Abstract
The eukaryotic translation initiation factor (eIF) 4G is required during protein synthesis to promote the assembly of several factors involved in the recruitment of a 40S ribosomal subunit to an mRNA. Although many eukaryotes express two eIF4G isoforms that are highly similar, the eIF4G isoforms in plants, referred to as eIF4G and eIFiso4G, are highly divergent in size, sequence, and domain organization but both can interact with eIF4A, eIF4B, eIF4E isoforms, and the poly(A)-binding protein. Nevertheless, eIF4G and eIFiso4G from wheat exhibit preferences in the mRNAs they translate optimally. For example, mRNA containing the 5′-leader (called Ω) of tobacco mosaic virus preferentially uses eIF4G in wheat germ lysate. In this study, the eIF4G isoform specificity of Ω was used to examine functional differences of the eIF4G isoforms in Arabidopsis. As in wheat, Ω-mediated translation was reduced in an eif4g null mutant. Loss of the eIFiso4G1 isoform, which is similar in sequence to wheat eIFiso4G, did not substantially affect Ω-mediated translation. However, loss of the eIFiso4G2 isoform substantially reduced Ω-mediated translation. eIFiso4G2 is substantially divergent from eIFiso4G1 and is present only in the Brassicaceae, suggesting a recent evolution. eIFiso4G2 isoforms exhibit sequence-specific differences in regions representing partner protein and RNA binding sites. Loss of any eIF4G isoform also resulted in a substantial reduction in reporter transcript level. These results suggest that eIFiso4G2 appeared late in plant evolution and exhibits more functional similarity with eIF4G than with eIFiso4G1 during Ω-mediated translation.
Keywords: eukaryotic translation initiation, eukaryotic translation initiation factor 4G (eIF4G), protein synthesis, RNA, translation, eIFiso4G, mRNA stability, translation initiation factors
Introduction
The synthesis of proteins from cellular mRNAs requires several translation initiation factors that assist in recruiting the 40S ribosomal subunit to an mRNA as well as in the recognition of the initiation codon and in the assembly of the 80S ribosome (1–3). Eukaryotic initiation factor (eIF) 4F is essential to promote 40S subunit binding to an mRNA and to assist in 40S subunit scanning of the 5′-leader in search of the initiation codon. eIF4F is a multisubunit factor composed of eIF4E, which binds the 5′-cap structure; the RNA helicase, eIF4A, which unwinds the secondary structure in a 5′-leader that would otherwise inhibit 40S subunit scanning; and the scaffolding protein, eIF4G, which interacts with eIF4E, eIF4A, eIF4B (which stimulates the RNA helicase activity of eIF4A), eIF3 (required for 40 S binding to an mRNA), and the poly(A)-binding protein (PABP)2 (2, 4–6). The interactions between eIF4G and eIF4E and between eIF4G and PABP results in mRNA circularization that stimulates translation by promoting 40S subunit recruitment (7, 8). An interaction between eIF4B and PABP further increases the affinity for poly(A) RNA of PABP (9–11). These factors, therefore, are necessary for the efficient translation of most mRNAs.
Although plants express two forms of eIF4G as do many eukaryotes (12), the plant isoforms are more divergent from each other than observed between eIF4G isoforms in other species. eIF4G in wheat is 168 kDa, whereas the second isoform, eIFiso4G, is only 83–86 kDa and they differ in sequence homology sufficiently that each interacts with a specific eIF4E isoform: eIF4G interacts with eIF4E, whereas eIFiso4G interacts with eIFiso4E (13). Despite their differences in size and sequence, both isoforms interact with eIF4B, eIF4A, and PABP, although eIF4G has two interaction domains for each of these partner proteins, whereas eIFiso4G has only one such domain for eIF4B and PABP (14, 15). The divergence between the two eIF4G isoforms has suggested functional specialization that allows them to discriminate between mRNAs that has been experimentally supported. For example, barley α-amylase and oat globulin mRNAs use eIF4G preferentially for their translation. In contrast, translation of Arabidopsis HSP21 and alfalfa mosaic virus RNA 4 is supported equally well by either eIF4G or eIFiso4G (16). eIF4G is used preferentially for the cap-independent translation mediated by the tobacco etch virus 5′-leader (17).
Although the activity of translation initiation factors might be expected to affect translational efficiency only, the translational status of an mRNA can influence the rate of mRNA decay (18). NPQ1 encodes violaxanthin deepoxidase (VDE), the enzyme responsible for the de-epoxidation of pigments in the xanthophyll cycle (19). Loss of eIFiso4G expression in Arabidopsis resulted in more than a 5-fold increase in the NPQ1 transcript level, which correlated with increases in VDE protein and VDE activity (20). The NPQ1 5′-leader was sufficient to confer this increase in transcript level in response to loss of eIFiso4G expression. This observation suggested that changes in eIF4G isoform expression can affect other aspects of gene expression in addition to translation.
Differential use of eIF4G isoforms has also been reported for the 5′-leader sequence from tobacco mosaic virus (TMV) (21). The TMV genome is a single strand, positive sense RNA that functions as the mRNA for the 5′-proximal cistron encoding the replicase. The 68-nucleotide TMV 5′-leader (known as Ω) facilitates the first round of translation of the genomic RNA to which the viral coat protein binds loosely releasing the RNA from the virion particle through a co-translational disassembly process (22). Translation in fractionated in vitro lysates demonstrated that eIF4F was 20–30-fold more active in translating mRNAs containing Ω as the leader than eIFiso4F (21).
In this study, the eIF4G isoform-specific preferences of the Ω 5′-leader were examined in vivo in Arabidopsis. The Arabidopsis genome contains a single gene encoding eIF4G, whereas two genes encode eIFiso4G isoforms (eIFiso4G1 and eIFiso4G2) (23). Loss of expression of any eIF4G isoform did not affect growth rate or size but did result in a substantial reduction in reporter gene expression in seedlings, adult leaves, and flowers, indicating that each eIF4G isoform contributes significantly to the protein synthetic capacity. Interestingly, much of the reduction in expression was a result of a decrease in reporter gene transcript level. The translational efficiency of Ω-containing mRNA, however, was reduced to a greater extent in an eif4g null mutant than in an eifiso4g1 null mutant. Surprisingly, the translational efficiency of Ω-containing mRNA was also reduced in an eifiso4g2 mutant. As eIFiso4G1 is expressed to a higher level than eIFiso4G2, this suggests that the two eIF4G isoforms are functionally different from one another and from eIF4G. Phylogenetic analysis demonstrated that eIFiso4G2 appears only in the Brassicaceae and is substantially divergent in sequence from eIFiso4G1 that is present in Arabidopsis. These results suggest that eIFiso4G2 is a Brassicaceae-specific isoform that exhibits functional differences from eIFiso4G1 present throughout the plant kingdom.
Experimental Procedures
Plant Growth and Transformation
Arabidopsis seeds were germinated on soil and grown in a plant growth room supplemented with Sylvania Gro-Lite fluorescent bulbs (Sylvania, Danvers, MA) at a photon flux density (PFD) of 100 μmol photons m−2 s−1. Wild-type Arabidopsis was transformed with the firefly luciferase (Luc) transgene under control of the cauliflower mosaic virus 35S promoter with or without the Ω 5′-leader using the binary vector, pBI121 as described (20). The primary inflorescences of Arabidopsis plants were removed, and the secondary inflorescences were allowed to initiate before infiltration. Inverted plants were dipped into the infiltration medium containing the Aglo1 strain of Agrobacterium containing a transgene. Infiltrated plants were kept on their side for 1 day and allowed to continue to flower in an upright position in the same growth room. Seeds of infiltrated plants were collected and screened on 0.25 × MS plates containing 50 μg/ml of kanamycin and 500 μg/ml of vancomycin.
Luciferase Assay
Plant samples were ground in liquid nitrogen and the soluble protein fraction resuspended in luciferase assay buffer (25 mm Tricine, pH 8, 5 mm MgCl2, 0.1 mm EDTA supplemented with 33.3 mm DTT, 270 μm coenzyme A, 500 μm ATP). Aliquots of total cell extract were assayed for luciferase activity following injection of 0.5 mm luciferin using a Monolight 2010 Luminometer (Analytical Luminescence Laboratory). Triplicate plant samples were prepared for each line and the average value reported. All luciferase expression data are normalized for equal amounts of total protein.
qPCR Analysis
Plant material was frozen in liquid nitrogen and ground to a fine powder, and 100 mg was resuspended in 1 ml of TRIzol® reagent (Invitrogen). Following centrifugation, the supernatant was extracted with 200 μl of chloroform and centrifuged to separate the phases. RNA was precipitated from the aqueous phase using isopropyl alcohol, and the RNA pellet was washed with 75% ethanol and resuspended in RNase-free H2O. 1 μg of RNA was used to obtain the first-strand cDNA by Omniscript RT kit (Qiagen) in a 20-μl reaction. The qPCR analysis was performed using a iQ5 real time PCR detection system (Bio-Rad) in 25-μl reactions containing 1× SYBR Green SuperMix 500 nm forward and reverse primers and 10 ng of cDNA. Reactions were carried out using the following conditions: 95 °C for 5 min (1 cycle) and 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s (35 cycles). To detect the presence of Luc, a forward primer, Luc-F1 (5′-CCGTTGTTGTTTTGGAGCACGGAAA-3′), and a reverse primer, Luc-R1 (5′-GATCTCTCTGATTTTTCTTGCGTCGAG-3′), were used. Protein phosphatase PP2A (At1g13320) was used as the reference gene for the quantitation of Luc expression in Arabidopsis leaves. To detect the expression of PP2A, a forward primer, PP2A-FW (5′-AGTATCGCTTCTCGCTCCAG-3′) and a reverse primer, PP2A-RV (5′-GTTCTCCACAACCGCTTGGT-3′), were used. The efficiency of PCR was determined by five 10-fold serial dilutions of the template DNAs in triplicate.
Sequence Alignment and Phylogenetic Analysis
The eIFiso4G1 and eIFiso4G2 amino acid sequences of Arabidopsis thaliana used in this study are available in the National Center for Biotechnology Information (NCBI) sequence database (www.ncbi.nlm.nih.gov) and were used as queries to perform BLAST searches of the Phytozome database (v10.3) (phytozome.jgi.doe.gov) for orthologs of the species used in the study. Amino acid sequence alignments for examining eIFiso4G2-specific sequence differences were performed by ClustalW2 with the following parameters: pairwise gap opening penalty 10, pairwise gap extension penalty 0.1, multiple gap opening penalty 10, multiple gap extension penalty 0.2, Gonnet protein weight matrix, and no end gap separation.
Using alignments obtained by MUSCLE, the evolutionary history was inferred by using the maximum likelihood method based on the JTT matrix-based model (24). The tree with the highest log likelihood (−6408.5683) is shown. Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using a JTT model. Similar trees were seen using the Whalen and Goldman substitution model and did not alter the findings. The tree is drawn to scale. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test is shown next to the branches in the figures with branch lengths measured in the number of substitutions per site. No branches were collapsed regardless of bootstrap values. The analysis involved 66 amino acid sequences. There were a total of 232 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (25).
Gene sequences used the analyses were from A. thaliana (AT2G24050 and AT5G57870); Arabidopsis lyrata (Al_481263, Al_495887, and Al_353746); Boechera stricta (Bs_27895s0080, Bs_26833s0467, and Bs_7867s0925); Capsella rubella (Cr_10022680m, Cr_10025916m, and Cr_10004122m); Capsella grandiflora (Cg_3181s0015, Cg_0380s0056, and Cg_2848s0070); Eutrema salsugineum (Es_10000058m and Es10012738m); Brassica rapa (Br_K00529, Br_H01394, Br_A00681, and Br_B01185); Carica papaya (Cp_supercontig_46.175); Gossypium raimondii (Gr_009G009800, Gr_004G273900, Gr_013G105700, and Gr_013G241300); Theobroma cacao (Tc_1EG036897 and Tc_1EG038403); Eucalyptus grandis (Eg_C00247 and Eg_C00248); Citrus clementina (Cc_10007494m and Cc_10007769m); Manihot esculenta (Me_1_001915m and Me_1_001934m); Ricinus communis (Rc_27455.m000039); Phaseolus vulgaris (Phv_003G154900 and Phv_009G207600); Glycine max (Glyma.17G072500.1, Glyma.02G205500.1, Glyma.06G225700.1, and Glyma.04G154100.1); Vitis vinifera (Vv_GSVIVT01023638001 and Vv_GSVIVT01035980001); Solanum lycopersicum (Sl_07g005810.2 and Sl_12g009960.1); Solanum tuberosum (St_PGSC0003DMT400029102, St_PGSC0003DMT400020464, and St_PGSC0003DMT400029103); Sorghum bicolor (Sb006G136400); Brachypodium distachyon (Bd_5g14687); Zea mays (Zm_GRMZM2G098577 and ZmGRMZM2G157061); Oryza sativa (Os02g39840 and Os04g42140); Panicum virgatum (Pv_Ab02269, Pv_Aa01373, Pv_J01984, and Pv_Gb01258); Setaria italica (Si_016412m and Si_009406m); Triticum aestivum (Ta_M95747); Selaginella moellendorffii (Sm_437322); Physcomitrella patens (Pp_014G077600, Pp_014G078000, Pp_017G069800, Pp_017G069500, and Pp_017G069100).
Results
Each eIF4G Isoform Contributes Substantially to Luc Expression Throughout Plant Development
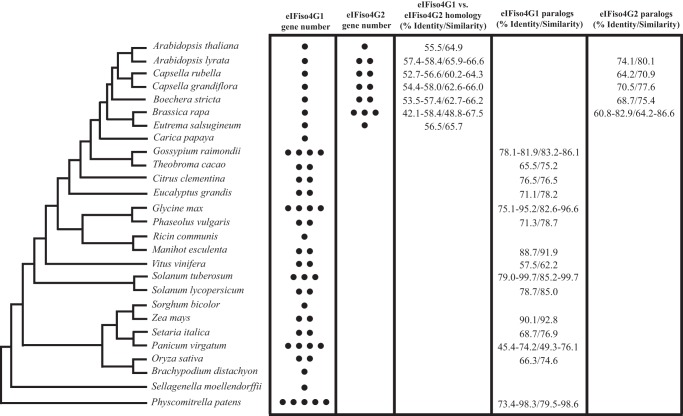
eIF4G is encoded by a single gene (i.e. At3g60240) in the A. thaliana genome. Other than a slight delay in flowering time, loss of eIF4G expression in A. thaliana results in no other readily observable phenotype (20). eIFiso4G is encoded by two genes, eIFiso4G1 and eIFiso4G2 (i.e. At5g57870 and At2g24050, respectively) (23), which share only 55.4% identity and 64.9% similarity. As in A. thaliana, eIFiso4G is encoded by two genes in maize (GRMZM2G157061 and GRMZM2G098577) but these are more similar (90.1% identity and 92.8% similarity). Loss of either eIFiso4G1 or eIFiso4G2 in A. thaliana does not result in a visible phenotype other than a slightly smaller stature in eifiso4g1 plants. However, the eIFiso4G double null mutant, i.e. eifiso4g1/2, is substantially smaller, grows slower, and contains less chlorophyll than WT plants (20, 23), demonstrating that both eIFiso4G genes contribute to vegetative growth and that they must share some functional similarities. Interestingly, eIFiso4E, the eIF4E isoform that interacts with the eIFiso4G isoforms, is encoded by a single gene (i.e. At5g35620) in A. thaliana but the eifiso4e null mutant has no visible phenotype (20), perhaps because eIF4E can interact with eIFiso4G as a less preferred partner in the absence of eIFiso4E (13). Although eIFiso4G and eIF4G exhibit functional differences (12, 16, 17, 20, 21), the limited homology between the eIFiso4G isoforms in A. thaliana suggests that they too may be functionally divergent in some respects.
To determine the extent to which each eIF4G and eIFiso4G gene products contribute to gene expression in Arabidopsis, the eifiso4e, eif4g, eifiso4g1, and eifiso4g2 mutants previously described (20) were crossed with Arabidopsis expressing the firefly luciferase (Luc) cDNA under the control of the CaMV promoter. As the eif4e triple null mutant is not viable (26), it could not be included. Progeny homozygous for the Luc transgene and null for the initiation factor were isolated. This approach avoided position effects of the transgene in the genome. Luciferase activity was then measured in different organs to determine the effect of loss of an initiation factor on its expression. Luciferase expression in 8-day-old eifiso4e null mutant seedlings, representing total steady-state luciferase activity normalized for equal amounts of total protein, was not significantly different from that in WT seedlings (Table 1). In contrast, loss of expression of either eIFiso4G1 or eIFiso4G2 in seedlings resulted in substantial reduction in luciferase activity although loss of eIFiso4G2 expression had a greater effect on reporter gene expression than did loss of eIFiso4G1 expression (Table 1). Luciferase activity in the eifiso4g1/2 double mutant was lower than in either single mutant. The greatest reduction in Luc expression, however, was observed in the eif4g mutant (Table 1).
TABLE 1.
Expression from Luc mRNA in WT and mutant lines
| Mutant line | Luciferase expressiona |
|||||
|---|---|---|---|---|---|---|
| Seedlings | Relative expression | Adult leaves | Relative expression | Flowers | Relative expression | |
| Luc/min/mg protein | ||||||
| WT | 1,025,659 ± 519,237 | 1 | 179,7961 ± 35249 | 1 | 4,479,681 ± 335,657 | 1 |
| eifiso4e | 1,122,767 ± 381,480 | 1.09 | 1,177,807 ± 97,045 | 0.655 | 2,682,324 ± 102,022 | 0.599 |
| eif4g | 4,404 ± 427 | 0.00429 | 32,955 ± 1,050 | 0.0183 | 92,614 ± 6,429 | 0.0207 |
| eifiso4g1 | 270,204 ± 107,473 | 0.263 | 172,935 ± 9,062 | 0.0961 | 249,183 ± 6,328 | 0.0556 |
| eifiso4g2 | 71,921 ± 7,613 | 0.0701 | 84,433 ± 5,200 | 0.0470 | 75,362 ± 6,679 | 0.0168 |
| eifiso4g1/2 | 28,821 ± 2,570 | 0.0281 | 164,344 ± 12,769 | 0.0914 | 231,244 ± 14,855 | 0.0516 |
a All luciferase expression data are normalized for equal amounts of total protein.
As in seedlings, loss of eIF4G expression in adult leaves resulted in the greatest reduction in steady-state Luc expression (Table 1). However, loss of expression of either eIFiso4G1, eIFiso4G2, or the double mutant also resulted in substantial reduction in Luc expression. Luc activity in eifiso4e adult leaves was reduced moderately relative to WT leaves (Table 1). Similar results were observed in flowers (Table 1). These results indicate that eIF4G and each of the eIFiso4G isoforms contribute substantially to expression throughout vegetative growth and in flowers.
To determine whether loss of one eIF4G isoform affects the expression of the remaining isoforms, qPCR was performed on the mutants to measure the relative levels of each eIF4G isoform. As expected, little expression of eIF4G was observed in the eif4g mutant and the level of eIF4G expression in the eifiso4g1 and eifiso4g2 single mutants was similar to WT, whereas a modest reduction in expression was observed in the eifiso4g1/2 double mutant (Fig. 1). Expression of eIFiso4G1 was not significantly different in the eif4g or eifiso4g2 mutants (Fig. 1). Similarly, eIFiso4G2 was not lower in the eif4g or eifiso4g1 mutants. These results suggest that the reduction in Luc expression following loss of expression of one eIF4G isoform was not a result of a substantial decrease in the expression of the remaining isoforms.
FIGURE 1.
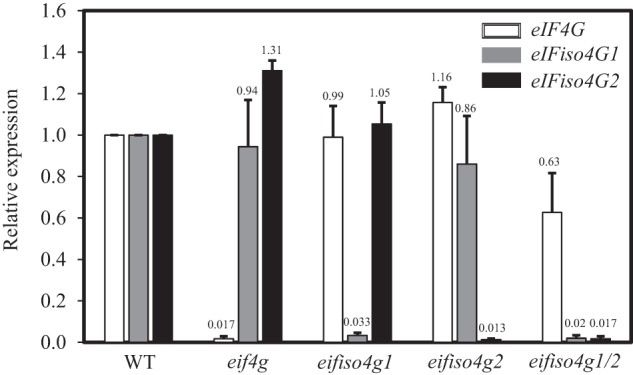
Expression of eIF4G isoforms in eIF4G mutants. qPCR was performed for eIF4G (white bars), eIFiso4G1 (gray bars), and eIFiso4G2 (black bars) in 8-day-old WT seedlings and eifiso4e, eif4g, eifiso4g1, eifiso4g2, and eifiso4g1/2 mutants. Expression is reported relative to the level in WT seedlings, which is set at a value of 1.
The Presence of Ω as the 5′-Leader Alters the Requirement for eIF4G Isoforms
To examine the eIF4G isoform requirement for a transcript containing Ω as the 5′-leader, the Ω sequence was introduced into the luciferase construct described above and plants containing the transgene were generated as previously described (20). The 35S::Ω-Luc transgene was then introduced into eIF4G isoform mutants through crosses to avoid position effects in the genome. Luciferase expression, representing total steady-state luciferase activity normalized for equal amounts of total protein, from the Ω-Luc construct was higher than from the Luc construct in WT seedlings (Table 2). A moderate reduction in expression was observed in the eifiso4e mutant relative to WT. As with the Luc construct, expression from the Ω-Luc construct was substantially lower in eif4g and eifiso4g mutants with expression being most affected following loss of eIF4G or eIFiso4G2 expression (Table 2). However, expression was also substantially reduced in the eifiso4g1 mutant. Expression in the eifiso4g1/2 double mutant was nearly as low as that observed for the eifiso4g2 mutant. In adult leaves and flowers, expression from the Ω-Luc construct was moderately higher in the eifiso4e mutant than in WT, in contrast to observations made in seedlings. However, as observed in seedlings, expression from the Ω-Luc construct was substantially reduced in the eif4g, eifiso4g1, eifiso4g2, and the eifiso4g1/2 double mutants and the reduction was more pronounced than that observed in seedlings.
TABLE 2.
Expression from Ω-Luc mRNA in WT and mutant lines
| Mutant line | Luciferase expressiona |
|||||
|---|---|---|---|---|---|---|
| Seedlings | Relative expression | Adult leaves | Relative expression | Flowers | Relative expression | |
| Luc/min/mg protein | ||||||
| WT | 1,484,868 ± 61880 | 1 | 2,113,573 ± 255,152 | 1 | 7,706,585 ± 324,332 | 1 |
| eifiso4e | 927,776 ± 146,235 | 0.625 | 3,961,323 ± 296,860 | 1.87 | 11,355,385 ± 396,134 | 1.47 |
| eif4g | 330 ± 274 | 0.000222 | 547 ± 67 | 0.000259 | 820 ± 132 | 0.000106 |
| eifiso4g1 | 202,298 ± 58,271 | 0.136 | 143,136 ± 4,312 | 0.0677 | 189,559 ± 5,766 | 0.0246 |
| eifiso4g2 | 1728 ± 147 | 0.00116 | 936 ± 63 | 0.000443 | 2,192 ± 40 | 0.00284 |
| eifiso4g1/2 | 14,640 ± 918 | 0.00986 | 106,912 ± 4,994 | 0.0506 | 108,006 ± 9,217 | 0.0140 |
a All luciferase expression data are normalized for equal amounts of total protein.
Loss of an eIF4G Isoform Affects Transcript Abundance
The luciferase expression data in Tables 1 and 3 represent contributions of the translatability and stability of an mRNA. Although eIF4G and its isoforms function to promote translation initiation, loss of an eIF4G isoform can affect the transcript level as shown by the increase in the level of Luc mRNA in the eifiso4g1/2 mutant when the VDE 5′-leader was present in the mRNA (20). To examine whether the reduced expression observed from the Ω-Luc and Luc constructs in eif4g, eifiso4g1, and eifiso4g2 mutants involves changes at the RNA level, the transcript level of each in each mutant was measured by qPCR. The transcript level of Luc was higher in the eifiso4e mutant than in WT plants but was substantially lower in eif4g, eifiso4g1, and eifiso4g2 mutants than in WT plants (Fig. 2A). It was also lower in the eifiso4g1/2 mutant, in good agreement with its level when expressed from the NPQ1 promoter in this same mutant (20). The Ω-Luc transcript level in eifiso4e was moderately lower than in WT plants (Fig. 2A), correlating with the modest reduction in Luc expression (Table 2). The Ω-Luc transcript level was substantially lower in eif4g, eifiso4g1, and eifiso4g2 mutants (Fig. 2A). It was also lower in eifiso4g1/2 plants as shown previously (20). When the transcript levels of Ω-Luc and Luc mRNAs were compared, Ω-Luc mRNA was more abundant in WT seedlings but was reduced to parity in the eifiso4e mutant (Fig. 2B). Although the transcript level of Ω-Luc mRNA relative to Luc mRNA was largely unchanged in eif4g and eifiso4g1, reductions in its relative level were observed in eifiso4g2 and eifiso4g1/2 seedlings (Fig. 2B). These results indicate that loss of eIFiso4E or eIFiso4G2 selectively reduces the level of Ω-Luc mRNA relative to the Luc transcript level but loss of any eIF4G isoform reduces the absolute level of Luc and Ω-Luc mRNAs. The reduction in their transcript level partially accounts for the reduction in Luc expression observed for these constructs.
TABLE 3.
eIF4G and eIFiso4G2 support Ω-mediated translation
| Mutant line | Luc mRNA |
Ω-Luc mRNA |
Ω dependence | ||
|---|---|---|---|---|---|
| Luciferase/qPCRa | Relative expression | Luciferase/qPCRa | Relative expression | ||
| WT | 1,025,659 | 1 | 331,629 | 1 | |
| eifiso4e | 1,122,767 | 1.09 | 1,181,881 | 3.56 | 0.307 |
| eif4g | 4,404 | 0.00429 | 56.2 | 0.000169 | 25.3 |
| eifiso4g1 | 270,204 | 0.263 | 45,562 | 0.137 | 1.92 |
| eifiso4g2 | 71,921 | 0.0701 | 883 | 0.00266 | 26.3 |
| eifiso4g1/2 | 28,821 | 0.0281 | 10,844 | 0.0327 | 0.859 |
FIGURE 2.
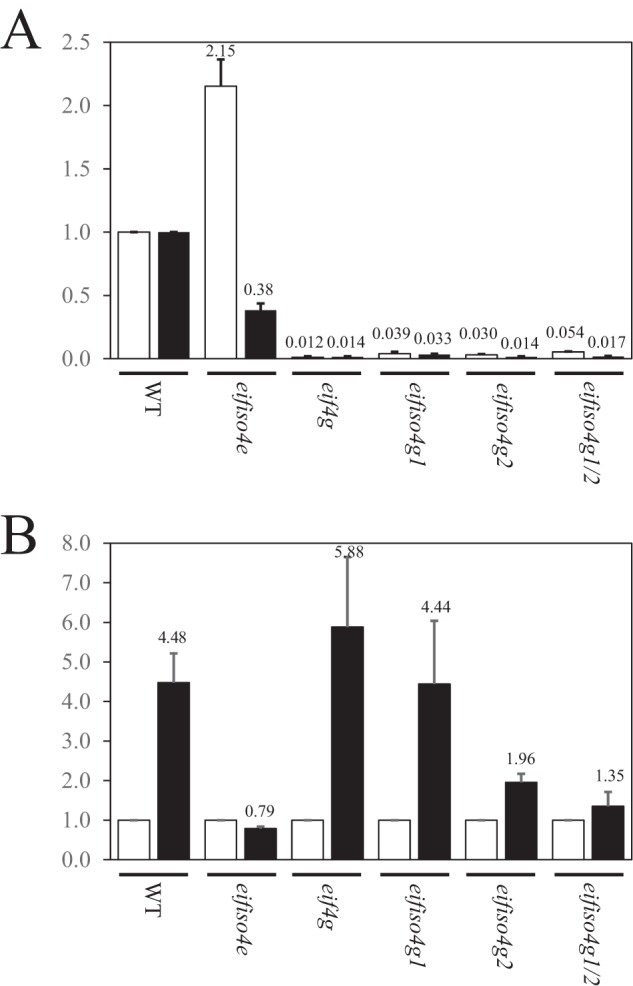
All eIF4G isoforms are required to support Luc and Ω-LUC transcript levels. qPCR was performed for expression from 35S::Luc and 35S::Ω-Luc transgenes in 8-day-old WT seedlings in WT plants and in eifiso4e, eif4g, eifiso4g1, eifiso4g2, and eifiso4g1/2 mutants into which each transgene was crossed. In A, expression is reported relative to the level of Luc (white bars) and Ω-Luc (black bars) mRNAs in WT seedlings, which is set at a value of 1. In B, the transcript level of Ω-Luc (black bars) is reported relative to the Luc (white bars) transcript level (set at a value of 1) for WT and mutant seedlings.
eIF4G2 Exhibits Functional Similarity with eIF4G during Ω-Mediated Translation
As noted above, loss of specific eIF4G isoforms affects Ω-Luc and Luc transcript abundance. To measure the relative translational efficiency of each mRNA, the Ω-Luc and Luc expression data obtained above in the eifiso4e, eif4g, eifiso4g1, and eifiso4g2 mutants can be normalized to RNA abundance using transcript levels determined by qPCR resulting in normalized values representing expression from a uniform amount of transcript (Fig. 2B). The degree to which the presence of Ω as the 5′-leader alters the translational efficiency of an mRNA can then be determined by the ratio of normalized luciferase expression from Ω-Luc mRNA to that from Luc mRNA. Relative comparisons between the normalized expression from the Ω-Luc and Luc constructs eliminates any indirect effect that loss of an eIF4G isoform may have on transcriptional activity, mRNA processing, or protein turnover. Thus the ratio of expression from Ω-Luc and Luc constructs normalized to mRNA levels enables the analysis of the effect that an eIF4G isoform has on translatability of an mRNA specifically.
This analysis revealed that translation from Ω-Luc mRNA was less dependent on eIFiso4E than Luc mRNA (Table 3), perhaps resulting from the unstructured nature of the Ω sequence and consistent with its ability to confer eIF4E-independent translation on an mRNA (27). In contrast, translation from Ω-Luc mRNA was ∼25-fold more dependent on eIF4G than Luc mRNA as translation from Ω-Luc mRNA was disproportionately reduced in the eif4g mutant (Table 3). Similarly, translation from Ω-Luc mRNA was ∼26-fold more dependent on eIFiso4G2 than Luc mRNA (Table 3). However, translation from Ω-Luc mRNA was significantly less dependent on the eIFiso4G1 isoform in that it was only 1.9-fold more dependent on this isoform than Luc mRNA (Table 3) indicating a significant difference between the two eIFiso4G isoforms in supporting translation from Ω-Luc mRNA. Interestingly, when both eIFiso4G isoforms were absent, the translation efficiency of Ω-Luc and Luc mRNAs was similar (Table 3), suggesting that a change in the relative abundance of the two eIFiso4G isoforms alters the translation efficiency of Ω-Luc mRNA.
eIFiso4G2 Is a Recently Evolved eIFiso4G Isoform Present Only in the Brassicaceae
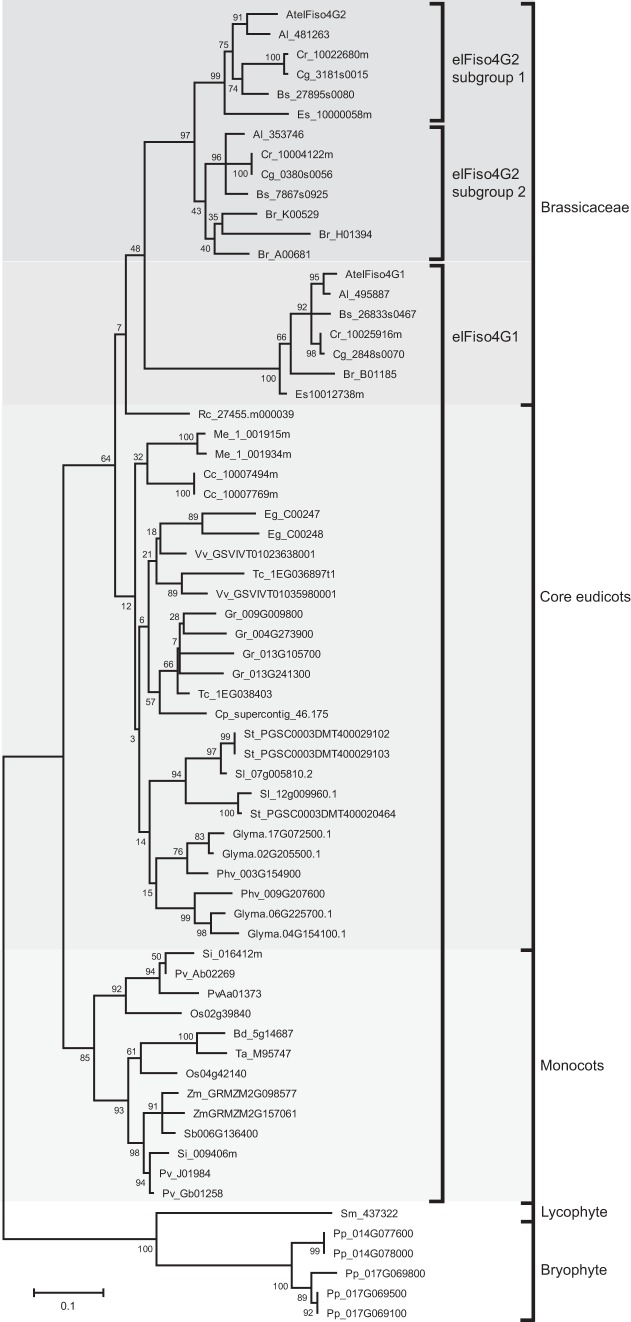
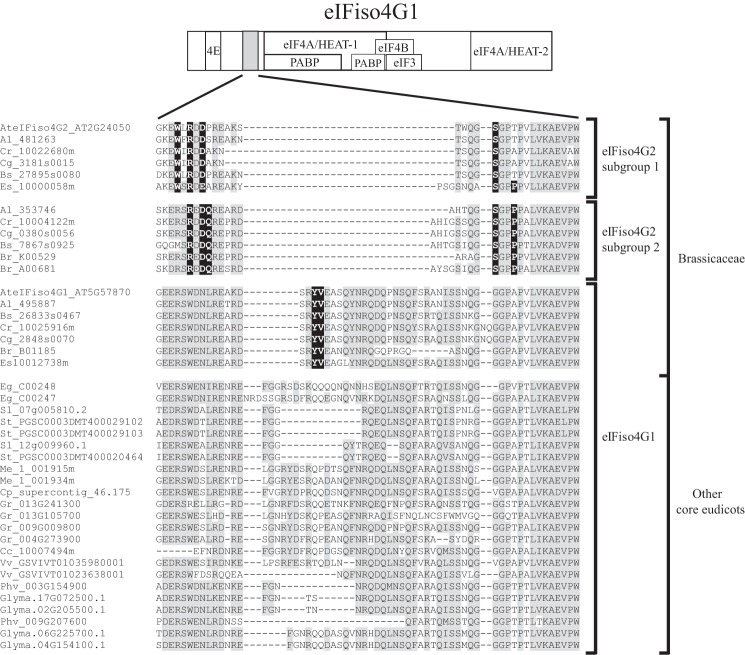
As noted above, although eIFiso4G1 and eIFiso4G2 are eIFiso4G isoforms, they exhibit substantial sequence divergence in Arabidopsis (Fig. 3). eIFiso4G1 and eIFiso4G2 orthologs in other species show similar low sequence conservation (Fig. 3). In species with multiple eIFiso4G2 paralogs, a higher level of sequence conservation was observed as it was among eIFiso4G1 paralogs (Fig. 3). To examine the relationship among eIFiso4G isoforms through land plant evolution, phylogenetic analysis was performed using eIFiso4G1 and eIFiso4G2 gene sequences representing non-vascular, vascular, and seed-bearing species. The tree was rooted using the eIFiso4G gene family from the non-vascular species, P. patens, which is composed of four members. As expected, the eIFiso4G isoforms from P. patens and the early vascular species, S. moellendorffii, cluster together and lie outside the eIFiso4G isoforms from flowering plants (Fig. 4). The eIFiso4G1 orthologs from monocotyledonous species also form a separate clade from that representing eIFiso4G1 orthologs of dicotyledonous species. The eIFiso4G isoforms in species of the Brassicaceae form a clade composed of a subclade of the eIFiso4G1 isoforms and two closely related subclades of eIFiso4G2 isoforms (Fig. 4). As eIFiso4G2 isoforms are present throughout the species of the Brassicaceae examined but are absent outside the Brassicaceae, these observations suggest that eIFiso4G2 arose early during the evolution of the Brassicaceae. The presence of multiple eIFiso4G2 isoforms in some species of the Brassicaceae that are present in each of the eIFiso4G2 subclades suggests gene duplication prior to speciation. The presence of an eIFiso4G2 isoform in each eIFiso4G2 subclade in Arabidopsis lyrata but only one eIFiso4G2 isoform in A. thaliana suggests that one eIFiso4G2 gene family member has been lost in the latter species. Together, these results indicate that eIFiso4G2 is a recently evolved eIFiso4G isoform not present outside the Brassicaceae.
FIGURE 3.
Evolution and sequence conservation of eIFiso4G1 and eIFiso4G2 in land plants. The presence of eIFiso4G1 and eIFiso4G2 genes in a species is indicated by a black dot and the number of genes for each type is indicated by the number of black dots. The evolutionary relationship of the species shown is indicated to the left. Sequence identity and similarity between eIFiso4G1 and eIFiso4G2 isoforms in each species is reported as are the identity and similarity among eIFiso4G1 isoforms and among eIFiso4G2 isoforms within a species when more than one isoform is present.
FIGURE 4.
Phylogenetic analysis of eIFiso4G1 and eIFiso4G2 isoforms in land plants. A phylogenetic tree of eIFiso4G1 and eIFiso4G2 isoforms was constructed using the maximum-likelihood method. The tree with the highest log likelihood is shown. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Numbers on each branch denote percentages of bootstrap support. eIFiso4G isoforms from the non-vascular species, P. patens, and the early vascular species, S. moellendorffii, were used to root the tree.
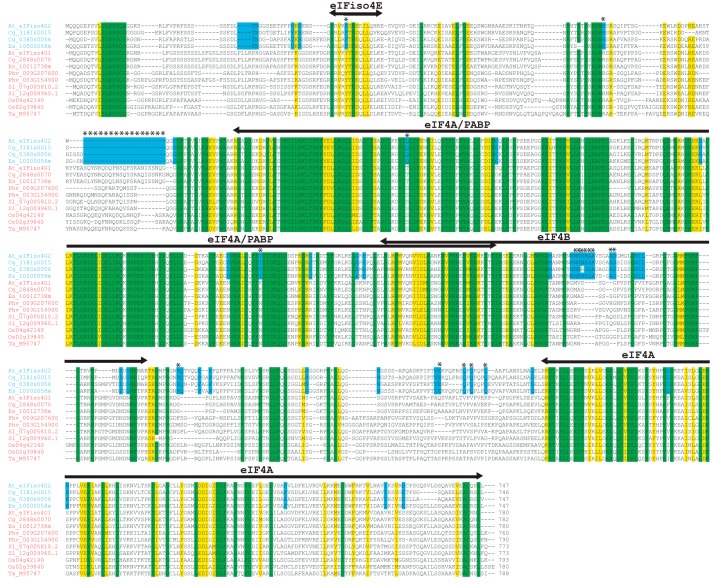
To examine in greater detail how eIFiso4G2 may differ from eIFiso4G1 at the amino acid level, selected eIFiso4G1 and eIFiso4G2 protein sequences representing Brassicaceae and other plant families were aligned. The amino acid sequence of eIFiso4G1 from wheat was included as its protein interaction domains and RNA-binding domains have been identified previously (15). Although the eIFiso4G1 and eIFiso4G2 isoforms exhibit sequence conservation throughout the protein, the regions of greatest homology represent the HEAT-1 and HEAT-2 domains (Fig. 5). eIF4A binds to both of these domains, whereas PABP binds within the HEAT-1 domain (15). eIF4B binds adjacent to this domain to a region also implicated in interaction with eIF3 in animal eIF4G (15, 28, 29). Analysis of those residues in eIFiso4G2, which differ from eIFiso4G1 isoforms but are conserved in eIFiso4G2 isoforms revealed a relatively small number of residues that are eIFiso4G2-specific (residues with asterisks in Fig. 5) and these are present largely in four regions of the protein.
FIGURE 5.
Sequence conservation among eIFiso4G1 and eIFiso4G2 isoforms. The sequence of eIFiso4G1 and eIFiso4G2 from A. thaliana (At), C. grandiflora (Cg), E. salsugineum (Es), P. vulgaris (Phv), S. lycopersicum (Sl), O. sativa (Os), and T. aestivum (Ta) were aligned. Amino acid identity is highlighted in green and amino acid similarity is highlighted in yellow. HEAT domains and binding sites for partner proteins of wheat eIFiso4G are indicated above the sequence as reported (15). eIFiso4G2-specific sequence differences that are conserved among eIFiso4G2 isoforms are indicated by asterisks.
One eIFiso4G2-specific residue is a tyrosine to phenylalanine change within the eIFiso4E binding site that occurs in both eIFiso4G2 subclades (Fig. 6). Although this represents a conserved change, the effects on eIFiso4E are unknown. An arginine to leucine change is also present in one of the eIFiso4G2 subclades (Fig. 6).
FIGURE 6.
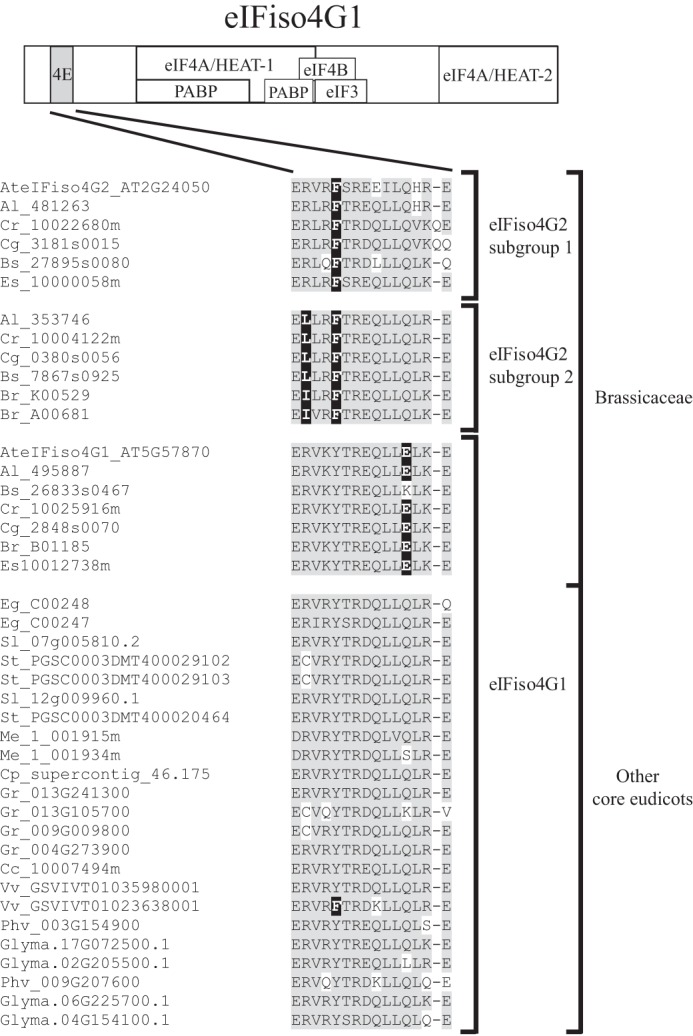
eIFiso4G2 isoforms differ from eIFiso4G1 isoforms within the eIFiso4E binding site. The sequences of the eIFiso4E binding site from eIFiso4G2 and eIFiso4G1 isoforms from core eudicot species were aligned and the conserved eIFiso4G2-specific sequence differences are shown. Amino acid identity is highlighted in gray and conserved eIFiso4G2-specific sequence differences are highlighted in black. The two eIFiso4G2 subgroups present in Brassicaceae are indicated to the right as are eIFiso4G1 isoforms present throughout core eudicots. The position of the eIFiso4E binding site is shown for wheat eIFiso4G at the top.
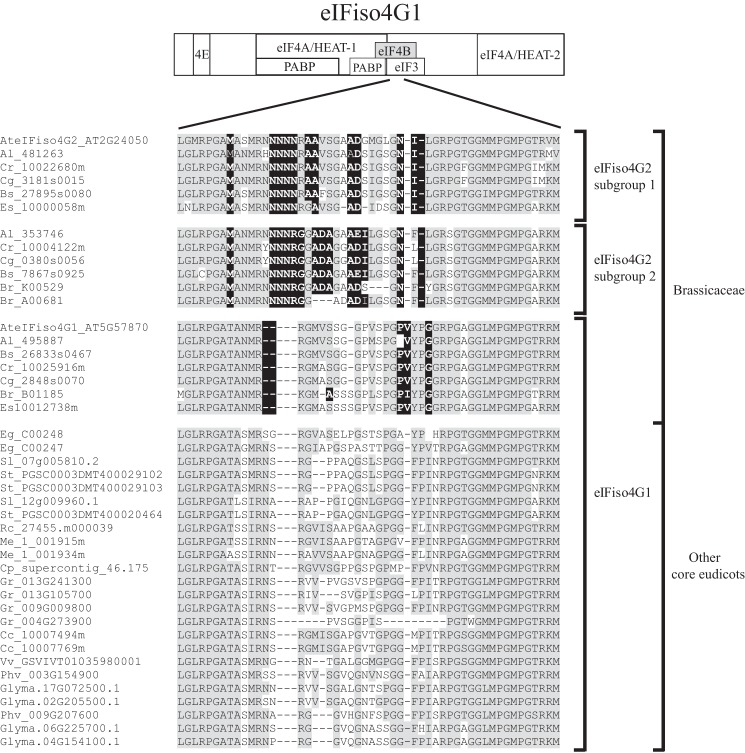
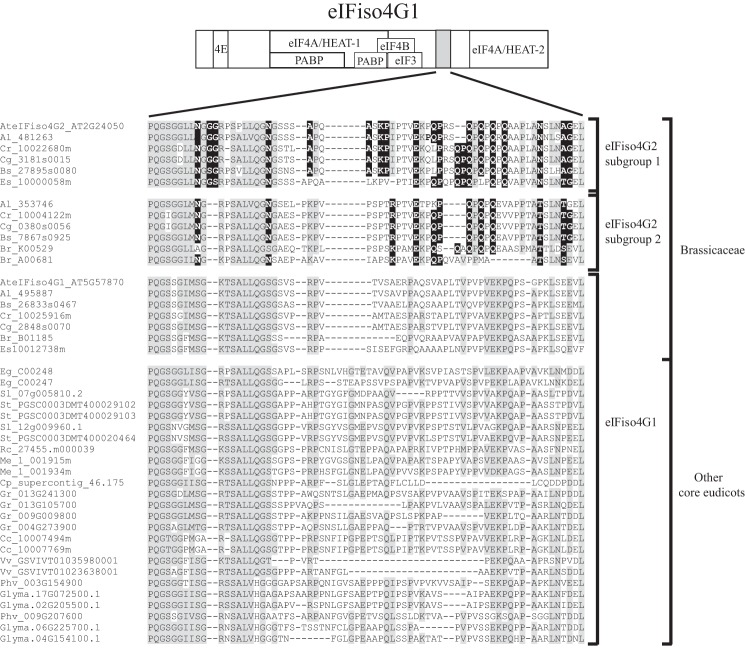
Several eIFiso4G2-specific residues are present within the eIF4B interaction domain that lies just C-proximal to the HEAT-1 domain. Analysis of all eIFiso4G2 isoforms confirmed the conservation of these eIFiso4G2-specific sequence differences (Fig. 7). As this region might also be involved in eIF3 binding, such sequence differences may affect interaction with one or both of these partners, although the eIF3 interaction with this region in eIFiso4G has not been examined. The region including the HEAT-1 domain also exhibits RNA binding activity as does the corresponding region in animal eIF4G (15, 30), although the role of this binding activity is unknown.
FIGURE 7.
eIFiso4G2 isoforms differ from eIFiso4G1 isoforms within the eIF4B binding site. Subsequences of the eIF4B binding site from eIFiso4G2 and eIFiso4G1 isoforms from core eudicot species were aligned and the conserved eIFiso4G2-specific sequence differences are shown. Amino acid identity is highlighted in gray and conserved eIFiso4G2-specific sequence differences are highlighted in black. The two eIFiso4G2 subgroups present in Brassicaceae are indicated to the right as are eIFiso4G1 isoforms present throughout core eudicots. The position of the eIF4B binding site is shown for wheat eIFiso4G at top.
The remaining two regions involve poorly conserved regions within eIFiso4G isoforms. All eIFiso4G2 isoforms examined contain a deletion with proximal sequence differences just upstream of the HEAT-1 domain in one such poorly conserved region (Fig. 8). A second poorly conserved sequence representing the region between the HEAT domains contains the last cluster of eIFiso4G2-specific changes that introduce a variable number of PQ repeats in addition to other sequence differences (Fig. 9). The function of this region has not been examined in detail but in animal eIF4G is subject to phosphorylation in response to the nutritional status of the cell, which regulates eIF4G activity (31). These results demonstrate that the recent evolution of eIFiso4G2 within the Brassicaceae has resulted in an isoform that differs from eIFiso4G1 in sequence including regions known to be involved in protein and RNA interactions.
FIGURE 8.
eIFiso4G2 isoforms differ from eIFiso4G1 isoforms within a region proximal to the eIF4A/HEAT-1 domain. The sequences of the region proximal to the eIF4A/HEAT-1 domain from eIFiso4G2 and eIFiso4G1 isoforms from core eudicot species were aligned and the conserved eIFiso4G2-specific sequence differences are shown. Amino acid identity is highlighted in gray and conserved eIFiso4G2-specific sequence differences are highlighted in black. The sequence absent in eIFiso4G2 isoforms is indicated by dashes. The two eIFiso4G2 subgroups present in Brassicaceae are indicated to the right as are eIFiso4G1 isoforms present throughout core eudicots. The region representing the sequence is shown for wheat eIFiso4G at the top.
FIGURE 9.
eIFiso4G2 isoforms differ from eIFiso4G1 isoforms within a region proximal to the eIF4A/HEAT-2 domain. The sequences of the region proximal to the eIF4A/HEAT-2 domain from eIFiso4G2 and eIFiso4G1 isoforms from core eudicot species were aligned and the conserved eIFiso4G2-specific sequence differences are shown. Amino acid identity is highlighted in gray and conserved eIFiso4G2-specific sequence differences are highlighted in black. Sequences absent in eIFiso4G2 and eIFiso4G1 isoforms are indicated by dashes. The two eIFiso4G2 subgroups present in Brassicaceae are indicated to the right as are eIFiso4G1 isoforms present throughout core eudicots. The region representing the sequence is shown for wheat eIFiso4G at the top.
Discussion
Plants are unusual in expressing two highly divergent eIF4G isoforms. Previous work has demonstrated that eIF4G and eIFiso4G from wheat exhibit clear differences in the types of mRNAs they translate optimally (12, 16, 17, 20, 21), which suggests that these two isoforms differ functionally as well as structurally. In addition to the divergence between eIF4G and eIFiso4G, some plant species express multiple eIFiso4G isoforms, which are significantly divergent from one another. The sequence divergence among eIFiso4G isoforms raises the question of whether they too may differ functionally. As the TMV 5′-leader was shown to discriminate between eIF4G and eIFiso4G during in vitro translation in wheat germ lysate, it was used in this study to interrogate the translational preference of each eIF4G isoform in vivo in Arabidopsis. This in vivo approach also made it possible to investigate how loss of a specific eIF4G isoform may affect expression in ways other than translational efficiency. As observed in wheat germ lysate (21), translation from mRNAs containing Ω as the 5′-leader exhibited a strong dependence on eIF4G in Arabidopsis. Surprisingly, Ω was also highly dependent on eIFiso4G2 but not on eIFiso4G1, the two eIFiso4G isoforms expressed in Arabidopsis. Loss of any eIF4G isoform did result in a substantial reduction in reporter transcript level regardless of whether Ω was present as the 5′-leader or not, suggesting that all eIF4G isoforms in Arabidopsis are functionally similar in supporting the abundance of the mRNA independent of these 5′-leaders.
The finding that eIFiso4G1 and eIFiso4G2 exhibit some functional difference was unexpected as loss of either isoform results in no visible phenotype in Arabidopsis but loss of both results in reduced stature, slow growth, and lower chlorophyll content (20, 23), suggesting functional overlap between the isoforms. Their functional overlap does not preclude, however, some degree of functional specialization. Clearly, the extent to which eIFiso4G1 and eIFiso4G2 are functionally different in supporting Ω-mediated translation must lie within their sequence differences. This raises the question, however, of why Ω-mediated translation in wheat germ lysate was not strongly dependent on eIFiso4G. Analysis of the evolution of eIFiso4G isoforms in land plants revealed that eIFiso4G2 is not present in early land plants and did not appear until after the divergence of monocots (e.g. wheat) from dicot species including Arabidopsis. In fact, eIFiso4G2 appears only in the Brassicaceae suggesting a late evolution of this eIFiso4G isoform. As an eIFiso4G2 homolog is not present in wheat, only eIF4G and eIFiso4G1 are available for translation in wheat germ lysate, which explains why Ω-mediated translation was dependent on eIF4G but not eIFiso4G in this earlier study (21). The finding that eIFiso4G2 does support Ω-mediated translation in Arabidopsis suggests that it and perhaps other species of the Brassicaceae have evolved a novel eIFiso4G isoform that exhibits some differences in the preference of mRNAs selected for translation.
As sequence differences between eIFiso4G1 and eIFiso4G2 must underlie their functional difference in supporting Ω-mediated translation, which are likely to be responsible for this functional specialization? The location of the sequence differences between eIFiso4G1 and eIFiso4G2 fall into two categories: those present in conserved domains known to be involved in interactions with partner proteins or RNA and those present in poorly conserved regions of the protein for which little to no function is known. The nature of these changes in eIFiso4G2 falls into two categories: those residues that differ from eIFiso4G1 but are conserved among eIFiso4G2 homologs and those that differ from eIFiso4G1 but are poorly conserved among eIFiso4G2 homologs. It is the former group of changes that are more likely to contribute to the observed functional differences between eIFiso4G1 and eIFiso4G2.
Conserved eIFiso4G2-specific sequence differences are found principally in four regions of the protein. A tyrosine to phenylalanine change within the eIFiso4E binding site is present in all eIFiso4G2 orthologs (Fig. 8). This is notable as this binding site in all plant eIF4G and eIFiso4G1 orthologs examined, with the exception of one of the two eIFiso4G1 isoforms in grape (Vv_GSVIVT01023638001) that shares no other eIFiso4G2-specific feature, contains a phenylalanine at this position. It is unknown how such a conserved change may alter interaction with eIFiso4E, but the conservation of such a change and the fact that eIFiso4E and eIF4E are responsible for binding the 5′-cap structure and therefore are involved in mRNA selection (32) suggests it could contribute in some way to the functional difference exhibited by eIFiso4G2.
A second region containing eIFiso4G2-specific amino acid differences lies C-proximal to the HEAT-1 domain where eIF4B interacts as determined in wheat eIFiso4G1 (15). It should be noted that eIF3 binds to the corresponding region in animal eIF4G but not in yeast eIF4G (29, 33, 34) and as such, it would be necessary to determine which is the case for plant eIFiso4G isoforms to consider whether the observed eIFiso4G2-specific sequence changes might affect this interaction. The eIFiso4G2-specific differences are clustered together in the central region of the eIF4B interaction domain and include an insertion of a stretch of asparagine residues (Fig. 7). eIF4B assists eIF4A in unwinding the RNA secondary structure and therefore would be expected to contribute most to the translation of those mRNAs containing a structured leader. Although the Ω 5′-leader is devoid of Watson-Crick base pairing, it may possess some degree of structure (35) and is still dependent on eIF4A (27). eIF4B also interacts with PABP, which may facilitate translation initiation (11, 36) so that changes to the interaction with eIF4B might be expected to affect multiple steps during translation initiation.
A region that lies just downstream of the HEAT-1 domain in eIF4G but within the N-terminal portion of the eIF4B interaction binding site has been implicated in permitting replication of cucumber mosaic virus in Arabidopsis as mutation of a proline inhibits viral accumulation (37). This region, designated H1-CT (38), is conserved in eukaryotes including eIFiso4G1 and eIFiso4G2, suggesting this region is not involved in the functional differences between eIFiso4G1 and eIFiso4G2.
The remaining two regions involving sequence differences that are highly conserved among eIFiso4G2 orthologs lie within poorly conserved regions within eIFiso4G isoforms. The first of these involves a deletion just N-proximal to the HEAT-1 domain in a region that is not well conserved (Fig. 8). The function of this region has not been investigated in any great detail although it lies within a region exhibiting RNA binding activity with preference for poly(A) and poly(G) RNA (15). If this region were involved in binding a 5′-leader to stabilize eIFiso4F binding to an mRNA during initiation, the eIFiso4G2-specific sequence differences might affect mRNA selection and therefore the degree to which a particular mRNA is dependent on eIFiso4G2 for translation.
A second poorly conserved sequence region lies between the HEAT-1 and HEAT-2 domains and contains a cluster of eIFiso4G2-specific sequence differences that introduce a variable number of PQ repeats (Fig. 9). In animal eIF4G, this region is subject to phosphorylation in response to the nutritional status of the cell (31) but this has not been examined for eIFiso4G.
Although eIFiso4G1 and eIFiso4G2 differ significantly in sequence, eIF4G differs even more substantially from eIFiso4G isoforms. eIF4G and eIFiso4G are similar in possessing two HEAT domains to which eIF4A binds and each interacts with eIF4B and PABP but they differ in how they interact with these partner proteins (14, 15). eIF4B and PABP bind within the eIFiso4G HEAT-1 domain at overlapping sites and bind eIFiso4G competitively (15). eIF4B and PABP only compete with eIF4A for binding the HEAT-1 domain in the absence of the HEAT-2 domain, suggesting the HEAT-2 domain stabilizes eIF4A binding in the presence of eIF4B or PABP.
PABP and eIF4B bind to overlapping sites near the HEAT-1 domain of eIF4G but their binding sites include only a portion of the HEAT-1 C-terminal region and as such, they bind competitively to this region but do not compete with eIF4A in binding to the HEAT-1-containing region (14). eIF4G differs most from eIFiso4G in that it contains a long N-terminal region to which PABP and eIF4B bind (14). Because their binding sites also overlap in this region, PABP and eIF4B bind competitively to this region as they do to the HEAT-1 proximal region. The HEAT-1-containing regions including the eIF4B and PABP binding sites in eIF4G and eIFiso4G also exhibit RNA binding activity. Although such activity in each was determined by binding to poly(A) and poly(G), their precise sequence preferences are unknown.
Despite the differences in domain organization and interaction with partner proteins, eIF4G and eIFiso4G2 support Ω-mediated translation to a greater extent than does eIFiso4G1. Examination of those sequence differences between eIFiso4G1 and eIFiso4G2 that are conserved among eIFiso4G2 orthologs revealed no similarity with the corresponding region in eIF4G, suggesting that it is not the eIFiso4G2-specific changes alone that are responsible for supporting Ω-mediated translation but rather it is likely their effect in the context of the protein in which they reside that enables this function.
Loss of any eIF4G or eIFiso4G isoform resulted in a substantial reduction in Luc transcript level. Similar reductions were observed whether Ω was present as the 5′-leader or not, indicating that Ω did not significantly influence the effect that loss of an eIF4G isoform had on the transcript level. The transcript level of Luc mRNA actually increased in eIFiso4G null Arabidopsis when the VDE 5′-leader was present, demonstrating that the 5′-leader can determine whether the Luc transcript level is affected by loss of an eIF4G isoform. Reduction in the Luc transcript level observed in this study could be a direct consequence of the loss of an eIF4G isoform, which renders the mRNA more susceptible to the degradation machinery in the absence of the factor binding the 5′-cap. It is also possible that loss of an eIF4G isoform could reduce the Luc transcript level indirectly through changes in transcription, nucleocytoplasmic transport, or expression of the mRNA decay machinery. The increase in transcript level of Luc containing the VDE 5′-leader following loss of eIFiso4G expression would argue against such a general and indirect effect on gene expression as would the absence of any visible phenotype in eif4g, eifiso4g1, or eifiso4g2 mutants. Instead, the contribution that an eIF4G or eIFiso4G isoform makes to the level of a specific mRNA under normal conditions may be determined by the strength of binding of eIF4E (or eIFiso4E) to the 5′-cap, which can be affected by the degree of secondary structure in a 5′-leader (30), the strength of binding of the RNA binding domains present in eIF4G (or eIFiso4G1 and eIFiso4G2), as well as those present in eIF4A, eIF4B, and PABP (11, 14, 15, 36) to sequences within the 5′-leader. The strength of interactions of these partner proteins with an eIF4G or eIFiso4G isoform might also be expected to influence mRNA selection.
The observation that eIFiso4G2 is functionally more similar to eIF4G than to eIFiso4G1 in supporting Ω-mediated translation raises the question of why such an isoform would be necessary in the Brassicaceae in which it appears. As eIFiso4G2 orthologs are present in all species of the Brassicaceae examined, it likely appeared early in the evolution of this family and must confer an advantage to these species as it has not been lost in any species of the Brassicaceae examined and in fact the gene family has expanded to more than one member in some species. Although the eIFiso4G1 orthologs in Arabidopsis and wheat showed relatively weak support for Ω-mediated translation, whereas Arabidopsis eIFiso4G2 showed strong support, to conclude that all eIFiso4G2 orthologs are alike in their support of Ω-mediated translation, whereas all eIFiso4G1 orthologs do not, may be too simple as there is sequence variation among eIFiso4G2 orthologs or among eIFiso4G1 orthologs that may influence the degree to which each functions in the translation of specific mRNAs. Moreover, in those species expressing more than one eIFiso4G1 or eIFiso4G2 ortholog, there may exist some degree of functional difference. Future work investigating differences among eIFiso4G1 or eIFiso4G2 orthologs in other species will be needed to show what possible range of functionality may exist for isoforms of this translation factor.
Author Contributions
D. R. G. conceived, designed, performed, and analyzed the experiments, and wrote the paper. The author has reviewed the results and approved the final version of the manuscript.
This work was supported by National Science Foundation Grant DBI-0820047 and the University of California Agricultural Experiment Station (to D. R. G.). The author declares that he has no conflicts of interest with the contents of this article.
- PABP
- poly(A)-binding protein
- HEAT
- Huntington, elongation factor 3, PR65/A, TOR
- TMV
- tobacco mosaic virus
- qPCR
- quantitative PCR
- VDE
- violaxanthin de-epoxidase
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- Ω
- ohm.
References
- 1.Preiss T., and W. Hentze M. (2003) Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays 25, 1201–1211 [DOI] [PubMed] [Google Scholar]
- 2.Kapp L. D., and Lorsch J. R. (2004) The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 73, 657–704 [DOI] [PubMed] [Google Scholar]
- 3.Pestova T. V., Lorsch J. R., and Hellen C.U.T. (2007) The mechanism of translation initiation in eukaryotes. in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., and Hershey J. W. B., eds) pp. 87–128, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 4.Gallie D. R. (2002) Protein-protein interactions required during translation. Plant Mol. Biol. 50, 949–970 [DOI] [PubMed] [Google Scholar]
- 5.Wei C.-C., Balasta M. L., Ren J., and Goss D. J. (1998) Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry 37, 1910–1916 [DOI] [PubMed] [Google Scholar]
- 6.Bi X., and Ren J., and Goss D. J. (2000) Wheat germ translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase activity by increasing the ATP binding affinity of eIF4A. Biochemistry 39, 5758–5765 [DOI] [PubMed] [Google Scholar]
- 7.Tarun S. Z. Jr., and Sachs A. B. (1995) A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 9, 2997–3007 [DOI] [PubMed] [Google Scholar]
- 8.Wells S. E., Hillner P. E., Vale R. D., and Sachs A. B. (1998) Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2, 135–140 [DOI] [PubMed] [Google Scholar]
- 9.Le H., Tanguay R. L., Balasta M. L., Wei C.-C., Browning K. S., Metz A. M., Goss D. J., and Gallie D. R. (1997) Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. J. Biol. Chem. 272, 16247–162559195926 [Google Scholar]
- 10.Le H., Browning K. S., and Gallie D. R. (2000) The phosphorylation state of poly(A)-binding protein specifies its binding to poly(A) RNA and its interaction with eukaryotic initiation factor (eIF) 4F, eIFiso4F, and eIF4B. J. Biol. Chem. 275, 17452–17462 [DOI] [PubMed] [Google Scholar]
- 11.Cheng S., and Gallie D. R. (2006) Wheat eukaryotic initiation factor 4B organizes assembly of RNA and eIFiso4G, eIF4A, and poly(A)-binding protein. J. Biol. Chem. 281, 24351–24364 [DOI] [PubMed] [Google Scholar]
- 12.Gallie D. R., and Browning K. S. (2001) eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J. Biol. Chem. 276, 36951–36960 [DOI] [PubMed] [Google Scholar]
- 13.Mayberry L. K., Allen M. L., Nitka K. R., Campbell L., Murphy P. A., and Browning K. S. (2011) Plant cap-binding complexes eukaryotic initiation factors eIF4F and eIFiso4F: molecular specificity of subunit binding. J. Biol. Chem. 286, 42566–42574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng S., and Gallie D. R. (2013) Eukaryotic initiation factor 4B and the poly(A)-binding protein bind eIF4G competitively. Translation 1, e24038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng S., and Gallie D. R. (2010) Competitive and noncompetitive binding of eIF4B, eIF4A, and the poly(A) binding protein to wheat translation initiation factor eIFiso4G. Biochemistry 49, 8251–8265 [DOI] [PubMed] [Google Scholar]
- 16.Mayberry L. K., Allen M. L., Dennis M. D., and Browning K. S. (2009) Evidence for variation in the optimal translation initiation complex: plant eIF4B, eIF4F, and eIF(iso)4F differentially promote translation of mRNAs. Plant Physiol. 150, 1844–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallie D. R. (2001) Cap-independent translation conferred by the 5′-leader of tobacco etch virus is eIF4G-dependent. J. Virol. 75, 12141–12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huch S., and Nissan T. (2014) Interrelations between translation and general mRNA degradation in yeast. WIREs RNA 5, 747–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niyogi K. K., Grossman A. R., and Björkman O. (1998) Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10, 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z., Jolley B., Caldwell C., and Gallie D. R. (2014) Eukaryotic translation initiation factor eIFiso4G is required to regulate violaxanthin de-epoxidase expression in Arabidopsis. J. Biol. Chem. 289, 13926–13936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallie D. R. (2002) The 5′-leader of tobacco mosaic virus promotes translation through enhanced recruitment of eIF4F. Nucleic Acids Res. 30, 3401–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mundry K. W., Watkins P. A., Ashfield T., Plaskitt K. A., Eisele-Walter S., and Wilson T. M. (1991) Complete uncoating of the 5′ leader sequence of tobacco mosaic virus RNA occurs rapidly and is required to initiate cotranslational virus disassembly in vitro. J. Gen. Virol. 72, 769–777 [DOI] [PubMed] [Google Scholar]
- 23.Lellis A. D., Allen M. L., Aertker A. W., Tran J. K., Hillis D. M., Harbin C. R., Caldwell C., Gallie D. R., and Browning K. S. (2010) Deletion of the eIFiso4G subunit of the Arabidopsis eIFiso4F translation initiation complex impairs health and viability. Plant Mol. Biol. 74, 249–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones D. T., Taylor W. R., and Thornton J. M. (1992) The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 [DOI] [PubMed] [Google Scholar]
- 25.Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patrick R. M., Mayberry L. K., Choy G., Woodard L. E., Liu J. S., White A., Mullen R. A., Tanavin T. M., Latz C. A., and Browning K. S. (2014) Two Arabidopsis loci encode novel eukaryotic initiation factor 4E isoforms that are functionally distinct from the conserved plant eukaryotic initiation factor 4E. Plant Physiol. 164, 1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altmann M., Blum S., Wilson T. M., and Trachsel H. (1990) The 5′-leader sequence of tobacco mosaic virus RNA mediates initiation-factor-4E-independent, but still initiation-factor-4A-dependent translation in yeast extracts. Gene 91, 127–129 [DOI] [PubMed] [Google Scholar]
- 28.LeFebvre A. K., Korneeva N. L., Trutschl M., Cvek U., Duzan R. D., Bradley C. A., Hershey J. W., and Rhoads R. E. (2006) Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J. Biol. Chem. 281, 22917–22932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villa N., Do A., Hershey J. W., and Fraser C. S. (2013) Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J. Biol. Chem. 288, 32932–32940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lomakin I. B., Hellen C. U., and Pestova T. V. (2000) Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20, 6019–6029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raught B., Gingras A. C., Gygi S. P., Imataka H., Morino S., Gradi A., Aebersold R., and Sonenberg N. (2000) Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 19, 434–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carberry S. E., and Goss D. J. (1991) Wheat germ initiation factors 4F and (iso)4F interact differently with oligoribonucleotide analogues of rabbit α-globin mRNA. Biochemistry 30, 4542–4545 [DOI] [PubMed] [Google Scholar]
- 33.He H., von der Haar T., Singh C. R., Ii M., Li B., Hinnebusch A. G., McCarthy J. E., and Asano K. (2003) The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol. Cell. Biol. 23, 5431–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh C. R., Watanabe R., Chowdhury W., Hiraishi H., Murai M. J., Yamamoto Y., Miles D., Ikeda Y., Asano M., and Asano K. (2012) Sequential eukaryotic translation initiation factor 5 (eIF5) binding to the charged disordered segments of eIF4G and eIF2b stabilizes the 48S preinitiation complex and promotes its shift to the initiation mode. Mol. Cell. Biol. 32, 3978–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovtun A. A., Shirokikh N. E., Gudkov A. T., and Spirin A. S. (2007) The leader sequence of tobacco mosaic virus RNA devoid of Watson-Crick secondary structure possesses a cooperatively melted, compact conformation. Biochem. Biophys. Res. Commun. 358, 368–372 [DOI] [PubMed] [Google Scholar]
- 36.Cheng S., and Gallie D. R. (2007) eIF4G, eIFiso4G, and eIF4B bind the poly(A)-binding protein through overlapping sites within the RNA recognition motif domains. J. Biol. Chem. 282, 25247–25258 [DOI] [PubMed] [Google Scholar]
- 37.Yoshii M., Nishikiori M., Tomita K., Yoshioka N., Kozuka R., Naito S., and Ishikawa M. (2004) The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 78, 6102–6111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marintchev A., and Wagner G. (2005) eIF4G and CBP80 share a common origin and similar domain organization: implications for the structure and function of eIF4G. Biochemistry 44, 12265–12272 [DOI] [PubMed] [Google Scholar]