Abstract
Iron is an essential nutrient for cells. It is unknown how iron, after its import into the cytosol, is specifically delivered to iron-dependent processes in various cellular compartments. Here, we identify an essential function of the conserved cytosolic monothiol glutaredoxins Grx3 and Grx4 in intracellular iron trafficking and sensing. Depletion of Grx3/4 specifically impaired all iron-requiring reactions in the cytosol, mitochondria and nucleus including the synthesis of Fe/S clusters, heme and di-iron centers. These defects were caused by impairment of iron insertion into proteins and iron transfer to mitochondria, indicating that intracellular iron is not bioavailable, despite highly elevated cytosolic levels. The crucial task of Grx3/4 is mediated by a bridging, glutathione-containing Fe/S center which functions both as an iron sensor and in intracellular iron delivery. Collectively, our study uncovers an important role of monothiol glutaredoxins in cellular iron metabolism with a surprising connection to cellular redox and sulfur metabolisms.
Keywords: Mitochondria, iron homeostasis, metallo proteins, glutathione, sulfur metabolism, ribonucleotide reductase
Introduction
Iron is essential for virtually all organisms, since it functions as a cofactor in central cellular processes such as respiration, DNA synthesis and repair, ribosome biogenesis, and metabolism. Research over the past decade has uncovered sophisticated systems facilitating the specific transport of iron across the plasma and various intracellular membranes (Hentze et al., 2004; Kaplan and Kaplan, 2009; Philpott and Protchenko, 2008; Vergara and Thiele, 2008). Despite its central metabolic function, little is known about the passage of iron through the eukaryotic cytosol to become incorporated into proteins and transported into various subcellular compartments. A soluble, low molecular-mass form of iron was described in the 1970s, but ever since the discovery of this ‘labile iron pool’ its physiological importance and composition has been under debate (Crichton and Charloteaux-Wauters, 1987; Richardson and Ponka, 1997). Presumably, iron may also be bound to dedicated proteins assuring specific delivery and insertion into iron-requiring sites. A metallo-chaperone function has been worked out for insertion of copper and nickel into respective metal-containing enzymes (Finney and O'Halloran, 2003; Lyons and Eide, 2007), but proteins performing a general role in iron trafficking or insertion are unknown. An iron donor function has been suggested for frataxin in mitochondrial Fe/S cluster biosynthesis (Bencze et al., 2006; Lill, 2009). In humans, the poly (rC) binding protein 1 (PCBP1) was shown to specifically deliver bound iron to ferritin, the major iron storage protein in higher eukaryotes (Shi et al., 2008). The apparently specific role of the PCBP1 iron chaperone and the fact that both ferritin and PCBP1 are not universally conserved leave open the possibility that other proteins with a general importance for iron trafficking exist. Clearly, the mode of specific iron delivery within the eukaryotic cytosol remains one of the fundamental unresolved problems of iron homeostasis.
Since iron is not only essential but also toxic at higher levels, cells have developed sophisticated systems for assuring a tightly regulated iron homeostasis (Hentze et al., 2004; Kaplan and Kaplan, 2009). While in mammals this process is executed by iron-regulatory proteins in a post-transcriptional fashion, the yeast S. cerevisiae uses the iron-sensing transcription factors Aft1 and Aft2. Under iron deprivation, Aft1-Aft2 activate transcription of genes of the iron regulon encoding cell surface iron transporters and proteins involved in intracellular iron utilization (Kaplan and Kaplan, 2009; Philpott and Protchenko, 2008). Sensing of intracellular iron by Aft1 also requires the regulatory proteins Fra1-Fra2, and the cytosolic-nuclear monothiol glutaredoxins Grx3 and Grx4, which are essential for the nuclear export of Aft1 in response to iron sufficiency (Kumanovics et al., 2008; Ojeda et al., 2006; Pujol-Carrion et al., 2006). The regulatory role of Grx3/4 is functionally conserved in fungi that utilize iron-regulated transcription systems unrelated to those from S. cerevisiae (Haas et al., 2008; Kaplan and Kaplan, 2009; Mercier and Labbe, 2009).
Grx3/4 belong to the large thioredoxin (Trx) fold family, and are composed of an N-terminal Trx and a C-terminal monothiol glutaredoxin (Grx) domain (Herrero and de la Torre-Ruiz, 2007; Lillig et al., 2008). Although the Grx3/4 subfamily of multi-domain monothiol glutaredoxins is conserved in eukaryotes, no universal function has been assigned to this family so far. In contrast to most members of the Grx protein family that catalyze dithiol-disulfide redox reactions, monothiol Grx proteins rarely possess oxidoreductase activity. Instead, after in vitro reconstitution or upon overexpression in E. coli they are capable of binding a bridging [2Fe-2S] cluster utilizing the active-site cysteine residue of the Grx domain and glutathione (GSH) as ligands (Li et al., 2009; Picciocchi et al., 2007). The existence of this unusual Fe/S center under physiological conditions, however, has not been demonstrated, and its functional role has remained unclear.
Here, we have used yeast as a model to define an essential role of Grx3/4 in intracellular iron trafficking. Depletion of Grx3/4 led to functional impairment of virtually all iron-dependent processes including heme biosynthesis, mitochondrial and cytosolic Fe/S protein biogenesis, and the formation of di-iron centers in mitochondria and the cytosol, eventually leading to the loss of cell viability. We provide evidence for the in vivo binding of a bridging Fe/S cluster to Grx3/4, and we assign a crucial physiological function to this cofactor both in cytosolic iron trafficking and as an iron sensor. Thus, the conserved cytosolic monothiol glutaredoxins use their bound Fe/S cofactor for a general role in intracellular iron trafficking.
Results
Deficiency in Grx3/4 is associated with defects in iron-dependent enzymes
To facilitate the functional analysis of Grx3/4, we constructed a regulatable yeast strain (Gal-GRX4; strain background W303-1A; Table S1) in which GRX3 was deleted and GRX4 was expressed under the control of the glucose-repressible GAL-L promoter. Upon Grx4 depletion Gal-GRX4 cells failed to grow on both fermentable and non-fermentable carbon sources (Figs. 1A and 3C). Likewise, double deletion of GRX3-GRX4 was lethal in the W303 strain background, distinguishing these cells from strain BY4742 grx3/4Δ which shows only severely retarded growth (Fig. 1A) (Ojeda et al., 2006). The strong effect of Grx3/4 deficiency on cell viability is not explained by their role in iron regulation, since Aft1 is not essential under iron-replete conditions (Kaplan and Kaplan, 2009). These data and the general conservation of Grx3/4 in eukaryotes suggest that these proteins perform a so far unknown, important function.
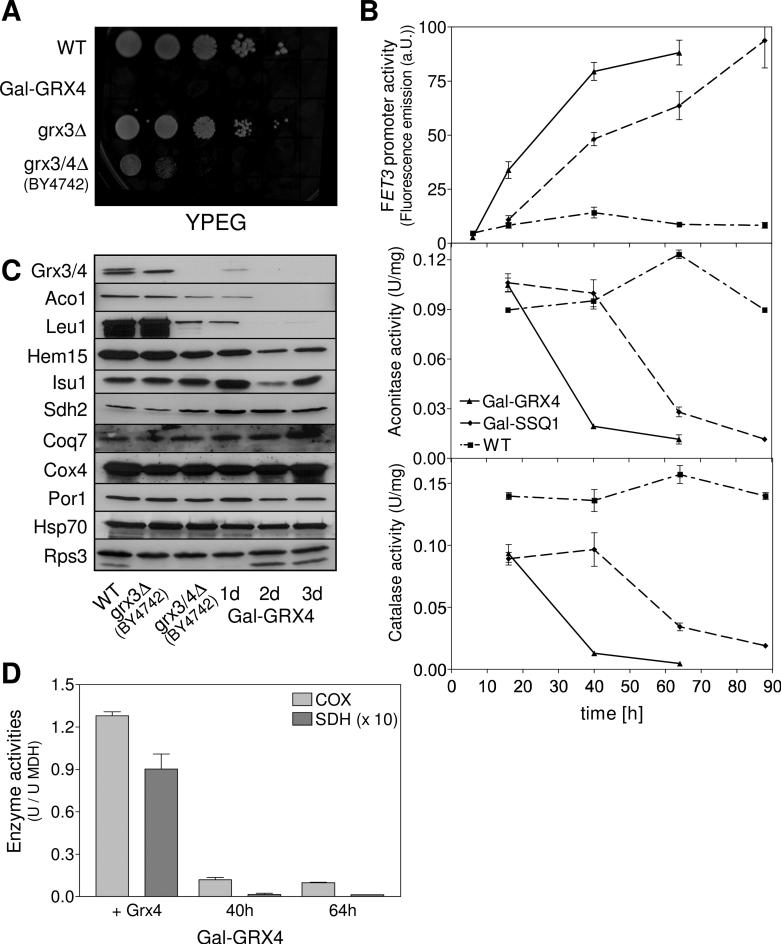
Fig. 1. Deficiency in Grx3/4 is associated with defects in iron-dependent enzymes.
(A) Wild-type (WT), Gal-GRX4, grx3Δ (strain background W303-1A), and grx3/4Δ (strain background BY4742) were grown in SD medium for 40 h. Tenfold serial dilutions were spotted onto YPEG plates and incubated for 2 days at 30°C. (B) WT, Gal-GRX4 and Gal-SSQ1 strains harbouring plasmid pFET3-GFP were grown in SD minimal medium. At the indicated times, FET3 promoter activities were determined by measuring the GFP-specific fluorescence emission of cells, and cell extracts were assayed for aconitase and catalase activities, or (C) analyzed for the indicated proteins by immunostaining. (D) Enzyme activities of respiratory complexes II (SDH) and IV (COX) were determined relative to malate dehydrogenase (MDH) in mitochondria isolated from Gal-GRX4 cells cultivated in rich glucose medium for 40 h and 64 h, and from Gal-GRX4 cells expressing GRX4 from vector pCM189 (+Grx4). Error bars indicate the SEM (n ≥ 4).
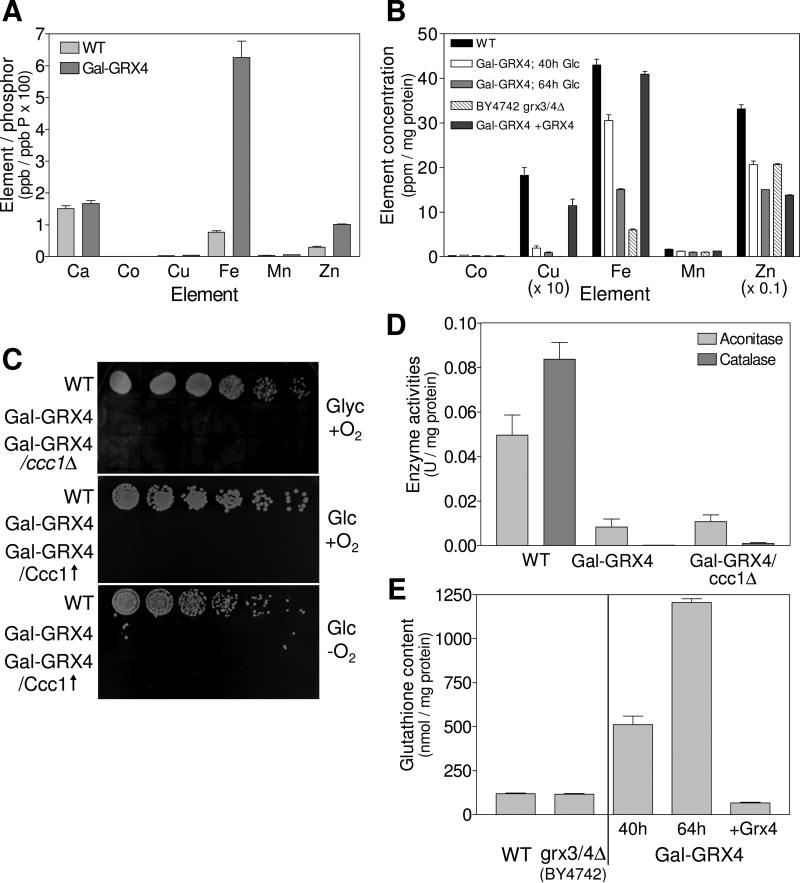
Fig. 3. Deficiency in Grx3/4 results in cytosolic iron and GSH accumulation.
The metal content of (A) wild-type (WT) and Gal-GRX4 cells (depleted for 64 h) and (B) mitochondria isolated from the indicated strains was determined by ICP-MS. (C) The indicated strains lacking (ccc1Δ) or overproducing Ccc1 (Ccc1↑) were cultivated in SD medium for 40 h. Tenfold serial dilutions were spotted onto SC agar plates containing glycerol (Glyc) or glucose (Glc), and cultivated at 30°C under aerobic (+O2) or anaerobic (−O2) conditions. (D) WT, Gal-GRX4 and Gal-GRX4/ccc1Δ cells were grown in SD medium for 64 h, and aconitase and catalase enzyme activities were determined. (E) GSH levels were determined in cell extracts from WT, BY4742 grx3/4Δ, Gal-GRX4 (depleted for 40 h or 64 h) and Gal-GRX4 cells expressing GRX4 from a plasmid (+Grx4).
Gal-GRX4 cells were used to investigate the immediate consequences of Grx3/4 deficiency. Gal-GRX4 cells were cultivated in minimal medium supplemented with glucose and iron chloride to gradually deplete Grx4 (Figs. 1B and 1C). A strong activation of the Aft1-dependent FET3 gene was observed using a FET3 promoter-GFP fusion as a reporter (Fig. 1B; (Ojeda et al., 2006)). Surprisingly, the activities of the mitochondrial Fe/S protein aconitase and cytosolic catalase, a heme-containing protein, drastically decreased, despite the presumed sufficient cellular iron supply. These effects resemble those upon depletion of Ssq1, a component the iron-sulfur cluster (ISC) assembly machinery, even though the changes occurred later due to slower depletion of Ssq1 (strain Gal-SSQ1). Grx3/4 deficiency was associated with a severe activity loss of respiratory complexes II (succinate dehydrogenase) and IV (cytochrome oxidase), but was fully complemented by expression of GRX4 from a plasmid (Fig. 1D). Likewise, low activities of both aconitase and respiratory complexes III and IV were observed in BY4742 grx3/4Δ cells (Fig. S1A), consistent with our earlier observation of an impaired 55Fe/S cluster incorporation into aconitase (Ojeda et al., 2006). Immunostaining of cell extracts from Grx4-depleted Gal-GRX4 cells and BY4742 grx3/4Δ cells further revealed changes in the levels of several iron-containing proteins including the aconitase-type Fe/S proteins Aco1 and Leu1, ferrochelatase Hem15, and the core mitochondrial ISC assembly protein Isu1 (Fig. 1C). These changes of protein levels correlate with those of the transcriptome of both iron-deprived and ISC machinery-compromised cells (Hausmann et al., 2008; Shakoury-Elizeh et al., 2004). In contrast, other iron-dependent proteins such as succinate dehydrogenase subunit 2 (Sdh2), and the ubiquinone biosynthesis enzyme Coq7 were hardly changed, and behaved similarly to the non-iron proteins mitochondrial cytochrome oxidase subunit 4 (Cox4) and porin (Por1), cytosolic Hsp70 and ribosomal subunit Rps3. Together, these findings indicate that Grx3/4-deficient cells develop severe defects in several mitochondrial and cytosolic iron-dependent proteins, despite the induction of the Aft1-dependent iron uptake system. Notably, these global iron-related defects are not detected upon the constitutive activation of Aft1 (Hausmann et al., 2008; Ihrig et al., 2009), suggesting that these consequences of Grx3/4 deficiency occur independently of Aft1 and a deregulated iron homeostasis.
Deficiency in Grx3/4 impairs the de novo synthesis of cellular Fe/S clusters and heme
We asked whether the decreased Fe/S protein activities in Grx3/4-depleted cells might be explained by an impaired de novo synthesis of their Fe/S clusters, and addressed this problem by using an established 55Fe radiolabelling and immunoprecipitation assay (Molik et al., 2007). First, the essential cytosolic Fe/S proteins Rli1, Dre2, and Nar1 were analyzed by expressing these proteins from a high-copy vector in wild-type and Gal-GRX4 cells. Fe/S cluster insertion into all three Fe/S protein targets was decreased 4-10-fold upon Grx4 depletion (Fig. 2A). The amount of Dre2 in Gal-GRX4 cells was comparable to that in wild-type cells, indicating a specific Fe/S cluster assembly defect (insert). In the case of Rli1 and Nar1, protein levels were diminished likely suggesting that the apoforms of these Fe/S proteins were degraded. Similar apoprotein instability is frequently observed upon strong defects in Fe/S protein biogenesis (Balk et al., 2004). Analysis of 55Fe incorporation into the mitochondrial Fe/S proteins Bio2 (biotin synthase) and Ilv3 (dihydroxyacid dehydratase), and the essential mitochondrial ISC scaffold protein Isu1 revealed an up to 4-fold lower 55Fe incorporation upon Grx4 depletion (Figs. 2B and 2C). Protein levels of Bio2, Ilv3, and Isu1 did not change upon depletion of Grx4 (inserts). These findings indicate a general impairment in the de novo assembly of Fe/S proteins upon depletion of Grx3/4.
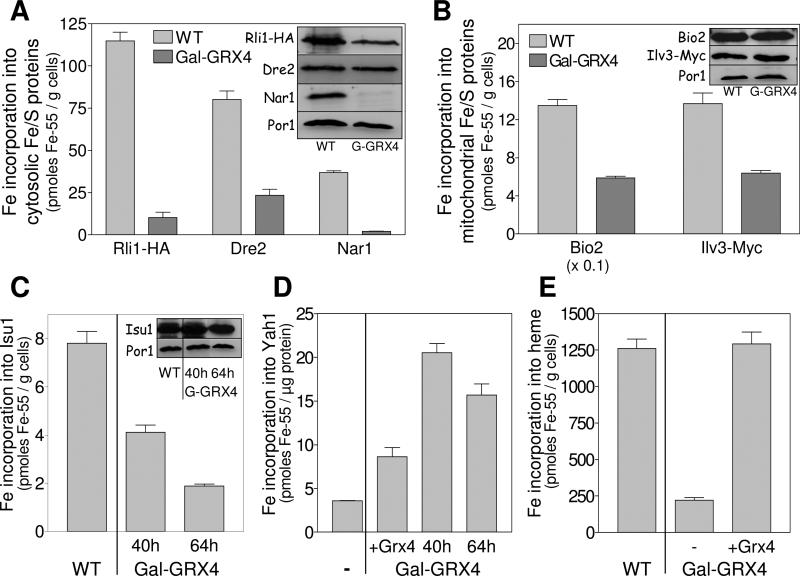
Fig. 2. Deficiency in Grx3/4 impairs the de novo synthesis of cellular Fe/S clusters and heme.
(A-C) Wild-type (WT) and Grx4-depleted Gal-GRX4 cells overproducing the cytosolic Fe/S proteins Rli1-HA, Dre2 or Nar1 (A), mitochondrial Bio2, Ilv3-Myc (B), or Isu1 (C) were radiolabeled with 10 μCi 55Fe for 2 h. The Fe/S proteins were immunoprecipitated and bound 55Fe was quantified by scintillation counting. Protein levels were assessed by immunostaining. Porin (Por1) served as a loading control. Gal-GRX4 cells were depleted for Grx4 by growth in SD medium for 40 h, and 64 h (in case of Isu1). (D) Purified apo-Yah1 was incubated under anaerobic conditions in the presence of 55Fe and cysteine either alone (−) or with detergent extracts of mitochondria isolated from 40 h or 64 h depleted Gal-GRX4 cells or Gal-GRX4 cells overproducing Grx4 (+Grx4). 55Fe/S cluster reconstitution on re-isolated Yah1 was quantified by scintillation counting. (E) WT and Gal-GRX4 cells (40 h depletion) harboring either vector pCM189 (−) or pCM189-GRX4 (+Grx4) were radiolabeled with 55Fe. 55Fe-heme was extracted with butyl-acetate and quantified by scintillation counting. Error bars indicate the SEM (n ≥ 4).
In principle, the defect of Fe/S protein maturation upon Grx3/4 depletion could be explained by a primary mitochondrial Fe/S protein assembly defect, since mitochondria are involved in generation of all cellular Fe/S proteins (Lill and Muhlenhoff, 2008). However, we note that the observed effects were less severe in mitochondria compared to the cytosol. To directly test the functionality of the mitochondrial ISC assembly machinery, we made use of an anaerobic in vitro system analyzing the capacity of mitochondrial detergent extracts to support Fe/S cluster insertion into the apoform of purified Yah1, a [2Fe-2S] ferredoxin (Molik et al., 2007). Apo-Yah1 was incubated with mitochondrial lysates and 55Fe, bound to Q-sepharose, and the amount of bound radioactivity was quantified by scintillation counting. Remarkably, extracts derived from Grx3/4-depleted mitochondria were even more competent in synthesizing the Fe/S cluster on Yah1 than Grx4-complemented Gal-GRX4 cells (Fig. 2D). This finding strongly suggests that the mitochondrial ISC assembly system is functional in Grx3/4-depleted cells rendering it likely that the decreased Fe/S cluster incorporation into apoproteins is explained by impaired iron supply.
Consistent with this idea, 55Fe insertion into heme was 5.5-fold lower in Grx4-depleted Gal-GRX4 cells compared to wild-type cells or Gal-GRX4 cells complemented by GRX4 (Fig. 2E). This diminished heme synthesis activity may explain the loss of function of heme-dependent enzymes such as catalase and cytochrome oxidase upon depletion of Grx3/4 (see above). In summary, Grx3/4-depleted cells are strongly impaired in both cellular Fe/S protein maturation and heme biosynthesis. Such defects are not observed in Aft1-activated cells (Hausmann et al., 2008; Ihrig et al., 2009).
Deficiency in Grx3/4 leads to impairment of di-iron enzymes despite cytosolic iron overload
The strong decrease of cellular Fe/S clusters and heme in Grx3/4-deficient cells is somewhat paradoxical, since these cells are expected to accumulate iron due to a constitutively activated cellular iron uptake system (Ojeda et al., 2006). To verify this, we measured the cellular iron content by ICP-MS analysis of wild-type and Grx4-depleted Gal-GRX4 cells grown in minimal medium supplemented with 100 μM FeCl3. Total cellular iron increased 6-fold upon depletion of Grx4 (Fig. 3A). The level of chelatable iron increased similarly and was mainly present as Fe2+ (Fig. S2). Cellular levels of other metals, with the exception of Zn (3-fold higher), were hardly changed. In contrast, mitochondrial iron levels were up to 2.3-fold lower in Grx4-depleted Gal-GRX4 cells compared to wild-type (Fig. 3B). Mitochondria from BY4742 grx3/4Δ cells contained even 7.5-fold less iron. Mitochondrial Mn, Co and Zn levels were hardly altered, but Cu changed in parallel to iron. The decrease in mitochondrial iron levels in Grx3/4-depleted cells is the more remarkable, as cells with mitochondrial Fe/S protein assembly defects usually display strongly elevated mitochondrial iron levels (Lill and Muhlenhoff, 2008). The fact that this was not observed, despite high levels of total cellular iron, indicates a defective delivery of iron to mitochondria in Grx3/4-deficient cells.
A reasonable explanation for these general defects in iron handling in the absence of Grx3/4 may be a sequestration of iron into the vacuole, the major iron storage compartment in fungi (Kaplan and Kaplan, 2009; Philpott and Protchenko, 2008). To test this, we varied the amount of Ccc1, the major importer of divalent metals into the vacuole. Deletion of CCC1 did not restore growth of Grx4-depleted Gal-GRX4 cells and did not increase the low enzyme activities of aconitase and catalase (Fig. 3C, top panel and Fig. 3D). Similarly, overproduction of Ccc1 failed to restore growth (Fig. 3C, middle). These data suggest that the accumulated iron is not stored in the vacuole.
Grx4-depleted Gal-GRX4 cells failed to grow under anaerobic conditions (Fig. 3C, bottom). Thus, reactive oxygen species (due to increased iron levels) are not responsible for the lethal phenotype of Grx3/4-depleted cells. Moreover, oxidized glutathione levels (GSSG; measured under anaerobic conditions) were below the detection limit (not shown). Rather, reduced glutathione (GSH) was strongly elevated in Grx4-depleted Gal-GRX4 cells, but not in BY4742 grx3/4Δ (Fig. 3E). Together, these results and the predominant presence of iron in its ferrous form (Fig. S2) indicate that reducing conditions prevail in Grx3/4-deficient cells.
The experiments presented above showed a maturation defect in cellular Fe/S and heme proteins in Grx3/4-deficient cells despite a cytosolic iron accumulation. Since this indicated a defective delivery of iron, we asked whether other iron-dependent enzymes were affected by Grx3/4 deficiency. First, the iron status of ribonucleotide reductase (Rnr), a cytosolic diferric-tyrosyl radical enzyme essential for deoxyribonucleotide synthesis, was analyzed (Perlstein et al., 2005). Upon depletion of Grx4 in Gal-GRX4 cells the protein levels of Rnr2 decreased slightly (Fig. S3). Nevertheless, the specific activity of Rnr was 6-fold lower compared to wild-type cells (Fig. 4A). This was likely due to inefficient metallation, since 55Fe insertion into Rnr in vivo (followed by immunoprecipitation of subunit Rnr2) was 5-7-fold decreased in Grx4-depleted cells (Fig. 4B). As a second di-iron protein, we analyzed the mitochondrial mono-oxygenase Coq7 which catalyses the hydroxylation of demethoxyubiquinol (DMQ6), the penultimate reaction of ubiquinone (CoQ6) biosynthesis (Tran et al., 2006). HPLC analysis of mitochondrial lipid extracts revealed diminished CoQ6 and increased DMQ6 levels upon Grx4 depletion (Fig. 4C). This effect was reversed to wild-type ratios by expression of GRX4 from a plasmid (Fig. 4D). The simultaneous accumulation of the substrate (DMQ6) and decrease of the product (CoQ6) of the enzyme Coq7 indicates a diminished activity of this di-iron mono-oxygenase upon depletion of Grx3/4. Collectively, these data demonstrate that a Grx3/4 deficiency causes a severe defect in cellular di-iron enzymes.
Fig. 4. Deficiency in Grx3/4 leads to functional impairment of di-iron enzymes.
(A) Permeabilised wild-type (WT) and Grx4-depleted Gal-GRX4 cells were assayed for specific ribonucleotide reductase activity (see also Fig. S3). (B) WT and Gal-GRX4 cells were grown in SD medium for 40 and 64 h, radiolabeled, and 55Fe binding to Rnr2 was analyzed by immunoprecipitation and scintillation counting. Protein levels of Rnr2 and Por1 were determined by immunoblotting (insert). (C) The substrate (demethoxyubiquinol DMQ6) and product (ubiquinone CoQ6) of mitochondrial mono-oxygenase Coq7 were analyzed by electrochemical detection coupled to HPLC separation of mitochondrial lipid extracts from WT and Grx4-depleted Gal-GRX4 cells. CoQ4 is a commercial standard. (D) Ratio of DMQ6 and CoQ6 levels in mitochondria isolated from WT, Gal-GRX4 cells (depleted for 40 h or 64 h) containing either vector pCM189 or pCM189-GRX4 (+Grx4). (E) WT and Grx4-depleted Gal-GRX4 cells overproducing Fe-only superoxide dismutase from E. coli (FeSod) were analyzed for superoxide dismutase in-gel activities and FeSod by immunoblotting.
Are the observed defects in Grx3/4-depleted cells specific for iron-dependent proteins? This question was addressed by analyzing the activities and protein levels of several metal-dependent enzymes. The in-gel activities of the endogenous Cu/Zn- and Mn-dependent superoxide dismutases (Sod1 and Sod2, respectively) remained unchanged in Grx4-depleted Gal-GRX4 cells (Fig. 4E). In marked contrast, both the level and activity of ectopically expressed iron-only superoxide dismutase (FeSod) from E. coli strongly declined. Further, the Zn-dependent alcohol dehydrogenase (ADH) was 3-fold more active upon Grx4 depletion (Fig. S1B). This increased ADH activity is characteristic for a switch towards fermentative metabolism and is typically observed upon iron deprivation. The normal function of several metal-reliant enzymes indicates that the described defects in Grx3/4-depleted cells are specific for iron-related processes. We conclude that Grx3/4 perform an essential role in cellular iron trafficking, in addition and independently of their non-essential function in iron uptake regulation.
Grx3/4 assemble a bridging Fe/S cluster independently of the CIA machinery
For a function of monothiol glutaredoxins in cellular iron trafficking, iron-binding may be a necessary prerequisite. Recent in vitro studies have shown that various glutaredoxins, including yeast Grx3/4, can bind a bridging, GSH-liganded [2Fe-2S] cluster (Bandyopadhyay et al., 2008; Berndt et al., 2007; Johansson et al., 2007; Li et al., 2009). We asked whether this unusual Fe/S cofactor is of physiological relevance and can be observed in a native environment using the 55Fe radiolabelling assay. Significant amounts of 55Fe were co-immunoprecipitated with anti-Grx4-antibodies from cell extracts derived from wild-type cells (strain BY4742), while only background levels were observed in grx3/4Δ cells (Fig. 5A). Similar amounts of 55Fe were co-immunoprecipitated from both grx3Δ and grx4Δ cells, indicating that the homologous Grx3 and Grx4 bind iron with similar efficiency and independently of each other. The amount of Grx4-bound 55Fe was unchanged both in wild-type cells of our standard strain W303-1A, and, remarkably, under anaerobic or oxidative stress conditions prevailing after addition of H2O2 or upon deletion of SOD1 (sod1Δ cells) indicating that iron binding to Grx3/4 was insensitive to oxidative stress (Fig. 5B).
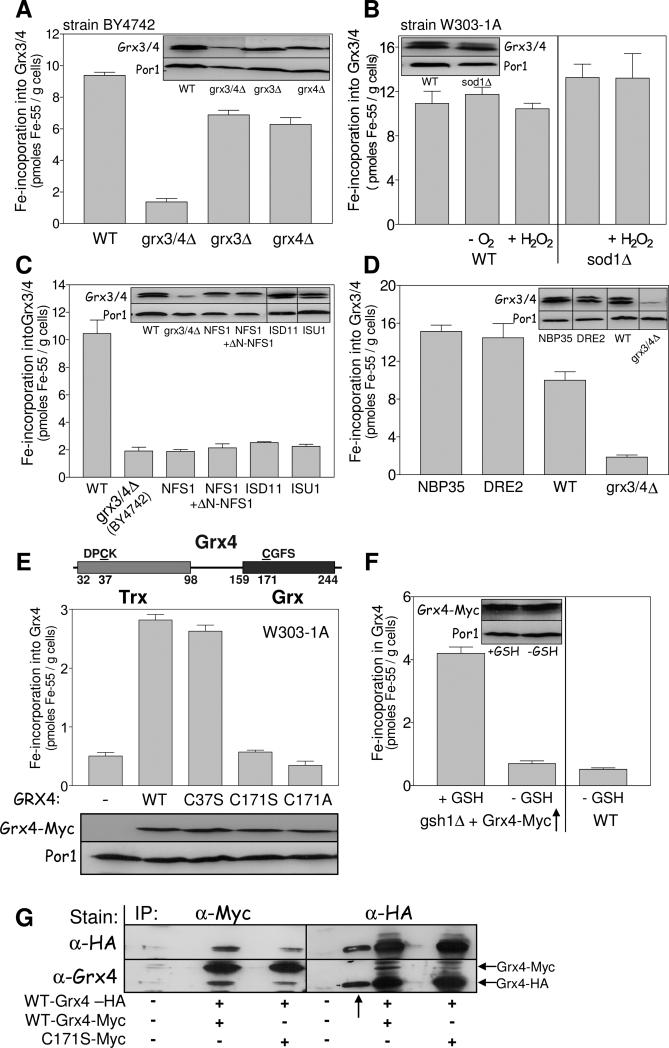
Fig. 5. Grx3/4 assemble a bridging Fe/S cluster independently of the CIA machinery.
(A) 55Fe binding to Grx3/4 was determined in wild-type (WT), grx3Δ, grx4Δ and grx3/4Δ cells (strain background BY4742). Grx3/4 protein levels were assessed by immunostaining. Por1 served as a loading control. The anti-Grx4 antiserum recognizes an unspecific band, in addition to Grx3/4. (B) 55Fe binding to Grx3/4 was determined in WT and sod1Δ cells (strain background W303-1A). Where indicated, cells were pretreated with 1 mM H2O2 for 10 min or examined under anaerobic (−O2) conditions. (C, D) 55Fe binding to Grx3/4 was determined in WT, Gal-NFS1 (with and without a plasmid expressing a cytosolic version of Nfs1 (+ΔN-NFS1)), Gal-ISD11, Gal-ISU1 (C) or Gal-NBP35 and Gal-DRE2 cells (D) after growth in SD medium for 40 h. BY4742 grx3/4Δ cells served as a background control. Grx3/4 and Por1 levels were assessed by immunostaining. (E) Top: Schematic presentation of the domain structure of Grx4. Middle: C-terminally Myc-tagged WT, C37S, C171S, or C171A mutant Grx4 were expressed in WT cells from vector pCM189, and 55Fe binding to Grx4 was determined by radiolabelling and immunoprecipitation with α-Myc beads. Bottom: Grx4-Myc and Por1 levels were assessed by immunostaining. (F) WT and gsh1Δ cells overproducing Grx4-Myc were cultivated in the presence or absence of 1 mM GSH for 64 h (Sipos et al., 2002). 55Fe binding to Grx4-Myc was determined by immunoprecipitation with α-Myc beads. (G) Extracts of WT cells expressing C-terminally HA-tagged Grx4 and wild-type or C171S Grx4-Myc as indicated were subjected to immunoprecipitation (IP) with α-Myc or α-HA-antibodies. The precipitate was analyzed by immunostaining with α-HA or α-Grx4 antibodies. The arrow (↑) indicates a lane with a molecular mass marker which is stained by the α-HA antiserum.
To analyze whether the iron bound to Grx3/4 is part of an Fe/S cluster, we investigated the requirement of 55Fe binding for components of Fe/S protein biogenesis (Lill and Muhlenhoff, 2008). We first analyzed the role of the cysteine desulfurase complex Nfs1-Isd11 which serves as the sulfur donor for Fe/S protein biogenesis. For regulated depletion of Nfs1 or Isd11 we used Gal-NFS1 or Gal-ISD11 cells, respectively. 55Fe binding to Grx4 (Fig. 5C) and to the cytosolic Fe/S protein Leu1 as a control (Fig. S4) declined to background levels upon depletion of either Nfs1 or Isd11. The same result was seen in Gal-ISU1Δisu2 cells depleted for the core ISC scaffold proteins Isu1/2. No recovery of iron binding to Grx3/4 was observed, when a cytosolic-nuclear version of Nfs1 (ΔN-Nfs1) was produced in Gal-NFS1 cells lacking mitochondrial Nfs1 (Fig. 5C). We found during these experiments that the Grx4 protein levels decreased upon Nfs1 depletion, while the Grx3 levels remained unchanged. While the reason for this specific decrease is unknown, we note that the remaining Grx3 did not bind 55Fe above background levels, unlike in grx4Δ cells (cf. Fig. 5A). The dependence of iron binding to Grx3/4 on core components of the mitochondrial ISC assembly system demonstrates that Grx3/4 bind an Fe/S cluster in vivo.
Does incorporation of the Fe/S cluster into Grx3/4 also require the cytosolic iron-sulfur protein assembly (CIA) system (Lill, 2009)? Depletion of the essential CIA components Nbp35 and Dre2 in the respective GAL promoter-exchange mutants significantly increased rather than impaired 55Fe binding to Grx3/4 (Fig. 5D), while hardly any 55Fe binding was observed to Leu1 as a control (Fig. S4). This surprising finding suggests that incorporation of the Fe/S cluster into Grx3/4, while requiring mitochondrial Nfs1-Isd11-Isu1, does not depend on the cytosolic CIA machinery.
The non-canonical pathway used for Fe/S cluster assembly on Grx3/4 may be due to the special nature of this cofactor, with a conserved Cys and GSH serving as ligands of a [2Fe-2S] cluster bridged between two Grx monomers, as observed after in vitro reconstitution or after overexpression in E. coli (Li et al., 2009; Picciocchi et al., 2007). We therefore sought to obtain in vivo evidence for this configuration. First, we determined which ligands might coordinate the Fe/S cluster. The importance of the conserved active-site cysteine residues C171 in the C-terminal Grx domain and C37 in the N-terminal Trx domain (Fig. 5E, top) for 55Fe binding was tested in wild-type cells expressing a plasmid-encoded Myc-tagged Grx4 (Grx4-Myc) in which these residues were mutated to serine (mutant proteins C171S and C37S) or alanine (C171A). Myc-tagged wild-type Grx4 was fully functional (see below), and was produced at similar levels as the mutant proteins (Fig. 5E, bottom). Mutant proteins C171S and C171A did not bind 55Fe above background levels (Fig. 5E, middle). In contrast, mutant protein C37S bound wild-type amounts of 55Fe suggesting that iron is bound via C171. Second, the importance of GSH for 55Fe binding to Grx3/4 in vivo was tested using the GSH synthesis-deficient mutant gsh1Δ that can be depleted for GSH upon growth in media lacking GSH. While significant amounts of 55Fe were bound to Grx4-Myc in the presence of exogenously added GSH, only background levels of 55Fe were found in GSH-deprived cells (Fig. 5F). Together, these findings suggest that the active-site C171 of the Grx domain and GSH serve as ligands of the Grx3/4-bound Fe/S cluster.
Finally, we tried to find in vivo evidence for the presence of a bridging Fe/S cluster on Grx3/4. A yeast strain was constructed that simultaneously expressed C-terminally HA- and Myc-tagged Grx4. Cell extracts were subjected to immunoprecipitation with anti-HA or anti-Myc immunobeads followed by immunoblotting. Immunoprecipitation with anti-HA antibodies led to co-isolation of Grx4-HA and a smaller amount of Grx4-Myc (Fig. 5G). The same result, yet with inversed intensities was observed using anti-Myc beads, while no cross-reacting bands were visible in wild-type cells. In cells expressing a C171S mutant Grx4-Myc and wild-type Grx4-HA, co-immunoprecipitation was far less efficient, but still detectable. These observations document the importance of residue C171 for efficient Grx4 dimer formation, and thus are consistent with the idea of a bridging Fe/S cluster between two Grx monomers. Collectively, our findings suggest that under physiological conditions Grx3/4 bind a bridging Fe/S cluster that is coordinated by the active-site cysteine and GSH consistent with the structure of glutaredoxins reconstituted in vitro.
The Grx3/4-bound Fe/S cluster is important for iron metabolism
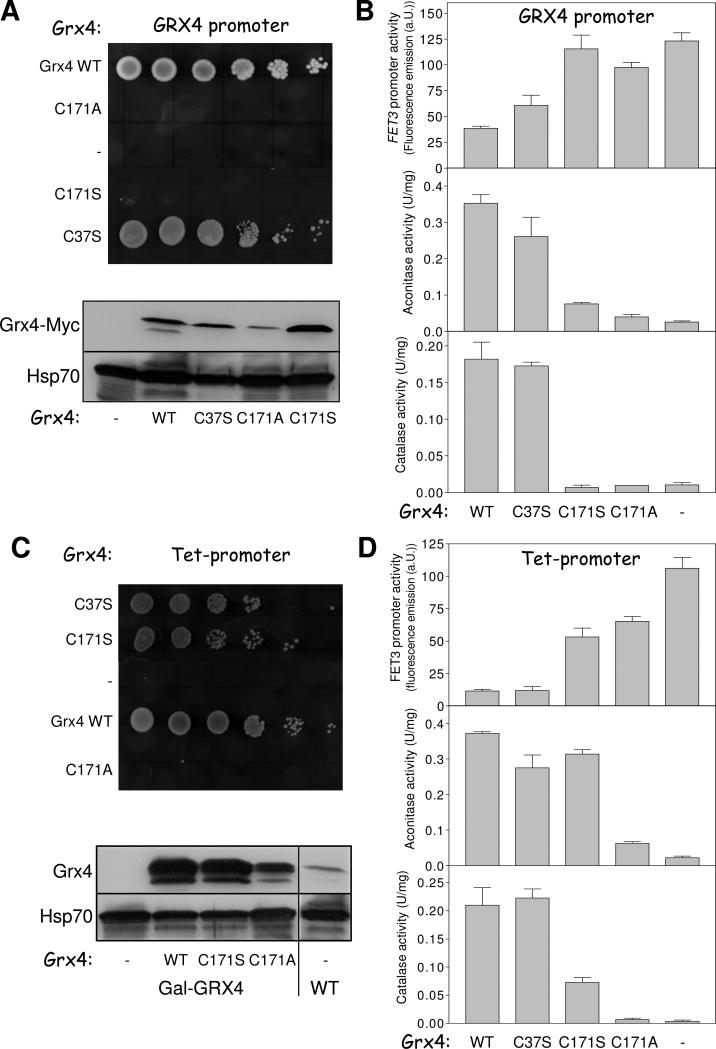
Is the Grx3/4-bound Fe/S cluster important for the function of Grx3/4 in iron trafficking and is this cofactor also required for iron sensing? As shown above the C171S and C171A Grx4-Myc mutant proteins have lost the ability to stably bind the Fe/S cluster (Fig. 5E). Grx4-depleted Gal-GRX4 cells expressing these mutant proteins from the endogenous GRX4 promoter failed to grow, whereas wild-type and C37S Grx4-Myc that retained normal iron binding supported wild-type growth (Fig. 6A, top). Aft1-dependent transcription of FET3 was fully activated and virtually no aconitase and catalase activities were observed in Gal-GRX4 cells producing C171S and C171A Grx4-Myc instead of wild-type Grx4 (Fig. 6B). In contrast, wild-type and C37S Grx4-Myc-expressing Gal-GRX4 cells showed wild-type signals in these assays. All Grx4-Myc proteins were present at similar levels, with the exception of C171A which apparently was less stable (Fig. 6A, bottom). These results demonstrate the crucial role of residue C171 and thus the Grx3/4-bound Fe/S cluster for both iron regulation and iron trafficking.
Fig. 6. The active-site cysteine of the Grx domain is essential for iron metabolism.
Gal-GRX4 cells lacking (−) or expressing wild-type (WT) Grx4-Myc or the Grx4-Myc mutant proteins C37S, C171S or C171A under the control of the GRX4 (A) or tetO7 (C) promoter from vector pCM189 were cultivated in SD medium. After 40 h, tenfold serial dilutions were spotted onto SC medium with glycerol. The levels of Grx4-Myc, endogenous Grx4, and cytosolic Hsp70 were assessed by immunostaining. (B, D) After 64 h cells from A and C were analyzed for FET3 promoter activities, or aconitase and catalase enzyme activities.
Formally, it is possible that the active-site C171 performs its essential role in Grx4 via thiol-dependent redox chemistry (Herrero and de la Torre-Ruiz, 2007; Lillig et al., 2008) rather than by coordination of the Fe/S cluster. Such a function can be excluded from the observation that overexpression (from a Tet promoter) of the C171S (but not C171A) Grx4-Myc mutant protein in Grx4-depleted Gal-GRX4 cells restored wild-type growth (Fig. 6C). We noted that Grx4 overexpression generally diminished FET3 induction (Fig. 6D). The corresponding C171S cells still displayed strong FET3 activation, yet wild-type aconitase and strongly increased catalase activities. The restored iron loading of these and presumably other iron-dependent proteins may explain why the C171S cells grow normally, despite a still disturbed iron regulation. Consistent with this interpretation, C171A cells did not show any recovery of enzyme activities. This striking result for C171S Grx4-Myc may be explained on the basis of earlier observations showing that, in some cases, Ser (but not Ala) may function as a (weak) ligand for iron (da Silva and Williams, 2001; Johnson et al., 2005). We therefore suppose that the C171S Grx4 still binds an Fe/S cluster, thus stabilizing the protein and maintaining its function in iron trafficking at high protein levels (Figs. 6A and 6C). However, the Fe/S cluster is bound in a fashion too labile to be isolated by our co-immunoprecipitation method, and to function properly in iron regulation. In summary, these data indicate that the essential function of residue C171 cannot be explained by thiol-dependent redox reactivity but rather by coordination of the bridging Fe/S cluster which was characterized here to perform an essential function in both intracellular iron trafficking and sensing.
Discussion
Our analysis of the cytosolic-nuclear monothiol glutaredoxins Grx3 and Grx4 provides strong evidence for an essential and general role of these proteins in intracellular iron trafficking, a so far poorly understood process (Fig. S5). This function of Grx3/4 is additional to their involvement in Aft1-dependent iron sensing, since the phenotypes of Grx3/4-depleted and Aft1-activated cells differ markedly in that only the former cells display global and severe biochemical defects in multiple iron-dependent enzymes (Hausmann et al., 2008; Ihrig et al., 2009). For instance, the function of cellular Fe/S-, heme- and other iron-containing proteins is strongly impaired upon depletion of Grx3/4, but not affected when Aft1 is activated. Many of these proteins are involved in essential processes such as DNA synthesis and ribosome function, explaining why Grx3/4-depleted cells eventually loose their viability. The two functions of Grx3/4 in iron trafficking and sensing are mediated by their crucial cofactor, a GSH-liganded and bridging [2Fe-2S] center. Destruction of this Fe/S center elicits a highly similar phenotype as the depletion of the Grx proteins. The observed biochemical consequences of Grx3/4 depletion are specific for iron, since the functions of other metal-dependent enzymes such as Cu,Zn- and Mn-dependent superoxide dismutases were not affected. Together, our findings demonstrate a crucial role of Grx3/4 in intracellular iron delivery from the cytosolic labile iron pool to virtually all iron-binding proteins or pathways in the cell (Fig. S5). Hence, Grx3/4 represent long-sought general factors that facilitate the proper assembly of various iron centers in proteins and cofactors. Grx3/4 are unlikely to make specific and direct contacts with the potential iron-dependent targets. Therefore, they may not function as classical metallo-chaperones that deliver their metal specifically to individual targets (Finney and O'Halloran, 2003; Lyons and Eide, 2007). Rather, the iron activated by Grx3/4 may be utilized by dedicated assembly factors such as components of the mitochondrial ISC or cytosolic CIA systems (Lill and Muhlenhoff, 2008) or the cytosolic ferritin-specific iron chaperone PCBP1 (Shi et al., 2008) for proper insertion into recipient apo-proteins.
Several observations make it unlikely that the severe iron trafficking defect in the absence of Grx3/4 is related to oxidative stress damage, even though, in principle, Fenton chemistry is possible in Grx3/4-depleted cells due to their massive accumulation of intracellular ferrous iron. First, the lethal phenotype of Grx3/4-depleted cells was not cured under anaerobic conditions, and the activities of iron-dependent enzymes were not recovered. Second, BY4742 grx3/4Δ cells display only weak signs of oxidative stress, possibly because the cytosolic redox balance in yeast is maintained by the dithiol glutaredoxins Grx1 and Grx2 (Rodriguez-Manzaneque et al., 1999). Third, binding of the essential Fe/S center to Grx3/4 was rather insensitive to oxidative stress. Finally, Grx3/4-depleted cells show hyper-accumulation of reduced GSH with no signs for an increase in oxidized GSSG.
The observed cytosolic iron trafficking defect in Grx3/4-deficient cells cannot be explained by low levels of iron in the cytosol. Rather, these cells accumulate high amounts of chelatable iron which in all likelihood is predominantly present in the cytosolic compartment. An iron efflux into the vacuole, the main storage organelle for excess iron in plants and fungi (Kaplan and Kaplan, 2009; Philpott and Protchenko, 2008), is unlikely since the deletion of the vacuolar divalent metal transporter gene CCC1 did not rescue any of the defects of Grx3/4-depleted cells. Mitochondria, which accumulate iron under conditions of hampered Fe/S protein biogenesis (Lill and Muhlenhoff, 2008), displayed diminished iron levels in Grx3/4-depleted cells. These data suggest that the surplus iron in Grx3/4-deficient cells is not bioavailable for efficient funneling into iron-dependent processes.
Our 55Fe radiolabelling experiments demonstrate that the Fe/S cluster of monothiol glutaredoxins, as initially characterized in vitro or after overexpression in E. coli (Li et al., 2009; Picciocchi et al., 2007), is also assembled in its native environment and is of utmost physiological relevance. The essential involvement of the core components of the mitochondrial ISC assembly system provides strong evidence that the bound iron is part of an Fe/S cluster, and that the sulfide ions of the [2Fe-2S] cluster are derived from the mitochondrial cysteine desulfurase complex Nfs1-Isd11. Our in vivo results support the unusual bridging character of the Fe/S cluster, which is coordinated by the Grx domain active-site Cys171 and GSH. Hence, GSH performs a dual role in cellular iron metabolism. As part of the Grx3/4 complex it is central for intracellular iron delivery and sensing, and as a component of the ISC export machinery it cooperates with the mitochondrial ABC transporter Atm1 both in the assembly of cytosolic-nuclear Fe/S proteins and in mitochondrial-cytosolic iron homeostasis (Lill, 2009; Sipos et al., 2002). Surprisingly, the Grx3/4 Fe/S cofactor is assembled independently of the CIA system, an observation which differs from all other known, non-scaffold cytosolic Fe/S proteins analyzed so far (Lill and Muhlenhoff, 2008). This unique assembly mode is likely related to the special type of Fe/S cluster on Grx3/4. The CIA machinery-independent maturation of Grx3/4 is consistent with earlier observations that this machinery is not important for cellular iron regulation in yeast (Hausmann et al., 2008; Kaplan and Kaplan, 2009).
While it is clear that the bridging Fe/S cluster of Grx3/4 is crucial for iron trafficking, the question remains how it may function. Currently, it cannot be discerned whether the iron moiety of the Fe/S cluster is mobilized and used for insertion into iron-dependent proteins, or whether the Fe/S cluster plays a more indirect role by activating cytosolic iron for specific trafficking. In any case, it is well-known for Fe/S clusters that they can reversibly loose one of their iron ions, the most prominent example being aconitase (Beinert et al., 1997). Thus, the Fe/S cluster of Grx3/4 may not be fully disassembled during its action in iron delivery. The role of the Grx3/4 Fe/S center in iron trafficking is clearly distinct from the proposed function of other monothiol glutaredoxins as Fe/S scaffold (or Fe/S carrier) proteins facilitating Fe/S cluster transfer to target apoproteins (Bandyopadhyay et al., 2008; Picciocchi et al., 2007). Thus, monothiol glutaredoxins may use diverse mechanisms for maturation of iron-containing proteins.
In principle, the essential requirement of the Grx active-site C171 for iron homeostasis could be explained by thiol-dependent redox chemistry. However, with few exceptions, monothiol glutaredoxins, including yeast Grx3/4, are inactive in GSH-dependent redox reactions in vitro, and there is little evidence that monothiol glutaredoxins carry out specific catabolic functions (Herrero and de la Torre-Ruiz, 2007). In addition, BY4742 grx3/4Δ cells display wild-type glutathione reductase activity (Rodriguez-Manzaneque et al., 1999). Most convincingly, the high-copy suppressor phenotype of the C171S mutation in the Grx domain excludes impaired thiol-related (redox) chemistry as an explanation for the functional defects prevailing in Grx3/4-depleted cells. High levels of the Cys171 mutant protein, but not the corresponding Ala variant, supported wild-type growth, and almost fully restored the activities of iron-dependent proteins. This suggests that the C171S protein is still capable of coordinating the Fe/S co-factor, as this is known for other proteins with Cys to Ser mutations (da Silva and Williams, 2001; Johnson et al., 2005). However, iron binding to the C171S mutant protein may be more labile thus precluding its detection by our in vivo 55Fe labeling and immunoprecipitation assay. As a consequence the amount of bound Fe/S cluster may be too low to support Grx4 function in Aft1 sensing. Consistent with this interpretation, in reconstituted human Grx2 and poplar GrxC1, the [2Fe-2S] cluster is coordinated in a binding pocket at the interface of two Grx monomers by the two active-site Cys residues and two non-covalently bound GSH molecules (Berndt et al., 2007; Johansson et al., 2007; Rouhier et al., 2007). The replacement of cysteine with serine may still provide a low-affinity iron binding site.
Grx3/4 play a role in iron-uptake regulation by cooperating with the iron-responsive transcription factor Aft1, the major regulator of cellular iron uptake in yeast (Philpott and Protchenko, 2008). In fact, the physical interaction between Grx3/4 and Aft1 was the initial evidence linking Grx3/4 and cellular iron homeostasis (Ojeda et al., 2006; Pujol-Carrion et al., 2006). Our study shows that the Grx3/4-bound Fe/S cluster functions as the long-sought iron sensor which communicates the cytosolic iron status to Aft1 (Fig. S5). Removal of the bound Fe/S cluster, e.g., by mutation of the active-site Cys of the Grx domain or by depletion of GSH abolished the ability of Grx3/4 to regulate Aft1. The iron-sensing role of Grx3/4 appears to be conserved in other fungi that utilize iron-responsive transcription factors, even if they are structurally unrelated to S. cerevisiae Aft1 (Kaplan and Kaplan, 2009; Mercier and Labbe, 2009).
The crucial function of Grx3/4 and its unique Fe/S center in intracellular iron trafficking and sensing opens an additional chapter of the intimate connection between the iron metabolism and Fe/S protein biogenesis. Not only is the iron metabolism regulated by (mitochondrial) Fe/S protein biogenesis (Kaplan and Kaplan, 2009), but conversely iron insertion into Fe/S proteins requires its activation by the Fe/S cluster-containing Grx3/4. The mutual dependence of these two important pathways is further linked to the cellular redox balancing systems via GSH which is crucial for both iron insertion into iron-dependent factors and regulation of cellular iron homeostasis (Lill and Muhlenhoff, 2008). These considerations reserve a central role for monothiol glutaredoxins Grx3/4 and their bound Fe/S center in cellular metabolism. The central function of Grx3/4 in intracellular iron trafficking may provide the explanation for the conservation of this protein family in eukaryotes. The discovery of this glutaredoxin function now paves the way towards mechanistic investigations of how iron is specifically activated for incorporation into iron-requiring processes.
Experimental procedures
Yeast strains, cell growth and plasmids
Yeast strains used in this study are listed in Table S1. If not stated otherwise, the strain background is W303-1A. Cells were cultivated in minimal medium containing all recommended supplements (SC), 2% w/v glucose (SD) or 3% w/v glycerol (Sherman, 2002). Iron-replete media were supplemented with 50 μM FeCl3. Media for anaerobic growth were supplemented with Tween80, ergosterol and methionine. Gal-GRX4 cells were depleted for Grx4 to critical levels by cultivation in SD medium for 40 to 64 h prior to analysis. Repression of other conditional Gal-strains was performed as described (Table S2). gsh1Δ cells were cultivated in the presence or absence of 1 mM GSH for 64 h prior to analysis (Sipos et al., 2002). Wild-type and site-directed mutant Grx4 proteins were expressed in yeast with a C-terminal Myc-tag from plasmid pCM189 (Ojeda et al., 2006) under the control of the tetO7 or the endogenous GRX4 promoter (Table S2).
Biochemical analyses
In vivo radiolabelling of yeast cells with 55FeCl3 (Perkin-Elmer) and measurement of 55Fe incorporation into proteins by immunoprecipitation and into heme was carried out as described (Pierik et al., 2009). Antibodies were raised in rabbits against recombinant proteins expressed in E. coli. 55Fe-labelling of added apo-ferredoxin in detergent extracts of isolated mitochondria was carried out as described (Molik et al., 2007). DMQ6 and CoQ6 were quantified according to (Tran et al., 2006), GSH and GSSG according to (Elledge and Davis, 1987).
Miscellaneous methods
The following published methods were used: manipulation of DNA and PCR (Sambrook and Russel, 2001); preparation of yeast mitochondria (Molik et al., 2007), immunological techniques (Harlow and Lane, 1988); FET3 promoter assays (Molik et al., 2007); determination of cellular and mitochondrial metal contents by ICP-MS (Muhlenhoff et al., 2003); enzyme activities of iron proteins, ADH, and MDH (Molik et al., 2007), ribonucleotide reductase (Wang et al., 2009), and superoxide dismutase (Flohe and Otting, 1984). Error bars represent the standard error of the mean (SEM) (n ≥ 4).
Supplementary Material
Highlights.
Glutaredoxins Grx3/4 are essential for iron insertion into proteins and cofactors
Deficiency in Grx3/4 functionally impairs all iron-requiring processes
Grx3/4-deficient cells accumulate iron but cannot biologically use it
The Fe/S center of Grx3/4 is critical for both iron trafficking and sensing
Acknowledgments
We are grateful to Dr. D.R. Winge for generous help and discussion. We acknowledge financial support from Deutsche Forschungsgemeinschaft (SFB 593, Gottfried-Wilhelm Leibniz program, and GRK 1216), von Behring-Röntgen Stiftung, Max-Planck Gesellschaft, Fonds der chemischen Industrie, NIH GM29295 (to J.S.), Ministerio de Ciencia e Innovacion (CSD2007-0020; to E.H.), Région Rhônes-Alpes program CIBLE 2009 (to F. P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balk J, Pierik AJ, Netz DJ, Muhlenhoff U, Lill R. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. Embo. 2004;J23:2105–2115. doi: 10.1038/sj.emboj.7600216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, Huynh BH, Herrero E, Jacquot JP, Johnson MK, Rouhier N. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. Embo J. 2008;27:1122–1133. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinert H, Holm RH, Munck E. Iron-sulfur clusters: nature's modular, multipurpose structures. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- Bencze KZ, Kondapalli KC, Cook JD, McMahon S, Millan-Pacheco C, Pastor N, Stemmler TL. The structure and function of frataxin. Crit Rev Biochem Mol Biol. 2006;41:269–291. doi: 10.1080/10409230600846058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt C, Hudemann C, Hanschmann EM, Axelsson R, Holmgren A, Lillig CH. How does iron-sulfur cluster coordination regulate the activity of human glutaredoxin 2? Antioxid Redox Signal. 2007;9:151–157. doi: 10.1089/ars.2007.9.151. [DOI] [PubMed] [Google Scholar]
- Crichton RR, Charloteaux-Wauters M. Iron transport and storage. Eur J Biochem. 1987;164:485–506. doi: 10.1111/j.1432-1033.1987.tb11155.x. [DOI] [PubMed] [Google Scholar]
- da Silva JJR, Williams RJP. Non-haem iron: redox reactions and controls, In The biological chemistry of the elements. In: da Silva JJR, Williams RJP, editors. Oxford University Press; Oxford, UK: 2001. pp. 340–369. [Google Scholar]
- Elledge SJ, Davis RW. Identification and isolation of the gene encoding the small subunit of ribonucleotide reductase from Saccharomyces cerevisiae: DNA damage-inducible gene required for mitotic viability. Mol Cell Biol. 1987;7:2783–2793. doi: 10.1128/mcb.7.8.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney LA, O'Halloran TV. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- Flohe L, Otting F. Superoxide dismutase assays. Methods Enzymol. 1984;105:93–104. doi: 10.1016/s0076-6879(84)05013-8. [DOI] [PubMed] [Google Scholar]
- Haas H, Eisendle M, Turgeon BG. Siderophores in fungal physiology and virulence. Annu Rev Phytopathol. 2008;46:149–187. doi: 10.1146/annurev.phyto.45.062806.094338. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Cold Spring Harbour Laboratory Press; Cold Spring Harbour, NY: 1988. [Google Scholar]
- Hausmann A, Samans B, Lill R, Muhlenhoff U. Cellular and Mitochondrial Remodeling upon Defects in Iron-Sulfur Protein Biogenesis. J Biol Chem. 2008;283:8318–8330. doi: 10.1074/jbc.M705570200. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- Herrero E, de la Torre-Ruiz MA. Monothiol glutaredoxins: a common domain for multiple functions. Cell Mol Life Sci. 2007;64:1518–1530. doi: 10.1007/s00018-007-6554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrig J, Hausmann A, Hain A, Richter N, Lill R, Muhlenhoff U. Iron regulation through the back door: Iron-dependent metabolite levels contribute to the transcriptional adaptation to iron deprivation in S. cerevisiae. Eukaryot Cell. 2009 doi: 10.1128/EC.00213-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson C, Kavanagh KL, Gileadi O, Oppermann U. Reversible sequestration of active site cysteines in a 2Fe-2S-bridged dimer provides a mechanism for glutaredoxin 2 regulation in human mitochondria. J Biol Chem. 2007;282:3077–3082. doi: 10.1074/jbc.M608179200. [DOI] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, Function, And Formation Of Biological Iron-Sulfur Clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Kaplan J. Iron acquisition and transcriptional regulation. Chem Rev. 2009;109:4536–4552. doi: 10.1021/cr9001676. [DOI] [PubMed] [Google Scholar]
- Kumanovics A, Chen OS, Li L, Bagley D, Adkins EM, Lin H, Dingra NN, Outten CE, Keller G, Winge D, et al. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J Biol Chem. 2008;283:10276–10286. doi: 10.1074/jbc.M801160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Mapolelo DT, Dingra NN, Naik SG, Lees NS, Hoffman BM, Riggs-Gelasco PJ, Huynh BH, Johnson MK, Outten CE. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry. 2009;48:9569–9581. doi: 10.1021/bi901182w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- Lillig CH, Berndt C, Holmgren A. Glutaredoxin systems. Biochim Biophys Acta. 2008;1780:1304–1317. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Lyons TJ, Eide DJ. Transport and storage of metal ions in biology, In Biological Inorganic Chemistry. In: Bertini I, Gray HB, Stiefel EI, Valentine JS, editors. University Science Books; Sausalito, CA: 2007. pp. 57–77. [Google Scholar]
- Mercier A, Labbe S. Both Php4 function and subcellular localization are regulated by iron via a multistep mechanism involving the glutaredoxin Grx4 and the exportin Crm1. J Biol Chem. 2009;284:20249–20262. doi: 10.1074/jbc.M109.009563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molik S, Lill R, Muhlenhoff U. Methods for studying iron metabolism in yeast mitochondria. Methods Cell Biol. 2007;80:261–280. doi: 10.1016/S0091-679X(06)80013-0. [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U, Stadler JA, Richhardt N, Seubert A, Eickhorst T, Schweyen RJ, Lill R, Wiesenberger G. A specific role of the yeast mitochondrial carriers MRS3/4p in mitochondrial iron acquisition under iron-limiting conditions. J Biol Chem. 2003;278:40612–40620. doi: 10.1074/jbc.M307847200. [DOI] [PubMed] [Google Scholar]
- Ojeda L, Keller G, Muhlenhoff U, Rutherford JC, Lill R, Winge DR. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J Biol Chem. 2006;281:17661–17669. doi: 10.1074/jbc.M602165200. [DOI] [PubMed] [Google Scholar]
- Perlstein DL, Ge J, Ortigosa AD, Robblee JH, Zhang Z, Huang M, Stubbe J. The active form of the Saccharomyces cerevisiae ribonucleotide reductase small subunit is a heterodimer in vitro and in vivo. Biochemistry. 2005;44:15366–15377. doi: 10.1021/bi051616+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott CC, Protchenko O. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:20–27. doi: 10.1128/EC.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picciocchi A, Saguez C, Boussac A, Cassier-Chauvat C, Chauvat F. CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry. 2007;46:15018–15026. doi: 10.1021/bi7013272. [DOI] [PubMed] [Google Scholar]
- Pierik AJ, Netz DJ, Lill R. Analysis of iron-sulfur protein maturation in eukaryotes. Nat Protoc. 2009;4:753–766. doi: 10.1038/nprot.2009.39. [DOI] [PubMed] [Google Scholar]
- Pujol-Carrion N, Belli G, Herrero E, Nogues A, de la Torre-Ruiz MA. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J Cell Sci. 2006;119:4554–4564. doi: 10.1242/jcs.03229. [DOI] [PubMed] [Google Scholar]
- Richardson DR, Ponka P. The molecular mechanisms of the metabolism and transport of iron in normal and neoplastic cells. Biochim Biophys Acta. 1997;1331:1–40. doi: 10.1016/s0304-4157(96)00014-7. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Manzaneque MT, Ros J, Cabiscol E, Sorribas A, Herrero E. Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8180–8190. doi: 10.1128/mcb.19.12.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Unno H, Bandyopadhyay S, Masip L, Kim SK, Hirasawa M, Gualberto JM, Lattard V, Kusunoki M, Knaff DB, et al. Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1. Proc Natl Acad Sci U S A. 2007;104:7379–7384. doi: 10.1073/pnas.0702268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular Cloning - A laboratory manual. 3rd edition, 3rd edn CSH Laboratory Press; ColdSpring Harbour, USA: 2001. [Google Scholar]
- Shakoury-Elizeh M, Tiedeman J, Rashford J, Ferea T, Demeter J, Garcia E, Rolfes R, Brown PO, Botstein D, Philpott CC. Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:1233–1243. doi: 10.1091/mbc.E03-09-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with Yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Shi H, Bencze KZ, Stemmler TL, Philpott CC. A cytosolic iron chaperone that delivers iron to ferritin. Science. 2008;320:1207–1210. doi: 10.1126/science.1157643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos K, Lange H, Fekete Z, Ullmann P, Lill R, Kispal G. Maturation of cytosolic iron-sulfur proteins requires glutathione. J Biol Chem. 2002;277:26944–26949. doi: 10.1074/jbc.M200677200. [DOI] [PubMed] [Google Scholar]
- Tran UC, Marbois B, Gin P, Gulmezian M, Jonassen T, Clarke CF. Complementation of Saccharomyces cerevisiae coq7 mutants by mitochondrial targeting of the Escherichia coli UbiF polypeptide: two functions of yeast Coq7 polypeptide in coenzyme Q biosynthesis. J Biol Chem. 2006;281:16401–16409. doi: 10.1074/jbc.M513267200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara SV, Thiele DJ. Post-transcriptional regulation of gene expression in response to iron deficiency: co-ordinated metabolic reprogramming by yeast mRNA-binding proteins. Biochem Soc Trans. 2008;36:1088–1090. doi: 10.1042/BST0361088. [DOI] [PubMed] [Google Scholar]
- Wang J, Lohman GJ, Stubbe J. Mechanism of inactivation of human ribonucleotide reductase with p53R2 by gemcitabine 5′-diphosphate. Biochemistry. 2009;48:11612–11621. doi: 10.1021/bi901588z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.