Abstract
Posttranslational lipid modifications mediate the membrane attachment of Rab GTPases, facilitating their function in regulating intracellular vesicular trafficking. In Arabidopsis, most Rab GTPases have two C-terminal cysteines and potentially can be double-geranylgeranylated by heterodimeric Rab geranylgeranyltransferases (Rab-GGTs). Genes encoding two putative α subunits and two putative β subunits of Rab-GGTs have been annotated in the Arabidopsis thaliana genome, but little is known about Rab-GGT activity in Arabidopsis. In this study, we demonstrate that four different heterodimers can be formed between putative Arabidopsis Rab-GGT α subunits RGTA1/RGTA2 and β subunits RGTB1/RGTB2, but only RGTA1·RGTB1 and RGTA1·RGTB2 exhibit bona fide Rab-GGT activity, and they are biochemically redundant in vitro. We hypothesize that RGTA2 function might be disrupted by a 12-amino acid insertion in a conserved motif. We present evidence that Arabidopsis Rab-GGTs may have preference for prenylation of C-terminal cysteines in particular positions. We also demonstrate that Arabidopsis Rab-GGTs can not only prenylate a great variety of Rab GTPases in the presence of Rab escort protein but, unlike Rab-GGT in yeast and mammals, can also prenylate certain non-Rab GTPases independently of Rab escort protein. Our findings may help to explain some of the phenotypes of Arabidopsis protein prenyltransferase mutants.
Keywords: Arabidopsis thaliana, post-translational modification (PTM), protein isoprenylation, Rab, small GTPase, Rab, Rab geranylgeranyltransferase, gene duplication, geranylgeranyl diphosphate
Introduction
Small GTPases serve as molecular switches that shuttle between active GTP-bound and inactive GDP-bound forms, providing transient signals to downstream effectors (1, 2). In plants, many membrane-localized small GTPases are important regulators of vesicular trafficking (1). They are typically anchored to membranes via posttranslational lipid modifications (3).
Rab GTPases constitute the largest family of the Ras superfamily of small GTPases (3–5). They are involved in regulating trafficking processes, such as vesicle formation, transport, membrane targeting, and docking (1, 5). The diversity and specific localization of Rab GTPases not only determine membrane identity, but also reflect the complexity of vesicle trafficking (1, 6).
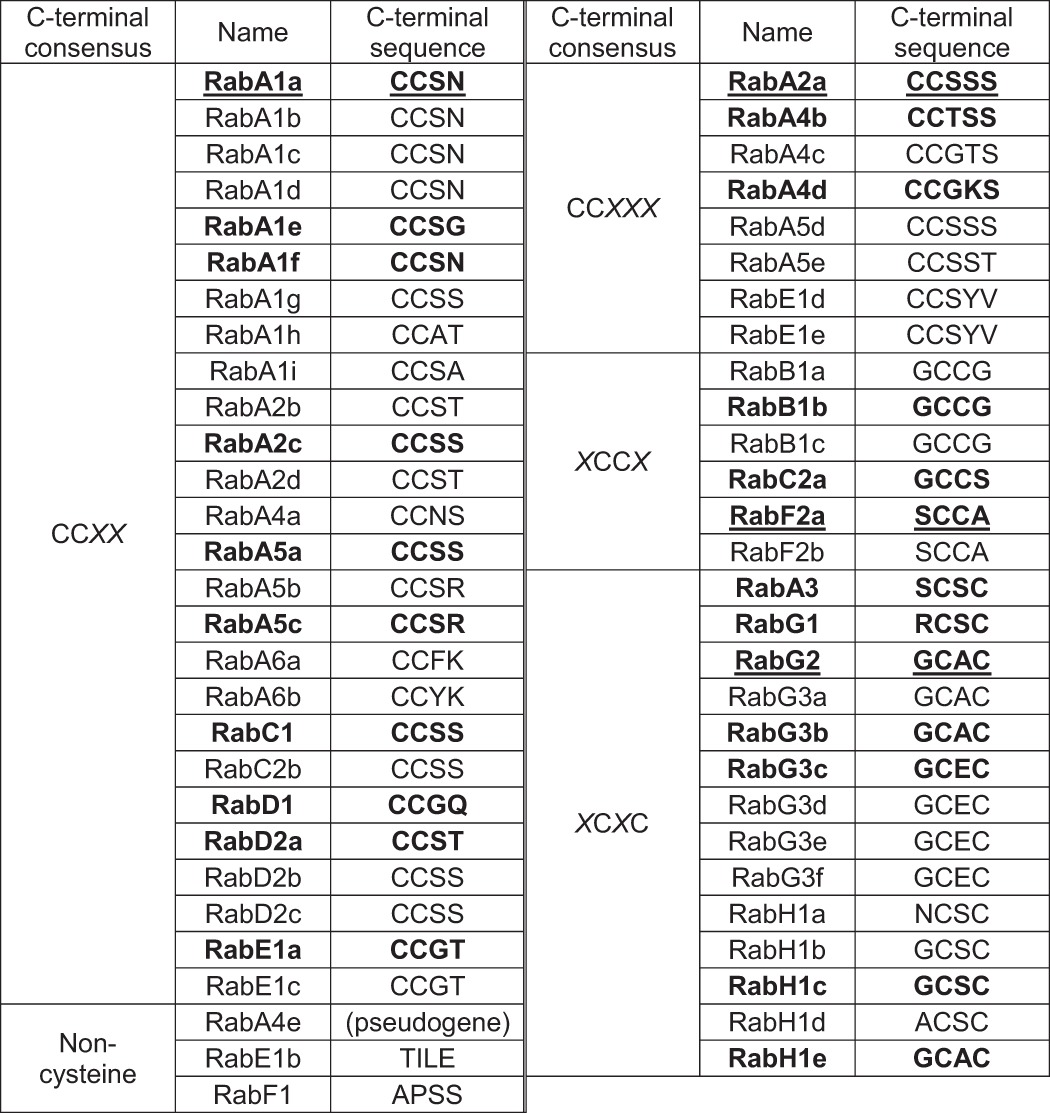
Phylogenetic analysis suggests that the 57 Arabidopsis Rab-encoding sequences fall into just eight subfamilies (3, 6), in contrast to ∼40 Rab subfamilies in mammals (7). Accordingly, the size of each Arabidopsis Rab subfamily is expanded. The distinct composition of the Arabidopsis Rab GTPase family suggests plant-specific functions (1, 6). Of these 57 members, 54 include two cysteines that are near the C terminus and are candidate prenylation sites (Table 1). At least one of the non-prenylated Rabs, RABF1/ARA6, is N-myristoylated and palmitoylated (8).
TABLE 1.
The members of the Arabidopsis Rab GTPase family
The 57 predicted members are grouped based on C-terminal sequences (-CCXX, -CCXXX, -XCCX, -XCXC, non-cysteine). The members highlighted in boldface type were chosen to test the target specificities of RGTA1·RGTB1, RGTA1·RGTB2, RGTA2·RGTB1, and RGTA2·RGTB2 (Table 2, Fig. 6). The members marked with underline were chosen to represent four types of C-terminal sequences in the cysteine substitution experiments (Fig. 4).
Protein prenylation irreversibly adds one 15-carbon isoprenoid (farnesylation), one 20-carbon isoprenoid (geranylgeranylation), or two 20-carbon isoprenoids (double geranylgeranylation) to one or two C-terminal cysteine residues of target proteins, by forming thioether bonds (9, 10). These three types of prenylation are respectively catalyzed by three distinct heterodimeric enzymes, collectively called protein prenyltransferases (11, 12). Protein farnesyltransferase (PFT)3 and protein geranylgeranyltransferase type I (PGGT-I) target a C-terminal CaaX box, in which C is the cysteine residue to be prenylated, and a is usually an aliphatic amino acid residue. For PFT, X is usually alanine, cysteine, glutamine, methionine, or serine; for PGGT-I, X is almost always leucine (10–12). In Arabidopsis, >250 proteins are predicted to be ideal targets for prenylation (10, 13). Among them, type I Rop GTPases and heterotrimeric G protein γ subunits AGG1 and AGG2 have a C-terminal CaaL box and can be geranylgeranylated by PGGT-I (9, 14, 15). Rab geranylgeranyltransferase (Rab-GGT, or protein geranylgeranyltransferase type II) has a broader spectrum of target sequences, most with two cysteine residues, including XCC, XCXC, XCCX, CCXX, and CCXXX (10, 12), where C is a cysteine residue that potentially can be prenylated, and X is a nonspecific amino acid residue. It is believed that Rab-GGT only prenylates Rab GTPases; hence the name (16).
All of the known protein prenyltransferases are heterodimeric enzymes that consist of a regulatory α subunit and a catalytic β subunit. PFT and PGGT-I share a common α subunit but have unique β subunits (10, 17). Complete loss of PFT and PGGT-I activities leads to lethality in yeast and animals (17–19). In contrast, mutations in the Arabidopsis PFT/PGGT-I α subunit PLP (pluripetala) cause significant developmental defects, but the mutants are viable and fertile (20). The viability of plp mutants suggests that additional prenylation mechanisms or other types of lipid modification in Arabidopsis might compensate for the loss of PFT/PGGT-I activities (10, 20). Mutations in the Arabidopsis PFT β subunit ERA1 (enhanced response to abscisic acid 1) result in only mild phenotypes (21–24), whereas mutations in the PGGT-I β subunit GGB (geranylgeranyltransferase-I β subunit) result in no detectable phenotypes under normal conditions (25), suggesting considerable target cross-specificity between Arabidopsis PFT and PGGT-I (10, 25).
Rab-GGT has a distinct set of subunits, and no shared subunits between Rab-GGT and PFT/PGGT-I have been reported. Despite sharing only 20–30% amino acid sequence similarity with their counterparts in PFT and PGGT-I, Rab-GGT subunits structurally resemble PFT and PGGT-I subunits (10, 16). The N-terminal helical domain of the Rab-GGT α subunit is structurally very similar to the PFT/PGGT-I α subunit, whereas the Rab-GGT β subunit forms an α-α barrel structure, as does the PFT β subunit (16, 26, 27). Mammalian and plant Rab-GGT α subunits have an additional immunoglobulin (Ig)-like domain and a leucine-rich repeat (LRR) domain, both of which all known PFT/PGGT-I α subunits and yeast Rab-GGT α subunits lack (12, 26). However, it appears that neither of these two domains is required for Rab-GGT activity (28). Remarkably, rather than recognizing target proteins by itself as PFT and PGGT-I do, Rab-GGT relies on a cofactor protein called Rab escort protein (REP) to recognize Rab GTPases (26, 29). According to different models, REP binds to unprenylated Rab GTPase, either before or after forming a complex with the Rab-GGT α·β heterodimer and helps load the C-terminal end of the unprenylated Rab GTPase into the catalytic site of Rab-GGT (30–32).
Very little is known about Rab-GGT activity in plants. Most early studies were carried out using total cell extracts (33–35), and only recently have functional studies on specific subunits been conducted. In the Arabidopsis genome, genes encoding two putative α subunits, RGTA1 and RGTA2, and two putative β subunits, RGTB1 and RGTB2, of Rab-GGT were annotated (36). rgtb1 mutants show a series of defects in shoot morphology, shoot gravitropism, tip growth of root hairs and pollen tubes, and light response (37, 38). rgtb2 mutants are broadly indistinguishable from wild-type plants under normal conditions but are also defective in tip growth (38). The rgtb1 rgtb2 double mutants are pollen-lethal, suggesting that they are at least partially genetically redundant (38). The effects of mutations in either RGTA1 or RGTA2 have not been reported. However, loss of the sole Physcomitrella patens Rab-GGT α subunit results in lethality (39), suggesting that Arabidopsis lacking a functional Rab-GGT α subunit might also be non-viable. The REP homolog in Arabidopsis has also been characterized biochemically. Recombinant AtREP can promote the prenylation of various Rab GTPases in Arabidopsis cell extracts but fails to complement a yeast REP mutation due to a change in AtREP of an arginine residue conserved in non-plant REPs to an asparagine (40).
Indirect evidence that RGTB1 is a bona fide Rab-GGT subunit comes from studies of rgtb1 mutants; specifically, the level of prenylated RABA2A is reduced in rgtb1 mutants, and rgtb1 total extracts cannot efficiently prenylate recombinant RABA2A in vitro (37). However, the biochemical activity of the other putative subunits and possible target specificity differences among different α·β combinations remain unknown. It is also unclear whether AtREP is required for Arabidopsis Rab-GGT activity. In this study, we used an isotope-based in vitro prenylation assay to address these questions. Here we report that RGTB1 and RGTB2 are biochemically redundant Rab-GGT β-subunits in Arabidopsis, whereas RGTA1 is the only active α subunit. Arabidopsis Rab-GGT also appears to show a preference for prenylation of cysteines in particular positions at the C terminus. Arabidopsis Rab-GGT not only prenylates a vast variety of Rab GTPases with various C-terminal sequences in vitro in an REP-dependent manner, but, unlike what has been reported for other eukaryotic Rab-GGTs, can also prenylate certain non-Rab small GTPases in vitro in an REP-independent manner. Our results help partially explain the survivability of Arabidopsis mutants lacking PFT/PGGT-I activity, the lack of phenotypes in PGGT-I mutants, and the observation of partial membrane localization of PGGT-I targets in PFT/PGGT-I mutants (15).
Experimental Procedures
Protein Sequence Analysis
The sequences of Arabidopsis genes and proteins were acquired from the Arabidopsis Information Resource (TAIR) online database (41). The sequences of yeast genes and proteins were acquired from the Saccharomyces Genome Database (42). The sequences of P. patens proteins were acquired from PlantGDB (43). The rice protein sequence was acquired from the Rice Genome Annotation Project website (44). The Drosophila melanogaster PTAR3 protein sequence was acquired from FlyBase (45). The sequences of genes and proteins of rat and human were acquired from the NCBI Reference Sequence (RefSeq) database (46). The pairwise alignments were performed with EMBOSS Needle (47). The multiple sequence alignments were performed with Clustal Omega (48). The conserved motif predictions were performed by InterPro version 51.0 (49) and Motif Scan (50). The alignment of RGTA1 and RGTA2 with rat RABGGTA (Protein Data Bank entry 1LTX) (31) was performed with the NCBI Cn3D application (51).
Expression of Rab-GGT in Yeast
The coding sequences of RGTA1, RGTA2, RGTB1, and RGTB2 were amplified by high-fidelity PCR using cDNA from Arabidopsis Col-0 wild-type plants as a template. The resulting PCR products were cloned into the yeast expression vector pESC-HIS (Agilent Technologies, Santa Clara, CA) in two steps. First, the coding sequences of RGTB1 and RGTB2 were cloned into MCS1 (multiple cloning site 1) of pESC-HIS by double digestion with EcoRI and ClaI or SpeI (New England Biolabs), followed by ligation with T4 DNA ligase (Promega, Madison, WI) to generate in-frame C-terminal fusions with the FLAG epitope tag. The resulting pESC-HIS-RGTB1-FLAG and pESC-HIS-RGTB2-FLAG plasmids were sequenced to verify the absence of PCR-induced mistakes and were used to separately express FLAG-tagged RGTB1 and RGTB2 proteins, respectively. Then the coding sequences of RGTA1 and RGTA2 were cloned into MCS2 of pESC-HIS-RGTB1-FLAG and pESC-HIS-RGTB2-FLAG by single digestion with XmaI (New England Biolabs), followed by calf intestinal phosphatase treatment (New England Biolabs) and ligation with T4 DNA ligase to generate in frame C-terminal fusions with the c-Myc epitope tag. The direction of the insert was checked by colony PCR using a GAL1 forward sequencing primer and RGTA1/2 gene-specific reverse primers. The resulting pESC-HIS-RGTB1-FLAG-RGTA1-c-Myc, pESC-HIS-RGTB1-FLAG-RGTA2-c-Myc, pESC-HIS-RGTB2-FLAG-RGTA1-c-Myc, and pESC-HIS-RGTB2-FLAG-RGTA2-c-Myc constructs were verified by sequencing and were used to co-express one α subunit with one β subunit.
The pESC constructs were transformed into S. cerevisiae YPH499 competent cells using a LiAc/SS carrier DNA/PEG method (52) with a modification that replaced a 42 °C heat shock with overnight incubation at room temperature. The preparation of YPH499 competent cells has also been described previously (52).
The expression of c-Myc-tagged RGTA1/2 and FLAG-tagged RGTB1/2 was driven by GAL1 and GAL10 promoters, respectively, and thus was inhibited by glucose but induced by galactose. The yeast cells containing the expression construct were first grown to A600 1.0 in SD−His medium with 2% glucose as a carbon source. The cells were then pelleted by centrifugation, washed with sterile water, and resuspended in SG − His medium with 2% galactose and 1% raffinose as carbon sources for induction. The cells were harvested after an 18-h induction.
Protein Purification, Pull-down, and Western Blot
The yeast cells harvested after expression induction were resuspended in an equal volume of prenyltransferase extraction buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm MgCl2, 20 μm ZnCl2, 2 mm DTT) supplemented with protease inhibitor mixture for fungal and yeast extracts (Sigma-Aldrich). The cell suspension was directly dropped into liquid nitrogen using a pipettor and snap-frozen into small droplets of 30–40 μl. The frozen cells were ground into fine powder by 5-mm steel beads in a Retsch M300 mixer mill (Qiagen, Valencia, CA). The powder was thawed on ice, and the resulting cell lysate was then separated by centrifugation. The supernatant contained soluble proteins from the cytosol and was used in pull-down experiments and for Western blots.
We used EZ-View Red FLAG M2 affinity gel beads (Sigma-Aldrich) to pull down FLAG-tagged RGTB1 and RGTB2, along with their respective interacting proteins. We followed the manufacturer's protocol for the equilibrating, binding, washing, and elution procedures, using 3× FLAG peptide to elute bound proteins.
To detect the tagged proteins by Western blot, protein samples (e.g. soluble fractions from cell extracts and pull-down eluates) were boiled with 2× Laemmli sample buffer (Bio-Rad) and then loaded onto 10% SDS-polyacrylamide gels. The separated proteins were transferred to a Whatman Protran nitrocellulose membrane (GE Healthcare). The membrane was incubated in blotto (1× PBS, pH 7.4, 0.05% Tween 20, 5% nonfat dry milk) at room temperature with shaking for 1 h. For detecting FLAG-tagged RGTB1/2, the washed membrane was incubated with monoclonal anti-FLAG M2-HRP antibody (Sigma-Aldrich) at 1:1000 dilution in 1× PBST (1× PBS, pH 7.4, 0.05% Tween 20), shaking at room temperature for 1 h. For detecting c-Myc-tagged RGTA1/2, the washed membrane was incubated with monoclonal anti-c-Myc antibody (clone 9E10, Sigma-Aldrich) at a 1:5000 dilution in 1× PBST and shaken at room temperature for 2 h, followed by another round of washes and incubation with anti-mouse IgG (Fab-specific)-peroxidase (Sigma-Aldrich) at 1:6500 dilution in 1× PBST while shaking at room temperature for 1 h. We used Pierce ECL Western blotting substrate (Thermo Scientific) for final detection, following the user manual. The chemiluminescence image was taken by an ImageQuant LAS4000 mini imager (GE Healthcare).
Expression of GST-tagged Rab GTPases and Substitution of C-terminal Cysteines
The coding sequences of selected Arabidopsis Rab GTPases (Table 1) were amplified by PCR using cDNA from Arabidopsis Col-0 wild-type plants as a template. The coding sequences were cloned into the pGEX-4T-1 vector (GE Healthcare) to generate inducible in frame N-terminal GST fusions by a single digest with BamHI or EcoRI, followed by calf intestinal phosphatase treatment and ligation with T4 DNA ligase. Three of the constructs, pGEX-4T-1 RABA4B, pGEX-6P-1 RABF2A, and pGEX-6P-1 RABG3C, were kindly provided by Dr. Erik Nielsen (University of Michigan) (53). All clones were sequenced to verify that they encoded wild-type proteins.
The pGEX RAB constructs were transformed into chemically prepared Escherichia coli BL21 competent cells using a heat-shock method. To express N-GST-tagged Rab GTPases, the BL21 cells containing the expression construct were grown in LB medium with 100 μg/ml ampicillin to A600 0.6–0.8. The expression was then induced by adding isopropyl β-d-1-thiogalactopyranoside into the culture to a final concentration of 0.4 mm. After a 6-h induction at 25 °C, the cells were harvested and resuspended in ice-cold 1× PBS with 100 μg/ml lysozyme, 10 μg/ml DNase I, and 5 mm DTT. After a 5-min incubation at room temperature, the cells were lysed by sonication. The GST-tagged proteins in the supernatant of the cell lysate were purified with GST SpinTrap columns (GE Healthcare), following the manufacturer's instructions.
The C-terminal cysteine substitution mutant proteins were generated by introducing point mutations into reverse primers for amplifying the coding sequence from the pGEX constructs of the corresponding wild-type proteins. The cloning, expression, and purification procedures were the same as those for wild-type proteins. The presence of the introduced point mutations was verified by DNA sequencing.
We used Precision Red advanced protein assay reagent (Cytoskeleton, Denver, CO) to quantify all protein concentrations.
In Vitro Prenylation Assay
The preparation of AtREP protein, procedures of desalting and concentrating the purified recombinant proteins, and procedures of the isotope-based in vitro prenylation assays have been described previously in great detail (54). Unless specified, we generally followed this protocol with one modification that added 0.2–0.3 μl, instead of 1 μl, of tritium-labeled geranylgeranyl diphosphate (3H-GGPP; American Radiolabled Chemicals, St. Louis, MO) per reaction. We minimized the volume of the 3H-GGPP added to the reactions to minimize inhibition of the prenylation reactions by isopropyl alcohol and ammonia present in the solvent.
Results
RGTA1/2 and RGTB1/2, Encoded by Two Pairs of Paralogous Genes, Are Putative α and β Subunits of Arabidopsis Rab-GGT
Based on the annotated full-length coding sequences in the TAIR database and our cDNA sequencing result, 75% of the aligned nucleotides are identical between RGTA1 (At4g24490) and RGTA2 (At5g41820), whereas 85%of the aligned nucleotides are identical between RGTB1 (At5g12210) and RGTB2 (At3g12070), suggesting that RGTA1/2 and RGTB1/2 are two pairs of paralogous genes. However, our search using the Plant Genome Duplication Database (55) does not map any of these four genes to chromosome regions that were duplicated during the most recent Arabidopsis whole genome duplication (56), contrary to a previous report that these two duplications resulted from the whole genome duplication (57).
Pairwise alignment of the protein sequences shows that RGTB1 (321 aa) and RGTB2 (317 aa) are almost identical to each other, with 85% amino acid identity and 91% similarity. Both RGTB1 and RGTB2 are highly conserved with mammalian Rab-GGT β subunits (RABGGTB) in protein sequences, sharing 72 and 70% similarity to rat (Rattus norvegicus) RABGGTB, respectively, suggesting that they are paralogous putative β subunits of Arabidopsis Rab-GGT.
The protein sequences of RGTA1 (678 aa) and RGTA2 (687 aa) are also highly similar to each other, although the similarity is not as high as that of the two putative β subunits, with 68% amino acid identity and 76% similarity (Fig. 1A). RGTA1 and RGTA2 share 39 and 41% similarity to rat RABGGTA, respectively (Fig. 1A).
FIGURE 1.
The protein sequence alignments of Arabidopsis RGTA1, RGTA2, and their orthologs. A, the alignment of the full-length protein sequences of Arabidopsis RGTA1 (AtRGTA1), Arabidopsis RGTA2 (AtRGTA2), and rat Rab-GGT α subunit (RnRABGGTA). The predicted PPTA repeats are marked as I, II, III, IV, and V. The 12-aa insertion (HQKQDDEKQDDP) in the third PPTA repeat (III) of RGTA2 is underlined. B, alignment of the third PPTA repeats of Arabidopsis RGTA1, Arabidopsis RGTA2, and Rab-GGT α subunits of yeast (ScBET4), P. patens (PpRGTA1), rice (Os06g0677500), D. melanogaster (DmPTAR3), rat (RnRABGGTA), and human (HsRABGGTA). Black background and asterisk, identical residues; gray background and dot, similar residues.
A search for conserved domains in RGTA1 predicts five protein prenyltransferase α subunit (PPTA) repeats in the N-terminal region based on both the Prosite (58) profile (PS51147) and the Pfam (59) profile (PF01239), whereas a similar search for RGTA2 predicts five PPTA repeats based on the Prosite profile but only three based on the Pfam profile. The first and third PPTA repeats in RGTA2 predicted by the Prosite profile are not recognized by the Pfam profile. The alignment of RGTA1, RGTA2, and rat RABGGTA shows high similarity in the N-terminal helical domain consisting of the PPTA repeats (Fig. 1A, I–V). The most noticeable difference within this domain is a 12-aa insertion in the middle of the third (III) PPTA repeat of RGTA2 predicted by the Prosite profile (RGTA2 aa 137–148, Fig. 1A), which apparently disrupts this very conserved motif and may be responsible for the discrepancy between the predictions by the Prosite and Pfam profiles for the third repeat. PPTA repeats have been identified only in known protein prenyltransferase α subunits (27, 60). Similar to RGTA1, both the Prosite and Pfam profiles recognize five PPTA repeats in most protein prenyltransferase α subunits, including mammalian and yeast Rab-GGT α subunits (not shown). With predicted PPTA repeats highly similar to mammalian RABGGTA, RGTA1 and RGTA2 appear to be paralogous putative α subunits of Arabidopsis Rab-GGT. Disruption in the third PPTA repeat is unique to RGTA2 among the Rab-GGT α subunits of various eukaryotic species from yeast to humans (Fig. 1B), and it might result in some variation in the secondary structure and possibly also in the biochemical activity of RGTA2.
The alignment also shows that RGTA1 and RGTA2 have extended C-terminal regions compared to rat RABGGTA that contain conserved LRR motifs (Fig. 1A). The intermediating regions of RGTA1 and RGTA2 are much less similar to the Ig-like domain of mammalian RABGGTA, which lies between the helical domain and the LRR domain, despite a few patches of similar sequences found in this region (Fig. 1A). However, previous studies in mammals have shown that the Ig-like domain and the LRR domain are not involved in prenyltransferase activity (28, 61). Therefore, the differences in these regions of RGTA1 and RGTA2 are not likely to affect their putative function as Rab-GGT α subunits.
Putative Rab-GGT α Subunits Form Heterodimers with Putative β Subunits
All known protein prenyltransferases function as heterodimers consisting of one α subunit and one β subunit (10, 12). To examine the hypothesis that putative Rab-GGT α subunits in Arabidopsis partner with putative β subunits as functional Rab-GGTs, we first performed a pull-down experiment to test all four combinations between RGTA1/2 and RGTB1/2 for physical interactions. Each of the subunit combinations was co-expressed in a yeast strain in which the α subunit was c-Myc-tagged and the β-subunit was FLAG-tagged. The β subunit and interacting proteins were pulled down with anti-FLAG beads, and then anti-c-Myc antibody was used to detect the α subunit in the resulting eluate. We used individually expressed and purified RGTA2-c-Myc as the negative control. No detectable RGTA2-c-Myc was present in the pull-down eluate, indicating that RGTA2-c-Myc could only be pulled down in a complex with FLAG-tagged RGTB1/2. The experiments revealed that the two subunits in all of the four combinations physically interact with each other (Fig. 2). Thus, there are four putative Rab-GGT heterodimers in Arabidopsis: RGTA1·RGTB1, RGTA1·RGTB2, RGTA2·RGTB1, and RGTA2·RGTB2.
FIGURE 2.
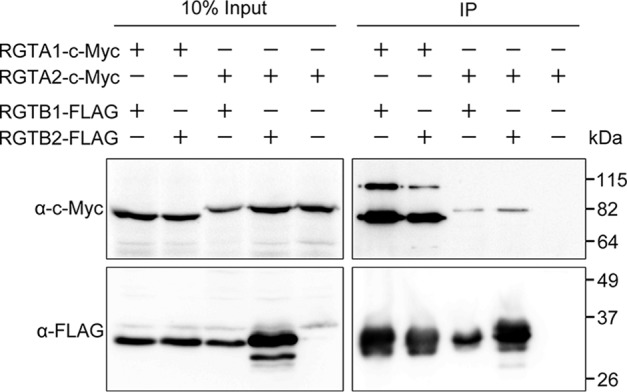
RGTA1 and RGTA2 interact with RGTB1 and RGTB2. RGTA1·c-Myc and RGTA2-c-Myc co-immunoprecipitated (IP) with RGTB1-FLAG and RGTB2-FLAG in the yeast cell extracts co-expressing the two subunits. The FLAG-tagged RGTB subunit was pulled down with anti-FLAG beads, and monoclonal anti-c-Myc antibody was used to detect whether the c-Myc-tagged RGTA was pulled down along with FLAG-tagged RGTB. RGTA2-c-Myc alone could not be pulled down by anti-FLAG antibody and thus did not present in the resulting eluate. Input and pull-down samples were resolved with two separate 10% SDS-polyacrylamide gels, and different epitope tags were detected with separate blots and one antibody at a time (see “Experimental Procedures”).
However, we noticed that RGTA1 and RGTA2 behaved differently in these experiments. We reproducibly obtained lower amounts of RGTA2 protein than RGTA1 protein in the eluates from pull-down experiments (Fig. 2). There are two possible explanations, which are not mutually exclusive, for this result: 1) the interaction between RGTA2 and either putative β subunit is weaker than that between RGTA1 and either putative β subunit, and 2) RGTA2, in our observation, is less stable than the other tested subunits during the processes of protein expression and purification (data not shown). These findings, together with the observation that a conserved PPTA motif in RGTA2 is disrupted by an insertion (Fig. 1), suggest that heterodimers with RGTA2 might be destabilized by the additional amino acids and/or might show altered biochemical activity in vitro. Given that RGTA1 better meets the criteria of a functional Rab-GGT α subunit compared with RGTA2 and that biochemical evidence has shown that RGTB1 is involved in Rab geranylgeranylation activity (37), we decided to use RGTA1·RGTB1 to initiate the following biochemical assays for testing Rab-GGT activity.
The RGTA1·RGTB1 Heterodimer Shows Rab-GGT Activity in Vitro in the Presence of AtREP
All known Rab-GGT heterodimers in animals and yeast require a third component, REP, for full activity (30, 62). In Arabidopsis, a single REP, AtREP (AT3G06540), has been identified. AtREP stimulates the prenylation of Rab GTPases in Arabidopsis cell extracts but not in cell extracts of the yeast REP mutant strain msr6, possibly due to a change of a conserved arginine residue to an asparagine (40). However, the necessity of AtREP for Arabidopsis Rab-GGT function has not been definitely determined.
To facilitate these studies, we developed an isotope-based in vitro prenylation assay consisting of purified recombinant enzymes, target and escort proteins, and 3H-GGPP (54). Similar to the interaction test above, RGTA1-c-Myc and RGTB1-FLAG were co-expressed in yeast and purified as a complex by pull-down with anti-FLAG beads. We then tested whether the purified RGTA1-c-Myc·RGTB1-FLAG complex has Rab-GGT activity, by combining it with GST-tagged target proteins and His-AtREP and assaying its ability to attach 3H-GGPP to the C-terminal cysteine residues of the target protein.
There are four classes of C-terminal putative prenylation target sequences found in Arabidopsis Rab GTPases: -CCXXX, -CCXX, -XCCX, and -XCXC (Table 1). We chose one Rab GTPase from each class as the representative to test Arabidopsis Rab-GGT activity (Table 1). Our results indicate that RGTA1·RGTB1 prenylates RABA1A, RABA2A, RABF2A, and RABG2 in vitro in the presence of AtREP (Fig. 3A). The requirement for AtREP was tested by omitting REP from otherwise identical reactions. Without AtREP, RGTA1·RGTB1 did not exhibit detectable Rab-GGT activity on any of the four Rab GTPases tested, indicating that AtREP is required for the Rab-GGT activity of RGTA1·RGTB1 (Figs. 3A and 4). The isotope-based detection of prenylated proteins was validated by altered migration of prenylated RABG2 in SDS-PAGE (Fig. 3B). This altered migration is consistent with that seen in previous studies of plant and human Rab GTPases (37, 63).
FIGURE 3.
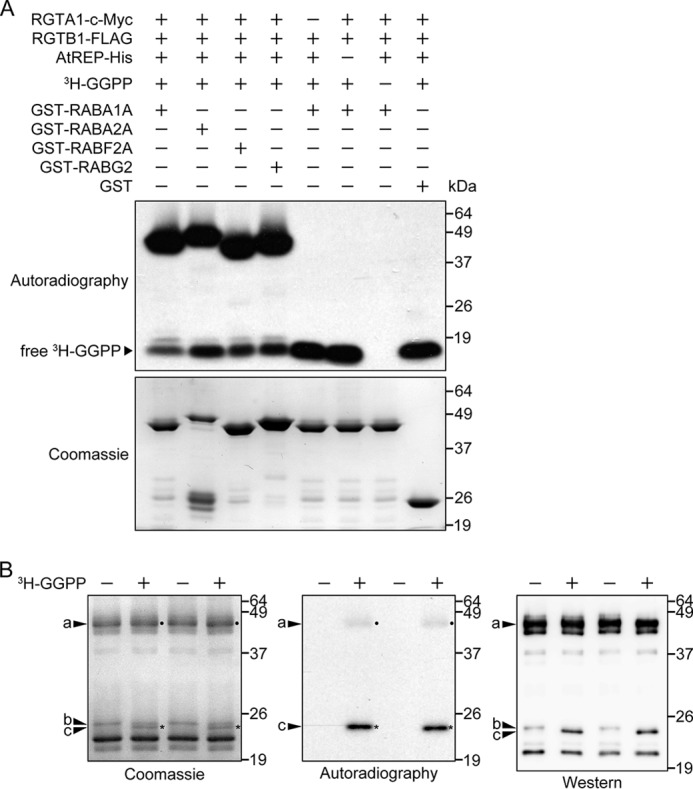
RGTA1·RGTB1 shows Rab-GGT activity in vitro. A, RGTA1·RGTB1 prenylates RABA1A, RABA2A, RABF2A, and RABG2 in vitro, in the presence of AtREP. The presence or absence of individual components in each reaction is indicated by a plus or minus sign, respectively. The reaction mixture was resolved by 10% SDS-PAGE. The x-ray film was exposed to the vacuum-dried SDS-PAGE gel at −80 °C for 48 h to detect radiolabeled 3H-GGPP. The bands of free, unincorporated 3H-GGPP at the bottom of the gels indicate that the labeled lipid substrate was always in excess in the reactions. B, prenylated RABG2 has altered SDS-PAGE migration. In vitro prenylation reactions, including RGTA1·RGTB1, AtREP, FLAG-RABG2, and with (+) or without (−) 3H-GGPP, were carried out and resolved on two 20% SDS-polyacrylamide gels. Proteins on one gel were visualized by Coomassie staining (left), and the gel was subsequently dried and exposed to an x-ray film for 12 h to detect radiolabeled prenylated proteins (center). The corresponding bands are marked with dots or asterisks on their right. Proteins on the other gel were transferred to a nitrocellulose membrane and probed with an anti-FLAG antibody (right). a, GST- and FLAG-double tagged RABG2 (GST-FLAG-RABG2), either prenylated or unprenylated; b, unprenylated FLAG-tagged RABG2 (FLAG-RABG2); c, prenylated FLAG-RABG2 (FLAG-RABG2-GG).
FIGURE 4.
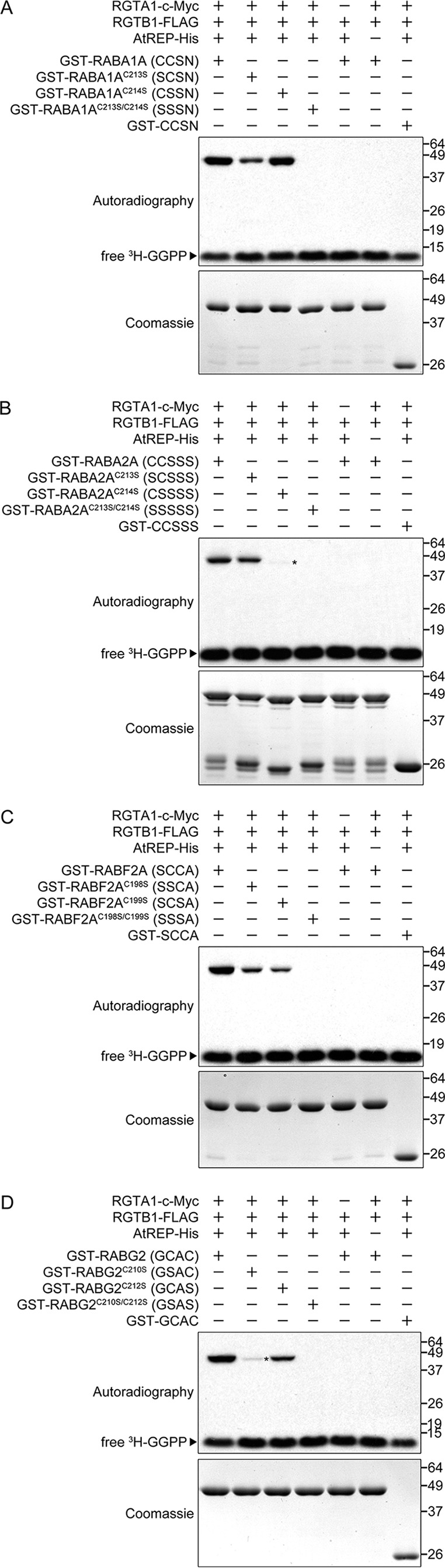
RGTA1·RGTB1 prenylates cysteine residues in a variety of C-terminal sequences in vitro. The presence or absence of individual components in each reaction is indicated by a plus or minus sign, respectively. Four proteins were chosen to represent the four different types of C-terminal sequences found in Arabidopsis Rab GTPases: RABA1A for -CCXX (A), RABA2A for -CCXXX (B); RABF2A for -XCCX (C); and RABG2 for -XCXC (D). For each representative protein, two single-cysteine substitution mutants and one double-cysteine substitution mutant were generated and tested in the in vitro prenylation assay. Exposure time for autoradiography was 24 h. The Coomassie Blue staining shows that equal amounts of target proteins were used in each reaction within each panel. Faint bands are marked with asterisk to help visualization.
To rule out the possibility that the prenylation activity may come from RGTB1 on its own or from yeast Rab-GGT subunits co-purified during the enzyme preparation, we used RGTB1-FLAG expressed in and purified from yeast as a negative control for Rab-GGT in the assay. RGTB1-FLAG on its own did not exhibit Rab-GGT activity either with (Fig. 3A) or without (data not shown) the addition of AtREP, indicating that it requires a partner α subunit for Rab-GGT activity. The result also indicates that RGTB1 does not form a functional Rab-GGT heterodimer with yeast Rab-GGT α subunit BET4. Therefore, in the above assay, RGTA1·RGTB1 exhibits bona fide Rab-GGT activity, and there is no detectable background activity from yeast Rab-GGT subunits.
RGTA1·RGTB1 Prenylates C-terminal Cysteine Residues at Different Positions in Vitro and May Exhibit Preference for Particular Positions
Unlike PFT and PGGT-I, which target more specific C-terminal CaaX sequences, Rab-GGT targets a wide variety of C-terminal sequences, most of which contain two cysteine residues for double geranylgeranylation (4, 16). In Arabidopsis, 54 of 57 identified Rab genes are predicted to encode Rab GTPases that have C-terminal cysteine residues and potentially can be prenylated (Table 1). To investigate which cysteine residues are prenylated by Rab-GGT, we use the same representative Rabs in the above activity assays to generate three cysteine-to-serine substitution mutants: two with only one cysteine residue substituted and one with both cysteine residues substituted. We then performed in vitro prenylation assays using the same quantity of wild-type and corresponding mutant Rab GTPase proteins across different reactions, in the presence of RGTA1·RGTB1 and AtREP.
Consistent with our earlier findings (Fig. 3), all wild-type target proteins tested (RABA1A, RABA2A, RABF2A, and RABG2) were prenylated by RGTA1·RGTB1 in the presence of AtREP. All single-substitution mutant proteins (RABA1AC213S, RABA1AC214S, RABA2AC213S, RABA2AC214S, RABF2AC198S, RABF2AC199S, RABG2C210S, and RABG2C212S) were also prenylated to some extent. In contrast, all double-substitution mutant proteins (RABA1AC213S/C214S, RABA2AC213S/C214S, RABF2AC198S/C199S, and RABG2C210S/C212S) remained unprenylated (Fig. 4, A–D). Altogether, these results suggest that 1) in all four types of C-terminal sequences, either cysteine residue can be geranylgeranylated by Rab-GGT, and 2) no other amino acid residue in Rab GTPases is geranylgeranylated (Fig. 4, A–D).
RGTA1·RGTB1 appears to have some preferences for prenylation of cysteine residues at different positions, as suggested by the isotope signal intensity from single-prenylated, single-substitution mutant proteins (Fig. 4, A–D). The cysteine residue at the fifth position from the C-terminal end of RABA2AC214S (C-terminal sequence: CSSSS) is very weakly prenylated (Fig. 4B), suggesting that the fifth amino acid residue from the C-terminal end is not a preferred prenylation site. Similarly, the very last residue at the C-terminal end also might not be preferred, because RABG2C210S (GSAC) is also weakly prenylated (Fig. 4D). RABA1AC214S (CSSN) and RABA2AC213S (SCSSS) are most strongly prenylated among all single-substitution mutant proteins, suggesting that the fourth residue from the C-terminal end might be the most preferred site for prenylation (Fig. 4, A and B). The second and third positions appear to be intermediately preferred, and the preferences of these two positions are not distinguishable from each other in our assays (Fig. 4, A, C, and D).
As controls for this experiment, we generated several artificial target proteins, GST-CCSSS, GST-CCSN, GST-SCCA, and GST-GCAC, by adding the C-terminal sequences from RABA1A, RABA2A, RABF2A, and RABG2, respectively, to the C-terminal end of GST protein. These proteins were not prenylated by RGTA1·RGTB1 in the presence of AtREP, suggesting that the target protein specificity of Arabidopsis Rab-GGT requires not only cysteine-containing C-terminal sequences but also other sequence or structural features of Rab GTPases for target recognition (Fig. 4, A–D).
RGTA1·RGTB1 Can Also Prenylate Certain PGGT-I Targets in Vitro Independently of AtREP
Rab-GGT is believed to exclusively prenylate Rab GTPases in animals and yeast (16, 26) because Rab-GGT relies on REP for target protein specificity (61). Given that AtREP also binds to Arabidopsis Rab GTPases (40) and our findings that 1) AtREP is required for Arabidopsis Rab-GGT activity and 2) artificial target proteins cannot be prenylated by Arabidopsis Rab-GGT, it is possible that Arabidopsis Rab-GGTs might have target protein specificity similar to that of their counterparts in animals and yeast. However, contrary to the lethality of PFT/PGGT-I α subunit loss-of-function mutants in animals and yeast, the Arabidopsis PFT/PGGT-I α subunit knockout mutant plp is viable and fertile (20), suggesting that some other prenyltransferase activity, possibly from Rab-GGT, can partially compensate for the loss of PFT/PGGT-I.
To test this hypothesis, we chose several non-Rab GTP-binding proteins reported to be prenylated by PGGT-I in Arabidopsis to perform in vitro prenylation assays: AGG1 and AGG2, which are two γ subunits of Arabidopsis heterotrimeric G proteins (15, 64, 65), and AtROP1, one of the Arabidopsis Rop family GTPases (9, 66). Our results indicate that RGTA1·RGTB1 can also prenylate AGG2 and AtROP1, but not AGG1, in vitro (Fig. 5). Moreover, the cross-specificity of RGTA1·RGTB1 on AGG2 and AtROP1 does not require AtREP, although the presence of AtREP appears to stimulate the prenylation of AtROP1 (Fig. 5). Therefore, RGTA1·RGTB1 can prenylate certain Arabidopsis PGGT-I target proteins in an REP-independent manner.
FIGURE 5.
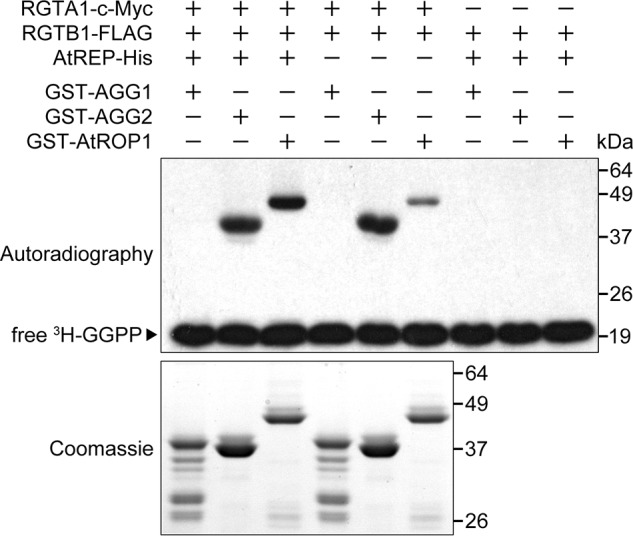
RGTA1·RGTB1 prenylates PGGT-I targets AGG2 and AtROP1 in vitro, independently of AtREP. The presence or absence of individual components in each reaction is indicated with a plus or minus sign, respectively. The amounts of AGG1 and AGG2 proteins were twice as much as that of AtROP1 protein used in each in vitro prenylation reaction. The prenylation of AtROP1 in the absence of AtREP is less efficient than that in the presence of AtREP. Exposure time for autoradiography was 48 h.
RGTB1 and RGTB2 Are Redundant Rab-GGT β Subunits, whereas RGTA2 Does Not Appear to Be a Functional Rab-GGT α Subunit in Vitro
By using in vitro prenylation assays, we have shown that RGTA1·RGTB1 is a bona fide Rab-GGT. However, as discussed earlier, the other three putative Rab-GGT heterodimers may have altered activities and/or target specificities due to differences between paralogous putative subunits.
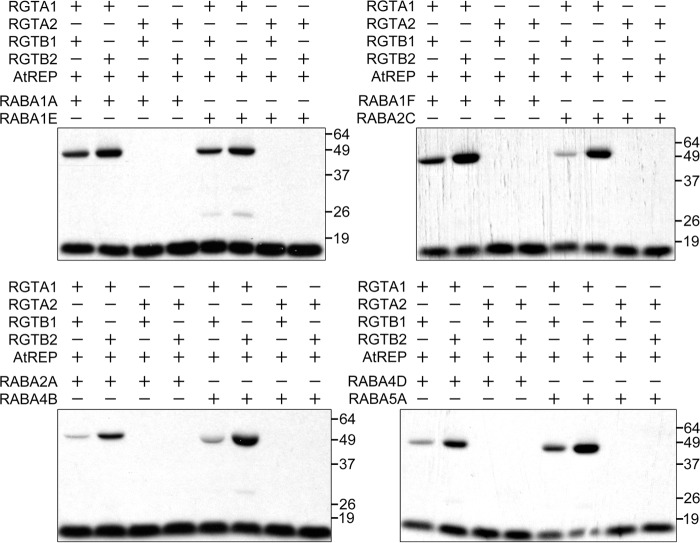
To investigate whether RGTA1·RGTB2, RGTA2·RGTB1, and RGTA2·RGTB2 are also functional and, if so, whether they have different target specificities, we performed in vitro prenylation assays using a subset of Arabidopsis Rab GTPases to represent the different subfamilies of the entire Rab family. The representative proteins were chosen based on their phylogenetic relationship (3, 6), expressed sequence tag availability, and C-terminal sequences (Table 1). AGG2 and AtROP1, which we showed above to be prenylated by RGTA1·RGTB1, were also included. For each target protein, four reactions were carried out, using the four possible α·β subunit heterodimers, respectively. The results, which are summarized in Table 2, demonstrate that RGTA1·RGTB1 and RGTA1·RGTB2 can prenylate every target protein tested in the presence of AtREP, suggesting that RGTB1 and RGTB2 are biochemically redundant when partnered with RGTA1 in vitro (Table 2 and Fig. 6). However, none of the target proteins tested were prenylated by RGTA2·RGTB1 or RGTA2·RGTB2, indicating that RGTA2 appears not to be a functional Rab-GGT α subunit when partnered with either RGTB1 or RGTB2 in vitro (Table 2 and Fig. 6). As discussed earlier, the loss of Rab-GGT α subunit function might result from the disrupted third PPTA repeat in the RGTA2 helical domain (Fig. 1).
TABLE 2.
Target specificities of RGTA1·RGTB1, RGTA1·RGTB2, RGTA2·RGTB1, and RGTA2·RGTB2
Shown is a summary of the results of the in vitro prenylation assay using the four Rab-GGT heterodimers, 25 different target proteins, and AtREP. A plus sign denotes that the target protein was prenylated by the Rab-GGT α·β heterodimer in the presence of AtREP. A minus sign indicates that the target protein was not prenylated. See Fig. 6 for the original autoradiography data from several representative experiments.
| Target protein |
Rab-GGT activity |
||||
|---|---|---|---|---|---|
| Name | C-terminal sequence | RGTA1·RGTB1 | RGTA1·RGTB2 | RGTA2·RGTB1 | RGTA2·RGTB2 |
| RABA1A | CCSN | + | + | − | − |
| RABA1E | CCSG | + | + | − | − |
| RABA1F | CCSN | + | + | − | − |
| RABA2A | CCSSS | + | + | − | − |
| RABA2C | CCSS | + | + | − | − |
| RABA3 | SCSC | + | + | − | − |
| RABA4B | CCTSS | + | + | − | − |
| RABA4D | CCKGS | + | + | − | − |
| RABA5A | CCSS | + | + | − | − |
| RABA5C | CCSR | + | + | − | − |
| RABB1B | GCCG | + | + | − | − |
| RABC1 | CCSS | + | + | − | − |
| RABC2A | GCCS | + | + | − | − |
| RABD1 | CCGQ | + | + | − | − |
| RABD2A | CCST | + | + | − | − |
| RABD2B | CCSS | + | + | − | − |
| RABE1A | CCGT | + | + | − | − |
| RABF2A | SCCA | + | + | − | − |
| RABG2 | GCAC | + | + | − | − |
| RABG3B | GCAC | + | + | − | − |
| RABG3C | GCEC | + | + | − | − |
| RABH1C | GCSC | + | + | − | − |
| RABH1E | GCAC | + | + | − | − |
| AGG2 | CSIL | + | + | − | − |
| ATROP1 | CSIL | + | + | − | − |
FIGURE 6.
Representative in vitro prenylation assays to test target specificities of RGTA1·RGTB1, RGTA1·RGTB2, RGTA2·RGTB1, and RGTA2·RGTB2. The presence or absence of individual components in each reaction is indicated by plus or minus signs, respectively. For the in vitro prenylation reactions testing the same Rab target, a mixture including reaction buffer, AtREP, Rab, and 3H-GGPP was prepared before adding different RGTA-RGTB heterodimers to each aliquot. The results are not quantitative. Exposure time for autoradiography was 24 h.
Discussion
Rab-GGT activity was detected in plants nearly 20 years ago (33–35). Two pairs of paralogous genes in Arabidopsis, RGTA1/2 and RGTB1/2, have long been annotated as genes encoding putative α and β subunits of Arabidopsis Rab-GGTs, respectively, based on homology (36). However, except for studies done with rgtb1 mutant plant extracts (37), the biochemical activities, partner/cofactor requirements, and substrate specificities of those putative Rab-GGT subunits had not been characterized. In this study, we present biochemical evidence that all four α·β combinations among RGTA1/2 and RGTB1/2 form heterodimers. Our assays indicate that RGTA1·RGTB1 and RGTA1·RGTB2 exhibit similar Rab-GGT activity and can prenylate a wide spectrum of Rab GTPases in vitro. In contrast, RGTA2·RGTB1 and RGTA2·RGTB2 did not show detectable Rab-GGT activity in our assays, possibly due to a 12-aa insertion that disrupts the third PPTA repeat in RGTA2. We also demonstrate that AtREP is required for the Rab-GGT activity of RGTA1·RGTB1 and RGTA1·RGTB2 prenylation of Rab GTPases.
By substituting the C-terminal cysteine residues, we demonstrate that the Arabdiopsis Rab-GGT RGTA1·RGTB1 can recognize and prenylate all four types of C-terminal sequences found in Arabidopsis Rab GTPases (-CCXX, -CCXXX, -XCCX, -XCXC), and both cysteine residues in the C-terminal sequences can be prenylated when GGPP is in abundance. However, the single-substitution mutant Rab GTPases show different degrees of prenylation, suggesting some preference in prenylation of cysteine residues at various positions. The fourth amino acid residue from the C-terminal end appears to be the most favored prenylation site, whereas the fifth and the first appear to be least favored. Previous work in mammals has shown that the double geranylgeranylation of Rab GTPases occurs in two sequential but independent steps, and the order of the two steps appears to be random (67). The proximal sequences on the N-terminal side of the cysteine residues are flexible in terms of prenylation target specificity (68). Therefore, the preference that we observe may solely rely on the position of the amino acid residue relative to the C-terminal end, although we cannot rule out the possibility that Arabidopsis Rab-GGTs show a greater preference for certain proximal amino acids. We hypothesize that the space limitation in the Rab-GGT catalytic site is responsible for the prenylation preference at different positions because the size or shape of the site might confine the C-terminal sequence in a certain conformation and only allow the cysteine residues close to the catalytic center to be efficiently prenylated.
Several target protein cross-specificities between PFT and PGGT-I have been reported (15, 19, 20, 69), but it has long been believed that Rab-GGT only prenylates Rab GTPases (16, 26). One of the novel findings in our study is that, in addition to Rab GTPases, Arabidopsis Rab-GGT can also prenylate certain PGGT-I targets in vitro, including the G-protein γ subunit AGG2 and the Rop family GTPase AtROP1. This finding may help to explain the viability and fertility of the Arabidopsis PFT/PGGT-I α subunit mutant plp as well as the mild phenotype of the Arabidopsis PGGT-I β subunit mutant ggb and residual membrane localization of the PGGT-I target AGG2 in plp (15, 20, 25), because Rab-GGT may at least partially compensate for the loss of PGGT-I in Arabidopsis. However, unidentified additional prenyltransferase components as well as other types of lipid modifications, such as S-acylation, myristoylation, and palmitoylation, may also potentially compensate for the loss of PFT/PGGT-I activity (10).
Previous studies of Rab-GGTs in mammals and yeast have shown that Rab-GGTs are completely dependent on AtREP for target specificity (i.e. the recognition and binding of Rab GTPases) (61). However, our results indicate that the prenyltransferase activities of Arabidopsis Rab-GGT in prenylation of AGG2 and AtROP1 are independent of AtREP, suggesting that Arabidopsis Rab-GGT can recognize and recruit certain target proteins, other than Rab GTPases, by itself. One similar case has been observed in C. elegans, in which the prenylation of some specific Rab GTPases is independent of REP (70). It has been proposed that an ancient Rab-GGT, once a PGGT-I-like protein, evolved to interact with an accessory protein over time and eventually gave up the specificity to the accessory protein, thus giving rise to the modern Rab-GGT and REP system (61). It is possible that Arabidopsis Rab-GGT retained or regained some specificity cues from a PGGT-I-like ancestor.
In animals and yeast, generally only one copy of each Rab-GGT subunit gene is present in the genome. In contrast, duplications of Rab-GGT subunits are found in multiple plant species (37, 39, 57). It has been suggested that duplications in different plant species have occurred independently, rather than having been inherited from a common ancestor (37). Some researchers have proposed that the two sets of Rab-GGT subunits in Arabidopsis were duplicated simultaneously in the recent whole genome duplication event (57). However, based on our analysis, none of the Arabidopsis Rab-GGT genes are found in any of the duplicated chromosome regions proposed to be involved in the whole genome duplication. Moreover, the flanking sequences of the Rab-GGT genes are not related to any sequence in other chromosome regions, suggesting that these genes have not been duplicated in large syntenic blocks.
The functional significance of having two copies of Rab-GGT subunits remains unclear (57). In yeast, loss of either of the single-copy Rab-GGT subunits leads to lethality (37). In Arabidopsis and P. patens, in which RGTB is duplicated, the rgtb1 and rgtb2 single knock-out mutants are viable, whereas the rgtb1 rgtb2 double mutants are non-viable, indicating genetic redundancy between the duplicated RGTB genes (38, 39). The duplicated RGTB genes appear to be completely redundant in P. patens, because neither single rgtb knock-out shows a detectable phenotype (39). However, each of the Arabidopsis rgtb single knockouts has a distinct set of mutant phenotypes (37, 38), suggesting that Arabidopsis RGTB1 and RGTB2 are only partially redundant. Our results show that Arabidopsis RGTB1 and RGTB2 are biochemically redundant in vitro, suggesting that there might be additional factors that differentiate RGTB1 and RGTB2 functions in vivo. Alternatively, the partial redundancy of RGTB1 and RGTB2 in Arabidopsis may result from differential expression.
In contrast, no Arabidopsis rgta1 or rgta2 mutants have been reported. Our results indicate that, although RGTA1 partners with both RGTB1 and RGTB2 to form a functional Rab-GGT, RGTA2 seems not to be functional in vitro. If this is also true in vivo, rgta1 mutants should be non-viable, similar to P. patens rgta1 mutants (39), whereas rgta2 mutants might exhibit no phenotype. It has been proposed that, despite possible redundancy, the duplicated Rab-GGT subunits in Arabidopsis may result in increased enzyme dosage and differential specificity in order to deal with the large family of Arabidopsis Rab GTPases (38, 57). However, our finding that RGTA1 is possibly the only functional Rab-GGT α subunit may make it the limiting factor in forming heterodimeric enzymes. Together with the observation that the transcript level of RGTA1 is much lower than that of either β subunit throughout the plant (37), the dosage effect hypothesis may not be supported in Arabidopsis.
Among numerous variations between the protein sequences of Arabidopsis RGTA1 and RGTA2, probably the strongest explanation for the loss of RGTA2 α subunit function is the 12-aa insertion in the third PPTA repeat, which is the longest stretch of continuous variation in the pairwise alignment between RGTA1 and RGTA2. This insertion may be unique to the Arabidopsis lineage, based on our search for RGTA homologs in various plant species (data not shown). We also noticed that some of the nucleotide sequences encoding the inserted amino acids contain some repetitive sequences (data not shown), suggesting that the insertion might have been introduced by replication slippage after RGTA was duplicated. By aligning RGTA2 protein sequence to the structure of rat RABGGTA in the Rab-GGT·REP complex (Protein Data Bank entry 1LTX) (31), we located the insertion at the C-terminal end of the α6 helix. The insertion might result in an extended linker between α6 and α7 helices or even more significant changes in the structural conformation that could impair function. For example, the C-terminal end of the α6 helix is facing and close to the Rab-GGT α·β interface; thus, the additional amino acids might interfere with dimerization, consistent with our observation that the interactions between RGTA2 and RGTB1/2 are weaker than that of RGTA1. It is also possible that the extension of the linker between helices caused by the insertion might block the access of Rab GTPase to the enzyme's catalytic center. It would be interesting to see whether removing the insertion from RGTA2 rescues its interaction with β subunits and its α subunit function.
Author Contributions
W. S. designed experiments, performed all experiments, prepared all figures and tables, and wrote the manuscript. Q. Z. provided select constructs and helped to develop in vitro prenylation assay protocols. M. P. R. provided intellectual framework and input, guided experimental design and direction, and revised the manuscript. B. N. K. provided intellectual and experimental input and laboratory facilities and revised the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Erik Nielsen for providing select Rab expression constructs; Dr. Hani Zaher for technical advice on yeast protein expression and purification; and Dr. Ram Dixit, Dr. Hani Zaher, Dr. Sona Pandey, and Dr. Kenneth Olsen for constructive discussion.
This study was supported by Kentucky Science and Engineering Foundation Grant KSEF-2841-RDE-016 and National Science Foundation Grants NSF-IIA-1355438 and NSF-IOS-1456884 (to M. P. R.). The authors declare that they have no conflicts of interest with the contents of this article.
- PFT
- protein farnesyltransferase
- PGGT-I
- protein geranylgeranyltransferase type I
- Rop
- Rho of plants
- Rab-GGT
- Rab geranylgeranyltransferase
- LRR
- leucine-rich repeat
- REP
- Rab escort protein
- 3H-GGPP
- tritium labeled geranylgeranyl diphosphate
- aa
- amino acid(s)
- PPTA
- protein prenyltransferase α subunit.
References
- 1.Molendijk A. J., Ruperti B., and Palme K. (2004) Small GTPases in vesicle trafficking. Curr. Opin. Plant Biol. 7, 694–700 [DOI] [PubMed] [Google Scholar]
- 2.Woollard A. A., and Moore I. (2008) The functions of Rab GTPases in plant membrane traffic. Curr. Opin. Plant Biol. 11, 610–619 [DOI] [PubMed] [Google Scholar]
- 3.Vernoud V., Horton A. C., Yang Z., and Nielsen E. (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131, 1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira-Leal J. B., Hume A. N., and Seabra M. C. (2001) Prenylation of Rab GTPases: molecular mechanisms and involvement in genetic disease. FEBS Lett. 498, 197–200 [DOI] [PubMed] [Google Scholar]
- 5.Grosshans B. L., Ortiz D., and Novick P. (2006) Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. U.S.A. 103, 11821–11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutherford S., and Moore I. (2002) The Arabidopsis Rab GTPase family: another enigma variation. Curr. Opin. Plant Biol. 5, 518–528 [DOI] [PubMed] [Google Scholar]
- 7.Pinheiro H., Samalova M., Geldner N., Chory J., Martinez A., and Moore I. (2009) Genetic evidence that the higher plant Rab-D1 and Rab-D2 GTPases exhibit distinct but overlapping interactions in the early secretory pathway. J. Cell Sci. 122, 3749–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ueda T., Yamaguchi M., Uchimiya H., and Nakano A. (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20, 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorek N., Bloch D., and Yalovsky S. (2009) Protein lipid modifications in signaling and subcellular targeting. Curr. Opin. Plant Biol. 12, 714–720 [DOI] [PubMed] [Google Scholar]
- 10.Running M. P. (2014) The role of lipid post-translational modification in plant developmental processes. Front. Plant Sci. 5, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casey P. J., and Seabra M. C. (1996) Protein prenyltransferases. J. Biol. Chem. 271, 5289–5292 [DOI] [PubMed] [Google Scholar]
- 12.Maurer-Stroh S., Washietl S., and Eisenhaber F. (2003) Protein prenyltransferases. Genome Biol. 4, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galichet A., and Gruissem W. (2003) Protein farnesylation in plants: conserved mechanisms but different targets. Curr. Opin. Plant Biol. 6, 530–535 [DOI] [PubMed] [Google Scholar]
- 14.Sorek N., Gutman O., Bar E., Abu-Abied M., Feng X., Running M. P., Lewinsohn E., Ori N., Sadot E., Henis Y. I., and Yalovsky S. (2011) Differential effects of prenylation and S-acylation on type I and II ROPS membrane interaction and function. Plant Physiol. 155, 706–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng Q., Wang X., and Running M. P. (2007) Dual lipid modification of Arabidopsis G γ-subunits is required for efficient plasma membrane targeting. Plant Physiol. 143, 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung K. F., Baron R., and Seabra M. C. (2006) Thematic review series: lipid posttranslational modifications. geranylgeranylation of Rab GTPases. J. Lipid Res. 47, 467–475 [DOI] [PubMed] [Google Scholar]
- 17.Mijimolle N., Velasco J., Dubus P., Guerra C., Weinbaum C. A., Casey P. J., Campuzano V., and Barbacid M. (2005) Protein farnesyltransferase in embryogenesis, adult homeostasis, and tumor development. Cancer Cell 7, 313–324 [DOI] [PubMed] [Google Scholar]
- 18.He B., Chen P., Chen S. Y., Vancura K. L., Michaelis S., and Powers S. (1991) RAM2, an essential gene of yeast, and RAM1 encode the two polypeptide components of the farnesyltransferase that prenylates a-factor and Ras proteins. Proc. Natl. Acad. Sci. U.S.A. 88, 11373–11377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trueblood C. E., Ohya Y., and Rine J. (1993) Genetic evidence for in vivo cross-specificity of the CaaX-box protein prenyltransferases farnesyltransferase and geranylgeranyltransferase-I in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 4260–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Running M. P., Lavy M., Sternberg H., Galichet A., Gruissem W., Hake S., Ori N., and Yalovsky S. (2004) Enlarged meristems and delayed growth in plp mutants result from lack of CaaX prenyltransferases. Proc. Natl. Acad. Sci. U.S.A. 101, 7815–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cutler S., Ghassemian M., Bonetta D., Cooney S., and McCourt P. (1996) A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273, 1239–1241 [DOI] [PubMed] [Google Scholar]
- 22.Running M. P., Fletcher J. C., and Meyerowitz E. M. (1998) The WIGGUM gene is required for proper regulation of floral meristem size in Arabidopsis. Development 125, 2545–2553 [DOI] [PubMed] [Google Scholar]
- 23.Bonetta D., Bayliss P., Sun S., Sage T., and McCourt P. (2000) Farnesylation is involved in meristem organization in Arabidopsis. Planta 211, 182–190 [DOI] [PubMed] [Google Scholar]
- 24.Yalovsky S., Kulukian A., Rodríguez-Concepción M., Young C. A., and Gruissem W. (2000) Functional requirement of plant farnesyltransferase during development in Arabidopsis. Plant Cell 12, 1267–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson C. D., Chary S. N., Chernoff E. A., Zeng Q., Running M. P., and Crowell D. N. (2005) Protein geranylgeranyltransferase I is involved in specific aspects of abscisic acid and auxin signaling in Arabidopsis. Plant Physiol. 139, 722–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H., Seabra M. C., and Deisenhofer J. (2000) Crystal structure of Rab geranylgeranyltransferase at 2.0 Å resolution. Structure 8, 241–251 [DOI] [PubMed] [Google Scholar]
- 27.Zhang H. (2006) Protein prenyltransferases. in Handbook of Metalloproteins, John Wiley & Sons, Inc., New York, 10.1002/0470028637.met002 [DOI] [Google Scholar]
- 28.Dursina B., Thomä N. H., Sidorovitch V., Niculae A., Iakovenko A., Rak A., Albert S., Ceacareanu A. C., Kölling R., Herrmann C., Goody R. S., and Alexandrov K. (2002) Interaction of yeast Rab geranylgeranyl transferase with its protein and lipid substrates. Biochemistry 41, 6805–6816 [DOI] [PubMed] [Google Scholar]
- 29.Andres D. A., Seabra M. C., Brown M. S., Armstrong S. A., Smeland T. E., Cremers F. P., and Goldstein J. L. (1993) cDNA cloning of component A of Rab geranylgeranyl transferase and demonstration of its role as a Rab escort protein. Cell 73, 1091–1099 [DOI] [PubMed] [Google Scholar]
- 30.Anant J. S., Desnoyers L., Machius M., Demeler B., Hansen J. C., Westover K. D., Deisenhofer J., and Seabra M. C. (1998) Mechanism of Rab geranylgeranylation: formation of the catalytic ternary complex. Biochemistry 37, 12559–12568 [DOI] [PubMed] [Google Scholar]
- 31.Pylypenko O., Rak A., Reents R., Niculae A., Sidorovitch V., Cioaca M. D., Bessolitsyna E., Thomä N. H., Waldmann H., Schlichting I., Goody R. S., and Alexandrov K. (2003) Structure of Rab escort protein-1 in complex with Rab geranylgeranyltransferase. Mol. Cell 11, 483–494 [DOI] [PubMed] [Google Scholar]
- 32.Baron R. A., and Seabra M. C. (2008) Rab geranylgeranylation occurs preferentially via the pre-formed REP-RGGT complex and is regulated by geranylgeranyl pyrophosphate. Biochem. J. 415, 67–75 [DOI] [PubMed] [Google Scholar]
- 33.Biermann B., Randall S. K., and Crowell D. N. (1996) Identification and isoprenylation of plant GTP-binding proteins. Plant Mol. Biol. 31, 1021–1028 [DOI] [PubMed] [Google Scholar]
- 34.Loraine A. E., Yalovsky S., Fabry S., and Gruissem W. (1996) Tomato Rab1A homologs as molecular tools for studying Rab geranylgeranyl transferase in plant cells. Plant Physiol. 110, 1337–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yalovsky S., Loraine A. E., and Gruissem W. (1996) Specific prenylation of tomato Rab proteins by geranylgeranyl type-II transferase requires a conserved cysteine-cysteine motif. Plant Physiol. 110, 1349–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lange B. M., and Ghassemian M. (2003) Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol. Biol. 51, 925–948 [DOI] [PubMed] [Google Scholar]
- 37.Hála M., Soukupová H., Synek L., and Zárský V. (2010) Arabidopsis RAB geranylgeranyl transferase β-subunit mutant is constitutively photomorphogenic and has shoot growth and gravitropic defects. Plant J. 62, 615–627 [DOI] [PubMed] [Google Scholar]
- 38.Gutkowska M., Wnuk M., Nowakowska J., Lichocka M., Stronkowski M. M., and Swiezewska E. (2015) Rab geranylgeranyl transferase β subunit is essential for male fertility and tip growth in Arabidopsis. J. Exp. Bot. 66, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thole J. M., Perroud P. F., Quatrano R. S., and Running M. P. (2014) Prenylation is required for polar cell elongation, cell adhesion, and differentiation in Physcomitrella patens. Plant J. 78, 441–451 [DOI] [PubMed] [Google Scholar]
- 40.Hála M., Eliás M., and Zárský V. (2005) A specific feature of the angiosperm Rab escort protein (REP) and evolution of the REP/GDI superfamily. J. Mol. Biol. 348, 1299–1313 [DOI] [PubMed] [Google Scholar]
- 41.Huala E., Dickerman A. W., Garcia-Hernandez M., Weems D., Reiser L., LaFond F., Hanley D., Kiphart D., Zhuang M., Huang W., Mueller L. A., Bhattacharyya D., Bhaya D., Sobral B. W., Beavis W., Meinke D. W., Town C. D., Somerville C., and Rhee S. Y. (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 29, 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cherry J. M., Hong E. L., Amundsen C., Balakrishnan R., Binkley G., Chan E. T., Christie K. R., Costanzo M. C., Dwight S. S., Engel S. R., Fisk D. G., Hirschman J. E., Hitz B. C., Karra K., Krieger C. J., Miyasato S. R., Nash R. S., Park J., Skrzypek M. S., Simison M., Weng S., and Wong E. D. (2012) Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40, D700–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duvick J., Fu A., Muppirala U., Sabharwal M., Wilkerson M. D., Lawrence C. J., Lushbough C., and Brendel V. (2008) PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res. 36, D959–D965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawahara Y., de la Bastide M., Hamilton J. P., Kanamori H., McCombie W. R., Ouyang S., Schwartz D. C., Tanaka T., Wu J., Zhou S., Childs K. L., Davidson R. M., Lin H., Quesada-Ocampo L., Vaillancourt B., Sakai H., Lee S. S., Kim J., Numa H., Itoh T., Buell C. R., and Matsumoto T. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.dos Santos G., Schroeder A. J., Goodman J. L., Strelets V. B., Crosby M. A., Thurmond J., Emmert D. B., Gelbart W. M., and FlyBase Consortium (2015) FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 43, D690–D697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruitt K. D., Brown G. R., Hiatt S. M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C. M., Hart J., Landrum M. J., McGarvey K. M., Murphy M. R., O'Leary N. A., Pujar S., Rajput B., Rangwala S. H., Riddick L. D., Shkeda A., Sun H., Tamez P., Tully R. E., Wallin C., Webb D., Weber J., Wu W., DiCuccio M., Kitts P., Maglott D. R., Murphy T. D., and Ostell J. M. (2014) RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 42, D756–D763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McWilliam H., Li W., Uludag M., Squizzato S., Park Y. M., Buso N., Cowley A. P., and Lopez R. (2013) Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 41, W597–W600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitchell A., Chang H. Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S., Sangrador-Vegas A., Scheremetjew M., Rato C., Yong S. Y., Bateman A., Punta M., Attwood T. K., Sigrist C. J., Redaschi N., Rivoire C., Xenarios I., Kahn D., Guyot D., Bork P., Letunic I., Gough J., Oates M., Haft D., Huang H., Natale D. A., Wu C. H., Orengo C., Sillitoe I., Mi H., Thomas P. D., and Finn R. D. (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43, D213–D221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pagni M., Ioannidis V., Cerutti L., Zahn-Zabal M., Jongeneel C. V., and Falquet L. (2004) MyHits: a new interactive resource for protein annotation and domain identification. Nucleic Acids Res. 32, W332–W335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Geer L. Y., Chappey C., Kans J. A., and Bryant S. H. (2000) Cn3D: sequence and structure views for Entrez. Trends Biochem. Sci. 25, 300–302 [DOI] [PubMed] [Google Scholar]
- 52.Gietz R. D., and Schiestl R. H. (2007) Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 35–37 [DOI] [PubMed] [Google Scholar]
- 53.Preuss M. L., Serna J., Falbel T. G., Bednarek S. Y., and Nielsen E. (2004) The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell 16, 1589–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi W., Zeng Q., and Running M. P. (2013) In vitro prenylation assay of Arabidopsis proteins. Methods Mol. Biol. 1043, 147–160 [DOI] [PubMed] [Google Scholar]
- 55.Lee T. H., Tang H., Wang X., and Paterson A. H. (2013) PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41, D1152–D1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 [DOI] [PubMed] [Google Scholar]
- 57.Rasteiro R., and Pereira-Leal J. B. (2007) Multiple domain insertions and losses in the evolution of the Rab prenylation complex. BMC Evol. Biol. 7, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sigrist C. J., de Castro E., Cerutti L., Cuche B. A., Hulo N., Bridge A., Bougueleret L., and Xenarios I. (2013) New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., Heger A., Hetherington K., Holm L., Mistry J., Sonnhammer E. L., Tate J., and Punta M. (2014) Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Charng W. L., Yamamoto S., Jaiswal M., Bayat V., Xiong B., Zhang K., Sandoval H., David G., Gibbs S., Lu H. C., Chen K., Giagtzoglou N., and Bellen H. J. (2014) Drosophila Tempura, a novel protein prenyltransferase α subunit, regulates notch signaling via Rab1 and Rab11. PLoS Biol. 12, e1001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo Z., Wu Y. W., Das D., Delon C., Cramer J., Yu S., Thuns S., Lupilova N., Waldmann H., Brunsveld L., Goody R. S., Alexandrov K., and Blankenfeldt W. (2008) Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation. EMBO J. 27, 2444–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seabra M. C., Brown M. S., Slaughter C. A., Südhof T. C., and Goldstein J. L. (1992) Purification of component A of Rab geranylgeranyl transferase: possible identity with the choroideremia gene product. Cell 70, 1049–1057 [DOI] [PubMed] [Google Scholar]
- 63.Sanford J. C., Foster L., Kapadia Z., and Wessling-Resnick M. (1995) Analysis of the stoichiometry of Rab protein prenylation. Anal. Biochem. 224, 547–556 [DOI] [PubMed] [Google Scholar]
- 64.Mason M. G., and Botella J. R. (2000) Completing the heterotrimer: isolation and characterization of an Arabidopsis thaliana G protein γ-subunit cDNA. Proc. Natl. Acad. Sci. U.S.A. 97, 14784–14788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mason M. G., and Botella J. R. (2001) Isolation of a novel G-protein γ-subunit from Arabidopsis thaliana and its interaction with Gβ. Biochim. Biophys. Acta 1520, 147–153 [DOI] [PubMed] [Google Scholar]
- 66.Lin Y., Wang Y., Zhu J. K., and Yang Z. (1996) Localization of a Rho GTPase implies a role in tip growth and movement of the generative cell in pollen tubes. Plant Cell 8, 293–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Durek T., Alexandrov K., Goody R. S., Hildebrand A., Heinemann I., and Waldmann H. (2004) Synthesis of fluorescently labeled mono- and diprenylated Rab7 GTPase. J. Am. Chem. Soc. 126, 16368–16378 [DOI] [PubMed] [Google Scholar]
- 68.Wu Y. W., Goody R. S., Abagyan R., and Alexandrov K. (2009) Structure of the disordered C terminus of Rab7 GTPase induced by binding to the Rab geranylgeranyl transferase catalytic complex reveals the mechanism of Rab prenylation. J. Biol. Chem. 284, 13185–13192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Armstrong S. A., Hannah V. C., Goldstein J. L., and Brown M. S. (1995) CAAX geranylgeranyl transferase transfers farnesyl as efficiently as geranylgeranyl to RhoB. J. Biol. Chem. 270, 7864–7868 [DOI] [PubMed] [Google Scholar]
- 70.Tanaka D., Kameyama K., Okamoto H., and Doi M. (2008) Caenorhabditis elegans Rab escort protein (REP-1) differently regulates each Rab protein function and localization in a tissue-dependent manner. Genes Cells 13, 1141–1157 [DOI] [PubMed] [Google Scholar]