Abstract Abstract
The taxonomic circumscription of the large and diverse leaf beetle genus Chrysolina Motschulsky is not clear, and its discrimination from the closely related genus Oreina Chevrolat has classically been controversial. In addition, the subgeneric arrangement of the species is unstable, and proposals segregating Chrysolina species into new genera have been recently suggested. In this context, the availability of a phylogenetic framework would provide the basis for a stable taxonomic system, but the existing phylogenies are based on few taxa and have low resolution. In the present study we perform a phylogenetic analysis based on mitochondrial (cox1 and rrnL) and nuclear (H3) DNA sequences from a sample of fifty-two Chrysolina species representing almost half of the subgeneric diversity of the group (thirty out of sixty-five subgenera) and most of the morphological, ecological and karyological variation in the genus. In addition, five Oreina species from two subgenera have also been analysed. The resulting phylogeny is used to evaluate some of the most relevant taxonomic hypotheses for Chrysolina, and also to reconstruct its ancestral host plant associations in a Bayesian framework. Our findings support the paraphyly of Chrysolina as currently defined due to the inclusion of Oreina, the monophyly of the Chrysolina (plus Oreina) species including the divergent Chrysolina (Polysticta) vigintimaculata (Clark, 1864), and enable inferences of deep-level evolutionary relationships among the studied subgenera. The plant family Lamiaceae is inferred as the ancestral host of the study group, whose evolution is characterized by continuous host-shifting among pre-existing host plant families. Some Chrysolina clades include mixtures of species with different levels of diet breadth, indicating that niche width has varied through time.
Keywords: Coleoptera, Chrysomelidae, Chrysolina, Oreina, Phylogeny, Insect-plant interaction, cox1, rrnL, H3
Introduction
The genus Chrysolina Motschulsky is a very large and diverse group of leaf-beetles that are mainly distributed in Europe, Asia and Africa (Bieńkowski 2001). Nearly 450 species belonging in 65 subgenera have been recognized (Bieńkowski 2001), and new species are still being described (e.g. Ge et al. 2011, Bourdonné et al. 2013, Lopatin 2011, 2014). However, despite the number of taxonomic studies focused on Chrysolina its taxonomy is chronically confused (Kippenberg 2010), and even the circumscription of the genus remains unclear. In fact, the most recent and updated taxonomic review (Bieńkowski 2007) does not contribute a comparative morphological diagnosis to differentiate Chrysolina from the closely related genera. In this sense the most controversial case is the one concerning the genera Chrysolina and Oreina Chevrolat, whose discrimination mainly relies in the ratio between the length of the metasternum and the length of the first abdominal sternite (Weise 1893). It has been suggested that this morphological attribute could be inconsistent (Bieńkowski 2007), thus reinforcing the inclusion of the genus Oreina within Chrysolina (Chapuis 1874, Bourdonné and Doguet 1991, Daccordi 1994) or conversely the recognition of Chrysolina as a subgenus of Oreina (Monrós and Bechyné 1956). In addition, taxonomic rearrangements are frequent in Chrysolina, including decisions splitting species into new genera (e.g. Craspeda Motschulsky [=Zeugotaenia Motschulsky]: Bourdonné 2005, Camerounia Jolivet: Bieńkowski 2007, Chalcoidea Motschulsky: Bourdonné 2012). Likewise, the subgeneric arrangement of the Chrysolina species is also unstable (Mikhailov 2000, 2002, Bieńkowski 2001, 2007, Bourdonné 2008, 2012, Kippenberg 2010). This taxonomic instability reflects the lack of a supraspecific systematic for the genus Chrysolina, due in part to the absence of a phylogenetic background.
Phylogenetic studies focused on Chrysolina are scarce and limited to a reduced number of taxa. Bourdonné and Doguet (1991) proposed the first evolutionary hypothesis for 10 groups of Palaearctic species attending to both their chromosome numbers and host-plant affiliations. From a molecular perspective, Garin et al. (1999) performed a phylogenetic analysis based on mtDNA sequences (cox1 and rrnL) from 30 Chrysolina species representing 22 subgenera plus two Oreina species. The resulting phylogenetic trees allowed for the identification of monophyletic lineages comprising few species each, but the deep level relationships were poorly resolved. On the other hand, the two Oreina species nested within the Chrysolina clade, but this relationship was unsupported. Simultaneously, Hsiao and Pasteels (1999) also inferred a molecular phylogeny based on mtDNA markers (12S and rrnL) from 16 Chrysolina species ascribed to 14 subgenera and 14 Oreina species, but the resulting topologies also had low resolution at the basal nodes. Oreina species were recovered as a monophyletic lineage that also included Chrysolina fastuosa (Scopoli, 1763), and all of them were nested in the Chrysolina clade. Both molecular studies highlighted the reciprocal monophyly of the subgenera Melasomoptera Bechyné and Synerga Weise, and of Hypericia Bedel and Sphaeromela Bedel, however discrepancies were observed regarding the systematic position of the subgenera Colaphodes Motschulsky and Taeniochrysea Bechyné.
Apart from taxonomic purposes, the availability of a phylogenetic hypothesis for the species of Chrysolina may allow for the study of evolutionary processes such as their ancestral host plant affiliations. In this regard, this leaf beetle genus constitutes a suitable and interesting study group as most of the species are oligophagous, each of them feeding on a narrow range of closely related plants (Jolivet and Petitpierre 1976, Bourdonné and Doguet 1991). Indeed, the taxonomic conservatism in host plant use found in Chrysolina is so high that host use has been frequently coupled with other systematic characters to circumscribe species assemblages (Petitpierre and Segarra 1985, Bourdonné and Doguet 1991, Petitpierre and Mikhailov 2009). The ancestral reconstruction of the trophic affiliations in Chrysolina and Oreina was addressed in the phylogenetic studies performed by Garin et al. (1999) and by Hsiao and Pasteels (1999), inferring the plant family Lamiaceae as the most likely ancestral host for Chrysolina + Oreina (Garin et al. 1999) and the Asteraceae for Oreina (Hsiao and Pasteels 1999). However, these reconstructions were based on poorly resolved phylogenetic trees from few taxa.
In this work we present the results of a phylogenetic study based on mitochondrial and nuclear DNA sequences from a sample of Chrysolina and Oreina species, using Bayesian and (ML) inference approaches. We expand the taxon sampling of previous molecular studies (Garin et al. 1999, Hsiao and Pasteels 1999) through the inclusion of representatives for nearly half of the Chrysolina subgenera comprising most of the morphologically defined groups and ecological variation of the genus. In addition, the inferred molecular phylogeny is used to test the validity of a number of taxonomic hypotheses derived from morphological, ecological, chemical and genetic data. Finally, we aim to investigate the evolution of the host plant associations in the genus Chrysolina.
Materials and methods
Taxon sampling
We have studied 52 Chrysolina species representing 30 out of the ca. 65 subgenera currently recognized for the genus (Bieńkowski 2001, Kippenberg 2010), plus five Oreina species from two subgenera. Our sampling includes type species representatives regarding 13 of the studied Chrysolina subgenera and one type species for Oreina. In addition, several representatives of other genera of the subfamily Chrysomelinae were analysed as outgroups, including a species from the early-divergent genus Timarcha Latreille (Gómez-Zurita et al. 2008) (Table 1). Beetles were collected by us in the field or received from colleagues in absolute ethanol and stored in the laboratory at -20 °C before processing. Voucher specimens are deposited for long-term storage at the DNA and tissue collection of the Biodiversity, Systematics and Evolution group (Bio6Evo) of the University of the Balearic Islands.
Table 1.
Studied taxa, sources, host plants and GenBank accession numbers. Species groups defined by Bourdonné and Doguet (1991) are also indicated. a: Baselga and Novoa (2006), b: Bieńkowski 2010, c: Bieńkowski 2011, d: Bourdonné 2005, e: Bourdonné and Doguet 1991, f: Cobos 1953, g: Garin et al. 1999, h: Jolivet and Petitpierre 1976, i: Jolivet et al. 1986, j: Koch 1992, k: Lopatin and Mikhailov 2010, l: Mikhailov 2006, m: Petitpierre 1981, n: Rizza and Pecora 1980, o: Vela and Bastazo 1999.
| Species | Source | Host(s) | Host(s) references | Bourdonné and Doguet’s (1991) group | cox1 | rrnL | H3 |
|---|---|---|---|---|---|---|---|
| Chrysolina aeruginosa (Faldermann, 1835) | SE Tuva, Siberia, Russia | Asteraceae (Artemisia), Lamiaceae (Thymus) | b | LN833682 | LN833808 | LN833745 | |
| Chrysolina baetica (Suffrian, 1851) | Murcia, Spain | Lamiaceae (Satureja, Thymus) | i | 2 | LN833683 | LN833809 | LN833746 |
| Chrysolina americana (Linnaeus, 1758) | Almuñecar, Spain | Lamiaceae (Lavandula, Rosmarinus) | b, h | 2 | LN833684 | LN833810 | LN833747 |
| Chrysolina aurichalcea (Gebler in Mannerheim, 1825) | Ticino, Switzerland | Apocynaceae (Vincetoxicum officinale), Asteraceae (Arctium, Artemisia, Aster, Kalimerus, Petasites) | b, j | 9 | LN833685 | LN833811 | LN833748 |
| Chrysolina banksi (Fabricius, 1775) | Balearic Islands, Spain | Lamiaceae, Plantaginaceae | h | 2 | LN833686 | LN833812 | LN833749 |
| Chrysolina bicolor (Fabricius, 1775) | Canary Islands, Spain | Lamiaceae (Saccocalyx, Salvia, Thymus) | h | 2 | LN833687 | LN833813 | LN833750 |
| Chrysolina carnifex (Fabricius, 1792) | Barcelona, Spain | Asteraceae (Artemisia, Santolina) | b | 9 | LN833688 | LN833814 | LN833751 |
| Chrysolina cerealis cyaneoaurata (Motschulsky, 1860) | Altai, Siberia, Russia | 2 | LN833689 | LN833815 | LN833752 | ||
| Chrysolina colasi (Cobos, 1952) | Granada, Spain | Lamiaceae (Sideritis glacialis) | o | 1 | LN833690 | LN833816 | LN833753 |
| Chrysolina convexicollis (Jakobson, 1901) | SE Tuva, Siberia, Russia | Asteraceae (Artemisia) | c | LN833691 | LN833817 | LN833754 | |
| Chrysolina costalis (Olivier, 1807) (=Chrysolina obsoleta Brullé, 1838 sensu Bieńkowski 2014 unpubl.) | Canary Islands, Spain | Ranunculaceae (Ranunculus) | e | 2 | LN833714 | LN833818 | LN833777 |
| Chrysolina diluta (Germar, 1824) | Granada, Spain | Plantaginaceae (Plantago) | h | 3 | LN833693 | LN833819 | LN833756 |
| Chrysolina eurina (Frivaldszky, 1883: 17) | Mundybash, Kemerovskaya oblast’, Russia | Asteraceae (Tanacetum vulgare) | b | 9 | LN833694 | LN833820 | LN833757 |
| Chrysolina fastuosa (Scopoli, 1763) | Lleida, Spain | Lamiaceae (Galeopsis, Lamium, Leonorus, Prunella) | h, i | 2 | LN833695 | LN833821 | LN833758 |
| Chrysolina femoralis (Olivier, 1790) | Girona, Spain | Lamiaceae (Satureja, Thymus) | h, i | 2 | LN833696 | LN833822 | LN833759 |
| Chrysolina fuliginosa (Olivier, 1807) | Lleida, Spain | Asteraceae (Centaurea) | h | 9 | LN833697 | LN833823 | LN833760 |
| Chrysolina gemina (Brullé, 1838) | Canary Islands, Spain | Lamiaceae (Lavandula) | h | 2 | LN833698 | LN833824 | LN833761 |
| Chrysolina geminata (Paykull, 1799) | Lleida, Spain | Hypericaceae (Hypericum) | b | 10 | LN833699 | LN833825 | LN833762 |
| Chrysolina haemochlora (Gebler, 1823) | Ust’-Koksa, Altai Republic, Russia | Apiaceae (Aegopodium, Angelica, Conioselinum, Heracleum, Pleurospermum) | c | LN833700 | LN833826 | LN833763 | |
| Chrysolina haemoptera (Linnaeus, 1758) | La Coruña, Spain | Plantaginaceae (Plantago) | m | 7 | LN833701 | LN833827 | LN833764 |
| Chrysolina helopioides (Suffrian, 1851) | Málaga, Spain | Apiaceae (Ferula) | h | 4 | LN833702 | LN833828 | LN833765 |
| Chrysolina herbacea (Duftschmid, 1825) | Teruel, Spain | Lamiaceae (Mentha) | b, h | 2 | LN833703 | LN833829 | LN833766 |
| Chrysolina hyperici (Forster, 1771) | Bragança, Portugal | Hypericaceae (Hypericum) | b | 10 | LN833704 | LN833830 | LN833767 |
| Chrysolina jakowlewi (Weise, 1894) | Sayan Mts., Tuva, Russia | LN833705 | LN833831 | LN833768 | |||
| Chrysolina janbechynei Cobos, 1953 [= Chrysolina curvilinea (Weise, 1884)] | Murcia, Spain | Asteraceae (Artemisia) | f | 9 | LN833692 | LN833832 | LN833755 |
| Chrysolina kocheri (Codina Padilla, 1961) | Smimou, Morocco | Plantaginaceae (Plantago coronopus) | d | 3 | LN833706 | LN833833 | LN833769 |
| Chrysolina kuesteri (Helliesen, 1912) | Tejeda, Granada, Spain | Lamiaceae, Scrophulariaceae (Linaria) | b, e | 1 | LN833707 | LN833834 | LN833770 |
| Chrysolina lepida (Olivier, 1807) | Huéscar, Granada, Spain | Asteraceae (Mantisalca salmantica) | e | 9 | LN833708 | LN833835 | LN833771 |
| Chrysolina lucida (Olivier, 1807) | Almería, Spain | Lamiaceae (Mentha) | h | 2 | LN833709 | LN833836 | LN833772 |
| Chrysolina lucidicollis grossepunctata (Lindberg, 1950) | Canary Islands, Spain | Scrophulariaceae (Linaria) | e | 1 | LN833710 | LN833837 | LN833773 |
| Chrysolina marginata (Linnaeus, 1758) | Girona, Spain | Asteraceae (Achillea) | b, e, h | 9 | LN833711 | LN833838 | LN833774 |
| Chrysolina affinis mesatlantica (Kocher, 1958) | Moyen Atlas, Morocco | 2 | LN833712 | LN833839 | LN833775 | ||
| Chrysolina obscurella (Suffrian, 1851) | Var, France | Apiaceae | e | 4 | LN833713 | LN833840 | LN833776 |
| Chrysolina oirota Lopatin, 1990 | Ivanovsky massif, Kazakhstan | Asteraceae (Saussurea latifolia), Lamiaceae (Lamium) | k | LN833715 | LN833841 | LN833778 | |
| Chrysolina pedestris (Gebler, 1823) | Sekisovka, Kazakhstan | Apiaceae (Seselis) | c | LN833716 | n.a. | LN833779 | |
| Chrysolina peregrina (Herrich-Schaeffer, 1839) | Balearic Islands, Spain | Apiaceae (Daucus, Phoeniculum) | g, h | 8 | LN833717 | n.a. | LN833780 |
| Chrysolina perforata (Gebler, 1830) | Erzin, Russia | Asteraceae, Lamiaceae | c | LN833718 | LN833842 | LN833781 | |
| Chrysolina petitpierrei Kippenberg, 2004 | Lleida, Spain | LN833719 | LN833843 | LN833782 | |||
| Chrysolina polita (Linnaeus, 1758) | Girona, Spain | Lamiaceae (Lycopus, Mentha, Origanum, Satureja) | b, h, i | 2 | LN833720 | LN833844 | LN833783 |
| Chrysolina quadrigemina (Suffrian, 1851) | Bragança, Portugal | Hypericaceae (Hypericum) | h | 10 | LN833721 | LN833845 | LN833784 |
| Chrysolina reitteri (Weise, 1884) | Susuz, Turkey | LN833722 | LN833846 | LN833785 | |||
| Chrysomela rossia (Illiger, 1802) | Torino, Italy | Lamiaceae (Mentha piperita), Scrophulariaceae (Linaria, Veronica) | b, n | 1 | LN833723 | LN833847 | LN833786 |
| Chrysolina rufoaenea (Suffrian, 1851) | Zamora, Spain | Apiaceae (Carum verticillatum) | a, i | 8 | LN833724 | LN833848 | LN833787 |
| Chrysolina soiota (Jakobson, 1924) | Kulumys range, Oisky pass, Russia | LN833726 | LN833849 | LN833789 | |||
| Chrysolina sturmi (Westhoff, 1882) | Chelyabinsk, Russia | Asteraceae (Cirsium), Lamiaceae (Glechoma), Scrophulariaceae (Linaria) | b | LN833727 | n.a. | LN833790 | |
| Chrysolina sylvatica (Gebler, 1823) | Kulumys range, Oisky pass, Russia | Ranunculaceae (Aquilegia glandulosa) | l | LN833728 | LN833850 | LN833791 | |
| Chrysolina timarchoides (Brisout de Barneville, 1882) | Girona, Spain | Apiaceae (Bupleurum, Heracleum) | h | 4 | LN833729 | LN833851 | LN833792 |
| Chrysolina tundralis (Jakobson, 1910) | Serebryansky Mount, Russia | Asteraceae (Arnica, Saussurea), Lamiaceae (Lamium purpureum) | c | LN833730 | LN833852 | LN833793 | |
| Chrysolina vernalis pyrenaica (Dufour, 1843) | Lleida, Spain | Plantaginaceae (Plantago) | m | 7 | LN833731 | LN833853 | LN833794 |
| Chrysolina vigintimaculata (Clark, 1864) | KwaZulu-Natal, South Africa | LN833732 | n.a. | LN833795 | |||
| Chrysolina viridana (Kuster, 1844) | Riofrio, Granada, Spain | Lamiaceae (Mentha) | h | 2 | LN833733 | LN833854 | LN833796 |
| Chrysolina wollastoni (Bechyné, 1957) [=Chrysolina rutilans (Wollaston, 1864)] | Canary Islands, Spain | Lamiaceae (Mentha) | h | 2 | LN833725 | LN833855 | LN833788 |
| Oreina cacaliae (Schrank, 1785) | Lleida, Spain | Asteraceae (Adenostyles, Petasites) | i | 6 | LN833735 | LN833857 | LN833798 |
| Oreina fairmairiana (De Gozis, 1882) [=Oreina splendidula (Fairmaire, 1865)] | Lleida, Spain | Apiaceae, Asteraceae (Senecio) | e | 6 | LN833739 | LN833858 | LN833802 |
| Oreina ganglbaueri (Jakob, 1953) | Lleida, Spain | Apiaceae (Angelica, Heracleum, Meum) | i | 5 | LN833736 | LN833859 | LN833799 |
| Oreina speciosa (Linnaeus, 1767) | Massif des Vosges, Haut-Rhin, France | Apiaceae (Angelica, Heliosiadium, Laserpitium, Peucedanum) | i | 5 | LN833737 | n.a. | LN833800 |
| Oreina speciosissima (Scopoli, 1763) | Lleida, Spain | Asteraceae (Adenostyles, Cirsinus, Petasites, Senecio) | i | 6 | LN833738 | LN833860 | LN833801 |
| Lamprolina aeneipennis (Boisduval, 1835) | Mount Keira, NSW, Australia | LN833734 | LN833856 | LN833797 | |||
| Paropsis atomaria Olivier, 1807 | Molonglo Gorge Nature Reserve, ACT, Australia | LN833740 | LN833862 | LN833803 | |||
| Paropsisterna liturata (Marsham, 1808) | Black Mountain, ACT, Australia | LN833741 | LN833861 | LN833804 | |||
| Phyllocharis cyanicornis (Fabricius, 1801) | Royal National Park, NSW, Australia | LN833742 | LN833863 | LN833805 | |||
| Poropteromela epipleuralis Lea, 1916 | Mount Moombil, NSW, Australia | LN833743 | LN833864 | LN833806 | |||
| Timarcha sinuatocollis Fairmaire 1861 | Lleida, Spain | LN833744 | LN833865 | LN833807 |
DNA isolation, PCR amplification and sequencing
Total DNA was purified from beetle head and pronotum using the DNeasy Tissue kit (Qiagen, West Sussex, UK) and following the manufacturer’s protocol. Elutions were done in 200 μL volume and one microliter was used in PCR reactions. Three different molecular markers were selected for the study, including a partial sequence of the mitochondrial 16S rDNA (rrnL; primers LR-N-13398 and LR-J-12887; Simon et al. 1994), a partial sequence of the mitochondrial cytochrome c oxidase subunit 1 gene (cox1; primers C1-J-2183 and TL2-N-3014; Simon et al. 1994), and a fragment from the nuclear histone 3 gene (H3; primers H3aF and H3aR; Colgan et al. 1998). PCR conditions used 0.2 μM of each primer and 3.5 mM MgCl2 using a standard protocol of 35 cycles with annealing temperature ranging from 50 to 45 °C (60s) depending on the sample, and denaturation (94 °C) and elongation (72 °C) lasted 30 and 60s, respectively. PCR products were visualized by 1% agarose gel electrophoresis and subsequently purified using MSB Spin PCRapace (Invitek, Berlin, Germany). Sanger sequencing was performed with the same primers as above using the BigDye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA). Sequences were edited and contigs were assembled using BIOEDIT v. 7 (Hall 1999), and deposited at GenBank under the accession numbers referred in Table 1.
Phylogenetic analyses
Heterogeneity in base composition across taxa was explored for each codon position of the protein-coding genes and for rrnL using the chi-square test for base frequency differences implemented in PAUP*4.0b10 (Swofford 2003). Multiple sequence alignment was performed using MAFFT 7 online version (http://mafft.cbrc.jp/alignment/server/, Katoh and Standley 2013) under default parameters. Molecular markers were checked for combinability using the (ILD) test (Farris et al. 1994) implemented in PAUP* v4.0b10 (Swofford 2002). The test was run using 100 random stepwise additions and 1000 replicates of heuristic search with tree bisection–reconnection (TBR) branch swapping. The optimal partitioning strategy and evolutionary models for the combined sequence matrix were assessed with PartitionFinder (Lanfear et al. 2012) under the (BIC) and using the implemented greedy algorithm.
Bayesian phylogenetic inference was conducted using MrBayes 3.2 (Ronquist et al. 2012). Two independent analyses consisting of four chains each were run for 5·106 generations specifying a sampling frequency every 100 generations, and setting a burn-in fraction of 10%. MCMC convergence and the (ESS) estimates were checked with TRACER v. 1.5 (Rambaut and Drummond 2007). Additionally, a maximum likelihood search was done using GARLI v.2.01 (Zwickl 2006) and performing 100 bootstrap replicates.
Taxonomic hypotheses testing
Specific hypotheses of monophyly were tested using a ML framework and the (AU test, Shimodaira 2000) as implemented in the CONSEL program (Shimodaira and Hasegawa 2001). We compared our molecular phylogenetic hypothesis with some of the most relevant systematic proposals for the genus Chrysolina (see results). Prior to the evaluation of each taxonomic scenario, a ML phylogenetic analysis was performed in GARLI v.2.01 using the same partitioning scheme and models as in the phylogenetic searches described above, but enforcing the monophyly of the taxa of interest. Once the resulting ML trees were obtained, their per site log-likelihoods were calculated using RAxML v8.0.X program (Stamatakis 2014) and used as input data in CONSEL.
Ancestral character reconstruction
Ancestral host plant affiliations were reconstructed using BayesTraits v. 2.0 (Pagel and Meade 2013) selecting the MCMC mode and the “multistate” model of evolution (Pagel et al. 2004). To take into account phylogenetic uncertainty, reconstructions were based on 1000 randomly selected post-burnin Bayesian trees from the phylogenetic analysis in MrBayes 3.2. Following the manual’s recommendations (http://www.evolution.rdg.ac.uk/BayesTraitsV2.0Files/TraitsV2Manual.pdf), the (RJ) MCMC with a hyperprior approach was chosen, and the interval of 0–30 for the RJ-hyperprior implementing an exponential distribution was applied. The “addMRCA” command was used to calculate the posterior distribution of ancestral character states at selected nodes in the Bayesian Chrysolina tree. A total of 10·106 generations were run, with samples taken every 100 iterations and discarding a burn-in fraction of 10%. Results of the MCMC runs including the ESS values were analysed in TRACER v. 1.5.
We also used BayesTraits to evaluate different ancestral host plant affiliations scenarios at the root of the Chrysolina tree. Analyses were conducted by enforcing the ancestral state of the (mrca) for the core Chrysolina node (excluding the divergent species Chrysolina vigintimaculata) to be one of the eight host plant families recorded for the studied Chrysolina species. MCMC was used to explore the samples and the space rate parameter of 1000 post-burnin trees generated in the MrBayes analysis. We performed two independent runs of 10·106 generations for each one of the constrained searches, and sampling rate parameters every 100 generations. The constrained runs were then compared by calculating the Bayes factors between the best and second best models based on the harmonic mean of the likelihood from each analysis as indicated in BayesTraits manual.
Results
Sequence data and phylogenetic analysis
Lengths of the amplified gene fragments ranged from 581 to 794 bp for cox1, 278 to 512 bp for rrnL, and 294 to 363 for H3. Total length of the concatenated DNA sequence matrix was 1682 bp. In cox1, 48.36% of the aligned positions were variable, indicating high divergence level among the studied sequences. Indeed, accumulation of mutations for cox1 was higher than for the other markers, as shown by the pairwise sequence divergence (p-distance), which ranged between 0.0063 and 0.2236 (average: 0.1331±0.0105) for cox1, 0.0012 and 0.1723 (average: 0.0924±0.0100) for rrnL, and 0.0027 and 0.1077 (average: 0.0641±0.0108) for H3. Also, cox1 and rrnL sequences showed the well-known A+T bias typical of insect mtDNA (69.9% and 76,4%, respectively), whereas base frequency was more balanced in the nuclear H3 marker (54,8%). Chi-squared tests for bias in base composition showed no significant heterogeneity in our datasets (P>0.99). On the other hand, ILD test revealed no evidence of incongruence among molecular markers (P= 0.24), and we therefore performed all subsequent phylogenetic analyses following a supermatrix approach.
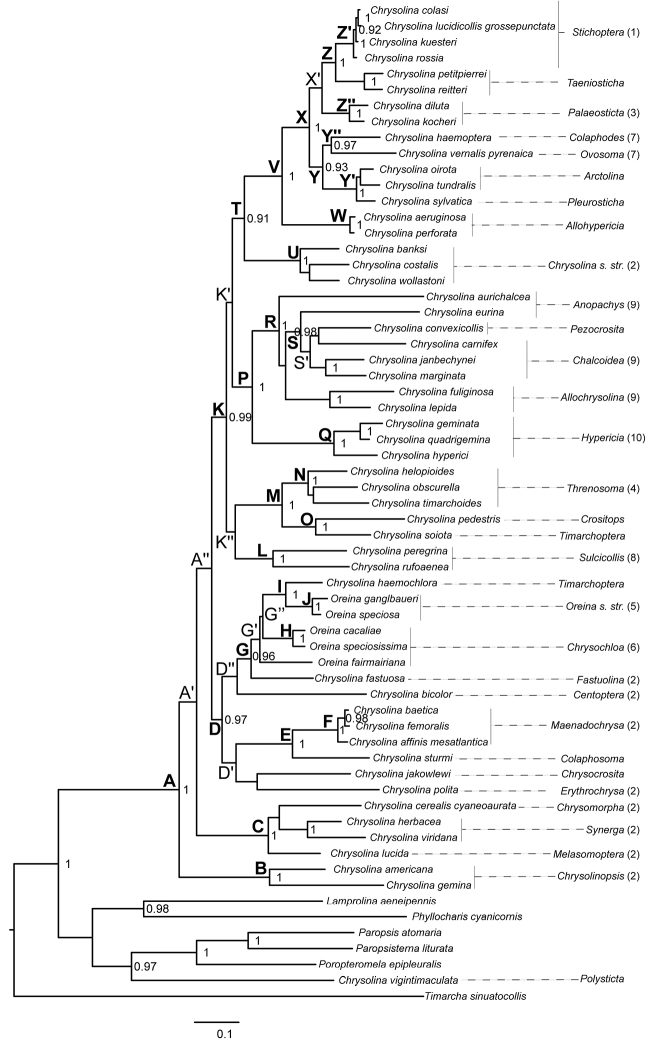
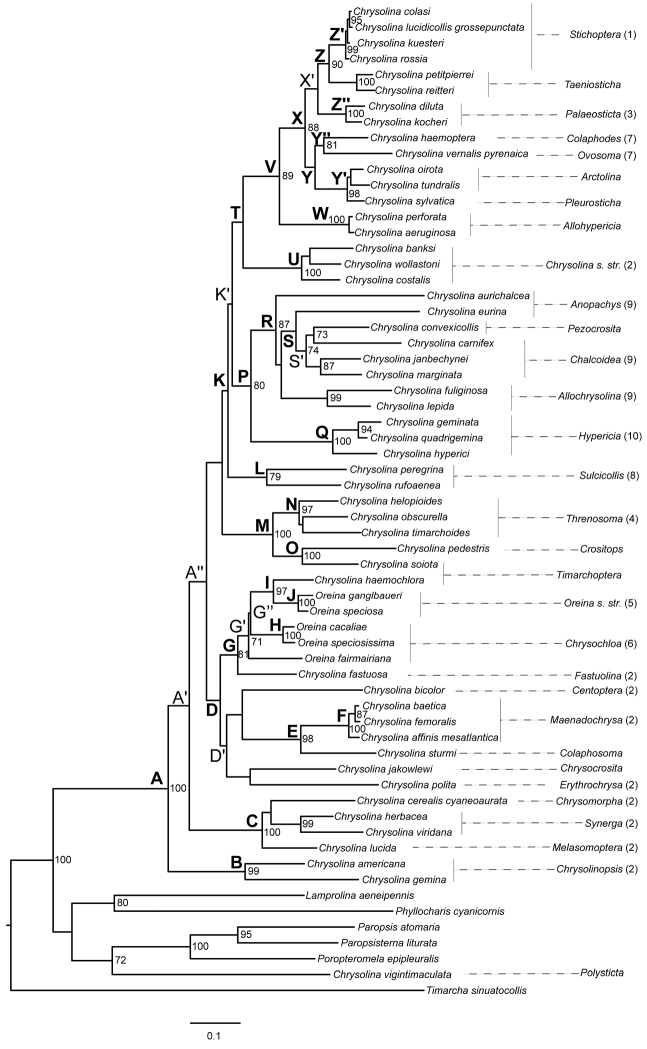
The best-fit partitioning scheme selected by PartitionFinder under BIC divided the data into seven subsets, each with its own model of molecular evolution (Table 2). The effective sample size value for each parameter sampled from the MCMC analysis was always >200. Bayesian and ML searches resulted in almost the same topology (Figures 1 and 2), with few discrepancies affecting only unsupported relationships such as the placement of the species Chrysolina bicolor (Fabricius, 1775), the position of the subgenus Sulcicollis (Fairmaire, 1887), and the internal branching pattern of the three species of the subgenus Chrysolina s. str. Motschulsky. Both phylogenetic approaches also yielded similar results in terms of nodal support, differing mainly in the values associated to some of the basal nodes of the core Chrysolina clade, which were higher in the Bayesian analysis (e.g. nodes K, D and T). The resulting phylogenetic trees revealed the paraphyly of the genus Chrysolina as currently described, due to the inclusion of the Oreina representatives within the Chrysolina clade (Figures 1 and 2). The genus Oreina is also recovered as a paraphyletic clade that includes the species Chrysolina haemochlora (Gebler, 1823). The results showed the monophyly of the studied Chrysolina (plus Oreina) species [clade A, Bayesian posterior probability (pp)=1, bootstrap=100] excepting the African taxa Chrysolina (Polysticta) vigintimaculata, which showed a higher affinity with outgroup taxa. In addition, the monophyletic status of the subgenera with more than one species sampled in the study was recovered in all cases excepting Anopachys Motschulsky, Chalcoidea, Timarchoptera Motschulsky and Oreina subgenus Chrysochloa Hope. The inferred topology allowed for the identification of four main monophyletic subgenera assemblages within the core Chrysolina clade with high support values in at least one of the resulting trees (clades B, C, D and K). Within these main lineages, it was also possible to identify systematic relationships among subgenera at different phylogenetic levels. The inferred groups of phylogenetically related subgenera and their statistical supports are summarized in Table 3.
Table 2.
Optimal partitioning strategy and evolutionary models selected using PartitionFinder under the Bayesian Information Criterion.
| Partition | Model |
|---|---|
| cox1 codon pos. 1 | GTR+I+G |
| cox1 codon pos. 2 | HKY+I+G |
| cox1 codon pos. 3 | GTR+G |
| rrnL | GTR+I+G |
| H3 codon pos. 1 | SYM+G |
| H3 codon pos. 2 | JC |
| H3 codon pos. 3 | HKY+I+G |
Figure 1.
Bayesian phylogenetic tree obtained from the combined analysis of cox1, rrnL and H3. Node numbers represent Bayesian posterior probability values. Only support values higher than 0.9 are shown. Numbers accompanying the subgeneric classification of the Chrysolina species on the right correspond to the systematic groups defined by Bourdonné and Doguet (1991). Clades mentioned in the text are highlighted.
Figure 2.
Maximum likelihood phylogenetic tree obtained from the combined analysis of cox1, rrnL and H3. Node numbers represent bootstrap support values. Only support values higher than 0.7 are shown. Numbers accompanying the subgeneric classification of the Chrysolina species on the right correspond to the systematic groups defined by Bourdonné and Doguet (1991). Clades mentioned in the text are highlighted.
Table 3.
Inferred phylogenetic relationships among Chrysolina and Oreina subgenera and their statistical supports. Nodes have been coded according to Figures 1 and 2.
| Node (Bayesian posterior probability; ML bootstrap) | Subgenera included | |||||
|---|---|---|---|---|---|---|
| B (1.00; 99) | Chrysolinopsis | |||||
| C (1.00; 100) | Chrysomorpha | |||||
| Melasomoptera | ||||||
| Synerga | ||||||
| D (0.97; <70) | Centoptera | |||||
| Chrysocrosita | ||||||
| Erythrochrysa | ||||||
| E (1.00; 98) | Colaphosoma | |||||
| Maenadochrysa | ||||||
| G (0.96; 81) | Fastuolina | |||||
| Oreina subgenus Chrysochloa | ||||||
| I (1.00; 97) | Oreina s. str. | |||||
| Timarchoptera partim. | ||||||
| K (0.99; <70) | Sulcicollis | |||||
| M (1.00; 100) | Threnosoma | |||||
| O (1.00; 100) | Crositops | |||||
| Timarchoptera partim. | ||||||
| P (1.00; 80) | Hypericia | |||||
| R (1.00; 87) | Anopachys | |||||
| Allochrysolina | ||||||
| S’ (<0.9; 74) | Chalcoidea | |||||
| Pezocrosita | ||||||
| T (0.91; <70) | Chrysolina s. str. | |||||
| V (1.00; 89) | Allohypericia | |||||
| X (1.00; 88) | Palaeosticta | |||||
| Y (0.93; <70) | Y’ (1.00; 98) | Arctolina | ||||
| Pleurosticha | ||||||
| Y’’ (0.97; 81) | Colaphodes | |||||
| Ovosoma | ||||||
| Z (1.00; 90) | Stichoptera | |||||
| Taeniosticha | ||||||
Testing for monophyly of key groups
Constrained ML searches were used to evaluate a number of taxonomic hypotheses for Chrysolina and Oreina using the AU test (Table 4). The phylogenetic scenarios that were rejected in the analyses included the systematic placement of Oreina as a different genus from Chrysolina (P=0.016), the synonymy of subgenera Paraheliostola L. N. Medvedev and Timarchoptera (Mikhailov 2002, P=0.001), the suggestion of a close relationship between Threnosoma Motschulsky and Chrysolina (Timarchoptera) haemochlora (Mikhailov 2005, P<0.001), the reciprocal monophyly of Colaphodes and Taeniochrysa (Hsiao and Pasteels 1999, P<0.001), the inclusion of Chrysolina timarchoides (Brisout, 1882) within the subgenera Maenadochrysa Bechyné (Bieńkowski 2001, P<0.001), the recognition of Craspeda sensu Bourdonné 2005 as a different genus from Chrysolina (P<0.01), the segregation from Chrysolina of the subgenera Allochrysolina Bechyné, Chalcoidea and Pezocrosita Jakobson (Bourdonné 2012, P<0.01), the monophyly of the Chrysolina species belonging to the “group 2” described by Bourdonné and Doguet (1991) (P<0.001) (Table 1), as well as the monophyly of the Chrysolina species feeding on hosts from the same plant family (Apiaceae, Asteraceae, Lamiaceae, Plantaginaceae, Ranunculaceae and Scrophulariaceae; P≤0.001 in all cases). Conversely, the molecular data could not reject the reciprocal monophyly of several taxa assemblages, such as Chrysolina vigintimaculata and the rest of the studied Chrysolina species (P=0.165), Chrysolina species belonging to the “group 6” described by Bourdonné and Doguet (1991) (P=0.527) (Table 1), subgenera Allochrysolina and Anopachys (Hsiao and Pasteels 1999, P=0.215), subgenera Chalcoidea and Hypericia (Pasteels et al. 2003, P=0.066), subgenera Allochrysolina, Chalcoidea and Pezocrosita (Bourdonné 2012, P=0.205), and the subgenera Palaeosticta Bechyné and Taeniosticha Motschulsky (Bourdonné 2005, P=0.198). Also, the monophyly of the sampled species concerning the subgenera Anopachys, Chalcoidea and Oreina subgenus Chrysochloa could not be rejected (P≥0.212 in all cases).
Table 4.
Results of the Approximately Unbiased test (AU test, Shimodaira 2000). Statistically significant P values are indicated in bold (P < 0.05).
| Hypothesis of monophyly | Authorship | AU test |
|---|---|---|
| Chrysolina timarchoides + Maenadochrysa | Bienkowski (2001) | 0.000 |
| Palaeosticta + Taeniosticha | Bourdonné (2005) | 0.198 |
| Craspeda as a different genus from Chrysolina | Bourdonné (2005) | 0.007 |
| Allochrysolina + Chalcoidea + Pezocrosita | Bourdonné (2012) | 0.205 |
| Allochyrsolina + Chalcoidea + Pezocrosita as a different genus from Chrysolina | Bourdonné (2012) | 0.003 |
| Species “group 2” | Bourdonné and Doguet (1991) | 0.000 |
| Species “group 6” | Bourdonné and Doguet (1991) | 0.527 |
| Allochrysolina + Anopachys | Hsiao and Pasteels (1999) | 0.215 |
| Colaphodes + Taeniochrysa | Hsiao and Pasteels (1999) | 0.000 |
| Paraheliostola + Timarchoptera | Mikhailov (2002) | 0.001 |
| Chrysolina haemochlora + Threnosoma | Mikhailov (2005) | 0.000 |
| Chalcoidea + Hypericia | Pasteels et al. (2003) | 0.066 |
| Anopachys species | 0.212 | |
| Chalcoidea species | 0.383 | |
| Chrysochloa species | 0.528 | |
| Oreina as a different genus from Chrysolina | 0.016 | |
| Chrysolina vigintimaculata + rest of the Chrysolina species + Oreina | 0.165 | |
| Species feeding on Apiaceae | 0.000 | |
| Species feeding on Asteraceae | 0.000 | |
| Species feeding on Lamiaceae | 0.000 | |
| Species feeding on Plantaginaceae | 0.000 | |
| Species feeding on Ranunculaceae | 0.001 | |
| Species feeding on Scrophulariaceae | 0.000 |
Ancestral character reconstruction
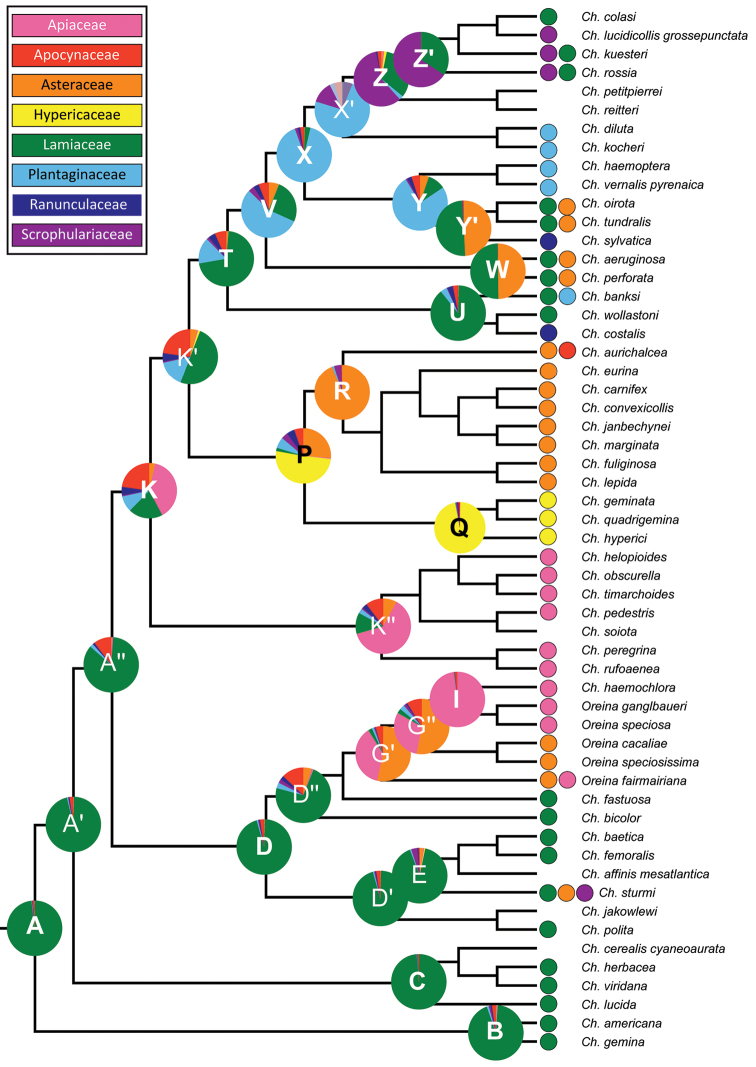
The Bayesian reconstruction of ancestral host plant associations showed an ancient affiliation with Lamiaceae at the root of the core Chrysolina clade (Figure 3, node A, P=0.98; Table 5). This plant family was also recovered as the most likely ancestral host for three of the main clades in our molecular phylogeny (nodes B, C and D; P=0.94, 0.99 and 0.95, respectively). Within clade D, a host shift from Lamiaceae towards Asteraceae (P=0.54) and/or Apiaceae (P=0.37) was detected for the mrca of Oreina and Chrysolina (Timarchoptera) haemochlora (clade G’). On the other hand, ancestral host plant reconstruction for node K was ambiguous, recovering associations with multiple families. However, it was possible to identify the occurrence of several host shifts for its derived lineages towards a new trophic association with (i) Apiaceae (node K’’, P=0.62), (ii) Hypericaceae (nodes P and Q, P=0.51 and 0.97, respectively), (iii) Asteraceae (node R, P=0.94), (iv) Plantaginaceae (node X, P=0.91), and (v) Scrophulariaceae (node Z’, PP=0.66). Nodes W and Y’ respectively showed a reversal shift from an ancestral Plantaginaceae host to the original Lamiaceae host family (P=0.5) as well as a new trophic link with Asteraceae (P=0.5).
Figure 3.
Ancestral reconstruction of host plant affiliations in the studied species of Chrysolina and Oreina. Terminal taxa are coded according to the available host plants records from the literature (Table 1). Pie charts at selected nodes show probabilities of each state from the Bayesian analysis in BayesTraits. Clades mentioned in the text are highlighted.
Table 5.
Posterior probability values of ancestral host-plant affiliations calculated in BayesTraits for the selected nodes in the Chrysolina-Oreina phylogeny. The highest probability value(s) for each node are highlighted in bold. Ast.=Asteraceae, Api.=Apiaceae, Hyp.=Hypericaceae, Lam.=Lamiaceae, Plant.=Plantaginaceae, Scro.=Scrophulariaceae, Ran.=Ranunculaceae, Apo.=Apocynaceae.
| Host-plant family | ||||||||
|---|---|---|---|---|---|---|---|---|
| Node | Ast. | Api. | Hyp. | Lam. | Plant. | Scro. | Ran. | Apo. |
| A | 0.000 | 0.001 | 0.000 | 0.980 | 0.003 | 0.002 | 0.002 | 0.010 |
| A’ | 0.001 | 0.002 | 0.001 | 0.959 | 0.006 | 0.003 | 0.006 | 0.022 |
| A’’ | 0.002 | 0.010 | 0.000 | 0.852 | 0.020 | 0.001 | 0.011 | 0.104 |
| B | 0.002 | 0.006 | 0.002 | 0.937 | 0.011 | 0.010 | 0.008 | 0.024 |
| C | 0.000 | 0.000 | 0.000 | 0.987 | 0.002 | 0.002 | 0.001 | 0.007 |
| D | 0.002 | 0.001 | 0.000 | 0.952 | 0.008 | 0.006 | 0.006 | 0.024 |
| D’ | 0.002 | 0.001 | 0.000 | 0.952 | 0.008 | 0.006 | 0.006 | 0.024 |
| D’’ | 0.048 | 0.010 | 0.001 | 0.732 | 0.033 | 0.023 | 0.024 | 0.129 |
| E | 0.022 | 0.005 | 0.006 | 0.910 | 0.008 | 0.036 | 0.008 | 0.006 |
| G’ | 0.536 | 0.374 | 0.001 | 0.023 | 0.012 | 0.008 | 0.002 | 0.044 |
| G’’ | 0.531 | 0.300 | 0.001 | 0.027 | 0.029 | 0.015 | 0.009 | 0.089 |
| I | 0.001 | 0.979 | 0.000 | 0.001 | 0.001 | 0.000 | 0.002 | 0.015 |
| K | 0.036 | 0.387 | 0.000 | 0.200 | 0.093 | 0.007 | 0.049 | 0.227 |
| K’ | 0.040 | 0.005 | 0.013 | 0.499 | 0.158 | 0.007 | 0.049 | 0.227 |
| K’’ | 0.080 | 0.624 | 0.001 | 0.124 | 0.028 | 0.009 | 0.029 | 0.104 |
| P | 0.262 | 0.005 | 0.511 | 0.019 | 0.064 | 0.039 | 0.047 | 0.053 |
| Q | 0.001 | 0.002 | 0.967 | 0.001 | 0.003 | 0.008 | 0.008 | 0.010 |
| R | 0.941 | 0.000 | 0.000 | 0.000 | 0.010 | 0.042 | 0.001 | 0.006 |
| T | 0.011 | 0.001 | 0.001 | 0.709 | 0.153 | 0.015 | 0.041 | 0.068 |
| U | 0.001 | 0.001 | 0.001 | 0.890 | 0.039 | 0.004 | 0.034 | 0.031 |
| V | 0.059 | 0.001 | 0.001 | 0.257 | 0.555 | 0.034 | 0.033 | 0.060 |
| W | 0.498 | 0.000 | 0.000 | 0.501 | 0.000 | 0.001 | 0.000 | 0.001 |
| X | 0.003 | 0.000 | 0.000 | 0.033 | 0.908 | 0.018 | 0.014 | 0.023 |
| X’ | 0.005 | 0.000 | 0.001 | 0.055 | 0.736 | 0.128 | 0.028 | 0.047 |
| Y | 0.052 | 0.000 | 0.000 | 0.103 | 0.757 | 0.011 | 0.026 | 0.050 |
| Y’ | 0.492 | 0.000 | 0.000 | 0.498 | 0.001 | 0.001 | 0.002 | 0.006 |
| Z | 0.009 | 0.008 | 0.016 | 0.327 | 0.023 | 0.586 | 0.009 | 0.023 |
| Z’ | 0.000 | 0.000 | 0.000 | 0.344 | 0.000 | 0.656 | 0.000 | 0.000 |
Results from Bayes factor comparisons of the constraint hypotheses for the ancestral plant family at the root of the core Chrysolina clade (node A) corroborated MCMC ancestral state reconstruction, offering positive to very strong statistical support for an ancestral trophic association with Lamiaceae (Table 6).
Table 6.
Comparing model support with the Bayes factor. Bayes factors were calculated as described in the BayesTraits manual: BF=2(ln LhA−ln LhB), where ln Lhx is the marginal likelihood from the harmonic mean of the post-convergence. The plant family Lamiaceae is the most likely ancestral host at the root of the core Chrysolina clade with the highest harmonic mean. The right column indicates the Bayes factor compared against Lamiaceae as the favoured ancestral host. * Indicates positive evidence, ** indicates strong evidence, and *** indicates very strong evidence for the favoured hypothesis.
| Host plant family | ln Lh | Bayes Factor |
|---|---|---|
| Apiaceae | -62.77 | 5.27** |
| Apocynaceae | -63.78 | 7.30** |
| Asteraceae | -65.71 | 11.16*** |
| Hypericaceae | -65.59 | 10.92*** |
| Lamiaceae | -60.13 | - |
| Plantaginaceae | -62.44 | 4.61* |
| Ranunculaceae | -62.57 | 4.86* |
| Scrophulariaceae | -63.24 | 6.20** |
Discussion
Molecular systematics of Chrysolina
The mitochondrial and nuclear genes used here provided an expanded and better-resolved tree topology for the genus Chrysolina, significantly improving previous phylogenetic hypotheses. Our results support the reciprocal monophyly of the studied species of Chrysolina (plus Oreina) including the divergent Chrysolina (Polysticta) vigintimaculata, whose relationship with the core Chrysolina-Oreina clade could not be rejected by the AU test. The inferred tree topologies recovered Chrysolina vigintimaculata as a well-differentiated lineage sister to the rest of the ingroup taxa. This species has been traditionally assigned to the subgenus Atechna Chevrolat (Bieńkowski 2001), a species of which was included in the phylogenetic analysis of Gómez-Zurita et al. (2008) based on three ribosomal genes and showing a clear divergence from the Chrysolina-Oreina clade. In addition, the same pattern was observed in a different phylogenetic study based on five molecular markers (Jurado-Rivera et al. in prep.) that included the species Chrysolina (Atechna) striata (Degeer, 1778). Although more data are needed, the available information indicates that these taxa may represent a lineage of early divergence within Chrysolina whose taxonomic status should be further investigated.
The inferred topology also supported most of the current subgeneric taxonomy of Chrysolina (Bieńkowski 2001, Kippenberg 2010), since the monophyly of the subgenera screened for more than one species could be demonstrated or alternatively could not be rejected by the AU test. The exceptions in this regard are the synonymy of the subgenus Paraheliostola with the subgenus Timarchoptera by Mikhailov (2002) and the combination of the species Chrysolina (Threnosoma) timarchoides with the subgenus Maenadochrysa by Bieńkowski (2001). In both cases the taxa in question were recovered with support as well-differentiated lineages, thus indicating that such taxonomic decisions could be wrong. Therefore, the subgenus Paraheliostola (type species Chrysolina soiota Jacobson, 1924) should be restored according to the present molecular phylogeny. Moreover, the available karyological evidence also conflicts with Bieńkowski’s (2001) proposal (Petitpierre 1975, 1981), and we thus agree with Daccordi and Ruffo (2005) and with Kippenberg (2010) in that Chrysolina timarchoides belongs in the subgenus Threnosoma.
The new molecular phylogeny also sheds light on the contentious issue of the taxonomic status of Oreina. Our analyses supported the inclusion of the studied Oreina species within the core Chrysolina clade, which was also backed up statistically in the AU test constraining these genera to be reciprocally monophyletic (Table 4). The sample included the type species of the genus, Oreina speciosa (Linnaeus, 1758), which further strengthens our findings and corroborates previous hypotheses that consider Oreina as part of the Chrysolina lineage (Chapuis 1874, Bourdonné and Doguet 1991, Daccordi 1994). Moreover, the species feeding on Apiaceae hosts, Oreina ganglbaueri (Jakob, 1953) and Oreina speciosa, were recovered as more closely related to the also Apiaceae feeding Chrysolina haemochlora than to the remainder of the Oreina species analysed here, reinforcing our conclusions and highlighting the need for a taxonomic revision for the group. On the other hand, the proposal of considering the genera Craspeda and Chalcoidea (sensu Bourdonné 2005 and 2012, respectively) as separate lineages from the remainder of the Chrysolina species is not supported in our phylogenetic framework, although the monophyly of the taxa included in each of them could not be statistically rejected (Table 4). Thus, the recognition of Craspeda and/or Chalcoidea as valid genera would render Chrysolina paraphyletic.
Excluding the divergent species Chrysolina vigintimaculata, Chrysolina could be subdivided into four major clades (Figures 1 and 2, clades B, C, D and K). The clades B and C comprised species from the “group 2” defined by Bourdonné and Doguet (1991), all of them feeding on host plants belonging to the family Lamiaceae and with a diploid chromosome number of 2n=24 (Petitpierre 1975, 1981, 1983). The hypothetical monophyly of the aforementioned “group 2” was statistically rejected by the AU test, thus reinforcing our finding that such an assemblage of species does not constitute a natural group. The clade B included two monotypic subgenera (Chrysolinopsis Bechyné and Taeniochrysea, sensu Bieńkowski 2001) that have been recently regarded as synonyms by Kippengberg (2010), a taxonomic decision that is strongly supported in our phylogenetic analyses. The monophyly of the species nested in clade C were also noted in the phylogenetic study of Garin et al. (1999), excepting the species Chrysolina cerealis (Linnaeus, 1767) that they recovered in a divergent clade as sister to Chrysolina fastuosa with maximum bootstrap support. Here we have analysed the subspecies Chrysolina cerealis cyaneoaurata (Motschulsky, 1860) inferring a clear relationship with the remainder of the members in clade C that is supported with maximum posterior probability and bootstrap values. Genetic distances (p-distance) between the sequences deposited in GenBank by Garin et al. (1999) regarding Chrysolina cerealis and our data for Chrysolina cerealis cyaneoaurata were unusually high for an intraspecific comparison (cox1: 0.14; rrnL: 0.08), thus suggesting that the taxa in question do not belong to the same species. It remains to be investigated whether their divergence is due to specimen misidentification or whether Chrysolina cerealis s. str. and Chrysolina cerealis cyaneoaurata really are different species. Meanwhile, the results about the systematic position of Chrysolina cerealis should be interpreted with caution.
Clade D defined the monophyletic origin of seven Chrysolina subgenera traditionally associated with the “group 2” proposed by Bourdonné and Doguet (1991) plus two Oreina subgenera included in “groups 5 and 6”, all of them with a karyotype 2n = 24 (Petitpierre 1975, 1981, 1983) excepting Chrysolina haemochlora (2n=27, Petitpierre and Mikhailov 2009). The affinity between the subgenera Colaphosoma Motschulsky and Maenadochrysa could be established with confidence agreeing with their shared feeding habits on Lamiaceae species of the tribe Mentheae (Jolivet and Petitpierre 1976, Jolivet et al. 1986, Bieńkowski 2010). On the other hand, the close relationship recovered in the present work among Chrysolina fastuosa and the studied Oreina species is consistent with the findings of Hsiao and Pasteels (1991) based on a different set of molecular markers. The authors concluded that such association was contradicted by strong morphological evidence, highlighting the need of further research on this issue. Our molecular phylogeny not only confirmed the monophyly of these taxa, but also revealed the inclusion of an additional Chrysolina species in this clade, Chrysolina haemochlora.
Interestingly, our results regarding the clade K were fully consistent with most species groupings established by Bourdonné and Doguet (1991) based on morphology, karyology and biology of the species (“groups 1, 3, 4, 7, 8, 9, 10 and 2 partim.”). Available molecular phylogenies of Chrysolina (Garin et al. 1999, Hsiao and Pasteels 1999) failed at recovering supported relationships among these groups, excepting the monophyletic origin of the species belonging in the “groups 1, 3 and 7” inferred by Garin et al. (1999). In contrast, our analyses allowed for the identification of their phylogenetic relationships at deep taxonomic level, and also extended the results to seven Chrysolina subgenera not studied by Bourdonné and Doguet (1991). The latter was the case of clade M, where the subgenera Crositops Marseul and Timarchoptera (more likely Paraheliostola, see above) were recovered as the sister lineage of the Threnosoma species regarded as “group 4”. Indeed, the subgenera Crositops and Threnosoma are known to share morphological attributes (Mikhailov 2005). Although no information is available for the species Chrysolina soiota, the remainder of the species in clade M feed on Apiaceae and also share a male karyotype 2n=47 (Petitipierre 1981, 1999, Petitpierre et al. 2004, Petitpierre and Mikhailov 2009), which is highly consistent with their close association recovered here. On the other hand, the existence of a relationship between the Mediterranean subgenus Threnosoma and the Siberian subgenus Timarchoptera proposed by Mikhailov (2005) was rejected by the AU test. Another subgenus that was not analysed by Bourdoneé and Doguet (1991) is represented in our sampling by the species Chrysolina (Pezocrosita) convexicollis (Jakobson, 1901), which appeared in the trees clearly nested within the species “group 9” (clade R) sharing with them a trophic link with Asteraceae. Our phylogenetic hypotheses also allowed for the identification of two main evolutionary lineages within “group 9”, on one hand the species belonging in the subgenera Anopachys [excluding Chrysolina aurichalcea (Gebler, 1825)], Chalcoidea and Pezocrosita, all of them feeding on closely related plant species in the family Asteraceae in the tribe Anthemideae (Achillea, Artemisia, Santolina, Tanacetum; Cobos 1953, Jolivet and Petitpierre 1976, Bieńkowski 2010, 2011, clade S) and sharing a karyotype of 2n=40 [cytogenetic data for Chrysolina eurina (Frivaldszky, 1883) and Chrysolina convexicollis are not available], and on the other hand the species in the subgenera Allochrysolina with a male karyotype 2n=42 (Petitpierre 1999) and feeding on closely related Asteraceae host plants in the subtribe Centaureinae (Centaurea, Mantisalca, Jolivet and Petitpierre 1976, Bourdonné and Doguet 1991). In turn, the species in “group 9” were recovered as the sister lineage of the species classified in the “group 10” (subgenus Hypericia; clade Q), thus contradicting Bourdonné and Doguet’s (1991) view that the subgenus Hypericia is so differentiated from the remainder of the Chrysolina subgenera that it deserves a generic status. Recognition of the genus Hypericia would render Chrysolina paraphyletic. Also regarding this lineage, Pasteels et al. (2003) found that the subgenera Hypericia, Chalcoidea and Sphaeromela are the only Chrysomelinae leaf beetles producing polyoxygenated steroids as defensive toxins, and suggested that they could be raised to a distinct genus. However, our inferred topologies were not compatible with this hypothesis, although the AU test could not reject the constrained monophyly of Chalcoidea and Hypericia. On the other hand, the well-supported and resolved clade T allowed for the identification of the phylogenetic relationships among four of the systematics groups defined by Bourdonné and Doguet (1991), and also expanded our knowledge regarding the systematic position of four subgenera not included before in any phylogenetic analysis. The species in the subgenera Chrysolina s. str. were placed in the “group 2” based on their trophic link with the plant family Lamiaceae but our results clearly contradict this association (clade U), agreeing with their unique male karyotype (2n=23; Petitpierre 1975, 1981, 1983). The common ancestry of Colaphodes, Ovosoma Motschulsky, Palaeosticta and Stichoptera Motschulsky demonstrated by Garin et al. (1999) was confirmed here, and in addition we show that the subgenera Allohypericia Bechyné, Arctolina Kontkanen, Pleurosticha Motschulsky and Taeniosticha also belong in this monophyletic lineage. The close relationship between the subgenera Arctolina and Pleurosticha has been previously proposed according to their morphology (Bieńkowski 2004) and their karyological resemblances [2n=26 (Xyp), Petitpierre and Mikhailov 2009]. In this regard, our study contributes additional evidence confirming their phylogenetic relatedness (clade Y’). The monophyly of the species adapted to the plant family Plantaginaceae (subgenera Palaeosticta, Colaphodes and Ovosoma) could not be rejected, indicating that they could conform to a natural group, thus expanding Bourdonné and Doguet’s (1991) “group 7”. On the other hand, the Stichoptera species of the “group 1” sensu Bourdonné and Doguet (1991) were demonstrated to be sister to the morphologically well-defined subgenus Taeniosticha (Bourdonné et al. 2013). Stichoptera species are characterized by their marked asymmetrical karyotypes (Petitpierre 1999) and their affiliation with Lamiaceae and Scrophulariaceae host plants, but unfortunately no data are available regarding the biology and the cytogenetics of the subgenus Taeniosticha to contrast with our molecular results.
Evolution of the host plant associations in Chrysolina
The initial stages of the evolutionary history of the genus Chrysolina were closely related to the plant family Lamiaceae (Figure 3, node A), which is in line with the pioneering studies based on the karyology and the ecology of the species (Petitpierre and Segarra 1985, Bourdonné and Doguet 1991) and also on mtDNA sequences (Garin et al. 1999). The inferred ancestral association with Lamiaceae was highly favoured in our analyses compared to the alternative hypotheses, including an original affiliation with the family Asteraceae suggested by Crowson (1981).
The most basal clades in our Chrysolina phylogeny are those living on Lamiaceae. However, the phylogenetic uncertainty affecting this region of the tree prevents us for drawing firm conclusions about the number of lineages that have adapted to this plant family at the early stages of the evolution of the genus. In contrast, our phylogenetic analyses allowed for the identification of a minimum of eight host plant family shifts in the Chrysolina tree, thus indicating that the feeding spectrum of the extant Chrysolina species is the result of frequent and abrupt host shifts in their evolutionary history. While some of these shifts are between plant families belonging to the same order (Lamiaceae, Plantaginaceae, Scrophulariaceae; order Lamiales; APG 2009), others are between distant plant families from different subclasses [shift from families in the subclass Asterids to Hypericaceae (subclass Rosids); APG 2009] or even from more divergent lineages [shifts from Asterids to Ranunculaceae (basal Eudicot); APG 2009]. Three main hypotheses have been proposed concerning the macroevolution of insect–plant associations (Nyman 2010): (i) the ‘cospeciation’ or ‘parallel cladogenesis’ model (Fahrenholz 1913): matching of speciation events between insects and their host plants; (ii) the ‘escape and radiate’ model (Ehrlich and Raven 1964): plants ‘escape‘ from herbivory due to novel defences and radiate, followed by colonization of new insect taxa that then radiate on them; and (iii) the ‘sequential evolution’ model (Jermy 1984): insects have little effect on the speciation of their hosts, whereas the diversification of hosts increases possibilities of ecological speciation in insects. The hypothesis of ‘parallel cladogenesis’ between Chrysolina lineages and their host plant families can be discarded as the temporal origin of the more closely related host plant families recorded for Chrysolina (Lamiaceae and Scrophulariaceae: mrca >65Ma, Bremer et al. 2004) clearly pre-dates the diversification of the Chrysolina lineage itself [mrca ca. 40Ma, (ca. 20Ma excluding the divergent subgenera Atechna), Gómez-Zurita et al. 2007]. Consistently, this pattern of asynchronous diversification has been found among other phytophagous insect groups and their host plants (Lopez-Vaamonde et al. 2006, McKenna et al. 2009). Regarding the ‘escape and radiate’ model, the existence of coincident radiations at a large scale among host families and the Chrysolina lineages is also not possible due to this time lag in their respective origins. Conversely, the ancestral host plant family affiliations inferred for Chrysolina seem to fit better the ‘sequential evolution’ model, as deduced from the continuous host-shifting among pre-existing host families that characterizes the evolution of the genus (Nyman 2010). Indeed, some Chrysolina clades have experienced multiple host shifts from the ancestral affiliation with Lamiaceae. As an example we could cite the case of the preference for Lamiaceae observed in the derived lineages Allohypericia (clade W), Stichoptera (clade Z’), Arctolina and Pleurosticha (clade Y’), which seems to be a back-colonization of this family from ancestors previously adapted to Plantaginaceae. Another case of multiple shifts is illustrated by the transition from Lamiaceae to Asteraceae and then to Apiaceae inferred for the Oreina clade, which is highly consistent with previous results based on allozyme data (Dobler et al. 1996) and mtDNA sequences (Hsiao and Pasteels 1999). In addition, convergent shifts to the same host plant family in different Chrysolina lineages have also occurred (Apiaceae: clades G’ and K’’; Asteraceae: clades G’, R, W’ and Y’, Chrysolina sturmi (Westhoff, 1882) and Chrysolina cerealis cyaneoaurata; Ranunculaceae: Chrysolina costalis (Olivier, 1807) and Chrysolina silvatica (Gebler, 1823); Scrophulariaceae: clade Z’ and Chrysolina sturmi), thus suggesting the existence of evolutionary constraints in host shifts as it has been described in other phytophagous insects including Chrysomelidae (Futuyma et al. 1993, Futuyma and Mitter 1996, Janz et al. 2001, Nosil 2002). A possible explanation for the continuous and convergent shifts among restricted sets of plant taxa is the phytochemical similarity among the alternative hosts (Feeny 1992), and indeed this seems to be the underlying mechanism in other herbivorous beetle groups (Becerra 1997, Kergoat et al. 2005). It also has been suggested that convergent shifts may not be independent, in the sense that an ancestral trait allowing the colonisation of a given plant group might have been already present in the insect lineages (Janz and Nylin 2008).
Chrysolina leaf beetles are highly specialized herbivores feeding on a narrow range of host plants (Jolivet and Petitpierre 1976, Bourdonné and Doguet 1991). However, despite the high level of specialization, their diet breadth ranges from species feeding on few plant species from the same genus or family (i.e., monophagous or oligophagous, respectively) to more generalist species exploiting few species but from different plant families (i.e., polyphagous). In this regard, Garin et al. (1999) reported the subgenus Chrysolina s. str. as the only lineage within the genus experiencing a shift to a generalist feeding habit at the plant family level. Now, our expanded taxon sampling coupled with the availability of a more complete host plant record shows that polyphagy is distributed across the Chrysolina tree, although it occurs at a lower frequency than mono- and oligophagy. Moreover, our results suggest that niche widths have varied through time, since some Chrysolina clades include mixtures of species with different levels of diet breadth (clades E, G’, R, U, Y’ and Z’). Oscillations in host range over evolutionary time are thought to play an important role in the diversification of the phytophagous insects (oscillation hypothesis, Janz et al. 2006, Janz and Nylin 2008). Under this model, speciation is driven by successive cycles of expansion of the host-plant range and generation of new species through specialization on different hosts. The oscillations are maintained through the ability to retain essential parts of the genetic “machinery” to utilize ancestral hosts, and therefore the probability of a major host shift seems to be positively influenced by polyphagy (Janz 2011). Our results on Chrysolina are still too preliminary to offer any scenario for the evaluation of this hypothesis. However, as it has been shown here, the evolutionary history of the genus is deeply associated with the occurrence of frequent and abrupt host shifts giving rise to the specialization on a restricted set of divergent host plant taxa, which is consistent with the model predictions. Optimizing niche width on the Chrysolina phylogeny would help in elucidating whether the diet breadth of the extant polyphagous species indeed represent an event of host range expansion from specialized ancestors, and whether polyphagy has been a transitional stage during host shifts. However, ancestral host range reconstruction will require very detailed information on host plant records and a well-resolved phylogeny for all Chrysolina species (Janz and Nylin 2008). In this respect, future research will be directed towards the expansion of the taxonomic sampling and the exploration of additional molecular markers in order to improve phylogenetic resolution. The implementation of DNA-based techniques for the taxonomic identification of the host plants (Jurado-Rivera et al. 2009) would also contribute to our understanding on the evolution of the ecological associations in this large and highly diversified leaf-beetle genus.
Conclusions
The combined phylogenetic analysis of mitochondrial (cox1 and rrnL) and nuclear (H3) DNA sequences allows for the identification of the main evolutionary lineages in a sample of Chrysolina species representing almost half of the subgeneric diversity and most of the morphological and ecological variation in the genus. Our results reveal the paraphyly of the genus Chrysolina as currently described, due to the inclusion of the Oreina representatives within the Chrysolina clade. In this regard, the recognition of the genera Craspeda and Chalcoidea (sensu Bourdonné 2005 and 2012, respectively) would also render Chrysolina paraphyletic. The molecular phylogeny support for the reciprocal monophyly of the studied species of Chrysolina (plus Oreina) including the divergent Chrysolina (Polysticta) vigintimaculata, whose relationship with the core Chrysolina clade cannot be statistically rejected. The molecular data are consistent with the current subgeneric arrangement of the species, excepting the synonymy of the subgenus Paraheliostola with the subgenus Timarchoptera by Mikhailov (2002) and the combination of the species Chrysolina (Threnosoma) timarchoides with the subgenus Maenadochrysa by Bieńkowski (2001). In addition, our hypothesized molecular phylogeny allows for the identification of deep-level evolutionary relationships among the studied Chrysolina subgenera. The Bayesian reconstruction of the host plant associations in the Chrysolina phylogeny points to the family Lamiaceae as the ancestral host of the genus, in agreement with previous studies. The feeding spectrum of the extant Chrysolina species has been shaped by continuous host-shifting among pre-existing host plant families throughout the evolution of the genus. Many clades include mixtures of species with different levels of diet breadth, indicating that niche width has varied through time.
Acknowledgements
We would like to thank the following colleagues who kindly provided us several specimens for this study: Dr. Yuri Mikhailov (Ural State Forestry Engineering University, Ekaterinburg, Russia; the species from Russia and Kazakhstan), Jean-Claude Bourdonné (Lesparrou, Ariège, France; two species from France) and Prof. José Serrano (Univ. Murcia, Spain; the species from Turkey).
Citation
Jurado-Rivera JA, Petitpierre E (2015) New contributions to the molecular systematics and the evolution of host-plant associations in the genus Chrysolina (Coleoptera, Chrysomelidae, Chrysomelinae). In: Jolivet P, Santiago-Blay J, Schmitt M (Eds) Research on Chrysomelidae 5. ZooKeys 547: 165–192. doi: 10.3897/zookeys.547.6018
References
- APG: Angiosperm Phylogeny Group (2009) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society 161: 105–121. doi: 10.1111/j.1095-8339.2009.00996.x [Google Scholar]
- Baselga A, Novoa F. (2006) Diversity of Chrysomelidae (Coleoptera) in Galicia, Northwest Spain: estimating the completeness of the regional inventory. Biodiversity & Conservation 15(1): 205–230. doi: 10.1007/s10531-004-6904-x [Google Scholar]
- Becerra JX. (1997) Insects on plants: macroevolutionary chemical trends in host use. Science 276(5310): 253–256. doi: 10.1126/science.276.5310.253 [DOI] [PubMed] [Google Scholar]
- Bieńkowski AO. (2001) A study on the genus Chrysolina Motschulsky, 1860, with a checklist of all the described subgenera, species, subspecies, and synonyms (Coleoptera: Chrysomelidae: Chrysomelinae). Genus 12(2): 105–235. [Google Scholar]
- Bieńkowski AO. (2004) A review of the subgenus Arctolina Kontkanen, 1959 of the genus Chrysolina Motschulsky, 1860 (Coleoptera: Chrysomelidae: Chrysomelinae). Genus 15(2): 187–233. [Google Scholar]
- Bieńkowski AO. (2007) A monograph of the genus Chrysolina Motschulsky, 1860 (Coleoptera: Chrysomelidae) of the world Part 1. Techpolygraphcentre Publ., Moscow. [Google Scholar]
- Bieńkowski AO. (2010) A review of the leaf-beetle genus Chrysolina Motschulsky (Coleoptera, Chrysomelidae) from Russia and European countries of the former USSR: I. A key to species with developed hind wings. Entomological Review 90(7): 885–902. doi: 10.1134/S0013873810070079 [Google Scholar]
- Bieńkowski AO. (2011) Review of the leaf-beetle genus Chrysolina Motschulsky (Coleoptera, Chrysomelidae) from Russia and the European countries of the former USSR: II. A key to species with the reduced hind wings. Entomological Review 91(5): 645–664. doi: 10.1134/S0013873811050095 [Google Scholar]
- Bourdonné JC. (2005) Révision du sous-genre Taeniosticha Motschulsky, 1860 du genre Craspeda Motschulsky, 1860 (Coleoptera, Chrysomelidae). 1ère partie. Nouvelle Revue d’Entomologie 21(4): 297–363. [Google Scholar]
- Bourdonné JC. (2008) Mise au point et compléments sur la révision des Taeniosticha du genre Craspeda (Coleoptera, Chrysomelidae, Chrysomelinae). Nouvelle Revue d’Entomologie 25: 17–26. [Google Scholar]
- Bourdonné JC. (2012) Révision du genre Chalcoidea Motschulsky, 1860 avec description d’une espèce nouvelle, Chalcoidea (s. str.) hollandei (1ère partie) (Coleoptera, Chrysomelidae, Chrysomelinae). Nouvelle Revue d’Entomologie 28: 99–181. [Google Scholar]
- Bourdonné JC, Doguet S. (1991) Données sur la biosystématique des Chrysolina l.s. (Coleoptera: Chrysomelidae: Chrysomelinae). Annales de la Société entomologique de France 27(1): 29–64. [Google Scholar]
- Bourdonné JC, Doguet S, Petitpierre E. (2013) Chrysolina (Stichoptera) oceanoripensis nova species, endémique des dunes françaises des Landes de Gascogne et considérations sur le sous-genre Stichoptera Motschulsky, 1860 (Coleoptera, Chrysomelidae). Nouvelle Revue d’Entomologie 29(1): 33–52. [Google Scholar]
- Bremer K, Friis EM, Bremer B. (2004) Molecular phylogenetic dating of flowering plants shows early Cretaceous diversification. Systematic Biology 53: 496–505. doi: 10.1080/10635150490445913 [DOI] [PubMed] [Google Scholar]
- Chapuis F. (1874) Famille des Phytophages. In: Lacordaire T, Chapuis F. Histoire naturelle des Insectes. Genera des Coléoptères ou exposé méthodique et critique de tous les genres proposés jusqu’ici dans cet ordre d’insectes. Tome dixième. Roret, Paris, 455 pp. [Google Scholar]
- Cobos A. (1953) Especies nuevas de Coleópteros de la provincia de Almeria. Archivos del Instituto de Aclimatación (Almería) 1: 128–134. [Google Scholar]
- Colgan DJ, McLauchlan A, Wilson GDF, Livingston SP, Edgecombe GD, Macaranas J, Cassis G, Gray MR. (1998) Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution. Australian Journal of Zoology 46: 419–437. doi: 10.1071/ZO98048 [Google Scholar]
- Crowson RA. (1981) The biology of the Coleoptera. Academic Press, London, 802 pp. [Google Scholar]
- Daccordi M. (1994) Notes for phylogenetic study of Chrysomelinae with descriptions of new taxa and a list of all the known genera (Coleoptera: Chrysomelidae: Chrysomelinae. In: Furth D. (Ed.) Proceedings of the Third International Symposium on the Chrysomelidae, Beijing, 1992. Backhuys Publishers, Leiden, 60–84.
- Daccordi M, Ruffo S. (2005) Considerazioni biogeografiche sulle Chrysolina delle province appenninica e sicula con descrizione di Chrysolina (Stichoptera) bourdonnei n. sp. (Coleoptera, Chrysomelidae). Studi trentini di Scienze naturali, Acta Biologica 81: 113–127. [Google Scholar]
- Dobler S, Mardulyn P, Pasteels JM, Rowell-Rahier M. (1996) Host-plant switches and the evolution of chemical defense and life history in the leaf beetle genus Oreina. Evolution 50(6): 2373–2386. doi: 10.2307/2410706 [DOI] [PubMed] [Google Scholar]
- Ehrlich PR, Raven PH. (1964) Butterflies and plants: a study in coevolution. Evolution 18: 586–608. doi: 10.2307/2406212 [Google Scholar]
- Fahrenholz H. (1913) Ectoparasiten und Abstammungslehre. Zoologischer Anzeiger 41: 371–374. [Google Scholar]
- Farris JS, Källersjö M, Kluge AG, Bult C. (1994) Testing significance of incongruence. Cladistics 10: 315–319. doi: 10.1111/j.1096-0031.1994.tb00181.x [Google Scholar]
- Feeny P. (1992) The evolution of chemical ecology: contributions from the study of herbivorous insects. In: Rosenthal GA, Berenbaum M. (Eds) Herbivores: their interactions with secondary plant metabolites – Evolutionary and Ecological Processes. Academic Press, San Diego, 1–44. doi: 10.1016/b978-0-08-092545-5.50006-7
- Futuyma DJ, Keese MC, Scheffer SJ. (1993) Genetic constraints and the phylogeny of insect-plant associations: responses of Ophraella communa (Coleoptera: Chrysomelidae) to host plants of its congeners. Evolution 47(3): 888–905. doi: 10.2307/2410192 [DOI] [PubMed] [Google Scholar]
- Futuyma DJ, Mitter C. (1996) Insect-plant interactions: the evolution of component communities. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 351(1345): 1361–1366. doi: 10.1098/rstb.1996.0119 [Google Scholar]
- Garin CF, Juan C, Petitpierre E. (1999) Mitochondrial DNA phylogeny and the evolution of host-plant use in Palearctic Chrysolina (Coleoptera, Chrysomelidae) leaf beetles. Journal of Molecular Evolution 48(4): 435–444. doi: 10.1007/PL00006488 [DOI] [PubMed] [Google Scholar]
- Ge S, Daccordi M, Li W, Yang XK. (2011) Seven new species of the genus Chrysolina Motschulsky from China (Coleoptera: Chrysomelidae: Chrysomelinae). Zootaxa 2736: 31–43. [Google Scholar]
- Gómez-Zurita J, Hunt T, Kopliku F, Vogler AP. (2007) Recalibrated tree of leaf beetles (Chrysomelidae) indicates independent diversification of angiosperms and their insect herbivores. PloS One 2: e360. doi: 10.1371/journal.pone.0000360 [DOI] [PMC free article] [PubMed]
- Gómez-Zurita J, Hunt T, Vogler AP. (2008) Multilocus ribosomal RNA phylogeny of the leaf beetles (Chrysomelidae). Cladistics 24: 34–50 doi: 10.1111/j.1096-0031.2007.00167.x [Google Scholar]
- Hsiao TH, Pasteels JM. (1999) Evolution of host-plant affiliation and chemical defense in Chrysolina-Oreina leaf beetles as revealed by mtDNA phylogenies. In: Cox ML. (Ed.) Advances in Chrysomelidae Biology 1. Backhuys, 321–342.
- Janz N. (2011) Ehrlich and Raven revisited: mechanisms underlying codiversification of plants and enemies. Annual Review of Ecology, Evolution, and Systematics 42(1): 71–89. doi: 10.1146/annurev-ecolsys-102710-145024 [Google Scholar]
- Janz N, Nyblom K, Nylin S. (2001) Evolutionary dynamics of host-plant specialization: a case study of the tribe Nymphalini. Evolution 55(4): 783–796. doi: 10.1554/0014-3820(2001)055[0783:EDOHPS]2.0.CO;2 [DOI] [PubMed]
- Janz N, Nylin S. (2008) The oscillation hypothesis of host-plant range and speciation. In: Tilmon KJ. (Ed.) Specialization, speciation, and radiation: the evolutionary biology of herbivorous insects. University of California Press, California, 203–215. doi: 10.1525/california/9780520251328.003.0015
- Janz N, Nylin S, Wahlberg N. (2006) Diversity begets diversity: host expansions and the diversification of plant-feeding insects. BMC Evolutionary Biology 6(1): . doi: 10.1186/1471-2148-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermy T. (1984) Evolution of insect/host plant relationships. American Naturalist 124: 609–630. doi: 10.1086/284302 [Google Scholar]
- Jolivet P, Petitpierre E. (1976) Les plantes hôtes connues des Chrysolina (Col. Chrysomelidae) – Essai sur les types de sélections trophiques. Annales de la Société entomologique de France 12(1): 123–149. [Google Scholar]
- Jolivet P, Petitpierre E, Daccordi M. (1986) Les plantes-hôtes des Chrysomelidae. Quelques nouvelles précisions et additions (Coleoptera). Nouvelle Revue Entomologique 3: 341–357. [Google Scholar]
- Jurado-Rivera JA, Vogler AP, Reid CAM, Petitpierre E, Gómez-Zurita J. (2009) DNA barcoding insect-host plant associations. Proceedings of the Royal Society B 276: 639–648. doi: 10.1098/rspb.2008.1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kergoat GJ, Delobel A, Fédière G, Le Rü B, Silvain JF. (2005) Both host-plant phylogeny and chemistry have shaped the African seed-beetle radiation. Molecular Phylogenetics and Evolution 35(3): 602–611. doi: 10.1016/j.ympev.2004.12.024 [DOI] [PubMed] [Google Scholar]
- Kippenberg H. (2010) Chrysomelinae. In: Löbl I, Smetana A. (Eds) Catalogue of Palaearctic Coleoptera. Vol. 6. Chrysomeloidea. Apollo Books, Stenstrup, 390–443.
- Koch K. (1992) Die Käfer Mitteleuropas. Ökologie 3 Goecke & Evers, Krefeld. [Google Scholar]
- Lanfear R, Calcott B, Ho SY, Guindon S. (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution 29(6): 1695–1701. doi: 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- Lopatin IK. (2011) New species of leaf-beetles (Coleoptera, Chrysomelidae) from China: X. Entomological review 91(7): 855–865. doi: 10.1134/S0013873811070074 [Google Scholar]
- Lopatin IK. (2014) New species of leaf-beetles (Coleoptera, Chrysomelidae) from China: XI. Entomological Review 94(3): 397–407. doi: 10.1134/S0013873814030105 [Google Scholar]
- Lopatin IK, Mikhailov YE. (2010) Contribution to the knowledge of the leaf-beetle fauna (Coleoptera, Chrysomelidae) of Eastern Kazakhstan. Entomological Review 90(7): 877–884. doi: 10.1134/S0013873810070067 [Google Scholar]
- Lopez-Vaamonde C, Wikström N, Labandeira C, Godfray HCJ, Goodman SJ, Cook JM. (2006) Fossil-calibrated molecular phylogenies reveal that leaf-mining moths radiated several million years after their host plants. Journal of Evolutionary Biology 19: 1314–1326. doi: 10.1111/j.1420-9101.2005.01070.x [DOI] [PubMed] [Google Scholar]
- McKenna DD, Sequeira AS, Marvaldi AE, Farrell BD. (2009) Temporal lags and overlap in the diversification of weevils and flowering plants. Proceedings of the National Academy of Sciences of the USA 106: 7083–7088. doi: 10.1073/pnas.0810618106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov YE. (2000) New and little known leaf beetles of the genus Chrysolina Motschulsky, 1860 from Altai and Sayany mountains in South Siberia (Coleoptera: Chrysomelidae). Genus 11(2): 129–146. [Google Scholar]
- Mikhailov YE. (2002) Consideration of the Subgenera Timarchoptera Motschulsky, 1860 and Paraheliostola L. Medvedev, 1992 of the Genus Chrysolina Motschulsky (Coleoptera, Chrysomelidae) after Description of Two New Forms from Khakassia. Evraziatskii Entomologicheskii Zhurnal 1(2): 219–228. [Google Scholar]
- Mikhailov YE. (2005) Contribution to the knowledge of Chrysolina (Crositops) pedestris (Gebler, 1823) (Coleoptera, Chrysomelidae): preimaginal stages and ecology. Contributions to Systematics and Biology of Beetles – Papers Celebrating the 80th Birthday of Igor Konstantinovich Lopatin, 143–151.
- Mikhailov YE. (2006) On the types and status of Chrysolina sylvatica (Gebler, 1823) and Chrysolina subcostata (Gebler, 1848) from the subgenus Pleurosticha Motschulsky, 1860 (Coleoptera, Chrysomelidae). Eurasian Entomological Journal 5(4): 292–302. [Google Scholar]
- Monrós F, Bechyné J. (1956) Über einige verkannte Chrysomeliden-Namen. Entomologische Arbeiten aus dem Museum Gg. Frey 7(3): 1118–1137. [Google Scholar]
- Nosil P. (2002) Transition rates between specialization and generalization in phytophagous insects. Evolution 56(8): 1701–1706. doi: 10.1111/j.0014-3820.2002.tb01482.x [DOI] [PubMed] [Google Scholar]
- Nyman T. (2010) To speciate, or not to speciate? Resource heterogeneity, the subjectivity of similarity, and the macroevolutionary consequences of niche‐width shifts in plant-feeding insects. Biological Reviews 85(2): 393–411. doi: 10.1111/j.1469-185X.2009.00109.x [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A. (2013) BayesTraits v. 2.0. University of Reading, Reading.
- Pagel M, Meade A, Barker D. (2004) Bayesian estimation of ancestral character states on phylogenies. Systematic Biology 53(5): 673–684. doi: 10.1080/10635150490522232 [DOI] [PubMed] [Google Scholar]
- Pasteels JM, Termonia A, Daloze D, Windsor DM. (2003) Distribution of toxins in chrysomeline leaf beetles: possible taxonomic inferences. In: Furth DG. (Ed.) Special Topics in Leaf Beetle Biology. Proceedings of the Fifth International Symposium on the Chrysomelidae, Iguassu Falls (Brazil), August 2000. Pensoft Publishers, Sofia-Moscow, 261–275. [Google Scholar]
- Petitpierre E. (1975) Notes on chromosomes of ten species of the genus Chrysolina Mots. (Coleoptera: Chrysomelidae). Genetica 45(3): 349–354. doi: 10.1007/BF01508309 [Google Scholar]
- Petitpierre E. (1981) New data on the cytology of Chrysolina Mots. and Oreina Chevr. (Coleoptera, Chrysomelidae). Genetica 54: 265–272 doi: 10.1007/BF00135045 [Google Scholar]
- Petitpierre E. (1983) Karyometric differences among nine species of the genus Chrysolina Mots. (Coleoptera, Chrysomelidae). Canadian Journal of Genetics and Cytology 25: 33–39. doi: 10.1139/g83-006 [Google Scholar]
- Petitpierre E. (1999) The cytogenetics and cytotaxonomy of Chrysolina Mots. and Oreina Chevr. (Coleoptera, Chrysomelidae). Hereditas 131: 55–62. doi: 10.1111/j.1601-5223.1999.00055.x [Google Scholar]
- Petitpierre E, Kippenberg H, Mikhailov Yu, Bourdonné JC. (2004) Karyology and cytotaxonomy of the genus Chrysolina Motschulsky (Coleoptera, Chrysomelidae). Zoologischer Anzeiger 242: 347–352. doi: 10.1078/0044-5231-00108 [Google Scholar]
- Petitpierre E, Mikhailov Y. (2009) Chromosomal evolution and trophic affiliation in the genus Chrysolina (Coleoptera: Chrysomelidae). In: Jolivet P, Santiago-Blay J, Schmitt M. (Eds) Research on Chrysomelidae 2. Brill, Leiden-Boston, 225–234. doi: 10.1163/ej.9789004169470.1-299.91
- Petitpierre E, Segarra C. (1985) Chromosomal variability and evolution of Chrysomelidae (Coleoptera) particularly that of Chrysomelinae and Palearctic Alticinae. Entomography 3: 403–426. [Google Scholar]
- Rambaut A, Drummond AJ. (2007) Molecular evolution, phylogenetics and epidemiology, Tracer v.1.5. http://tree.bio.ed.ac.uk/software/tracer/
- Rizza A, Pecora P. (1980) Biology and host specificity of Chrysomela rossia, a candidate for the biological control of dalmatian toadflax, Linaria dalmatica. Annals of the Entomological Society of America 73(1): 95–99. doi: 10.1093/aesa/73.1.95 [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. doi: 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. (2000) Another calculation of the p-value for the problem of regions using the scaled bootstrap resamplings. Technical report No. 2000–35, Department of Statistics, Stanford University. [Google Scholar]
- Shimodaira H, Hasegawa M. (2001) CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17: 1246–1247. doi: 10.1093/bioinformatics/17.12.1246 [DOI] [PubMed] [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Annals of the entomological Society of America 87(6): 651–701. doi: 10.1093/aesa/87.6.651 [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 1312–1313. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. (2003) PAUP*. Phylogenetic Analysis Using Parsimony (and other Methods), Version 4.0. Sinauer Associates, Sunderland, Massachusetts. [Google Scholar]
- Vela JM, Bastazo G. (1999) Ecological and biogeographic aspects of the Andalusian leaf beetle endemisms. In: Cox ML. (Ed.) Advances in Chrysomelidae Biology 1, Backhuys Publishers, Leiden, 137–158. [Google Scholar]
- Weise J. (1893) Chrysomelidae. Lieferung 6 Naturgeschichte der Insecten Deutschland – Erste Abtheilung Coleoptera – Sechster Band [1893]. Nicolaische Verlags-Buchhandlung R. Stricker, Berlin, 961–1161. [Google Scholar]
- Zwickl D. (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis, University of Texas, Austin. [Google Scholar]