Abstract
Amyloid propagation requires high levels of sequence specificity so that only molecules with very high sequence identity can form cross-β-sheet structures of sufficient stringency for incorporation into the amyloid fibril. This sequence specificity presents a barrier to the transmission of prions between two species with divergent sequences, termed a species barrier. Here we study the relative effects of protein sequence, seed conformation, and environment on the species barrier strength and specificity for the yeast prion protein Sup35p from three closely related species of the Saccharomyces sensu stricto group; namely, Saccharomyces cerevisiae, Saccharomyces bayanus, and Saccharomyces paradoxus. Through in vivo plasmid shuffle experiments, we show that the major characteristics of the transmission barrier and conformational fidelity are determined by the protein sequence rather than by the cellular environment. In vitro data confirm that the kinetics and structural preferences of aggregation of the S. paradoxus and S. bayanus proteins are influenced by anions in accordance with their positions in the Hofmeister series, as observed previously for S. cerevisiae. However, the specificity of the species barrier is primarily affected by the sequence and the type of anion present during the formation of the initial seed, whereas anions present during the seeded aggregation process typically influence kinetics rather than the specificity of prion conversion. Therefore, our work shows that the protein sequence and the conformation variant (strain) of the prion seed are the primary determinants of cross-species prion specificity both in vivo and in vitro.
Keywords: amyloid, prion, protein conformation, protein evolution, yeast, Hofmeister series, species barrier
Introduction
Amyloidogenic proteins form ordered self-seeding fibrous aggregates that are known to be associated with a variety of neurodegenerative diseases in humans and other mammals, such as Alzheimer disease, Parkinson disease, and prion diseases, including sheep scrapie, mad cow disease, elk and deer chronic wasting disease, and human Creutzfeldt-Jakob disease, kuru, and fatal familial insomnia (1–5). The amyloid form of an amyloidogenic protein can convert the cellular form of a protein of the same or very similar amino acid sequence into an amyloid conformation, usually via cross-β interactions (6–9). Transmissible amyloids called prions can spread the amyloid state between organisms. Cross-species transmission of the prion state is impaired by sequence divergence within the prion proteins, resulting in a “species barrier” to transmission of a prion from one species to another (10). However, the species barrier can be overcome in some species combinations. For example, bovine spongiform encephalopathy, which possibly originated from transmission of a scrapie prion from sheep to cattle, resulted in the widely known “mad cow” disease epidemic that greatly affected the United Kingdom in the 1990s (11, 12). Bovine spongiform encephalopathy has also been found to be transmitted from cattle to humans, manifesting itself as a new variant of Creutzfeldt-Jakob disease called variant Creutzfeldt-Jakob disease (13–16). Therefore, understanding the mechanisms of species barrier and cross-species prion transmission is crucial for preventing future outbreaks of prion diseases. However, the rules governing species barriers as well as the effects of the physiological and environmental conditions on the barrier are poorly understood to date.
Yeast prions are cytoplasmic elements heritable in a non-Mendelian fashion (for a review, see Refs. 17, 18). Because many of them control phenotypically detectable traits, yeast prions provide a useful model for studying the molecular basis of prion phenomena. One of the best studied yeast prion proteins is translation termination factor Sup35. Prion formation by Sup35 causes translational readthrough (nonsense suppression), a phenotypically detectable trait in specifically designed yeast strains (17, 18). Previously, we (19, 20) and others (21) have reported observations of the species barrier and cross-species prion transmission between Sup35 proteins of three closely related yeast species, Saccharomyces cerevisiae, Saccharomyces paradoxus, and Saccharomyces bayanus, containing Sup35p with high levels of sequence similarity.
The Sup35 protein can be divided into three segments: the N-terminal prion domain (Sup35N); the linker middle domain (Sup35M); and the functional C-terminal domain (Sup35C), responsible for translation termination and cell viability. Levels of similarity between the Sup35N fragments of the Sup35 proteins from Saccharomyces sensu stricto are close to those observed in mammalian prion proteins (22, 23), which renders this system a useful model for studying the species barrier. The Sup35N domain alone is difficult to express because it is poorly soluble and aggregates too rapidly, whereas the addition of the Sup35M domain resolves all of the abovementioned issues. Therefore, Sup35NM is used widely as a model protein for studying amyloid aggregation in vitro and has been shown to transmit prion properties to full-size Sup35 after transfection into yeast cells (24, 25). The amino acid similarities between the Sup35NM regions of the three different species are 95% and 93% (S. cerevisiae/S. paradoxus combination), 85% and 78% (S. cerevisiae/S. bayanus combination), and 85% and 79% (S. paradoxus/S. bayanus combination) (19, 20). Our previous experiments, performed with divergent Sup35 proteins (or proteins containing divergent or chimeric Sup35N domains) in S. cerevisiae cells, have demonstrated that the prion species barrier depends on (but is not arithmetically proportional to) the level of divergence of Sup35N sequences and that different subregions (modules) of Sup35N play a primary role in determining the barrier in different cross-species combinations (19, 20, 26). A prion species barrier has also been observed with divergent Sup35NM fragments in vitro (19).
Prion proteins, including mammalian PrP and yeast Sup35, not only can fold into alternative prion and non-prion forms but can also adopt multiple distinct amyloid conformations, known as “strains” or “variants” (17, 27–33). Different strains are associated with distinct disease patterns in mammals and different stringencies of phenotypic effects in fungi (because of this, yeast prion strains can be termed “strong,” “intermediate,” “weak,” etc.). When formed, the prion strain is typically reproduced faithfully, although strain “mutations” may occur with a low frequency (34–38). In both mammals and yeast, prion strain properties influence the species barrier (26, 39). For example, transmission of the specific strong prion strain from the S. cerevisiae to S. paradoxus Sup35 prion domain (PrD)7 in S. cerevisiae cells is relatively efficient, whereas transmission of the specific weak prion strain in the same direction is rare (20). Notably, underlying S. cerevisiae prion strain patterns were maintained during propagation through the S. paradoxus protein but altered irreversibly during propagation through the S. bayanus protein, which could be detected as a “switch” of prion strain after reverse transmission back to the S. cerevisiae sequence (20). This shows that not only the efficiency of prion transmission but also the fidelity of reproduction of prion conformation during transmission is controlled by the level of identity of interacting protein sequences.
Previous work from our group has also demonstrated that the ion type present in solution greatly influences not only processes such as deactivation of enzymes (40–42) but also in vitro amyloid formation by S. cerevisiae Sup35NM and that ion effects are determined by their position in the Hofmeister series in the fashion of an “inverse” Hofmeister effect. Specifically, strongly hydrated anions (kosmotropes) initiate nucleation quickly and promote rapid fiber elongation, whereas poorly hydrated anions (chaotropes) delay nucleation and mildly affect the elongation rate (43, 44). A similar effect of kosmotropes has also been observed by another group for the mammalian prion protein PrP (45). Moreover, amyloid formation by Sup35NM in the presence of different anions resulted in the generation of different spectra of prion strains, with kosmotropes favoring the formation of strong strains (characterized by smaller aggregate size and higher efficiency of fragmentation and proliferation), and chaotropes favoring the formation of weak strains (characterized by larger aggregate size and lower efficiency of fragmentation and proliferation) (43, 44).
In this work, we specifically address the contribution of the cellular composition and the conditions of the aggregation reaction to the specificity of prion transmission. The results of the in vivo experiments are compared in cells of different yeast species to determine whether species-specific patterns of intracellular environment influence the parameters of the prion species barrier. Further, in vitro aggregation experiments are performed to determine whether anions of the Hofmeister series influence cross-species specificity of prion transmission. Our results are consistent with the notion that protein sequence and conformation remain the primary determinants of cell specificity, whereas environmental conditions influence specificity primarily via favoring the formation of different prion strains.
Experimental Procedures
Yeast Strains and Plasmids
Saccharomyces cerevisiae strains
The S. cerevisiae strain GT17 (MATa ade1–14 his3 leu2 trp1 ura3 [psi− pin−]) was used for transfection of in vitro generated aggregates (19, 44). The strain GT256-23C (MATα ade1–14 his3Δ (or 11,15) lys2 leu2–3,112 trp1 ura3–52 sup35::HIS3 [PSI+ PIN+] [CEN LEU2 SUP35Sc]) was used for all shuffle experiments performed in the S. cerevisiae species. This strain harbors a strong prion variant. The S. cerevisiae strains GT797 (MATa ade1–14 his3Δ (or 11,15) lys2 ura3–52 leu2–3,112 trp1 sup35::HIS3 [CEN URA3 SUP35Sp] [psi− pin−]) or GT987 (MATa ade1–14 his3Δ (or 11,15) lys2 ura3–52 leu2–3,112 trp1 sup35::HIS3 [URA3 SUP35Sb] [psi− pin−]), bearing, respectively, the SUP35 genes from S. paradoxus or S. bayanus, were used as recipients for transfection with in vitro generated S. paradoxus or S. bayanus Sup35NM amyloids.
Plasmids
Centromeric (low-copy) vectors having URA3, LEU2, or LYS2 markers and bearing the divergent or chimeric SUP35 genes under the control of the endogenous S. cerevisiae SUP35 promoter (PSUP35) were employed in the plasmid shuffle experiments. Construction of the chimeric SUP35 derivatives has been described previously (19, 20). Plasmids pFA6a-kanMX6 (46), pBluescript-URA3 I (constructed by J. Kumar in the Chernoff laboratory), pRS303N (47), and pRS303H (47) were used as a source of marker genes to be introduced into S. paradoxus (see below). Plasmid pRS316 was used in the S. paradoxus ade1–14Sc construction (as described below) for co-transformation with the PCR product.
S. paradoxus Strain Construction
Natural strains of S. paradoxus are non-mating homothallic diploids lacking genetic markers that can be used for transformation and prion detection. To perform prion studies in S. paradoxus, we constructed genetically marked haploid heterothallic S. paradoxus strains bearing the ade1–14 (UGA) reporter (see above), which allows for [PSI+] detection. This construction included a multistep process, outlined below.
Generation of lys2 and ura3-P2 Mutations
The lys2 and ura3 auxotrophic mutations were introduced into the homothallic diploid S. paradoxus strain SP7-1D (provided by G. Naumov) by irradiation with UV light for 15–45 s and selecting mutants on α-aminoadipic- or 5-fluoroorotic acid-containing medium, respectively. Mutant alleles with the lowest reversion frequencies were chosen for further work. Diploid strains containing each allele were sporulated, and the resulting haploid spores were mated to generate double-heterozygous diploids. These diploids were sporulated and dissected, and spore clones unable to grow on both the medium lacking uracil (-Ura) and medium lacking lysine (-Lys) were identified. The respective mutant strains were sporulated, and spores were mated to obtain diploids heterozygous by both lys2 and ura3 alleles. These diploids were, in turn, sporulated to obtain homozygous double lys2 ura3 mutant strains.
Generation of Heterothallic S. paradoxus Strains
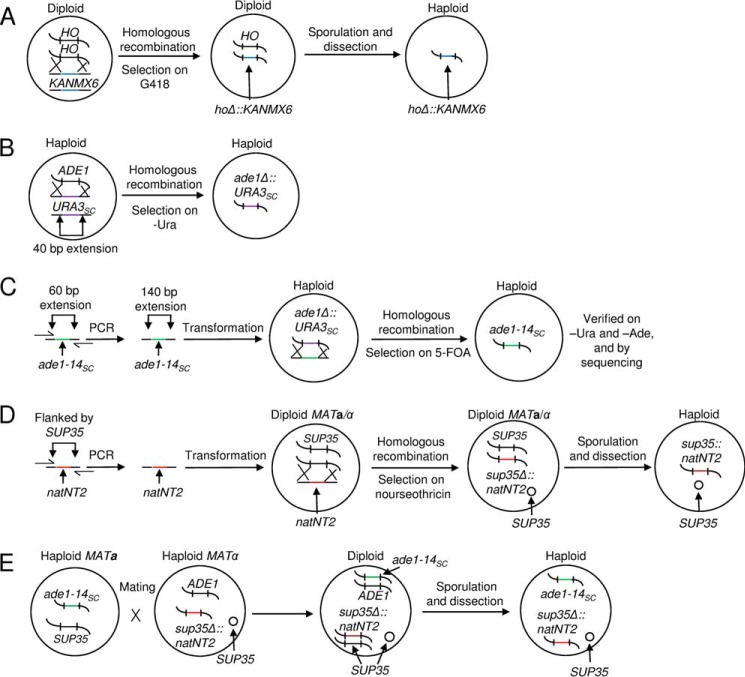
The initially homothallic lys2 ura3 S. paradoxus strain GT749-1B, generated as described previously, was used to produce heterothallic strains via replacement of the HO gene with the bacterial kanMX6 gene (Fig. 1A). For this purpose, the kanMX6 gene was PCR-amplified from plasmid pFA6a-kanMX6 (46), with primers having 50-bp extensions homologous to flanking regions of the S. paradoxus HO gene on both sides. This PCR fragment was used to transform the GT749-1B S. paradoxus strain. Replacement of HO by kanMX6 as a result of homologous recombination conveyed G418 resistance to the yeast cells and was verified by PCR. The resulting heterozygous HO/ho::KANMX6 strain was sporulated and dissected to produce heterothallic haploid ho::KANMX6 lys2 ura3 strains of both mating types.
FIGURE 1.
The steps in construction of the S. paradoxus strains. A, the HO gene was disrupted by replacement with the bacterial KANMX6 gene. B, the ADE1 gene in S. paradoxus was disrupted by the URA3 gene from S. cerevisiae (URA3Sc). C, the ade1–14 and ura3 mutations were engineered by replacing URA3Sc (previously inserted to disrupt ADE1) with ade1–14Sc. D, the SUP35 gene in a S. paradoxus chromosome was replaced by natNT2 (conveying resistance to nourseothricin). E, an S. paradoxus strain having both ade1–14 and sup35Δ::natNT2 in its genotype was obtained by crossing two S. paradoxus yeast strains (one containing ade1–14, and another containing sup35Δ::natNT2 and a wild-type copy of SUP35 on a plasmid) of the opposite mating type. This was followed by sporulation and dissection.
Generation of the ade1–14Sc Allele in S. paradoxus
Next, the endogenous S. paradoxus wild-type ADE1 allele was replaced by the S. cerevisiae ade1–14 (UGA) mutant allele (ade1–14Sc). This enabled us to detect the presence of [PSI+] by read-through of ade1–14 (17) and compare the [PSI+] strains obtained in S. cerevisiae and S. paradoxus by using one and the same reporter. This construction was performed in two steps: replacement of endogenous S. paradoxus ADE1 by the S. cerevisiae URA3 gene (Fig. 1B) followed by replacement of URA3 by the S. cerevisiae ade1–14 allele (Fig. 1C). For the initial ADE1 replacement with URA3, the S. cerevisiae URA3 gene was PCR-amplified from the pBluescript-URA3 I plasmid (see above) using primers with 40-bp 5′ extensions homologous to the flanks of S. paradoxus ADE1 and transformed into a haploid S. paradoxus ho::KANMX6 lys2 ura3 strain constructed as described above. The transformants with ade1Δ::URA3 replacement were selected on -Ura medium and confirmed to become Ade−. For the subsequent replacement of URA3 with ade1–14Sc, the ade1–14Sc allele was PCR-amplified from the S. cerevisiae genome using primers with 60-bp 5′ extensions homologous to the flanking regions of previously inserted URA3 in the S. paradoxus genome, followed by a second round of PCR aimed at further increasing the length of the homologous flanking regions by an additional 80 bp on each side for more efficient homologous recombination. The resulting PCR product containing 140 bp flanking extensions was co-transformed together with the LYS2 plasmid pRS317 (used to enrich the sample by transformants versus revertants) into a haploid ade1Δ::URA3 lys2 ura3 S. paradoxus strain constructed as described above. Transformants that were selected on -Lys medium with 5-fluoroorotic acid media and subsequently tested on -Ade and -Ura media. Ura- Ade- transformants were cured of the LYS2 plasmid and verified by PCR amplification and sequencing of the ade1–14Sc allele.
Generation of the S. paradoxus Strains with SUP35 Genes of Different Origins
For this purpose, the chromosomal SUP35 allele of the S. paradoxus strain was replaced by natNT2, conveying nourseothricin resistance, and PCR amplified from plasmid pRS303N (47) by using primers with 40-bp 5′ extensions homologous to flanking regions of the S. paradoxus SUP35 gene (Fig. 1D). The PCR fragment was transformed into a diploid HO/ho::kanMX6 lys2 ura3 S. paradoxus strain homozygous for wild-type SUP35 and incorporated by homologous recombination in place of one of the alleles. The diploid also contained an additional copy of SUP35 on a centromeric plasmid. Transplacement was followed by sporulation and dissection to obtain haploid sup35Δ::natNT2 spores kept alive by the SUP35 plasmid. Such a spore clone was mated to the ade1–14Sc lys2 ura3 strain constructed as described earlier, followed by sporulation and dissection (Fig. 1E). The resulting haploid strain contained the only functional SUP35 copy on a plasmid. Therefore, derivatives with the SUP35 genes of various origins could be generated by introducing the plasmid with the respective SUP35 gene and subsequently losing the original SUP35 plasmid (this procedure is known as plasmid shuffle and is described in more detail below). As the final result of these manipulations, the S. paradoxus GT1320-36A strain containing the S. cerevisiae SUP35 gene was produced with the following genotype: MATa ade1Δ::ade1–14Sc (UGA) lys2 ura3-P2 Δsup35::natN2 ho::kanMX6 [SUP35Sc LYS2] [psi− pin−]. To generate a [PSI+] version of this strain, it was transfected with the cell extracts of S. cerevisiae strain GT256-23C as described below.
Yeast Cultivation and Genetic Techniques
Cultivation
Standard yeast media and growth conditions were employed (48). Yeast cultures were incubated at 30 °C. Standard procedures (49) were used for [PSI+] detection, characterization, and curing by guanidine hydrochloride. All strains have an ade1–14 nonsense mutation in the ADE1 gene that contains a premature UGA stop codon, allowing detection of [psi−] and [PSI+] through the read-through or nonsense suppression assay (49).
Yeast Transformation and Transfection
Standard techniques for yeast transformation were employed (48, 49). For transfection with yeast extracts, cells of the [PSI+] donor strain were disrupted via a standard glass bead lysis procedure, and the resulting lysate was transfected into the spheroplasts of the [psi−] S. paradoxus strain GT1320-36A using a modified protocol described by Rubin et al. (44) and originally adapted from Tanaka et al. (24). To prepare spheroplasts, the cell wall of a [psi−] recipient cell was fragmented with zymolase. An empty URA3 vector was co-transfected into the recipient cells as an indicator that material had passed across the cell membrane (Fig. 2A). Transfectants were obtained on the medium lacking both uracil and tryptophane, and counterselecting against the Trp− donor strain (-Ura-Trp) to avoid contamination by donor cells. Both small and large colonies were observed on -Ura-Trp. Only large colonies contained the URA3 plasmid, whereas smaller colonies (without the plasmid) resulted from a background growth, possibly because of a low concentration of YPD present in the transfectant selection medium. The large Ura+ colonies were tested on the medium lacking adenine (-Ade) to check for [PSI+] (Fig. 2B).
FIGURE 2.
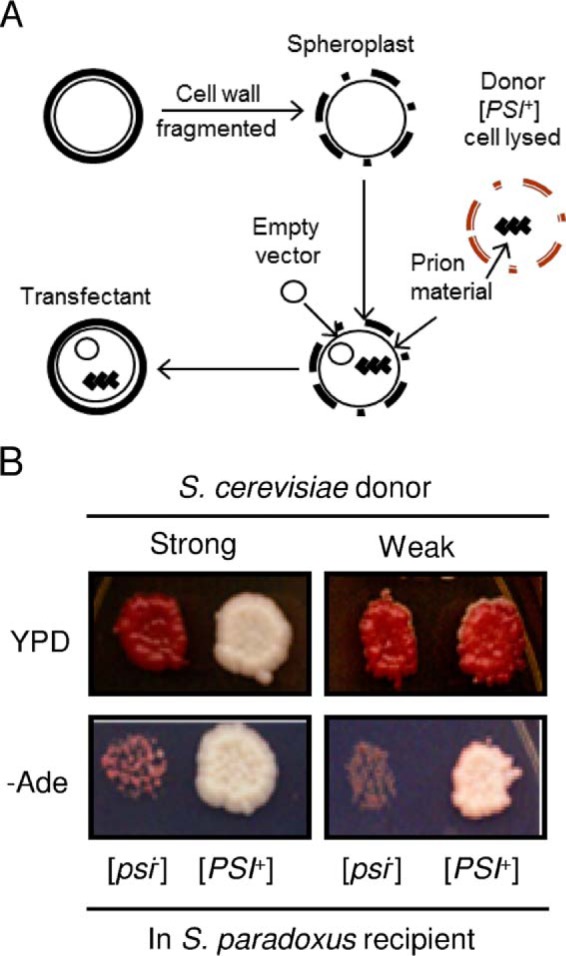
Cross-species prion transfection. A, summary of the transfection procedure. B, cellular extract was transfected from either a strong or weak [PSI+] S. cerevisiae strain into a [psi−] S. paradoxus strain, expressing the Sup35 protein from S. cerevisiae. Representative [PSI+] S. paradoxus transfectants obtained from either the strong (left) or weak (right) [PSI+] donor strains are shown. Yeast cells were grown on YPD (for the color assay) and on -Ade medium (for the suppression assay) at 30 °C for 8 days.
Transfection of in vitro generated aggregates into S. cerevisiae strains GT797 and GT987 was performed as described previously (44), with optional sonication to increase the frequency of transfection and using an empty plasmid with the LYS2 marker for selection of transfectants.
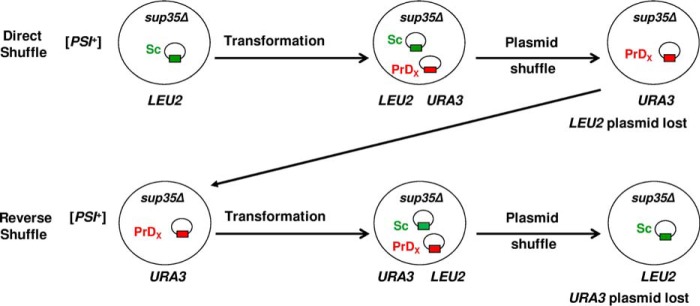
Direct and Reverse Shuffle Procedures
The SUP35 genes of various origins were exchanged in the S. paradoxus strain, constructed as described above, by using a plasmid shuffle procedure (Fig. 3) performed as described previously for S. cerevisiae (19, 20), except that LYS2 and URA3 markers were used on plasmids instead of LEU2 and URA3. For direct shuffle, cells containing a SUP35Sc LYS2 plasmid were transformed with a plasmid containing the SUP35 gene of the same or different origin and an URA3 marker, followed by loss of the original LYS2 plasmid. From each individual [PSI+] transformant, only a single Ura+ Lys− colony was analyzed. To perform a reverse shuffle, cells obtained from the direct shuffle and containing the SUP35Sp URA3 plasmid were transformed with the SUP35Sc LYS2 plasmid and cured of the original URA3 plasmid.
FIGURE 3.
Plasmid shuffle schematic. Shown is a schematic of direct and reverse plasmid shuffle. Sc, SUP35 from S. cerevisiae; PrDX, SUP35 genes of various origins or chimeric constructs.
In Vitro Techniques
Expression and Purification of Sup35NM
Plasmid constructs containing the SUP35NM coding regions of different origins with the attached C-terminal His6 tags were generated as described previously (50) and expressed in Escherichia coli host strain HMS174 (DE3) pLysS (Novagen) to produce the Sup35NM-(His6) proteins. The cell pellets were stored at −80 °C until purification. Purification of Sup35NM proteins from E. coli was carried out as described previously (51). The purified protein was precipitated using methanol at −20 °C and stored at −80 °C in 80% methanol.
Kinetic Assays Using Thioflavin T
The protein pellet stored at −80 °C was collected by centrifugation. The supernatant was discarded, and the protein was resuspended in 8 m urea. Sup35NM was then concentrated by a 10-kDa centrifugal filter and diluted 100-fold with PBS (pH 7.4). The samples were boiled for about 10 min before starting the aggregation experiments to break down any preformed aggregates. 1 mm thioflavin T (Sigma-Aldrich) solution was prepared fresh in PBS. Aggregation experiments were conducted in triplicates in a 96-well plate with final thioflavin T and Sup35NM concentrations of 100 and 10 μm, respectively, in the presence of 0.4 m sodium salt. The seeded experiments contained 5% by volume of sonicated preformed amyloid. Polymerization was carried out at 25 °C in a 96-well plate by shaking it linearly at 17 Hz in a Synergy H4 hybrid multimode microplate reader (BioTek, Winooski, VT). Fluorescence readings were taken every minute for about 13 h using an excitation wavelength of 440 nm and emission wavelength of 480 nm.
Preparation of Amyloid Seeds
The respective sodium salt was added to Sup35NM, purified, and prepared as described above to a final concentration of 0.5 m salt and 10 μm protein in a microcentrifuge tube. The samples were allowed to rotate at 20 rpm at room temperature for 2 days. After polymerization, the amyloid samples were stored at −80 °C until use.
Results
[PSI+] Transfection into Saccharomyces paradoxus Cells
To transfer a S. cerevisiae prion into the cell of a different Saccharomyces species, we transfected the S. paradoxus strain bearing the SUP35Sc gene that was generated as described above with the extracts of [PSI+] S. cerevisiae cells (Fig. 2A). The Ade+ colonies were identified among transfectants and shown to be curable of the Ade+ phenotype by guanidine hydrochloride, an agent eliminating yeast prions. Therefore, we showed that the prion formed in the S. cerevisiae donor species can be transfected into a different recipient species, S. paradoxus. Moreover, the major properties of specific prion variants were maintained independently of the cell environment (Fig. 2B). For example, transfection with the cellular extract from the strong [PSI+] S. cerevisiae strain GT256-23C generated strong [PSI+] isolates in S. paradoxus, as judged from growth on -Ade medium, color on YPD medium (Fig. 2B), and an almost complete lack of spontaneous [PSI+] loss in mitotic divisions (only 3 of 1624 colonies obtained from 5 independent cultures lost [PSI+]). In contrast, transfection with the cellular extract from the weak [PSI+] S. cerevisiae strain GT988-1A generated weak [PSI+] isolates in S. paradoxus (Fig. 2B). Therefore, we were able to use S. paradoxus cells containing the same strong [PSI+] strain that had been studies previously in the S. cerevisiae cell environment for our shuffle experiments, which allowed a direct assessment of the effect of the cell environment on the species barrier.
Comparison of Cross-species Prion Transmission in S. cerevisiae and S. paradoxus Cells
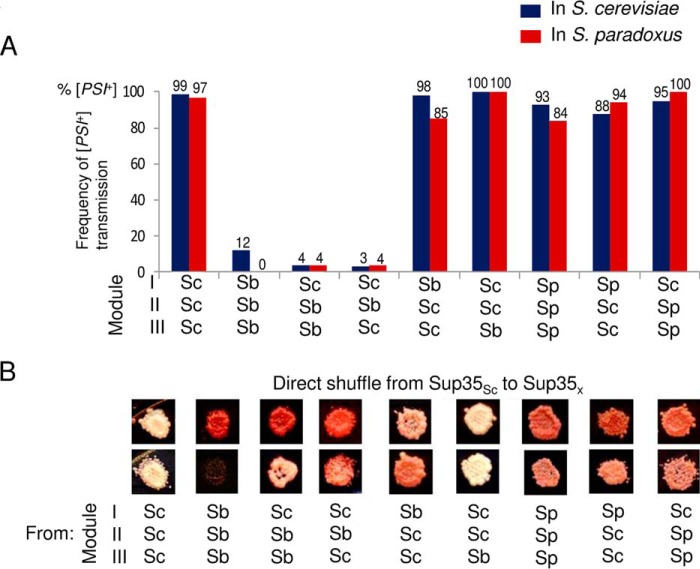
To study the extent of influence of the cell environment on the species barrier, cross-species transmission of a strong Sup35Sc [PSI+] strain to Sup35 proteins with a prion domain (PrDs) of different origin or with a chimeric PrD was measured in S. paradoxus cells by using a plasmid shuffle procedure (Fig. 3) and compared with previous results obtained in S. cerevisiae cells (20). Constructs with chimeric Sup35 PrDs were produced by reshuffling modules I (positions 1–33 in S. cerevisiae nomenclature), II (positions 34–96), and III (positions 97–123) in all possible combinations, as described earlier (20). Modules I and II roughly correspond to the QN-rich stretch and region of oligopeptide repeats, respectively. These are subregions of Sup35 PrD that have been shown previously to influence the specificity and fidelity of cross-species prion transmission (20). The results and comparison with the previous data are shown in Fig. 4A. Overall, similar patterns of the species barrier were observed in both S. cerevisiae and S. paradoxus cells, although some numerical differences were found. In both cases, the strongest barrier was detected between the S. cerevisiae and S. bayanus PrDs (in the S. paradoxus cells, no transmission was detected for this combination at all). By contrast, there was only a slight decrease in prion transmission from Sup35Sc to proteins with the S. paradoxus PrD or its QN-rich stretch (module I). Constructs with chimeric PrDs described previously (20) were used to determine the roles of various regions of Sup35 in the species barrier. Similar to the experiments in S. cerevisiae cells, the region of oligopeptide repeats (module II) was a primary determinant of the species specificity of prion conversion in the combination of S. cerevisiae and S. bayanus. Notably, transmission of the prion state from Sup35Sc to any protein with the QN-rich stretch (module I) and/or the region of oligopeptide repeats (module II) coming from a species other than S. cerevisiae resulted in the appearance of a phenotypically weaker prion (Fig. 4B), as observed previously in S. cerevisiae (20).
FIGURE 4.
Comparison of direct cross-species prion transmission in the S. cerevisiae and S. paradoxus cell environments. A, frequency of [PSI+] transmission from S. cerevisiae Sup35 proteins to proteins with divergent or chimeric prion domains (as indicated). The data obtained from the S. cerevisiae cell environment (blue) have been published previously by Chen et al. (20) I, II, and III refer to the exchangeable modules of the PrD. B, phenotypes of [PSI+] produced from direct transmission in S. paradoxus. -Ade and YPD plates were photographed after 8 days.
In further experiments, prions generated by transmission from Sup35Sc to proteins with divergent or chimeric PrDs were transmitted back to Sup35Sc by using reverse shuffle (Fig. 5). In most cases, the results followed the same pattern as detected previously in S. cerevisiae (20). As in S. cerevisiae, reverse transmission of the prion state to Sup35Sc from the constructs containing module II of S. bayanus was high in S. paradoxus, confirming the asymmetry of the species barrier in the S. cerevisiae/S. bayanus combination. On the other hand, although proteins containing the S. paradoxus PrD or at least its module I efficiently transmitted the [PSI+] state to Sup35Sc, a noticeable decrease was detected in S. paradoxus compared with S. cerevisiae cells.
FIGURE 5.
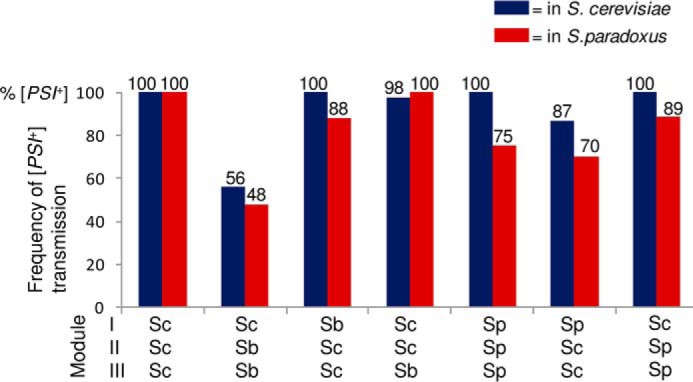
Comparison of reverse cross-species prion transmission in S. cerevisiae and S. paradoxus. The frequencies of [PSI+] transmission from the divergent or chimeric prion domains (as indicated) to the S. cerevisiae protein are shown. Notably, the reverse shuffle constructs containing the S. paradoxus PrD or its module I restored the original strong [PSI+] variant. In contrast, reverse shuffle from constructs containing module II of S. bayanus produced weak [PSI+] variants. I, II, and III refer to the exchangeable modules of the PrD.
For most combinations, the strong [PSI+] phenotype was restored after reverse transmission to Sup35Sc, confirming that, to the extent detectable by the resolution of our approach, prion weakening during its propagation by a divergent or chimeric protein was due to changes in its phenotypic expression rather than in the molecular “properties” of the prion strain. However, the Sup35Sc prion strain became weak after the reverse transmission from the protein containing at least module II from S. bayanus (Fig. 6). Therefore, the underlying properties of the prion variant were changed irreversibly in this combination, again confirming previous observations made in S. cerevisiae cells (20).
FIGURE 6.
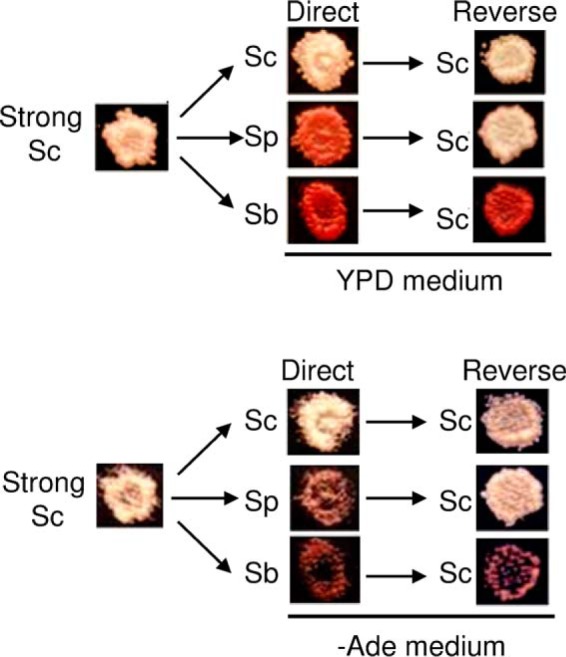
Reproduction and switch of prion variants in cross-species transmission in the S. paradoxus cell environment on reverse shuffle. Shown are representatives of the direct shuffle experiment where [PSI+] isolates obtained from a strong S. cerevisiae prion variant in the S. paradoxus cell environment went through direct shuffle to the control S. cerevisiae Sup35 protein or chimeric proteins with either S. paradoxus PrD or S. bayanus module II PrDs, followed by reverse shuffle back to S. cerevisiae Sup35.
Overall, our data show that the major rules of a transmission barrier, applying to both the efficiency of prion transmission and fidelity of the reproduction of prion strain patterns by a divergent protein, are generally invariant in S. cerevisiae and S. paradoxus host cells, even though some quantitative differences were detected. Major species barrier patterns are also apparently not influenced by the presence of the Rnq1 prion ([PIN+]) because the S. paradoxus strain used in this work was [pin−], whereas the S. cerevisiae strain used previously was [PIN+].
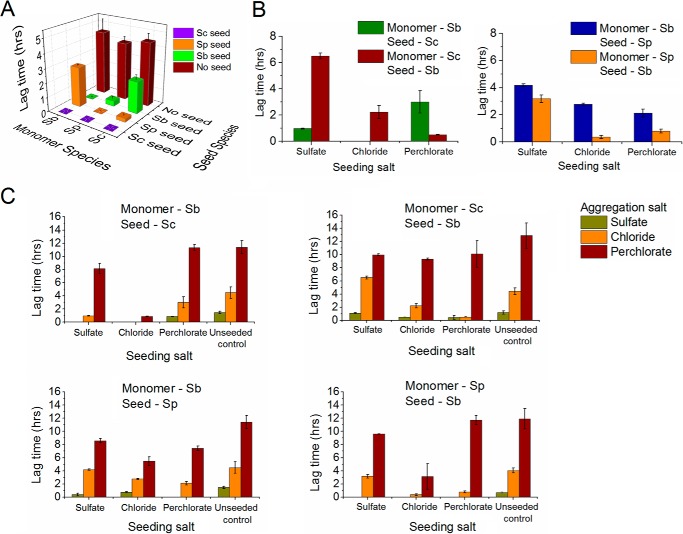
Amyloid Formation by Sup35NM of Various Origins in the Presence of Different Salts along the Hofmeister Series
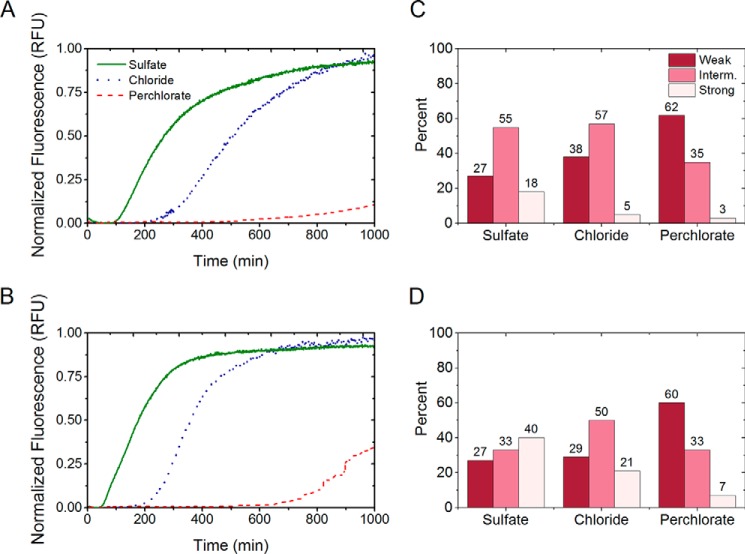
To obtain materials for studying the cross-species prion transmission in vitro, we first examined the unseeded amyloid formation of Sup35NMSb and Sup35NMSp in the presence of the salts (Fig. 7, A and B) and compared the results with our previous data for Sup35NMSc (44). We determined that the parameters of in vitro aggregation of Sup35NMSb and Sup35NMSp are influenced by anions of the Hofmeister series in the same way as observed previously for Sup35NMSc. For both proteins, the lag time of amyloid formation was shortened in the presence of sulfate (strong kosmotrope) and prolonged in the presence of perchlorate (strong chaotrope) compared with chloride (mild chaotrope). This establishes the generality of the inverse Hofmeister effect on the amyloid formation by each of the three Sup35NM proteins from the Saccharomyces species used in this work.
FIGURE 7.
Aggregation profiles of Sup35NMSb and Sup35NMSp in the presence of 0.4 m salts monitored by Thioflavin T fluorescence assay and phenotypic characterization of Sup35NMSb and Sup35NMSp prion strains. A and B, amyloid formation by Sup35NMSb (A) and Sup35NMSp (B) in the presence of 0.4 m sulfate, chloride, or perchlorate showing the effect of Hofmeister ions on their aggregation. In the presence of a strong kosmotrope (sulfate), the lag times are short, and elongation rates are fast. The opposite is true of aggregation in a chaotropic solution (perchlorate), whereas the aggregation plots show intermediate lag times and elongation rates in the presence of the mildly chaotropic chloride ions. RFU, relative fluorescence units. C and D, distribution of weak, intermediate (Interm.), and strong [PSI+] colonies obtained after the transfection of the yeast strains GT987 (expressing Sup35Sb) and GT797 (expressing Sup35Sp) with Sup35NMSb (C) and Sup35NMSp (D) amyloids, respectively, obtained in the presence of sulfate, chloride, or perchlorate salts. For both proteins, amyloids formed in sulfate resulted in a higher number of strong strains that appeared white or faintly pink in color on YPD, and the amyloids formed in perchlorate formed more weak strains appearing dark pink on YPD. Amyloids formed in chloride showed intermediate strain spectra compared with those formed in sulfate and perchlorate.
We also determined whether patterns of amyloids generated by Sup35NMSp and Sup35Sb in vitro are influenced by ionic composition. For this purpose, amyloids generated by Sup35NMSp or Sup35NMSb in the presence of various salts were transfected into the [psi−] S. cerevisiae strain bearing either Sup35Sp or Sup35Sb protein instead of Sup35Sc. For both proteins, a higher proportion of weaker [PSI+] isolates was detected after transfection with amyloids obtained in the presence of chloride or perchlorate compared with amyloids obtained in the presence of sulfate (Fig. 7, C and D). This confirms that a kosmotrope favors the formation of stronger prion strains, whereas a chaotrope favors the formation of weaker prion strains by Sup35NMSp and Sup35NMSb, similar to what has been described previously for Sup35NMSc (44).
Cross-species Amyloid Seeding in the Presence of a Mild Chaotrope
Next we investigated the effect of sequence divergence on the species barrier between Sup35NM proteins from the three species. For this purpose, we studied cross-species amyloid seeding in an intermediate environment; that is, in the presence of a mild chaotrope (chloride) that is positioned between sulfate and perchlorate in the Hofmeister series. Amyloids used as seeds were also formed in the presence of chloride. The monomeric proteins (referred to as “monomers”) and the sonicated amyloid fibers (referred to as “seeds”) used as seeds for the aggregation assays can be considered in vitro analogues of the in vivo [psi−] recipients and [PSI+] donors, respectively. Our results (Fig. 8A) show that Sup35NMSc and Sup35NMSp are highly efficient in seeding each other in the presence of chloride because we detect zero (or near zero) lag times for cross-seeding of Sup35NMSc with Sup35NMSp and vice versa. In contrast, Sup35NMSb is less efficient in seeding the other two proteins or being seeded by them and, therefore, exhibits a stronger species barrier (Fig. 8A). This generally agrees with in vivo data (Refs. 19, 20 and above) and can be explained by a higher degree of homology between the PrDs of Sup35NMSc and Sup35NMSp and a lower degree of homology between the PrDs of any of these proteins and Sup35NMSb. Also, in accordance with previous reports, we observed an asymmetry in the species barrier (19, 20, 34, 52). For example, seeding of the Sup35NMSc monomer with the Sup35NMSb seed results in a lag time of 2.22 h, whereas seeding of the Sup35NMSb monomer with the Sup35NMSc seed shows essentially no lag time. A similar asymmetric effect is seen in the Sp/Sb heterologous seeding combination.
FIGURE 8.
Effects of salts on the lag time of intraspecies and cross-species aggregation. A, lag time in hours for seeding experiments performed in 0.4 m sodium chloride. Monomer species, seed species, and lag time are shown along the y axis, x axis, and z axis of the three-dimensional plot, respectively. Unseeded aggregation data (red columns) are shown for comparison. The seeds were formed in the presence of chloride salt. Homologous seeding is highly efficient with zero lag times. Sc/Sp combination results in zero or close to zero lag times. Larger lag times are observed in some cases involving Sb as a seed or monomer. B, effect of a salt present during seed formation on heterologous co-aggregation in the Sup35NMSc/Sup35NMSb (left panel) and Sup35NMSp/Sup35NMSb (right panel) combinations. The salt present during the seed formation (seeding salt) is shown on the x axis. The different colors of the columns depict different monomer-seed combinations. Amyloid seeds were formed in the presence of sulfate, chloride, and perchlorate salts, and aggregation was carried out in the presence of chloride salt. Salts present during seed formation affect co-aggregation kinetics, as seen in a variation of the lag time. C, effect of a salt present during the aggregation reaction on heterologous co-aggregation in the Sup35NMSc/Sup35NMSb and Sup35NMSp/Sup35NMSb combinations. The seeding salt is shown on the x axis, whereas the salt present during the aggregation reaction (aggregation salt) is shown by different colors of the columns. Unseeded aggregation data are shown for comparison. Amyloid seeds were formed in the presence of sulfate, chloride, and perchlorate salts. Irrespective of seed origin, aggregation is the fastest in the presence of the strongly kosmotropic sulfate and slowest in the presence of the strongly chaotropic perchlorate.
Notably, the asymmetry pattern observed in the presence of chloride was different from the asymmetry patterns observed in vivo as well as in the previous in vitro data (19). Those results uncovered a stronger barrier for Sup35NMSb seeding with the Sup35NMSc seeds rather than in the opposite direction. To further investigate this discrepancy, we attempted both seed formation and cross-seeding in PBS, as in our previous work. Indeed, the asymmetry pattern observed in PBS resembled the in vivo data and our previous in vitro results rather than the asymmetry pattern observed in chloride (Fig. 9). Therefore, the addition of chloride appears to reverse an asymmetry pattern of the species barrier, indicating that the ionic composition of the solution influences the barrier. To decipher the mechanism of this phenomenon, we performed a more systematic study aimed at determining whether ionic composition acts via influencing seed properties or directly affects cross-seeding interactions.
FIGURE 9.
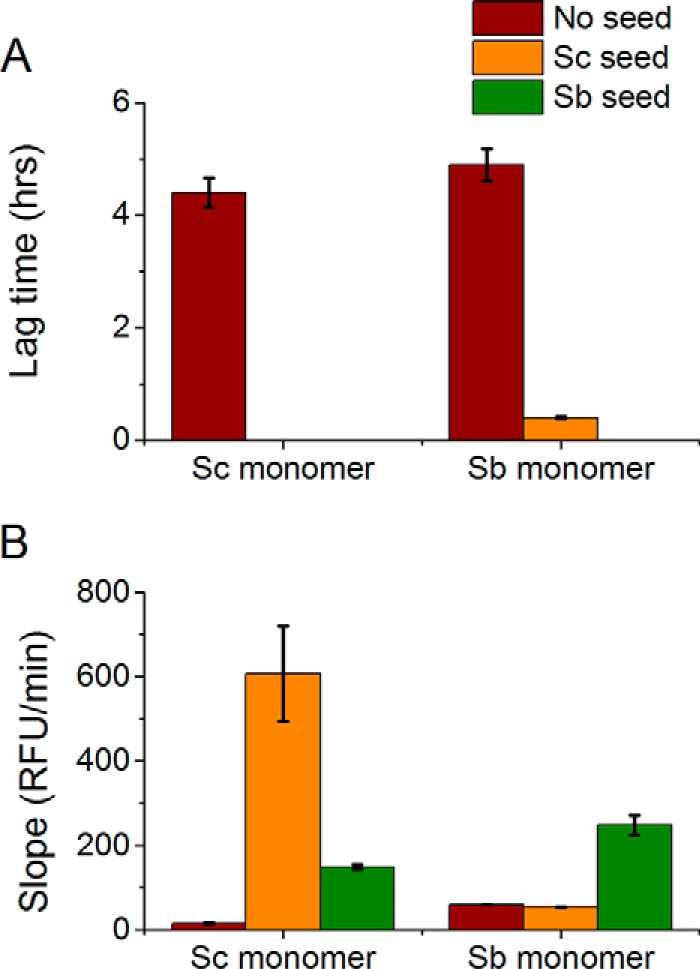
Lag time (A) and slope in RFU/minute (B) for unseeded and cross-seeding aggregation reactions between S. cerevisiae and S. bayanus in PBS. The monomer species is shown along the x axis, and the seeds species (or no seed control) is depicted by different colors of the columns. The seeds were also formed in PBS. Seeding the Sup35NMSc monomer with Sup35NMSb seeds is more efficient (zero lag time and higher slope) than seeding the Sup35NMSb monomer with Sup35NMSc seeds (non-zero lag time and lower slope).
Comparison of the Cross-seeding Capabilities of Seeds Obtained in the Presence of Different Salts
First we determined whether cross-seeding is influenced by the conditions under which a seed is generated. Hofmeister ions have been shown to affect both the kinetics of amyloid aggregation and the structural parameters of the resulting aggregates, reflecting distinct protein conformations (43, 44). Kosmotropic ions such as sulfate lead to faster kinetics and stronger strains, and chaotropic ions such as perchlorate lead to slower kinetics and weaker strains (43, 44, 51, 53). We confirmed this for Sup35NMSp and Sup35NMSb (see above). Because strain patterns influence cross-species prion conversion (20, 23), we compared cross-seeding by seeds obtained in different salts. In all reactions, we saw that amyloid seeds of same protein formed in different salts have different lag times and slopes depending on the salt in which they are formed (Figs. 8C and 10). Interestingly, seeds formed in sulfate do not always result in the shortest lag times, and seeds formed in perchlorate do not always lead to the longest lag times. Fig. 8B shows the effect of salt present during seed formation on the species barrier in the combinations Sup35NMSc/Sup35NMSb and Sup35NMSp/Sup35NMSb. One can see that, in the case of Sup35NMSc/Sup35NMSb (Fig. 8B, left panel), the seed obtained in chloride shows the shortest lag time in one of the combinations, whereas the seed obtained in perchlorate shows the shortest lag time in the other combinations. This agrees with our previous observation that, for the Sup35Sc/Sup35Sb combination in vivo, the species barrier is more severe for the strong strain compared with the weak strain. The combination Sup35NMSp/Sup35NMSb shows an even stronger trend in this direction, exhibiting the longest lag period for the sulfate-generated seeds in both reciprocal combinations (Fig. 8B, right panel). Overall, our data demonstrate that salts of the Hofmeister series influence species barriers via altering the structural parameters of the seed produced in the presence of a respective salt.
FIGURE 10.
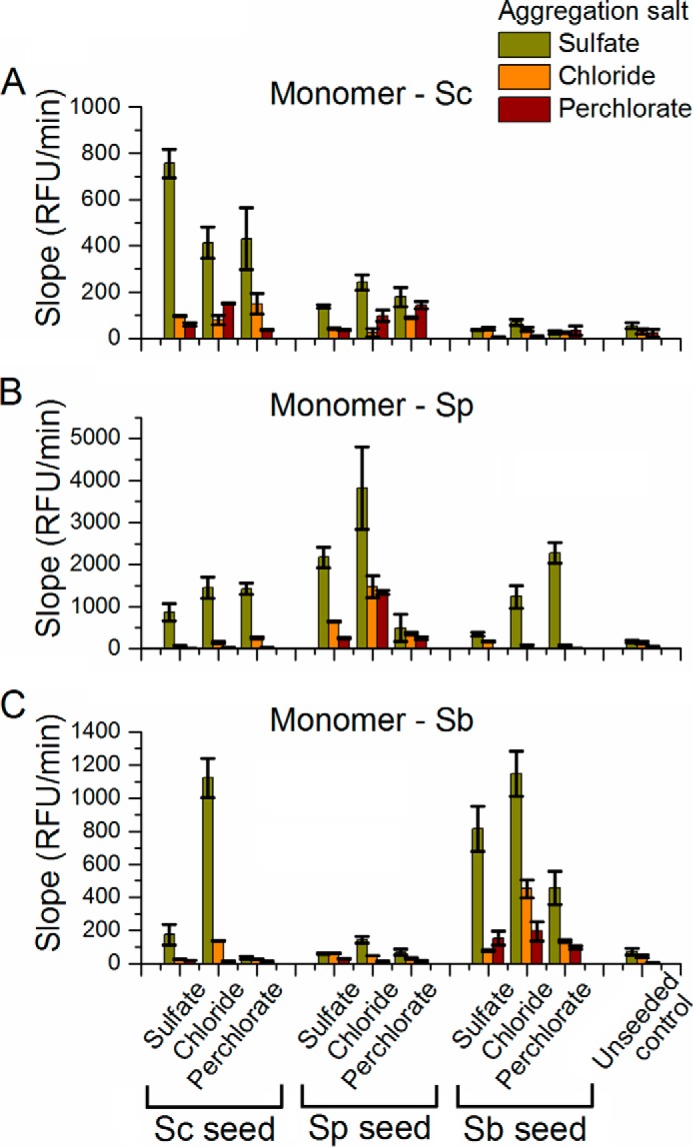
Slope in RFU/min for unseeded and seeded aggregation reactions. A–C, slopes obtained for aggregation assays performed with the monomeric proteins Sup35NMSc (A), Sup35NMSp (B), and Sup35NMSb (C) in both unseeded and seeded (with homologous and heterologous amyloid seeds) aggregation reactions. Homologous aggregation is more efficient than heterologous aggregation. Co-aggregation in the presence of sulfate leads to the highest slopes, and aggregation in perchlorate results in the smallest slopes.
Comparison of Cross-seeding Reactions Performed in the Presence of Different Salts
To determine whether the environment of the in vitro reaction influences the specificity of cross-seeding, we investigated the effect of salts present in the aggregation medium on cross-seeding between Sup35NM proteins from the three Saccharomyces species. For the Sup35NMSc/Sup35NMSp combination, zero or close to zero lag times were observed (supplemental Figs. S1 and S2), which can be explained by the higher degree of homology between the prion domains of Sup35NMSc and Sup35NMSp. For the Sup35NMSb/Sup35NMSc and Sup35NMSb/Sup35NMSp combinations, aggregation in sulfate, which is a strong kosmotrope, resulted in faster aggregation kinetics with shorter lag times (Fig. 8C) and steeper slopes (Fig. 10). On the other hand, aggregation in perchlorate, which is a strong chaotrope, resulted in longer lag times and smaller slopes. The aggregation kinetics in the slightly chaotropic chloride salt fell in between the kinetics observed for sulfate and perchlorate as the aggregation salts. Despite numerical differences, this overall pattern was detected independently of the salt in which the initial seed was obtained (for the effect on the slope, see Fig. 10). Therefore, the salt in which the cross-seeding reaction is performed has a strong and systematic influence on the kinetics of the cross-seeding reaction but does not affect the specificity of reaction, in contrast to the salt in which the initial seed is generated.
Discussion
By constructing a unique set of S. paradoxus strains allowing for prion detection and using the transfection protocol, we were able to obtain S. cerevisiae and S. paradoxus cultures with one and the same strain of prion that helped us differentiate the effects of prion protein properties and intracellular environment on cross-species prion conversion. Our data show that both transmission barrier and conformational fidelity in vivo are primarily determined by the protein itself rather than by the environment. Therefore, differences between the S. cerevisiae and S. paradoxus intracellular environments do not affect major rules of [PSI+] transmission, although they might influence the quantitative characteristics of the process.
The relative stability of the species barrier characteristics in the [PSI+] strain used in our studies differs somewhat from the results of Bateman and Wickner (35), who observed that parameters of [PSI+] transmission specificity could fluctuate reversibly during mitotic propagation of one and the same original prion strain even in the same S. cerevisiae intracellular environments. Authors interpreted these data as evidence for strain instability and existence of a “strain cloud.” However, other researchers (54) did not see such fluctuations in the same experimental system for [PSI+] strains used in their studies, indicating that the existence of the readily detectable cloud is not a general rule. It should be noted that, in our experiments, we analyzed only one colony from each original transformant. Therefore, even if rare clonal variations in transmission specificity occurred, then they would have a minimal effect in our case. Notably, Bateman and Wickner (35) used the model of an “intraspecies” barrier controlled by variations within module III of Sup35 PrD. This module does not play a significant role in the cross-species transmission differences detected in our work. It is therefore possible that intraspecies transmission barriers are based on the different molecular foundations compared with the cross-species barriers studied in our work. Moreover, it is not known whether the differences between the components of a cloud (that is, differences between interconvertible “substrains”) are determined by the same structural mechanisms that control differences between prion strains that are firmly established and faithfully reproduced at the phenotypic level.
Because S. cerevisiae and S. paradoxus species are relatively closely related to each other, it would be of interest to determine whether more drastic differences in the intracellular environment could influence the specificity of prion transmission. We engineered the system for [PSI+] detection in S. bayanus and showed that the [PSI+] prion can be transfected from the S. cerevisiae to S. bayanus strains.8 However, the systematic analysis of cross-species conversion in S. bayanus cells turned out to be impossible because of the overall low efficiency of the nonsense suppression assay in this species (data not shown). While continuing the optimization of the S. bayanus-based prion detection system, we moved the cross-species prion transmission experiments to an entirely different environment by performing in vitro experiments in solutions of various ionic compositions.
We have reported previously that anions of the Hofmeister series influence the kinetics and strain preference during in vitro amyloid formation by Sup35NMSc (44, 51), and now we show that Sup35NM proteins from the other species of the Saccharomyces sensu stricto group, Sup35NMSb and Sup35NMSp, also respond to the presence of kosmotropic or chaotropic ions in the same way as Sup35NMSc. We confirmed previous data by showing that, in vitro, in agreement with previous research, the more similar Sc/Sp proteins exhibit a relatively low barrier in the transmission of the prion state, whereas the more divergent Sb protein shows a high barrier with the other two proteins (19). We also performed systematic studies of the effects of the Hofmeister series anions on cross-species prion transmission. Overall, our data show that the salt present during amyloid formation can alter the parameters of the species barrier. The simplest explanation for such a result is that salts present during seed formation determine the seed conformation, which, in turn, influences the efficiency of the conformational adaptation of the added monomer to the preformed nuclei provided by this seed. This agrees with our previous in vivo data showing that the prion strain pattern controls the specificity of transmission of the prion state to the newly immobilized protein (20). In contrast, salts present during the process of cross-seeding exhibit a strong and systematic influence on the kinetics of cross-species aggregation in accordance with the inverse Hofmeister trend that has been reported previously for homologous aggregation (44), but they do not alter the transmission specificity.
Interestingly, we observed that seeds formed in chloride are more efficient in promoting both homologous and cross-species aggregation than seeds formed in sulfate or perchlorate (Figs. 8C and 10). We hypothesize that this might be because the seed nucleation in chloride produces a most diverse mix of various prion strains. This diversity helps to provide a pool of seed conformations, some of which are more amenable for the monomer to template onto. Therefore, it is always possible to select a fraction of strains that can act as highly efficient templates under any given conditions. By contrast, aggregation in the presence of either highly kosmotropic (e.g. sulfate) or highly chaotropic (e.g. perchlorate) salt exhibits a more pronounced bias toward one particular strain. Therefore, the type of the strain “preadapted” to the changed environment might be lacking.
Previous work by the Weissman group (24, 55, 56) has shown that temperature can be used to affect the specificity of the species barrier between S. cerevisiae and Candida albicans. Tanaka et al. (24) have observed that, when Sup35NMSc is aggregated at low (4 °C) temperature, more “promiscuous” aggregates with a stronger phenotype and higher seeding capabilities can be generated compared with aggregates produced at a high (37 °C) temperature. Tanaka et al. (55) have shown that the seeds of the S. cerevisiae Sup35NM protein formed at different temperatures show a variable ability to cross-seed the Sup35NM protein from C. albicans. Chien et al. (56) have shown that polymerization of a chimeric protein, combining the regions from the Sup35 prion domains of both S. cerevisiae and C. albicans, can produce distinct prion strains with different seeding specificities depending on temperature. These results agree with our observations that the differences in ionic composition influence the species barrier by promoting the formation of different strains. However, our work represents the first systematic comparison of the effects of aggregation conditions at the stages of initial aggregate formation and cross-species seeding. Our data confirm that protein sequence and conformation play a central role in determining the specificity of prion transmission and show that external factors influence transmission specificity primarily by altering the nature of the initial seed, whereas the conditions of the actual cross-seeding reaction itself have an effect only on the kinetics of the process.
Author Contributions
A. S. B. and Y. O. C. designed the study. A. S. and K. L. B. performed the experiments. B. C., S. G., and K. L. B. constructed the strains. A. S., K. L. B., S. H. B., A. S. B., and Y. O. C. analyzed the results and wrote and edited the manuscript.
Supplementary Material
Acknowledgments
We thank D. Deng, K. Jang, and A. Tippur for help with some experiments and A. Grizel, G. Newnam, and A. Romanyuk for helpful discussions.
The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figures S1 and S2.
K. L. Bruce, B. Chen, S. Gyoneva, and Y. O. Chernoff, unpublished data.
- PrD
- prion domain
- Sc
- Saccharomyces cerevisiae
- Sp
- Saccharomyces paradoxus
- Sb
- Saccharomyces bayanus
- YPD
- yeast extract/peptone/dextrose rich organic medium.
References
- 1.Chiti F., and Dobson C. M. (2006) Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 [DOI] [PubMed] [Google Scholar]
- 2.Eisenberg D., and Jucker M. (2012) The amyloid state of proteins in human diseases. Cell 148, 1188–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisenberg D., Nelson R., Sawaya M. R., Balbirnie M., Sambashivan S., Ivanova M. I., Madsen A. Ø., and Riekel C. (2006) The structural biology of protein aggregation diseases: fundamental questions and some answers. Acc. Chem. Res. 39, 568–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregersen N., Bross P., Vang S., and Christensen J. H. (2006) Protein misfolding and human disease. Annu. Rev. Genomics Hum. Genet. 7, 103–124 [DOI] [PubMed] [Google Scholar]
- 5.Jackson G. S., and Clarke A. R. (2000) Mammalian prion proteins. Curr. Opin. Struct. Biol. 10, 69–74 [DOI] [PubMed] [Google Scholar]
- 6.Prusiner S. B. (1998) Prions. Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan K.-M., Baldwin M., Nguyen J., Gasset M., Serban A., Groth D., Mehlhorn I., Huang Z., Fletterick R. J., and Cohen F. E. (1993) Conversion of aα-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. U.S.A. 90, 10962–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen J., Baldwin M. A., Cohen F. E., and Prusiner S. B. (1995) Prion protein peptides induce α-helix to β-sheet conformational transitions. Biochemistry 34, 4186–4192 [DOI] [PubMed] [Google Scholar]
- 9.Ross E. D., Minton A., and Wickner R. B. (2005) Prion domains: sequences, structures and interactions. Nat. Cell Biol. 7, 1039–1044 [DOI] [PubMed] [Google Scholar]
- 10.Pattison I. H. (1965) Experiments with scrapie with special reference to the nature of the agent and the pathology of the disease: Slow, latent and temperate virus infections. NINDB Monograph 2, 249–257 [Google Scholar]
- 11.Prusiner S. B. (1997) Prion diseases and the BSE crisis. Science 278, 245–251 [DOI] [PubMed] [Google Scholar]
- 12.Wilesmith J. W., Ryan J. B., and Atkinson M. (1991) Bovine spongiform encephalopathy: epidemiological studies on the origin. Vet. Rec. 128, 199–203 [DOI] [PubMed] [Google Scholar]
- 13.Ghani A. C., Donnelly C. A., Ferguson N. M., and Anderson R. M. (2002) The transmission dynamics of BSE and vCJD. C. R. Biol. 325, 37–47 [DOI] [PubMed] [Google Scholar]
- 14.Hill A. F., Desbruslais M., Joiner S., Sidle K. C., Gowland I., Collinge J., Doey L. J., and Lantos P. (1997) The same prion strain causes vCJD and BSE. Nature 389, 448–450, 526 [DOI] [PubMed] [Google Scholar]
- 15.Hueston W. D. (2013) BSE and variant CJD: emerging science, public pressure and the vagaries of policy-making. Prev. Vet. Med. 109, 179–184 [DOI] [PubMed] [Google Scholar]
- 16.Ferguson N. M., Donnelly C. A., Ghani A. C., and Anderson R. M. (1999) Predicting the size of the epidemic of the new variant of Creutzfeldt-Jakob disease. Brit. Food J. 101, 86–98 [Google Scholar]
- 17.Liebman S. W., and Chernoff Y. O. (2012) Prions in yeast. Genetics 191, 1041–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chernova T. A., Wilkinson K. D., and Chernoff Y. O. (2014) Physiological and environmental control of yeast prions. FEMS Microbiol. Rev. 38, 326–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen B., Newnam G. P., and Chernoff Y. O. (2007) Prion species barrier between the closely related yeast proteins is detected despite coaggregation. Proc. Natl. Acad. Sci. U.S.A. 104, 2791–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B., Bruce K. L., Newnam G. P., Gyoneva S., Romanyuk A. V., and Chernoff Y. O. (2010) Genetic and epigenetic control of the efficiency and fidelity of cross-species prion transmission. Mol. Microbiol. 76, 1483–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Afanasieva E. G., Kushnirov V. V., Tuite M. F., and Ter-Avanesyan M. D. (2011) Molecular basis for transmission barrier and interference between closely related prion proteins in yeast. J. Biol. Chem. 286, 15773–15780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wopfner F., Weidenhöfer G., Schneider R., von Brunn A., Gilch S., Schwarz T. F., Werner T., and Schätzl H. M. (1999) Analysis of 27 mammalian and 9 avian PrPs reveals high conservation of flexible regions of the prion protein. J. Mol. Biol. 289, 1163–1178 [DOI] [PubMed] [Google Scholar]
- 23.Šandula J., and Vojtková-Lepšíková A. (1974) Immunochemical studies on mannans of the genus Saccharomyces. Folia Microbiol. 19, 94–101 [DOI] [PubMed] [Google Scholar]
- 24.Tanaka M., Chien P., Naber N., Cooke R., and Weissman J. S. (2004) Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 [DOI] [PubMed] [Google Scholar]
- 25.Tanaka M., and Weissman J. S. (2006) An efficient protein transformation protocol for introducing prions into yeast. Methods Enzymol. 412, 185–200 [DOI] [PubMed] [Google Scholar]
- 26.Bruce K. L., and Chernoff Y. O. (2011) Sequence specificity and fidelity of prion transmission in yeast. Semin. Cell Dev. Biol. 22, 444–451 [DOI] [PubMed] [Google Scholar]
- 27.Derkatch I. L., Chernoff Y. O., Kushnirov V. V., Inge-Vechtomov S. G., and Liebman S. W. (1996) Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144, 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toyama B. H., Kelly M. J., Gross J. D., and Weissman J. S. (2007) The structural basis of yeast prion strain variants. Nature 449, 233–237 [DOI] [PubMed] [Google Scholar]
- 29.Stein K. C., and True H. L. (2014) Prion strains and amyloid polymorphism influence phenotypic variation. PLoS Pathog. 10, e1004328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kretzschmar H., and Tatzelt J. (2013) Prion disease: a tale of folds and strains. Brain Pathol. 23, 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parchi P., Strammiello R., Giese A., and Kretzschmar H. (2011) Phenotypic variability of sporadic human prion disease and its molecular basis: past, present, and future. Acta Neuropathol. 121, 91–112 [DOI] [PubMed] [Google Scholar]
- 32.Gambetti P., Cali I., Notari S., Kong Q., Zou W.-Q., and Surewicz W. K. (2011) Molecular biology and pathology of prion strains in sporadic human prion diseases. Acta Neuropathol. 121, 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Surewicz W. K., and Apostol M. I. (2011) Prion protein and its conformational conversion: a structural perspective. Top. Curr. Chem. 305, 135–167 [DOI] [PubMed] [Google Scholar]
- 34.Bateman D. A., and Wickner R. B. (2012) [PSI+] prion transmission barriers protect Saccharomyces cerevisiae from infection: intraspecies 'species barriers'. Genetics 190, 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bateman D. A., and Wickner R. B. (2013) The [PSI+] prion exists as a dynamic cloud of variants. PLoS Genet. 9, e1003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J., Browning S., Mahal S. P., Oelschlegel A. M., and Weissmann C. (2010) Darwinian evolution of prions in cell culture. Science 327, 869–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahal S. P., Browning S., Li J., Suponitsky-Kroyter I., and Weissmann C. (2010) Transfer of a prion strain to different hosts leads to emergence of strain variants. Proc. Natl. Acad. Sci. 107, 22653–22658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghaemmaghami S., Watts J. C., Nguyen H.-O., Hayashi S., DeArmond S. J., and Prusiner S. B. (2011) Conformational transformation and selection of synthetic prion strains. J. Mol. Biol. 413, 527–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill A. F., and Collinge J. (2004) Prions. A Challenge for Science, Medicine and the Public Health System, pp. 33–49, Karger, Basel, Switzerland [Google Scholar]
- 40.Broering J. M., and Bommarius A. S. (2005) Evaluation of Hofmeister effects on the kinetic stability of proteins. J. Phys. Chem. B 109, 20612–20619 [DOI] [PubMed] [Google Scholar]
- 41.Broering J. M., and Bommarius A.S. (2007) Cation and strong co-solute effects on protein kinetic stability. Biochem. Soc. Trans. 35, 1602–1605 [DOI] [PubMed] [Google Scholar]
- 42.Broering J. M., and Bommarius A. S. (2008) Kinetic model for salt-induced protein deactivation. J. Phys. Chem. B 112, 12768–12775 [DOI] [PubMed] [Google Scholar]
- 43.Hofmeister F. (1888) On the understanding of the effects of salts. Arch. Exp. Pathol. Pharmacol. 24, 247–260 [Google Scholar]
- 44.Rubin J., Khosravi H., Bruce K. L., Lydon M. E., Behrens S. H., Chernoff Y. O., and Bommarius A. S. (2013) Ion-specific effects on prion nucleation and strain formation. J. Biol. Chem. 288, 30300–30308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diaz-Espinoza R., Mukherjee A., and Soto C. (2012) Kosmotropic anions promote conversion of recombinant prion protein into a PrPSc-like misfolded form. PLoS ONE 7, e31678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Longtine M. S., McKenzie A. III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 47.Taxis C., and Knop M. (2006) System of centromeric, episomal, and integrative vectors based on drug resistance markers for Saccharomyces cerevisiae. BioTechniques 40, 73–78 [DOI] [PubMed] [Google Scholar]
- 48.Kaiser C., Michaelis S., and Mitchell A. (1994) Methods in Yeast Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 49.Chernoff Y. O., Uptain S. M., and Lindquist S. L. (2002) Analysis of prion factors in yeast. Methods Enzymol. 351, 499–538 [DOI] [PubMed] [Google Scholar]
- 50.Allen K. D., Wegrzyn R. D., Chernova T. A., Müller S., Newnam G. P., Winslett P. A., Wittich K. B., Wilkinson K. D., and Chernoff Y. O. (2005) Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 169, 1227–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeh V., Broering J. M., Romanyuk A., Chen B., Chernoff Y. O., and Bommarius A. S. (2010) The Hofmeister effect on amyloid formation using yeast prion protein. Protein Sci. 19, 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edskes H. K., McCann L. M., Hebert A. M., and Wickner R. B. (2009) Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics 181, 1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klement K., Wieligmann K., Meinhardt J., Hortschansky P., Richter W., and Fändrich M. (2007) Effect of different salt ions on the propensity of aggregation and on the structure of Alzheimer's aβ(1–40) amyloid fibrils. J. Mol. Biol. 373, 1321–1333 [DOI] [PubMed] [Google Scholar]
- 54.Huang Y.-W., Chang Y.-C., Diaz-Avalos R., and King C.-Y. (2015) W8, a new Sup35 prion strain, transmits distinctive information with a conserved assembly scheme. Prion 9, 207–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanaka M., Chien P., Yonekura K., and Weissman J. S. (2005) Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell 121, 49–62 [DOI] [PubMed] [Google Scholar]
- 56.Chien P., DePace A. H., Collins S. R., and Weissman J. S. (2003) Generation of prion transmission barriers by mutational control of amyloid conformations. Nature 424, 948–951 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.