Abstract
Propionibacterium freudenreichii belongs to the class Actinobacteria (Gram positive with a high GC content). This “Generally Recognized As Safe” (GRAS) species is traditionally used as (i) a starter for Swiss-type cheeses where it is responsible for holes and aroma production, (ii) a vitamin B12 and propionic acid producer in white biotechnologies, and (iii) a probiotic for use in humans and animals because of its bifidogenic and anti-inflammatory properties. Until now, only strain CIRM-BIA1T had been sequenced, annotated and become publicly available. Strain CIRM-BIA129 (commercially available as ITG P20) has considerable anti-inflammatory potential. Its gene content was compared to that of CIRM-BIA1 T. This strain contains 2384 genes including 1 ribosomal operon, 45 tRNA and 30 pseudogenes.
Keywords: GRAS, QPS, probiotic, anti-inflammatory, immunomodulation, surface proteins
Introduction
Propionibacterium freudenreichii belongs to the class of Actinobacteria (Gram positive bacteria with a high GC content). This ‘Generally Recognized As Safe’ species is traditionally used as (i) a starter for Swiss-type cheeses where it is responsible for holes and aroma production (ii) a vitamin B12 and propionic acid producer in white biotechnologies, and (iii) a probiotic for use in humans and animals because of its bifidogenic properties (enhancing intestinal transit). A recent screening of 23 strains belonging to this species revealed the considerable anti-inflammatory properties of strain CIRM-BIA129 (ITG-P20) [1]. Its anti-inflammatory potential is superior to that of the previously sequenced CIRM-BIA1 T strain. CIRM-BIA129 (ITG-P20) is currently used to make cheese at an industrial scale and indeed is consumed in large quantities because one gram of Swiss-type cheese contains at least 109 CFU. The genetic basis for the anti-inflammatory properties of P. freudenreichii are still poorly understood. Sequencing CIRM-BIA129 (ITG-P20) would enable investigation of the genomic determinants responsible for the important anti-inflammatory properties of this strain. To our knowledge, CIRM-BIA129 is the second strain in this species to be sequenced, annotated and made publicly available.
Organism information
Classification and features
P. freudenreichii CIRM-BIA129 (ITG P20) cells are Gram-positive, microaerophilic, pleiomorphic (coccoid to rod shape forming ‘Chinese characters’) bacillae (1.0–1.5 μm × 0.5–0.8 μm wide) forming creamy-white colonies on YEL agar plates (Table 1). Cells grow at the bottom of liquid medium tubes (to escape oxygen) and tend to clot in liquid culture at the beginning of the stationary phase. Transmission electron microscopy pictures of liquid-grown cultures revealed a thick cell wall (Fig. 1) made of peptidoglycan. No exopolysaccharides were observed at the surface of the bacteria, unlike what is seen in numerous strains of P. freudenreichii, including the type strain CIRM-BIA1 T (Fig. 2).
Table 1.
Classification and general features of P. freudenreichii CIRM-BIA129 (ITG-P20) according to MIGS guidelines and the Catalogue of Life database
| MIGS ID | Property | Term | Evidence code (a) |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [15] | |
| Phylum Actinobacteria | TAS [16, 17] | ||
| Class Actinobacteria | TAS [16, 17] | ||
| Order Actinomycetales | TAS [16] | ||
| Family Propionibacteriaceae | TAS [18] | ||
| Genus Propionibacterium | TAS [19] | ||
| Species Propionibacterium freundenreichii | TAS [20] | ||
| Strain CIRM-BIA 129 alias ITG P20 | NAS | ||
| Gram staining | Positive | NAS | |
| Cell shape | Pleiomorph (coccoids to rods) | NAS | |
| Motility | Non-motile | NAS | |
| Sporulation | Non-sporulating | NAS | |
| Temperature range | Not tested | ||
| Temperature optimum | 32 °C | NAS | |
| pH range; Optimum | Not tested | ||
| Carbon source | glycerol, erythritol, L-arabinose, adonitol, galactose, D-glucose, D-fructose, D-mannose, inositol, arbutine, esculine, lactose, lactate and gluconate | IDA | |
| MIGS-6 | Habitat | Unknown | |
| MIGS-6.3 | Salinity | Tolerate up to 1 M NaCl | IDA |
| MIGS-22 | Oxygen requirement | Microaerophilic | NAS |
| MIGS-15 | Biotic relationship | Free-living | NAS |
| MIGS-14 | Pathogenicity | Non-pathogen, GRAS, QPS | NAS |
| MIGS-4 | Geographic location | Not reported | |
| MIGS-5 | Sample collection time | Not reported | |
| MIGS-4.1 | Latitude | Not reported | |
| MIGS-4.2 | Longitude | Not reported | |
| MIGS-4.3 | Depth | Not reported | |
| MIGS-4.4 | Altitude | Not reported |
(a) Evidence codes – IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are those of the Gene Ontology project [21]
Fig. 1.
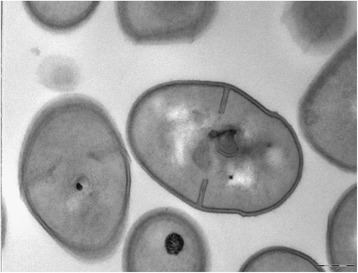
Transmission electron microscopy pictures of liquid-grown cultures of CIRM-BIA129 (ITG P20)
Fig. 2.

The evolutionary history of the strain was inferred using the Neighbor-Joining method [12]. The optimal tree with a sum of branch lengths = 0.80 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method [13] and are in the units of the number of base substitutions per site. The analysis involved 18 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1376 positions in the final dataset. Evolutionary analyses were performed under MEGA5 [14]
A BLASTn of the 16S sequence of type strain CIRM-BIA1 T against the contigs of CIRM-BIA129 (ITG-P20) confirmed the affiliation of this strain to the species (100 % identity, 100 % coverage).
Representative genomic 16S rRNA sequences of the strains were compared with those of other type strains belonging to Actinobacteria present in the Ribosomal Database Project. (Figure 2) shows the phylogenetic tree.
CIRM-BIA129 (ITG P20) was found to be able to utilize glycerol, erythritol, L-arabinose, adonitol, galactose, D-glucose, D-fructose, D-mannose, inositol, arbutine, esculine, lactose, lactate and gluconate as carbon sources according to the gallery-API results.
CIRM-BIA129 (ITG P20) was shown to grow with up to 1 M NaCl in a chemically defined medium in presence of osmoprotectant (G. Jan, personal communication).
Genome sequencing information
Genome project history
Propionibacterium freudenreichii CIRM-BIA 129 (ITG P20) genome was sequenced to obtain information regarding mechanism(s) or molecule(s) responsible for anti-inflammatory properties of the strain. Project information and associated MIGS are shown in Table 2.
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Improved high quality draft |
| MIGS-28 | Libraries used | 300 bp-insert Illumina library |
| MIGS-29 | Sequencing platforms | 36 nucleotide paired-end sequencing (Illumina genome analyser II). |
| MIGS-31.2 | Fold coverage | 681x |
| MIGS-30 | Assemblers | Velvet version 1.1.03 k-mer |
| MIGS-32 | Gene calling method | Show program |
| Locus Tag | PFCIRM129 | |
| EMBL ID | CCBE010000001-CCBE010000111 | |
| EMBL Date of Release | 20-Jun-2014 | |
| BIOPROJECT | PRJEB4826 | |
| MIGS-13 | Source Material Identifier | CIRM-BIA 129 |
| Project relevance | Probiotic, anti-inflammatory |
Growth conditions and genomic DNA preparation
The P. freudenreichii strain CIRM-BIA129 (ITG-P20), isolated from Emmental cheese by Actalia Dairy Products (Institut Technique du Gruyère, Actalia, Rennes, France), was provided by the CIRM-BIA Biological Resource Centre (Centre International de Ressources Microbiennes-Bactéries d’Intérêt Alimentaire, INRA, Rennes, France). It was cultivated at 30 °C without shaking in YEL [2]. Growth was monitored spectrophotometrically at 650 nm, as well as by counting colony-forming units in YEL medium containing 1.5 % agar.
A cell pellet (equivalent to 2 × 1010 CFU) was obtained by centrifugation for 10 min at 5000 × g from a one-day exponentional phase culture of CIRM-BIA129 (ITG P20). DNA was extracted using the Blood & Cell Culture DNA Midi Kit (Qiagen) according to the manufacturer’s recommendations with the following modifications. Briefly, complete bacterial lysis was obtained by adding 220 mg of lysozyme powder (Qbiogene) to 3.5 ml of B1 buffer (Qiagen) followed by 2.5 h of incubation at 37 °C. High molecular weight genomic DNA was purified by gravity flow and anion exchange chromatography, eluted in 5 ml QF buffer (Qiagen) and precipitated with 3.5 ml of isopropanol. DNA was collected by centrifugation for 10 min at 4 °C and 15000 g and then air dried. DNA was resuspended in 100 μL TE 1X buffer at pH8 (Sigma) for two hours at 55 °C. DNA integrity was checked on an 0.8 % agarose gel. The OD260/280nm was 1.9.
Genome sequencing and assembly
The genome of the CIRM-BIA129 (ITG-P20) strain was obtained using a whole-genome strategy based on Illumina 36 nucleotide paired-end sequencing (Illumina genome analyser II) (Table 2). After filtration and trimming, the number of reads was downsized to 50 million of 34.8 bases mean read length, representing a 681-fold genome coverage. The reads were assembled using Velvet version 1.1.03 [3] with a k-mer size of 31. The assembly resulted in 59 scaffolds of a length greater than 1 kb, with an N50 of 123 kb.
Genome annotation
Automatic and manual annotations were carried out with the AGMIAL platform [4], using P. freudenreichii CIRM-BIA 1 T as the reference [5]. Sub-cellular localization was predicted using SurfG+ [6], a software specific to sub-cellular prediction in Gram-positive bacteria. A comparison of the predicted proteome of CIRM-BIA129 with that of the sequenced type strain CIRM-BIA1 was performed with Koriblast software (Korilog, France) with its default parameters. Genes present in CIRM-BIA129 and not in the type strain CIRM-BIA1 were searched by tblastn of the CIRM-BIA129 predicted proteome against the complete genome of CIRM-BIA1. No hit proteins were specific to CIRM-BIA129. In order to search for genomic islands specific to CIRM-BIA129, a blastp of the CIRM-BIA129 proteome against the CIRM-BIA1 proteome was performed. Proteins filtered for a ‘hit coverage’ of less than 75 % and those corresponding to locus-tags colocalized along the chromosome were attributed to genomic islands. To search for probable pseudogenes in CIRM-BIA1, a blastp analysis of CIRM-BIA1 protein against CIRM-BIA129 proteins filtered on a ‘hit coverage’ of less than 75 % was performed. Among them, the corresponding genes were declared as pseudogenes in the event of two adjacent genes having the same ‘best hit’. S-layer homology domains inside the proteins were sought from the SLH.hmm pattern using hmmer with default parameters.
Genome properties
The genome was found to be composed of 111 contigs for a total size of 2,588,969 bp (67.3 % GC content). The contig length was encompassed between 158 bp and 179,656 bp with an average at 23,324 bp. The N50 is 46,655 bp. One ribosomal operon (containing one 16S rRNA gene and one 23S rRNA gene) was present, on contig 24, A total of 2354 complete genes were predicted, 2,307 of them protein-coding genes and 45 tRNA encoding genes. Thirty genes were pseudogenes. According to the COG results, 1,236 protein coding genes were assigned to a putative function, the remainder being annotated as hypothetical proteins. The properties and statistics of the genome are summarized in (Tables 3 and 4).
Table 3.
Nucleotide content and gene count levels of the genome
| Attribute | Value | % of total(a) |
|---|---|---|
| Genome size (bp) | 2,588969 | 100 |
| DNA coding (bp) | 2,209128 | 85.3 |
| DNA G + C (bp) | 1,741869 | 67.3 |
| DNA scaffolds | 59 | 100 |
| Total genes | 2,388 | 100 |
| Protein-coding genes | 2,307 | 96.6 |
| RNA genes | 3 | 0.2 |
| Pseudo genes | 30 | 1.2 |
| Genes in internal clusters | Not reported | Not reported |
| Genes with function prediction | 1190 | 49.8 |
| Genes assigned to COGs | 1,325 | 55.5 |
| Genes with Pfam domains | 1148 | 48.1 |
| Genes with signal peptides | 247 | 10.3 |
| Genes with transmembrane helices | 521 | 21.8 |
| CRISPR repeats | Not reported | Not reported |
(a)The total is based on the size of the genome in base pairs
Table 4.
Number of genes associated with 25 general COG functional categories (2337 proteins)
| Code | Value | % age | Description |
|---|---|---|---|
| J | 116 | 4.96 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.04 | RNA processing and modification |
| K | 83 | 3.55 | Transcription |
| L | 76 | 3.25 | Replication and repair |
| B | 0 | 0 | Chromatin structure and dynamics |
| D | 14 | 0.60 | Cell cycle control and mitosis |
| V | 30 | 1.28 | Defence mechanisms |
| T | 32 | 1.37 | Signal transduction mechanisms |
| M | 58 | 2.48 | Cell wall/membrane biogenesis |
| N | 0 | 0 | Cell motility |
| U | 16 | 0.68 | Intracellular trafficking and secretion |
| O | 51 | 2.18 | Post-translational modification, protein turnover, chaperone functions |
| C | 88 | 3.77 | Energy production and conversion |
| G | 119 | 5.09 | Carbohydrate metabolism and transport |
| E | 121 | 5.18 | Amino acid metabolism and transport |
| F | 47 | 2.01 | Nucleotide metabolism and transport |
| H | 88 | 3.77 | Coenzyme metabolism |
| I | 38 | 1.63 | Lipid metabolism |
| P | 88 | 3.77 | Inorganic ion transport and metabolism |
| Q | 21 | 0.90 | Secondary structure |
| R | 149 | 6.38 | General function prediction only |
| S | 89 | 3.81 | Function unknown |
| - | 1012 | 43.3 | Not in COG |
The total is based on the total number of protein coding genes in the genome
Insights into the genome sequence
Surface proteins encoding genes
In a previous research study [7], the authors observed that the removal of surface-associated proteins led to a marked decrease in anti-inflammatory properties. Numerous studies dealing with the anti-inflammatory properties of food or probiotic bacteria have also identified the S-layer proteins or other surface compounds responsible for this trait [8, 9].
The S-layer proteins were found to be paracrystalline mono-layered assemblies of proteins which coat the surface of bacteria. S-layer proteins were associated with the cell wall via an SLH domain, with a cell wall polymer serving as the anchoring structure. This SLH domain comprised about 40–50 amino-acids, and could be found as one or more copies in the protein. Proteins other than S-layer proteins are known to be anchored by SLH domains to the cell wall of bacteria and are called S-layer associated proteins [10]. As P. freudenreichii CIRM-BIA129 is known for its anti-inflammatory properties [7, 11], it was necessary to determine the presence of protein sequences containing SLH domains. This search led to the identification of eight genes: slpB (PFCIRM129_00700) that contained five SLH domains; slpE, (PFCIRM129_05460) containing four SLH domains; slpF (PFCIRM129_01545), slpG (PFCIRM129_09890), slpA (PFCIRM129_09350) and inlA (PFCIRM129_12235) that contained three SLH domains at the C-terminal part, and slh2 (PFCIRM129_03800) and slpD (PFCIRM129_11775), containing two SLH domains. Interestingly, CIRM-BIA129 (ITG-P20) did not present a typical, thick S-layer at its surface, so these genes most probably code for SLAPs.
Comparison with the reference strain
A tblastn analysis revealed 77 new genes (no hit) that were not present in the type strain CIRM-BIA1. Most of them were of unknown function except for four, which encoded two transporters PFCIRM129_07475 (pseudogene) and PFCIRM129_08355, an amidohydrolase PFCIRM129_03910 and a transcription factor PFCIRM129_03595. The blastp results revealed three genomic islands that were present in CIRM-BIA129 but absent from CIRM-BIA1: (i) from PFCIRM129_09365 to 09660 encompassed by conjugal transfer protein gene PFCIRM_09215 and conjugative transfer gene complex protein PFCIRM129_09665, probably corresponding to an integrative conjugal element, (ii) from _10870 to _10910 comprising several relaxase genes and an exopolyphosphatase gene PFCIRM129_10900, and (iii) from PFCIRM129_10830 to 10905 containing three alpha, beta hydrolase genes (PFCIRM129_10860, 10890, 10895).
A blastp analysis revealed a gene encoding a ribulokinase PFCIRM129_07785 which was pseudogenized in CIRM-BIA129 but appeared to be functional in CIRM-BIA1.
By contrast, the glucokinase gene PFREUD_00950 involved in gluconate degradation was pseudogenized in CIRM-BIA1 but complete in CIRM-BIA129. This difference may explain the ability of CIRM-BIA129 to degrade gluconate. In the same way, inlA (containing three SLH domains, see above) was pseudogenized in CIRM-BIA1 but complete in CIRM-BIA129. This suggests a better ability to interact with eukaryotic cells, as internalin A was described as a bacterial adhesin.
Conclusion
The genome of CIRM-BIA129 revealed new genes that had never previously been described in the species. Some of them, which probably encode surface exposed proteins, may be of considerable importance to the adaptation of the bacterium to the intestinal tract. Its ability to degrade gluconate may enable it to survive in the intestine where this sugar is abundant. Genes including the SLH domain may be candidates for the immune properties of the strain because SLH domains enable the protein to be anchored in the cell wall.
Acknowledgements
We would like to thank Actalia for supplying the CIRM-BIA129 (ITG P20) strain. Work on sequencing was funded by INRA through the “AIP-Bioressource” call for projects.
Abbreviations
- YEL
yeast extract lactate
Footnotes
Competing interests
Authors do not disclose any competing interests.
Authors’ contributions
HF extracted the DNA, annotated the genome, supervised the project and wrote the paper. SMD annotated surface proteins and provided pictures. VL cured the database and deposited the genome in the EMBL database. AH, JB and JMA assembled the genome. SP phenotyped the strain. VC stored the strain and checked its identity. VB sequenced the strain. GJ and YLL participated to the writing and provided funding for the research. All authors read and approved the final manuscript.
References
- 1.Foligne B, Breton J, Mater D, Jan G. Tracking the microbiome functionality: focus on Propionibacterium species. Gut. 2013;62:1227–1228. doi: 10.1136/gutjnl-2012-304393. [DOI] [PubMed] [Google Scholar]
- 2.Malik AC, Reinbold GW, Vedamuthu ER. Evaluation of the taxonomy of the Propionibacterium. Can J Microbiol. 1968;11:1185–1191. doi: 10.1139/m68-199. [DOI] [PubMed] [Google Scholar]
- 3.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryson K, Loux V, Bossy R, Nicolas P, Chaillou S, van de Guchte M, et al. AGMIAL: implementing an annotation strategy for prokaryote genomes as a distributed system. Nucleic Acids Res. 2006;34:3533–45. doi: 10.1093/nar/gkl471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falentin H, Deutsch S-M, Jan G, Loux V, Thierry A, Parayre S, et al. The Complete Genome of Propionibacterium freudenreichii CIRM-BIA1T, a Hardy Actinobacterium with Food and Probiotic Applications. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0011748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barinov A, Loux V, Hammani A, Nicolas P, Langella P, Ehrlich D, et al. Prediction of surface exposed proteins in Streptococcus pyogenes, with a potential application to other Gram-positive bacteria. Proteomics. 2009;9:61–73. doi: 10.1002/pmic.200800195. [DOI] [PubMed] [Google Scholar]
- 7.Foligne B, Deutsch SM, Breton J, Cousin FJ, Dewulf J, Samson M, et al. Promising immunomodulatory effects of selected strains of dairy propionibacteria evidenced in vitro and in vivo. Appl Environ Microbiol. 2010;76:8259–64. doi: 10.1128/AEM.01976-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci U S A. 2008;105:19474–8. doi: 10.1073/pnas.0810305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taverniti V, Stuknyte M, Minuzzo M, Arioli S, De Noni I, Scabiosi C, et al. S-layer protein mediates the stimulatory effect of Lactobacillus helveticus MIMLh5 on innate immunity. Appl Environ Microbiol. 2013;79:1221–31. doi: 10.1128/AEM.03056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson B, Selle K, O’Flaherty S, Goh YJ, Klaenhammer T. Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiology. 2013;159:2269–2282. doi: 10.1099/mic.0.070755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Marechal C, Peton V, Ple C, Vroland C, Jardin J, Briard-Bion V, et al. Surface proteins of Propionibacterium freudenreichii are involved in its anti-inflammatory properties. J Proteomics. 2015;113:447–61. doi: 10.1016/j.jprot.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 12.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a New Hierarchic Classification System, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 1997;47:479–491. doi: 10.1099/00207713-47-2-479. [DOI] [Google Scholar]
- 17.Whitman WB, Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Ludwig W, et al. Bergey’s Manual of Systematic Bacteriology: Volume 5a: The Actinobacteria. 2nd ed. Springer; 2012.
- 18.Zhi X-Y, Li W-J, Stackebrandt E. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int. J. Syst. Evol. Microbiol. 2009;59:589–608. doi: 10.1099/ijs.0.65780-0. [DOI] [PubMed] [Google Scholar]
- 19.Charfreitag O, Stackebrandt E. Inter- and Intrageneric Relationships of the Genus Propionibacterium as Determined by 16S rRNA Sequences. Microbiology. 1989;135:2065–2070. doi: 10.1099/00221287-135-7-2065. [DOI] [PubMed] [Google Scholar]
- 20.Judicial Commission of the International Committee on Systematics of Prokaryotes Necessary corrections to the Approved Lists of Bacterial Names according to Rule 40d (formerly Rule 46). Opinion 86. Int. J. Syst. Evol. Microbiol. 2008;58:1975. doi: 10.1099/ijs.0.2008/006015-0. [DOI] [PubMed] [Google Scholar]
- 21.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]


