Abstract
MFAP4 (microfibrillar-associated protein 4) is an extracellular glycoprotein found in elastic fibers without a clearly defined role in elastic fiber assembly. In the present study, we characterized molecular interactions between MFAP4 and elastic fiber components. We established that MFAP4 primarily assembles into trimeric and hexameric structures of homodimers. Binding analysis revealed that MFAP4 specifically binds tropoelastin and fibrillin-1 and -2, as well as the elastin cross-linking amino acid desmosine, and that it co-localizes with fibrillin-1-positive fibers in vivo. Site-directed mutagenesis disclosed residues Phe241 and Ser203 in MFAP4 as being crucial for type I collagen, elastin, and tropoelastin binding. Furthermore, we found that MFAP4 actively promotes tropoelastin self-assembly. In conclusion, our data identify MFAP4 as a new ligand of microfibrils and tropoelastin involved in proper elastic fiber organization.
Keywords: analytical ultracentrifugation, calcium-binding protein, elastin, site-directed mutagenesis, surface plasmon resonance (SPR), fibrillin, microfibrillar-associated protein 4
Introduction
Elastic fibers are key extracellular matrix structural elements of connective tissues that undergo repeated stretch, such as large arteries and the lung (1). The fibers consist of two major components: an amorphous elastin core surrounded by a sheath of fibrillin-rich microfibrils (2). Elastin is a highly hydrophobic polymer of the soluble precursor tropoelastin (3). Tropoelastin is known to undergo a self-assembly process known as coacervation (4), often believed to be a first step in the process of elastic fiber maturation. Because of the high content of lysine residues within the tropoelastin sequence, its assembly into a polymeric form is stabilized by formation of desmosine cross-links, catalyzed by the lysyl oxidase (LOX)3 enzyme family (5).
Microfibrils, the other major component of elastic fibers, provide the structural scaffold for the deposition of elastin globules. They consist primarily of fibrillin-1 and fibrillin-2, large glycoproteins with a high degree of homology (6). Apart from fibrillins, numerous accessory proteins have been shown to associate with microfibrils or elastin and promote formation of mature fibers, including fibulins and microfibril-associated glycoproteins (MAGPs) (7–10). The importance of proper elastogenesis has been underscored by gene deficiency studies: mice lacking elastin, LOX, or fibrillin-1 die shortly after birth because of vascular abnormalities (11–13).
MFAP4 (microfibrillar-associated protein 4) is an extracellular matrix protein belonging to the fibrinogen-related domain (FReD) family. The family includes several proteins engaged in tissue homeostasis and innate immunity, such as FIBCD1 (fibrinogen C domain-containing 1), ficolins, and angiopoietins (14–16). The crystal structure of the FReD of several family members has been solved (17–19). The ligand-binding site, designated S1, is described in all the proteins and is located in close proximity to the calcium-binding site. MFAP4 has been reported to form homodimeric structures that further oligomerize, but its definite oligomerization pattern has not been established (20).
MFAP4 is considered to be the human homolog of MAGP-36 (36-kDa microfibril-associated glycoprotein). MAGP-36 has been found to bind elastin and collagen in a calcium-dependent manner and to co-localize with elastin microfibrils (21, 22). Human MFAP4 has been localized to elastic fibers in a variety of elastic tissues, including aorta (23), skin (24), and lung (23, 25). Immunogold electron microscopy studies have shown that MFAP4 localizes to elastin-associated microfibrils but not microfibrils away from the elastin bundle (24). Together, these observations suggest that MFAP4 may contribute to the elastic fiber assembly and/or maintenance. Indeed, MFAP4 has been found to interact with fibrillin-1 and promote elastic fiber formation in in vitro dermal skin fibroblast cultures (26).
In this study, we aimed to verify and further characterize the oligomerization pattern of MFAP4 and its molecular interactions with well established elastic fiber components such as tropoelastin, fibrillin-1 and -2, and LOX. We also used site-directed mutagenesis to determine amino acid positions critical for the ligand-binding properties of MFAP4 based on the sequence alignments of the predicted S1 binding site of MFAP4 and structural homologs. Finally, we explored the role of MFAP4 in the process of tropoelastin coacervation.
Experimental Procedures
Sequence Analysis, Alignment, and Molecular Modeling
DNA and protein sequence analyses were performed using the DNASTAR Lasergene v6 package. The sequence alignment of the FReD in MFAP4, l-ficolin, FIBCD1, and tachylectin 5A (TL5A) was conducted using the ClustalW method within the package.
Molecular modeling of MFAP4 was performed using the fully automated protein structure homology-modeling server SWISS-MODEL 8.05 (27–31). The structure of l-ficolin (Protein Data Bank code 2J3O) was used as a template to model FReD of MFAP4. An initial model was made based on the sequence alignment of l-ficolin and MFAP4, which was further adjusted to match the structural properties. A homology model of FReD was built by the server, and the figures were prepared using the MacPyMol software (DeLano Scientific).
Production of rMFAP4 Constructs
First-strand synthesis was performed using human lung total RNA (Clontech) as described previously (25). The coding sequence for human full-length recombinant MFAP4 (rMFAP4) without a His6 and V5 tag was inserted into the pcDNA5/FRT/V5-His TOPO®TA (Invitrogen) expression vector as described previously (32).
In addition, several genetically modified versions of rMFAP4 containing selected point mutations within the FReD sequence were created using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) and the plasmid containing the rMFAP4 as a template (Table 1). Expression vectors containing the desired mutations were transformed into competent cells using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer's instructions. The purification of the rMFAP4-coding plasmids was performed using GeneJETTM plasmid miniprep kit (Thermo Fisher Scientific, Waltham, MA). All the constructs were sequenced using primers T7 and bovine growth hormone terminator reverse primer (both provided by Eurofins MWG Operon) and the online web service supplied by Eurofins MWG Operon. The constructs were further analyzed for presence of desired mutations and absence of additional mutations using SeqManTM II DNASTAR Lasergene v6.
TABLE 1.
Primer sequences used for site-directed mutagenesis of human recombinant MFAP4
Complementary sets of oligonucleotide primers of ∼30 bp each containing the mutagenic substitutions of interest (S203A, S203Y, F241A, F241W, D191A, D193A, and D191A/D193A) were constructed using the QuikChange Primer Design program (Agilent Technologies).
| Base change | Base position | Amino acid change | Primer sequences |
|---|---|---|---|
| T/G | 632 | S203A | Forward: 5′-CTGCGCAGCTCTCGCCTCAGGAGCCTT-3′ |
| Reverse: 5′-AAGGCTCCTGAGGCGAGAGCTGCGCAG-3′ | |||
| C/A | 633 | S203Y | Forward: 5′-CTGCGCAGCTCTCTACTCAGGAGCCTTCT-3′ |
| Reverse: 5′-AGAAGGCTCCTGAGTAGAGAGCTGCGCAG-3′ | |||
| TT/GC | 746–747 | F241A | Forward: 5′-GGCCCAGTGGAAGGGCGCCTACTACTCCCTCAAA-3′ |
| Reverse: 5′-TTTGAGGGAGTAGTAGGCGCCCTTCCACTGGGCC-3′ | |||
| TC/GG | 746–747 | F241W | Forward: 5′-GCCCAGTGGAAGGGCTGGTACTACTCCCTCAAAC-3′ |
| Reverse: 5′-GTTTGAGGGAGTAGTACCAGCCCTTCCACTGGGC-3′ | |||
| A/C | 597 | D191A | Forward: 5′-TACCTTCGACCGGGCCCAGGACCTCTTTG-3′ |
| Reverse: 5′-CAAAGAGGTCCTGGGCCCGGTCGAAGGTA-3′ | |||
| A/C | 603 | D193A | Forward: 5′-GACCGGGACCAGGCCCTCTTTGTGCAG-3′ |
| Reverse: 5′-CTGCACAAAGAGGGCCTGGTCCCGGTC-3′ | |||
| A/C, A/C | 597, 603 | D191A/D193A | Forward: 5′-CCTTCGACCGGGCCCAGGCCCTCTTTGTGCA-3′ |
| Reverse: 5′-TGCACAAAGAGGGCCTGGGCCCGGTCGAAGG-3′ |
Expression and Purification of rMFAP4 WT and Mutant Forms
The expression and purification of MFAP4 variants was performed as described previously (25, 32).
Concentration of WT rMFAP4
Purified WT rMFAP4 was concentrated using a Vivaspin 6 ultrafiltration spin column, 10-kDa cut-off (GE Healthcare), and the buffer was changed to 50 mm Tris, 150 mm NaCl, pH 7.5. The purity of the concentrated protein was tested by SDS-PAGE followed by Coomassie staining using SimplyBlueTM SafeStain (Invitrogen). The concentration of WT rMFAP4 was ∼0.7 mg/ml.
Gel Permeation Chromatography
Gel permeation chromatography of WT rMFAP4 and MFAP4 mutant forms was conducted on a Superdex 200 10/30 column (Amersham Biosciences, GE Healthcare) in 50 mm Tris, pH 7.5, containing 150 mm NaCl. The elution profile was fitted with two Gaussian curves. The Stokes radius of WT rMFAP4 was determined from its elution position relative to the elution position of proteins of known Stokes radius (thyroglobulin, ferritin, aldolase, conalbumin, and ovalbumin) (33).
Analytical Ultracentrifugation
Sedimentation velocity experiments were carried out at 40,000 rpm and 20 °C using aluminum centerpieces in a Beckman XL-I analytical ultracentrifuge (Beckman Coulter). Scans were collected at 3-min intervals at 230 nm (0.18 mg/ml) and 1.5-min intervals at 280 nm (0.7 mg/ml) in separate experiments. The data were analyzed by using the program Sedfit with 68 and 45 scans, respectively (34). The proportion of each component of the mixture was very similar at each concentration, suggesting that oligomers do not interconvert appreciably over the concentration range examined. The partial specific volume of WT rMFAP4 was calculated as 0.702 ml/g based on its amino acid and carbohydrate composition (35).
Determination of MFAP4 Molecular Mass
The molecular mass of the WT rMFAP4 oligomers were calculated from their sedimentation coefficients (s20,w) and the Stokes radius derived from the gel filtration experiment (36) and separately in the sedimentation experiment from the spreading of the protein boundary over time.
Pulldown Assay
5 mg of type I collagen from bovine Achilles tendon (Sigma-Aldrich), bovine lung or aortic elastin (Elastin Products Company, Owensville, MO), and chitin from shrimp cells (Sigma-Aldrich) were washed in 1 ml of TBS, 0.05% Tween, 5 mm CaCl2 (TBST-Ca) containing 1 mg/ml BSA (Sigma-Aldrich) and hydrated in TBST-Ca overnight at 4 °C. Macromolecules were pelleted and incubated with 31 ng/ml WT rMFAP4 or purified mutant MFAP4 variants (diluted in TBST-Ca or TBST containing 10 mm EDTA, 1 ml of total volume) overnight at 4 °C. The supernatants were removed, and the pellets were washed with TBST-Ca containing 1 mg/ml BSA. Bound protein was eluted from the insoluble fraction by boiling the samples in SDS-PAGE buffer (40% (v/v) glycerol, 8% (w/v) SDS, 25% (v/v) 4× lower Tris, pH 8.8, 0.002% bromophenol blue) and subsequently analyzed by SDS-PAGE and Western blotting.
SDS-PAGE and Western Blotting
The samples were mixed with SDS-PAGE buffer with 0.6 m DTT in the reduced samples or 0.02 m iodoacetamide in the nonreduced samples. The samples were boiled for 1 min and finally alkylated by the addition of 1.4 m iodoacetamide to a final concentration of 90 mm. Proteins were separated by SDS-PAGE and transferred to a PVDF membrane (Amersham Biosciences). The membrane was blocked with 5% nonfat dry milk (Bio-Rad) in TBST for 2 h at room temperature and incubated with 0.5 μg/ml HG-HYB 7–5 anti-MFAP4 antibody at 4 °C overnight. The membrane was washed in TBST and incubated with rabbit anti-mouse Ig-HRP (Dako, Glostrup, Denmark), diluted 1:20,000 for 1 h at room temperature. Proteins were detected using ECL reagents (Amersham Biosciences).
Production of Recombinant Fibrillin Fragments
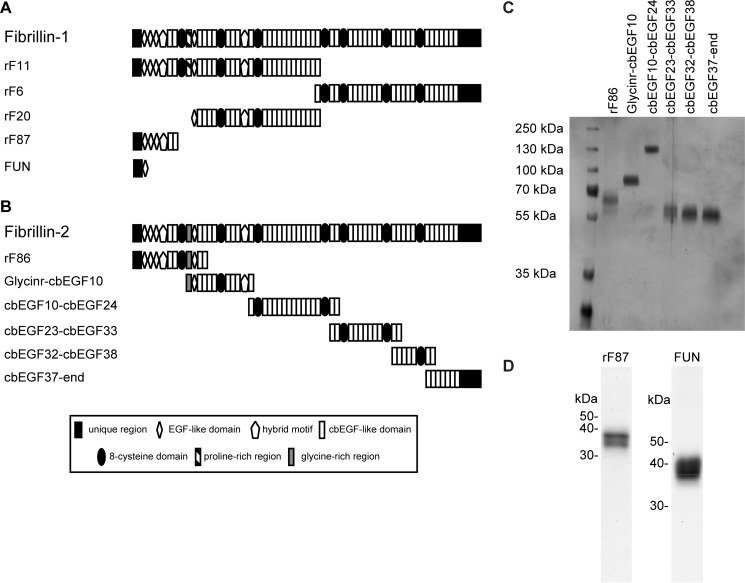
Recombinant fragments of human fibrillin-1 were overexpressed in HT1080 or HEK293/EBNA cells and purified using anion exchange chromatography as previously described (37, 38). The fibrillin-1 N-terminal unique region including the first epidermal growth factor (EGF)-like domain fused together with cbEGF19–22 and a tandem C-terminal Strep tag was expressed and purified as previously described (39) (Fig. 1A and Table 2).
FIGURE 1.
Structure of recombinant fibrillin fragments. A, domain structure of full-length fibrillin-1 and recombinant fibrillin-1 fragments used in this study. B, domain structure of full-length fibrillin-2 and recombinant fibrillin-2 fragments used in this study. Details of the domains are given in the legend. C, purity of recombinant fibrillin-2 fragments was analyzed by SDS-PAGE and silver staining. D, purity of recombinant fibrillin-1 fragments rF87 and fibrillin-1 N-terminal unique region (FUN) was analyzed by SDS-PAGE and Coomassie blue staining.
TABLE 2.
Recombinant fragments spanning full-length fibrillin-1 and -2
| Fibrillin fragment | Origin protein | Spanned region |
|---|---|---|
| rF11 | Fibrillin-1 | Ser19--Val1527 |
| rF6 | Asp1487--His2871 | |
| rF20 | Pro402--Val1527 | |
| rF87 | Ser19--Ile329 | |
| FUN | Ser19--His118 fused to Asp1363--Val1527 | |
| rF86 | Fibrillin-2 | Gln29--Asp535 |
| Glycinr-cbEGF10 | Glu412--Asp997 | |
| cbEGF10-cbEGF24 | Val956--Asp1734 | |
| cbEGF23-cbEGF33 | Ile1651--Asp2253 | |
| cbEGF32-cbEGF38 | Val2172--Asp2532 | |
| cbEGF37-end | Ile2450--Tyr2912 |
Recombinant fragments spanning human full-length fibrillin-2 were overexpressed with a tandem C-terminal StrepII-tag followed by purification using affinity chromatography as previously described (39) (Fig. 1B and Table 2). After transfection of HEK293 cells, stably transfected clones were selected in the presence of puromycin (1 μg/ml). Proteins were purified directly from serum-free cell culture medium. After filtration and centrifugation (25 min, 10,000 × g) culture supernatants were applied to a Streptactin column (1 ml; IBA, Goettingen, Germany) and eluted with 2.5 mm desthiobiotin, 10 mm Tris-HCl, pH 8.0.
Solid Phase Binding Assays
Plates were coated with tropoelastin (Sigma-Aldrich; 100 ng/ml), fibrillin-2 fragments (100 ng/ml), MFAP4 (500 ng/ml), full-length LOX (500 ng/ml; Origene, Rockville, MD), ovalbumin-desmosine conjugate (500 ng/ml; Elastin Products Company), or ovalbumin (500 ng/ml; Sigma-Aldrich) diluted in coating buffer (CO32+/HCO3−, pH 9.6) and incubated at 4 °C overnight. The next day the plates were washed three times with TBST-Ca and blocked with TBST-Ca containing 10 mg/ml BSA for at least 1 h at room temperature. Plates were then incubated with MFAP4 variants or recombinant LOX (OriGene) at the indicated concentrations diluted in TBST-Ca or TBST containing 10 mm EDTA for 2 h at room temperature. The plates were washed in TBST-Ca and incubated with 0.5 μg/ml biotinylated HG-HYB 7–5 anti-MFAP4 antibody or 1 μg/ml biotinylated anti-LOX (OriGene) for 1 h at room temperature, washed again, and subsequently incubated with 0.625 μg/ml streptavidin-HRP conjugate (Zymed; Thermo Fisher Scientific) for 30 min at room temperature. After the final wash, the plates were developed using 2 mg O-phenylenediamine dihydrochloride (Kem-En-Tec, Taastrup, Denmark) dissolved in 5 ml of citrate phosphate buffer, pH 5.0, with 35% (w/v) H2O2. The reaction was stopped with 1 m H2SO4, and the absorbance was read at 492 nm versus 620 nm. The signal from background binding to ovalbumin (for ovalbumin-desmosine) or BSA (all other proteins) was subtracted from the results. In some experiments, WT rMFAP4 was preincubated with increasing concentrations of tropoelastin or deoxypyridinoline cross-links (Santa Cruz Biotechnology; WT rMFAP4 concentrations: 125 or 500 ng/ml, respectively) for 2 h at room temperature before adding to the blocked plate.
In a separate set of experiments, plates were coated with 0.1 μm MFAP4 in PBS containing 5 mm CaCl2. Wells were blocked with 5% nonfat dry milk in TBS for 1 h at room temperature and incubated with recombinant fibrillin-2 fragments (serial dilutions starting from 500 nm) in 2% milk in TBS for 3 h at room temperature. Bound C-terminally double Strep-tagged fibrillin-2 variants were detected using a monoclonal anti-Strep tag antibody (StrepMAB-Classic; IBA) diluted in 2% milk in TBS, after a final incubation with enzyme-conjugated secondary antibodies. Color reaction was achieved using 3,3′,5,5′-tetramethylbenzidine (Sigma-Aldrich). The reaction was stopped with 0.1 n HCl, and the absorbance was read at 450 nm.
Surface Plasmon Resonance (SPR)
SPR experiments were performed using a BIAcore 2000 (BIAcore AB, Uppsala, Sweden). MFAP4 was covalently coupled to CM5 sensor chips at 2500 Rus and fibrillin-1 or -2 fragments were flown over at 100 nm in HBS-P buffer (BIAcore AB) containing either 5 mm CaCl2 or 3 and 10 mm EDTA.
In a separate set of experiments, ovalbumin and ovalbumin-desmosine conjugate were flown over immobilized MFAP4 at 50 nm. Alternatively, tropoelastin was coupled to sensor chips and WT rMFAP4 and MFAP4 mutant variants were flown over at 1000 nm.
Immunofluorescence Microscopy
Skin biopsies from male C57BL/6 WT and GT-8 mice (40) sacrificed at postnatal day 1 (P1), P7, and P14 were snap frozen in liquid nitrogen and then embedded in Tissue-Tek O.C.T. compound (Sakura Finetek Europe, Alphen aan Den Rijn, the Netherlands). Seven-μm-thick cryosections were cut using a Leica CM1850 cryostat. Sections were air dried for 15 min at room temperature and were then fixed for 10 min in cold acetone/methanol, blocked with 1% BSA in PBS for 45 min, and incubated with primary antibodies rabbit anti-fibrillin-1 pAb 9543 (41) and monoclonal mouse anti-MFAP4 (32) diluted 1:1000 in PBS/1% BSA overnight at 4 °C in a humidified chamber. After washing in PBS, sections were incubated with FITC-conjugated goat anti-rabbit antibody diluted 1:1000 in PBS, 1% BSA and Cy3-conjugated goat anti-mouse antibody (both from Life Technologies, Thermo Fisher Scientific) diluted 1:100 in PBS, 1% BSA for 60 min at room temperature. The sections were washed thoroughly, coverslipped using the Dako Fluorescence Mounting Medium (Dako), and visualized with a Leica SP5 confocal laser microscope. Micrographs were processed using the ImageJ software (42).
Coacervation Assay
Soluble tropoelastin and WT rMFAP4 or BSA were mixed in TBST-Ca to final concentrations of 1 mg/ml and 40 μg/ml, respectively, and kept on ice. The reaction mixtures (50 μl) were transferred to a 96-well plate, and the absorbance at 405 nm was measured at different temperatures ranging from 19 to 45 °C using Victor3 plate reader (PerkinElmer Life Sciences).
Statistics
Quantitative data were analyzed using Prism 5 software (GraphPad, La Jolla, CA) and are presented as means ± S.E. unless stated otherwise. The results were analyzed using unpaired Student's t test or two-way analysis of variance, with p values of <0.05 considered significant.
Results
MFAP4 Forms Trimers and Hexamers of Homodimers
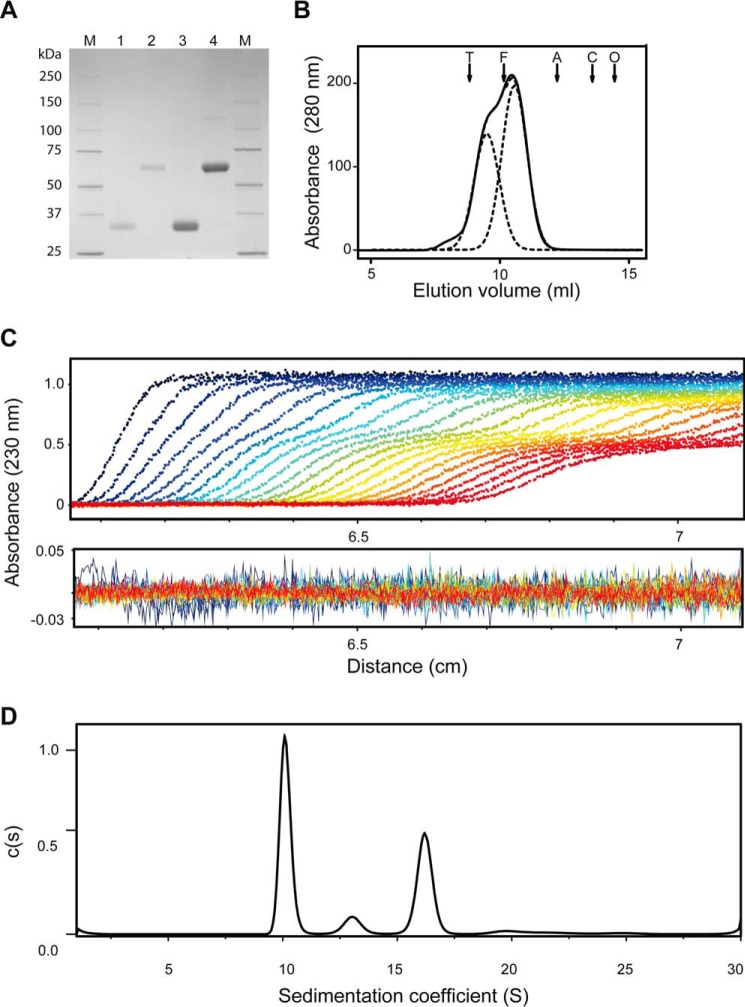
To investigate the structural organization and oligomerization of human MFAP4, we produced full-length rMFAP4. On SDS-PAGE, WT rMFAP4 migrated equivalent to a monomeric form (∼36 kDa) in the reduced state and to a homodimer (∼66 kDa) in the nonreduced state (Fig. 2A) as previously described (25). Further gel filtration chromatography analysis revealed that WT rMFAP4 eluted as two overlapping peaks corresponding to Stokes radii of 5.4 and 7.0 nm (Fig. 2B).
FIGURE 2.
Oligomerization of human MFAP4. A, purified and concentrated recombinant WT MFAP4 (WT rMFAP4). Lanes 1 and 2 show the purified protein in reduced and nonreduced state, respectively. Lanes 3 and 4 show concentrated WT rMFAP4 in reduced and nonreduced state, respectively. B, gel filtration elution profile of WT rMFAP4. Protein eluted as two overlapping peaks, and the elution profile was fitted with two Gaussian curves (dotted lines) corresponding to Stokes radii of 5.4 and 7 nm. Elution positions of bovine thyroid thyroglobulin (arrow T; 669 kDa), horse spleen apoferritin (arrow F; 440 kDa), rabbit muscle aldolase (arrow A; 158 kDa), conalbumin (arrow C; 75 kDa), and ovalbumin (arrow O; 43 kDa) are indicated. C, sedimentation of WT rMFAP4 (every third scan is shown). D, distribution of sedimenting species c(s) of WT rMFAP4. The data were fitted using continuous c(s) distribution using SEDFIT package. Four species were detected: 10.1 S (51%), 13.1 S (7%), 16.2 S (35%), and 20.2 S (3%).
Analytical ultracentrifugation was used to determine the sedimentation coefficient of WT rMFAP4. Four species were detected by sedimentation velocity experiments of WT rMFAP4: 10.1 S, 51%; 13.1 S, 7%; 16.2 S, 35%; and 20.2 S, 3% (Fig. 2D). The same proportion of each size of oligomers was observed with dilution of the protein, suggesting that the oligomers do not interconvert. Combining the values from the gel filtration with the sedimentation coefficients, s20,w, gives molecular masses of 208 and 432 kDa for the major species, thus suggesting trimeric and hexameric structures of homodimers (6- and 12-mer, respectively). Similar values (197 and 399 kDa) were calculated directly by Sedfit, which estimates the diffusion coefficient, and hence the Stokes radius, from the spreading of the protein boundary over time. Minor species of 289 and 553 kDa probably comprise tetrameric and octameric structures of homodimers (8- and 16-mer, respectively).
Binding of Human MFAP4 to Elastin and Collagen
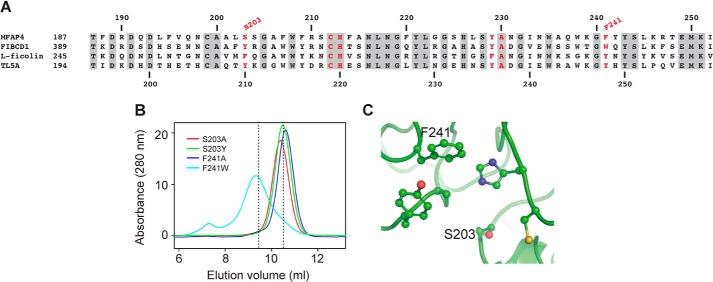
We used a site-directed mutagenesis strategy based on sequence alignments to compare the S1 binding site of MFAP4 and FIBCD1. Despite different ligand specificities, binding in all well characterized FReD family members is mediated at least in part via the S1 pocket. Comparison with FIBCD1, l-ficolin, and TL5A shows that of the six residues that are likely to form the S1 pocket in MFAP4, three residues are identical in all other family members (Cys212, His213 and Ala230), one is conserved in two of the three members (Tyr229), and the other two residues are different (Fig. 3A). Notably, Ser203 is a hydrophobic residue in FIBCD1, l-ficolin, and TL5A, whereas Phe241 is a tryptophan or a tyrosine. For mutagenesis, the serine in position 203 and the phenylalanine in position 241 were mutated to the corresponding amino acids in FIBCD1 or to the simple amino acid alanine. The objective of these experiments was to change the S1 binding pocket in MFAP4 and thereby disrupt binding toward MFAP4-specific ligands but to retain the integrity of the fibrinogen-like fold via conservative substitutions to the corresponding residues in FIBCD1. All four variant forms of rMFAP4 (S203A, S203Y, F241A, and F241W) were secreted normally and eluted at positions similar to WT rMFAP4 when analyzed by gel filtration chromatography (Fig. 3B), indicating that the mutations did not compromise biosynthesis or folding of the proteins. Fig. 3C shows the modeling of the predicted S1 binding pocket of WT MFAP4. We further attempted to investigate the involvement of calcium binding in MFAP4 interactions by mutating two conserved aspartate residues in positions 191 and 193 to alanine (D191A, D193A, and D191A/D193A). However, we were not able to obtain functional proteins for any of the three mutations (data not shown), indicating that the introduced changes compromised correct biosynthesis and/or folding of the proteins.
FIGURE 3.
Structure of the S1 binding site of MFAP4. A, sequence alignment of the S1 site in MFAP4, FIBCD1, l-ficolin, and TL5A. The numbers on top and bottom refer to MFAP4 and TL5A sequences, respectively. Conserved residues are highlighted in gray. Residues in the S1 site are marked in red. The mutations performed in this study are indicated. The accession numbers were as follows: MFAP4, AAH62415.1; l-ficolin, NP_001994.2; FIBCD1, NP_116232.3; and TL5A, 1JC9_A. B, gel filtration elution profile of MFAP4 mutant variants. Elution positions of WT rMFAP4 are indicated with dashed lines. C, modeling of S1 binding site in WT MFAP4, with the indicated amino acids mutated in this study. The structure of l-ficolin (Protein Data Bank code 2J3O) was used as a template for modeling by SWISS-MODEL 8.05, and the figures were prepared using MacPyMOL software.
Human WT rMFAP4 bound elastin and type I collagen in a calcium-dependent manner (Fig. 4A) as previously demonstrated for a bovine homolog (21). MFAP4 variants exhibited altered binding pattern to both ligands. Mutation S203A did not influence MFAP4 binding capacity. Mutation S203Y moderately lowered binding to elastin but potently attenuated binding to type I collagen. The mutations in position Phe241 had the strongest influence on MFAP4 binding properties, with both F241A and F241W variants showing strong attenuation or complete inhibition of interactions with elastin and type I collagen, respectively (Fig. 4, B and C).
FIGURE 4.
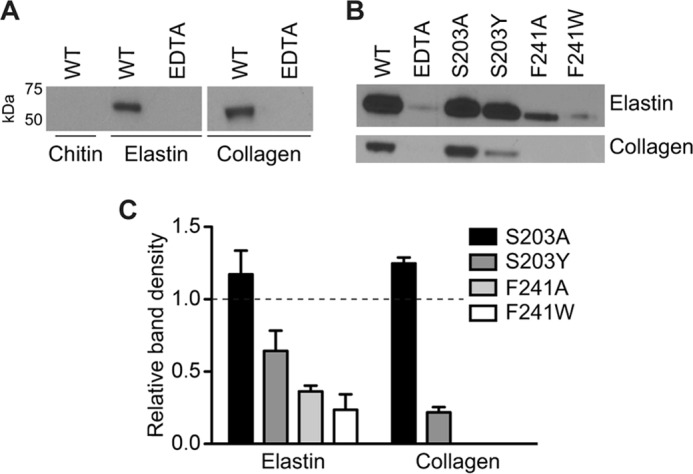
Binding of MFAP4 to insoluble elastin and type I collagen. A, WT MFAP4 binds to elastin and type I collagen, but not chitin (negative control), in a calcium-dependent manner. MFAP4 is pulled down by insoluble collagen or type I collagen but remains in solution in the presence of EDTA. B, point mutations within S1 site influence binding of MFAP4 to elastin and type I collagen. Although mutation S203A does not impact MFAP4 binding capacity, mutation S303Y lowers and mutations F241A and F241W strongly reduce elastin and collagen binding properties of MFAP4. C, densitometric analysis of (B) normalized to WT rMFAP4 (indicated with a dashed line). The data are representative (A and B) or means ± S.E. (C) of at least two independent experiments.
MFAP4 Interacts with Tropoelastin
Because MFAP4 is known to co-localize with elastic fibers, we investigated its molecular interactions with tropoelastin. Specific binding between WT rMFAP4 and immobilized tropoelastin was detected in a solid phase binding assay (Fig. 5A). The interaction was crucially dependent on calcium ions, because addition of EDTA fully inhibited observed effects (Fig. 5A). Moreover, the interaction was attenuated by WT rMFAP4 preincubation with soluble tropoelastin, confirming the specificity of our results (Fig. 5B). Mutant MFAP4 forms S203Y, F241W, and F241A exhibited significantly diminished binding to tropoelastin, whereas mutation S203A had no impact on the binding (Fig. 5C). Furthermore, SPR measurements showed a clear WT rMFAP4 binding to immobilized tropoelastin, which was undetectable in case of all four MFAP4 mutant variants (Fig. 5D).
FIGURE 5.
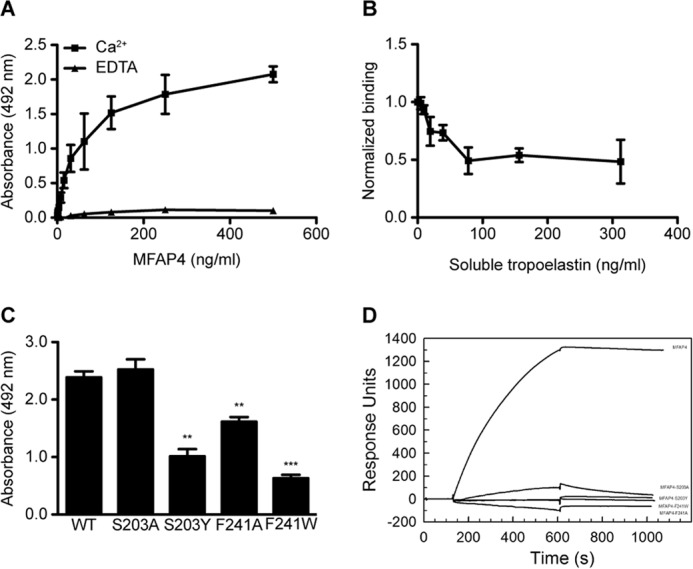
Molecular interactions between MFAP4 and tropoelastin. A, MFAP4 binds immobilized tropoelastin in a calcium-dependent manner in solid phase binding assays. B, MFAP4 interaction with immobilized tropoelastin is attenuated by preincubating MFAP4 with soluble tropoelastin. C, mutations of the S1 binding site modify MFAP4-tropoelastin binding capacity. D, BIAcore analysis of binding between immobilized tropoelastin and soluble MFAP4 variants. One representative experiment is shown. The data are means ± S.E. of three independent experiments. **, p < 0.01; ***, p < 0.001, calculated by Student's t test.
MFAP4 Interacts with Fibrillin-1 and -2 in Vitro and Co-localizes with Fibrillin-1 Fibers in Vivo
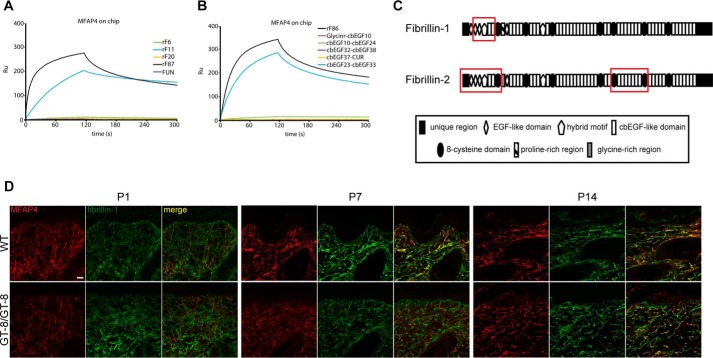
To test direct interaction of MFAP4 with fibrillins, a direct SPR interaction screen was performed using fibrillin truncation variants spanning full-length fibrillin-1 and -2 (Fig. 1, A and B). These binding experiments revealed distinct binding epitopes for MFAP4 within fibrillin-1 and fibrillin-2. Fibrillin-1 variants rF11 and rF87 (Figs. 1A and 6A) specifically interacted with immobilized WT rMFAP4, suggesting that the MFAP4 binding site lies within the N-terminal part of fibrillin-1 containing the second and third EGF-like domains, the first hybrid motif, and the first two cbEGF-like domains (Fig. 6C). A fragment covering the N-terminal unique region and the first EGF-like domain (fibrillin-1 N-terminal unique region) showed no binding (Fig. 6A), indicating that these domains are not involved in MFAP4-fibrillin-1 interaction.
FIGURE 6.
Molecular interactions between MFAP4 and fibrillins. A, BIAcore analysis of binding between immobilized MFAP4 and soluble fibrillin-1 fragments flown over at 100 nm in the presence of 5 mm CaCl2. One representative experiment is shown. B, BIAcore analysis of binding between immobilized MFAP4 and soluble fibrillin-2 fragments flown over at 100 nm in the presence of 5 mm CaCl2. One representative experiment is shown. C, structure of full-length fibrillin-1 and fibrillin-2 with identified MFAP4 binding sites (red rectangles). D, confocal immunofluorescence microscopy shows co-localization of MFAP4 (red) and fibrillin-1 (green) in the skin of WT and GT-8 mice (GT-8/GT-8) at postnatal days P1, P7, and P14. Representative pictures are shown. Original magnification, 126×. Scale bar, 20 μm.
Among fibrillin-2 variants, fragments rF86 and EGF23–33 bound to immobilized WT rMFAP4 (Fig. 6B). We confirmed our results using solid phase binding assays, where fragments rF86 and cbEGF23–33 bound WT rMFAP4 in a dose-dependent manner (data not shown). Thus, MFAP4 is able to bind fibrillin-2 at two distinct sites (Fig. 6C). The binding of both fibrillin-1 and fibrillin-2 fragments to WT rMFAP4 was not calcium-dependent, because the interaction was unchanged in the presence of EDTA (data not shown).
To confirm our in vitro observations, we performed co-localization studies in murine skin sections of WT mice and GT-8 mice, with the latter expressing a dominant negative mutant form of fibrillin-1 that causes progressive elastic fiber fragmentation (40). At day P1, when fibrillin-1 fibers in GT-8 mice look normal, MFAP4 and fibrillin-1 show only partially overlapping staining patterns (Fig. 6D). However, we detected strong co-localization of MFAP4 and fibrillin-1 at P7 and P14 time points, both in WT and GT8 tissues. In GT-8 skin at P14, MFAP4 exhibited a punctate staining pattern of the fragmented fibers, similar to fibrillin-1 (Fig. 6D).
MFAP4 Binds LOX but Does Not Tether LOX to Tropoelastin
In solid phase binding experiments, WT rMFAP4 was able to bind immobilized LOX (Fig. 7A). We then checked whether MFAP4 could act as a bridging protein and facilitate the binding of LOX to immobilized tropoelastin. However, we detected only a weak binding between LOX and tropoelastin, and the presence of WT rMFAP4 had no effect on this interaction (Fig. 7B). Thus, MFAP4 does not assist in tethering LOX to tropoelastin.
FIGURE 7.
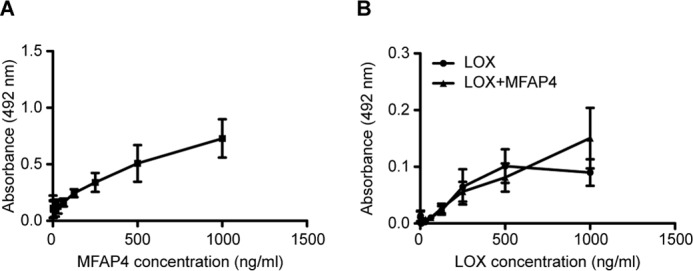
Molecular interactions between MFAP4 and LOX. A, MFAP4 binds to immobilized LOX. B, the interaction between LOX and immobilized tropoelastin is not influenced by the presence of MFAP4 in the solution. The data are means ± S.E. of three independent experiments.
Binding of MFAP4 to Desmosine
We detected calcium-dependent binding between WT rMFAP4 and ovalbumin-conjugated desmosine using solid phase binding setup (Fig. 8A). We confirmed our observations by SPR measurements, where we detected binding between immobilized WT rMFAP4 and ovalbumin-conjugated desmosine but not the ovalbumin control (Fig. 8B). Preincubation of MFAP4 with collagen-specific deoxypyridinoline cross-links did not influence MFAP4 binding to desmosine (data not shown).
FIGURE 8.
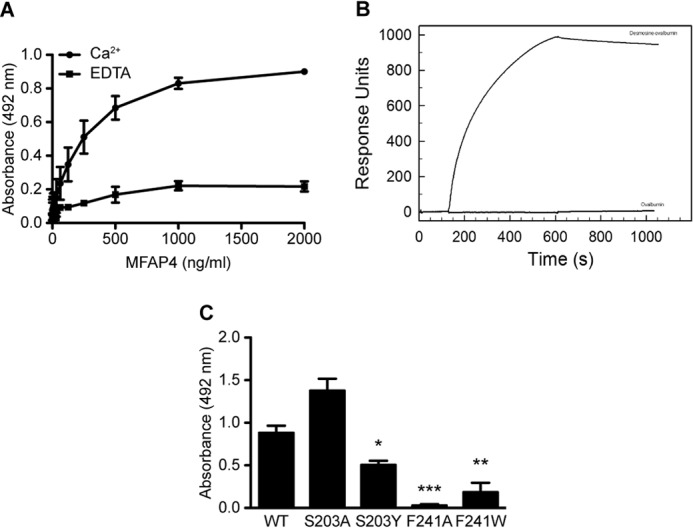
Molecular interactions between MFAP4 and desmosine. A, MFAP4 binds immobilized ovalbumin-desmosine conjugate in a calcium-dependent manner in solid phase binding assays. The background binding to ovalbumin was subtracted from the results. B, BIAcore analysis of binding between immobilized MFAP4 and soluble ovalbumin-desmosine conjugate and ovalbumin control. One representative experiment is shown. C, mutations of the S1 binding site modify MFAP4-desmosine binding capacity. The data are means ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001, calculated by Student's t test.
We also investigated whether binding to desmosine is disrupted by point mutations in the S1 binding site of MFAP4. Indeed, both Phe241 mutants and the S203Y mutant exhibited abolished or attenuated binding, underlying the importance of these two positions in mediating proper interactions with the ligand (Fig. 8C).
MFAP4 Promotes Coacervation of Tropoelastin
Finally, we investigated whether MFAP4 can influence the ability of tropoelastin to coacervate. Recombinant tropoelastin began to coacervate spontaneously at ∼37 °C. In the presence of WT rMFAP4, tropoelastin started to coacervate at lower temperatures, showing that MFAP4 promotes tropoelastin self-assembly (Fig. 9). Tropoelastin coacervation was not induced in the presence of negative control protein BSA (Fig. 9). No change in turbidity was observed for the buffer or WT rMFAP4 alone.
FIGURE 9.
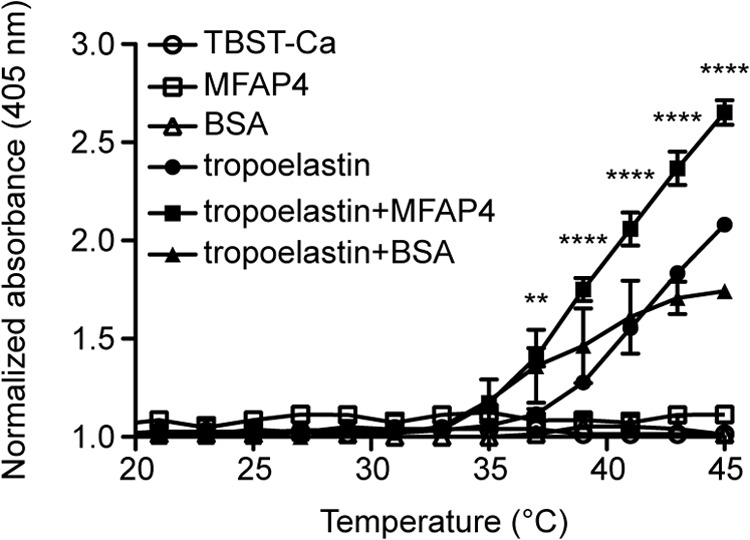
MFAP4 promotes tropoelastin coacervation. Self-aggregation of 1 mg/ml of soluble tropoelastin was induced in the presence or absence of recombinant WT MFAP4 by the increase in temperature, and the turbidity of the solution was measured. TBST-Ca, TBS/0.05% Tween containing 5 mm CaCl2. The data are means ± S.E. of two independent experiments. **, p < 0.01; ****, p < 0.0001, calculated by two-way analysis of variance.
Discussion
Although elastic fiber assembly is a critical molecular process responsible for proper function of elastic tissues, it still remains poorly understood. In the present study, we investigated molecular composition of MFAP4 and its interactions with elastic fiber components. We identified MFAP4 as a tropoelastin- and fibrillin-binding protein and pinpointed the specific residues in the MFAP4 structure responsible for its binding capacities. We also showed that MFAP4 facilitates tropoelastin coacervation. Our results revealed new details of elastic fiber assembly process, thus providing a better understanding of elastic fiber biology (Fig. 10).
FIGURE 10.
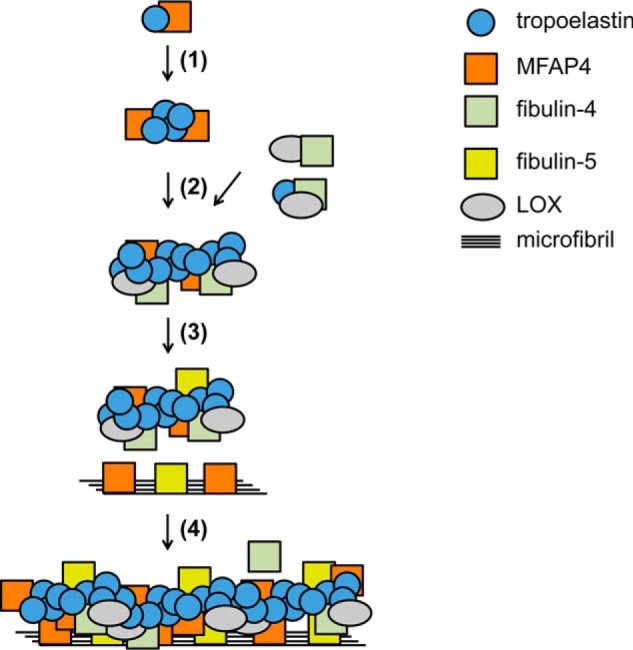
A model describing the proposed role of MFAP4 in elastic fiber assembly. MFAP4 binds to tropoelastin and promotes tropoelastin coacervation (step 1). Tropoelastin is further cross-linked by LOX, recruited by fibulin-4 (step 2). MFAP4 and other proteins such as fibulin-5 interact with fibrillin-1 as well as tropoelastin (step 3) and guide tropoelastin complex deposition onto microfibrils, resulting in a formation of a mature elastic fiber (step 4). Fibulin interactions have been described previously (8).
The multimerization and oligomeric state of FReD-containing proteins is considered to be important to establish a conformation that matches the spatial organization of their ligands, such as branched oligosaccharides, and thereby to increase the binding strength (15, 43). Using chemical cross-linking, it has been suggested that MFAP4 homodimers assemble into higher oligomeric forms of ∼280 kDa, corresponding to an 8-meric structure (25). This has been supported by gel permeation chromatography of rMFAP4, which eluted as a single peak corresponding to a size between 158 and 335 kDa (32). However, our results from gel permeation chromatography and ultracentrifugation suggest that MFAP4 primarily forms 6- and 12-meric structures with molecular masses of 208 and 432 kDa, respectively, together with minor species of 8- and 16-mers. Observed differences might be caused by the choice of column; we used the Superdex 200 with higher resolution than the Superose 6 used in the previous studies. In addition, we have previously observed that in physiologically relevant concentrations in human serum, MFAP4 elutes as a single peak that possibly represents the 6-meric form (32). This suggests that the 6-mer is the functionally important form of MFAP4 and, importantly, remains the main oligomeric form of purified WT rMFAP4. The higher 12-meric oligomer might appear only in concentrated MFAP4 solutions.
We detected specific binding between MFAP4 and tropoelastin, as well as between MFAP4 and both fibrillins. This partially contradicts previous observations of Kasamatsu et al. (26), who suggested interaction between MFAP4 and fibrillin-1 but not tropoelastin. These discrepancies might result from the choice of method and technical conditions, because we showed that although fibrillin binding to MFAP4 is calcium-independent, MFAP4 interactions with tropoelastin require calcium ions. We confirmed our SPR binding data in solid phase binding assays leading to similar results. Moreover, the coacervation-promoting properties of MFAP4 further confirm our findings, identifying MFAP4 as a novel tropoelastin ligand.
We extended the knowledge of the MFAP4-fibrillin-1 interaction by identifying that the MFAP4 binding site resides within five domains on the fibrillin-1 N terminus, containing the second and third EGF-like domains, the first hybrid motif, and the first and second calcium-binding EGF-like domains (Cys119–Ile329). Our interaction studies with fragments spanning full-length fibrillin-2 suggest that in addition to the N-terminal binding site localized to the corresponding region identified in fibrillin-1, an additional MFAP4 binding site is located on the C-terminal half within the region encompassed by the fifth and sixth 8-cysteine domain (Met1735–Asp2171). Fibrillin-1 and fibrillin-2 have both distinct and overlapping biological roles. Fibrillin-2 is mostly produced during embryogenesis, and its expression is turned off shortly after birth, whereas fibrillin-1 production continues throughout adulthood (44). Although both fibrillins can be found in equal amounts in fetal, heteropolymeric microfibrils, fibrillin-2 is largely absent or obscure in most postnatal tissues (41). The presence of an additional MFAP4 binding site on fibrillin-2 suggests that MFAP4 might be important during embryonic development.
GT-8/GT-8 homozygous mice exhibit fragmented microfibrils from around day P7 and die within 2–3 weeks after birth (40). Importantly, we observed that MFAP4 co-localizes with fibrillin-1 in vivo in healthy skin and remains bound to fibrillin-1 also in degenerating elastic fibers. This might indicate a role for MFAP4 in diseases characterized by fibrillin degradation, such as Marfan syndrome. Marfan syndrome is caused by mutations in fibrillin-1 gene, and its most serious manifestation is aortic aneurysm and dissection of the ascending aorta (45). Importantly, MFAP4 protein is up-regulated in ascending aortic samples from patients suffering from Marfan syndrome (46), suggesting that MFAP4 might play an important role in Marfan syndrome pathology and aneurysm formation.
Calcium independence of the MFAP4-fibrillin interaction suggests that fibrillins may interact with MFAP4 at a binding site distinct from the S1 site. Interestingly, the MFAP4 binding site is located around the first hybrid motif in the N-terminal part of fibrillin-1, the region recognized also by MAGP-1 and fibulin-5, known for facilitating tropoelastin-fibrillin interactions (7, 47). Considering the ultrastructural localization of MFAP4 at the elastin-microfibril interface, it may be hypothesized that MFAP4 acts as a bridging molecule that guides tropoelastin deposition onto microfibrils.
FIBCD1 is a FreD protein with the highest homology to MFAP4. Phylogenetically, the FReDs of MFAP4 and FIBCD1 are closely related (48), and the homologous S1 ligand-binding site can be predicted within the FReD of MFAP4. Analysis of the crystal structure of FIBCD1 revealed that the hydrophobic binding pocket of FIBCD1 is created by the aromatic side chains of Tyr405 (MFAP4, Ser203), His415 (MFAP4, His213), Tyr431 (MFAP4, Tyr229), and Trp443 (MFAP4, Phe241) (17). All of these residues were found to be important for FIBCD1 binding to acetylated structures, with slight binding impairment for Trp443 mutant and total inhibition for the remaining three mutants (49).
We chose to mutate two residues in the predicted S1 site of MFAP4 that are not conserved with regards to FIBCD1 sequence, namely Ser203 and Phe241. Variants S203A, S203Y, and F241A oligomerized toward a 6-mer, similarly to WT rMFAP4, whereas variant F241W assembled mainly into a 12-mer. This suggests that the substitution of Phe241 with tryptophan promotes formation of a larger oligomer. We showed that single amino acid change in either of these positions influences MFAP4 binding properties. Overall, MFAP4 interactions were compromised the most by mutations of Phe241. Phe241 is most likely involved in maintaining the hydrophobicity of the binding pocket. However, replacing it with another hydrophobic amino acid (F241W) also resulted in binding deficiency, implying that this substitution might have introduced an unacceptable steric hindrance. Because variant F241W, unlike other investigated mutants, polymerized predominantly toward a 12-mer, it is also possible that the 12-meric form of MFAP4 is relatively inactive compared with the main, 6-meric form. Interestingly, MFAP4 ligand binding was attenuated in S203Y but not S203A variant. This suggests that the hydroxyl group of Ser203 is not essential for binding, but the proper size of the amino acid in this position might ensure the proper conformation of the binding site or maintenance of contacts between other residues involved in the ligand recognition.
To our knowledge, MFAP4 is the first described desmosine-binding molecule. The desmosine cross-link is derived from the condensation of four lysine residues and occurs only in mature elastin (1). The presence of soluble desmosine in body fluids is an indicator of elastic fiber degradation and has been suggested as a biomarker for lung damage (50, 51). Interestingly, we have recently shown that plasma MFAP4 is associated to chronic obstructive pulmonary disease (COPD) severity and may serve as a stable COPD biomarker (52). The interaction between MFAP4 and desmosine seems specific, as shown by lack of competition by collagen-specific deoxypyridinoline cross-links. We speculate that the interaction of MFAP4 and desmosine may contribute to mature elastic fiber homeostasis and stability.
Cross-linking of tropoelastin aggregates is an important step in the development of mature elastic fibers, catalyzed by LOX and LOX-like proteins (LOXL1–4). Because of sequence differences in their pro-peptide regions, functional differences between LOX family members have been suggested (53). Indeed, at least LOXL1 has been shown to play a nonredundant role in elastic fiber homeostasis with ligand specificities distinct from LOX (54). Despite a detectable binding between LOX and MFAP4, our data did not support a role for MFAP4 in recruiting LOX to tropoelastin. However, we cannot exclude the possibility that MFAP4 interacts with LOXLs and thus contributes to cross-link formation.
Tropoelastin-binding proteins can exhibit coacervation-promoting properties, as previously demonstrated for fibulin-5 or fibrillin-1 (55, 56). We showed that MFAP4 not only binds strongly to tropoelastin but also promotes tropoelastin coacervation. Coacervation is believed to be crucial for initial concentration and proper aligning of tropoelastin molecules before cross-linking (4, 57, 58). Importantly, impaired coacervation is a cause of abnormal elastic fiber assembly in the patients with supravalvular aortic stenosis in whom mutant tropoelastin secretion is not impaired (59, 60). Thus, molecular interactions between tropoelastin and MFAP4 might positively affect the alignment and maturation of tropoelastin. Because we confirmed previously suggested interactions between MFAP4 and fibrillin-1 (26), it is also possible that MFAP4 together with fibrillin-1 cooperatively promote coacervation.
MFAP4 has been hypothesized to share functions with another extracellular matrix protein, fibulin-5 (21). Fibulin-5 also contains an RGD sequence, exhibits elastin-binding properties, and is essential for elastogenesis in vivo (61, 62). The functional overlap between MFAP4 and fibulin-5 was also predicted in the recent genome-wide gene expression study, where gene expression of MFAP4 and fibulin-5 in the lung tissue strongly correlated with each other and were both highly up-regulated in patients with COPD (63). Moreover, the overlap between MFAP4 and fibulin-5 binding sites within fibrillin-1 further strengthens the hypothesis that at least some functions of MFAP4 are shared with fibulin-5.
We have recently generated MFAP4-deficient mice and shown that they exhibit attenuated airway smooth muscle responses when subjected to an allergic asthma model (64). We have further investigated the steady state phenotype of MFAP4 deficiency and observed that MFAP4-null mice develop a spontaneous loss of lung function with moderate emphysema-like airspace enlargement, which progresses with age (65). However, the elastin protein content and elastic fiber ultrastructure were unchanged between genotypes, and we have not detected any obvious defects in other elastic organs in naïve MFAP4-deficient mice.4 This suggests that the role of MFAP4 in elastic fiber formation might be redundant in vivo and implies involvement of compensatory mechanisms.
In summary, we have shown that MFAP4 is an important elastic fiber component. Our study provides a novel molecular explanation for an active role of MFAP4 in elastic fiber formation.
Author Contributions
B. P. and A. T. H. conducted most of the experiments, analyzed the results, and wrote the manuscript. A. S. provided assistance throughout the study and participated in experiments shown in Fig. 4. J. B. M. participated in experiments shown in Fig. 3. A. P. W., A. V. Z., S. E. H., and G. S. conducted and analyzed the experiments shown in Fig. 6, and G. S. revised the manuscript. R. W. conducted the experiments shown in Figs. 2 (B–D) and 3B. S. K. M. conducted SPR experiments shown in Figs. 5 and 8. U. H. participated in study conception and supervision. G. L. S. conceived and supervised the study and revised the manuscript. All authors reviewed the paper and approved the final version of the manuscript.
Acknowledgments
We thank technician Ida Tornoe (Institute of Molecular Medicine, Faculty of Health Sciences, University of Southern Denmark, Odense, Denmark) for technical support and Dr. Lynn Sakai for providing the GT-8 mutant mice used for colony establishment; expression plasmids for rF6, rF11, rF20, and rF87; and anti-fibrillin-1 antibody.
This work was supported by the Danish Medical Research Council, Th. Maigaards Benthine Lunds Fond af 1.6.1978, Familien Hede Nielsens Fond, Fonden til Lægevidenskabens Fremme, Deutsche Forschungsgemeinschaft SFB829/Project B12, and Köln Fortune (to G. S.). The authors declare that they have no conflicts of interest with the contents of this article.
B. Pilecki, A. Schlosser, U. Holmskov, and G. L. Sorensen, unpublished observations.
- LOX
- lysyl oxidase
- COPD
- chronic obstructive pulmonary disease
- FReD
- fibrinogen-related domain
- MAGP
- microfibril-associated glycoprotein
- Pn
- postnatal day n
- SPR
- surface plasmon resonance
- TBST
- TBS with 0.05% Tween
- TL5A
- tachylectin 5A.
References
- 1.Wagenseil J. E., and Mecham R. P. (2007) New insights into elastic fiber assembly. Birth Defects Res. C. Embryo Today 81, 229–240 [DOI] [PubMed] [Google Scholar]
- 2.Baldwin A. K., Simpson A., Steer R., Cain S. A., and Kielty C. M. (2013) Elastic fibres in health and disease. Expert Rev. Mol. Med. 15, e8. [DOI] [PubMed] [Google Scholar]
- 3.Vrhovski B., and Weiss A. S. (1998) Biochemistry of tropoelastin. Eur. J. Biochem. 258, 1–18 [DOI] [PubMed] [Google Scholar]
- 4.Yeo G. C., Keeley F. W., and Weiss A. S. (2011) Coacervation of tropoelastin. Adv. Colloid Interface Sci. 167, 94–103 [DOI] [PubMed] [Google Scholar]
- 5.Bedell-Hogan D., Trackman P., Abrams W., Rosenbloom J., and Kagan H. (1993) Oxidation, cross-linking, and insolubilization of recombinant tropoelastin by purified lysyl oxidase. J. Biol. Chem. 268, 10345–10350 [PubMed] [Google Scholar]
- 6.Ramirez F., and Sakai L. Y. (2010) Biogenesis and function of fibrillin assemblies. Cell Tissue Res. 339, 71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rock M. J., Cain S. A., Freeman L. J., Morgan A., Mellody K., Marson A., Shuttleworth C. A., Weiss A. S., and Kielty C. M. (2004) Molecular basis of elastic fiber formation: critical interactions and a tropoelastin-fibrillin-1 cross-link. J. Biol. Chem. 279, 23748–23758 [DOI] [PubMed] [Google Scholar]
- 8.Choudhury R., McGovern A., Ridley C., Cain S. A., Baldwin A., Wang M. C., Guo C., Mironov A. Jr., Drymoussi Z., Trump D., Shuttleworth A., Baldock C., and Kielty C. M. (2009) Differential regulation of elastic fiber formation by fibulin-4 and -5. J. Biol. Chem. 284, 24553–24567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemaire R., Bayle J., Mecham R. P., and Lafyatis R. (2007) Microfibril-associated MAGP-2 stimulates elastic fiber assembly. J. Biol. Chem. 282, 800–808 [DOI] [PubMed] [Google Scholar]
- 10.Horiguchi M., Inoue T., Ohbayashi T., Hirai M., Noda K., Marmorstein L. Y., Yabe D., Takagi K., Akama T. O., Kita T., Kimura T., and Nakamura T. (2009) Fibulin-4 conducts proper elastogenesis via interaction with cross-linking enzyme lysyl oxidase. Proc. Natl. Acad. Sci. U.S.A. 106, 19029–19034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D. Y., Faury G., Taylor D. G., Davis E. C., Boyle W. A., Mecham R. P., Stenzel P., Boak B., and Keating M. T. (1998) Novel arterial pathology in mice and humans hemizygous for elastin. J. Clin. Invest. 102, 1783–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mäki J. M., Räsänen J., Tikkanen H., Sormunen R., Mäkikallio K., Kivirikko K. I., and Soininen R. (2002) Inactivation of the lysyl oxidase gene Lox leads to aortic aneurysms, cardiovascular dysfunction, and perinatal death in mice. Circulation 106, 2503–2509 [DOI] [PubMed] [Google Scholar]
- 13.Carta L., Pereira L., Arteaga-Solis E., Lee-Arteaga S. Y., Lenart B., Starcher B., Merkel C. A., Sukoyan M., Kerkis A., Hazeki N., Keene D. R., Sakai L. Y., and Ramirez F. (2006) Fibrillins 1 and 2 perform partially overlapping functions during aortic development. J. Biol. Chem. 281, 8016–8023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlosser A., Thomsen T., Moeller J. B., Nielsen O., Tornøe I., Mollenhauer J., Moestrup S. K., and Holmskov U. (2009) Characterization of FIBCD1 as an acetyl group-binding receptor that binds chitin. J. Immunol. 183, 3800–3809 [DOI] [PubMed] [Google Scholar]
- 15.Thomsen T., Schlosser A., Holmskov U., and Sorensen G. L. (2011) Ficolins and FIBCD1: soluble and membrane bound pattern recognition molecules with acetyl group selectivity. Mol. Immunol. 48, 369–381 [DOI] [PubMed] [Google Scholar]
- 16.Procopio W. N., Pelavin P. I., Lee W. M., and Yeilding N. M. (1999) Angiopoietin-1 and -2 coiled coil domains mediate distinct homo-oligomerization patterns, but fibrinogen-like domains mediate ligand activity. J. Biol. Chem. 274, 30196–30201 [DOI] [PubMed] [Google Scholar]
- 17.Shrive A. K., Moeller J. B., Burns I., Paterson J. M., Shaw A. J., Schlosser A., Sorensen G. L., Greenhough T. J., and Holmskov U. (2014) Crystal structure of the tetrameric fibrinogen-like recognition domain of fibrinogen C domain containing 1 (FIBCD1) protein. J. Biol. Chem. 289, 2880–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kairies N., Beisel H. G., Fuentes-Prior P., Tsuda R., Muta T., Iwanaga S., Bode W., Huber R., and Kawabata S. (2001) The 2.0-A crystal structure of tachylectin 5A provides evidence for the common origin of the innate immunity and the blood coagulation systems. Proc. Natl. Acad. Sci. U.S.A. 98, 13519–13524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garlatti V., Belloy N., Martin L., Lacroix M., Matsushita M., Endo Y., Fujita T., Fontecilla-Camps J. C., Arlaud G. J., Thielens N. M., and Gaboriaud C. (2007) Structural insights into the innate immune recognition specificities of L- and H-ficolins. EMBO J. 26, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lausen M., Lynch N., Schlosser A., Tornoe I., Saekmose S. G., Teisner B., Willis A. C., Crouch E., Schwaeble W., and Holmskov U. (1999) Microfibril-associated protein 4 is present in lung washings and binds to the collagen region of lung surfactant protein D. J. Biol. Chem. 274, 32234–32240 [DOI] [PubMed] [Google Scholar]
- 21.Toyoshima T., Nishi N., Kusama H., Kobayashi R., and Itano T. (2005) 36-kDa microfibril-associated glycoprotein (MAGP-36) is an elastin-binding protein increased in chick aortae during development and growth. Exp. Cell Res. 307, 224–230 [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi R., Tashima Y., Masuda H., Shozawa T., Numata Y., Miyauchi K., and Hayakawa T. (1989) Isolation and characterization of a new 36-kDa microfibril-associated glycoprotein from porcine aorta. J. Biol. Chem. 264, 17437–17444 [PubMed] [Google Scholar]
- 23.Wulf-Johansson H., Lock Johansson S., Schlosser A., Trommelholt Holm A., Rasmussen L. M., Mickley H., Diederichsen A. C., Munkholm H., Poulsen T. S., Tornøe I., Nielsen V., Marcussen N., Vestbo J., Saekmose S. G., Holmskov U., and Sorensen G. L. (2013) Localization of microfibrillar-associated protein 4 (MFAP4) in human tissues: clinical evaluation of serum MFAP4 and its association with various cardiovascular conditions. PLoS One 8, e82243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toyoshima T., Yamashita K., Furuichi H., Shishibori T., Itano T., and Kobayashi R. (1999) Ultrastructural distribution of 36-kD microfibril-associated glycoprotein (MAGP-36) in human and bovine tissues. J. Histochem. Cytochem. 47, 1049–1056 [DOI] [PubMed] [Google Scholar]
- 25.Schlosser A., Thomsen T., Shipley J. M., Hein P. W., Brasch F., Tornøe I., Nielsen O., Skjødt K., Palaniyar N., Steinhilber W., McCormack F. X., and Holmskov U. (2006) Microfibril-associated protein 4 binds to surfactant protein A (SP-A) and colocalizes with SP-A in the extracellular matrix of the lung. Scand. J. Immunol. 64, 104–116 [DOI] [PubMed] [Google Scholar]
- 26.Kasamatsu S., Hachiya A., Fujimura T., Sriwiriyanont P., Haketa K., Visscher M. O., Kitzmiller W. J., Bello A., Kitahara T., Kobinger G. P., and Takema Y. (2011) Essential role of microfibrillar-associated protein 4 in human cutaneous homeostasis and in its photoprotection. Sci. Rep. 1, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 28.Kiefer F., Arnold K., Künzli M., Bordoli L., and Schwede T. (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwede T., Kopp J., Guex N., and Peitsch M. C. (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guex N., and Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 31.Peitsch M. C., Wells T. N., Stampf D. R., and Sussman J. L. (1995) The Swiss-3DImage collection and PDB-Browser on the World-Wide Web. Trends Biochem. Sci. 20, 82–84 [DOI] [PubMed] [Google Scholar]
- 32.Saekmose S. G., Schlosser A., Holst R., Johansson S. L., Wulf-Johansson H., Tornøe I., Vestbo J., Kyvik K. O., Barington T., Holmskov U., and Sørensen G. L. (2013) Enzyme-linked immunosorbent assay characterization of basal variation and heritability of systemic microfibrillar-associated protein 4. PLoS One 8, e82383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laue T. M., Bhairavi D. S., Ridgeway T. M., and Pelletier S. L. (1992) Analytical Ultracentrifugation in Biochemistry and Polymers Sciences, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 34.Wallis R., and Drickamer K. (1997) Asymmetry adjacent to the collagen-like domain in rat liver mannose-binding protein. Biochem. J. 325, 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girija U. V., Dodds A. W., Roscher S., Reid K. B., and Wallis R. (2007) Localization and characterization of the mannose-binding lectin (MBL)-associated-serine protease-2 binding site in rat ficolin-A: equivalent binding sites within the collagenous domains of MBLs and ficolins. J. Immunol. 179, 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reinhardt D. P., Keene D. R., Corson G. M., Pöschl E., Bächinger H. P., Gambee J. E., and Sakai L. Y. (1996) Fibrillin-1: organization in microfibrils and structural properties. J. Mol. Biol. 258, 104–116 [DOI] [PubMed] [Google Scholar]
- 38.Sengle G., Charbonneau N. L., Ono R. N., Sasaki T., Alvarez J., Keene D. R., Bächinger H. P., and Sakai L. Y. (2008) Targeting of bone morphogenetic protein growth factor complexes to fibrillin. J. Biol. Chem. 283, 13874–13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch M., Veit G., Stricker S., Bhatt P., Kutsch S., Zhou P., Reinders E., Hahn R. A., Song R., Burgeson R. E., Gerecke D. R., Mundlos S., and Gordon M. K. (2006) Expression of type XXIII collagen mRNA and protein. J. Biol. Chem. 281, 21546–21557 [DOI] [PubMed] [Google Scholar]
- 40.Charbonneau N. L., Carlson E. J., Tufa S., Sengle G., Manalo E. C., Carlberg V. M., Ramirez F., Keene D. R., and Sakai L. Y. (2010) In vivo studies of mutant fibrillin-1 microfibrils. J. Biol. Chem. 285, 24943–24955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charbonneau N. L., Dzamba B. J., Ono R. N., Keene D. R., Corson G. M., Reinhardt D. P., and Sakai L. Y. (2003) Fibrillins can co-assemble in fibrils, but fibrillin fibril composition displays cell-specific differences. J. Biol. Chem. 278, 2740–2749 [DOI] [PubMed] [Google Scholar]
- 42.Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanio M., Kondo S., Sugio S., and Kohno T. (2007) Trivalent recognition unit of innate immunity system: crystal structure of trimeric human M-ficolin fibrinogen-like domain. J. Biol. Chem. 282, 3889–3895 [DOI] [PubMed] [Google Scholar]
- 44.Zhang H., Hu W., and Ramirez F. (1995) Developmental expression of fibrillin genes suggests heterogeneity of extracellular microfibrils. J. Cell Biol. 129, 1165–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson P. N., Arteaga-Solis E., Baldock C., Collod-Béroud G., Booms P., De Paepe A., Dietz H. C., Guo G., Handford P. A., Judge D. P., Kielty C. M., Loeys B., Milewicz D. M., Ney A., Ramirez F., Reinhardt D. P., Tiedemann K., Whiteman P., and Godfrey M. (2006) The molecular genetics of Marfan syndrome and related disorders. J. Med. Genet. 43, 769–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pilop C., Aregger F., Gorman R. C., Brunisholz R., Gerrits B., Schaffner T., Gorman J. H. 3rd, Matyas G., Carrel T., and Frey B. M. (2009) Proteomic analysis in aortic media of patients with Marfan syndrome reveals increased activity of calpain 2 in aortic aneurysms. Circulation 120, 983–991 [DOI] [PubMed] [Google Scholar]
- 47.El-Hallous E., Sasaki T., Hubmacher D., Getie M., Tiedemann K., Brinckmann J., Bätge B., Davis E. C., and Reinhardt D. P. (2007) Fibrillin-1 interactions with fibulins depend on the first hybrid domain and provide an adaptor function to tropoelastin. J. Biol. Chem. 282, 8935–8946 [DOI] [PubMed] [Google Scholar]
- 48.Doolittle R. F., McNamara K., and Lin K. (2012) Correlating structure and function during the evolution of fibrinogen-related domains. Protein Sci. 21, 1808–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomsen T., Moeller J. B., Schlosser A., Sorensen G. L., Moestrup S. K., Palaniyar N., Wallis R., Mollenhauer J., and Holmskov U. (2010) The recognition unit of FIBCD1 organizes into a noncovalently linked tetrameric structure and uses a hydrophobic funnel (S1) for acetyl group recognition. J. Biol. Chem. 285, 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma S., Turino G. M., and Lin Y. Y. (2011) Quantitation of desmosine and isodesmosine in urine, plasma, and sputum by LC-MS/MS as biomarkers for elastin degradation. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 879, 1893–1898 [DOI] [PubMed] [Google Scholar]
- 51.Lindberg C. A., Engström G., de Verdier M. G., Nihlén U., Anderson M., Forsman-Semb K., and Svartengren M. (2012) Total desmosines in plasma and urine correlate with lung function. Eur. Respir. J. 39, 839–845 [DOI] [PubMed] [Google Scholar]
- 52.Johansson S. L., Roberts N. B., Schlosser A., Andersen C. B., Carlsen J., Wulf-Johansson H., Saekmose S. G., Titlestad I. L., Tornoe I., Miller B., Tal-Singer R., Holmskov U., Vestbo J., and Sorensen G. L. (2014) Microfibrillar-associated protein 4: a potential biomarker of chronic obstructive pulmonary disease. Respir. Med. 108, 1336–1344 [DOI] [PubMed] [Google Scholar]
- 53.Thomassin L., Werneck C. C., Broekelmann T. J., Gleyzal C., Hornstra I. K., Mecham R. P., and Sommer P. (2005) The Pro-regions of lysyl oxidase and lysyl oxidase-like 1 are required for deposition onto elastic fibers. J. Biol. Chem. 280, 42848–42855 [DOI] [PubMed] [Google Scholar]
- 54.Liu X., Zhao Y., Gao J., Pawlyk B., Starcher B., Spencer J. A., Yanagisawa H., Zuo J., and Li T. (2004) Elastic fiber homeostasis requires lysyl oxidase-like 1 protein. Nat. Genet. 36, 178–182 [DOI] [PubMed] [Google Scholar]
- 55.Hirai M., Ohbayashi T., Horiguchi M., Okawa K., Hagiwara A., Chien K. R., Kita T., and Nakamura T. (2007) Fibulin-5/DANCE has an elastogenic organizer activity that is abrogated by proteolytic cleavage in vivo. J. Cell Biol. 176, 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clarke A. W., Wise S. G., Cain S. A., Kielty C. M., and Weiss A. S. (2005) Coacervation is promoted by molecular interactions between the PF2 segment of fibrillin-1 and the domain 4 region of tropoelastin. Biochemistry 44, 10271–10281 [DOI] [PubMed] [Google Scholar]
- 57.Bellingham C. M., Lillie M. A., Gosline J. M., Wright G. M., Starcher B. C., Bailey A. J., Woodhouse K. A., and Keeley F. W. (2003) Recombinant human elastin polypeptides self-assemble into biomaterials with elastin-like properties. Biopolymers 70, 445–455 [DOI] [PubMed] [Google Scholar]
- 58.Narayanan A. S., Page R. C., Kuzan F., and Cooper C. G. (1978) Elastin cross-linking in vitro: studies on factors influencing the formation of desmosines by lysyl oxidase action on tropoelastin. Biochem. J. 173, 857–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wachi H., Sato F., Nakazawa J., Nonaka R., Szabo Z., Urban Z., Yasunaga T., Maeda I., Okamoto K., Starcher B. C., Li D. Y., Mecham R. P., and Seyama Y. (2007) Domains 16 and 17 of tropoelastin in elastic fibre formation. Biochem. J. 402, 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu W. J., and Weiss A. S. (1999) Deficient coacervation of two forms of human tropoelastin associated with supravalvular aortic stenosis. Eur. J. Biochem. 266, 308–314 [DOI] [PubMed] [Google Scholar]
- 61.Nakamura T., Lozano P. R., Ikeda Y., Iwanaga Y., Hinek A., Minamisawa S., Cheng C. F., Kobuke K., Dalton N., Takada Y., Tashiro K., Ross J. Jr., Honjo T., and Chien K. R. (2002) Fibulin-5/DANCE is essential for elastogenesis in vivo. Nature 415, 171–175 [DOI] [PubMed] [Google Scholar]
- 62.Yanagisawa H., Davis E. C., Starcher B. C., Ouchi T., Yanagisawa M., Richardson J. A., and Olson E. N. (2002) Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature 415, 168–171 [DOI] [PubMed] [Google Scholar]
- 63.Brandsma C. A., van den Berge M., Postma D. S., Jonker M. R., Brouwer S., Paré P. D., Sin D. D., Bossé Y., Laviolette M., Karjalainen J., Fehrmann R. S., Nickle D. C., Hao K., Spanjer A. I., Timens W., and Franke L. (2015) A large lung gene expression study identifying fibulin-5 as a novel player in tissue repair in COPD. Thorax 70, 21–32 [DOI] [PubMed] [Google Scholar]
- 64.Pilecki B., Schlosser A., Wulf-Johansson H., Trian T., Moeller J. B., Marcussen N., Aguilar-Pimentel J. A., de Angelis M. H., Vestbo J., Berger P., Holmskov U., and Sorensen G. L. (2015) Microfibrillar-associated protein 4 modulates airway smooth muscle cell phenotype in experimental asthma. Thorax 70, 862–872 [DOI] [PubMed] [Google Scholar]
- 65.Holm A. T., Wulf-Johansson H., Hvidsten S., Jorgensen P. T., Schlosser A., Pilecki B., Ormhøj M., Moeller J. B., Johannsen C., Baun C., Andersen T., Schneider J. P., Hegermann J., Ochs M., Götz A. A., Schulz H., de Angelis M. H., Vestbo J., Holmskov U., and Sorensen G. L. (2015) Characterization of spontaneous air space enlargement in mice lacking microfibrillar-associated protein 4. Am. J. Physiol. Lung. Cell Mol. Physiol. 308, L1114–L1124 [DOI] [PubMed] [Google Scholar]