Abstract
Cockayne syndrome (CS) is a recessive disorder that results in deficiencies in transcription-coupled nucleotide excision repair (TC-NER), a subpathway of nucleotide excision repair, and cells from CS patients exhibit hypersensitivity to UV light. CS group B protein (CSB), which is the gene product of one of the genes responsible for CS, belongs to the SWI2/SNF2 DNA-dependent ATPase family and has an ATPase domain and an ubiquitin-binding domain (UBD) in the central region and the C-terminal region, respectively. The C-terminal region containing the UBD is essential for the functions of CSB. In this study, we generated several CSB deletion mutants and analyzed the functions of the C-terminal region of CSB in TC-NER. Not only the UBD but also the C-terminal 30-amino acid residues were required for UV light resistance and TC-NER. This region was needed for the interaction of CSB with RNA polymerase II, the translocation of CS group A protein to the nuclear matrix, and the association of CSB with chromatin after UV irradiation. CSB was modified by small ubiquitin-like modifier 2/3 in a UV light-dependent manner. This modification was abolished in a CSB mutant lacking the C-terminal 30 amino acid residues. However, the substitution of lysine residues in this region with arginine did not affect SUMOylation or TC-NER. By contrast, substitution of a lysine residue in the N-terminal region with arginine decreased SUMOylation and resulted in cells with defects in TC-NER. These results indicate that both the most C-terminal region and SUMOylation are important for the functions of CSB in TC-NER.
Keywords: chromatin, DNA damage, nucleotide excision repair, protein domain, small ubiquitin-like modifier (SUMO), Cockayne syndrome, transcription-coupled repair
Introduction
Nucleotide excision repair (NER)4 is a versatile DNA repair pathway that removes bulky, helix-distorting lesions, including UV light-induced cyclobutane pyrimidine dimers and pyrimidine-pyrimidone (6–4) photoproducts as well as chemical carcinogen-induced lesions (1). There are two subpathways in NER: global genome NER and transcription-coupled NER (TC-NER). Global genome NER operates throughout the entire genome. On the other hand, TC-NER specifically removes lesions from the transcribed strands of actively transcribed genes (2, 3). In TC-NER, RNA polymerase II (Pol II) stalled at a lesion on the transcribed strand is thought to serve as a damage recognition signal.
Cockayne syndrome (CS) is an autosomal recessive disorder that results in deficiencies in TC-NER. The major clinical features of CS are photosensitivity, growth failure, a progressive neurodevelopmental disorder, and premature aging (4). These features appear within a few years after birth, and most CS patients die before they reach adulthood. Cells from CS patients exhibit hypersensitivity to UV light and a reduction in the recovery of RNA synthesis after UV irradiation (5, 6). There are two genetic complementation groups in CS, namely, CS-A and CS-B (7, 8), and the causative genes are CSA (9) and CSB (10), respectively.
CSB is a multifunctional protein that works in transcription and TC-NER (11). CSB consists of 1493 amino acid residues and belongs to the SWI2/SNF2 DNA-dependent ATPase family (10). CSB has an ATPase domain in the central region (Fig. 1A). ATP hydrolysis is essential for chromatin remodeling after UV irradiation (12, 13). CSB interacts with Pol II (14–16), and the catalytic activity of CSB is hypothesized to be involved in remodeling of the Pol II-DNA interface (17). In the C-terminal region, CSB has an ubiquitin-binding domain (UBD), and this is essential for CSB function in TC-NER (18). CSB1–1220, which lacks the C-terminal 273 amino acid residues containing the UBD, has a similar level of ATPase activity as WT CSB and can interact with Pol II after UV irradiation. However, cells expressing CSB1–1220 exhibit hypersensitivity to UV light and reduced recovery of RNA synthesis after UV irradiation (18). CSB is a key factor in the recruitment of other repair factors to stalled Pol II (19). In addition, CSB is involved in the translocation of CSA to the nuclear matrix (20, 21). UV-sensitive syndrome (UVSS) is another TC-NER-deficient disorder. CSB is remarkably degraded by the proteasome after UV irradiation in UVSS group A (UVSS-A) cells (22–25). UVSSA, the product of the UVSS-A causative gene, forms a complex with USP7 (22, 23) and interacts with CSB and Pol II in a CSA- and UV light-dependent manner (22, 25).
FIGURE 1.
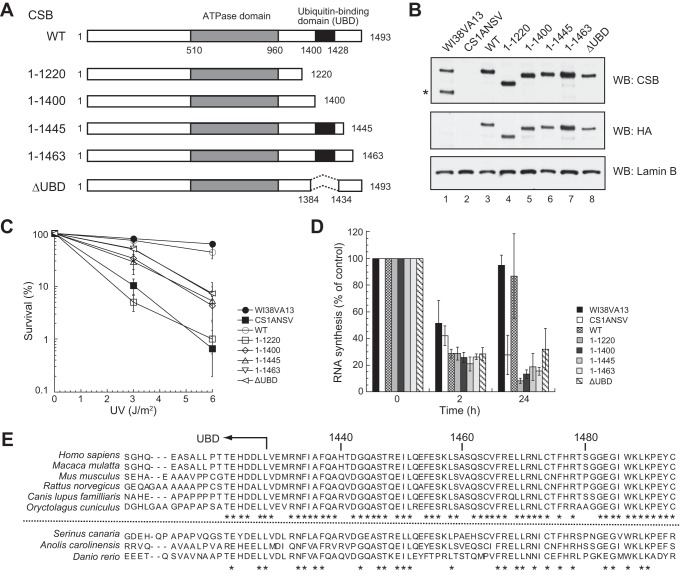
The C-terminal region of CSB is essential for TC-NER. A, schematic of the CSB mutants. The ATPase domain and UBD are indicated by the dark gray and black boxes, respectively. B, expression levels of CSB mutants. Each CSB mutant was tagged with FLAG and HA epitopes at the N terminus and expressed in CS1ANSV cells. Whole cell lysates were analyzed by Western blotting (WB) with anti-CSB and anti-HA antibodies. Lamin B is a loading control. The asterisk denotes the CPFP protein (a fusion protein consisting of N-terminal CSB1–465 and the piggyback transposon (30)). C, colony-forming ability of cells expressing each CSB mutant after UV irradiation. The points are the average of at least three independent experiments, and vertical bars indicate mean ± S.E. D, RNA synthesis of cells expressing each CSB mutant after UV irradiation. Cells were irradiated with 10 J/m2 UV light, and incorporation of [3H]uridine was measured 2 and 24 h later. The relative incorporation of [3H]uridine in UV-irradiated cells was compared with that in non-irradiated cells. The points are the average of at least three independent experiments, and vertical bars indicate mean ± S.E. E, alignment of amino acid sequences of CSB homologs in vertebrates. Sequences after 1410 amino acid residues are shown. Sequence information was obtained from the NCBI web site. The accession numbers are NP_000115.1 (Homo sapiens), AFH31457.1 (Macaca mulatta), AAI32448.1 (Mus musculus), NP_001100766.1 (Rattus norvegicus), XP_005637540.1 (Canis lupus familiaris), XP_008268053 (Oryctolagus cuniculus), XP_009085572 (Serinus canaria), XP_008115961.1 (Anolis carolinensis), and XP_688972 (Danio rerio). The asterisks denote conserved amino acids among species.
Posttranslational modifications play important roles in the functions of CSB (26). It has been reported that CSB is ubiquitinated and degraded in a UV light- and CSA-dependent manner (27). However, another study has reported that CSB is ubiquitinated by BRCA1 even in the absence of CSA (28). Therefore, it is unclear which ubiquitin ligase is responsible for the ubiquitination of CSB. Not only ubiquitination but also deubiquitination of CSB by UVSSA-USP7 might be essential for the progression of TC-NER (22, 23). There are at least eight candidate phosphorylation sites in CSB (26). CSB is phosphorylated by c-Abl kinase, and this modification is relevant to the subcellular localization of CSB (29).
Here we showed that the most C-terminal region of CSB affects its function in TC-NER and the UV light sensitivity of cells. In addition, we revealed that CSB is modified by small ubiquitin-like modifier (SUMO)-2/3 in a UV light-dependent manner and that the most C-terminal region is related to this modification. Moreover, the amino acid substitution of Lys-205 with Arg in CSB suppressed SUMOylation and resulted in a lack of TC-NER in cells. These results indicate the importance of the C-terminal region and SUMOylation of CSB in TC-NER.
Experimental Procedures
Expression Constructs and Stable Cell Lines
To generate epitope-tagged CSB expression constructs, WT and mutant CSB cDNA fragments were amplified by PCR and cut with XhoI at the 5′ end and with XbaI at the 3′ end. The fragments were cloned in-frame and downstream of the sequence encoding the FLAG epitope, followed by the HA epitope in pcDNA3.1 (Invitrogen). The CSB2K→R, CSB3K→R, and CSBK205R cDNAs were generated using the QuikChange II-E site-directed mutagenesis kit (Agilent Technologies) according to the instructions of the manufacturer. DNA sequencing of the plasmids ruled out the presence of PCR-derived mistakes. An expression construct for C-terminal FLAG- and HA-tagged WT CSB was also generated. CSB-FLAG-HA cDNA was cut out of pCAGGS-CSB-FLAG-HA (30) using XhoI and NotI and inserted into pcDNA3.1.
To isolate stable transfectants, CS1ANSV cells were transfected with the CSB expression constructs using Effectene transfection regent (Qiagen) according to the procedure of the manufacturer. Stable transfectants were selected in the presence of 500 μg/ml G418 (Nacalai Tesque).
Cell Culture
The cell lines used in this study were SV40 immortalized human fibroblasts and WI38VA13 (normal), CS1ANSV (CS-B), Kps3 (UVSS-A), CS3BESV (CS-A), and CS1ANSV cells stably expressing WT CSB or each CSB mutant. Kps3 cells stably expressing FLAG-HA-UVSSA and CS3BESV cells stably expressing CSA-FLAG-HA have been generated previously (21, 22). All cell lines were cultured in DMEM containing 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in an incubator containing 5% CO2.
UV Irradiation
Cells were washed once with PBS and irradiated with the indicated dose of UV-C light. Culture medium was added immediately after UV irradiation. For the UV light survival assay, 1.0 × 103 cells were seeded into 10-cm dishes and incubated overnight. After UV irradiation, cells were cultured for 10 days, fixed with 3% formaldehyde prepared in PBS, and stained with 2.5 mm crystal violet solution. Colonies were counted using a stereo microscope.
Preparation of Whole Cell Extracts
Cells (1.0 × 106) were washed once with PBS and lysed with 100 μl of SDS-PAGE sample buffer (62.5 mm Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 0.01% bromphenol blue, and 2.5% mercaptoethanol) by boiling for 5 min.
Recovery of RNA Synthesis after UV Irradiation
Cells were inoculated into 35-mm dishes at a density of 1.0 × 105 cells/dish and incubated overnight. Cells were washed once with PBS, irradiated with 10 J/m2 UV-C light or not irradiated, and incubated in 1 ml of culture medium. To measure RNA synthesis, [3H]uridine (PerkinElmer Life Sciences) was added to each dish to a concentration of 370 kBq/ml at each time point. After incubation for 1 h, labeling was terminated by adding NaN3 to a final concentration of 200 μg/ml. Cells were solubilized with 0.8% SDS. Then an equal volume of 10% TCA containing 0.1 m sodium diphosphate was added to the lysates and incubated on ice. Acid-insoluble material was collected on glass microfiber filters (Whatman GF/C). Radioactivity was measured in Insta-Fluor Plus mixture (PerkinElmer Life Sciences) with a liquid scintillation counter (PerkinElmer Life Sciences Tri-Carb 2810TR). The ratio of the radioactivity of UV-irradiated cells to that of non-irradiated cells was considered to reflect the recovery of RNA synthesis after UV irradiation.
Interaction of CSB with Pol II
After irradiation with 20 J/m2 UV light, cells were incubated for 30 min and harvested. In total, 1.0 × 106 cells were resuspended in micrococcal nuclease buffer (20 mm Tris-HCl (pH 7.5), 100 mm KCl, 2 mm MgCl2, 1 mm CaCl2, 300 mm sucrose, 0.1% Triton X-100, 1 mm DTT, and protease inhibitors) and incubated on ice for 10 min. Lysates were fractionated by centrifugation (5000 rpm, 5 min, 4 °C). Pellets were resuspended in micrococcal nuclease buffer containing 15 units/ml micrococcal nuclease and incubated at 25 °C for 15 min. The reaction was terminated by adding EDTA to a final concentration of 5 mm and centrifugation at 5000 rpm for 5 min at 4 °C. The supernatants were recovered. The pellets were washed with micrococcal nuclease buffer, and the supernatants were combined with the aforementioned supernatants and used as the solubilized chromatin fractions. The solubilized chromatin fractions were incubated with 20 μl of anti-FLAG M2-agarose beads (Sigma) overnight at 4 °C. The beads were washed five times with NETN buffer (50 mm Tris-HCl (pH 7.8), 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, and 0.5 mm PMSF) and then boiled in an equal volume of 2× SDS-PAGE sample buffer (125 mm Tris-HCl (pH 6.8), 4% SDS, 20% glycerol, 0.02% bromphenol blue, and 5% mercaptoethanol). SDS-PAGE was performed, and separated proteins were transferred to a PVDF membrane. Immunoblot detection was performed with anti-Pol II and anti-CSB antibodies using Pierce Western blotting Substrate Plus (Thermo Scientific).
Subcellular Fractionation
After irradiation with 20 J/m2 UV light, cells were harvested at each time point. In total, 1.0 × 106 (Fig. 2) or 4.0 × 106 (Fig. 4) cells were resuspended in CSK-Triton buffer (10 mm PIPES (pH 6.8), 100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 0.5% Triton X-100, 1 mm EGTA, 1 mm DTT, and protease inhibitors) with gentle vortexing and incubated on ice for 10 min. The lysates were fractionated by centrifugation (6000 rpm, 3 min, 4 °C). The supernatants were mixed with an equal volume of 2× SDS-PAGE sample buffer, boiled for 5 min, and used as the soluble fractions. The pellets were washed once with CSK-Triton buffer, boiled in 1× SDS-PAGE sample buffer, and used as the insoluble fractions.
FIGURE 2.
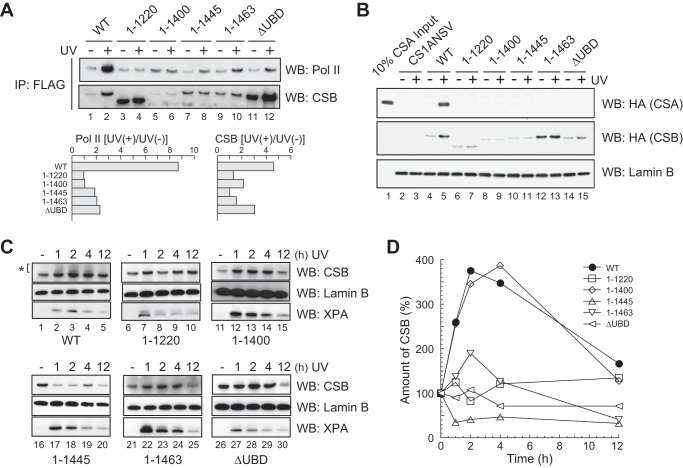
Effects of the deletion in CSB on TC-NER. A, interaction of CSB with Pol II. Top panel, solubilized chromatin fractions were prepared from UV-irradiated and non-irradiated cells, and CSB was immunoprecipitated with anti-FLAG-agarose. Immunoprecipitated samples were analyzed by Western blotting (WB) using anti-Pol II and anti-CSB antibodies. Bottom panel, the intensities of Pol II and CSB bands were quantified using ImageQuant TL software (GE Healthcare). The intensities in UV-irradiated cells wer compared with those in non-irradiated cells. B, UV-induced translocation of CSA to the nuclear matrix. CSK-ppt fractions were prepared from UV-irradiated and non-irradiated cells expressing CSB mutants, incubated with CSK-sup fraction containing FLAG-HA epitope-tagged CSA, and treated with DNase I. CSA retained in DNase I-insoluble fractions was detected by Western blotting with an anti-HA antibody. Lamin B is a loading control. C, association of CSB with chromatin. Insoluble fractions were prepared from cells at the indicated times after UV irradiation and analyzed by Western blotting with an anti-CSB antibody. Samples from the same number of cells were applied to each lane of the gel. Lamin B is a loading control. XPA is associated with chromatin after UV irradiation. The asterisk represents shifted bands of CSB. D, quantification of CSB bands in C. The intensities of CSB bands were quantified using ImageQuant TL software. The intensity in UV-irradiated cells was compared with that in non-irradiated cells. Points are the averages of two independent experiments.
FIGURE 4.
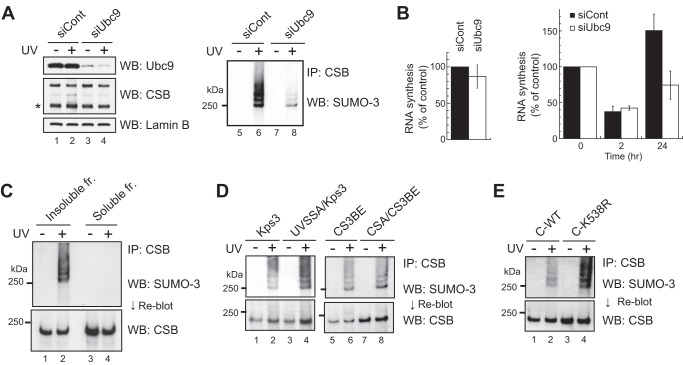
Analysis of UV-induced SUMOylation of CSB. A, effects of Ubc9 knockdown by siRNA. Left panel, WI38VA13 cells were transfected with siRNA against Ubc9 or control siRNA. Whole cell lysates were analyzed by Western blotting (WB) with anti-Ubc9 and anti-CSB antibodies. Lamin B is a loading control. The asterisk denotes the CPFP protein. Right panel, immunoprecipitated (IP) CSB was analyzed by Western blotting with an anti-SUMO-3 antibody. B, left panel, effects of Ubc9 knockdown on RNA synthesis. Incorporation of [3H]uridine was measured in knockdown cells. The points are the average of at least three independent experiments, and vertical bars indicate mean ± S.E. Right panel, effects of Ubc9 knockdown on the recovery of RNA synthesis after UV irradiation. Knockdown cells were irradiated with 10 J/m2 UV light, and incorporation of [3H]uridine was measured 2 and 24 h later. The incorporation of [3H]uridine in UV-irradiated cells was compared with that in non-irradiated cells. The points are the average of at least three independent experiments, and vertical bars indicate mean ± S.E. C, subcellular localization of SUMOylated CSB. Soluble and insoluble fractions were prepared with CSK-Triton buffer. CSB was immunoprecipitated from the fractions and analyzed by Western blotting with an anti-SUMO-3 antibody. After detection of SUMO-3, the membrane was reblotted with an anti-CSB antibody. D, SUMO-3 modification of CSB in UVSSA- and CSA-deficient cells. Kps3 and CS3BE cells are deficient in UVSSA and CSA, respectively. CSB was immunoprecipitated from lysates of these cells and of cells transfected with the responsible cDNA and analyzed by Western blotting with an anti-SUMO-3 antibody. After detection of SUMO-3, the membrane was reblotted with an anti-CSB antibody. E, SUMO-3 modification of ATPase-defective CSB. Lysates were prepared from cells expressing C-terminally FLAG-HA epitope-tagged CSAWT and CSAK538R. CSB was immunoprecipitated from the lysates and analyzed by Western blotting with an anti-SUMO-3 antibody. After detection of SUMO-3, the membrane was reblotted with an anti-CSB antibody. All samples were prepared from the same number of cells.
Toranslocation of CSA to the Nuclear Matrix after UV Irradiation
UV light-induced translocation of CSA in a cell-free system was examined as described previously (21). CS1ANSV cells expressing CSB were irradiated with 20 J/m2 UV light, incubated for 1 h, and then treated with CSK-Triton buffer to prepare the insoluble fractions (CSK-ppt fraction). The soluble fractions (CSK-sup fraction) were prepared from CS3BESV cells stably expressing CSA-FLAG-HA by treatment with CSK-Triton buffer. The CSK-sup fraction containing CSA-FLAG-HA was incubated with the CSK-ppt fraction on ice for 1 h. After centrifugation (6000 rpm, 3 min, 4 °C), the pellet was washed twice with CSK-Triton buffer, incubated with 0.1 units/μl DNase I (Takara) prepared in CSK-Triton buffer at 30 °C for 10 min, and centrifuged (6000 rpm, 3 min, 4 °C). After washing three times with CSK-Triton buffer, 1× SDS-PAGE sample buffer was added to the pellet and boiled for 5 min. CSA retained in the DNase I-insoluble fractions was detected by immunoblotting with an anti-HA antibody.
Detection of SUMOylated Proteins
SUMOylated proteins were detected as described previously (31) with some modifications. After irradiation with 20 J/m2 UV light, cells were incubated for 1 h and harvested. In total, 2.0 × 106 cells were solubilized by adding SDS lysis buffer (62.5 mm Tris-HCl (pH 6.8), 10% glycerol, 2% SDS, and protease inhibitors) and boiling for 10 min. The lysates were passed through a 25-gauge needle to shear DNA and diluted 10-fold with radioimmune precipitation assay buffer (50 mm Tris-HCl (pH 7.4), 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, and 1 mm EDTA) supplemented with 20 mm N-ethylmaleimide, 20 mm PR-619 (Life Sensors), 1 mm sodium fluoride, 1 mm sodium orthovanadate (V), and 2 mm β-glycerophosphate immediately before use. The diluted lysates were incubated with 20 μl of protein G-Sepharose 4 Fast Flow (GE Healthcare) and 2 μg of an anti-CSB antibody overnight at 4 °C. The beads were washed five times with radioimmune precipitation assay buffer and then boiled in an equal volume of 2× SDS-PAGE sample buffer. SDS-PAGE was performed, and separated proteins were transferred to a PVDF membrane. Immunoblot detection was performed with anti-ubiquitin, anti-SUMO 1, 2, and 3, and anti-CSB antibodies using Pierce Western blotting Substrate Plus.
RNA Interference
Ubc9 silencing was performed by transfection of siRNA (Santa Cruz Biotechnology, catalog no. sc-36773) using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the procedure of the manufacturer. Reverse transfection was performed first, and forward transfection was performed 24 h later. Cells were irradiated with UV light 24–48 h after forward transfection, incubated for the indicated amount of time, and harvested.
Antibodies
Polyclonal anti-CSB (catalog no. sc-10459), anti-SUMO-1 (catalog no. sc-9060), anti-ubiquitin (catalog no. sc-9133), anti-lamin B (catalog no. sc-6216), anti-CSA (catalog no. sc-10997), anti-XPA (catalog no. sc-853), and anti-Ubc9 (catalog no. sc-10759) antibodies and monoclonal anti-SUMO-3 (catalog no. sc-130884) and anti-Pol II (catalog no. sc-17798) antibodies were from Santa Cruz Biotechnology. A polyclonal anti-SUMO-2 antibody was from GeneTex. Monoclonal anti-ubiquitin (catalog no. D058-3) and anti-DDDDK (FLAG) tag (catalog no. M185-3) antibodies were from MBL. A monoclonal anti-HA tag (catalog no. 11867423001) antibody was from Roche.
Results
UV Sensitivity and Recovery of RNA Synthesis after UV Irradiation of Cells Expressing Mutant CSB Proteins with Deletions of the C-terminal Region
To analyze the functions of the C-terminal region of CSB in TC-NER, we generated several C-terminally truncated CSB mutants, including CSB1–1220 (Fig. 1A). CSB1–1400 had a deletion from just the N terminus of the UBD to the C terminus. CSB1–1445 and CSB1–1463 contained the UBD, and CSB1–1463 lacked only the C-terminal 30 amino acid residues. There is no report of the functional domains and amino acid residues in the region corresponding to amino acid residues 1221–1400, 1446–1463, and 1464–1493 of CSB, except for target sites for phosphorylation (Ser-1348 (32) and Ser-1461 (33)) and ubiquitination (Lys-1457 (34)). CSBΔUBD lacked 49 amino acids residues (1385–1433) containing the UBD. All CSB mutants have two nuclear localization signals (amino acid residues 466–481 and 1038–1055 (10)). In addition to the expression constructs for the CSB mutants, an expression construct for WT CSB (CSBWT) was also generated. All CSB proteins had FLAG and HA epitope tags at their N termini. These epitope-tagged CSBs were stably expressed in CS1ANSV cells. CS1ANSV cells are derived from a CS-B patient, theoretically only express short CSB (35) and do not have functional CSB. Immunoblot analysis using anti-CSB and anti-HA antibodies was performed to examine the expression of CSB in each transfectant (Fig. 1B). Expression levels of CSB in the transfectants were similar to those in normal (WI38VA13) cells. Transfectants expressing each CSB protein were designated CSBWT cells, CSB1–1220 cells, CSB1–1400 cells, CSB1–1445 cells, CSB1–1463 cells, and CSBΔUBD cells for convenience.
First, the colony-forming ability of UV light-irradiated transfectants was measured (Fig. 1C). CSBWT cells exhibited almost the same level of UV light sensitivity as normal (WI38VA13) cells. CSB1–1220 cells showed UV light hypersensitivity comparable with that of CS1ANSV cells, as described previously (18). Cells expressing other deletion mutants were more sensitive to UV irradiation than CSBWT cells but were less sensitive to UV irradiation than CS1ANSV cells regardless of whether the CSB mutant protein contained the UBD. Therefore, all cell lines expressing CSB deletion mutants exhibited hypersensitivity to UV irradiation.
Next, the recovery of RNA synthesis after UV irradiation, which is an index of TC-NER, was measured. RNA synthesis was decreased 2 h after UV irradiation, but, in WI38VA13 and CSBWT cells, recovered to the level of non-irradiated cells 24 h after UV irradiation (Fig. 1D). No such recovery was observed in CS1ANSV cells. All cell lines expressing CSB deletion mutants had little ability to recover RNA synthesis after UV irradiation. CSB1–1220 cells showed the most severe reduction in RNA synthesis. On the other hand, the reduction in RNA synthesis in CSBΔUBD cells was slightly less than that in cells expressing other deletion mutants but was almost the same as that in CS1ANSV cells. Regardless, all cell lines expressing CSB deletion mutants were deficient in TC-NER. These results indicate that not only the UBD but also the C-terminal region downstream of the UBD (1446–1493) affected TC-NER. Even deletion of only the C-terminal 30 amino acid residues had the same effect.
Therefore, we compared the C-terminal amino acid sequences of CSB homologs (Fig. 1E). Approximately 75% of amino acid residues in the C-terminal region (1446–1493) are identical among six mammals (Fig. 1E, top). At non-conserved amino acid residues, most residues are substituted for similar residues. Amino acid residues in this region of other animals are relatively conserved (Fig. 1E, bottom; ∼40% identical). In the frog (Xenopus tropicalis) CSB homolog and Rad26 (Saccharomyces cerevisiae CSB homolog), there are no amino acid residues corresponding to this region.
The Effects of C-terminal Deletion of CSB on the Function in TC-NER
Because CSB interacts with Pol II in a UV light-dependent manner (14–16), we examined whether the mutant CSB proteins interact with Pol II after UV irradiation. N-terminal FLAG-HA epitope-tagged CSB was immunoprecipitated from solubilized chromatin fractions using anti-FLAG-agarose, and Western blotting was performed with an anti-Pol II antibody (Fig. 2A). The amount of Pol II co-precipitated with CSBWT increased after UV irradiation (Fig. 2A, lanes 1 and 2), whereas the amount co-precipitated with CSB1–1220 and CSB1–1400 was the same before and after UV irradiation (Fig. 2A, lanes 3–6). The amount of Pol II co-precipitated with CSB1–1445, CSB1–1463, and CSBΔUBD increased slightly after UV irradiation (Fig. 2A, lanes 7–12). These results indicate that both the UBD and C-terminal region of CSB are involved in the UV-induced interaction with Pol II.
CSB is also needed for the translocation of CSA to the nuclear matrix after UV irradiation (20, 21). This translocation is relevant to TC-NER. Therefore, we next investigated the ability of each CSB mutant to translocate CSA to the nuclear matrix using a cell-free system (21). Mutant CSB cells were irradiated with UV light or not irradiated and incubated for 1 h. CSK-ppt fractions were prepared from the cells. CSK-sup fractions were prepared from CS-A (CS3BESV) cells expressing FLAG and HA epitope-tagged CSA. This fraction contained FLAG-HA-tagged CSA. Each CSK-ppt fraction was incubated with the CSK-sup fraction. After DNase I treatment, the CSK-ppt fractions were analyzed by Western blotting with an anti-HA antibody (Fig. 2B). When CSA translocation occurs, HA-tagged CSA is detected in a UV light-dependent manner. As reported previously, CSA was detected when the CSK-ppt fraction was prepared from CSBWT cells after UV irradiation (Fig. 2B, lanes 4 and 5) but was not detected when CS1ANSV cells were used (Fig. 2B, lanes 2 and 3). When the CSK-ppt fractions were prepared from mutant CSB cells, CSA was not detected (Fig. 2B, lanes 6–15). These results indicate that the UBD and C-terminal region of CSB are required for the translocation of CSA to the nuclear matrix.
Association of the Mutant CSB Proteins with Chromatin after UV Irradiation
TC-NER factors are thought to associate with chromatin for progression of TC-NER. Therefore, we examined the association of mutant CSB proteins with chromatin. Insoluble fractions containing chromatin were prepared from mutant CSB cells after UV irradiation and analyzed by Western blotting using an anti-CSB antibody (Fig. 2C). More CSBWT was detected in the insoluble fractions after UV irradiation. The intensities of the bands were quantified and plotted (Fig. 2D). The amount of CSBWT in the insoluble fraction increased until 2 h after UV irradiation and decreased to the level of non-irradiated cells by 12 h after UV irradiation. The level of CSB1–1400 in the insoluble fraction also increased in a UV light-dependent manner. The levels of CSB1–1220 and CSB1–1463 in the insoluble fraction increased slightly until 2 h after UV irradiation and then decreased. The level of CSBΔUBD in the insoluble fraction did not increase after UV irradiation. The amount of CSB1–1445 in the insoluble fraction was decreased by UV irradiation. These data show that the association of CSB with chromatin after UV irradiation was affected by deletion of the UBD or C-terminal region.
UV-dependent Posttranslational Modification of CSB by SUMO-2/3
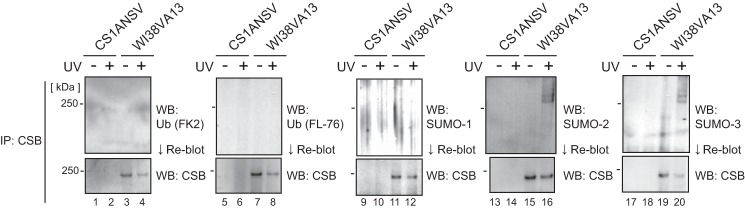
In Fig. 2C we show that some high molecular weight bands were observed after UV irradiation in the CSBWT panel (lanes 2–4, asterisk). In the panel of CSBΔUBD, the shifted bands were extremely faint but visible (Fig. 2C, lanes 27–29). In the panels of other CSB mutants, only a single band of CSB (i.e. no shifted bands) was detected. It has been reported that CSB is modified by ubiquitin and phosphate and that these posttranslational modifications affect the function of CSB in TC-NER (26–29). In addition, the SUMOplot analysis program (Abgent) showed that Lys-1489 in the C-terminal region has a high potential to be modified by SUMO. The shifted bands were presumed to reflect modifications of CSB, and we hypothesized that there is a relationship between the C-terminal deletion of CSB and specific posttranslational modifications. Therefore, we first examined which modifications of CSB were detected after UV irradiation. Cells were treated with SDS lysis buffer, which disrupts protein interactions, and then CSB was immunoprecipitated from the lysate. Modifications by ubiquitin and SUMO were examined by Western blotting using specific antibodies (Fig. 3). We could not detect ubiquitinated CSB regardless of UV irradiation (Fig. 3, lanes 1–8). No bands were detected using an anti-SUMO-1 antibody (Fig. 3, lanes 9–12), whereas, remarkably, three bands higher than 250 kDa and some even higher faint bands were detected with anti-SUMO-2 (Fig. 3, lanes 13–16) and anti-SUMO-3 (Fig. 3, lanes 17–20) antibodies specifically after UV irradiation. The amino acid sequences of SUMO-2 and SUMO-3 are 97% identical, and differences in their function have not been found. Therefore, they are usually denoted as SUMO-2/3. Almost the same band patterns were detected with anti-SUMO-2 and anti-SUMO-3 antibodies in Fig. 3 (lanes 16 and 20). Therefore, we subsequently only used an anti-SUMO-3 antibody to detect the shifted bands.
FIGURE 3.
UV-induced posttranslational modifications of CSB. CSB immunoprecipitated (IP) from cell lysates was analyzed by Western blotting (WB) with two anti-ubiquitin (Ub) antibodies and three anti-SUMO antibodies. After detection of the modifications, the membrane was reblotted with an anti-CSB antibody. Immunoprecipitated samples from the same number of cells were applied to each lane of the gel.
Analysis of CSB SUMOylation
Ubc9 is the only SUMO-conjugating enzyme (E2) in eukaryotes. Ubc9 was knocked down by transfection of siRNA (Fig. 4A). The amount of Ubc9 was extremely low after the transfection but that of CSB was not (Fig. 4A, lanes 1 and 3). Then CSB was immunoprecipitated from the cell lysate. The bands detected with an anti-SUMO-3 antibody became faint (Fig. 4A, lane 8). These data indicate that the bands are due to SUMO modification.
Next, soluble and insoluble fractions were prepared with CSK-Triton buffer, and SUMOylation of CSB was analyzed. Bands were detected in the insoluble fraction but not in the soluble fraction (Fig. 4C). The insoluble fraction contained chromatin, suggesting that SUMOylated CSB is associated with chromatin.
We then tested whether the SUMO modification of CSB is affected in TC-NER-deficient cells (Fig. 4D). UVSSA-deficient Kps3 cells and CSA-deficient CS3BE cells were used. CSB SUMOylation following UV irradiation was detected in both cell lines (Fig. 4D, lanes 1, 2, 5, and 6) as well as in cells transfected with the responsible cDNA (Fig. 4D, lanes 3, 4, 7, and 8).
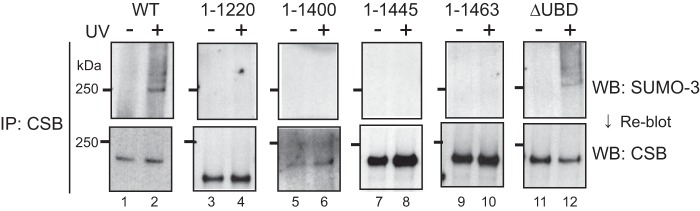
The ATPase activity of CSB is essential for its function in TC-NER. One amino acid substitution in the ATPase domain (K538R) disrupts the ATPase activity and causes cells to become hypersensitive to UV irradiation (36). In cells expressing CSBK538R, this dysfunctional CSB was also SUMOylated after UV irradiation (Fig. 4E). We next examined whether the CSB deletion mutants described above are SUMOylated (Fig. 5). CSBΔUBD was SUMOylated after UV irradiation, similar to CSBWT (Fig. 5, lanes 1, 2, 11, and 12), but no other CSB mutants were SUMOylated.
FIGURE 5.
SUMO-3 modification of CSB deletion mutants. Lysates were prepared from cells expressing CSB mutants. CSB was immunoprecipitated (IP) from the lysates and analyzed by Western blotting (WB) with an anti-SUMO-3 antibody. After detection of SUMO-3, the membrane was reblotted with an anti-CSB antibody. Samples from the same number of cells were applied to each lane of the gel.
Taken together, UV light-dependent SUMO modification of CSB occurred independently of UVSSA, CSA, and the ATPase activity of CSB. In addition, the C-terminal region of CSB was involved in the SUMOylation of CSB but the UBD was not.
Effects of Substitution of Lysine to Arginine in the C-terminal Region of CSB
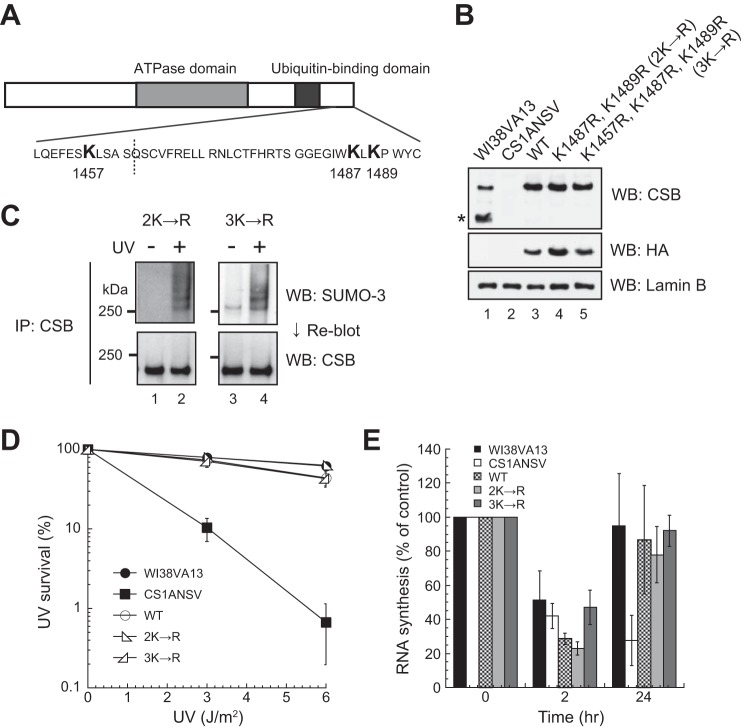
As shown in Fig. 5, the C-terminal truncated CSB mutants (CSB1–1220, CSB1–1400, CSB1–1445, and CSB1–1463) were not SUMOylated after UV irradiation. CSB1–1463, which has the smallest deletion, lacks only 30 amino acid residues. Therefore, it was assumed that lysine residues of a SUMO acceptor site are present among the most C-terminal 30 amino acid residues of CSB. In this region, there are two lysine residues, Lys-1487 and Lys-1489 (Fig. 6A). To investigate whether these lysine residues were responsible for SUMOylation of CSB, we generated two CSB mutants with FLAG-HA epitope tags at their N termini: CSBK1487R, K1489R (2K→R) and CSBK1457R, K1487R, K1489R (3K→R). Lys-1457 is located outside of the 30 amino acid residues but just precedes this region. Transition of a SUMO-modified site to a nearby lysine residue can occur when an authentic lysine for SUMOylation is substituted. Therefore, Lys-1457 was substituted with arginine in conjunction with the substitutions at Lys-1487 and Lys-1489. Lys-1457 is also reportedly modified by ubiquitin independently of UV irradiation (34). In immunoblot analysis, the expression levels of both mutant CSB proteins in the transfectants were similar to those in normal (WI38VA13) and CSAWT cells (Fig. 6B). SUMOylation of the substitution mutants was examined as described above. Substitutions of Lys-1457, Lys-1487, and Lys-1489 with arginine did not abolish SUMOylation of CSB after UV irradiation (Fig. 6C). We noted that the band pattern of CSB3K→R in non-irradiated cells was different from that of the other CSB mutants. A single band was detected in CSB3K→R cells (Fig. 6C, lane 3), but no bands were detected in the other mutant cells.
FIGURE 6.
Effects of lysine-to-arginine substitutions in the C-terminal region of CSB. A, schematic of the positions of lysine residues in the C-terminal region of CSB. B, expression levels of CSB mutants. N-terminally FLAG-HA epitope-tagged CSB with two and three amino acid substitutions (K1487R, K1489R and K1457R, K1487R, K1489R, respectively) was expressed in CS1ANSV cells. Whole cell lysates were analyzed by Western blotting (WB) with anti-CSB and anti-HA antibodies. Lamin B is a loading control. The asterisk denotes the CPFP protein. C, SUMO-3 modification of the lysine-substituted CSB mutants. Lysates were prepared from cells expressing CSB mutants. CSB was immunoprecipitated (IP) from the lysates and analyzed by Western blotting with an anti-SUMO-3 antibody. After detection of SUMO-3, the membrane was reblotted with an anti-CSB antibody. Samples from the same number of cells were applied to each lane of the gel. D, colony-forming ability of cells expressing the lysine-substituted CSB mutants after UV irradiation. The points are the average of at least three independent experiments, and vertical bars indicate mean ± S.E. E, RNA synthesis of cells expressing the lysine-substituted CSB mutants after UV irradiation. Cells were irradiated with 10 J/m2 UV light, and incorporation of [3H]uridine was measured 2 and 24 h later. The incorporation of [3H]uridine in UV-irradiated cells was compared with that in non-irradiated cells. The points are the average of at least three independent experiments, and vertical bars indicate mean ± S.E.
In addition, the colony-forming ability and recovery of RNA synthesis after UV irradiation of the substitution mutants were examined. The substitutions did not affect UV light sensitivity (Fig. 6D) or RNA synthesis recovery following UV irradiation (Fig. 6E). These results indicate that the three lysine residues in the C-terminal region are not SUMO acceptor sites after UV irradiation and that the substitution of these residues with arginine has no effect on the function of CSB in TC-NER.
Substitution of Lys-205 with Arginine Represses SUMOylation of CSB
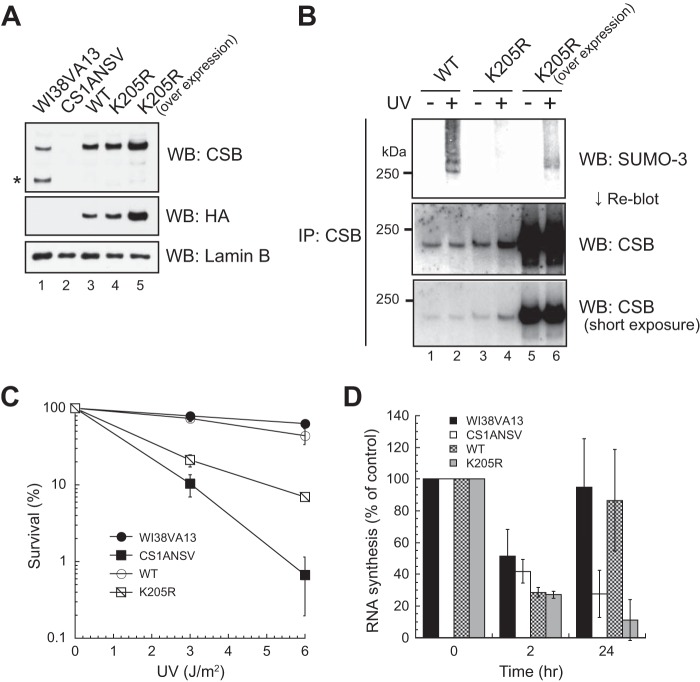
There is no lysine residue for SUMOylation in the C-terminal region of CSB. The SUMOplot analysis program (Abgent) indicated that Lys-205 has a high probability of being SUMOylated. In addition, according to PhosphoSitePlus (Cell Signaling Technology), many amino acid residues near Lys-205 are modified by posttranslational modifications. Therefore, CSB with Lys-205 substituted (CSBK205R) and tagged with the FLAG-HA epitope at its N terminus was generated and stably expressed in CS1ANSV cells (Fig. 7A). SUMOylation of CSBK205R was not detected with an anti-SUMO3 antibody even after UV irradiation (Fig. 7B, lanes 3 and 4). When cells overexpressing CSBK205R were used, faint bands were detected, dependent on UV light, but the unique shifted bands were not (Fig. 7B, lanes 5 and 6).
FIGURE 7.
Effects of the K205R substitution of CSB. A, expression levels of CSB mutants. N-terminally FLAG-HA epitope-tagged CSB with the K205R substitution was expressed in CS1ANSV cells. Whole cell lysates were analyzed by Western blotting (WB) with anti-CSB and anti-HA antibodies. Lamin B is a loading control. The asterisk denotes the CPFP protein. B, SUMO-3 modification of CSBK205R. Lysates were prepared from CSBK205R cells. CSB was immunoprecipitated (IP) from the lysates and analyzed by Western blotting with an anti-SUMO-3 antibody. After detection of SUMO-3, the membrane was reblotted with an anti-CSB antibody. Samples from the same number of cells were applied to each lane of the gel. C, colony-forming ability of CSBK205R cells after UV irradiation. The points are the average of at least three independent experiments, and vertical bars indicate mean ± S.E. D, RNA synthesis of CSBK205R cells after UV irradiation. Cells were irradiated with 10 J/m2 UV light, and incorporation of [3H]uridine was measured 2 and 24 h later. The incorporation of [3H]uridine in UV-irradiated cells was compared with that in non-irradiated cells. The points are the average of at least three independent experiments, and vertical bars indicate mean ± S.E.
CSBK205R cells were more sensitive to UV irradiation than CSBWT cells but less sensitive to UV irradiation than CS1ANSV cells (Fig. 7C), similar to CSB1–1400, CSB1–1445, CSB1–1463, and CSBΔUBD cells. Moreover, the recovery of RNA synthesis after UV irradiation was not detected in CSBK205R cells, similar to all cell lines expressing CSB deletion mutants (Fig. 7D).
To investigate the effects of SUMOylation on TC-NER, Ubc9 knockdown cells were used (Fig. 4B). RNA synthesis in knockdown cells not irradiated with UV was almost equal to that in control cells. Next, the recovery of RNA synthesis after UV irradiation was examined. RNA synthesis was decreased in both cell types 2 h after UV irradiation. RNA synthesis was recovered in control cells 24 h after UV irradiation, whereas RNA synthesis in Ubc9 knockdown cells was only 70% of that in non-irradiated cells. These results indicate that TC-NER is hampered by Ubc9 knockdown. There are no reports showing that any TC-NER-specific factors are SUMOylated in eukaryotes. Regardless of UV irradiation, neither CSA nor UVSSA was modified by any SUMO isoforms (data not shown). These data suggest that SUMOylation of CSB is relevant to TC-NER.
Discussion
When UV light-induced lesions occur at the transcribed strands of actively transcribed genes, Pol II is arrested at the damaged sites (37, 38), and TC-NER is initiated. CSB interacts with Pol II, and this interaction is increased after UV irradiation (15). CSB is a key factor to recruit repair factors to stalled Pol II at damaged sites (19, 22, 39). In this study, we characterized CSB mutants with deletions of various C-terminal regions. All cells expressing deletion mutants showed hypersensitivity to UV light (Fig. 1C) and did not recover RNA synthesis after UV irradiation (Fig. 1D), indicating that both the UBD and C-terminal region are necessary for TC-NER. Although the recovery of RNA synthesis in all mutant cells was as defective as that in CS1ANSV cells, the mutant cells, except CSB1–1220 cells, were less sensitive to UV irradiation than CS1ANSV cells. These results suggest that the region corresponding to amino acid residues 1221–1400 has functions for cell viability after UV irradiation; for example, suppression of apoptosis. It is noteworthy that Ser-1348 is phosphorylated (32). It is likely that phosphorylation at this site is important for the functions of CSB.
The interaction between CSB with deletion of either the UBD or C-terminal region and Pol II was increased slightly after UV irradiation in contrast to the large increase in the interaction between WT CSB and Pol II (Fig. 2A), even when CSB was associated with chromatin (Fig. 2, C and D). These results indicate that both regions of CSB are required for its complete interaction with Pol II after UV irradiation. The interaction between Pol II and CSB is thought to be critical for TC-NER. If the interaction is incomplete after UV irradiation, then it is expected that the TC-NER reaction is not initiated.
Translocation of CSA to the nuclear matrix after UV irradiation is related to TC-NER and is dependent on CSB (20, 21). Because the translocation did not occur with the deletion mutants (Fig. 2B), both the UBD and C-terminal regions are necessary for the translocation. It is considered that complete function of CSB to initiate the TC-NER reaction is required for the translocation.
It has been reported that CSB1–1220 can interact with Pol II after UV irradiation (18). However, our results did not correspond with this. This difference is likely due to experimental conditions. A cross-linking reagent was used in the previous study but not in this study. It is possible that CSB1–1220 interacts with Pol II after UV irradiation but that the interaction is too weak to be detected under the conditions we used. Transplantation of the UBA domain from S. cerevisiae Rad23 to CSB1–1220 (CSBdel in the report) restores CSB function in TC-NER. Although UBA domain-transplanted CSB is considered to be equivalent to CSB1–1463 used in this study, CSB1–1463 exhibited dysfunction similar to CSBΔUBD. It is possible that the UBA domain of Rad23 has some functions that can compensate for the absence of the most C-terminal region of human CSB.
CSB was modified by SUMO-2/3 in a UV light-dependent manner (Fig. 3), and the most C-terminal region was needed for this modification (Fig. 5). However, amino acid substitutions of lysine residues with arginine in this region did not affect its SUMOylation (Fig. 6), indicating that a SUMO acceptor site does not exist in this region. The most C-terminal region is possibly required for the interaction with SUMOylation machinery, for example, Ubc9, or is required for positioning in the location where CSB is SUMOylated. Because the UBD was dispensable for SUMOylation, the complete interaction between CSB and Pol II is not essential for the modification. SUMOylation of CSB was repressed by substitution of Lys-205, a residue in the N-terminal region, with arginine, and this substitution led to cells becoming hypersensitive to UV radiation and exhibiting defects in TC-NER (Fig. 7), indicating that the SUMOylation plays a role in TC-NER. A certain amount of CSB was associated with chromatin without UV irradiation, and SUMOylated CSB was also associated with chromatin after UV irradiation (Fig. 4C). So far, we cannot discriminate between whether chromatin-bound CSB is modified or modified CSB binds to chromatin. In any case, SUMOylated CSB is thought to function in chromatin. SUMOylation of CSB occurred even in the absence of CSA and UVSSA (Fig. 4D), suggesting that the modification is upstream of CSA and UVSSA in the TC-NER reaction. However, we cannot exclude the possibility that SUMOylated CSB functions with CSA and UVSSA or downstream of them. It has been suggested that both the N- and C-terminal regions of CSB are required for its proper function in TC-NER (40). The involvement of the N- and C-terminal regions in SUMOylation might give a clue regarding the function of CSB.
In the global genome NER pathway, XPC is modified by SUMO-1 after UV irradiation (31, 41). This SUMOylation is related to UV light-induced and DNA damage-binding protein-dependent ubiquitination of XPC and is assumed to protect XPC from degradation by the proteasome (31). On the other hand, in the absence of SUMOylation, XPC is normally recruited to sites with DNA lesions but is then immobilized by the DNA damage-binding protein complex, resulting in a deficiency in NER (41). SUMO-2 or -3 chains reportedly act as a signal for the recruitment of an E3 ubiquitin ligase and lead to SUMOylated substrates undergoing proteasomal degradation (42). Although the role of CSB modification by SUMO-2/3 is unclear, our findings provide a new point of view regarding the function of CSB in TC-NER.
Author Contributions
Y. S., K. T., and M. S. designed the study and wrote the manuscript. Y. S. and M. S. performed the experiments. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank all members of the Tanaka laboratory for discussions and encouragement.
This work was supported by Grant-in-Aid for Scientific Research on Innovative Areas 22131009 (to K.T.) and Grant-in-Aid for Scientific Research (B) 15H02820 (to M.S.) from the Ministry of Education, Cultures, Sports, Science, and Technology (MEXT). The authors declare that they have no conflicts of interest with the contents of this article.
- NER
- nucleotide excision repair
- TC-NER
- transcription-coupled nucleotide excision repair
- Pol II
- RNA polymerase II
- CS
- Cockayne syndrome
- UBD
- ubiquitin-binding domain
- UVSS
- UV-sensitive syndrome
- SUMO
- small ubiquitin-like modifier
- CSK-ppt
- CSK-Triton buffer insoluble
- CSK-sup
- CSK-Triton buffer soluble.
References
- 1.Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., and Ellenberger T. (2005) DNA Repair and Mutagenesis, 2nd Ed., ASM Press, Washington, D. C. [Google Scholar]
- 2.Hanawalt P. C., and Spivak G. (2008) Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 [DOI] [PubMed] [Google Scholar]
- 3.Marteijn J. A., Lans H., Vermeulen W., and Hoeijmakers J. H. (2014) Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 15, 465–481 [DOI] [PubMed] [Google Scholar]
- 4.Nance M. A., and Berry S. A. (1992) Cockayne syndrome: review of 140 cases. Am. J. Med. Genet. 42, 68–84 [DOI] [PubMed] [Google Scholar]
- 5.Mayne L. V., and Lehmann A. R. (1982) Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 42, 1473–1478 [PubMed] [Google Scholar]
- 6.Lehmann A. R., Francis A. J., and Giannelli F. (1985) Prenatal diagnosis of Cockayne's syndrome. Lancet 325, 486–488 [DOI] [PubMed] [Google Scholar]
- 7.Tanaka K., Kawai K., Kumahara Y., Ikenaga M., and Okada Y. (1981) Genetic complementation groups in Cockayne syndrome. Somatic Cell Genet. 7, 445–455 [DOI] [PubMed] [Google Scholar]
- 8.Lehmann A. R. (1982) Three complementation groups in Cockayne syndrome. Mutat. Res. 106, 347–356 [DOI] [PubMed] [Google Scholar]
- 9.Henning K. A., Li L., Iyer N., McDaniel L. D., Reagan M. S., Legerski R., Schultz R. A., Stefanini M., Lehmann A. R., Mayne L. V., and Friedberg E. C. (1995) The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell 82, 555–564 [DOI] [PubMed] [Google Scholar]
- 10.Troelstra C., van Gool A., de Wit J., Vermeulen W., Bootsma D., and Hoeijmakers J. H. (1992) ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell 71, 939–953 [DOI] [PubMed] [Google Scholar]
- 11.Vélez-Cruz R., and Egly J.-M. (2013) Cockayne syndrome group B (CSB) protein: at the crossroads of transcriptional networks. Mech. Ageing Dev. 134, 234–242 [DOI] [PubMed] [Google Scholar]
- 12.Citterio E., Van Den Boom V., Schnitzler G., Kanaar R., Bonte E., Kingston R. E., Hoeijmakers J. H., and Vermeulen W. (2000) ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 20, 7643–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lake R. J., Geyko A., Hemashettar G., Zhao Y., and Fan H.-Y. (2010) UV-Induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol. Cell 37, 235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tantin D., Kansal A., and Carey M. (1997) Recruitment of putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complex. Mol. Cell. Biol. 17, 6803–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van den Boom V., Citterio E., Hoogstraten D., Zotter A., Egly J. M., van Cappellen W. A., Hoeijmakers J. H., Houtsmuller A. B., and Vermeulen W. (2004) DNA damage stabilizes interaction of CSB with the transcription elongation machinery. J. Cell Biol. 166, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Gool A. J., Citterio E., Rademakers S., van Os R., Vermeulen W., Constantinou A., Egly J. M., Bootsma D., and Hoeijmakers J. H. (1997) The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 16, 5955–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svejstrup J. Q. (2003) Rescue of arrested RNA polymerase II complex. J. Cell Sci. 116, 447–451 [DOI] [PubMed] [Google Scholar]
- 18.Anindya R., Mari P. O., Kristensen U., Kool H., Giglia-Mari G., Mullenders L. H., Fousteri M., Vermeulen W., Egly J. M., and Svejstrup J. Q. (2010) A ubiquitin-binding domain in Cockayne syndrome B required for transcription-coupled nucleotide excision repair. Mol. Cell 38, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fousteri M., Vermeulen W., van Zeeland A. A., and Mullenders L. H. (2006) Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell 23, 471–482 [DOI] [PubMed] [Google Scholar]
- 20.Kamiuchi S., Saijo M., Citterio E., de Jager M., Hoeijmakers J. H., and Tanaka K. (2002) Translocation of Cockayne syndrome group A protein to the nuclear matrix: possible relevance to transcription-coupled DNA repair. Proc. Natl. Acad. Sci. U.S.A. 99, 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saijo M., Hirai T., Ogawa A., Kobayashi A., Kamiuchi S., and Tanaka K. (2007) Functional TFIIH is required for UV-induced translocation of CSA to the nuclear matrix. Mol. Cell. Biol. 27, 2538–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X., Horibata K., Saijo M., Ishigami C., Ukai A., Kanno S., Tahara H., Neilan E. G., Honma M., Nohmi T., Yasui A., and Tanaka K. (2012) Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat. Genet. 44, 593–597 [DOI] [PubMed] [Google Scholar]
- 23.Schwertman P., Lagarou A., Dekkers D. H., Raams A., van der Hoek A. C., Laffeber C., Hoeijmakers J. H., Demmers J. A., Fousteri M., Vermeulen W., and Marteijn J. A. (2012) UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat. Genet. 44, 598–602 [DOI] [PubMed] [Google Scholar]
- 24.Nakazawa Y., Sasaki K., Mitsutake N., Matsuse M., Shimada M., Nardo T., Takahashi Y., Ohyama K., Ito K., Mishima H., Nomura M., Kinoshita A., Ono S., Takenaka K., Masuyama R., Kudo T., Slor H., Utani A., Tateishi S., Yamashita S., Stefanini M., Lehmann A. R., Yoshiura K., and Ogi T. (2012) Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat. Genet. 44, 586–592 [DOI] [PubMed] [Google Scholar]
- 25.Fei J., and Chen J. (2012) KIAA1530 protein is recruited by Cockayne syndrome complementation group protein A (CSA) to participate in transcription-coupled repair (TCR). J. Biol. Chem. 287, 35118–35126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lake R. J., and Fan H.-Y. (2013) Structure, function and regulation of CSB: a multi-talented gymnast. Mech. Ageing Dev. 134, 202–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Groisman R., Kuraoka I., Chevallier O., Gaye N., Magnaldo T., Tanaka K., Kisselev A. F., Harel-Bellan A., and Nakatani Y. (2006) CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 20, 1429–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei L., Lan L., Yasui A., Tanaka K., Saijo M., Matsuzawa A., Kashiwagi R., Maseki E., Hu Y., Parvin J. D., Ishioka C., and Chiba N. (2011) BRACA1 contributes to transcription-coupled repair of DNA damage through polyubiquitination and degradation of Cockayne syndrome B protein. Cancer Sci. 102, 1840–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imam S. Z., Indig F. E., Cheng W. H., Saxena S. P., Stevnsner T., Kufe D., and Bohr V. A. (2007) Cockayne syndrome protein B interacts with and is phosphorylated by c-Abl tyrosine kinase. Nucleic Acids Res. 35, 4941–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horibata K., Saijo M., Bay M. N., Lan L., Kuraoka I., Brooks P. J., Honma M., Nohmi T., Yasui A., and Tanaka K. (2011) Mutant Cockayne syndrome group B protein inhibits repair of DNA topoisomerase I-DNA covalent complex. Genes Cells 16, 101–114 [DOI] [PubMed] [Google Scholar]
- 31.Wang Q. E., Zhu Q., Wani G., El-Mahdy M. A., Li J., and Wani A. A. (2005) DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 33, 4023–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., and Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R. 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., and Elledge S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 34.Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., and Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laugel V., Dalloz C., Durand M., Sauvanaud F., Kristensen U., Vincent M. C., Pasquier L., Odent S., Cormier-Daire V., Gener B., Tobias E. S., Tolmie J. L., Martin-Coignard D., Drouin-Garraud V., Heron D., Journel H., Raffo E., Vigneron J., Lyonnet S., Murday V., Gubser-Mercati D., Funalot B., Brueton L., Sanchez Del Pozo J., Muñoz E., Gennery A. R., Salih M., Noruzinia M., Prescott K., Ramos L., Stark Z., Fieggen K., Chabrol B., Sarda P., Edery P., Bloch-Zupan A., Fawcett H., Pham D., Egly J. M., Lehmann A. R., Sarasin A., and Dollfus H. (2010) Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum. Mutat. 31, 113–126 [DOI] [PubMed] [Google Scholar]
- 36.Citterio E., Rademakers S., van der Horst G. T., van Gool A. J., Hoeijmakers J. H., and Vermeulen W. (1998) Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J. Biol. Chem. 273, 11844–11851 [DOI] [PubMed] [Google Scholar]
- 37.Mei Kwei J. S., Kuraoka I., Horibata K., Ubukata M., Kobatake E., Iwai S., Handa H., and Tanaka K. (2004) Blockage of RNA polymerase II at a cyclobutane pyrimidine dimer and 6-4 photoproduct. Biochem. Biophys. Res. Commun. 320, 1133–1138 [DOI] [PubMed] [Google Scholar]
- 38.Fousteri M., and Mullenders L. H. (2008) Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 18, 73–84 [DOI] [PubMed] [Google Scholar]
- 39.Sarker A. H., Tsutakawa S. E., Kostek S., Ng C., Shin D. S., Peris M., Campeau E., Tainer J. A., Nogales E., and Cooper P. K. (2005) Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne syndrome. Mol. Cell 20, 187–198 [DOI] [PubMed] [Google Scholar]
- 40.Wang L., Limbo O., Fei J., Chen L., Kim B., Luo J., Chong J., Conaway R. C., Conaway J. W., Ranish J. A., Kadonaga J. T., Russell P., and Wang D. (2014) Regulation of the Rhp26ERCC6/CSB chromatin remodeler by a novel conserved leucine latch motif. Proc. Natl. Acad. Sci. U.S.A. 111, 18566–18571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akita M., Tak Y.-S., Shimura T., Matsumoto S., Okuda-Shimizu Y., Shimizu Y., Nishi R., Saitoh H., Iwai S., Mori T., Ikura T., Sakai W., Hanaoka F., and Sugasawa K. (2015) SUMOylation of xeroderma pigmentosum group C protein regulates DNA damage recognition during nucleotide excision repair. Sci. Rep. 10.1038/srep10984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geoffroy M.-C., and Hay R. T. (2009) An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 10, 564–568 [DOI] [PubMed] [Google Scholar]