its ability to enhance the yield of transcripts. This result further demonstrates that the binding of δ to DNA is essential for δ-mediated transcription regulation.
FIGURE 4.
Effect of salt on the activity of δ. A, in vitro transcription assay. 200 nm of RNAP holo, 50 nm of abrB promoter DNA in the absence and presence of δ at 10 and 100 mm NaCl concentration. Run-off transcript of 144 nucleotides in length. B, same as in A, but for the rrnBP1 promoter DNA. Run-off transcript of 47 nucleotides in length. C, binding of δ to RNAP core at different NaCl concentration. For EMSA, 40 nm of TMR-labeled δ protein was incubated with 400 nm of RNAP core in transcription buffer at different NaCl concentration as indicated at 37 °C for 30 min, and the products were separated on 5% TBE PAGE. The bands were visualized by scanning the gel using fluorescence scanning. D, binding of RNAP holo to promoter at different salt concentration. For EMSA, 100 or 200 nm RNAP holo were incubated with 20 nm of 32P-labeled abrB promoter DNA in transcription buffer containing either 10 or 100 mm NaCl at 37 °C for 30 min. The products were challenged with 400 nm of unlabeled DNA and separated on 5% TBE PAGE. The bands were visualized by phosphorimaging. E, binding of δ to promoter DNA fragment at 100 mm NaCl. For fluorescence anisotropy assay, 20 nm TMR-labeled δ was added to the abrB promoter DNA fragment (length −95/+144) in transcription buffer containing 100 mm NaCl. Each data set represents mean of three replicates.
δ Affects Transcription Initiation at the Open Complex Formation, Not at the Promoter Escape State
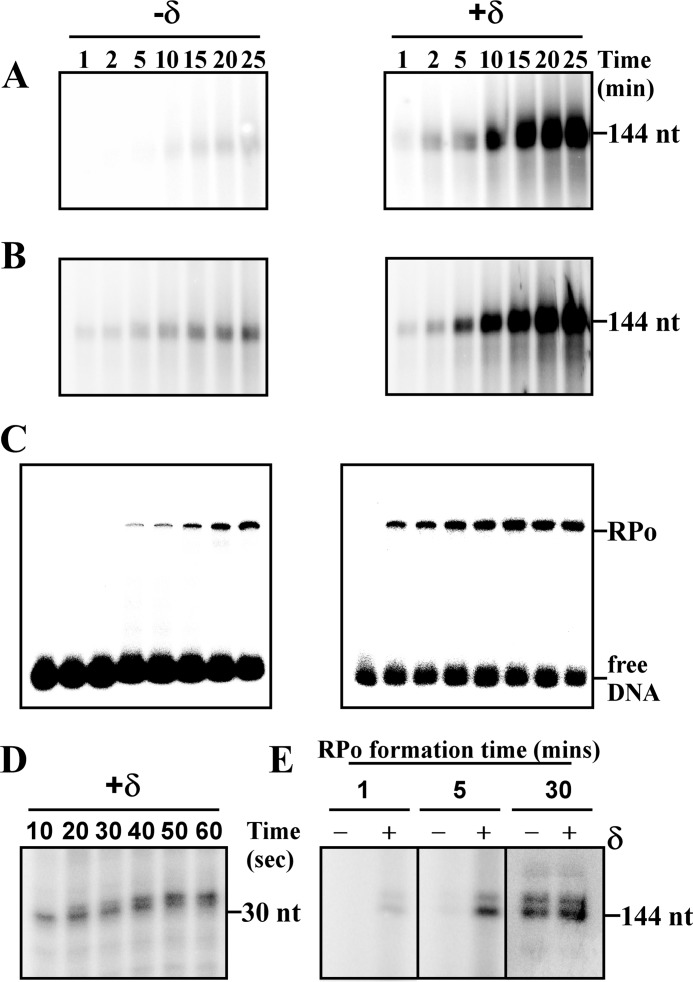
From the above results, it is apparent that δ functions as a transcriptional regulator. We therefore tested the possibility whether the protein has any role in transcription initiation. Because transcription initiation involves two rate-limiting steps, namely, open complex formation and promoter escape, we wished to monitor the effect of δ on both of these steps and therefore performed two types of in vitro transcription assays. In the first assay, we incubated all the components of the transcription reaction, e.g. RNAP, promoter DNA, NTP, and δ (or no δ), except σA and initiated transcription reactions were with the addition of σA (Fig. 5A). This reaction involved open complex formation, as well as promoter escape steps. In the second assay, we first formed the open complex by incubating RNAP holo and promoter DNA for 20 min at 37 °C and subsequently initiated the reactions with NTP. δ (or no δ) was added to the reactions along with NTP (Fig. 5B). Previously it had been shown that the multiround transcription with Ec RNAP does not occur successfully on linear DNA template containing no transcriptional terminator (22, 23). It is plausible that the level of transcripts in the presence of δ is higher than that in the absence of δ at each time point, because of the occurrence of multiround transcription in the presence of δ (Fig. 5B). However, a remarkable observation of this assay is the appearance of the first bands of transcripts within 1 min, both in the presence and in the absence of δ. Because the open complex was formed prior to transcription initiation in both the cases, these results indicate that δ does not have any role in the promoter escape. On the other hand, in the first assay, the first band of transcripts appeared at 1 min in the presence of δ and at 10 min in the absence of δ (Fig. 5A). In this assay, the time taken by RNAP to synthesize run-off transcripts reflects the time taken by RNAP to form open complex as well as for promoter escape. Because δ has no role in promoter escape, the difference in the time taken by RNAP to synthesize transcripts may be attributed to the difference in the time taken by RNAP to form open complex in the presence or absence of δ. This result allows us to conclude that δ may facilitate the open complex formation, possibly by reducing the DNA melting time. This was further confirmed by EMSA, as well as single round transcription assay. In the EMSA, we used 20 nm radiolabeled abrB promoter DNA fragment (−66/+30). After forming the open complex, the complexes were challenged with the same unlabeled DNA fragment at 400 nm concentration to remove any nonspecific complexes. The EMSA data showed that the open complex was formed within 1 min in the presence of δ but required ∼15 min to reach the same level in the absence of δ (Fig. 5C). Identical results were observed with the rrnBP1 promoter (data not shown). With the present experimental setup, we were unable to study the open complex formation in less than 1 min. To monitor the minimum time required by RNAP to synthesize transcripts from a promoter, a single round transcription assay was performed. Heparin was added to the reaction at the time of transcription initiation by NTP to prevent recycling of RNAP. We also used 1.5-fold higher concentrations of RNAP and DNA in this assay to increase the amount of transcripts for easy visualization. The data showed that RNAP is able to synthesize transcripts as early as 10 s in the presence of δ (Fig. 5D). Thus, the presence of δ reduces the time of transcription initiation by RNAP to the second scale, compared with the minute scale as observed in the absence of δ. When single-round transcription assays were further carried out by forming the open complex for 1, 5, and 30 min in the presence and absence of δ, transcription activation by δ were observed with the open complexes of 1 and 5 min, not significant with the open complex of 30 min (Fig. 5E). At 30 min, the amount of the open complex in the presence and absence of δ were the same, and at the saturating level, as a result, there was no change in the amount of transcript.
FIGURE 5.
Effect of δ on the open complex formation and promoter escape. A, effect of δ on open complex formation. For in vitro transcription assay, 200 nm RNAP core, 50 nm abrB promoter DNA, and NTP were incubated in the absence or presence of 200 nm δ. Transcription reactions were initiated by addition of 800 nm of σA. Reactions were stopped by FLB at given time points as indicated. Products were run on a same gel and visualized by phosphorimaging with the same contrast. B, effect of δ on promoter escape. For in vitro transcription assay, first open complex was formed by incubating 200 nm RNAP holo with 50 nm abrB promoter. Transcription reaction was initiated with addition of NTP. δ (200 nm) was added to the reaction at the time of NTP addition. 10 μl of the reaction samples were aliquoted at given time points, mixed with FLB, and denatured by heating at 95 °C for 5 min. The obtained products were separated by running in 8% urea PAGE gel. Run-off transcripts sizes were 144 nucleotides. Products were run on a same gel and visualized by phosphorimaging with same contrast. C, effect of δ on open complex formation. For EMSA, 200 nm of RNAP holo samples were incubated with 25 nm 32P-labeled abrB promoter DNA fragments (length −66/+30) in the absence and presence of 200 nm δ for the time points indicated in the figure to form complexes, the products were challenged with 400 nm unlabeled DNA fragments. The samples were separated on 5% native PAGE. D, single-round in vitro transcription assay. 300 nm of RNAP holo and 75 nm of abrB promoter DNA were incubated at 37 °C for 30 min in the presence of 300 nm δ. The reactions were initiated by addition of NTP and 0.25 μg/μl heparin and stopped by FLB at time indicated. Products were separated on 12% urea PAGE. Run-off transcripts sizes were 30 nucleotides. E, single-round in vitro transcription assay. 100 nm RNAP holo samples were incubated with 25 nm abrB promoter DNA fragments in the absence or presence of 100 nm δ at 37 °C for the time as indicated in the figure. Single-round transcription reactions were initiated by addition of NTP and 0.25 μg/μl heparin at the time indicated points. The reactions were stopped by addition of FLB after 30 min and resolved on 8% urea-PAGE. Run-off transcripts sizes were 144 nucleotides.
δ Involves in Recycling of RNAP from Stalled Elongation Complex
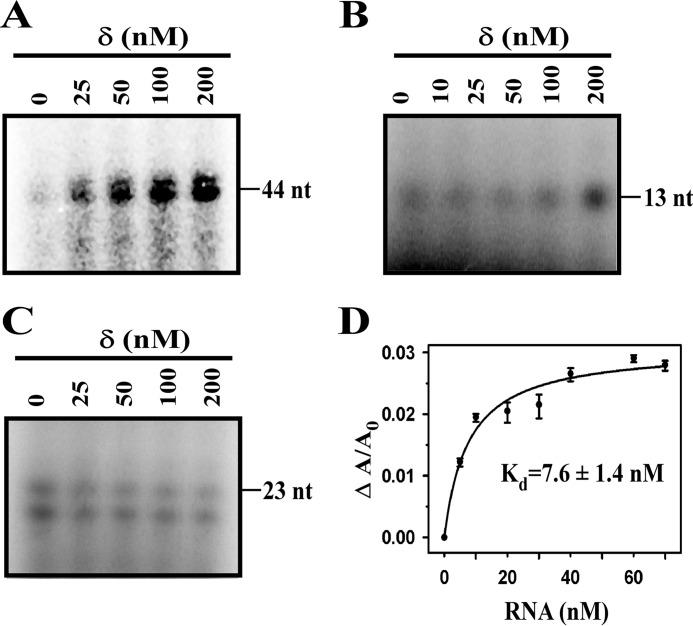
The previous study (10), as well as our observation, shows that δ is involved in recycling of RNAP in transcription. To test whether the protein is able to recycle RNAP in the stalled elongation complex, we used three sets of complexes stalled at positions +13, +23, and +44 on the abrB promoter derivatives. To our surprise, we observed an increase in the transcript yield in multiround transcription assay with a stalled elongation complex at +44 (Fig. 6A). Ideally, because RNAP does not dissociate from the stalled elongation complex, recycling of RNAP does not occur under normal condition. Therefore, the increase in the transcription yield can only be explained by the recycling of RNAP. However, when we formed stalled elongation complexes with short nucleotides (13 or 23 nucleotides), we did not find any increase in the transcript yield (Fig. 6, B and C) in the presence of δ. Next, we tested whether δ binds to RNA, using fluorescence anisotropy assay with 20 nm labeled δ and 144-nucleotides RNA (Fig. 6D). The apparent Kd value of δ binding of RNA is ∼8 nm. Because we used the hyperbolic function to fit the data, the actual Kd value should be less than estimated value. Helmann and co-workers (10) suggested that the recycling of RNAP occurs through δ-mediated release of RNA and a concomitant release of RNAP from the elongation complex. Our observation additionally suggests that δ-mediated release of RNA is possible only if the length of the RNA reaches a critical length at least greater than 23 nucleotides.
FIGURE 6.
Effect of δ on recycling of RNAP in stalled elongation complex. A, effect of δ on stalled elongation complex. For in vitro transcription assay, 100 nm RNAP holo were incubated with 25 nm abrB promoter DNA fragment in which the first C base appear at +45 of the template strand, in the presence of δ. Transcription reactions were initiated by addition of ATP, UTP, and GTP and kept for 15 min before the reactions were stopped by the addition of FLB and resolved on 12% urea-PAGE. Transcripts sizes were 44 nucleotides. B, same as A except using abrB promoter DNA fragment in which first C base appear at +14. Transcripts sizes were 13 nucleotides. C, same as A except using abrB promoter DNA fragment in which first C base appear at +24. Transcripts sizes were 23 nucleotides. D, binding of δ to RNA. For fluorescence anisotropy assay, 20 nm TMR-labeled δ was titrated with RNA. Each data set represents the means of three replicates. The Kd values of δ to RNA is 7.6 ± 1.4 nm.
Discussion
The δ protein is present in most Gram-positive bacteria (especially in fermicutes). Because δ copurifies with RNAP, it has been thought to be a subunit of RNAP (1, 5–10). Using several biochemical assays, we elucidate that δ binds to RNAP core, but not to RNAP holo. We further show that δ functions as a transcriptional regulator and binds to DNA upstream of the promoter region. Structural studies on the N-terminal domain of δ revealed that it contains an HARE-HTH motif (10, 24, 25), known to interact with DNA (26). The upstream DNA to the −35 element of the two promoters reported in this study contains AT-rich sequences, as found in most σA-dependent promoters in B. subtilis (27). Replacement of A bases around −40 by C-rich ones or removal of the AT sequences from the promoter DNA fragments impaired transcription activation by δ, indicating that A-rich sequences immediately upstream of the −35 element are required for the binding of δ to DNA, as well as its function. Because δ does not bind RNAP holo and because δ-mediated transcription assay was performed with RNAP holo, the possibility that δ binds to DNA as a subunit of RNAP is unlikely.
The binding of δ to DNA region upstream of the promoter allows RNAP to form the open complex much faster. As a result, the synthesis of transcripts is found to occur in the second scale in contrast to an order of magnitude slower (minute scale) rate observed in the absence of δ. The location of binding of δ is immediately upstream of the −35 element, where RNAP binds to the promoter DNA. Thus, the protein may facilitate the open complex formation possibly by interacting with RNAP.
Our observations that δ facilitates open complex formation and activates transcription from certain promoters including rrnBP1 do not support the proposition by Rabatinová et al. (12) that δ destabilizes the open complex at the rrnBP1 promoter and mediates changes in the requirement of iNTP by RNAP to stabilize the formation of open complexes. We further observed that not only transcription initiating nucleotide (iNTP, GTP for rrnBP1) but increasing concentrations of ATP also resulted in a higher amount of transcript from the same promoter (data not shown). Thus, the observed rise in the level of transcript in the presence of higher GTP amount is an effect of nucleotide concentration on the transcript yield, not caused by stabilization of the open complex by iNTP in the presence of δ. On the other hand, their conclusion that δ is required for competitive fitness of the cell could be explained by the involvement of δ in the transcriptional regulation of essential genes under stress conditions.
The fact that δ is involved in both up-regulation and down-regulation of genes can be explained by its role as a transcriptional regulator (13, 28–30). In this study, δ is found to act as an activator of transcription on the rrnBP1 and abrB promoters. However, it is also possible that δ binds to other promoters and interacts with RNAP in a way that inhibits transcription initiation, as has been observed in the case of certain repressors (31, 32). We have observed that δ acts as a repressor on the spo0B promoter (33), although the mechanism of repression by the protein needs to be ascertained.4
Because δ was able to release RNA from the RNAP-RNA binary complex, it was suggested that δ is also involved in the release of RNAP from the RNAP-DNA-RNA ternary complex by releasing RNA from the complex (10). This leads to the suggestion that δ was involved in recycling of RNAP in multiround transcription yielding higher amounts of transcripts. However, this recycling of RNAP was only observed when a linear DNA fragment was used in the assay without having any transcription terminator. δ was unable to enhance the transcript yield when a promoter DNA fragment contained a terminator. This was observed in a previous study (5), as well as in our study (data not shown). Because recycling of RNAP occurs upon transcription termination, the effect of δ was not apparent on the DNA template that contains a terminator sequence. However, in the linear template without a terminator sequence, Ec RNAP forms a dead end complex upon synthesis of full-length RNA, and no recycling of RNAP occurs (22, 23). It is likely that Bs RNAP also forms dead end complex like Ec RNAP. In the event, δ releases the RNAP from the dead end complex and therefore allows it to rebind to the promoter for multiround transcription. This explains the higher level of transcript in the presence of δ in Fig. 5 (A and B). When the interaction of δ with DNA was abrogated by mutating or removing the upstream region of the promoter, the protein was unable to increase the level of transcripts. Although RNAP is available to rebind to the promoter DNA because of recycling of RNAP by δ in this assay, the polymerase is unable to initiate rapid synthesis of transcript without the binding of δ at the promoter DNA and therefore is unable to increase the yield of transcript. Thus, this observation further indicates that δ-mediated recycling of RNAP could not be the sole reason for δ function.
The results of the single-round transcription assay show that δ has no effect on the overall yield of transcripts (Fig. 5E), when sufficient time (30 min) was available for the formation of the open complex. However, δ increases the yield of transcripts by severalfold in multiround transcription assays even if the open complex formation was allowed for 30 min (Fig. 1D). This observation could be explained by the effect of δ on the rate of open complex formation. Because δ facilitates the open complex formation, the protein drastically reduces the time to initiate RNA synthesis: from the minute scale to the second scale. Thus, after each round of transcription, an RNAP molecule could rebind to the promoter, quickly form an open complex, and synthesize a transcript. Therefore, recycling of RNAP occurs rapidly in multiround transcription and increases the yield of transcripts.
Interestingly, we also observed δ-mediated recycling of RNAP in the stalled elongation complex. Because no recycling of RNAP is expected from the stalled elongation complex, our observation of δ-mediated increase in the level of transcripts from the complex provides direct evidence for the recycling of RNAP from the ternary complex. The increase of transcript yield was observed only when the stalled complex contained large transcripts (44 nucleotides) and not when the transcripts were shorter (13 or 23 nucleotides). Because the nascent transcripts (>15 nucleotides) emerge from the RNA exit channel in RNAP (34–36), it is likely that the elongation complex containing 13-nucleotide RNA remains within the RNA exit channel and thus remains inaccessible to δ. For the complex containing the 23-nucleotide RNA, ∼9-nucleotide-long RNA would lie outside the exit channel and accessible to the solution. However, it may not be fully accessible to δ. Therefore, the above results suggest that δ-mediated recycling of RNAP in the stalled elongation complex is possible only when the nascent RNA is fully accessible to δ, and the interaction of the transcript with δ is essential for the release of RNA (and the concomitant release of RNAP) from the ternary complex. This is consistent with our observation that δ binds to RNA with high affinity. However, whether the interaction of δ with RNAP is required during δ-mediated release of RNA from stalled ternary complex, further study is required. Previously, a single strand nucleic acid binding protein from E. coli, SSB, has been shown to recycle N4 virion RNAP by releasing the nascent transcript from the elongation complex when its size is greater than 32 nucleotides (37). Because δ has no effect on DNA template with a terminator sequence as recycling of RNAP occurs even without δ, the utility of δ-mediated recycling is not clearly understood at this point. The only plausible explanation could be the involvement of δ in releasing RNAP from the unwanted paused complex for efficient transcription. Overall, we propose a model of transcription activation by δ in which δ binds to DNA at A-rich sequence immediately upstream of the −35 element of promoter DNA and facilitates the open complex formation that leads to rapid synthesis of transcripts.
Author Contributions
J. M. conceived and designed the experiments; R. K. P. performed purification of RNAP, in vitro transcription assays, EMSAs, fluorescence anisotropy assays, and site-directed mutagenesis; S. S. performed cloning and purification of δ; P. R. performed fluorescence based in vitro transcription assays; R. K. P. and J. M. analyzed the data; and J. M. wrote the paper.
Acknowledgments
We thank K. Murakami (Penn State University), R. Sur (University of Calcutta), A. B. Datta, and P. Parrack (Bose Institute) for critically reading the manuscript and comments.
This work was supported by Research Grants BT/PR 5345/MED/29/648/2012 and BT/PR 5270/BRB/10/1066/2012 from the Department of Biotechnology, Ministry of Science and Technology, India. The authors declare that they have no conflicts of interest with the contents of this article.
R. K. Prajapati, S. Sengupta, and J. Mukhopadhyay, unpublished results.
- RNAP
- RNA polymerase
- Bs
- B. subtilis
- IPTG
- isopropyl β-d-thiogalactopyranoside
- TMR
- tetramethyl rhodamine
- Ec
- E. coli.
References
- 1.Pero J., Nelson J., and Fox T. D. (1975) Highly asymmetric transcription by RNA polymerase containing phage-SP01-induced polypeptides and a new host protein. Proc. Natl. Acad. Sci. U.S.A. 72, 1589–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tjian R., Losick R., Pero J., and Hinnebush A. (1977) Purification and comparative properties of the delta and sigma subunits of RNA polymerase from Bacillus subtilis. Eur. J. Biochem. 74, 149–154 [DOI] [PubMed] [Google Scholar]
- 3.Achberger E. C., and Whiteley H. R. (1981) The role of the delta peptide of the Bacillus subtilis RNA polymerase in promoter selection. J. Biol. Chem. 256, 7424–7432 [PubMed] [Google Scholar]
- 4.Dobinson K. F., and Spiegelman G. B. (1987) Effect of the delta subunit of Bacillus subtilis RNA polymerase on initiation of RNA synthesis at two bacteriophage phi 29 promoters. Biochemistry 26, 8206–8213 [DOI] [PubMed] [Google Scholar]
- 5.Juang Y. L., and Helmann J. D. (1994) The delta subunit of Bacillus subtilis RNA polymerase. An allosteric effector of the initiation and core-recycling phases of transcription. J. Mol. Biol. 239, 1–14 [DOI] [PubMed] [Google Scholar]
- 6.Williamson V. M., and Doi R. H. (1979) Sigma factor is not released during transcription in Bacillus subtilis. Mol. Gen. Genet. 174, 47–52 [DOI] [PubMed] [Google Scholar]
- 7.Dickel C. D., Burtis K. C., and Doi R. H. (1980) Delta factor increases promoter selectivity of Bacillus subtilis vegetative cell RNA polymerase. Biochem. Biophys. Res. Commun. 95, 1789–1795 [DOI] [PubMed] [Google Scholar]
- 8.Hyde E. I., Hilton M. D., and Whiteley H. R. (1986) Interactions of Bacillus subtilis RNA polymerase with subunits determining the specificity of initiation: sigma and delta peptides can bind simultaneously to core. J. Biol. Chem. 261, 16565–16570 [PubMed] [Google Scholar]
- 9.López de Saro F. J., Yoshikawa N., and Helmann J. D. (1999) Expression, abundance, and RNA polymerase binding properties of the delta factor of Bacillus subtilis. J. Biol. Chem. 274, 15953–15958 [DOI] [PubMed] [Google Scholar]
- 10.López de Saro F. J., Woody A. Y., and Helmann J. D. (1995) Structural analysis of the Bacillus subtilis delta factor: a protein polyanion which displaces RNA from RNA polymerase. J. Mol. Biol. 252, 189–202 [DOI] [PubMed] [Google Scholar]
- 11.Gao H., and Aronson A. I. (2004) The delta subunit of RNA polymerase functions in sporulation. Curr. Microbiol. 48, 401–404 [DOI] [PubMed] [Google Scholar]
- 12.Rabatinová A., Ŝanderová H., Jirát Matêjĉková J., Korelusová J., Sojka L., Barvík I., Papouŝková V., Sklenár V., Zídek L., and Krásný L. (2013) The delta subunit of RNA polymerase is required for rapid changes in gene expression and competitive fitness of the cell. J. Bacteriol. 195, 2603–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiss A., Ibarra J. A., Paoletti J., Carroll R. K., and Shaw L. N. (2014) The delta subunit of RNA polymerase guides promoter selectivity and virulence in Staphylococcus aureus. Infect. Immun. 82, 1424–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X., and Lewis P. J. (2008) Overproduction and purification of recombinant Bacillus subtilis RNA polymerase. Protein Expr. Purif. 59, 86–93 [DOI] [PubMed] [Google Scholar]
- 15.Mukhopadhyay J., Mekler V., Kortkhonjia E., Kapanidis A. N., Ebright Y. W., and Ebright R. H. (2003) Fluorescence resonance energy transfer (FRET) in analysis of transcription-complex structure and function. Methods Enzymol. 371, 144–159 [DOI] [PubMed] [Google Scholar]
- 16.Kim Y., Ho S. O., Gassman N. R., Korlann Y., Landorf E. V., Collart F. R., and Weiss S. (2008) Efficient site-specific labeling of proteins via cysteines. Bioconjug. Chem. 19, 786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudra P., Prajapati R. K., Banerjee R., Sengupta S., and Mukhopadhyay J. (2015) Novel mechanism of gene regulation: the protein Rv1222 of Mycobacterium tuberculosis inhibits transcription by anchoring the RNA polymerase onto DNA. Nucleic Acids Res. 43, 5855–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banerjee R., Rudra P., Saha A., and Mukhopadhyay J. (2015) Recombinant reporter assay using transcriptional machinery of Mycobacterium tuberculosis. J. Bacteriol. 197, 646–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daubendiek S. L., Ryan K., and Kool E. T. (1995) Rolling circle RNA synthesis: circular oligonucleotides as efficient substrates for T7 RNA-polymerase. J. Am. Chem. Soc. 117, 7818–7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strauch M. A., Perego M., Burbulys D., and Hoch J. A. (1989) The transition state transcription regulator AbrB of Bacillus subtilis is autoregulated during vegetative growth. Mol. Microbiol. 3, 1203–1209 [DOI] [PubMed] [Google Scholar]
- 21.Stewart G. C., and Bott K. F. (1983) DNA sequence of the tandem ribosomal RNA promoter for B. subtilis operon rrnB. Nucleic Acids Res. 11, 6289–6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arndt K. M., and Chamberlin M. J. (1990) RNA chain elongation by Escherichia coli RNA polymerase: factors affecting the stability of elongating ternary complexes. J. Mol. Biol. 213, 79–108 [DOI] [PubMed] [Google Scholar]
- 23.Nudler E., Gusarov I., Avetissova E., Kozlov M., and Goldfarb A. (1998) Spatial organization of transcription elongation complex in Escherichia coli. Science 281, 424–428 [DOI] [PubMed] [Google Scholar]
- 24.Motácková V., Sanderová H., Zídek L., Novácek J., Padrta P., Svenková A., Korelusová J., Jonák J., Krásný L., and Sklenár V. (2010) Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase and its classification based on structural homologs. Proteins 78, 1807–1810 [DOI] [PubMed] [Google Scholar]
- 25.Demo G., Papouŝková V., Komárek J., Kader̂ávek P., Otrusinová O., Srb P., Rabatinová A., Krásný L., Zídek L., Sklenár̂ V., and Wimmerová M. (2014) X-ray vs. NMR structure of N-terminal domain of delta-subunit of RNA polymerase. J. Struct. Biol. 187, 174–186 [DOI] [PubMed] [Google Scholar]
- 26.Aravind L., and Iyer L. M. (2012) The HARE-HTH and associated domains: novel modules in the coordination of epigenetic DNA and protein modifications. Cell Cycle 11, 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helmann J. D. (1995) Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23, 2351–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue X., Tomasch J., Sztajer H., and Wagner-Döbler I. (2010) The delta subunit of RNA polymerase, RpoE, is a global modulator of Streptococcus mutans environmental adaptation. J. Bacteriol. 192, 5081–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue X., Sztajer H., Buddruhs N., Petersen J., Rohde M., Talay S. R., and Wagner-Döbler I. (2011) Lack of the delta subunit of RNA polymerase increases virulence related traits of Streptococcus mutans. PLoS One 6, e20075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue X., Li J., Wang W., Sztajer H., and Wagner-Döbler I. (2012) The global impact of the delta subunit RpoE of the RNA polymerase on the proteome of Streptococcus mutans. Microbiology 158, 191–206 [DOI] [PubMed] [Google Scholar]
- 31.Choy H. E., Park S. W., Aki T., Parrack P., Fujita N., Ishihama A., and Adhya S. (1995) Repression and activation of transcription by Gal and Lac repressors: involvement of alpha subunit of RNA polymerase. EMBO J. 14, 4523–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinones M., Kimsey H. H., Ross W., Gourse R. L., and Waldor M. K. (2006) LexA represses CTXphi transcription by blocking access of the alpha C-terminal domain of RNA polymerase to promoter DNA. J. Biol. Chem. 281, 39407–39412 [DOI] [PubMed] [Google Scholar]
- 33.Varughese K. I., Madhusudan, Zhou X. Z., Whiteley J. M., and Hoch J. A. (1998) Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase. Mol. Cell 2, 485–493 [DOI] [PubMed] [Google Scholar]
- 34.Komissarova N., and Kashlev M. (1998) Functional topography of nascent RNA in elongation intermediates of RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 95, 14699–14704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolb K. E., Hein P. P., and Landick R. (2014) Antisense oligonucleotide-stimulated transcriptional pausing reveals RNA exit channel specificity of RNA polymerase and mechanistic contributions of NusA and RfaH. J. Biol. Chem. 289, 1151–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murakami K. S., and Darst S. A. (2003) Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 13, 31–39 [DOI] [PubMed] [Google Scholar]
- 37.Davydova E. K., and Rothman-Denes L. B. (2003) Escherichia coli single-stranded DNA-binding protein mediates template recycling during transcription by bacteriophage N4 virion RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 100, 9250–9255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J., and Russell D. W. (2001) Molecular cloning: a laboratory manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]