Abstract
Many pectin methylesterases (PMEs) are expressed in plants to modify plant cell-wall pectins for various physiological roles. These pectins are also attacked by PMEs from phytopathogens and phytophagous insects. The de-methylesterification by PMEs of the O6-methyl ester groups of the homogalacturonan component of pectin, exposing galacturonic acids, can occur processively or non-processively, respectively, describing sequential versus single de-methylesterification events occurring before enzyme-substrate dissociation. The high resolution x-ray structures of a PME from Aspergillus niger in deglycosylated and Asn-linked N-acetylglucosamine-stub forms reveal a 10⅔-turn parallel β-helix (similar to but with less extensive loops than bacterial, plant, and insect PMEs). Capillary electrophoresis shows that this PME is non-processive, halophilic, and acidophilic. Molecular dynamics simulations and electrostatic potential calculations reveal very different behavior and properties compared with processive PMEs. Specifically, uncorrelated rotations are observed about the glycosidic bonds of a partially de-methyl-esterified decasaccharide model substrate, in sharp contrast to the correlated rotations of processive PMEs, and the substrate-binding groove is negatively not positively charged.
Keywords: carbohydrate processing, crystal structure, electrostatics, molecular dynamics, processivity, capillary electrophoresis, pectin methylesterase
Introduction
The heterogeneous polysaccharide pectin (1–4) is a key component of the plant cell wall (5). A host of enzymes, including pectin methylesterase (PME)3 as well as endo- and exo-polygalacturonases, modifies pectin fine-structure locally in space and time for purposes of plant-cell differentiation, cell adhesion/separation, growth, and development from roots to meristem, morphogenesis, seed and fruit development, and defense against pathogens (6–16). PMEs hydrolyze the O6-methyl ester groups of the homogalacturonan (HG) chains that form the backbone of pectin (Fig. 1). As well as being ubiquitous in plants, and frequently manifesting in a staggering number of isoforms (more than 66 for Arabidopsis thaliana) (8, 10), PMEs are found in phytopathogenic bacteria and fungi (17–26) and in Archaea (especially Halobacteriaceae (27)), and they have also been observed in the genomes of several phytophagous insects (28–30) and in human gut microflora (31). Recently, plant pollen PMEs have been identified as a key human allergen (32, 33).
FIGURE 1.
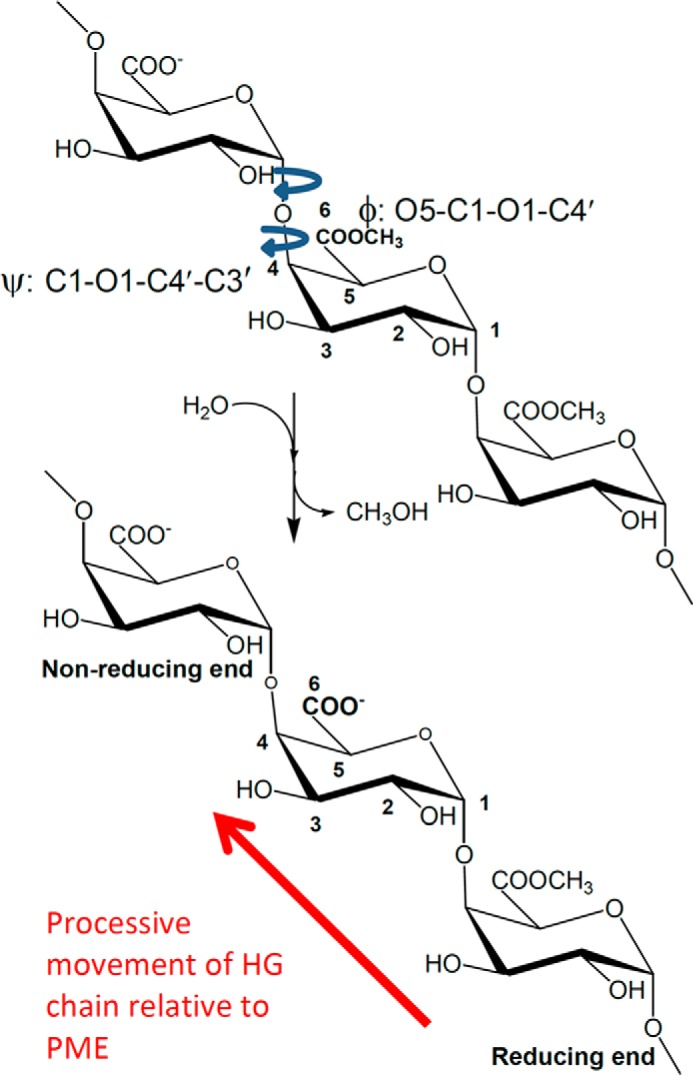
Hydrolysis (de-methylesterification) of a short homogalacturonan oligomer. For processive PMEs, the direction of de-methylesterification is indicated by the arrow. The reducing and non-reducing ends of the HG oligomer are annotated. The φ and ψ torsion angles linking pairs of residues are shown; the primed atoms belong to the glycolytically linked adjacent residue toward the reducing end of the HG chain.
Bacterial and plant PMEs that have been functionally characterized are generally observed to be processive (34–55), i.e. the enzyme binds to the HG substrate and then successively hydrolyzes a block of methyl ester groups before dissociating. In all cases characterized to date, the direction of processivity is from the non-reducing end of the HG chain toward the reducing end, as shown in Fig. 1. The degree of processivity (also called the action pattern) has been reported to be dependent on ionic strength and pH (56), and various classifications of PMEs exist (48, 49, 57–60). In general, processive PMEs have their isoelectric points, pI, and pH for optimum activity at near-neutral to basic pH, although one exception, a ficus (jelly fig) PME, has been reported (61, 62). Where salt sensitivity and the pH optimum for activity have been reported, generally, the pH of optimum activity has decreased as salt concentration increases.
To date, the fungal PMEs from Aspergillus (Emericella) nidulans (63) and Trichoderma reesei (Hypocrea jecorina) are reported to be processive (50, 64), whereas others are reported to be non-processive with the pattern of de-methylesterification of HG chains being near-random, indicating that these enzymes release their substrate after each de-methylesterification event (38, 42, 50, 52, 65–67). This mode of action is also referred to as multiple attack (56).
Sequence identity in pairwise comparisons of PMEs between plants and bacteria, between bacteria and fungi, and between plants and fungi is generally on the order of 30%. Between PMEs from phytophagous insects and other organisms, the identity is somewhat lower. Fig. 2 shows a ClustalW-based (68) alignment of all structurally characterized PMEs, including the fungal PME of this study.
FIGURE 2.
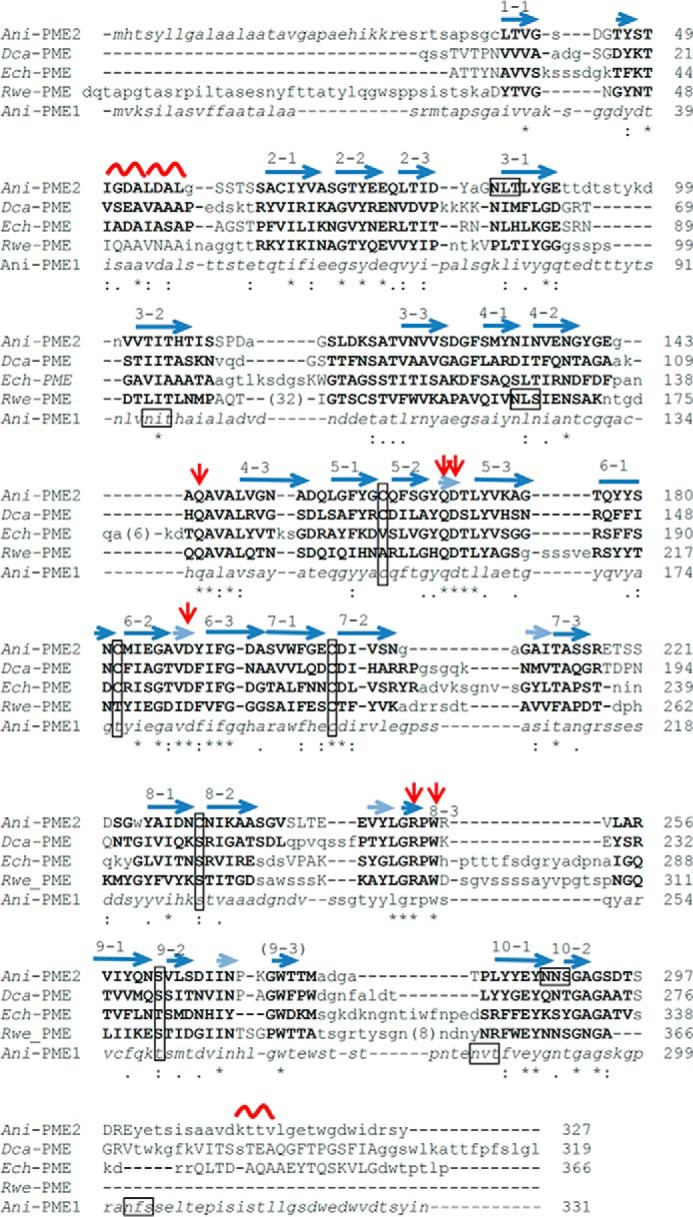
Adjusted ClustalW alignment of selected PME sequences. The raw sequence-based alignment has been adjusted to show structural alignment, where three-dimensional structural characterization has been made. Residues structurally homologous for fungal, plant, bacterial, and insect PMEs are highlighted in bold capitals; where structural homology is limited to two of the four structures light-face capitals are used. Residues in lowercase are not structurally homologous, and those in italics were not structurally characterized or observed. The secondary structure elements are noted above the pile-up of sequences. The β-helical turns are denoted n (n = 1–10); the strands comprising each turn are appended as n·1, n·2, and n·3; a series of very short β-strands, which form a 5-stranded β-sheet and encompass several active-site residues, are shown in a semi-transparent representation. The active-site Asp and Gln residues are flagged by red arrows; potential N-glycosylation motifs (NX(S/T)) of eukaryotic PMEs are boxed, and the ladder of cysteine and serine/threonine residues are boxed (vertical boxes). For clarity, extended insertions have been excised and the number of missing residues placed in parentheses.
Plant and bacterial PMEs have been structurally characterized, and recently so has an insect PME. They all share a distinctive parallel β-helix structure, shown in Fig. 3. A pair of absolutely conserved Asp residues, Asp-178 and Asp-199 in the Erwinia chrysanthemi PME, forms the active site, along with a pair of Gln residues (Gln-153 and Gln-177) that potentially form an oxyanion hole for the putative negatively charged acyl intermediate. The structure of a carrot PME, Dca-PME, has been determined (69) as well as the structure of tomato PME, the latter with a tightly associated kiwi fruit inhibitor (70). Bacterial PMEs from the soft-rot plant pathogen E. chrysanthemi (Dickeya dadantii), Ech-PME, and from the zoonotic diarrhea-causing Yersinia enterocolica, Yen-PME, have also been structurally characterized (46, 71, 72). The PME from the rice weevil shares the same distinctive β-helix structure and key active-site residues observed for other PMEs (73).
FIGURE 3.
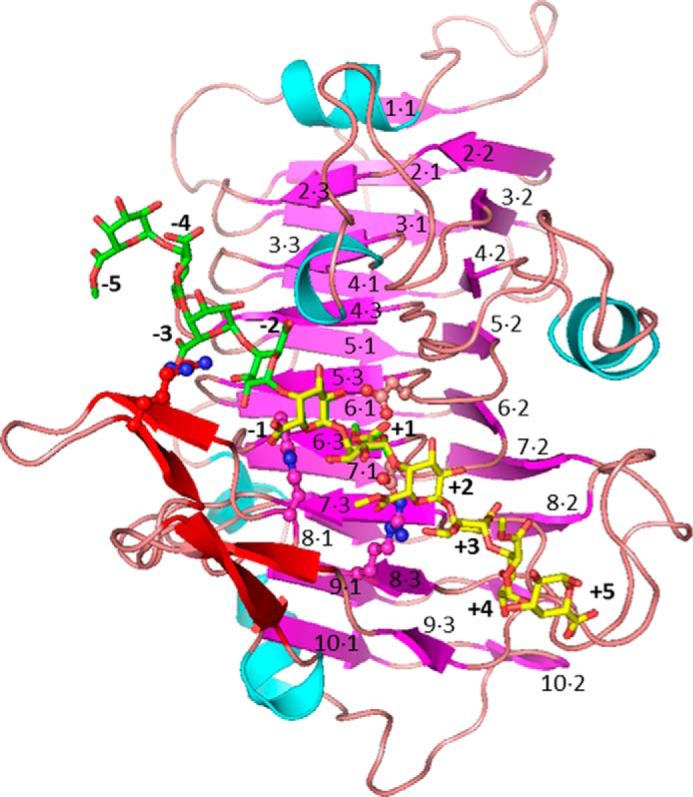
Structure of the PME from E. chrysanthemi. The PME is colored according to secondary structure (magenta for β-strands of the parallel core β-helix, red for non-core β-strands, cyan for helices, and pink for loops). The decasaccharide used for molecular dynamics simulations and formed by combining hexasaccharide oligomers (yellow, methyl-esterified except at the reducing end; green, de-methyl-esterified except at the non-reducing end) is modeled into the binding groove. Note: saccharide residues at −1 and +1 sites are overlapped, and the orientation of the galacturonic acid/methylgalacturonate residues alternates. The labeling scheme for the core parallel β-helix is shown. Key active-site residues are shown as sticks.
In the case of the PME from E. chrysanthemi, complexes of the enzyme with a variety of hexasaccharide HG oligomers with different patterns of methylesterification have been characterized (46). This study established that the oligosaccharide binding groove is ∼10 residues long. The active site is denoted as +1, and it was established that galacturonic acid moieties prefer sites −1 and −2, whereas methyl-esterified residues prefer sites +1 and +3. Thus, the direction of processivity was rationalized. Binding at sites −3, −2, −1, +1, +2, and +3 is well conserved among the different hexasaccharides, whereas binding at the outer sites of the binding groove is less well defined. Additionally, a key feature of the oligosaccharide conformations was a pseudo-21 screw axis that placed neighboring methyl ester groups alternately facing into and away from the binding groove. This creates a topological problem as processivity cannot be accomplished by simple translation of substrate with respect to enzyme (or vice versa).
Recently, we performed extensive molecular dynamics simulations on decasaccharide Ech-PME species, derived from hexasaccharide Ech-PME complexes (46), which deciphered the mechanism of processivity (74, 75). Concerted rotations of residues spanning the active site were observed, but only for a decasaccharide oligomer that represented the transition state where a just de-methyl-esterified residue was at the active site +1, galacturonate groups were present at sites −5 to −1, and methyl-esterified residues at sites +2 to +5 (denoted X5XM4). These rotations oriented the methyl ester group of the monosaccharide at site +2 toward the active site at +1, in the correct orientation for a second de-methylesterification event after the translation of the chain (74). In one simulation, this translation was also observed. The processivity of this PME can therefore be considered as a Brownian ratchet, where the free energy that rectifies thermal fluctuations is carried endogenously in the methyl ester groups, in contrast to classical molecular motors that are powered by exogenous sources, such as hydrolysis of ATP, or by relaxation of proton or ion gradients (75).
To date, no structure has been determined of a fungal or of a non-processive PME, or for that matter of a PME where the pI and the pH of optimum activity are <4.5. The black rot fungus Aspergillus niger expresses three PMEs, for lack of better names, Ani-PME1 (UniProt G3Y7R7, pmeA), Ani-PME2 (Uniprot G3YAL0, pmeB), and Ani-PME3 (UniProt G3Y257) (76). Ani-PME1 and Ani-PME2 share identity of only 42% (Fig. 2), much lower than that among pairs of isoforms for a given plant or even between plants. Ani-PME1 has been shown to have three glycosylation sites (77), whereas Ani-PME2 is predicted to have just two (Fig. 2). Ani-PME1 has been expressed in Pichia pastoris, and its activity and lack of processivity have been widely characterized (78–80). Here, we present the first structure of a fungal PME, Ani-PME2, which, while preserving key active-site residues (Fig. 2), has distinctly different loop structures and surface electrostatic potential compared with plant, bacterial, and insect PMEs. Using highly methyl-esterified pectin samples, we have examined in parallel the mode of action of Ani-PME1 and Ani-PME2 alongside that of a PME from orange (Citrus sinensis), denoted here as Csi-PME. We also present results of molecular dynamics simulations on Ani-PME2, conducted with the same methodology as our calculations on Ech-PME. These serve two purposes as follows: first, to understand the basis of non-processivity in this fungal enzyme, and second, to validate our calculations on the processive Ech-PME by providing a point of comparison.
With this work on fungal PME, we now have, for the first time, both detailed biochemical characterizations and structures for the following: (i) processive and non-processive PMEs; (ii) PMEs from three distantly related organisms; (iii) PMEs that have activity from acidic to basic conditions; and (iv) PMEs that vary in their requirement for ionic strength (salt concentration). Our results provide then a key addition to the voluminous attention paid by Nature to pectin and pectin methylesterases in plant cell-wall construction, modification, and deconstruction by researchers seeking to understand the myriad facets of the biochemistry and biophysics of pectin and PME and by technologists applying pectin methylesterases in numerous industrial scale processes and products (5).
Experimental Procedures
Sources of Chemicals
All chemicals and reagents used were AR grade or higher, sourced from Sigma (New Zealand), Merck KGaA, and Thermo Fisher (Australia Pty. Ltd.). Deglycosylation and cloning enzymes, purification kits, and media were from Bio-Rad, GE Healthcare, Life Technologies, Inc., New England Biolabs, Roche Diagnostics (New Zealand), and Zymo Research Corp. Crystallization solutions and plates were obtained from Art Robbins Instruments, Hampton Research, and Molecular Dimension Ltd. Custom primers and gBlock were supplied by Integrated DNA Technologies Pty. Ltd. Orange PME (EC 3.1.1.11) (Csi-PME), obtained from Sigma (P5400, >150 units/mg), and apple pectin, obtained from Fluka and from Sigma (catalog no. 76282, degree of methylation (DM), ∼75%) were used as received. All water used was of MilliQ standard.
Vector Modification for Synthesis and Cloning of Ani-PME1 and Ani-PME2
The pYES2 vector (Life Technologies, Inc.) (Scheme 1) was digested with XhoI and confirmed by 1% agarose gel electrophoresis. All restriction digests were done in 1× SuRE Cut buffer H or B, as appropriate (Roche Diagnostics), overnight at 37 °C the enzyme was heat-inactivated at 65 °C for 20 min, and the fragments were purified with DNA Clean & ConcentratorTM-5 (Zymo Research Corp.). The purified fragment was further digested with HindIII and dephosphorylated with 1 unit of thermosensitive alkaline phosphatase (Promega) in 1× Multi-core buffer at 37 °C for 15 min and heat-inactivated at 74 °C. The gBlock was digested simultaneously with XhoI and HindIII. The resulting DNA products were ligated overnight at 4 °C and then for 3 h at 10 °C in 1× T4 DNA ligase buffer (Roche Diagnostics) with 0.5 units of T4 DNA ligase. Escherichia coli XL-1 Blue competent cells were transformed with 8 μl of the ligation mixture by heat shock, and cells were plated on SOB/ampicillin agar plates and grown for 20 h at 30 °C. Several colonies were picked and used to inoculate separate cultures of LB/ampicillin medium. The cultures were grown at 37 °C overnight, and colony PCRs were run using custom primers (Integrated DNA Technologies) to select isolates with the correct length. Plasmids were isolated using a High Pure Plasmid Isolation kit (Roche Diagnostics). The sequences of the resulting plasmid products were confirmed by DNA sequencing (Massey Genome Service, Massey University, New Zealand). The construct is shown in Scheme 1.
SCHEME 1.
Ani-PME2 construct showing vector pYES2 (gray) with insertions including gBlock (bold, with S. cerevisiae consensus sequence AAAAAAATG immediately followed by S. cerevisiae α-mating factor) and Ani-PME2 (beginning with the added Kex cleavage site AAAAGA). Ani-PME2 primers are in italics and utilized restriction sites are underlined.
RNA Extraction
Material from a frozen vial of A. niger van Tieghem, anamorph (ATCC 11414) obtained from the Massey University Culture Collection, was streaked onto a maltose extract agar plate. A 1-cm2 of growth was cut from the plate and vortexed in a 0.01% Triton X-100 solution. This was used to inoculate a modified Czapek-Dox media (sucrose replaced with 1% w/v apple pectin (Sigma, DM 70–75), pH 6.8) and grown at 30 °C for 6–7 days. Activity assay was performed by adding media filtered through a 0.2-μm syringe filter to 0.5% w/v highly methyl-esterified homogalacturonan (degree of methylesterification of 96%, gift from Marie-Christine Ralet) and incubating at 35 °C. De-methylesterification was visualized with capillary electrophoresis (see under “Functional Analyses of Ani-PME2 and Csi-PME” for more detail). Upon confirmation of PME activity, total RNA was isolated by grinding cells in liquid nitrogen with a pestle and mortar and extracted using Illustra RNAspin mini kit (GE Healthcare) with the DNase step repeated once.
cDNA Synthesis and PME Cloning
cDNA was synthesized from total RNA in a reverse transcriptase reaction containing 4 μl of SuperScript® VILOTM MasterMix (Life Technologies, Inc.) and 0.3 μg of RNA. The annealing reaction was incubated at 25 °C for 10 min, followed by elongation at 42 °C for 120 min, and termination at 85 °C for 5 min. PME genes were PCR-amplified from the resulting cDNA and then modified by PCR to add restriction sites and a Kex cleavage site using custom primers (Integrated DNA Technologies, see Scheme 1). The DNA fragments and modified pYES2 vector were digested in separate reactions containing XhoI and XbaI for 16 h at 37 °C. The amplifications and digests were confirmed by 1% agarose gel electrophoresis. Vector de-phosphorylation, ligation, transformation, growth, PCR, and sequence confirmation were performed as described above (see under “Vector Modification for Synthesis and Cloning of Ani-PME1 and Ani-PME2”). The clones were transformed into uracil-auxotrophic INVSc1 yeast expression host by heat shock according to the pYES2 protocol. Several colonies of INVSc1 containing the pYES2_αMF_Ani-PME construct were inoculated into individual 8 ml of SC-U medium (synthetic minimal defined medium minus uracil supplement) containing 2% glucose. The cultures were grown at 30 °C with shaking overnight and were made into glycerol stocks for −80 °C storage.
Expression and Purification of Ani-PME1 and Ani-PME2
One vial of frozen stock was used to inoculate a starter culture of 200 ml of SC-U medium in a 1-liter flask and grown at 30 °C for 24 h with shaking. A volume of cells sufficient to make a final starting density (A600) of 0.4 were spun down at 1500 × g for 10 min and resuspended in 10 ml of induction medium (SC medium containing 2% galactose instead of glucose). The suspension was divided equally and placed into two 5-liter flasks containing 1 liter of induction medium. Cells were grown at 30 °C with vigorous shaking for 72 h. Cells were harvested by centrifugation at 5000 × g for 20 min, and the cell pellets were discarded. The supernatant was filtered through a 0.8-μm filter paper (Merck Millipore), and the pH was adjusted to 5.5 with NaOH. A bed volume of 40 ml of DEAE-Sephadex A-25 (GE Healthcare) equilibrated with buffer B (20 mm BisTris-HCl, 0.5 m NaCl, pH 6.5) was added and stirred gently at room temperature overnight. After binding, the resin slurry was packed into an XK26/200 column (GE Healthcare) by gravity and washed with 20 column volumes (CV) of 80% buffer A (20 mm BisTris-HCl, pH 6.5), 20% buffer B. PME was eluted in a linear gradient from 20 to 100% buffer B in 6 CV at a flow rate of 1.5 ml/min delivered by an ÄKTAexplorer 10S (GE Healthcare). The fractions were checked for PME activity, and purity was assessed by SDS-12% PAGE. Active fractions of similar purity were pooled, buffer-exchanged into storage buffer (10 mm sodium acetate, 50 mm NaCl, pH 4.5), and concentrated with Vivaspin 20 concentrators with 10,000 molecular weight cutoff (GE Healthcare).
Deglycosylation of Ani-PME1 and Ani-PME2
Removal of N-linked high mannose glycans from Ani-PME was performed by incubation with maltose-binding protein-tagged EndoHf (New England Biolabs) or His-tagged PNGaseF (81). For EndoHf reactions, the sample was deglycosylated in 1× G5 Reaction Buffer with 40,000 units of EndoHf per ml of sample and incubated for 20 h at 37 °C. The resulting mixture was applied to a 500-μl amylose column (New England Biolabs), and the deglycosylated PME was recovered in the flow-through fraction and stored at −80 °C. Similarly for PNGaseF reactions, concentrated buffer was added to the PME sample in storage buffer for a final concentration of 50 mm Na3PO4, 10 mm sodium acetate, 50 mm NaCl, 1 mm EDTA, pH 7.5; PNGaseF was then added for a final concentration of 15 μg/ml. The reaction was incubated at 37 °C for 60 h. Imidazole-HCl was added to a final concentration of 25 mm at the end of incubation and applied to a 500-μl Chelating Fast Flow column (GE Healthcare) charged with Ni2+. The deglycosylated PME was recovered in the flow-through fraction and stored at −80 °C.
Mass Spectrometry of Deglycosylated Ani-PME1 and Ani-PME2 and Csi-PME
The identity of the PME was confirmed by in-gel digestion of excised SDS-polyacrylamide gel bands (82) and tandem MS/MS experiments. Mass spectrometric measurements were performed using a 6520 series Q-TOF mass spectrometer and an HPLC-Chip Cube interface equipped with a ProtID-Chip-43 (II) chip (Agilent Technologies). The peptides were eluted from the chip using a linear gradient of MeCN with 0.1% formic acid from 5 to 55% in 15 min at a flow rate of 4 nl/min delivered by a 1260 Infinity Nano LC system (Agilent Technologies). Capillary voltage of 1830 V, fragmentor voltage of 175 V, and skimmer voltage of 65 V were used for LC-MS/MS experiments. The spectra were acquired in the m/z range from 400 to 1700 at six and four spectra/s for parent and fragmented ions, respectively. Data acquisition, processing, and exportation were performed using Mass Hunter Workstation version B6.0 SP1 (Agilent Technologies). The data were searched against the non-redundant database under the subset of A. niger combined with a contaminant database using the Trans Proteome Pipeline (83) and MASCOT search engine. Search parameters took into account modifications such as carboxymethyl on Cys, deamidation of Gln and Asn, and methionine sulfoxide. A cutoff score based on p < 0.05 was used to filter the searches, and each reported protein hit has at least two unique peptides following “Paris Guidelines” (84). The intact deglycosylated protein did not fly on either Q-TOF or Orbi-trap instruments, due to large negative surface charge, but an approximate mass of 32.0 (6) kDa was obtained by MALDI-TOF.
Functional Analyses of Ani-PME2 and Csi-PME
To analyze for processivity, three different protocols were used. In all cases, capillary electrophoresis, which separates components on the basis of hydrodynamic size and charge density, was used. First, this allowed the de-methylesterification process to be monitored via increases in the negative charge density of an apple pectin substrate. This was conducted at different pH values and ionic strength to determine the pH and ionic strength for optimal activity. Second, the digest patterns of the modified substrates could be interrogated after incubation with degrading enzymes to infer the blockiness of the charge distribution of the pre-digested polymer from the number of completely unmethyl-esterified fragments produced.
Capillary Electrophoresis
Experiments carried out in this work used an automated CE system (HP 3D), equipped with a diode array detector. Electrophoresis was carried out in a fused-silica capillary of 50 μm internal diameter and a 46.5-cm total length (40 cm from inlet to detector). The capillary incorporated an extended light path detection window (150 μm) and was thermostatically controlled at 25 °C. Phosphate buffer at pH 7.0 was used as a CE background electrolyte (BGE) and was prepared by mixing 0.2 m Na2HPO4 and 0.2 m NaH2PO4 in appropriate ratios and subsequently reducing the ionic strength to that desired (typically 50 or 90 mm). All new capillaries were conditioned by rinsing for 30 min with 1 m NaOH, 30 min with a 0.1 m NaOH solution, 15 min with water, and 30 min with BGE. Between runs, the capillary was washed for 2 min with 1 m NaOH, 2 min with 0.1 m NaOH, 1 min with water, and 2 min with BGE. Detection was carried out using the far-UV absorbance at 192 nm (reporting on the presence of the carboxyl groups present on each GalA residue). Samples were loaded hydrodynamically (various injection times at 5000 pascal, typically giving injection volumes of the order of 10 nl) and typically electrophoresed across a potential difference of 25 kV. All experiments were carried out at normal polarity (inlet anodic) unless otherwise stated.
First, 2 μl of Ani-PME2 in glycosylated and EndoHf-treated (NAG stub) forms (2 and 12 mg/ml, respectively) were each added to 150 μl of 0.5% w/v of highly methyl-esterified (∼75%) apple pectin in 50 mm acetate buffer at pH 4.2 with 100 mm NaCl. CE was used to follow hydrolysis of the methyl ester groups of the pectin HG chains to galacturonate residues as a function of incubation time. The evolution of the methylesterification distribution, relative to the starting pectin, was inferred from the CE profile as described elsewhere (47, 85). Second, aliquots of 0.5% w/v apple pectin were incubated with (i) glycosylated Ani-PME2 (pH 4.5, 100 mm NaCl), (ii) orange PME (pH 7.5), and (ii) base (pH 11.5), respectively, without buffering. The plant PME and the base were included as reference points for processive and non-processive de-methylesterification, respectively. In all cases as de-methylesterification proceeds, the pH drops as pectic acid groups are liberated. In response, 10-μl aliquots of 0.1 m NaOH were periodically added to the solutions to maintain the pH. When the total amount of NaOH added corresponded to the liberation of sufficient carboxyl protons so that the degree of methylesterification of the pectin was inferred to have been reduced to around 35%, the reactions were quenched by dropping the pH to 3 with acetic acid and subsequently denaturing the enzymes by heating (95 °C for 5 min) where appropriate. Again, the degree of methylesterification and the intermolecular DM distribution of these substrates were obtained by CE. The advantage of this batch-method is that three low DM substrates (all ∼(35 ± 2)%) can be obtained from the same mother pectin in a facile manner, performing the de-methylesterification in three different ways. Third, the samples generated under the second protocol were treated with endopolygalacturonase II (EndoPG II) from A. niger. This enzyme preferentially cuts the HG chain in unmethyl- or de-methyl-esterified regions, and it has a strict requirement for non-methyl-esterified GalA residues on either side of the scission point (86). Hence, the digest pattern (the relative amount of different non-esterified or partially methyl-esterified oligosaccharides of different degrees of polymerization liberated) is rendered sensitive to the patterning of methyl ester moieties along the chain and therefore to the processive nature or otherwise of the reaction that removed the methyl ester groups from the highly esterified starting material. These digests were performed in 50–100 mm acetate buffer at pH 4.2, and the resulting oligogalacturonides were examined after a 24-h incubation at 25 °C by CE.
Crystallization of Ani-PME2
The sitting-drop vapor diffusion method was used for initial crystallization trials, with solutions dispensed by means of a liquid-handling robot Mosquito (TTP Labtech) into 96-well Shallow-well Intelli plates (Art Robbins Instruments). The screening conditions used were Structure Screen I+II HT-96 and JCSG-plus HT-96 (Molecular Dimensions) at 25 °C. 800-nl drops were made with equal volumes of protein at 6.5 mg/ml and reservoir solutions in subwell 2 and 800 nl of reservoir solution alone in subwell 3. Crystallization conditions yielding micro-crystals were further optimized by the hanging-drop vapor-diffusion method. Suitable crystals were grown at 21 °C in drops of 2 μl, made with protein and reservoir solutions in the ratios of 1:1 and 2:3, the latter containing 1.9 m (NH4)2SO4, 100 mm sodium acetate, pH 3.6 (EndoHf-deglycosylated Ani-PME2), or 1.8 m (NH4)2SO4, 100 mm sodium acetate, pH 4.1 (PNGaseF-deglycosylated Ani-PME2). Crystals grew as clusters of very fine flattened needles, up to 0.6 mm long and 0.015 × 0.010 mm in cross-section over a period of a week. Single crystals of fully glycosylated protein have not been obtained at this stage.
Single Crystal X-ray Diffraction Data Collection, Structure Solution, and Refinement of Ani-PME2
Single crystals were briefly soaked in mother liquor containing 20% glycerol as cryoprotectant and flash-frozen in liquid nitrogen. Single crystal x-ray diffraction data to resolutions of 1.8 Å (PNGaseF-treated enzyme) and 1.75 Å (EndoHf-treated enzyme), harvested from an RAxisIV++ detector using x-rays generated by a Rigaku MM007 microfocus rotating copper-anode x-ray generator that were monochromated and focused by an AXCo PX70 capillary optic, were processed using CRYSTALCLEAR and d*TREK (87). The structure of the PNGaseF-treated enzyme was solved by molecular replacement using MrBump, (88–91) as embedded in the CCP4i suite of programs (92, 93). A composite model was based on the PMEs from tomato (Solanum lycopersicum; PDB code 1xg2) and carrot (Daucus carota, PDB code 1gq8), with side chains trimmed by CHAINSAW to a common stub shared by Ani-PME2. After refinement by means of REFMAC5 (94) of the PHASER-produced ensemble solution, Rcryst and Rfree were 0.409 and 0.445, respectively. Notwithstanding the rather high Rfree value, the electron density maps, displayed and manipulated via COOT (95), readily allowed rethreading of many loops, adjustment of rotamers for side chains common to search and target molecules, and addition of all side chains. No significant electron density before residue 29 (Ani-PME2 numbering) was observed. Several rounds of refinement and inspection of electron density maps with COOT led to additional adjustments and corrections to main chain and side chain conformations along with the addition of sulfate ions, water molecules, and glycerol molecules to complete the structural model. The validation tools of COOT were used to assess and check the structure for infelicities.
Data collection and structure refinement of the EndoHf-treated Ani-PME proceeded very similarly to the PNGaseF-treated protein. Because the structure is isomorphous to the PNGaseF-treated enzyme, the set of reflections hkl set aside for calculation of Rfree for the structure of the PNGaseF-treated enzyme was transferred to the data set for the EndoHf-treated enzyme, and additional reflections were added, because resolution of this x-ray data extended to 1.75 Å. Refinement for the latter structure began with rigid-body refinement of the model for the former structure, stripped of non-proteinaceous moieties. Salient details of data processing and structure refinement are given for both structures in Table 1. All molecular diagrams were prepared using PyMOL (Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC).
TABLE 1.
Data processing and structure refinement statistics for Ani-PME2
| PNGaseF-treated PME2 | EndoHf-treated PME2 | |
|---|---|---|
| Data collection and processing | ||
| Crystal system | Orthorhombic | Orthorhombic |
| Space group | C2221 | C2221 |
| Unit cell: a, b, c (Å) | 75.25, 113.84, 88.74 | 74.61, 113.54, 88.77 |
| Resolution range(highest shell) (Å) | 88.74–1.80 (1.86–1.80) | 62.35–1.75 (1.81–1.75) |
| No. of reflections | 32,510 | 34,821 |
| Rmerge | 0.075 (0.313) | 0.084 (0.366) |
| I/σ(I) (post-averaging) | 9.8 (3.7) | 8.3 (3.1) |
| Completeness | 95.7 (91.4) | 95.6% (89.6) |
| Redundancy | 3.79 (3.75) | 3.94 (3.71) |
| Wilson B value (Å2) | 19.4 | 19.0 |
| Structure refinement | ||
| Z′ | 1 | 1 |
| Rcryst | 0.170 | 0.171 |
| Rfree | 0.203 | 0.201 |
| Amino acid residues | 29–327 | 29–327 |
| No. of amino acid atom sites | 2280 | 2335 |
| No. of water molecules | 312 | 323 |
| Other atoms/groups | 2.25 glycerol/3.5 SO42−/1 Cl−/0.5 CH3COO− | 3 glycerol/4.5 SO42−/1 Cl−/0.5CH3COO−/1 NAG |
| Average B value: protein/water/other (Å2) | 18.9/30.4 /33.9 | 19.8/31.1/44.3 |
| Root mean square deviation bond distances (Å) | 0.0069 | 0.0082 |
| Root mean square deviation bond angles (o) | 1.26 | 1.31 |
| Ramachandran | ||
| % Favored/allowed/forbidden | 95.5/4.2/0.3 (Met276) | 95.2/4.5/0.3 (Met276) |
| PDB code | 5c1c | 5c1e |
Calculation and Comparison of PME Electrostatic Properties
The comparison of electrostatic potentials has been performed on the basis of similarity indices describing the quantitative differences of the proteins' electrostatic potentials in three-dimensional space. The protonation state of each protonatable amino acid has been adjusted to that calculated at pH 4. The pKa values for each amino acid have been calculated using the software PROPKA Version 3.0 (96). Hydrogens, atomic charges, and radii were placed using the software PDB 2PQR Version 1.9 (97) using the parameters derived by the PARSE force field, which has been optimized for calculations in implicit solvent (98). Electrostatic potentials have then been calculated by solving the linearized form of the Poisson-Boltzmann equation using the software APBS Version 1.3 (97). Similarity indices were calculated using the webserver PIPSA (99). PIPSA calculates similarity indices using Equation 1,
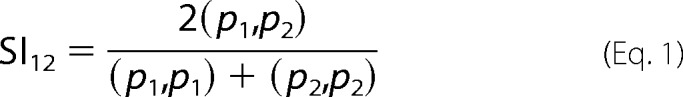 |
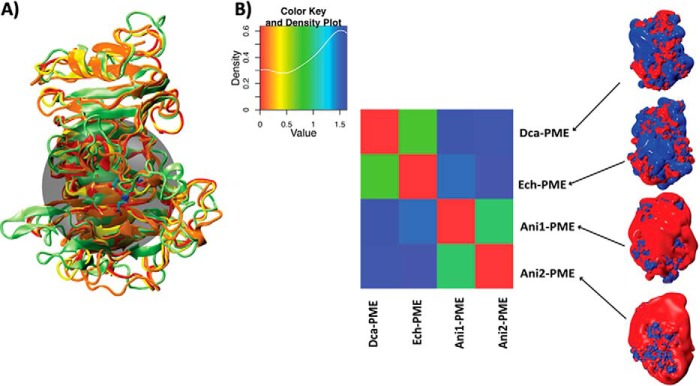
where (p1,p2), (p1,p1), and (p2,p2) are the scalar products of the electrostatic potentials calculated by discretizing the solutions of the Poisson-Boltzmann equation on a three-dimensional grid. After superimposing the structures, the electrostatic potentials have been calculated over the whole Ech-PME, Dca-PME, Ani-PME1, and Ani-PME2, but the similarity indices described here are relative to the binding groove of the enzyme, defined as the region described by a sphere (see Fig. 13A) having as its center the coordinates of Asp-199_OD2 of Ech-PME and the radius 15 Å. A matrix, reporting the differences between the electrostatic potentials of the PMEs expressed by the fourth species, was obtained and is shown in Fig. 13B.
FIGURE 13.
Comparison of the electrostatic potentials for processive and non-processive PMEs. A, structural alignment of the PMEs from D. carota (Dca-PME, orange, PDB code 1gq8), E. chrysanthemi (Ech-PME, light green, PDB code 2nt6 and 2nt9), A. niger isoform 1 (Ani-PME1, red, homology-modeled structure), and A. niger isoform 2 (Ani-PME2, yellow). The transparent black sphere, centered on atom OD2 atom of Ech-PME Asp-199 and having a radius of 15 Å, shows the region chosen for the quantitative comparison of the electrostatic potentials. B, diagonalized matrix colored according to similarity indices calculated for the comparison of the electrostatic potentials of the PMEs shown in A. The color plot and the co-respective values of the similarity index are reported in the inset above. In the right-hand panel, the electrostatic potential densities of the four analyzed PMEs are shown and colored between −1 (red) and +1 kBT/e.
Modeling of Ani-PME2·HG Complexes and Molecular Dynamics Simulations
HG decasaccharides were modeled in complex with Ani-PME2, using as starting coordinates those from two x-ray crystal structures of Ech-PME in complex with hexasaccharides (46). The two structures chosen showed the hexasaccharides docked along different subsites of the binding groove. In one case (PDB code 2nsp), the hexasaccharide occupied subsites −5 to +1 and in the other (PDB code 2nt9) −1 to +5. Coordinate merging of the two hexasaccharides and the deletion of one of the two pairs of overlapping monosaccharides (subsites −1 and +1) provided a decasaccharide spanning the entire length of the binding groove. The complex between such a decasaccharide and Ani-PME2 has been modeled through the structural alignment of Ech-PME, docked with an HG decasaccharide, and Ani-PME2. The decasaccharide selected to test for processivity was unmethyl-esterified on residues docked in the subsites −5 to +1 and methyl-esterified on residues docked in the subsites +2 to +5, denoted X5XM4, and models the product of the reaction, whereby the active site (+1) accommodates a de-methyl-esterified residue. Energy minimizations in vacuo removed steric clashes between the protein and the oligosaccharide. The preparation of the system for production of molecular dynamics runs and the analysis and visualization of the collected trajectories were performed as described previously (74). Each system was simulated for a total time of 250 ns in five independent simulations of 50 ns for which the set of initial particle velocities was different.
Results
Expression, Purification, Deglycosylation, and Activity of Ani-PME1 and Ani-PME2
The genes for Ani-PME1 and Ani-PME2 had the A. niger export tag replaced by one optimized for the expression host, Saccharomyces cerevisiae. The protein was expressed and exported into the growth medium in good yield and then readily purified. Sequence analysis revealed two potential motifs for asparagine N-linked glycosylation (see Fig. 2). To obtain fully deglycosylated protein, Ani-PME2 was treated with PNGaseF. To obtain protein bearing N-linked N-acetylglucosamine (NAG) stub(s), the protein was treated with EndoHf. The enzyme in all three forms is active. Mass spectrometry analysis of pepsin-digested fragments established that the export tag was cleaved as expected, and relative to the full-length native protein, the recovered Ani-PME2 began at residue 29. X-ray crystallographic analysis confirmed that only Asn-84 was N-glycosylated (see below).
The activity of the protein was monitored over the pH range 3–7, in the presence or absence of NaCl. Maximum activity was observed in the pH range 4.0–4.5 and with concentration of NaCl of 0.8 m. Without added NaCl, protein activity is highly reduced. At pH 7 and above in the presence of 100 mm NaCl, Ani-PME2 is nearly inactive. A similar activity profile has been established for Ani-PME1 (79, 100).
Processivity of Ani-PME1, Ani-PME2, and Csi-PME
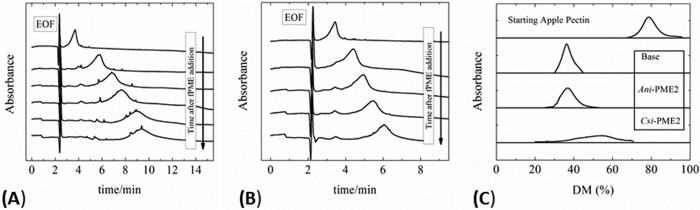
Three experimental protocols on highly methyl-esterified apple pectin establish that under conditions close to their pH optima Ani-PME2 is non-processive, whereas Csi-PME is processive as expected. In the first protocol, the de-methylesterification of apple pectin by Ani-PME2 in glycosylated and Endo-Hf-treated (N-acetylglucosamine stub) forms was followed as a function of time, as summarized in Fig. 4, A and B. In both cases, CE reveals that the Gaussian peak shape is preserved, which is indicative of random de-methylesterification of the apple pectin by the Ani-PME2 species (47). The glycan appears to have minimal effect on the resultant charge distribution and only a mild effect on the activity so that the EndoHf-treated enzyme achieves a similar extent of de-methylesterification of pectin in about two-thirds the time of the fully glycosylated Ani-PME2.
FIGURE 4.
Capillary electrophoresis of de-methyl-esterified pectin. CE results of de-methylesterification of Fluka apple pectin in 50 mm acetate buffer at pH 4.2 containing 100 mm NaCl by Ani-PME2 (glycosylated) (A) and Ani-PME2 (EndoHf-treated) (B). The pectin peak (carboxyl group absorbance at 192 nm) moves to the right as its negative charge density increases. (The large spike at around 2 min results from the electro-osmotic flow.) C, capillary electrophoresis of batch de-methyl-esterified of pectin. Pectin was de-methyl-esterified to generate substrates of similar degree (35%) of methylesterification (DM), maintaining pH at a constant value by titration with 0.10 m NaOH solution. Orange PME (Csi-PME at pH 7.5), Ani-PME2 (at pH 4.5), and base (at pH 11.5) were used to de-methyl-esterify 0.5% w/v apple pectin. The degree of methylesterification and the molecular distribution were obtained by CE.
In the second protocol, batch de-methylesterification, apple pectin samples were hydrolyzed to a constant average degree of methylesterification of ∼35%. Under strongly basic conditions (pH 11.5), apple pectin undergoes non-enzymatic, non-processive (random) de-methylesterification; the Gaussian distribution of charged forms of unmodified apple pectin is preserved upon treatment with a strong base. A Gaussian distribution is also maintained upon treatment of apple pectin by glycosylated Ani-PME2 at pH 4.5 and 100 mm NaCl, indicative of a lack of processivity. However, for apple pectin treated with Csi-PME, CE shows a broadened and unsymmetrical distribution, which is consistent with processive action by this enzyme, as illustrated in Fig. 4C.
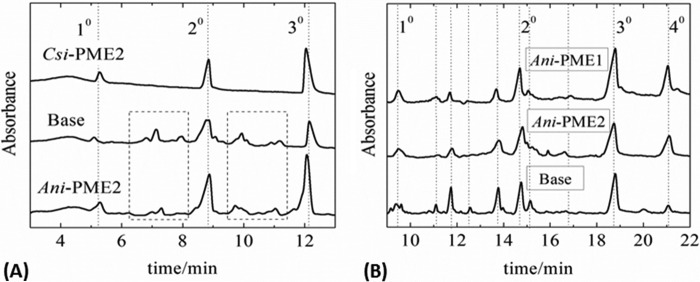
In the third protocol, the samples, each having an approximately sample-averaged 35% degree of methylesterification as prepared under the above second protocol, were digested with EndoPG II, which preferentially cleaves the glycosidic bonds of the HG chain at sites containing a de-methyl-esterified or non-methyl-esterified galacturonic acid residue. That Csi-PME is highly processive is confirmed by the presence of just three peaks in the CE chromatogram, corresponding simply to mono-, di-, and tri-galacturonic acid species (101), as shown in Fig. 5A. Moreover, non-processive (or near-random) de-methylesterification by Ani-PME2 at pH 4.2 or by strong base at pH 11.5 is confirmed by a plethora of fragments of varying length and charge, as shown in Fig. 5A (101). Although the distributions of EndoPG II-generated fragments for base-treated and Ani-PME2-treated samples are slightly different, this difference is probably attributable to imperfectly random de-methylesterification by Ani-PME2, as hinted by the slight tailing of the Gaussian distribution for the Ani-PME2-treated apple pectin, apparent on close inspection of Fig. 4C.
FIGURE 5.
Capillary electrophoresis of EndoHf-digested partly de-methyl-esterified pectin. A, CE of apple pectin samples digested with EndoPG II and run with 50 mm background electrolyte at 20 kV applied voltage. Highly methyl-esterified apple pectin has first been treated with strong base (labeled Base), Csi-PME, or Ani-PME2 to a constant degree of methylesterification of ∼35%. EndoPG II digestion of the de-methyl-esterified sample obtained by using Csi-PME produces only mono-, di-, and tri-galacturonic acid fragments of upon digestion with EndoPG II. For the base-generated and Ani-PME2-treated substrate, both of similar DM, however, many more fragments (see dotted boxes) can be identified as being partially methyl-esterified oligogalacturonides, consistent with the more random placement of the unmethyl-esterified residues by base or Ani-PME2. B, capillary electrophoresis of EndoHf-digested partly de-methyl-esterified pectin. CE of apple pectin samples digested with EndoPG II and run with 100 mm phosphate running buffer at 30 kV applied voltage. Highly methyl-esterified apple pectin was first treated with strong base (labeled Base) or glycosylated Ani-PME2, or Ani-PME1 to a constant degree of methylesterification of ∼40%. Unmethyl-esterified peaks are labeled, and additional peaks corresponding to methyl-esterified fragments appearing in all three digests are indicated by dotted lines (101).
Finally, samples of a second highly methyl-esterified apple pectin (to investigate the reproducibility of the data with a different starting substrate) that had undergone prior treatment with either strong base, or Ani-PME1, or glycosylated Ani-PME2 to a constant DM of ∼40% were then treated with EndoPG II, and the products were examined by CE (Fig. 5B) under conditions designed to give higher resolution than for the previous runs (different ionic strength of the background electrolyte and voltage). Both Ani-PME1 and Ani-PME2 share a similar lack of processivity.
Structures of Ani-PME2
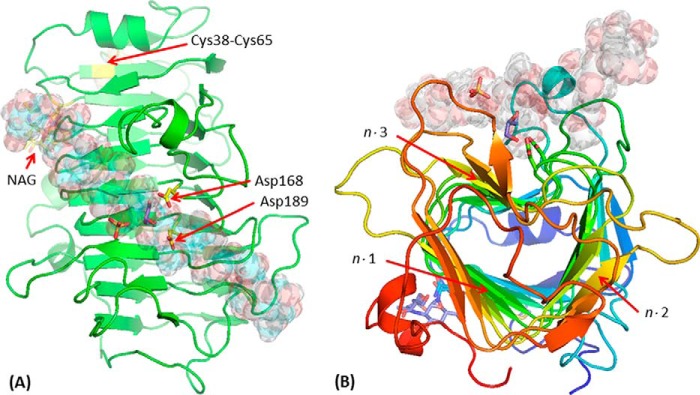
The structures of Ani-PME2 in PNGaseF-treated (fully deglycosylated) and EndoHf-treated (N-acetylglucosamine, NAG, stub) forms have been determined to 1.80 and 1.75 Å resolution, respectively. Both forms are isomorphous and monomeric. Other structurally characterized PMEs are also monomeric and all share a distinctive parallel β-helix in which three strands, with a minimum number of residues of 20, complete one turn of the helix, and the clockwise (right-handed) helix includes 10⅔ turns, although parts of the first and last turns do not have three β-strands. The entire helix undergoes a left-handed super-helical twist of ∼60°. Ignoring the extended loops, the β-helix has a somewhat triangular aspect when viewed end-on. Denoting each helical turn with the number n (n = 1,10), each β-sheet face of the three distinct faces of the helix can be denoted as n·m (m = 1,3). The putative substrate-binding groove, with reference to the Ech-PME structure with various hexasaccharide moieties bound, lies diagonally along the n·3 face. The structure of EndoHf-treated Ani-PME is shown in Fig. 6. The root-mean-square deviation between these two structures, superimposing Cα atoms by means of LSKAB, is 0.081 Å, indicating that the NAG stub, and by extrapolation the entire glycan, has essentially no influence on the core protein structure.
FIGURE 6.
Structure of EndoHf-treated Ani-PME2. The active-site Asp residues (here Asp-168 and Asp-189) are highlighted in a ball-and-stick representation (carbon atoms in yellow); the disordered NAG stub is similarly highlighted (carbon atoms in yellow). Other atoms, including those of a sulfate and a glycerol near the active site, and the disulfide bridge Cys-38–Cys-65, are in standard CPK coloring. A decasaccharide (de-methyl-esterified for residues labeled −5, −4, −3, −2, −1, and +1 and methyl-esterified for residues +2, +3, +4, and +5) is modeled in the binding groove and shown in semi-transparent form. A, view looking down on the active site. Note that the carboxylate group of the decasaccharide residue at the site −1 coincides closely with one of the sulfate anions, and a glycerol is located close to the active-site aspartate residues. B, view looking approximately down the β-helix axis of Ani-PME2, rainbow colored from the N terminus in blue to the C terminus in red. The n·m faces of the β-helix are labeled (helix turn n = 1–12; face m = 1–3).
High quality electron density is apparent for protein main chain and almost all side chains from residue 29 through to the final residue at 327. Asn-84 is revealed as the sole site of N-linked glycosylation by the following: (i) electron density for the NAG stub at Asn-84 and none at Asn-288 for EndoHf-treated Ani-PME2, and (ii) atomic displacement parameters consistent with Asp-84 (the product of hydrolysis of the Asn-linked glycan) and Asn-288.
For both structures, oxidation of a pair of exposed cysteines leads to a disulfide link between exposed Cys-38 and Cys-65. These cysteine residues are not present in plant, bacterial, or insect PMEs or in Ani-PME1.
Except for the N- and C-terminal regions, the parallel β-helix forms a remarkably rigid framework to which the substrate HG chains bind. Fig. 7 shows the relative atomic displacement parameters for the structurally characterized processive PMEs from the bacterium E. chrysanthemi and the plant D. carota (carrot) and the non-processive fungal PME, Ani-PME2, as reported here. The rigidity of the long loops surrounding the active site for Ech-PME is also observed in the molecular dynamics simulations published previously. (74) These simulations revealed that the motions of the substrate and of the loops surrounding it were correlated in the case of the substrate capable of undergoing processive de-methylesterification. In Fig. 8, the superposition of the structures of Ani-PME2, Ech-PME, Dca-PME, and rice weevil PME is shown, highlighting the very different loops among these PMEs and the highly conserved core parallel β-helix. Indeed, notwithstanding sequence identity of only ∼30%, in pairwise comparisons of this fungal PME with plant (Dca-PME, PDB code 1gq8) and bacterial (Ech-PME PDB code 1qjv) PMEs, the root-mean-square deviations by secondary structure matching with a 5-Å threshold for inclusion (102) are 1.42 Å (271 residues aligned) and 1.56 Å (259 residues aligned), respectively.
FIGURE 7.
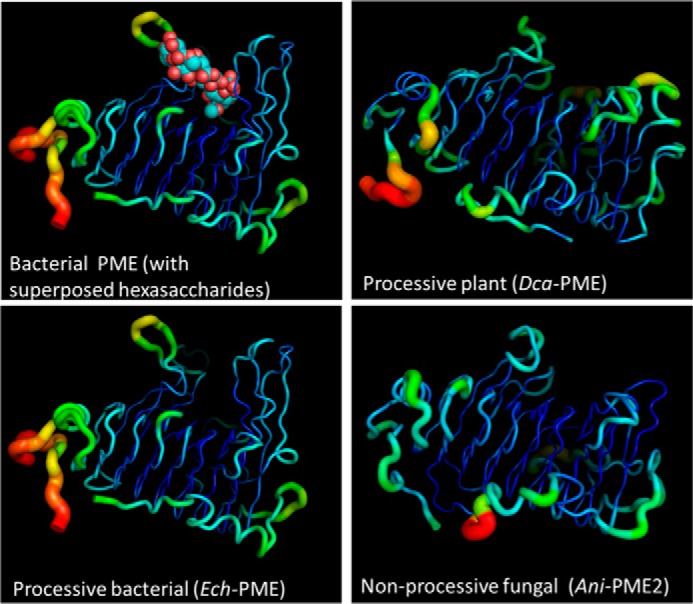
Flexibility of Ani-PME2 reported by crystallographic atomic displacement parameters (B values). “Putty” diagrams (PyMOL) of the relative atomic displacement parameters, in rainbow colors, for two processive plant and bacterial PMEs and for the non-processive fungal PME, Ani-PME2. The largest atomic displacement parameters are colored red; the smallest dark blue. The top left frame shows the decasaccharide derived from superposition of two hexasaccharides (PDB codes 2nsp and 2ntp). Note that the loops surrounding the substrate-binding groove are relatively inflexible in all structures, notwithstanding greatly varying lengths, and note the absence of bound oligosaccharide in the case of Dca-PME and Ani-PME.
FIGURE 8.
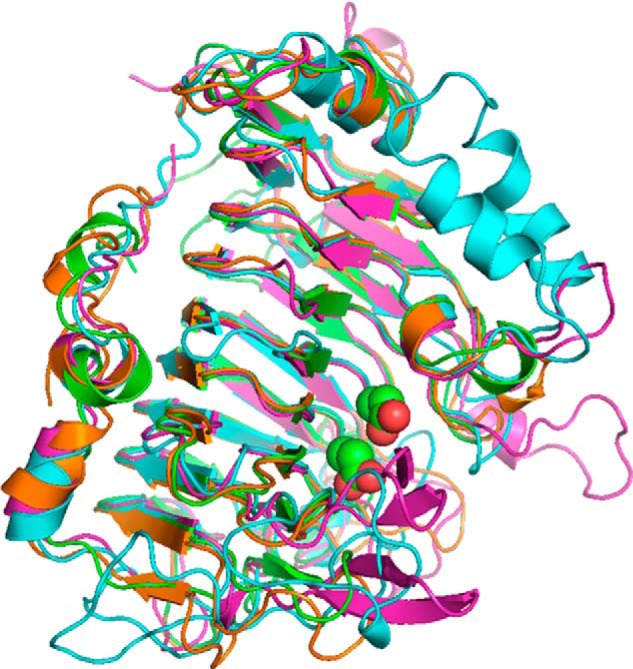
Schematic representation of the superposition of PME structures. The PME2 from A. niger is shown in green, D. carota in orange (PDB code 1gq8), rice weevil (Sitophilus oryzae) in cyan (PDB code 4pmh), and E. chrysanthemi in magenta (PDB codes 2nt6 and 2nt9). The active-site Asp residues are shown as spheres. Relative to Fig. 6A, the view here is rotated ∼90° anti-clockwise about an axis running south-north. The N and C termini of the rice weevil PME are cyan-highlighted; the others are shown in black.
Cys Ladder
For Ani-PME2, there is a distinctive quartet of buried cysteine residues, Cys-161, Cys-182, Cys-202, and Cys-231, that sit at the turn between the strands n·1 and n·2 (n = 5–8) and face internally into the core of the β-helix (Fig. 9). In both structures of Ani-PME2, these cysteines all adopt the same rotamer, and there are no disulfide linkages. For Ani-PME1, these residues alternate as Cys/Thr/Cys/Ser, and in other PME structures only one or two of these cysteines are conserved (46, 69, 71).
FIGURE 9.
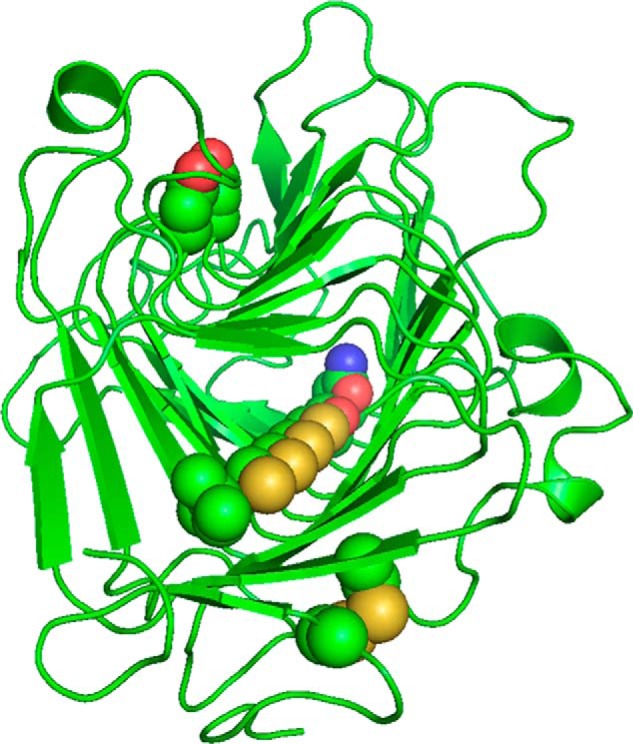
Cysteine ladder comprising Cys-161, Cys-182, Cys-202, and Cys-231. These Cys and Ser-262 share a common orientation and lie in the turn between β-helix faces n·1 and n·2. The Asn and Ile bookends are shown. Also shown are the Cys-38–Cys-64 disulfide bridge and the active-site aspartate residues, Asp-168 and Asp-189. For clarity, residues 49–57 are omitted.
Homology Modeling of the Structure of Ani-PME1
A ClustalW alignment (103) of Ani-PME1 and Ani-PME2 reveals about 40% identity (Fig. 2). Active-site residues and their immediate surroundings are conserved between these two fungal PMEs. Loop lengths are also conserved in general; the exception is the loop between strands 9·3 and 10·1, which is three residues longer for Ani-PME1 compared with Ani-PME2. Thus, the secondary and tertiary structure of Ani-PME1 is expected to resemble very closely that of Ani-PME2. Fig. 13A shows a superposition of the structures of Ani-PME1 (model), Ani-PME2, Ech-PME, and Dca-PME. Note that a rethreading of the loop and strand consisting of residues 91–115 of Ani-PME2 would place this loop for Ani-PME1 as lining the substrate-binding groove, in a manner similar to that for the plant and bacterial PME structures. Moreover, this rethreading would place a putative glycosylation motif (NIT) in a loop, rather than in a β-strand.
Molecular Dynamics Simulations on Ani-PME2
MD simulations performed on Ani-PME2 reveal a substantially different behavior compared with that previously reported for Ech-PME (74, 75). The motions of the decasaccharide featuring de-methyl-esterified residues docked at subsites −5 to +1 (X5XM4) are shown in Fig. 10 for the non-processive Ani-PME2 and the processive Ech-PME. The sliding motions of the decasaccharide along the substrate-binding groove (parallel to the z direction) are similar for Ani-PME2 and the processive Ech-PME. However, motions in the y direction (approximately across the substrate-binding groove) and especially in the x directions (away from the enzyme) are noticeably greater for Ani-PME2 than for the processive Ech-PME. Collectively, these motions, shown as root-mean-square fluctuations in Fig. 10B and as the derived diffusion coefficients in Fig. 10C, are entirely consistent with the lack of processivity for Ani-PME2 experimentally observed by capillary electrophoresis methods.
FIGURE 10.
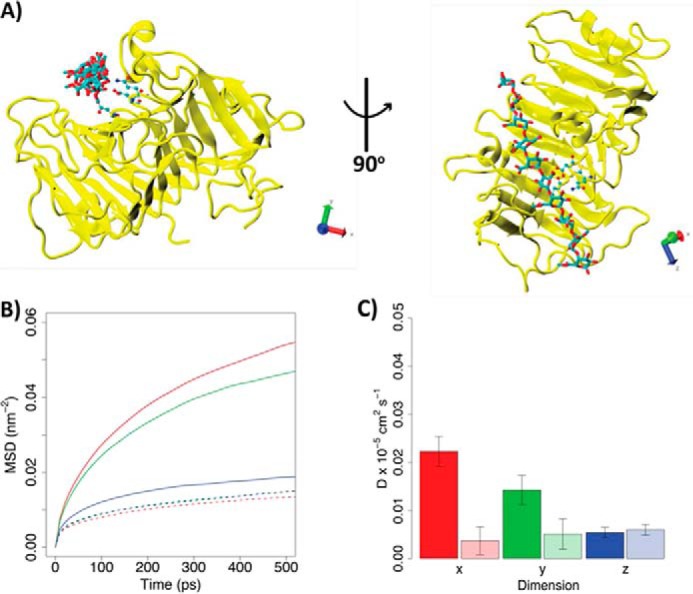
Diffusion of HG decasaccharide (X5XM4) docked into the binding grooves of processive Ani-PME2 and processive Ech-PME. A, structure of Ani-PME2 in complex with the X5XM4 decasaccharide modeled into the enzyme binding groove. Ani-PME2 is colored in yellow, and the oligosaccharide is shown in licorice and colored by atom type. The binding of X5XM4 to Ech-PME is generally similar and is shown in Ref. 74. The Cartesian axes show the alignment of the oligosaccharide along the z-dimension. B, mean-square displacements of the oligosaccharide during the first 500 ps from the start of the simulation in the x (red), y (green), and z (blue) dimensions. The solid line refers to Ani-PME2; the dotted line shows Ech-PME. The mean-square displacement in the z direction is very similar for both enzymes. C, diffusion coefficients calculated from the fitting of the mean-square displacements shown in B between 100 and 400 ps. Diffusion coefficients describe the movements of the oligosaccharide along the x (red), y (green), and z (blue) dimensions. The z-direction represents the direction of oligosaccharide sliding in the case of a processive activity. The fully colored boxes refer to Ani-PME2; the pale-shaded boxes show Ech-PME. The diffusion of the oligosaccharide is markedly greater in the x-direction, away from the substrate-binding groove, for the non-processive Ani-PME2 than for the processive Ech-PME.
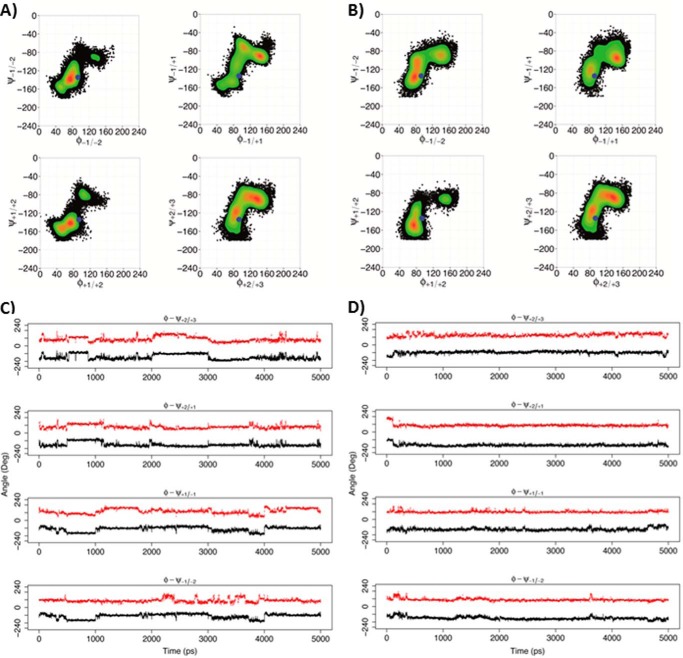
But what of the dynamic behavior of the decasaccharide X5XM4 at the level of the individual residues? The concerted rotations about the glycosidic bonds for residues in the vicinity of the active site that were observed for the processive Ech-PME2 are absent for the non-processive Ani-PME2. Fig. 11A shows a Ramachandran-like plot of the distribution of the φ/ψ torsion angles (defined in Fig. 1) for the glycosidic linkages upstream and downstream of the residue bound in the active site for the non-processive Ani-PME2 and processive Ech-PME enzymes. Fig. 11C shows the time evolution of these angles and their highly correlated changes. In contrast, for the non-processive Ani-PME2, the distribution of these φ/ψ torsion angles is noticeably different from that for Ech-PME for most of the linkages (Fig. 11B), and the absence of correlated rotations is clearly apparent in Fig. 11D.
FIGURE 11.
Rotations around the glycosidic linkages for the X5XM4 substrate. A and C, non-processive Ani-PME2. B and D, processive Ech-PME. A and B, φ/ψ dihedral angles for glycosidic linkages +2/+3, +2/+1, +1/−1, and −1/−2, where +1 is the residue X at the active site (+1). The φ and ψ angles are defined, respectively, as O5-C1-O1-C4′ and C1-O1-C4′-C3′ (see Fig. 1). The blue dots mark the starting point for the MD simulations. The scale on the right reports the relative probability of the φ/ψ values sampled during the MD simulations. C, plot of φ (red) and ψ (black) dihedral angles as a function of time for the non-processive Ani-PME2, showing uncorrelated and non-concerted rotations. D, plot of φ (red) and ψ (black) dihedral angles as a function of time for the processive Ech-PME, showing correlated and concerted rotations. The plots in B and D were obtained from the MD trajectories as published previously (74).
In our previous study, the sliding of an HG decasaccharide (X5XM4) along the binding groove of the processive Ech-PME was observed in association with a sharp drop of the enzyme-substrate electrostatic binding energy, indicating that charge neutralization between enzyme and substrate may be occurring upon sliding (75). We discuss below the implications on processivity of the very different electrostatic potential presented to the substrate by Ani-PME2 compared with that presented by plant and bacterial PMEs.
Discussion
Similarities and Differences among PME Structures
The active-site residues identified in the structures of Ech-PME, Dca-PME, and rice weevil PME are well conserved in the Ani-PME2 structure, with one exception; there is for Ani-PME2 no equivalent to Thr-109 of Ech-PME, Thr-83 of Dca-PME, and Thr-146 of rice weevil PME (Fig. 2 and Table 2). This threonine, which appears to be highly conserved in bacterial, plant, and insect PMEs, binds to substrate at the −1 and −2 sites through both main chain and side chain moieties. Moreover, the loop on which this threonine residue resides in the plant, bacterial, and insect PME structures, between strands 3·2 and 3·3, occupies a different region of space in the fungal structure. This residue appears also to be absent in the Ani-PME1 sequence.
TABLE 2.
Correspondence table for structurally conserved residues
Most functions are inferred from the structures of Ech-PME (as the D178A mutant) in complex with variously decorated hexasaccharides and from the changes in enzyme activity on mutation of selected residues (46). The PROPKA3-calculated pKa values of active-site residues are given in parentheses (96).
| Residue (structural element) | Ani-PME2 | Dca-PME | Ech-PME | Rice weevil PME | Function/comment |
|---|---|---|---|---|---|
| Thr (loop 3-2/3-3) | 83 | 109 | 146 | Substrate binding | |
| Gln (loop 4-2/4-3) | 145 | 115 | 153 | 177 | Substrate binding |
| Gln (loop 5-2/5-3) | 167 | 135 | 177 | 199 | Substrate binding/stabilization of anionic acyl intermediate |
| Asp (loop 5-2/5-3) | 168 (8.17) | 136 (9.06) | 178 (9.00) | 200 (8.51) | General acid/base |
| Asp (loop 6-2/6-3) | 189 (4.36) | 157 (4.89) | 199 (4.58) | 226 (4.39) | Positioned for nucleophilic attack on methyl ester moiety |
| Arg (strand 8-3) | 249 (13.87) | 225 (13.39) | 267 (14.70) | 290 (13.89) | Substrate binding |
| Trp (loop 8-8/9-1) | 251 | 227 | 269 | 292 | Substrate binding (H-bond of substrate to NE1) |
| Met (strand 9-3) | 276 | Trp252 | 306 | Ala332 | Dispersion interactions with substrate |
Table 3 shows the predicted intermolecular contacts between a decasaccharide and PME, based on the intermolecular contacts observed for differently patterned hexasaccharide moieties bound to Ech-PME (46). By and large these contacts appear to be well conserved between bacterial and fungal PMEs, with the exception of the Thr noted above.
TABLE 3.
Residues forming hydrogen-bonding contacts with an HG substrate for Ech-PME and their structural equivalents for Ani-PME2 and Dca-PME
Interactions are inferred from the structures of Ech-PME (as the D178A mutant) in complex with variously decorated hexasaccharides (46).
| Subsite | Ani-PME2 | Dca-PME | Ech-PME |
|---|---|---|---|
| +5 | –a | –a | Asn-226 |
| +4 | No contacts | (Lys-182, Gly-180_O)b | No contacts |
| +3 | No contacts | Gln-181c | Arg-219 |
| +2 | Arg-249, Gln-167 | Arg-255, Gln-135 | Arg-267, Gln-177 |
| +1 | Gln-167, Asp-168, Asp-189 | Gln-135, Asp-136, Asp-157 | Gln-177, Asp-178, Asp-199 |
| −1 | Gln-145, Trp-251, –, Arg-252d | Gln-115, Trp-227, Thr-83, Lys-228d | Gln-153, Trp-269, Thr-109, Thr-272_OG/N |
| −2 | –, Leu-116_N,e Arg-252d | Thr-83_N, Phe-84_N, Lys-228d | Thr-109_N(weak), Ala-110_N, Thr-272_N/O |
| −3, −4, −5 | No direct contacts | No direct contacts | No direct contacts |
a No structural equivalent or inequivalent was in the vicinity to Ech-PME (D178A) with bound hexasaccharides.
b Data were inferred from superposition; no structural equivalent or inequivalent for Ech-PME (D178A) with bound hexasaccharides was in the vicinity.
c Data were structurally inequivalent but approximately positioned at Ech-PME Arg-219.
d Data were structurally inequivalent but approximately positioned at Ech-PME Thr-272_N.
e Data were structurally inequivalent but approximately positioned at Ech-PME Ala-110_N and Dca-PME Phe-84_N.
Loops and Processivity
The β-helix forms a very rigid framework from which extends an array of loops. The substrate-binding groove, which lies diagonally along the face formed by strands n·3, is lined by five loops of variable length and mobility (see Figs. 2, 7, and 8). For Ech-PME, few changes in protein flexibility are seen upon binding hexasaccharides, except for the loop between strands 7·2 and 7·3, which decreases in mobility on substrate binding; Asn-229 from this loop makes contact with hexasaccharide residue binding at site +5. These loops are much longer for Ech-PME than for Dca-PME and Ani-PME2; moreover, several loops for Dca-PME are several residues longer than those for Ani-PME2 (Fig. 2). In the case of Ani-PME2, the loops all appear more rigid than those for the plant and bacterial PMEs, and the loop linking strands 9·3 and 10·1, although of similar length to that in the plant structure, is particularly rigid (see Fig. 7).
Nonetheless, the plant PME with loops of similar length to those for Ani-PME2 and much shorter than those of Ech-PME (Fig. 10) is processive, suggesting that factors additional to loop movements may be critical for determining processivity. Electrostatic forces, having a longer range of action with respect to other interatomic interactions, may play a crucial role considering the highly charged nature of the substrate and enzyme. To test this hypothesis, a comparison of the electrostatic properties of PMEs that have different processive abilities has been carried out.
Surface Charge and Processivity
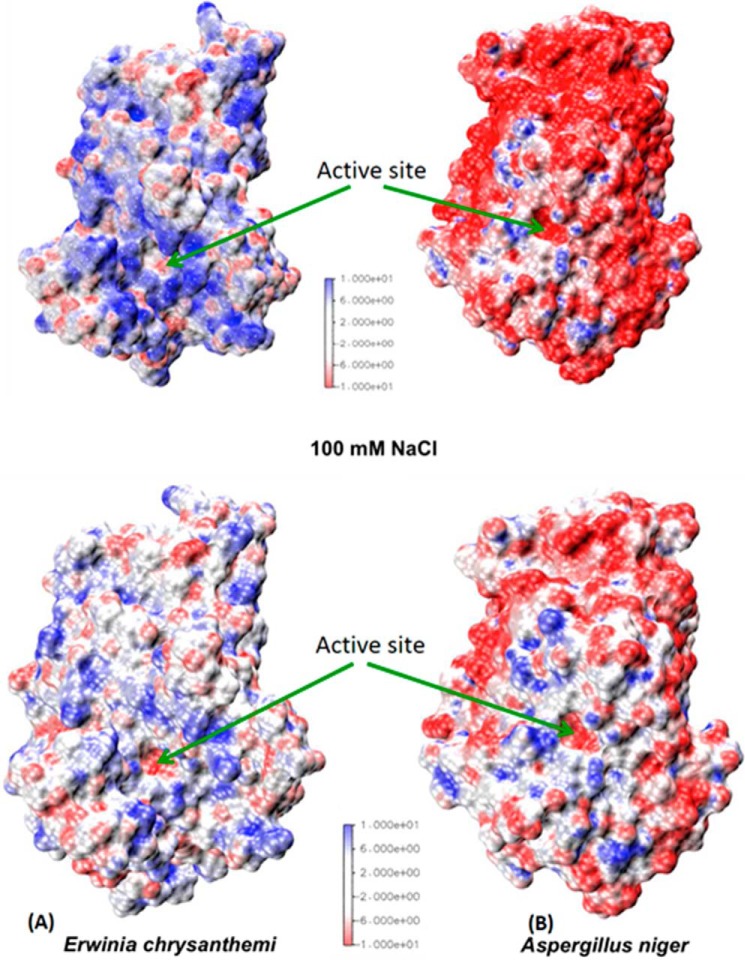
The surface electrostatic potential of Ani-PME2 (and also that predicted for Ani-PME1) is markedly different from that for plant and bacterial PMEs, especially when calculated at pH 7.4, the pH at which bacterial and plant enzymes are the most active. Whereas the plant and bacterial PMEs show a strongly positive potential (except for the active-site Asp residue), Ani-PME2 shows a very strongly negative electrostatic potential (Fig. 12). Calculations have also been performed in the presence of 100 mm NaCl, for which the screening effect substantially reduces the absolute values of the surface potentials.
FIGURE 12.
Comparison of surface electrostatic potentials at pH 7. 4. A, Ech-PME. B, Ani-PME2 at 0 mm NaCl (top frames) and 100 mm NaCl (bottom frames). The color scale is in multiples of kBT/e (T = 298.15 K). The electrostatic potential for Dca-PME is similar to that for Ech-PME.
At pH 7.4 and low salt, Ani-PME2 is only weakly active. This may be correlated with repulsion of substrate, which, unless fully methyl-esterified, is negatively charged. Lowering the pH to 4.2, the pH at which Ani-PME2 is most active, still leaves the protein surface partially negatively charged. Nonetheless, intrinsic activity of the orange PME (Csi-PME) and the fungal PMEs (in the presence of 100 mm NaCl) at their respective pH optima are similar. Thus, the positive electrostatic potential extended by the bacterial and most plant PMEs may be responsible for providing overall electrostatic attraction that retains the increasingly negatively charged substrate in the binding groove, thereby favoring processivity. In contrast, these two fungal PMEs provide an overall electrostatic repulsion, favoring dissociation of substrate and random non-processive enzymatic action. Note that galacturonic acid moieties have a pKa of ∼3.5 and hence are substantially deprotonated, when exposed, at the pH optimum of 4.2 for Ani-PME2.
Table 4 summarizes the number and type of residues that are usually charged at pH ∼7, in essence the electrostatic density of the structurally characterized plant, bacterial, insect, and fungal PMEs. The plant and fungal enzymes carry a similar number of charged residues, but the fungal enzymes carry many more aspartate and glutamate residues and many fewer lysine and arginine residues than the plant PME, leading to a deficit of −25 and −26 for the fungal enzymes compared with a surplus of +5 for the plant Dca-PME at pH 7. Although the bacterial Ech-PME carries a larger number of negatively charged residues than the fungal enzymes, it carries an even larger surplus of positively charged residues leading to a surplus of positively over negatively charged residues of +5, the same as the plant PME. Thus, compared with Dca-PME and Ani-PME2, the surface charge of Ech-PME changes more rapidly with changes in pH about its isoelectric point.
TABLE 4.
Basic and acidic residues for PMEs from A. niger, D. carota, and E. chrysanthemi
| Ani-PME2 | Ani-PME1 | Dca-PME | Ech-PME | Rice weevil PME | |
|---|---|---|---|---|---|
| Asp | 23 | 20 | 17 | 27 | 18 |
| Glu | 15 | 19 | 6 | 6 | 8 |
| Total acidic | 38 | 39 | 23 | 33 | 26 |
| Lys | 6 | 5 | 15 | 22 | 14 |
| Arg | 7 | 8 | 15 | 16 | 10 |
| Total basic | 13 | 13 | 30 | 38 | 24 |
| His | 1 | 6 | 3 | 3 | 2 |
| Total charged (excluding His) | 51 | 52 | 53 | 71 | 50 |
| Difference (no. of basic − no. of acidic) | −25 | −26 | +7 | +5 | −2 |
| Calculated pIa | 3.87 | (4.27) | 9.90 | 9.51 | 7.09 |
| Calculated charge at pH 7a | −23.50 | − | 7.61 | 11.25 | 0.16 |
| Calculated charge at pH 4a | −3.57 | − | 21.26 | 44.26 | 12.74 |
| pH optimum | 4.2–4.5 | 4.2–4.5 | ∼7.5 | ∼7.5 | ? |
| Salt sensitivity | Yes | Yes | No | No | ? |
a Data were calculated from the folded structure by PROPKA3 (96).
Taking into account the different pKa values of aspartate compared with glutamate and lysine compared with arginine, and the local structural environment of the side chains, leads to calculated values of the isoelectric point for the folded protein of 9.90 for Dca-PME compared with 9.51 for Ech-PME and for Ani-PME2 of 3.87 (96). The rice weevil PME has a similar total number of charged residues as the fungal PMEs, but the number of basic and acidic residues is nearly balanced leading to a calculated pI of 7.09. The pI calculated for unfolded PMEs is lower by ∼0.3 than those calculated for these PMEs when folded.
Focusing on electrostatics in the substrate-binding groove, we have calculated the similarity index for the electrostatic potential over the area within a 15-Å radius of the active site (see “Experimental Procedures” for details). Fig. 13B reports the diagonalized matrix of the electrostatic potential similarity index in the region of the binding groove (Fig. 13A). Notwithstanding low conservation of residues in this region (Fig. 2), Dca-PME and Ech-PME, which are both processive, show a positively charged electrostatic potential and share a high degree of similarity in their electrostatic potentials in the region that binds HG oligomers, as a result of conservative substitutions near, but not at, the structurally equivalent sites. In contrast, the two fungal PMEs show an overall negative potential, with Ani-PME1 showing a stronger negative density in this region than Ani-PME2.
Among the structurally characterized PMEs, only one positively charged residue lining the substrate-binding groove is conserved, Arg-249 in Ani-PME2 on strand 8·3, which is part of the conserved LGRPW motif of PMEs. For Ech-PME, eight positively charged side chains line the substrate-binding groove, congregated toward the end that favors binding of negatively charged galacturonate residues. However, the plant and fungal PMEs, Dca-PME and Ani-PME2, have just three positively charged residues lining this groove. At the end favoring binding of methyl-esterified groups, several aspartate residues are found for Ech-PME that are not conserved in plant and fungal structures. For Ani-PME2, six carboxylic acid residues, mostly glutamic acid and hence partly protonated at the pH optimum of ∼4.2, also line the putative substrate-binding groove as follows: Glu-142, Asp-194, Glu-218, Glu-243, Glu-244, and Asp-278.
Provided that a minimal suite of positive charges are lining the substrate-binding groove, enzyme activity does not appear to be influenced by the presence of negatively charged residues elsewhere on the surface. However, an overall negative charge along the binding groove appears inimical to processivity in PMEs characterized to date.
Focusing now on the active site, the nearly buried active-site aspartate, Asp-168 for Ani-PME2, has an anomalously high PROPKA3-calculated pKa value of 8.16 (96). This value is nonetheless smaller than the very high values calculated for Dca-PME, Ech-PME, and rice weevil PME (respectively, 9.06, 9.00, and 8.51), not inconsistent with the lower pH optimum for Ani-PME2 (Table 2). This then allows Asp-168 of Ani-PME2 (and the equivalent residues for Dca-PME, Ech-PME, and rice weevil PME) to function as a general acid/base. The other active-site aspartate residue, the nucleophile Asp-189 for Ani-PME2, and the structurally equivalent residues for plant and bacterial PMEs (Table 2) all have slightly elevated pKa values of ∼1.5–2.0 greater than that of a free aspartate side chain (pKa of 3.80). The putative substrate-binding arginine, Arg-249 in the case of Ani-PME2, which forms a strong salt bridge with Asp-189, also shows elevated pKa values of >14 (reference point of 12.50).
On the Nature of Processivity
Factors determining processivity are clearly subtle. The presence of a deep binding groove alone is insufficient to confer processivity. Indeed, the long loops of bacterial and insect PMEs may serve to deter the binding of PME inhibitors. Moreover, favorable electrostatics by themselves do not determine processivity. The role of surface electrostatic potential, explored in part in our molecular dynamics simulations following the evolution of the structure of the enzyme and substrate subsequent to the in silico de-methylesterification event, appears to be a key factor in determining processivity, overriding specific interactions of substrate and enzyme, which are largely preserved among the various PMEs studied (Tables 2 and 3). The degree of processivity is highly sensitive to the strengths of substrate and product binding, with respect to thermal energy, that is to the Boltzmann factor exp(−E/RT). A continuum of processivity is therefore to be expected. Indeed, it is remarkable that only vestiges of processivity are apparent in the de-methylesterification by Ani-PME2 (and Ani-PME1) relative to that by a strong base, given the high concentration of base with respect to substrate (and PME concentration for enzymatic attack) and the vastly greater mutual diffusion of base and pectin compared with pectin and PME. Thus, the chances of PME rebinding to the same pectin molecule and binding to a site adjacent to that just de-methyl-esterified are better than random.
In the case of Ani-PME2, the pI and pH of optimum activity approximately coincide. This is true for many other PMEs, both those with a pH optimum that is acidic and those with a pH optimum that that is near-neutral to basic. However, for the jelly fig and T. reesei PMEs, the pH of optimum activity (∼4.5) and the calculated pI differ by more than 4 log units. Moreover, both proteins are processive. Processivity is therefore not correlated with pH, but at this point it does appear to be correlated with pI.
Biological Implications
One intriguing feature of Ani-PME1 and -PME2 is that the pH optimum for activity is mildly acidic, and a moderate ionic strength (100 mm NaCl) considerably enhances activity. Both A. niger and E. chrysanthemi are plant pathogens. Whereas E. chrysanthemi attacks the vascular tissue of plants, in particular seeking access to the xylem, and operates intracellularly, A. niger appears to attack fruiting bodies, seeds and bulbs, and operates extracellularly. Although most fruits are acidic and hence a PME operating at acidic pH, such as Ani-PME2, is optimal for pectin hydrolysis, seeds and bulbs generally have a pH value close to neutral. A 31-kDa PME (one of three) from the fungus Fusarium asiaticum has been found to have a pH optimum of 6.5 (104), and there is a third putative PME for A. niger that awaits characterization.
Conclusions
The first structures of a fungal, acidophilic, halophilic PME, Ani-PME2, have been determined in fully deglycosylated and N-acetylglucosamine-stub forms. The enzyme has been shown, by using capillary electrophoresis, to observe pectin de-methylesterification and the subsequent methylesterification state of the EndoPG II digests to be non-processive. Molecular dynamics simulations of this enzyme in complex with a partly de-methyl-esterified decasaccharide representing the putative transition state immediately after de-methylesterification independently suggest that Ani-PME2 is non-processive, with uncorrelated rotations of the decasaccharide residues and large diffusion coefficients for movement of the substrate away from the active site. In so doing, these simulations provide further validation, as a negative control, of our previous MD work, which revealed that the mechanism of processivity in Ech-PME involves correlated rotations of residues about the glycosidic linkages before sliding of the reoriented substrate into position for the next round of de-methylesterification.
PMEs are ubiquitous in plants and are widely distributed across animal, fungal, and bacterial domains. Generalities prominent in the literature concerning the mode of action (processive/non-processive) with respect to pH, salt dependence, and source of PME may need to be revisited as exceptions to these generalities are revealed.
Author Contributions
L. M. K., T. S. L., D. M., M. A. K. W., and G. B. J. carried out the experiments. M. A. K. W., G. B. J., and D. M., analyzed the results. L. D. M., M. A. K. W., D. M., and G. B. J. conceived the study and devised the experiments. G. B. J. wrote the paper with substantial contributions from all co-authors. All authors approved this version of the manuscript.
Acknowledgments
We thank Brett Savary for fruitful and enlightening discussions and Gill Norris for general support of this project.
This work was supported by the Riddet Institute, a Centre of Research Excellence. All authors reviewed the results and approved the final version of the manuscript.
The atomic coordinates and structure factors (codes 5c1c and 5c1e) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PME
- pectin methylesterase
- BGE
- background electrolyte
- CE
- capillary electrophoresis
- DM
- degree of methylesterification, percentage of methyl-esterified galacturonic acid residues in HG chains of pectin
- EndoHf
- endoglycosidase H fused to maltose-binding protein
- EndoPG II
- endopolygalacturonase II from A. niger
- HG
- homogalacturonan
- NAG
- N-acetylglucosamine
- PNGaseF
- peptide-N4-(N-acetyl-β-glucosaminyl)asparagine amidase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PDB
- Protein Data Bank.
References
- 1.Atmodjo M. A., Hao Z., and Mohnen D. (2013) Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 64, 747–779 [DOI] [PubMed] [Google Scholar]
- 2.Caffall K. H., and Mohnen D. (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344, 1879–1900 [DOI] [PubMed] [Google Scholar]
- 3.Mohnen D. (2008) Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11, 266–277 [DOI] [PubMed] [Google Scholar]
- 4.Markovic O., and Janecek S. (2004) Pectin methylesterases: sequence-structural features and phylogenetic relationships. Carbohydr. Res. 339, 2281–2295 [DOI] [PubMed] [Google Scholar]
- 5.Albersheim P., Darvill A., Roberts K., Sederoff R., and Staehelin A. (2010) Plant Cell Walls: From Chemistry to Biology, Garland Science, New York [Google Scholar]
- 6.Hongo S., Sato K., Yokoyama R., and Nishitani K. (2012) Demethylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the Arabidopsis stem. Plant Cell 24, 2624–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jolie R. P., Duvetter T., Van Loey A. M., and Hendrickx M. E. (2010) Pectin methylesterase and its proteinaceous inhibitor: a review. Carbohydr. Res. 345, 2583–2595 [DOI] [PubMed] [Google Scholar]
- 8.Pelloux J., Rustérucci C., and Mellerowicz E. J. (2007) New insights into pectin methylesterase structure and function. Trends Plant Sci. 12, 267–277 [DOI] [PubMed] [Google Scholar]
- 9.Liu Q., Talbot M., and Llewellyn D. J. (2013) Pectin methylesterase and pectin remodelling differ in the fibre walls of two gossypium species with very different fibre properties. PLoS ONE 8, e65131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang M., Yuan D., Gao W., Li Y., Tan J., and Zhang X. (2013) A comparative genome analysis of PME and PMEI families reveals the evolution of pectin metabolism in plant cell walls. PLoS ONE 8, e72082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J., Shiu S. H., Thoma S., Li W. H., and Patterson S. E. (2006) Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 7, R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogawa M., Kay P., Wilson S., and Swain S. M. (2009) Arabidopsis dehiscence zone polygalacturonase1 (ADPG1), ADPG2, and quartet2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Ovidio R., Mattei B., Roberti S., and Bellincampi D. (2004) Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant-pathogen interactions. Biochim. Biophys. Acta 1696, 237–244 [DOI] [PubMed] [Google Scholar]
- 14.De Lorenzo G., and Ferrari S. (2002) Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr. Opin. Plant Biol. 5, 295–299 [DOI] [PubMed] [Google Scholar]
- 15.Lionetti V., Cervone F., and Bellincampi D. (2012) Methylesterification of pectin plays a role during plant-pathogen interactions and affects plant resistance to diseases. J. Plant Physiol. 169, 1623–1630 [DOI] [PubMed] [Google Scholar]
- 16.Lionetti V., Francocci F., Ferrari S., Volpi C., Bellincampi D., Galletti R., D'Ovidio R., De Lorenzo G., and Cervone F. (2010) Engineering the cell wall by reducing de-methyl-esterified homogalacturonan improves saccharification of plant tissues for bioconversion. Proc. Natl. Acad. Sci. U.S.A. 107, 616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li P., Feng B., Wang H., Tooley P. W., and Zhang X. (2011) Isolation of nine Phytophthora capsici pectin methylesterase genes which are differentially expressed in various plant species. J. Basic Microbiol. 51, 61–70 [DOI] [PubMed] [Google Scholar]
- 18.Abo-El-Dahab M. K. (1964) Production of pectic and cellulolytic enzymes by Xanthomonas malvacearum. Phytopathology 54, 597–601 [Google Scholar]
- 19.Jia Y. J., Feng B. Z., Sun W. X., and Zhang X. G. (2009) Polygalacturonase, pectate lyase and pectin methylesterase activity in pathogenic strains of Phytophthora capsici incubated under different conditions. J. Phytopathol. 157, 585–591 [Google Scholar]
- 20.Knoesel D. (1967) Occurrence and activity of pectolytic and cellulolytic enzymes in phytopathogenic bacteria. Z. Pflanzenkr. Pflanzenschutz 74, 150–155 [Google Scholar]
- 21.Marei G. I., Abdel R. M., and Abdelgaleil S. A. (2012) Comparative antifungal activities and biochemical effects of monoterpenes on plant pathogenic fungi. Pestic. Biochem. Physiol. 103, 56–61 [Google Scholar]
- 22.Ortega J. (1998) Production of pectolytic enzymes by Curvularia senegalensis, a phytopathogenic fungus. Texas J. Sci. 50, 327–332 [Google Scholar]
- 23.Zaki M. M., Mahmoud S. A., and Fawzi F. G. (1979) Enzymic activities of soft rot bacteria in relation to pathogenesis. Egypt. J. Microbiol. 13, 1–7 [Google Scholar]
- 24.Khanh N. Q., Ruttkowski E., Leidinger K., Albrecht H., and Gottschalk M. (1991) Characterization and expression of a genomic pectin methyl esterase-encoding gene in Aspergillus niger. Gene 106, 71–77 [DOI] [PubMed] [Google Scholar]
- 25.Benoit I., Coutinho P. M., Schols H. A., Gerlach J. P., Henrissat B., and de Vries R. P. (2012) Degradation of different pectins by fungi: correlations and contrasts between the pectinolytic enzyme sets identified in genomes and the growth on pectins of different origin. BMC Genomics 13, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boccara M., and Chatain V. (1989) Regulation and role in pathogenicity of Erwinia chrysanthemi 3937 pectin methylesterase. J. Bacteriol. 171, 4085–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waterhouse R. M., Tegenfeldt F., Li J., Zdobnov E. M., and Kriventseva E. V. (2013) OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 41, D358–D365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen Z., Pappan K., Mutti N. S., He Q.-J., Denton M., Zhang Y., Kanost M. R., Reese J. C., and Reeck G. R. (2005) Pectin methylesterase from the rice weevil, Sitophilus oryzae: cDNA isolation and sequencing, genetic origin, and expression of the recombinant enzyme. J. Insect. Sci. 5, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keeling C. I., Henderson H., Li M., Yuen M., Clark E. L., Fraser J. D., Huber D. P., Liao N. Y., Docking T. R., Birol I., Chan S. K., Taylor G. A., Palmquist D., Jones S. J., and Bohlmann J. (2012) Transcriptome and full-length cDNA resources for the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major insect pest of pine forests. Insect Biochem. Mol. Biol. 42, 525–536 [DOI] [PubMed] [Google Scholar]
- 30.Pauchet Y., Wilkinson P., Chauhan R., and Ffrench-Constant R. H. (2010) Diversity of beetle genes encoding novel plant cell wall degrading enzymes. PLoS ONE 5, e15635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martens E. C., Lowe E. C., Chiang H., Pudlo N. A., Wu M., McNulty N. P., Abbott D. W., Henrissat B., Gilbert H. J., Bolam D. N., and Gordon J. I. (2011) Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLos Biol. 9, e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salamanca G., Rodríguez R., Quiralte J., Moreno C., Pascual C. Y., Barber D., and Villalba M. (2010) Pectin methylesterases of pollen tissue, a major allergen in olive tree. FEBS J. 277, 2729–2739 [DOI] [PubMed] [Google Scholar]
- 33.Jimenez-Lopez J. C., Kotchoni S. O., Rodríguez-García M. I., and Alché J. D. (2012) Structure and functional features of olive pollen pectin methylesterase using homology modeling and molecular docking methods. J. Mol. Model. 18, 4965–4984 [DOI] [PubMed] [Google Scholar]
- 34.Tanhatan-Nasseri A., Crepeau M.-J., Thibault J.-F., and Ralet M.-C. (2011) Isolation and characterization of model homogalacturonans of tailored methylesterification patterns. Carbohydr. Polym. 86, 1236–1243 [Google Scholar]
- 35.Cameron R. G., Luzio G. A., Goodner K., and Williams M. A. K. (2008) Demethylation of a model homogalacturonan with a salt-independent pectin methylesterase from citrus: I. Effect of pH on demethylated block size, block number and enzyme mode of action. Carbohydr. Polym. 71, 287–299 [Google Scholar]
- 36.Cameron R. G., Luzio G. A., Kim Y., Vasu P., Savary B. J., and Williams M. A. K. (2011) Characterization of nanostructural modifications introduced into a model pectic homogalacturonan by esterases or chemical saponification and modeling of enzyme mode of action. NSTI-Nanotech 3, 275–278 [Google Scholar]
- 37.Dedeurwaerder S., Menu-Bouaouiche L., Mareck A., Lerouge P., and Guerineau F. (2009) Activity of an atypical Arabidopsis thaliana pectin methylesterase. Planta 229, 311–321 [DOI] [PubMed] [Google Scholar]
- 38.Dongowski G., and Bock W. (1984) Effects of molecular parameters of pectin substrates on the activities of pectinesterases from Aspergillus niger and from higher plants. Nahrung 28, 507–516 [Google Scholar]
- 39.Kim Y., Cameron R. G., Luzio G. A., Vasu P., Savary B. J., and Willams M. A. (2011) 242nd ACS National Meeting & Exposition, Denver, CO, August 28 to September 1, 2011, ACS Meeting Abstr. AGFD-102 [Google Scholar]
- 40.Kim Y., Williams M. A. K., Galant A. L., Luzio G. A., Savary B. J., Vasu P., and Cameron R. G. (2013) Nanostructural modification of a model homogalacturonan with a novel pectin methylesterase: effects of pH on nanostructure, enzyme mode of action and substrate functionality. Food Hydrocolloids 33, 132–141 [Google Scholar]
- 41.Lee H., Rivner J., Urbauer J. L., Garti N., and Wicker L. (2008) De-esterification pattern of Valencia orange pectin methylesterases and characterization of modified pectins. J. Sci. Food Agric. 88, 2102–2110 [Google Scholar]
- 42.Limberg G., Korner R., Buchholt H. C., Christensen T. M., Roepstorff P., and Mikkelsen J. D. (2000) Analysis of different de-esterification mechanisms for pectin by enzymatic fingerprinting using endopectin lyase and endopolygalacturonase II from A. niger. Carbohydr. Res. 327, 293–307 [DOI] [PubMed] [Google Scholar]
- 43.Luzio G. A., and Cameron R. G. (2008) Demethylation of a model homogalacturonan with the salt-independent pectin methylesterase from citrus: part II. Structure-function analysis. Carbohydr. Polym. 71, 300–309 [Google Scholar]
- 44.Markovic O., Cederlund E., Griffiths W. J., Lipka T., and Jörnvall H. (2002) Characterization of carrot pectin methylesterase. Cell. Mol. Life Sci. 59, 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ralet M.-C., Williams M. A. K., Tanhatan-Nasseri A., Ropartz D., Quéméner B., and Bonnin E. (2012) Innovative enzymatic approach to resolve homogalacturonans based on their methylesterification pattern. Biomacromolecules 13, 1615–1624 [DOI] [PubMed] [Google Scholar]
- 46.Fries M., Ihrig J., Brocklehurst K., Shevchik V. E., and Pickersgill R. W. (2007) Molecular basis of the activity of the phytopathogen pectin methylesterase. EMBO J. 26, 3879–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams M. A. K., Cucheval A., Nasseri A. T., and Ralet M.-C. (2010) Extracting intramolecular sequence information from intermolecular distributions: highly nonrandom methyl ester substitution patterns in homogalacturonans generated by pectin methylesterase. Biomacromolecules 11, 1667–1675 [DOI] [PubMed] [Google Scholar]
- 48.Savary B. J., Hotchkiss A. T., and Cameron R. G. (2002) Characterization of a salt-independent pectin methylesterase purified from Valencia orange peel. J. Agric. Food Chem. 50, 3553–3558 [DOI] [PubMed] [Google Scholar]
- 49.Cameron R. G., Savary B. J., Hotchkiss A. T., Fishman M. L., Chau H. K., Baker R. A., and Grohmann K. (2003) Separation and characterization of a salt-dependent pectin methylesterase from Citrus sinensis var. Valencia fruit tissue. J. Agric. Food Chem. 51, 2070–2075 [DOI] [PubMed] [Google Scholar]
- 50.Markovic O., Jarosova A., Machova E., and Slezarik A. (1996) Effect of oligomeric and polymeric d-galacturonans on the activity of pectinesterases from tomato, Aspergillus foetidus, and Trichoderma reesei. Biologia (Bratislava) 51, 293–297 [Google Scholar]
- 51.Cameron R. G., Luzio G. A., Vasu P., Savary B. J., and Williams M. A. K. (2011) Enzymatic modification of a model homogalacturonan with the thermally tolerant pectin methylesterase from Citrus: 1. Nanostructural characterization, enzyme mode of action, and effect of pH. J. Agric. Food Chem. 59, 2717–2724 [DOI] [PubMed] [Google Scholar]
- 52.Duvetter T., Fraeye I., Sila D. N., Verlent I., Smout C., Hendrickx M., and Van Loey A. (2006) Mode of de-esterification of alkaline and acidic pectin methylesterases at different pH conditions. J. Agric. Food. Chem. 54, 7825–7831 [DOI] [PubMed] [Google Scholar]
- 53.Catoire L., Pierron M., Morvan C., du Penhoat C. H., and Goldberg R. (1998) Investigation of the action patterns of pectin methylesterase isoforms through kinetic analyses and NMR spectroscopy. Implications in cell wall expansion. J. Biol. Chem. 273, 33150–332156 [DOI] [PubMed] [Google Scholar]
- 54.Goldberg R., Pierron M., Bordenave M., Breton C., Morvan C., and du Penhoat C. H. (2001) Control of mung bean pectin methylesterase isoform activities. Influence of pH and carboxyl group distribution along the pectic chains. J. Biol. Chem. 276, 8841–8847 [DOI] [PubMed] [Google Scholar]
- 55.Williams M. A. K., Cucheval A., Ström A., and Ralet M.-C. (2009) Electrophoretic behavior of copolymeric galacturonans, including comments on the information content of the intermolecular charge distribution. Biomacromolecules 10, 1523–1531 [DOI] [PubMed] [Google Scholar]
- 56.Denès J. M., Baron A., Renard C. M., Péan C., and Drilleau J. F. (2000) Different action patterns for apple pectin methylesterase at pH 7.0 and 4.5. Carbohydr. Res. 327, 385–393 [DOI] [PubMed] [Google Scholar]
- 57.Christensen T. M., Nielsen J. E., Kreiberg J. D., Rasmussen P., and Mikkelsen J. D. (1998) Pectin methyl esterase from orange fruit. Characterization and localization by in situ hybridization and immunohistochemistry. Planta 206, 493–503 [DOI] [PubMed] [Google Scholar]
- 58.Gaffe J., Tieman D. M., and Handa A. K. (1994) Pectin methylesterase isoforms in tomato (Lycopersicon esculentum) tissues (effects of expression of a pectin methylesterase antisense gene). Plant Physiol. 105, 199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warrilow A. G., and Jones M. G. (1995) Different forms of tomato pectinesterase have different kinetic properties. Phytochemistry 39, 277–282 [Google Scholar]
- 60.Versteeg C., Rombouts F. M., and Pilnik W. (1978) Purification and some characteristics of two pectinesterase isoenzymes from oranges. Lebensm.-Wiss. Technol. 11, 267–274 [Google Scholar]
- 61.Hsiao E. S., Chen J. C., Tsai H.-Y., Khoo K.-H., Chen S.-T., and Tzen J. T. (2009) Determination of N-glycosylation site and glycan structures of pectin methylesterase in jelly fig (Ficus awkeotsang) Achenes. J. Agric. Food Chem. 57, 6757–6763 [DOI] [PubMed] [Google Scholar]
- 62.Lin T. P., Liu C. C., Chen S. W., and Wang W. Y. (1989) Purification and characterization of pectin methylesterase from Ficus awkeotsang Makino Achenes. Plant Physiol. 91, 1445–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller J. W., Vasu P., and Savary B. J. (2011) in 241st ACS National Meeting and Exposition, Anaheim, CA, ACS Meeting Abstr. AGFD20 [Google Scholar]
- 64.Markovic O., and Kohn R. (1984) Mode of pectin deesterification by Trichoderma reesei pectinesterase. Experientia 40, 842–843 [Google Scholar]
- 65.Ralet M.-C., and Thibault J.-F. (2002) Interchain heterogeneity of enzymatically deesterified lime pectins. Biomacromolecules 3, 917–925 [DOI] [PubMed] [Google Scholar]
- 66.Videcoq P., Garnier C., Robert P., and Bonnin E. (2011) Influence of calcium on pectin methylesterase behaviour in the presence of medium methylated pectins. Carbohydr. Polym. 86, 1657–1664 [Google Scholar]
- 67.Kester H. C., Benen J. A., Visser J., Warren M. E., Orlando R., Bergmann C., Magaud D., Anker D., and Doutheau A. (2000) Tandem mass spectrometric analysis of Aspergillus niger pectin methylesterase: mode of action on fully methyl-esterified oligogalacturonates. Biochem. J. 346, 469–474 [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson J. D., Gibson T. J., and Higgins D. G. (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics Chapter 2, Unit 2.3 [DOI] [PubMed] [Google Scholar]
- 69.Johansson K., El-Ahmad M., Friemann R., Jörnvall H., Markovic O., and Eklund H. (2002) Crystal structure of plant pectin methylesterase. FEBS Lett. 514, 243–249 [DOI] [PubMed] [Google Scholar]
- 70.Di Matteo A., Giovane A., Raiola A., Camardella L., Bonivento D., De Lorenzo G., Cervone F., Bellincampi D., and Tsernoglou D. (2005) Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 17, 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jenkins J., Mayans O., Smith D., Worboys K., and Pickersgill R. W. (2001) Three-dimensional structure of Erwinia chrysanthemi pectin methylesterase reveals a novel esterase active site. J. Mol. Biol. 305, 951–960 [DOI] [PubMed] [Google Scholar]
- 72.Boraston A. B., and Abbott D. W. (2012) Structure of a pectin methylesterase from Yersinia enterocolitica. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 129–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teller D. C., Behnke C. A., Pappan K., Shen Z., Reese J. C., Reeck G. R., and Stenkamp R. E. (2014) The structure of rice weevil pectin methylesterase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 70, 1480–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mercadante D., Melton L. D., Jameson G. B., Williams M. A. K., and De Simone A. (2013) Substrate dynamics in enzyme action: rotations of monosaccharide subunits in the binding groove are essential for pectin methylesterase processivity. Biophys. J. 104, 1731–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mercadante D., Melton L. D., Jameson G. B., and Williams M. A. K. (2014) Processive pectin methylesterases: the role of electrostatic potential, breathing motions and bond cleavage in the rectification of Brownian motions. PLoS ONE 9, e87581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martens-Uzunova E. S., and Schaap P. J. (2009) Assessment of the pectin degrading enzyme network of Aspergillus niger by functional genomics. Fungal Genet. Biol. 46, S170–S179 [DOI] [PubMed] [Google Scholar]
- 77.Warren M. E., Kester H., Benen J., Colangelo J., Visser J., Bergmann C., and Orlando R. (2002) Studies on the glycosylation of wild-type and mutant forms of Aspergillus niger pectin methylesterase. Carbohydr. Res. 337, 803–812 [DOI] [PubMed] [Google Scholar]
- 78.van Alebeek G. J., van Scherpenzeel K., Beldman G., Schols H. A., and Voragen A. G. (2003) Partially esterified oligogalacturonides are the preferred substrates for pectin methylesterase of Aspergillus niger. Biochem. J. 372, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang X., Chen P., Yin M., and Yang Q. (2013) Constitutive expression, purification and characterisation of pectin methylesterase from Aspergillus niger in Pichia pastoris for potential application in the fruit juice industry. J. Sci. Food Agric. 93, 375–381 [DOI] [PubMed] [Google Scholar]
- 80.Duwe B., and Khanh N. Q. (1996) Site-directed mutagenesis of the active site of pectin methylesterase from Aspergillus niger RH5344. Biotechnol. Lett. 18, 621–626 [Google Scholar]
- 81.Loo T., Patchett M. L., Norris G. E., and Lott J. S. (2002) Using secretion to solve a solubility problem: high yield expression in Escherichia coli and purification of the bacterial glycoamidase PNGase F. Protein Expr. Purif. 24, 90–98 [DOI] [PubMed] [Google Scholar]
- 82.Shevchenko A., Tomas H., Havlis J., Olsen J. V., and Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 83.Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., and Aebersold R. (2010) A guided tour of the trans-proteomic pipeline. Proteomics 10, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rodriguez H., Snyder M., Uhlén M., Andrews P., Beavis R., Borchers C., Chalkley R. J., Cho S. Y., Cottingham K., Dunn M., Dylag T., Edgar R., Hare P., Heck A. J., Hirsch R. F., et al. (2009) Recommendations from the 2008 International Summit on Proteomics Data Release and Sharing Policy: the Amsterdam principles. J. Proteome Res. 8, 3689–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Williams B., Ralet M., and Cameron R. (2010) in Pacifichem 2010, International Chemical Congress of Pacific Basin Societies, Honolulu, HI, Abstr. BIOL-241 [Google Scholar]
- 86.Kester H. C., Magaud D., Roy C., Anker D., Doutheau A., Shevchik V., Hugouvieux-Cotte-Pattat N., Benen J. A., and Visser J. (1999) Performance of selected microbial pectinases on synthetic monomethyl-esterified di- and trigalacturonates. J. Biol. Chem. 274, 37053–37059 [DOI] [PubMed] [Google Scholar]
- 87.Pflugrath J. W. (1999) The finer things in x-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 88.Keegan R. M., and Winn M. D. (2008) MrBUMP: an automated pipeline for molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 64, 119–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stein N. (2008) CHAINSAW: a program for mutating PDB files used as templates in molecular replacement. J. Appl. Crystallogr. 41, 641–643 [Google Scholar]
- 90.Vagin A., and Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 91.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 93.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Emsley P., and Cowtan K. (2004) Coot: model-rebuilding tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 96.Olsson M. H., Sondergaard C. R., Rostkowski M., and Jensen J. H. (2011) PROPKA3: consistent treatment of internal and surface residues in empirical pKa Predictions. J. Chem. Theory Comput. 7, 525–537 [DOI] [PubMed] [Google Scholar]
- 97.Unni S., Huang Y., Hanson R. M., Tobias M., Krishnan S., Li W. W., Nielsen J. E., and Baker N. A. (2011) Web servers and services for electrostatics calculations with APBS and PDB2PQR. J. Comput. Chem. 32, 1488–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sitkoff D., Sharp K. A., and Honig B. (1994) Accurate calculation of hydration free energies using macroscopic solvent models. J. Phys. Chem. 98, 1978–1988 [Google Scholar]
- 99.Richter S., Wenzel A., Stein M., Gabdoulline R. R., and Wade R. C. (2008) webPIPSA: a web server for the comparison of protein interaction properties. Nucleic Acids Res. 36, W276–W280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maldonado M. C., Strasser D. S., and Callieri D. (1994) Purification and characterization of pectinesterase produced by a strain of Aspergillus niger. Curr. Microbiol. 28, 193–196 [Google Scholar]
- 101.Ström A., and Williams M. A. K. (2004) On the separation, detection and quantification of pectin derived oligosaccharides by capillary electrophoresis. Carbohydr. Res. 339, 1711–1716 [DOI] [PubMed] [Google Scholar]
- 102.Krissinel E., and Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 103.Thompson J. D., Higgins D. G., and Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Glinka E. M., and Liao Y. C. (2011) Purification and partial characterisation of pectin methylesterase produced by Fusarium asiaticum. Fungal Biol. 115, 1112–1121 [DOI] [PubMed] [Google Scholar]