Abstract
Background & Aims
Hepatic stellate cells (HSCs) play a key role in liver fibrosis in various chronic liver disorders including nonalcoholic fatty liver disease (NAFLD). The development of liver fibrosis requires a phenotypic switch from quiescent to activated HSCs. The trigger for HSC activation in NAFLD remain poorly understood. We investigated the role and molecular mechanism of extracellular vesicles (EVs) released by hepatocytes during lipotoxicity in modulation of HSC phenotype.
Methods
EVs were isolated from fat-laden hepatocytes by differential centrifugation and incubated with HSCs. EV internalization and HSC activation, migration, and proliferation were assessed. Loss- and gain-of-function studies were performed to explore the potential role of peroxisome proliferator-activated receptor-γ (PPAR-γ)-targeting microRNAs (miRNAs) carried by EVs into HSC.
Results
Hepatocyte-derived EVs released during lipotoxicity are efficiently internalized by HSCs resulting in their activation, as shown by marked up-regulation of profibrogenic genes (collagen-I, α-smooth muscle actin, and tissue inhibitor of metalloproteinases-2), proliferation, chemotaxis, and wound-healing responses. These changes were associated with miRNAs shuttled by EVs and suppression of PPAR-γ expression in HSCs. The hepatocyte-derived EV miRNA content included various miRNAs that are known inhibitors of PPAR-γ expression, with miR-128-3p being the most efficiently transferred. Furthermore, loss- and gain-of-function studies identified miR-128-3p as a central modulator of the effects of EVs on PPAR-γ inhibition and HSC activation.
Conclusions
Our findings demonstrate a link between fat-laden hepatocyte-derived EVs and liver fibrosis and have potential implications for the development of novel antifibrotic targets for NAFLD and other fibrotic diseases.
Keywords: Extracellular Vesicles, Hepatic Stellate Cell, Lipotoxicity, Liver Fibrosis, miRNAs
Abbreviations used in this paper: B2M, β2-microglobulin; CDAA, choline-deficient l-amino acid; CSAA, choline supplemented l-amino acid; CTGF, connective tissue growth factor; fsDMEM, Dulbecco’s modified Eagle’s medium; EV, extracellular vesicle; FBS, fetal bovine serum; FFAs, free fatty acids; GFAP, glial fibrillary acidic protein; Hep-EV, HepG2-derived extracellular vesicles; HSC, hepatic stellate cell; LX2, human immortalized hepatic stellate cells; mHSC, mouse primary hepatic stellate cell; miRNA, microRNA; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; PCR, polymerase chain reaction; PPAR-γ, peroxisome proliferator-activated receptor-γ; PMH, primary mouse hepatocytes; PMH-EV, primary mouse-derived extracellular vesicles; qRT-PCR, quantitative real-time polymerase chain reaction; scRNA, scramble RNA; siRNA, silencing RNA; α-SMA, α-smooth muscle actin; TGF-β, transforming growth factor-β; TIMP-2, tissue inhibitor of metalloproteinases-2; VNN1, Vanin-1; WT, wild type
Summary.
Extracellular vesicles released by hepatocytes during lipotoxicity carry and shuttle specific microRNA-targeting peroxisome proliferator-activated receptor-γ into hepatic stellate cells and induce a phenotypical switch from quiescent to activated cells.
Nonalcoholic fatty liver disease (NAFLD) has emerged as a serious public health problem in the United States and many other countries.1, 2 It affects both adults and children and may progress to cirrhosis and end-stage liver disease.3, 4, 5, 6 An increased fat deposit in the liver is an early event and a prerequisite for the development of NAFLD.7, 8 Considerable evidence supports the concept that certain lipids, such as saturated free fatty acids (FFAs), contribute to disease progression through toxic effects on hepatocytes. Indeed, lipotoxicity may result in hepatocyte damage, triggering an inflammatory reaction and abnormal wound-healing response that results in the development of nonalcoholic steatohepatitis (NASH) and fibrosis.6, 9, 10 Patients with fibrotic NASH are at significant risk for disease progression to cirrhosis with significant increase in liver-related morbidity and mortality.8, 11, 12
Hepatic stellate cells (HSCs) play a crucial role during liver fibrosis in various chronic liver disorders, including NAFLD.13 During liver fibrogenesis, HSCs undergo a phenotypical switch from quiescent vitamin A–storing cells to activated and proliferative myofibroblast-like cells in a process called activation, which involves up-regulation of various genes, including α-smooth muscle actin (α-SMA), collagen-1α1, tissue inhibitor of metalloproteinases (TIMP-1 and 2), and transforming growth factor-β (TGF-β).14 Activated HSCs play a crucial role in sustaining and promoting extracellular matrix deposition, and they acquire the ability to proliferate and migrate toward the area of injury.13, 15 Currently, the molecular mechanisms linking fat-laden hepatocytes to HSC activation remain incompletely understood.
We have recently demonstrated that extracellular vesicles (EVs) membrane-bound vesicles released during cell stress or death that are key cell-to-cell communicators are released both in vitro in cultured hepatocytes exposed to saturated free fatty acids and in vivo in animal models of NASH.16, 17 In these studies, the levels of circulating EVs strongly correlated with the severity of liver fibrosis, and treatment with a neutralizing antibody for Vanin-1 (VNN1) a surface protein identified in proteomic analysis of EVs played a crucial role in modulating the internalization of EVs into target cells. Based on this evidence, we tested the hypothesis that EVs released by hepatocytes during lipotoxicity carry and transfer microRNAs (miRNA, miR) that regulate fibrogenesis by inducing a phenotypical switch from quiescent to activated HSCs.
Materials and Methods
Animal Studies
Male C57BL/6 wild-type (WT) mice, 20 to 25 g of body weight, 7 weeks old, were placed on a choline-deficient l-amino acid (CDAA) diet or control diet of choline supplemented l-amino acid (CSAA) (n = 12) (Dyets, Bethlehem, PA) for 20 weeks to induce NASH.18 In addition, six mice were placed on a high-fat diet (45% kcal from fat, 18.8 kJ/g) (Research Diets, New Brunswick, NJ) or normal chow for 12 weeks as an alternative model of diet-induced NAFLD.19 The mice were sacrificed, and the liver and blood were collected under anesthesia achieved by injecting intraperitoneally, with a 21G needle, a mixture of 100 mg/kg of ketamine and 10 mg/kg of xylazine dissolved in a 0.9% saline solution.20
The studies were approved by the University of California San Diego Institutional Animal Care and Use Committee and followed the National Institutes of Health guidelines outlined in “Guide for the Care and Use of Laboratory Animals.” Collection of liver specimens, H&E staining, and NAFLD activity score analysis were performed as previously described elsewhere.17
Cell Culture
Human hepatoma cell line (HepG2) was maintained in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Grand Island, NY), supplemented with 10% fetal bovine serum (FBS; CellGro, Manassas, VA), 5000 U/mL penicillin, and 5000 μg/mL streptomycin sulfate in 0.85% NaCl and 5% sodium pyruvate (Sigma-Aldrich, St. Louis, MO). Long-chain FFAs, palmitic acid (Sigma-Aldrich) was dissolved in 95% ethanol (stock solution 100 mM) and stored at −20° C before the experiments. Human immortalized hepatic stellate cells (LX2) were kindly provided by Prof. Scott Friedman; they were cultured in DMEM, supplemented with 1% FBS (CellGro) and 5000 U/mL penicillin and 5000 μg/mL streptomycin sulfate in 0.85% NaCl.
Hepatocyte Isolation
Primary murine hepatocytes were isolated by using a two-step method previously described elsewhere.21 Briefly, WT C57/B6 mice were deeply anesthetized, the abdominal cavity was opened and livers were perfused in situ with EGTA for 5 minutes and collagenase D (Roche Diagnostics, Indianapolis, IN) for 10 minutes at a flow rate of 10 mL/min. After perfusion, the partially digested liver was excised, and the digest was passed through a 70-μm nylon mesh to remove undigested materials. Purified hepatocytes were seeded on collagen-coated culture dishes and cultured in William’s E media (Life Technologies) supplemented with 10% FBS and 5000 U/mL penicillin and 5000 μg/mL streptomycin sulfate in 0.85% NaCl. Hepatocyte vitality was determined by Tripan blue staining and counting on a hemacytometer. Experiments were performed the day after the isolation.
Hepatic Stellate Cell Isolation
Primary mouse hepatic stellate cells (mHSCs) were isolated from WT C57/B6 mice by previously published protocols.21, 22 Briefly, livers were perfused in situ with EGTA for 5 minutes, with pronase E (0.4 mg/mL, Roche Diagnostics) for 5 minutes, and collagenase D (0.5 mg/mL; Roche Diagnostics) for 8 minutes at a flow rate of 5 mL/min. After perfusion, the liver was excised from the body and fully digested in collagenase D, pronase E, and DNAse I (2 mg/mL, Roche Diagnostics) for 15 minutes. The digested livers were filtered through a cell strainer and washed with Gey’s balanced salt solution. HSCs were purified from other parenchymal and nonparenchymal liver cells by flotation on 6.4% (w/v) Nicodenz (Axis-Shield PoC AS, Oslo, Norway)/Gey’s balanced salt solution (without NaCl).
The purity of the HSCs was assessed by detecting vitamin A autofluorescence. The cells were counted, and approximately 0.2 × 106 cells were seeded onto a 12-well plate in high-glucose DMEM media containing 10% FBS, 1% penicillin/streptomycin, and 1% HEPES (Sigma-Aldrich) for up to 7 days. Cells were imaged every day with an Olympus contrast phase microscope (Olympus America, Center Valley, PA), and they were treated with primary hepatocyte-derived microparticles on day 2 from isolation and for 48 hours before performing the experiments.
Extracellular Vesicle Isolation
HepG2 and primary mouse hepatocytes (PMH) were seeded onto a 100-mm dish or six-well plate, respectively, and cultured until reaching 80% to 85% confluence. Cells were incubated with 0.25 mM palmitic acid in serum-free DMEM supplemented with 1.1% penicillin and streptomycin, and 1% endotoxin-free bovine serum albumin for up to 24 hours. Extracellular vesicles were isolated by differential centrifugation, as previously described elsewhere.16
Briefly, collected medium was centrifuged twice at 1700g for 15 minutes to remove cell debris and aggregates. The supernatant was then transferred to new tubes and ultracentrifuged at 100,000g for 90 minutes at 10°C.23 The supernatant was collected in new tubes and used as the EV-free control, and the pelleted EVs (13–15 × 103/mL medium/100-mm petri dish) were resuspended in 500 μL of serum-free DMEM for subsequent in vitro studies.
To trace the hepatocyte-derived EVs (Hep-EV), PKH26 dye (Sigma-Aldrich) was used according to the manufacturer’s instructions. A complete characterization of Hep-EVs, including size, composition, and distribution, was performed by dynamic light scattering, transmission electron microscope, liquid chromatography with tandem mass spectrometry, and fluorescence-activated cell sorting as reported previously elsewhere.16 In selected studies, Hep-EV were incubated with 10 μg/mL of RNase (Roche Diagnostics) for 30 minutes at 37°C to remove any RNA adhering to the external leaflet.
Cell Transfection
Transfection of miR-128-3p MISSION microRNA MIMIC (Sigma-Aldrich) and mirVana miR-128-3p inhibitor in LX2 was obtained by using Lipofectamin RNAiMAX transfection reagent (Life Technologies). Briefly, LX2 cells were plated at a density of 0.5–4 × 105 cell/well in a 12-well or 24-well plate and transfected with 75 nM of miR-128-MIMIC or 30 nM of AntagomiR-128-3p. Transfected cells were incubated at 37°C with serum-free and antibiotic-free Opti-MEM for 6 hours, then the medium was replaced with standard culture medium supplemented with 1% FBS. After 24 hours the cells were treated with hepatocyte-derived EVs or controls for an additional 48 hours and then harvested or used for subsequent experiments. Anti-miR Negative Control (Life Technologies) and miRIDIAN microRNA Mimic Negative Control (Sigma-Aldrich) were used as the negative controls.
For HepG2 transfection with Dicer1 and Drosha silencing RNA (siRNA) mixture, we used Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. Briefly, HepG2 (3 × 105 cell/well in a six-well plate) were transfected with 40 nM of Silencer Select Dicer1 and Drosha siRNA (IDs: s23756 and s26492; Life Technologies) in Opti-MEM and incubated at 37°C for up to 6 hours. After 6 hours Opti-MEM was replaced with regular medium with 10% FBS. After 48 hours from transfection, the cells were treated with or without 0.25 μM of palmitic acid (Sigma-Aldrich) or vehicle (1% endotoxin-free, low-FFA bovine serum albumin) for 24 hours. After the incubation with palmitic acid, the EVs were isolated, and the cells were harvested for protein and RNA isolation.
Silencing efficacy was assessed by quantitative polymerase chain reaction (qPCR) and Western blot analyses. Transfection of mirVana miR-128-3p inhibitor in HepG2 was obtained using Lipofectamine RNAiMAX transfection reagent (Life Technologies) according to the manufacturer’s instructions. Briefly, HepG2 were seeded at a density of 0.5–1 × 106 in a six-well plate and transfected with 30 nM of AntagomiR-128-3p. Transfected cells were incubated at 37°C with serum-free and antibiotic-free Opti-MEM for 6 hours and then the medium was replaced with serum-free DMEM with 0.25 mM of palmitic acid and 1% low-endotoxin FFA-free bovine serum albumin for 24 hours. After 24 hours of incubation, the EVs were isolated. Isolation of RNA and miRNAs was performed as described herein.
Migration Assays
In vitro migration studies have been performed by using LX2 cells and primary mouse HSCs. The wound-healing assay was performed to analyze HSC nonoriented migration (chemokinesis) by using Radius 24-well Cell Migration Assay (Cell Biolabs, San Diego, CA) according to the manufacturer’ instructions. The HSC were seeded onto the radius of a 24-well plate and treated with Hep-EV or controls for 24 hours. Analysis of the number of migrated cells onto the radius was performed and quantification reported in histogram. Mitomycin (1 μg/mL) was used to inhibit cell proliferation during the wound-healing assay. Boyden’s chamber assay was performed to analyze the HSC-oriented migration (chemotaxis) by using cell culture inserts of 8-μm pore size (Millipore, Billerica, MA) and a 24-well plate. Each well was filled with 500 μL of serum-free DMEM with Hep-EVs or controls.
For some selected studies, HSCs were incubated with Hep-EVs with anti-Vanin-1 neutralizing antibody, miR-128-3p mimic, anti-miR-128-3p, or Hep-EVs isolated from HepG2 transfected with Dicer/Drosha siRNA. The inserts were placed on top of each well, and 150 μL of cell suspension (5 × 104 cells) was added. The plates were incubated overnight at 37°C, and then the filters were removed and stained with Vectashield mounting medium with 4′,6-diamino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA). Migrated cells were detected with a fluorescence microscope, and the number per field of their nuclei was counted.
Proliferation Assay
The HSC proliferation assay was performed by detecting DNA synthesis via incorporation of 5-bromo-2-deoxyuridine (BrdU) using a CyQuant Direct Cell Proliferation Assay (Life Technologies) according to the manufacturer’ instructions. The HSCs were plated onto a 96-well plate (0.15 × 105 cell/well) and treated with controls or testing compounds for 48 hours. A bottom-read plate reader was used to detect fluorescein isothiocyanate-positive signals. The relative fluorescence was reported in the quantification graphs as the fold-change to the control.
Western Blot Analysis
The HSCs were digested in 400 μL of radioimmunoprecipitation assay buffer containing Phosphatase and Protease Inhibitor Cocktail (Roche Diagnostics). After cellular lysis, 30–50 μg of protein was solubilized in Laemmli buffer, resolved by a 4%–20% Criterion Tris-HCl gel electrophoresis system (Bio-Rad Laboratories, Hercules, CA) and transferred to a 0.2-μm nitrocellulose membrane (Bio-Rad Laboratories). Primary rabbit polyclonal or mouse monoclonal antibody anti-human TGF-β (Cell Signaling Technology, Beverly, MA), α-SMA, peroxisome proliferator-activated receptor-γ (PPAR-γ), Dicer1, tubulin, connective tissue growth factor (CTGF), glial fibrillary acidic protein (GFAP) (1:1000; Genetex, Irvine, CA), and actin (1:5000; Genetex) were incubated overnight at 4°C. Primary antibodies were detected by using appropriate horseradish peroxidase–secondary antibodies (Cell Signaling Technology). Proteins were visualized by Supersignal West Pico chemiluminescence reagents (Pierce Biotechnology, Rockford, IL). Western blot data were obtained from three independent experiments, and a representative gel is shown in the figures.
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction
Total RNA was isolated using RNeasy kit (Qiagen, Valencia, CA) and reverse transcribed by iScript cDNA synthesis kit (Bio-Rad Laboratories) according to the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed on a BioRad Cycler (Bio-Rad Laboratories) by use of SYBRGreen real-time PCR master mix (Kapabiosystem, Woburn, MA) according to the manufacturer’s instructions. The housekeeping gene β2-microglobulin (B2M) was used as the internal control. The PCR primers used to amplify each gene are listed in Supplementary Table 1.
MiRNAs were isolated from hepatocytes, Hep-EVs, HSCs, or murine liver tissue by miRNeasy Mini kit (Qiagen), and miRNA expression was analyzed by using the TaqMan microRNA Reverse Transcription kit and TaqMan Universal PCR kit (Life Technologies) on 7300 Real-time PCR system (Life Technologies). Identification of miRNAs targeting PPAR-γ was assessed by combining computational data from three prediction algorithms: miRanda, TargetScan, and mirWalk. We selected three of the most conserved miRNAs: miR-130b, miR-128-3p, and miR-27b. Specific primers for selected miRNAs (Life Technologies) were used in separate reactions, and the fold-change expression with respect to control was calculated for all samples. The U6 small-nuclear RNA was used as a control (Life Technologies).
Incorporation of EVs in Hepatic Stellate Cells and Transfer of RNA
Incorporation of Hep-EVs into HSCs was evaluated by confocal microscope after the incubation of 0.15 × 105 HSC/well with PKH26-positive EVs for up to 6 hours in a four-well tissue culture slide. In selected studies, Hep-EVs were incubated overnight at 4°C with Vanin-1 (Genetex) neutralizing antibody (4 μg/mL) to inhibit the internalization of EVs into HSCs. To analyze the RNA transfer into HSCs by Hep-EVs, we incubated Hep-EVs with RNA fluorescein isothiocyanate-positive stain SYTO RNASelect (Life Technologies) and then with PKH26 (Sigma-Aldrich) to label the EV membrane, according to the manufacturer’s instructions. HSCs were incubated with PKH26/SYTO-positive EVs for up to 6 hours, and the fluorescence intensity was evaluated by Olympus FV1000 Spectral Confocal microscope. The miRNA transfer to HSCs was also evaluated by qRT-PCR at different time points (3 hours, 6 hours, 16 hours, and 24 hours). We used 4′,6-diamidino-2-phenylindole stain (DAPI) to label cell nuclei. A 40× magnification was used for the microphotographs.
Statistical Analysis
All data are expressed as mean ± standard deviation (SD) unless otherwise indicated. Differences between three or more groups were compared by a nonparametric Kruskal-Wallis analysis of variance (ANOVA) test. If a statistically significant effect was detected, post hoc pairwise comparisons were performed using Mann-Whitney tests with Bonferroni correction. Differences between two groups were compared with a two-sided Student t test if the data were normal or a Mann-Whitney test if the data deviated from the normal distribution. P < .05 was considered statistically significant. All statistical analyses were performed using GraphPad Prism 4.0c (La Jolla, CA) or R v3.0.2 (www.r-project.org).
Results
Hepatocyte-Derived Extracellular Vesicles Released During Lipotoxicity Are Internalized Into Hepatic Stellate Cells and Induce Their Activation
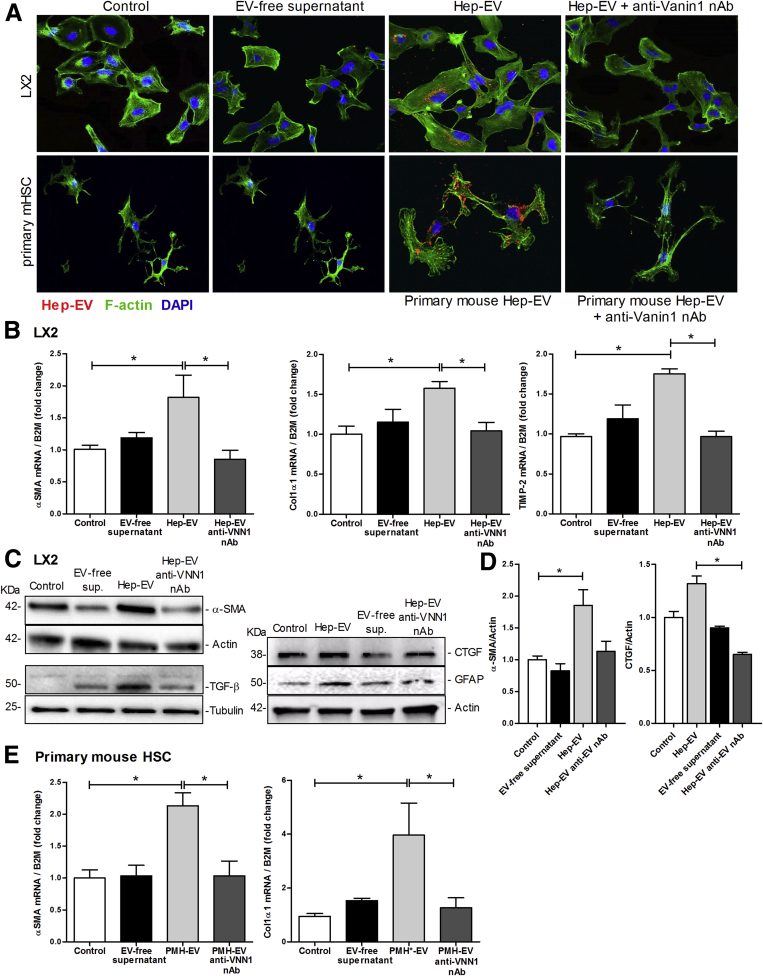
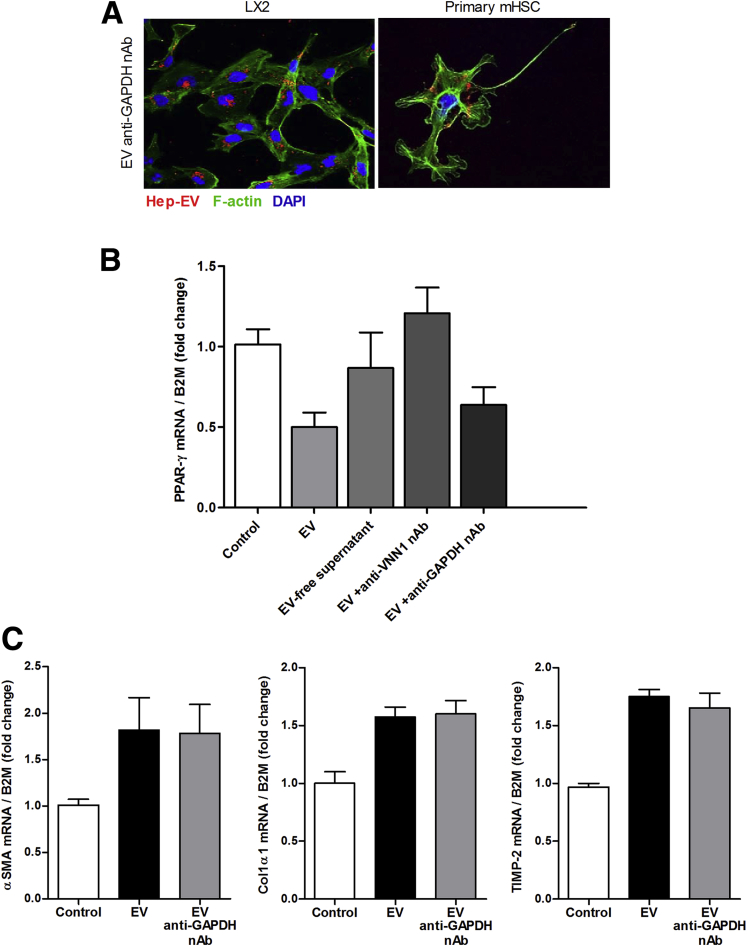
We have recently demonstrated that while cultured hepatocytes released a small number of EVs into the supernatant, treatment of these cells with lipotoxic fatty acids results in a several-fold increase in the number of EVs released.16 The EVs released by lipotoxic hepatocytes (Hep-EV) are enriched in VNN1 protein.16 To study the effects of Hep-EVs on HSCs, we initially assessed whether Hep-EVs released by HepG2 (hepatoma cells) or PMH exposed to palmitic acid for 24 hours were internalized into HSCs. To address this, HSCs were exposed to Hep-EVs labeled with PKH26 (a lipophilic dye binding the cell membrane) for 1, 3, or 6 hours. We observed that Hep-EVs (HepG2-EVs and primary mouse hepatocyte-EVs) were internalized into both human immortalized HSCs (LX2) and primary mouse HSCs (primary mHSC) particularly after 6 hours of incubation (Figure 1A). To assess whether the internalization was dependent on Vanin-1 (VNN1), as previously demonstrated,16 we preincubated Hep-EVs with 4 μg/mL of VNN1 neutralizing antibody (VNN1 nAb). We observed that the Hep-EV uptake was dramatically reduced (Figure 1A) by blocking VNN1 on the EV external leaflet. Treatment of Hep-EVs with a blocking antibody against GAPDH had no effect on Hep-EV internalization into both human and mouse HSCs, suggesting a key role of VNN1 in Hep-EV internalization into HSCs (Supplementary Figure 1).
Figure 1.
Hepatocyte-derived extracellular vesicles (EVs) are internalized into hepatic stellate cells (HSC) and induce up-regulation of profibrogenic markers. (A) Internalization of HepG2-derived extracellular vesicles (HepG2-EVs) and primary mouse hepatocyte EVs (PMH-EVs) labeled with PKH26 into immortalized human hepatic stellate cells (LX2) and primary mouse hepatic stellate cells (mHSC) assessed by indirect immunofluorescence after 6 hours of incubation with EVs and imaged by confocal microscope using 40× magnification. A Vanin-1 neutralizing antibody (VNN1 nAb) was used to block EV internalization. (B) Quantitative polymerase chain reaction analyses for profibrogenic genes α-smooth muscle actin (α-SMA), collagen type 1, and tissue inhibitor of metalloproteinases-2 (TIMP-2) in LX2 exposed to Hep-EVs for 16 hours with or without a Vanin-1 neutralizing antibody. (C) Western blot analysis of markers of HSC activation α-smooth muscle actin (α-SMA), TGF-β (transforming growth factor-β), (connective tissue growth factor) CTGF, and (glial fibrillary acidic protein) GFAP in LX2 exposed to Hep-EVs for 24 hours with or without VNN1 nAb. (D) Quantification of α-SMA and CTGF protein levels normalized to actin. (E) Quantitative qRT-PCR analyses for profibrogenic genes α-SMA and collagen type 1 in primary mouse hepatic stellate cells (mHSC) to PMH-EVs for 16 hours with or without VNN1 nAb. β2-Microglobulin (B2M) was used as housekeeping gene for qRT-PCR, and actin and tubulin were used as loading controls for Western blotting. Values represent mean ± SD from three independent experiments. *P < .05, **P < .005, ***P < .0005, Kruskal-Wallis test with post–hoc Mann-Whitney test and Bonferroni correction.
Supplementary Figure 1.
A control neutralizing antibody against GAPDH does not influence extracellular vesicle (EV) internalization or EV-induced hepatic stellate cell (HSC) activation. (A) Representative microphotographs of internalization of hepatocyte-derived, PKH26-positive extracellular vesicles (EV, red) into LX2 and primary mouse HSCs (F-actin fibers, green and nuclei, DAPI) after 6 hours of incubation with EVs incubated with GAPDH neutralizing antibody (GAPDH nAb). (B) Quantitative PCR analysis of PPAR-γ in human immortalized HSCs (LX2) treated with HepG2-derived EVs (Hep-MP) and EVs incubated with GAPDH nAb for 16 hours. (C) Quantitative PCR analyses for profibrogenic genes α-SMA, Collagen type 1 and TIMP-2 in LX2 treated with Hep-EVs and EVs incubated with glyceraldehyde-3-phosphate dehydrogenase neutralizing antibody (GAPDH nAb) for 16 hours. β2-Microglobulin was used as the control. Values represent mean ± standard deviation from three independent experiments. *P < .05, **P < .005, ***P < .0005, Kruskal-Wallis test with post hoc Mann-Whitney test and Bonferroni correction.
To analyze the effect of internalized Hep-EVs on the modulation of HSC phenotype, we exposed human (LX2) and mouse primary HSCs to Hep-EVs for up to 16 hours. Subsequently, we analyzed mRNA expression for various markers of HSC activation, including α-smooth muscle actin (α-SMA), collagen-1α1, and tissue inhibitor growth factor-2 (TIMP-2) by qRT-PCR. LX2 cells incubated with Hep-EVs showed a significant up-regulation of α-SMA, collagen-1α1, and TIMP-2 compared with untreated LX2 (control) and LX2 treated with EV-free supernatant (Figure 1B). These results matched with increased protein levels of α-SMA, TGF-β, CTGF, and GFAP, markers of HSC activation (Figure 1C and D).13
To confirm these findings in primary cells, we exposed primary mouse HSCs (primary mHSC) to primary hepatocyte-derived EVs (PMH-EV) released during lipotoxicity. Similar to the findings observed for LX2, α-SMA and collagen-1α1 were both markedly up-regulated in primary mHSC by the exposure to EVs compared with controls (Figure 1E). Notably, internalization of Hep-EVs and up-regulation of HSC activation were dramatically reduced by blocking Vanin-1 with VNN1 nAb (Figure 1A–E).
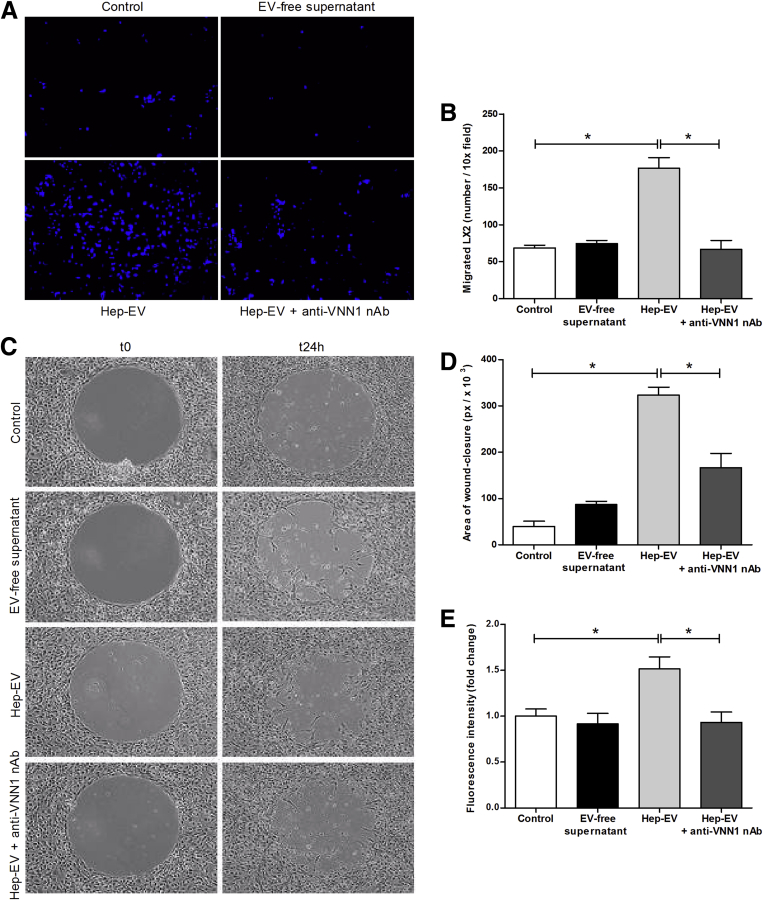
Extracellular Vesicles Released by Lipotoxic Hepatocytes Induce Hepatic Stellate Cell Migration and Proliferation
During hepatic fibrosis, the activation, migration, and proliferation of HSCs occur rapidly in response to various stimuli present in the extracellular environment and released during liver injury. To investigate Hep-EVs released during lipotoxicity-induced HSC profibrogenic responses, we exposed LX2 to Hep-EVs and tested oriented migration (chemotaxis), nonoriented migration (chemokinesis), and proliferation. The exposure of LX2 to Hep-EVs resulted in a marked stimulation of cell chemotaxis (Figure 2A and B), chemokinesis (Figure 2C and D), and DNA synthesis as an index of proliferation (Figure 2E) compared with untreated cells or cells treated with EV-free supernatant. To further analyze whether blocking Hep-EV internalization could reduce HSC activation, we neutralized Vanin-1 on the Hep-EVs surface. Notably, we observed a significant reduction of LX2 migration (chemotaxis and chemokinesis) and proliferation (Figure 2A–E). Taken together, these findings demonstrate that EVs released by hepatocytes exposed to lipotoxic palmitic acid are efficiently internalized into HSCs in a process that, at least in part, depends on the expression of VNN1 on the EV surface and that they induce HSC activation.
Figure 2.
Hepatocyte-derived extracellular vesicles (EVs) induce migration, wound-healing response, and proliferation of hepatic stellate cells. (A) Representative microphotographs (10× magnification) and (B) corresponding quantification histogram of Boyden’s chamber assay of human immortalized hepatic stellate cells (LX2) exposed to HepG2-derived EVs (Hep-EV) for 16 hours. (C) Representative microphotographs and (D) corresponding quantification graph of wound-healing assay of LX2 treated with Hep-EVs for 24 hours. (E) Quantification graph of proliferation assay of LX2 treated with Hep-EVs for 48 hours. A Vanin-1 neutralizing antibody was used to block the internalization of Hep-EVs. Values represent mean ± standard deviation from three independent experiments. *P < .05, **P < .005, ***P < .0005, Kruskal-Wallis test with post hoc Mann-Whitney test and Bonferroni correction.
Extracellular Vesicles Mediate the Profibrogenic Effects of Hepatic Stellate Cells by Transferring Specific Peroxisome Proliferator-Activated Receptor-γ-Targeting MicroRNAs
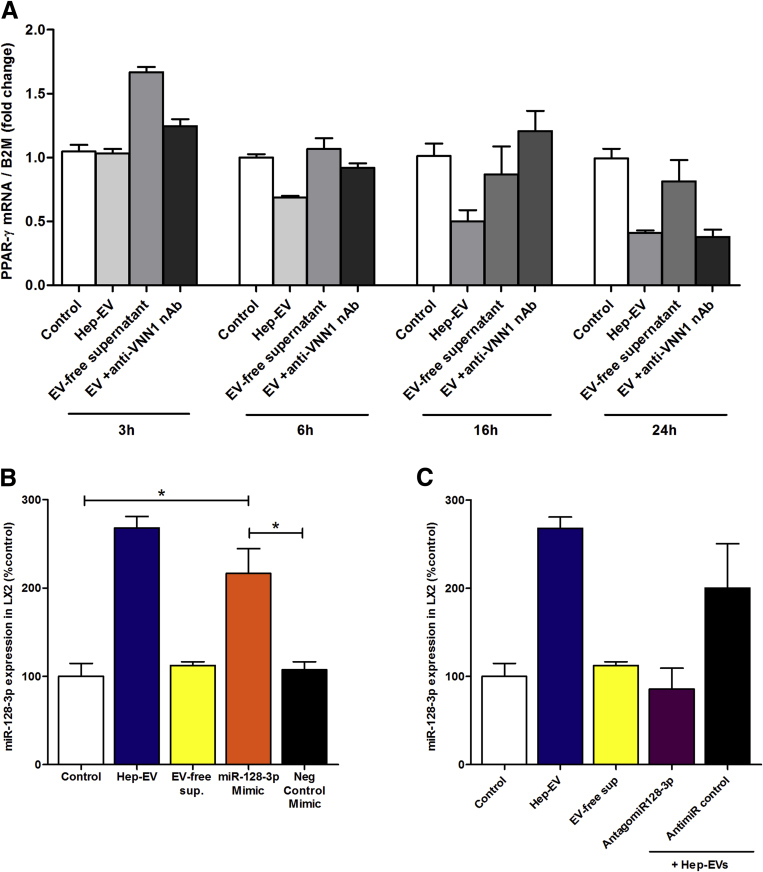
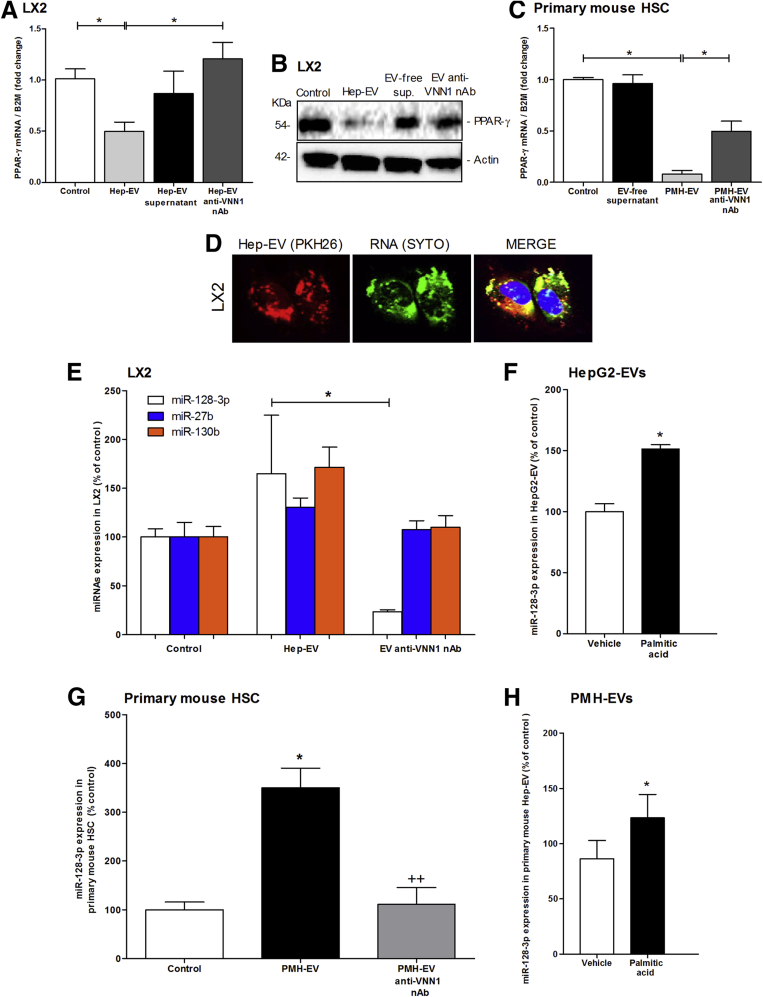
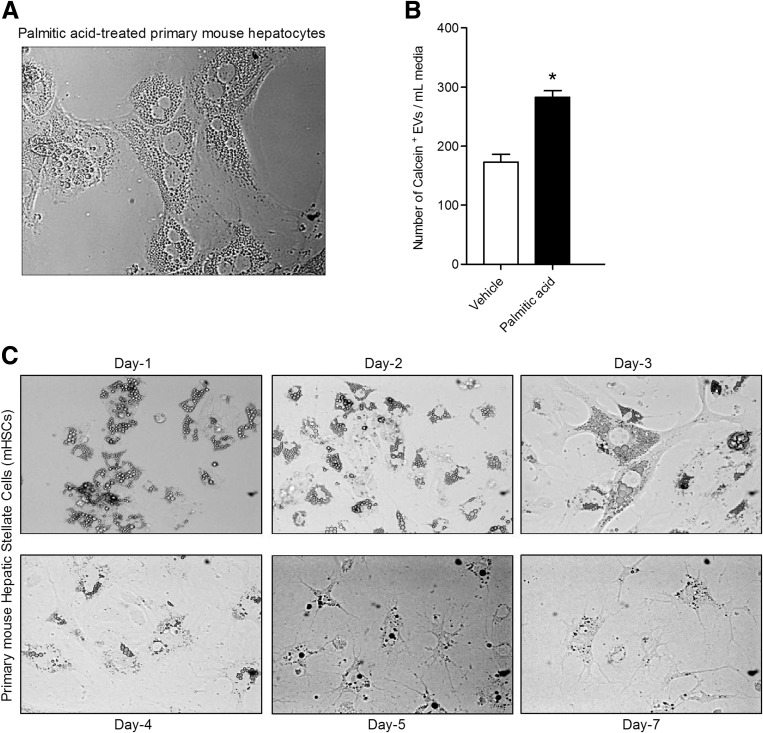
Several studies support a central role of PPAR-γ as key mediator in maintaining a quiescent HSC phenotype in normal liver.24, 25, 26 Notably, it has been shown that PPAR-γ progressively decreases during primary HSC activation in vitro and that it is completely depleted in fully activated HSCs.25, 27 The molecular mechanisms responsible for the changes in PPAR-γ expression during HSC activation remain incompletely understood. To investigate whether hepatocyte-derived EVs induce a phenotypical switch from quiescent to activated HSCs by regulating PPAR-γ expression, we exposed LX2 cells to Hep-EVs, and determined the mRNA and protein expression level of PPAR-γ. Exposure of LX2 to Hep-EVs, particularly for 16 hours (Supplementary Figure 3A), resulted in a 50% reduction of PPAR-γ mRNA expression compared with untreated cells and cells treated with EV-free supernatant (Figure 3A). The decrease of the PPAR-γ mRNA level was concomitant with a marked reduction of the PPAR-γ protein level after 24 hours of exposure to Hep-EVs (Figure 3B). To further confirm these findings, we exposed PMH to palmitic acid and collected and quantified PMH-derived EVs (Supplementary Figure 2A and B).
Supplementary Figure 3.
PPAR-γ mRNA expression time-course and miR-128-3p level in hepatic stellate cells (HSCs) transfected with miR-128-3p mimic or antagomiR-128-3p. (A) Quantitative polymerase chain reaction (qPCR) analysis of peroxisome proliferator-activated receptor-γ (PPAR-γ) time-course in LX2 exposed to HepG2-derived extracellular vesicles (EVs) or controls for 3, 6, 16, or 24 hours. A Vanin-1 neutralizing antibody (VNN1 nAb) was used to block EV internalization. (B) MiR-128-3p level in LX2 transfected with miR-128-3p MIMIC or exposed to EVs and controls for 16 hours. (C) MiR-128-3p level in LX2 transfected with AntagomiR-128-3p or exposed to EV and controls for 16 hours. Values represent mean ± standard deviation from three independent experiments. *P < .05, **P < .005, ***P < .0005, Kruskal-Wallis test with post hoc Mann-Whitney test and Bonferroni correction.
Figure 3.
Hepatocyte-derived extracellular vesicles (EVs) down-regulate peroxisome proliferator-activated receptor-γ (PPAR-γ) by shuttling specific microRNAs. (A) Quantitative polymerase chain reaction (qPCR) analysis of PPAR-γ in human immortalized hepatic stellate cells (LX2) treated with HepG2-derived EVs (Hep-EVs) for 16 hours with or without Vanin-1 neutralizing antibody (VNN1 nAb). (B) Western blot analysis of PPAR-γ protein level in LX2 treated with Hep-EVs for 24 hours in the presence or absence of VNN1 nAb. (C) Quantitative PCR analysis of PPAR-γ in primary mouse hepatic stellate cells (mHSCs) treated with primary mouse hepatocyte-derived EVs (PMH-EVs) for 16 hours with or without VNN1 nAb. β2-Microglobulin (B2M) was used as housekeeping gene for quantitative PCR, and actin was used as loading control for Western blotting. (D) Representative confocal microphotographs of internalization (60× magnification) of PKH26-positive Hep-EVs (red) and EV-RNA content labeled with SYTO RNA (green). (E) Quantitative PCR of PPAR-γ-targeting miR-128-3p, miR-27b, and miR-130b in LX2 treated with Hep-EVs for 16 hours with or without VNN1 nAb. (F) Quantitative PCR analysis of miR-128-3p in EVs isolated from HepG2 treated with 0.25 μM of palmitic acid or vehicle for 24 hours. (G) Quantitative PCR of PPAR-γ-targeting miR-128-3p in (mHSC) treated with Hep-EVs for 16 hours with or without VNN1 nAb. (H) Quantitative PCR analysis of miR-128-3p in EVs isolated from PMH-EVs treated with 0.25 μM of palmitic acid or vehicle for 24 hours. U6 was used as the control. Values represent mean ± standard deviation from three independent experiments. *P <.05, **P < .005, ***P < .0005, Kruskal-Wallis test with post hoc Mann-Whitney test and Bonferroni correction.
Supplementary Figure 2.
Extracellular vesicles (EVs) isolated from primary mouse hepatocytes and incubated with quiescent mouse primary hepatic stellate cells (HSCs). (A) Representative microphotographs of primary mouse hepatocytes exposed to 0.25 μM of palmitic acid for 24 hours. (B) Flow cytometry analysis of the amount of calcein + EVs isolated from palmitic acid-treated or vehicle-treated primary mouse hepatocytes. (C) Representative microphotographs of primary mouse HSCs cultured in vitro from day-1 (quiescent HSCs) to day-7 (activated HSC). Values represent mean ± standard deviation from three independent experiments. *P < .05, **P < .005, ***P < .0005, Kruskal-Wallis test with post–hoc Mann-Whitney test and Bonferroni correction.
Primary mouse quiescent HSCs isolated from WT mice were incubated with PMH-EVs on day 2 from isolation and for 24 hours, when they still have a quiescent phenotype (Supplementary Figure 2C). As previously observed for LX2 cells, PPAR-γ mRNA expression resulted in a 90% down-regulation in EV-treated primary mHSCs compared with untreated cells (control) and mHSC treated with EV-free supernatant (Figure 3C). In both human immortalized and primary mouse HSCs, PPAR-γ mRNA and protein levels were unchanged or not significantly changed, compared with controls, by blocking EVs internalization with Vanin-1 neutralizing antibody (VNN1 nAb) (Figure 3A–C).
It has been extensively reported that EVs carry and transfer a variety of different bioactive molecules, such as mRNA, miRNAs, proteins, and lipids16, 28, 29, 30, 31 from the cell of origin to the target cell. Currently, miRNAs have been found to play essential roles in HSC differentiation, proliferation, apoptosis, and migration. Studies have shown that up-regulation of miR-126 in HSCs promoted hepatic fibrosis by down-regulating IkBα.32 In other studies, overexpression of miR-150 and miR-194 resulted in cell proliferation inhibition in LX-2 as well as reduction in collagen type 1 and α-SMA via inhibition of c-myb and rac1 expression.33, 34 Furthermore, previous reports have identified and validated several microRNAs that target PPAR-γ.35, 36 Here, we tested whether Hep-EVs carry some specific microRNAs targeting PPAR-γ and transfer them into HSC, enhancing their profibrogenic activation. EVs isolated from fat-laden hepatocytes were double-labeled with SYTO green fluorescent nucleic acid stain for EV-RNA and PKH26 for EV membrane. Confocal microscopic imaging identified EVs containing RNA incorporated into HSCs after 6 hours of exposure (Figure 3D).
We further identified the level of three validated miRNAs targeting PPAR- γ: miR-128-3p, miR-27b, and miR-130b.35, 37, 38, 39 We identified all three miRNAs in LX2 exposed to Hep-EVs. We observed that LX2 cells were enriched in all three miRNAs compared with the untreated cells, but only the amount of miR-128-3p was dramatically reduced by neutralizing Vanin-1 on EVs, suggesting that EV internalization is a key mechanism for shuttling miR-128-3p into target cells (Figure 3E).
To confirm these results, we quantified the amount of miR-128-3p in EVs isolated from palmitic acid-treated HepG2, and observed that EVs were particularly enriched in this miRNA compared with control-treated HepG2 (Figure 3F). These results were confirmed by similar results in primary mouse HSCs incubated with palmitic acid-treated PMH-EV (Figure 3G and H). These findings suggest that Hep-EVs carry and shuttle PPAR-γ-targeting miRNAs in HSCs, thus promoting their activation.
In Vitro Manipulation of MicroRNA-128-3p With Gain- and Loss-of-Function Approaches Results in Differential Modulation of Hepatic Stellate Cell Phenotype
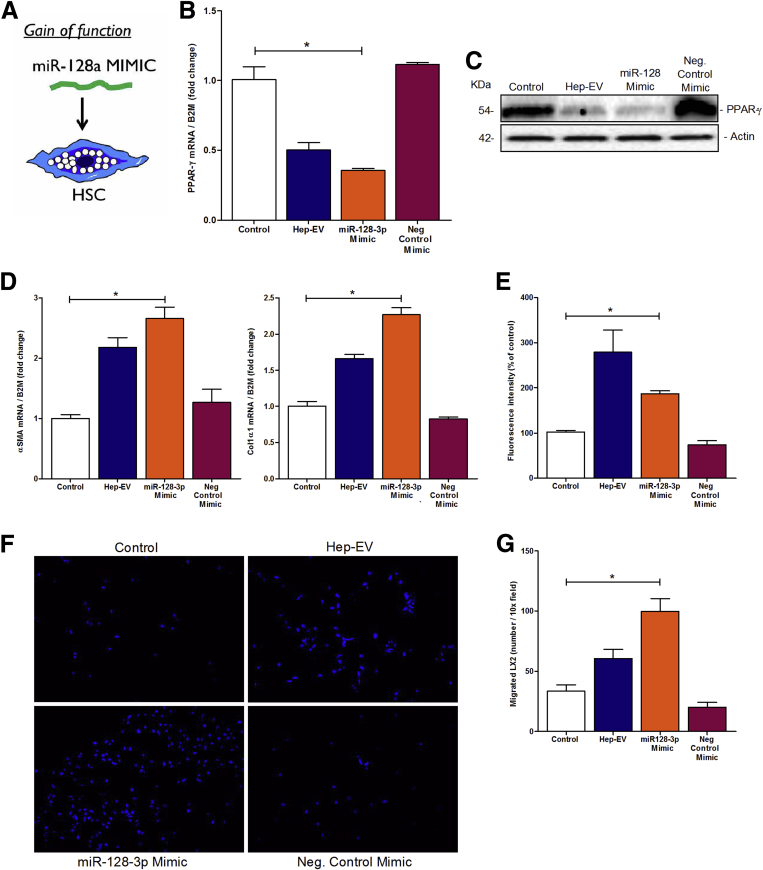
To evaluate the profibrogenic role of miR-128-3p encapsulated in Hep-EVs, we used different gain- and loss-of-function approaches. First, we treated LX2 with miR-128-3p mimic, which reproduces mature endogenous miR-128-3p, or negative control mimic (Figure 4A). LX2 cells transfected with miR-128-3p Mimic showed a significant down-regulation of PPAR-γ mRNA, as well as protein, compared with LX2 transfected with negative control mimic, or cells treated with EV-free supernatant (Figure 4B and C). Additionally, miR-128-3p mimic induced a significant up-regulation in the levels of miR-128-3p (Supplementary Figure 3B) and profibrogenic genes α-SMA and collagen-1α1 compared with controls (Figure 4D). We then analyzed whether miR-128-3p mimic also modulated LX2 profibrogenic responses. In this regard, we observed a dramatic increase of LX2 cell mitogenic activity (Figure 4E), as well as chemotaxis (Figure 4F and G), similar to the effects induced by Hep-EVs.
Figure 4.
An artificial exposure of hepatic stellate cells (HSC) to miR-128-3p mimic showed similar profibrogenic responses induced by miR-128-3p shuttled by HepG2-derived extracellular vesicles (Hep-EVs). (A) Graphical summary of gain-of-function approach in HSCs. (B) Quantitative polymerase chain reaction (qPCR) analysis of peroxisome proliferator-activated receptor-γ (PPAR-γ) in human immortalized hepatic stellate cells (LX2) treated with HepG2-EVs, miR-128-3p mimic or negative control mimic for 16 hours. (C) Western blot analysis of PPAR-γ protein level in LX2 treated with HepG2-EVs, miR-128-3p mimic, or negative control mimic for 24 hours. (D) Quantitative PCR analyses for profibrogenic genes α-smooth muscle actin (α-SMA) and collagen type 1 in LX2 treated with HepG2-EVs, miR-128-3p mimic or negative control mimic for 16 hours. β2-Microglobulin (B2M) was used as housekeeping gene for qPCR, and actin was used as loading control for Western blotting. (E) Proliferation assay of 5-bromo-2-deoxyuridine–positive LX2 after exposure to HepG2-EVs, miR-128-3p mimic or negative control mimic for 48 hours. (F) Representative microphotographs and (G) corresponding quantification graph of Boyden’s chamber assay of LX2 incubated with HepG2-EVs, miR-128-3p mimic, and negative control mimic for 16 hours. Values represent mean ± standard deviation from three independent experiments. *P < .05, **P < .005, ***P < .0005, Kruskal-Wallis test with post hoc Mann-Whitney test and Bonferroni correction.
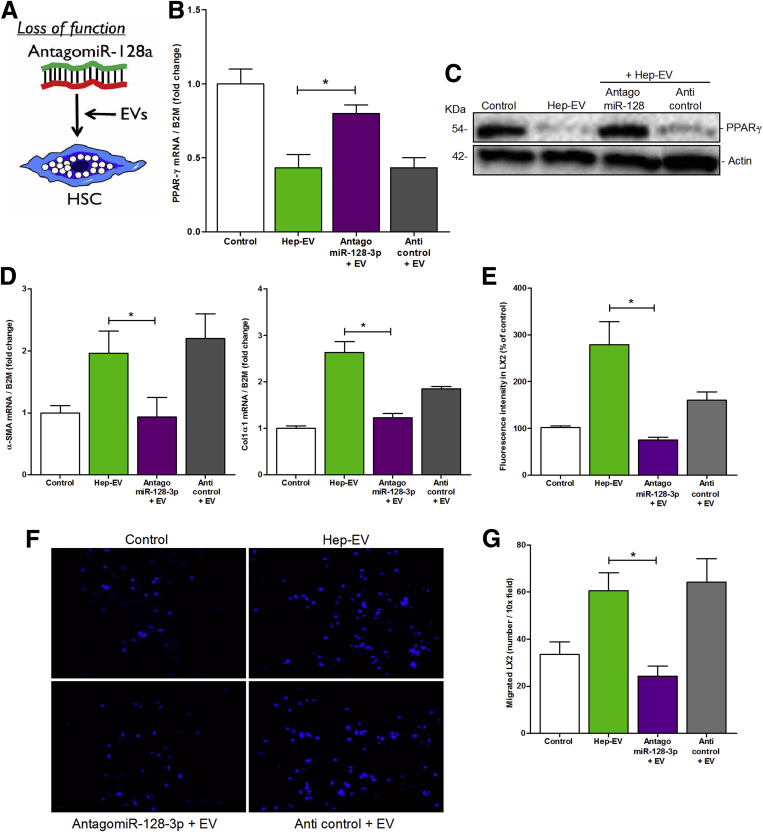
Second, we transfected LX2 cells with a selected miR-128-3p antagonist (antagomiR-128-3p) or with negative control (anticontrol) (Figure 5A). By inhibiting miR-128-3p in LX2 cells exposed to Hep-EVs (Supplementary Figure 3C), we rescued both mRNA and protein levels of PPAR-γ which resembled those of untreated cells or cells treated with EV-free supernatant (Figure 5B and C). The expression of PPAR-γ remained unchanged in LX2 treated with anticontrol. Inhibition of miR-128-3p also resulted in a significant abrogation of the profibrogenic markers α-SMA and collagen-1α1 (Figure 5D) as well as a marked decrease in HSC mitogenic activity (Figure 5F) and oriented migration (Figure 5F and G) compared with anticontrol.
Figure 5.
Loss of function of miR-128-3p in hepatic stellate cells (HSC) rescued PPAR-γ expression and reduced activation, migration and proliferation. (A) Graphical summary of loss-of-function approach in hepatic stellate cells (HSCs). (B) Quantitative polymerase chain reaction (qPCR) analysis of peroxisome proliferator-activated receptor-γ (PPAR-γ) in human immortalized hepatic stellate cells (LX2) treated with HepG2-derived extracellular vesicles (Hep-EVs), AntagomiR-128-3p, or anticontrol for 16 hours. (C) Western blot analysis of PPAR-γ protein level in LX2 treated with Hep-EVs, antagomiR-128-3p, or anticontrol for 24 hours. (D) Quantitative polymerase chain reaction (qPCR) analyses for profibrogenic genes α-smooth muscle actin (α-SMA) and collagen type 1 in LX2 treated with Hep-EVs, antagomiR-128-3p or anticontrol for 16 hours. β2-Microglobulin (B2M) was used as housekeeping gene for quantitative PCR, and actin was used as loading control for Western blotting. (E) Proliferation assay of 5-bromo-2-deoxyuridine–positive LX2 after exposure to Hep-EVs, AntagomiR-128-3p or anticontrol for 48 hours. (F) Representative microphotographs and (G) corresponding quantification graph of Boyden’s chamber assay of LX2 incubated with Hep-EVs, AntagomiR-128-3p or anticontrol for 16 hours. Values represent mean ± SD from three independent experiments. *P < .05, **P < .005, ***P < .0005, Kruskal-Wallis test with post hoc Mann-Whitney test and Bonferroni correction.
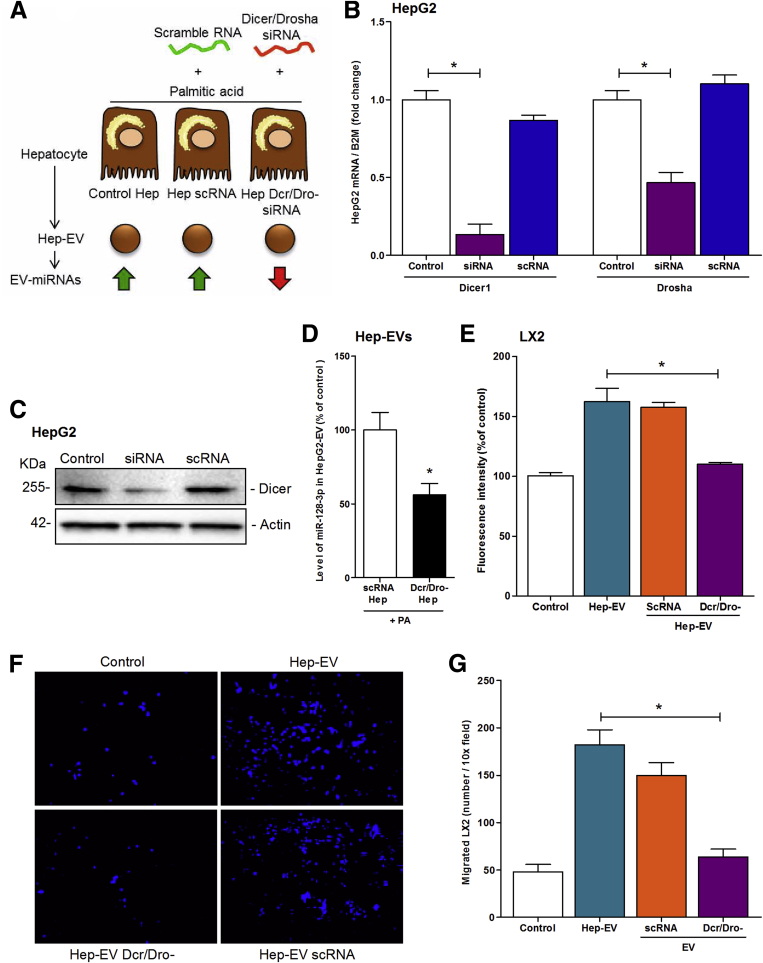
To further confirm the role of microRNAs shuttled by Hep-EVs in HSC activation, we used an alternative loss-of-function approach by double-silencing Dicer1 and Drosha (Dcr/Dro-siRNA) in HepG2 followed by exposure to palmitic acid to generate Hep-EVs with reduced miRNA content compared with EVs released by HepG2 transfected with scramble RNA (scRNA) (Figure 6A). The double-silencing approach resulted in an approximately 90% suppression of mRNA level of Dicer and 60% of Drosha compared with control and scRNA cells (Figure 6B). Western blot analysis confirmed that Dicer protein synthesis was blocked by the siRNA approach (Figure 6C). Knocking-down Dicer/Drosha in fat-laden HepG2 reduced the level of miR-128-3p of 50% in EVs compared with EVs released by fat-laden HepG2 transfected with scRNA (Figure 6D).
Figure 6.
Depletion of the microRNA machinery in HepG2 resulted in HepG2-derived extracellular vesicles (Hep-EVs) with reduced profibrogenic effect on HSC. (A) Graphical summary of double-silencing of Dicer/Drosha in HepG2 to generate miRNA-reduced EVs released during lipotoxicity. (B) Quantitative PCR analysis of Dicer1 (Dcr) and Drosha (Dro) in HepG2 treated with Dcr/Dro silencing RNA (siRNA) or scramble RNA (scRNA) for 48 hours and exposed to 0.25 μM of palmitic acid or vehicle for 24 hours for EV release. (C) Western blot analysis of Dicer in HepG2 treated with or without silencing RNA (siRNA) and scramble RNA (scRNA) for 72 hours. β2-Microglobulin (B2M) was used as housekeeping gene for quantitative polymerase chain reaction (PCR), and actin was used as loading control for Western blot analysis. (D) Quantitative PCR analysis of miR-128-3p amount in Hep-EVs released from HepG2 treated with Dcr/Dro siRNA or scRNA. (E) Proliferation assay of 5-bromo-2-deoxyuridine–positive LX2 exposed to EV isolated from palmitic acid-treated HepG2, scRNA-treated HepG2 and Dcr/Dro siRNA-treated HepG2 (Dcr/Dro-) followed by palmitic acid exposure. (F) Representative microphotographs and (G) corresponding quantification histogram of Boyden’s chamber assay of LX2 exposed to EVs derived from palmitic acid-treated HepG2, scRNA-treated HepG2 and Dcr/Dro siRNA-treated HepG2 (Dcr/Dro-) followed by plamitic acid exposure. Values represent mean ± standard deviation from three independent experiments. *P < .05, **P < .005, ***P < .0005, Kruskal-Wallis test with post hoc Mann-Whitney test and Bonferroni correction.
We further tested whether reducing EV-miRNAs, including miR-128-3p, could reduce HSC activation. Indeed, the exposure of LX2 cells to miRNA-reduced EVs resulted in a significant decrease of HSC mitogenic activity (Figure 6E) and chemotaxis (Figure 6F and G). This effect was not observed after the exposure of HSCs to EVs isolated from HepG2 transfected with scramble RNA (scRNA) (Figure 6E–G).
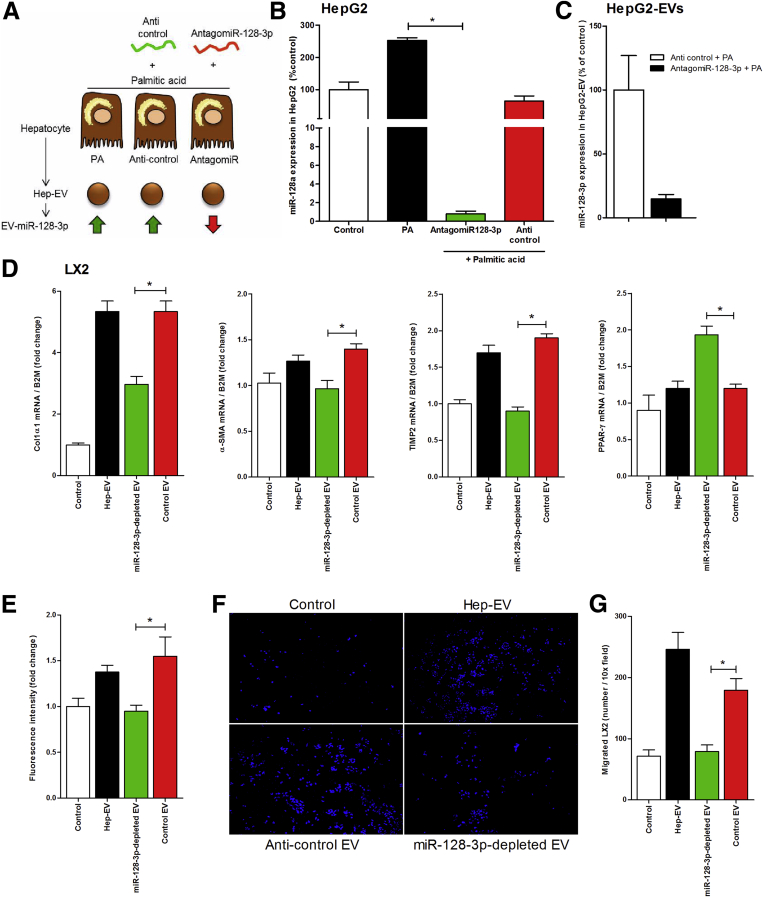
To further confirm the role of miR-128-3p in HSC activation, we transfected palmitic acid-treated HepG2 with antagomiR-128-3p or with anticontrol (Figure 7A). This approach resulted in a 90% reduction of miR-128-3p in both fat-laden HepG2 as well as Hep-derived EVs (Figure 7B and C). The exposure of miR-128-3p-depleted EVs to LX2 cells resulted in a down-regulation of profibrogenic markers α-SMA, collagen-1α1, and TIMP-2 and up-regulation of the HSC quiescent marker, PPAR-γ compared with EVs isolated from fat-laden HepG2 transfected with anticontrol (Figure 7D). Furthermore, miR-128-3p-depleted Hep-EVs showed reduced HSC proliferation (Figure 7E) and oriented-migration compared with control EVs (Figure 7F and G).
Figure 7.
Specific depletion of miR-128-3p in HepG2-derived extracellular vesicles (Hep-EVs) significantly reduced hepatic stellate cell (HSC) profibrogenic responses. (A) Graphical summary of approach used to inhibit miR-128-3p in palmitic acid-treated HepG2 and generate miR-128-3p-depleted EVs. (B) Quantitative polymerase chain reaction (qPCR) analysis of miR-128-3p expression in HepG2 transfected with antagomiR-128-3p, or anticontrol, and exposed to 0.25 mM of palmitic acid. (C) Quantitative PCR analysis of miR-128-3p level in Hep-EV released by HepG2 transfected with anticontrol and antagomiR-128-3p. Mean values were normalized to U6 small-nuclear RNA. (D) Quantitative PCR analyses of expression of profibrogenic genes α-SMA, TIMP-1, and collagen1α1 as well as of the HSC quiescent marker PPAR-γ in LX2 treated with miR-128-3p-depleted Hep-EV, Hep-EV, or Hep-EV isolated from anticontrol transfected HepG2. β2-Microglobulin (B2m) was used as housekeeping gene. (E) Proliferation assay of 5-bromo-2-deoxyuridine–positive LX2 after exposure to miR-128-3p-depleted Hep-EV, Hep-EV, or Hep-EV isolated from anticontrol transfected HepG2, for 48 hours. (F) Representative microphotographs and (G) corresponding quantification graph of Boyden’s chamber assay of LX2 incubated with miR-128-3p-depleted Hep-EV, Hep-EV, or Hep-EV isolated from anticontrol transfected HepG2, for 16 hours. Values represent mean ± standard deviation from three independent experiments. *P < .05, **P < .005, ***P < .0005, Kruskal-Wallis test with post hoc Mann-Whitney test and Bonferroni correction.
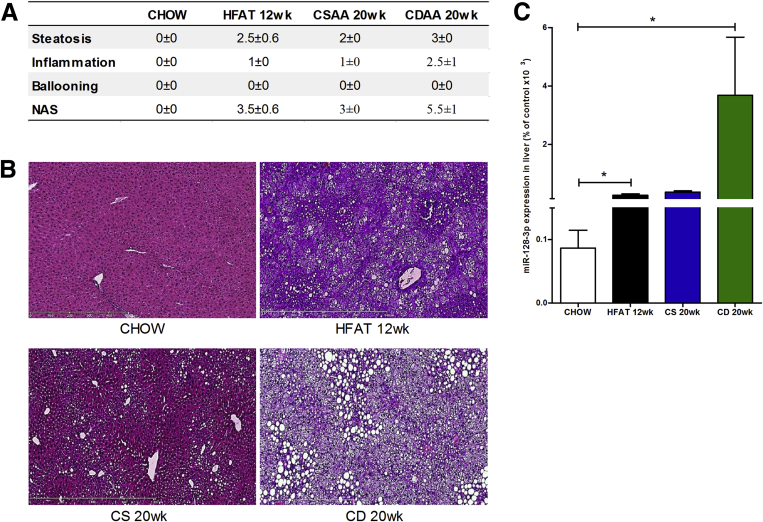
To test whether miR-128-3p was differentially expressed in the liver during lipotoxicity, we measured the level of this miRNA in liver samples isolated from mice fed high-fat diet for 12 weeks and mice fed CDAA diet for 20 weeks, which developed mild and severe NASH, respectively (Figure 8A and B). Mice fed high-fat or CDAA diets showed significant up-regulation of miR-128-3p in the liver compared with mice that received the respective control diets (Figure 8C). These findings demonstrate that EV-miR-128-3p contributes to HSC activation, which may be inhibited or reduced by blocking miR-128-3p.
Figure 8.
Up-regulation of hepatic miR-128-3p in two experimental models of diet-induced nonalcoholic fatty liver disease (NAFLD)/nonalcoholic steatohepatitis (NASH). (A) H&E liver specimens were analyzed in a blinded manner by a trained pathologist for steatosis, inflammation, and ballooning, determining the NAFLD activity score analysis score in mice fed chow, high-fat, choline supplemented l-amino acid (CSAA), or choline-deficient l-amino acid (CDAA). (B) Representative microphotographs of H&E staining of liver specimens harvested from mice fed a high-fat diet for 12 weeks, CSAA or CDAA diet for 20 weeks, or normal chow. (C) Expression level of miR-128-3p in liver samples harvested from mice fed a high fat diet for 12 weeks, CSAA or CDAA diet for 20 weeks or normal chow. Mean values were normalized to U6 small-nuclear RNA. Values represent mean ± standard deviation from three independent experiments. *P < .05, **P < .005, ***P < .0005, Kruskal-Wallis test with post hoc Mann-Whitney test and Bonferroni correction.
Discussion
The principal findings of this study relate to the mechanisms linking overloading of hepatocytes with lipids and the activation of HSCs the key fibrogenic cells in the liver. Our data demonstrate that EVs released by hepatocytes exposed to lipotoxic FFAs are efficiently internalized in HSCs in a process that depends at least in part on the expression of VNN1 on the surface of EVs. EVs internalized into HSCs not only induce a phenotypic switch from quiescent to activated HSCs, but also shuttle miRNAs in to the target cells—that is, miR-128-3p, a specific PPAR-γ-targeting miRNA. Gain- and loss-of-function experiments identified miR-128-3p as a key mediator of HSC activation induced by fat-laden hepatocyte-derived EVs.
NAFLD has grown to become the most common chronic liver disease in both adults and children.40, 41 The early stages of the disease are characterized by the overaccumulation of fat mainly in the form of triglycerides in the liver resulting in hepatic steatosis.42 Although this condition appears to be benign, some patients develop features of hepatocellular damage and inflammation in a condition termed steatohepatitis.8, 43 Like other chronic liver disorders, this process may trigger an abnormal wound-healing response with development of liver fibrosis; this single most important feature of disease severity in patients with NAFLD44 can lead to cirrhosis and the need for liver transplantation.45 Recent research has provided significant information regarding the molecular and cellular basis for the development of hepatic fibrosis in NAFLD. In particular, studies have focused on the crosstalk between inflammatory cells, mainly the resident liver macrophages, or Kupffer cells, and HSCs, the key cell responsible for liver scar formation.13, 46
The links between damaged parenchymal cells in the liver, modulation of HSC phenotype, and the development of liver fibrosis in NAFLD remain incompletely understood. Previous studies have suggested that hepatocyte death may be an important signal for the activation of HSCs. Indeed, engulfment of apoptotic bodies by HSCs stimulates the fibrogenic activity of these cells and may be one mechanism by which hepatocyte apoptosis promotes fibrosis.47 Previous data have also demonstrated that DNA from apoptotic hepatocytes acts as an important mediator of HSC activation and differentiation by providing a stop signal when they have reached an area of apoptotic hepatocytes and inducing a stationary phenotype-associated up-regulation of collagen production.48
Lipotoxicity, a process by which an accumulation of certain toxic lipids, such as saturated FFAs, in hepatocytes triggers various molecular pathways of cell stress and eventually results in cell death, has evolved as a key event during NAFLD progression. We have recently demonstrated that during lipotoxicity hepatocytes release EVs, which are enriched in Vanin-1 (VNN1) on the external leaflet, and are internalized into endothelial cells where they induce proangiogenic effects.16 Through various approaches we identified VNN1 as an important mediator of the process of internalization. EVs are a heterogeneous population of small membrane-bound structures released mainly by stressed or dying cells via exocytosis from the cytosol or ectocytosis from the plasma membrane of the parenteral cells.49, 50 EVs carry a variety of different bioactive molecules from the parenteral cells, including proteins, mRNA, miRNAs, and lipids.51, 52, 53 The wide spectrum of biological activities promoted by EVs makes them efficient cell-to-cell communicators.
Circulating levels of EVs are increased in animal models of NASH and in patients with cirrhosis.17, 54 In the former, the levels of circulating EVs strongly correlated with the severity of liver fibrosis. In our present study, we observed that EVs are released in large amounts by hepatocytes exposed to the lipotoxic fatty acid palmitic acid and are efficiently internalized into HSCs. Similar to what we previously observed in endothelial cells, the internalization of Hep-EVs into HSCs required the presence of VNN1 on their surface. More importantly, we found that the internalization of vesicles induced a significant phenotypic switch in HSCs from quiescent to activated.
A large body of evidence supports a central role of PPAR-γ, a member of the nuclear hormone-receptor superfamily, as a key modulator of HSC quiescence.18, 55 Notably, it has been shown that PPAR-γ progressively decreases during HSC activation and that it is completely depleted in fully activated HSCs.25, 27 The findings that the internalization of EVs into HSCs resulted in the transfer of their RNA content into the HSCs, including various miRNAs that are specific modulators of PPAR-γ expression, led us to investigate their potential role in the activation of HSCs. We focused our studies on miR-128-3p, which inhibits PPAR-γ expression, because it was recently identified as being differentially expressed in the livers of patients with NASH.56 Our findings show that this miRNA was enriched in EVs derived from fat-laden hepatocytes and that it was dramatically up-regulated in liver samples isolated from two murine diet-induced NAFLD/NASH models. Additionally, miR-128-3p was selectively transferred to HSCs by Hep-EVs. By using both gain- and loss-of-function approaches, we identified miR-128-3p as a key mediator of HSC activation induced by EVs in a process dependent, at least in part, on the modulation of PPAR-γ expression.
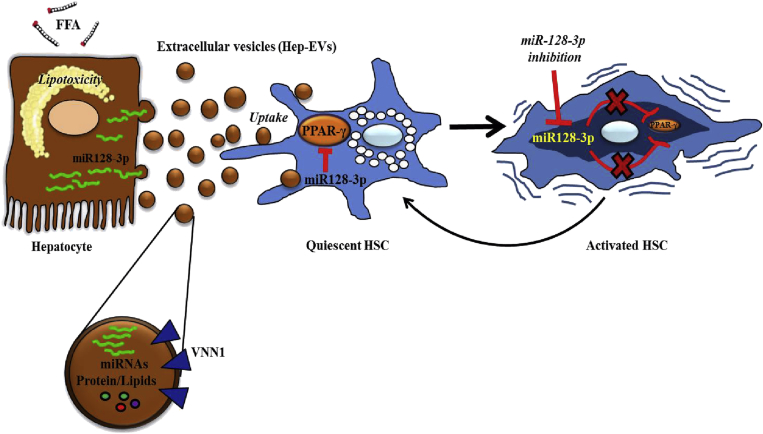
In summary, our study uncovers a novel pathway linking lipotoxicity in liver parenchymal cells to activation of the main fibrogenic cells in the liver, identifying a potential key mechanism of liver fibrosis in NASH. The results support a model in which overloading of hepatocytes with toxic lipids results in the release of EVs that can be efficiently internalized by HSCs and induce a phenotypic switch to profibrogenic HSCs. This process involves the delivery of miR-128-3p and suppression of PPAR-γ expression (Figure 9). These findings provide further insight into the pathogenesis of liver fibrosis in NASH and identify potential molecular targets for antifibrotic therapeutic interventions.
Figure 9.
Hepatocyte-derived extracellular vesicles (EVs) are released during lipotoxicity and induce a phenotypical switch from quiescent to activated hepatic stellate cells (HSC). During liver steatosis, free fatty acids accumulate in the hepatocytes leading to lipid-induced toxicity (lipotoxicity) and cell death. Dying hepatocytes produce and release EVs that carry a variety of different bioactive molecules, particularly microRNAs such as miR-128-3p, which is highly up-regulated in fat-laden hepatocytes. EVs shuttle miR-128-3p into HSCs, which leads to an inhibition of peroxisome proliferator-activated receptor-γ (PPAR-γ) and consequent phenotypical switch from quiescent to activated HSCs. A selective inhibition of miR-128-3p rescues PPAR-γ expression and results in HSC inactivation.
Acknowledgments
The authors thank the UCSD Neuroscience Core and especially Jennifer Santini for microscopy assistance, and Dr Hongying Li of the UCSD Moores Cancer Center, Division of Biostatistics and Bioinformatics, for the statistical consultation.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was funded by National Institutes of Health grants R01 DK082451, U01 AA022489 (to A.E.F.), American Liver Foundation Postdoctoral Research Fellowship (to D.P.), Gilead Sciences Research Scholars Program in Liver Disease (to A.E.), and grant NS047101 for the UCSD Microscopy Facility (to Jennifer Santini).
Supplementary Material
Supplementary Table 1.
List of primers used for quantitative polymerase chain reaction
| Gene | Primers |
|---|---|
| hTIMP-2 | |
| Forward | 5-GGGGCCGTGTAGATAAACTCTAT-3 |
| Reverse | 5-AAGCGGTCAGTGAGAAGGAAG-3 |
| hCOL1A1 | |
| Forward | 5-GAGGGCCAAGACGAAGACATC-3 |
| Reverse | 5-CAGATCACGTCATCGCACAAC-3 |
| hα-SMA | |
| Forward | 5-CTATGAGGGCTATGCCTTGCC-3 |
| Reverse | 5-GCTCAGCAGTAGTAACGAAGGA-3 |
| h18S | |
| Forward | 5-CAGCCACCCGAGATTGAGCA-3 |
| Reverse | 5-TAGTAGCGACGGGCGGTGTG-3 |
| hPPAR-γ | |
| Forward | 5-TTCGCAATCCATCGGCGAG-3 |
| Reverse | 5-CCACAGGATAAGTCACCGAGG-3 |
| mPPAR-γ | |
| Forward | 5-CGAGGGCGATCTTGACAGGA-3 |
| Reverse | 5-GGCCACCTCTTTGCTCTGCT-3 |
| hGAPDH | |
| Forward | 5-ACAACTTTGGTATCGTGGAAGG-3 |
| Reverse | 5-GCCATCACGCCACAGTTTC-3 |
| m18S | |
| Forward | 5-ACGGAAGGGCACCACCAGGA-3 |
| Reverse | 5-CACCACCACCCACGGAATCG-3 |
| hDICER | |
| Forward | 5-GAGCTGTCCTATCAGATCAGGG-3 |
| Reverse | 5-ACTTGTTGAGCAACCTGGTTT-3 |
| hDROSHA | |
| Forward | 5-ATCCTTTAGGCCCCAAAATCTG-3 |
| Reverse | 5-GGGTACAAAGTCTGGTCGTGG-3 |
References
- 1.Roberts E.A. Non-alcoholic steatohepatitis in children. Clin Liver Dis. 2007;11:155–172. doi: 10.1016/j.cld.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Bellentani S., Saccoccio G., Masutti F. Prevalence of and risk factors for hepatic steatosis in Northern Italy. Ann Intern Med. 2000;132:112–117. doi: 10.7326/0003-4819-132-2-200001180-00004. [DOI] [PubMed] [Google Scholar]
- 3.Angulo P., Keach J.C., Batts K.P. Independent predictors of liver fibrosis in patients with nonalcoholic steatohepatitis. Hepatology. 1999;30:1356–1362. doi: 10.1002/hep.510300604. [DOI] [PubMed] [Google Scholar]
- 4.Knobler H., Schattner A., Zhornicki T. Fatty liver—an additional and treatable feature of the insulin resistance syndrome. QJM. 1999;92:73–79. doi: 10.1093/qjmed/92.2.73. [DOI] [PubMed] [Google Scholar]
- 5.Ratziu V., Giral P., Charlotte F. Liver fibrosis in overweight patients. Gastroenterology. 2000;118:1117–1123. doi: 10.1016/s0016-5085(00)70364-7. [DOI] [PubMed] [Google Scholar]
- 6.Vernon G., Baranova A., Younossi Z.M. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment Pharmacol Ther. 2011;34:274–285. doi: 10.1111/j.1365-2036.2011.04724.x. [DOI] [PubMed] [Google Scholar]
- 7.Marchesini G., Brizi M., Morselli-Labate A.M. Association of nonalcoholic fatty liver disease with insulin resistance. Am J Med. 1999;107:450–455. doi: 10.1016/s0002-9343(99)00271-5. [DOI] [PubMed] [Google Scholar]
- 8.Angulo P. Nonalcoholic fatty liver disease. New Engl J Med. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 9.Andersen T., Gluud C., Franzmann M.B. Hepatic effects of dietary weight loss in morbidly obese subjects. J Hepatol. 1991;12:224–229. doi: 10.1016/0168-8278(91)90942-5. [DOI] [PubMed] [Google Scholar]
- 10.Teli M.R., James O.F., Burt A.D. The natural history of nonalcoholic fatty liver: a follow-up study. Hepatology. 1995;22:1714–1719. [PubMed] [Google Scholar]
- 11.Ratziu V., Bonyhay L., Di Martino V. Survival, liver failure, and hepatocellular carcinoma in obesity-related cryptogenic cirrhosis. Hepatology. 2002;35:1485–1493. doi: 10.1053/jhep.2002.33324. [DOI] [PubMed] [Google Scholar]
- 12.Bugianesi E., Leone N., Vanni E. Expanding the natural history of nonalcoholic steatohepatitis: from cryptogenic cirrhosis to hepatocellular carcinoma. Gastroenterology. 2002;123:134–140. doi: 10.1053/gast.2002.34168. [DOI] [PubMed] [Google Scholar]
- 13.Friedman S.L. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rippe R.A., Brenner D.A. From quiescence to activation: gene regulation in hepatic stellate cells. Gastroenterology. 2004;127:1260–1262. doi: 10.1053/j.gastro.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 15.Olaso E., Friedman S.L. Molecular regulation of hepatic fibrogenesis. J Hepatol. 1998;29:836–847. doi: 10.1016/s0168-8278(98)80269-9. [DOI] [PubMed] [Google Scholar]
- 16.Povero D., Eguchi A., Niesman I.R. Lipid-induced toxicity stimulates hepatocytes to release angiogenic microparticles that require Vanin-1 for uptake by endothelial cells. Sci Signa. 2013;6:ra88. doi: 10.1126/scisignal.2004512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Povero D., Eguchi A., Li H. Circulating extracellular vesicles with specific proteome and liver MicroRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PLoS One. 2014;9:e113651. doi: 10.1371/journal.pone.0113651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miura K., Yang L., van Rooijen N. Hepatic recruitment of macrophages promotes nonalcoholic steatohepatitis through CCR2. Am J Physio Gastrointest Liver Physiol. 2012;302:G1310–G1321. doi: 10.1152/ajpgi.00365.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buettner R., Scholmerich J., Bollheimer L.C. High-fat diets: modeling the metabolic disorders of human obesity in rodents. Obesity (Silver Spring) 2007;15:798–808. doi: 10.1038/oby.2007.608. [DOI] [PubMed] [Google Scholar]
- 20.Li Z., Berk M., McIntyre T.M. The lysosomal-mitochondrial axis in free fatty acid-induced hepatic lipotoxicity. Hepatology. 2008;47:1495–1503. doi: 10.1002/hep.22183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Ambrosio D.N., Walewski J.L. Distinct populations of hepatic stellate cells in the mouse liver have different capacities for retinoid and lipid storage. PLoS One. 2011;6:e24993. doi: 10.1371/journal.pone.0024993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seki E., De Minicis S., Osterreicher C.H. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med. 2007;13:1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- 23.Kornek M., Popov Y., Libermann T.A. Human T cell microparticles circulate in blood of hepatitis patients and induce fibrolytic activation of hepatic stellate cells. Hepatology. 2011;53:230–242. doi: 10.1002/hep.23999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marra F., Efsen E., Romanelli R.G. Ligands of peroxisome proliferator-activated receptor gamma modulate profibrogenic and proinflammatory actions in hepatic stellate cells. Gastroenterology. 2000;119:466–478. doi: 10.1053/gast.2000.9365. [DOI] [PubMed] [Google Scholar]
- 25.Miyahara T., Schrum L., Rippe R. Peroxisome proliferator-activated receptors and hepatic stellate cell activation. J Biol Chem. 2000;275:35715–35722. doi: 10.1074/jbc.M006577200. [DOI] [PubMed] [Google Scholar]
- 26.Sharvit E., Abramovitch S., Reif S., Bruck R. Amplified inhibition of stellate cell activation pathways by PPAR-gamma, RAR and RXR agonists. PLoS One. 2013;8:e76541. doi: 10.1371/journal.pone.0076541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazra S., Xiong S., Wang J. Peroxisome proliferator-activated receptor gamma induces a phenotypic switch from activated to quiescent hepatic stellate cells. J Biol Chem. 2004;279:11392–11401. doi: 10.1074/jbc.M310284200. [DOI] [PubMed] [Google Scholar]
- 28.Fonsato V., Collino F., Herrera M.B. Human liver stem cell-derived microvesicles inhibit hepatoma growth in SCID mice by delivering antitumor microRNAs. Stem Cells. 2012;30:1985–1998. doi: 10.1002/stem.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garzetti L., Menon R., Finardi A. Activated macrophages release microvesicles containing polarized M1 or M2 mRNAs. J Leukocyte Biol. 2014;95:817–825. doi: 10.1189/jlb.0913485. [DOI] [PubMed] [Google Scholar]
- 30.Li J., Zhang Y., Liu Y. Microvesicle-mediated transfer of microRNA-150 from monocytes to endothelial cells promotes angiogenesis. J Biol Chem. 2013;288:23586–23596. doi: 10.1074/jbc.M113.489302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jansen F., Yang X., Hoelscher M. Endothelial microparticle-mediated transfer of MicroRNA-126 promotes vascular endothelial cell repair via SPRED1 and is abrogated in glucose-damaged endothelial microparticles. Circulation. 2013;128:2026–2038. doi: 10.1161/CIRCULATIONAHA.113.001720. [DOI] [PubMed] [Google Scholar]
- 32.Guo C.J., Pan Q., Xiong H. Dynamic expression of miR-126* and its effects on proliferation and contraction of hepatic stellate cells. FEBS Lett. 2013;587:3792–3801. doi: 10.1016/j.febslet.2013.09.047. [DOI] [PubMed] [Google Scholar]
- 33.Venugopal S.K., Jiang J., Kim T.H. Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am J Physio Gastrointest Liver Physiol. 2010;298:G101–G106. doi: 10.1152/ajpgi.00220.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y., Huang C., Zhang S.P. The potential of microRNAs in liver fibrosis. Cell Signal. 2012;24:2268–2272. doi: 10.1016/j.cellsig.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 35.Jennewein C., von Knethen A., Schmid T. MicroRNA-27b contributes to lipopolysaccharide-mediated peroxisome proliferator-activated receptor gamma (PPARgamma) mRNA destabilization. J Biol Chem. 2010;285:11846–11853. doi: 10.1074/jbc.M109.066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji J., Zhang J., Huang G. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS letters. 2009;583:759–766. doi: 10.1016/j.febslet.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 37.Pan S., Yang X., Jia Y. Microvesicle-shuttled miR-130b reduces fat deposition in recipient primary cultured porcine adipocytes by inhibiting PPAR-g expression. J Cell Physiol. 2014;229:631–639. doi: 10.1002/jcp.24486. [DOI] [PubMed] [Google Scholar]
- 38.Motohashi N., Alexander M.S., Casar J.C. Identification of a novel microRNA that regulates the proliferation and differentiation in muscle side population cells. Stem Cells Dev. 2012;21:3031–3043. doi: 10.1089/scd.2011.0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J., Yu X., Fries J.W. MicroRNA function in the profibrogenic interplay upon chronic liver disease. Int J Mol Sci. 2014;15:9360–9371. doi: 10.3390/ijms15069360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bellentani S., Scaglioni F., Marino M. Epidemiology of non-alcoholic fatty liver disease. Dig Dis. 2010;28:155–161. doi: 10.1159/000282080. [DOI] [PubMed] [Google Scholar]
- 41.Loomba R., Sirlin C.B., Schwimmer J.B. Advances in pediatric nonalcoholic fatty liver disease. Hepatology. 2009;50:1282–1293. doi: 10.1002/hep.23119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z.Z., Berk M., McIntyre T.M. Hepatic lipid partitioning and liver damage in nonalcoholic fatty liver disease: role of stearoyl-CoA desaturase. J Biol Chem. 2009;284:5637–5644. doi: 10.1074/jbc.M807616200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alkhouri N., Dixon L.J., Feldstein A.E. Lipotoxicity in nonalcoholic fatty liver disease: not all lipids are created equal. Expert Rev Gastroenterol Hepatol. 2009;3:445–451. doi: 10.1586/egh.09.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhala N., Angulo P., van der Poorten D. The natural history of nonalcoholic fatty liver disease with advanced fibrosis or cirrhosis: an international collaborative study. Hepatology. 2011;54:1208–1216. doi: 10.1002/hep.24491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Browning J.D., Szczepaniak L.S., Dobbins R. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 46.Friedman S.L. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Canbay A., Friedman S., Gores G.J. Apoptosis: the nexus of liver injury and fibrosis. Hepatology. 2004;39:273–278. doi: 10.1002/hep.20051. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe A., Hashmi A., Gomes D.A. Apoptotic hepatocyte DNA inhibits hepatic stellate cell chemotaxis via toll-like receptor 9. Hepatology. 2007;46:1509–1518. doi: 10.1002/hep.21867. [DOI] [PubMed] [Google Scholar]
- 49.Cocucci E., Racchetti G., Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 50.Ratajczak J., Wysoczynski M., Hayek F. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia. 2006;20:1487–1495. doi: 10.1038/sj.leu.2404296. [DOI] [PubMed] [Google Scholar]
- 51.Diehl P., Fricke A., Sander L. Microparticles: major transport vehicles for distinct microRNAs in circulation. Cardiovasc Res. 2012;93:633–644. doi: 10.1093/cvr/cvs007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan A., Farber E.L., Rapoport A.L. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS One. 2009;4:e4722. doi: 10.1371/journal.pone.0004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kornek M., Lynch M., Mehta S.H. Circulating microparticles as disease-specific biomarkers of severity of inflammation in patients with hepatitis C or nonalcoholic steatohepatitis. Gastroenterology. 2012;143:448–458. doi: 10.1053/j.gastro.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deregibus M.C., Cantaluppi V., Calogero R. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110:2440–2448. doi: 10.1182/blood-2007-03-078709. [DOI] [PubMed] [Google Scholar]
- 56.Cheung O., Puri P., Eicken C. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]