SUMMARY
The transition from transcription initiation to elongation at promoters of primary response genes (PRGs) in metazoan cells is controlled by inducible transcription factors, which utilize P-TEFb to phosphorylate RNA polymerase II (Pol II) in response to stimuli. Prior to stimulation, a fraction of P-TEFb is recruited to promoter-proximal regions in a catalytically inactive state bound to the 7SK small nuclear ribonucleoprotein (snRNP) complex. However, it remains unclear how and why the 7SK snRNP is assembled at these sites. Here we report that the transcriptional regulator KAP1 continuously tethers the 7SK snRNP to PRG promoters to facilitate P-TEFb recruitment and productive elongation in response to stimulation. Remarkably, besides PRGs, genome-wide studies revealed that KAP1 and 7SK snRNP co-occupy most promoter-proximal regions containing paused Pol II. Collectively, we provide evidence of an unprecedented mechanism controlling 7SK snRNP delivery to promoter-proximal regions to facilitate “on-site” P-TEFb activation and Pol II elongation.
Graphical Abstract
INTRODUCTION
Metazoan cells are capable of rapidly activating transcriptional cascades in response to intrinsic and extrinsic insults (stimuli). This capacity is mediated by a specialized transcriptional program of primary response genes (PRGs) (Fowler et al., 2011; Hargreaves et al., 2009; Smale, 2010), which is essential for tissue homeostasis, cell-fate choice, establishment of innate immune responses, and avoidance of malignancy. PRGs are rapidly induced in the absence of new protein synthesis and without chromatin remodeling and encode proteins necessary for the cell to immediately respond to a stimulus, thereby facilitating resolution of the insult (Hargreaves et al., 2009; Ramirez-Carrozzi et al., 2009).
Transcriptional regulation involves a network of inducible transcription factors (TFs) and cofactors that orchestrate the precise spatiotemporal synthesis of the proper set of genes in response to a stimulus. Prior to stimulation, the transcription preinitiation complex (PIC) assembles at promoters to recruit RNA polymerase II (Pol II) and initiate transcription at most PRGs (Diamant and Dikstein, 2013; Hargreaves et al., 2009; Smale, 2010), but Pol II pauses shortly after transcribing ~20–65 nt downstream of the transcription start site (TSS) by the action of negative elongation factors (Adelman and Lis, 2012; Peterlin and Price, 2006; Yamaguchi et al., 1999). In response to stimuli such as proinflammatory cytokines (tumor necrosis factor alpha, henceforth referred to as TNF) and environmental stresses, PRGs are rapidly activated by inducible TFs that mediate recruitment of coactivators such as positive elongation factors to help relieve Pol II pausing (Barboric et al., 2001; Rahl et al., 2010). One such elongation factor that has received considerable attention in the last two decades is the P-TEFb kinase, a heterodimer of Cyclin T1 or T2 (CycT1/2) and Cyclin-dependent kinase 9 (Cdk9). P-TEFb phosphorylates the C-terminal domain of Pol II and several negative elongation factors to promote the transcriptional pause release (Mancebo et al., 1997; Zhou et al., 2012).
To properly regulate its activity, the majority of P-TEFb is held in a catalytically inactive state reversibly bound to the 7SK small nuclear ribonucleoprotein (snRNP), composed of the 7SK RNA, the kinase inhibitor Hexim1, the 5′-RNA methyl capping enzyme MePCE, and the 3′-RNA stability protein Larp7 (He et al., 2008; Jeronimo et al., 2007; Krueger et al., 2008; Li et al., 2005; Michels et al., 2004; Nguyen et al., 2001; Yik et al., 2003). Although the majority of the 7SK snRNP complex exists in the soluble nucleoplasmic fraction, previous studies have shown that it is recruited to HIV and cellular promoter-proximal regions (Cherrier et al., 2013; D’Orso and Frankel, 2010; Ji et al., 2013; McNamara et al., 2013). It remains unclear, however, how and why the cell evolved a mechanism to selectively position the 7SK snRNP on promoter-proximal regions. We therefore sought to (1) identify factors that mediate 7SK snRNP recruitment to promoters and (2) characterize the biological significance of this complex in the activation of inducible transcriptional programs.
Here we present the unexpected findings that the Kruppel-associated box (KRAB)-interacting protein 1 (KAP1), also referred to as tripartite motif containing 28 (TRIM28) and transcription intermediary factor 1-beta (TIF1β) (Iyengar and Farnham, 2011), directly recruits the 7SK snRNP complex to most genes containing promoter-proximal paused Pol II, particularly at inducible genes. Recruitment of the KAP1-7SK snRNP complex to promoter-proximal regions is tightly correlated with Pol II pausing and transcriptional activity, and its recruitment regulates the rapid and robust response to stimulation. Although loss of KAP1 does not alter the recruitment of inducible TFs (nuclear factor κB; NF-κB) to target gene promoters in response to stimulation nor PIC assembly, it strongly antagonizes P-TEFb recruitment, slowing down Pol II pause release and largely attenuating gene activation. Our findings define a mechanism by which the 7SK snRNP complex is recruited to most promoter-proximal regions containing transcriptionally engaged Pol II, and explains the importance of this localized placement for rapid gene induction in response to stimulation. Moreover, our findings further illustrate that HIV evolved its genome to ensure that the virus is readily and robustly activated during infection through the placement of KAP1-7SK snRNP close to promoter-proximal paused Pol II.
RESULTS
The 7SK snRNP Complex Interacts with the Transcriptional Regulator KAP1/TRIM28/TIF1β
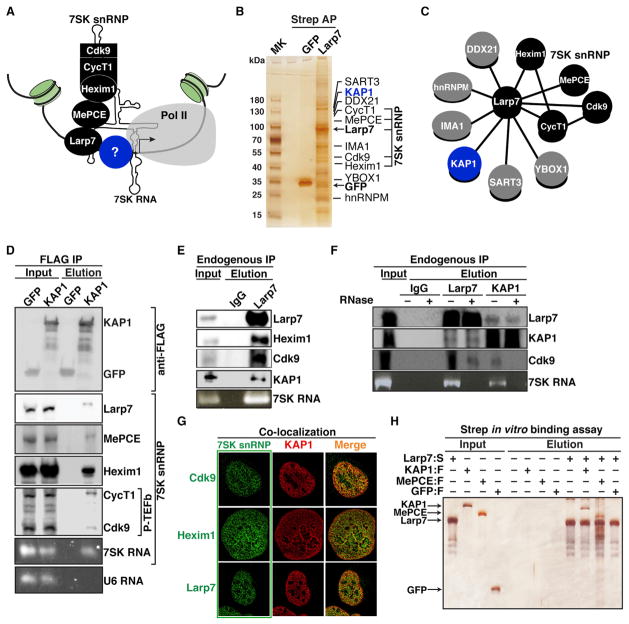
Previous studies have shown that the inhibitory 7SK snRNP complex is recruited to gene promoters (Figure 1A). However, the molecular basis of this recruitment mechanism and its precise role in the transcriptional cycle have yet to be understood. To identify factors that interact with the 7SK snRNP to potentially recruit it to promoters, we employed an affinity purification (AP) and mass spectrometry (MS) approach, which has proven to be an efficient unbiased and comprehensive method to discover the nature and composition of protein complexes. We generated HEK293 T-REx cell lines inducibly expressing a Strep-tagged version of the 7SK snRNP component Larp7 (Larp7:S) and GFP:S (negative control). Strep AP of Larp7:S from nuclear fractions revealed several specific interactors not present in the control sample (Figure 1B). To identify their identity, we subjected the affinity-purified products to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. High-confidence Larp7 interactors (based on their quantitative enrichment relative to control samples) are shown in Tables S1 and S2, along with their corresponding hypothetical bands in the silver-stained gel of Figure 1B. In addition to the core 7SK snRNP components (CycT1, Cdk9, MePCE, and Hexim1) and reversibly bound heterogeneous nuclear ribonucleoprotein (hnRNP) proteins, YBOX1 and DDX21 (Calo et al., 2015; Jeronimo et al., 2007; Krueger et al., 2008), we also identified one high-confidence interactor, the KAP1 protein (also known as TRIM28/TIF1β) (Iyengar and Farnham, 2011) (Figures 1B and 1C). In contrast to our AP-MS dataset, KAP1 was not identified in a previous Larp7 proteomics study (Krueger et al., 2008), probably attributable to the higher affinity of Strep AP (compared with regular antibody-mediated immunoprecipitation; IP), thereby leading to more sensitivity.
Figure 1. Identification of KAP1 as an Interactor of the 7SK snRNP Complex.
(A) Model for the tethering of the 7SK snRNP complex to a gene promoter containing paused Pol II through an as-yet unidentified factor(s) (blue).
(B) Strep affinity purification (AP) of GFP and Larp7 (arrows) from HEK293 T-REx cell lines. The top ten high-confidence Larp7 interactors were identified by mass spectrometry analysis and are indicated in the silver-stained gel. See also Tables S1 and S2.
(C) A network representation of protein-protein interactions between Larp7 and the factors identified in (B).
(D) Stably expressed FLAG-tagged GFP or KAP1 proteins were affinity purified from HEK293 T-REx and interactions were analyzed by western blot. Bottom: agarose gels showing qRT-PCR amplifications. Input and elution represent 0.5% and 5% of the initial material, respectively.
(E) Lysates from HEK293T cells were immunoprecipitated and protein and RNA interactions were analyzed by western blot or qRT-PCR, respectively. Input and elution represent 0.5% and 10% of the initial material, respectively.
(F) IPs were performed in the absence (−) or presence (+) of RNaseA. Input and elution represent 0.5% and 10% of the initial material, respectively. Ethidium bromide stain of PCR-amplified 7SK RNA demonstrates the efficiency of RNase treatment.
(G) Full 3D deconvoluted confocal microscopy images showing protein colocalizations in U2OS cells.
(H) FLAG-tagged proteins were incubated with immobilized Larp7:S (+) or empty beads (−). Note that both KAP1 and Larp7 protein purifications are devoid of any copurifying 7SK RNA (as judged based on qRT-PCR), thus indicating that the interaction is direct and not 7SK RNA mediated. Input and elution represent 20% of the initial material.
See also Figure S1.
To validate that KAP1 interacts with Larp7 as part of the 7SK snRNP complex, we generated HEK293 T-REx cell lines inducibly expressing FLAG-tagged KAP1 (KAP1:F), which expresses at similar levels to endogenous KAP1, and GFP:F (Figure S1A). Indeed, using nuclear extracts, we detected the primary components of the 7SK snRNP after FLAG IP of KAP1:F but not GFP:F, including 7SK RNA but not U6 (another abundant snRNA) (Figure 1D). Because we detected the KAP1-Larp7 protein interaction using epitope-tagged components, we next performed an IP of endogenous Larp7 (which is constitutively bound to the 7SK snRNP; Krueger et al., 2008) to demonstrate that the association occurs in a more biologically relevant system. Consistent with the previous results, we detected KAP1 in the IP of endogenous Larp7, as well as core 7SK snRNP components, but not when using a normal serum (IgG) (Figure 1E). Based on the material loaded, we estimated that about 10%–15% of the 7SK snRNP interacts with KAP1 in the conditions tested. Importantly, whereas the KAP1-Larp7 interaction was largely insensitive to RNase, as expected for a direct protein-protein interaction, the KAP1-Cdk9 interaction was RNA dependent (Figure 1F), indicating that KAP1 recruits P-TEFb through the Larp7 subunit of the 7SK snRNP complex.
To further validate the relevance of these findings, we performed high-resolution confocal microscopy of endogenous KAP1 and components of the 7SK snRNP and observed statistically significant colocalization indices (p value < 0.01) for all protein pair combinations in all cells examined (Figure 1G; Figures S1B–S1D), in contrast to noninteracting partners (Figure S1E).
To further test whether KAP1 associates with the 7SK snRNP complex through direct interaction with Larp7, we performed an in vitro binding assay using affinity-purified proteins from HEK293T cells under high-salt conditions. Using this method, we observed that Larp7:S directly bound KAP1:F in vitro, as well as MePCE:F, but not to GFP:F (Figure 1H). Although in the conditions used the KAP1-Larp7 interaction appears not to be stoichiometric, it is possible that Larp7 (in the absence of 7SK RNA) is biochemically poorly behaved or that the protein-protein interaction requires posttranslational modifications to achieve a higher binding affinity. Nonetheless, we have demonstrated using various approaches that KAP1 associates with the 7SK snRNP through the Larp7 subunit.
Given the relevance of P-TEFb in the transcriptional cycle, below we focus on studying the function of KAP1 in recruiting the 7SK snRNP to promoter-proximal regions to control Pol II pause release.
KAP1 and the 7SK snRNP Complex Co-occupy the HIV Promoter-Proximal Region
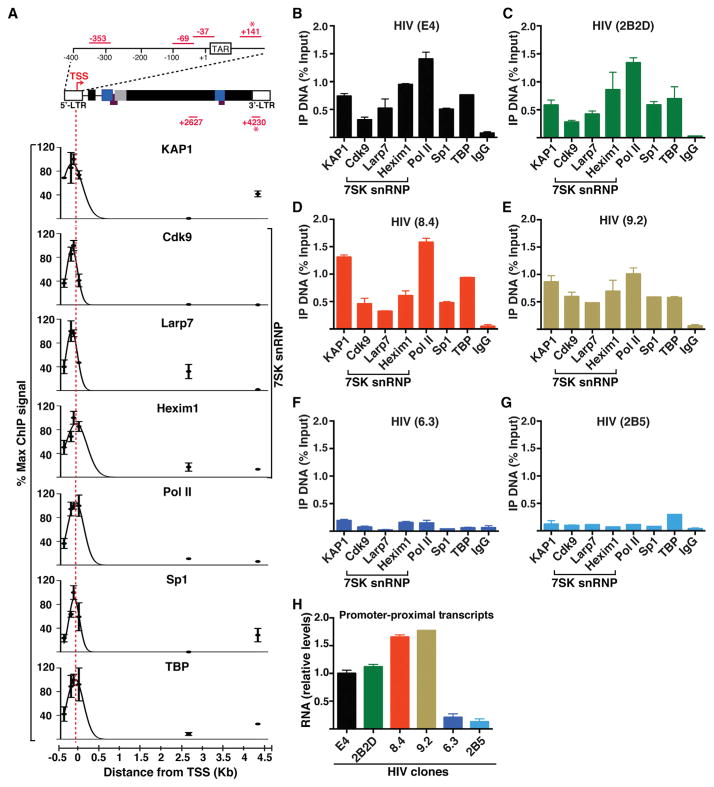
Given that KAP1 physically associates with the 7SK snRNP and that KAP1 is also recruited to gene promoters (Bunch et al., 2014; Hu et al., 2009), we asked whether both are corecruited to the promoter of HIV integrated into a Jurkat CD4+ T cell line known as clone E4 (Pearson et al., 2008). Using chromatin immunoprecipitation (ChIP) assays, we detected both KAP1 and the 7SK snRNP (Cdk9, Larp7, and Hexim1) at the HIV promoter-proximal region surrounding the TSS (5′ long terminal repeat; LTR) but not inside the genome (at least with the amplicons used) (Figure 2A).
Figure 2. KAP1 and the 7SK snRNP Complex Co-occupy the HIV Promoter-Proximal Region along with Paused Pol II.
(A) ChIP assays on the Jurkat HIV E4 clone followed by qPCR using primer pairs amplifying six different genomic regions (dots). Maximal ChIP signal was arbitrarily set to 100%, and all other amplifications were standardized to it (mean ± SEM; n = 3). Amplicons marked with an asterisk indicate 5′-LTR or 3′-LTR specific. From each dataset, best-fit nonlinear regression curves were generated using Prism (GraphPad) to approximate the ChIP signal throughout the HIV genome at a given location. TAR, transactivation response element.
(B) ChIP assays with the indicated antibodies including a normal serum (IgG) in the HIV E4 clone followed by qPCR assays using the 5′-LTR-specific amplicon. Percentage of immunoprecipitated DNA respective to input was calculated (mean ± SEM; n = 3).
(C) Same as in (B) for 2B2D.
(D) Same as in (B) for 8.4.
(E) Same as in (B) for 9.2.
(F) Same as in (B) for 6.3.
(G) Same as in (B) for 2B5.
(H) Quantitation of HIV promoter-proximal transcript levels (+141) normalized to ACTB by qRT-PCR (mean ± SEM; n = 3).
The presence of duplicated LTRs makes it difficult to assign ChIP signal to individual LTRs. Therefore, to distinguish PCR amplifications of the 5′-LTR from the 3′-LTR, we used LTR-specific primer sets (+141 and +4230, respectively) (Jadlowsky et al., 2014). We noted that KAP1 displayed a preference for the 5′-LTR, consistent with previous reports of it occupying the LTR of endogenous retroelements (Iyengar and Farnham, 2011; Rowe et al., 2010). A minor fraction of KAP1 was also detected at the 3′-LTR of the integrated provirus. However, none of the 7SK snRNP components displayed significant enrichment at the 3′-LTR, demonstrating that the inhibitory snRNP, and the majority of KAP1, primarily co-occupy the 5′-LTR of HIV. The peak intensity of the 7SK snRNP and KAP1 closely mirrors the occupancy profile of promoter-proximal paused Pol II and the PIC (as revealed by the occupancy of basal transcription factors Sp1 and TBP) at the promoter (Figure 2A).
Given that HIV integrates randomly and that integration site placement dictates basal transcriptional activity (Jordan et al., 2001), we asked whether the occupancy of KAP1 and the 7SK snRNP at the HIV promoter was dependent on the integration site, chromatin context, and promoter activity. To evaluate this, we performed ChIP-quantitative (q)PCR with the primer pair that specifically amplifies the HIV promoter-proximal region and measured transcription initiation levels across several cell-based models containing a single HIV integrant. Notably, we observed that KAP1 and the 7SK snRNP are recruited only to promoters containing a PIC (Sp1 and TBP) and transcriptionally engaged Pol II such as in the E4, 2B2D, 8.4, and 9.2 HIV clones (Figures 2B–2E). Conversely, KAP1 and the 7SK snRNP do not appear to occupy the promoter of a provirus integrated in chromatin-dense regions without noticeable PIC and paused Pol II (6.3 clone; Figure 2F) or a promoter containing a mutation that impairs PIC assembly and Pol II recruitment (2B5 clone; Figure 2G).
Given the correlation between PIC assembly, Pol II pausing, and the presence of the KAP1-7SK snRNP complex at promoter-proximal regions, we asked whether the HIV genomes in these cell lines are transcriptionally active. To test this, we performed qRT-PCR assays to measure levels of promoter-proximal transcripts (indicative of transcription initiation) and observed that higher transcript levels (in the E4, 2B2D, 8.4, and 9.2 clones) directly correlated with the presence of a promoter-assembled PIC, transcriptionally engaged Pol II, and the KAP1-7SK snRNP complex at promoter-proximal regions (Figure 2H). In contrast, the 6.3 and 2B5 clones showed fewer promoter-proximal transcripts (at least 7- to 10-fold less compared with active promoters). These results are consistent with previous evidence demonstrating that 6.3 exhibits very low transcript levels and 2B5 contains a nonfunctional promoter (Jordan et al., 2001; Pearson et al., 2008). Taken together, our findings indicate that KAP1-7SK snRNP occupies promoter-proximal regions of proviruses that show evidence of transcription initiation.
KAP1 Mediates Recruitment of the 7SK snRNP Complex to the HIV and Cellular Promoter-Proximal Regions
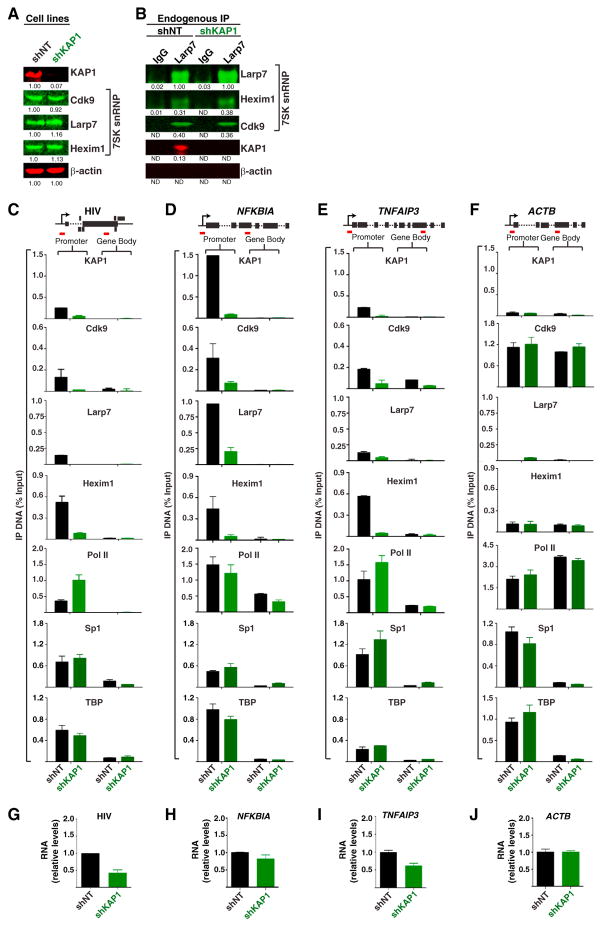
Given that KAP1 associates with the 7SK snRNP and that both are recruited to chromatin, we hypothesized that KAP1 recruits the inhibitory snRNP to promoters to control transcription elongation. To test this model, we generated Jurkat HIV E4 cell lines expressing nontarget (shNT) or KAP1 (shKAP1) small hairpin (sh) RNAs. Importantly, this approach yielded efficient knockdown (KD) of KAP1 without significantly affecting the stability of 7SK snRNP components (Figure 3A). Given that fluctuations in their levels, such as increased Hexim1, have been previously correlated with the release of P-TEFb from the 7SK snRNP (Liu et al., 2014), we asked whether 7SK snRNP stability is disrupted when KAP1 expression is lost. To test this, we performed IP of endogenous Larp7 in the two cell lines and quantified levels of coimmunoprecipitated 7SK snRNP components, and observed no clear alterations in the levels of Cdk9 and Hexim1 bound to Larp7 in response to KAP1 KD (Figure 3B). Although we cannot exclude minor fluctuations in the levels of competent 7SK snRNP, its stability appears for the most part to be KAP1 independent.
Figure 3. KAP1 Mediates Recruitment of the 7SK snRNP Complex to Promoter-Proximal Regions.
(A) Western blots of the E4 shNT and shKAP1 cell lines. Values represent relative protein levels normalized to β-actin.
(B) Lysates from (A) were immunoprecipitated to monitor levels of 7SK snRNP components. Values represent the amount of immunoprecipitated proteins relative to input. ND denotes below detection limit.
(C) ChIP assay at the HIV locus in the cell lines from (A). Values represent the average of three independent biological replicates minus a nonspecific IgG control (mean ± SEM; n = 3). Short red lines denote the relative locations of promoter and gene body amplicons.
(D) Same as in (C) for NFKBIA.
(E) Same as in (C) for TNFAIP3.
(F) Same as in (C) for ACTB.
(G) Relative HIV transcripts (+2,627) quantified by qRT-PCR and normalized to ACTB (mean ± SEM; n = 3).
(H) Same as in (G) for NFKBIA.
(I) Same as in (G) for TNFAIP3.
(J) ACTB transcript levels were standardized to 1 for each cell line.
See also Figure S2.
Using these cell lines, we then assessed whether KAP1 plays any role in recruiting the 7SK snRNP complex to the HIV and cellular gene promoters by ChIP. Importantly, because loss of KAP1 resulted in minimal 7SK snRNP disruption, this strategy allowed us to measure direct effects of KAP1 loss on the promoter-bound 7SK snRNP pool. Expectedly, KAP1 and 7SK snRNP components co-occupy the HIV promoter but not intragenic domains (gene body) in the control cell line (shNT). However, the occupancy of KAP1 and the 7SK snRNP at the HIV promoter was reduced by a factor of ~3- to 5-fold (depending on the subunit) upon KAP1 KD (Figure 3C). Although Pol II occupancy at the HIV promoter increased ~2-fold in response to KAP1 KD, we did not detect any significant increase in Pol II levels at the gene body (Figure 3C), probably indicating reduced pause release in the absence of KAP1. Notably, PIC assembly (Sp1 and TBP) at the HIV promoter remained unaffected.
Because different HIV clones (E4 and 2B2D) showed evidence of KAP1-7SK snRNP recruitment to the promoter-proximal region (Figure 2), we also tested the effect of KAP1 KD on 7SK snRNP recruitment in another Jurkat HIV clone (2B2D). Remarkably, we observed that the effect of KAP1 KD on 7SK snRNP occupancy at the HIV promoter in the 2B2D clone matches the effect observed on E4 (Figure S2A), suggesting that the results are clone independent.
The findings that KAP1 and the 7SK snRNP have been detected at promoter-proximal regions of NF-κB-regulated genes (McNamara et al., 2013) prompted us to test whether KAP1 mediates 7SK snRNP recruitment to cellular genes as observed with HIV. We chose two inflammatory-responsive PRGs, NF-κB inhibitor alpha (NFKBIA, encoding IκBα) and TNF alpha-induced protein 3 (TNFAIP3, encoding A20), due to the presence of a fully assembled PIC at their promoters, P-TEFb dependence, Pol II pausing, and a strong and rapid induction in the presence of TNF through NF-κB (RelA family member) (Diamant and Dikstein, 2013; Smale, 2010). Notably, both PRGs showed evidence of KAP1 and 7SK snRNP recruitment to promoters but not to gene bodies, as well as promoter-proximal paused Pol II prior to stimulation (Figures 3D and 3E). Strikingly, KAP1 KD produced a sharp reduction (~3- to 10-fold depending on the subunit) in the occupancy of the 7SK snRNP at both gene promoters. Similar to the HIV promoter, PIC assembly (Sp1 and TBP) at both promoters remains mostly unaffected upon KAP1 KD (Figures 3D and 3E), potentially indicating that KAP1 functions at a step downstream of PIC assembly to control transcription activation.
We also examined whether KAP1-7SK snRNP is recruited to an actively transcribed gene such as ACTB (encoding β-actin). We found high levels of Cdk9 but much lower levels of KAP1, Larp7, and Hexim1 at the promoter, but no significant effect of KAP1 KD on Cdk9 or Pol II levels at this gene (Figure 3F). This potentially indicates that paused PRGs are more sensitive than actively transcribed genes to the loss of KAP1.
Moreover, we observed that KAP1 and the 7SK snRNP also occupied the promoter-proximal region of the well-known Pol II paused gene HSPA1B (encoding HSP70) (Figure S2B) but not at the transcriptionally inactive gene regulated by estrogen in breast cancer (GREB1, encoding GREB-1) (Figure S2C) (Bunch et al., 2014; Lis et al., 2000). GREB1 served as a negative control because it contains neither an assembled PIC nor a transcriptionally engaged Pol II, similar to the virtually inactive HIV promoter in the 6.3 and 2B5 clones (Figures 2F and 2G, respectively).
To further examine how the observed changes in KAP1 and 7SK snRNP occupancy at the HIV and cellular genes affect RNA levels from these promoters, we performed qRT-PCR assays and observed no significant alterations in NFKBIA or a slight reduction in HIV and TNFAIP3 RNA steady-state levels in response to KAP1 KD (Figures 3G–3I) without alterations in ACTB levels (Figure 3J).
Taken together, our results demonstrate that recruitment of the 7SK snRNP to promoter-proximal regions of NF-κB-regulated PRGs is dictated by KAP1.
KAP1-Mediated Recruitment of the 7SK snRNP Complex to Promoter-Proximal Regions Enables Efficient Transcription Activation in Response to Stimulation
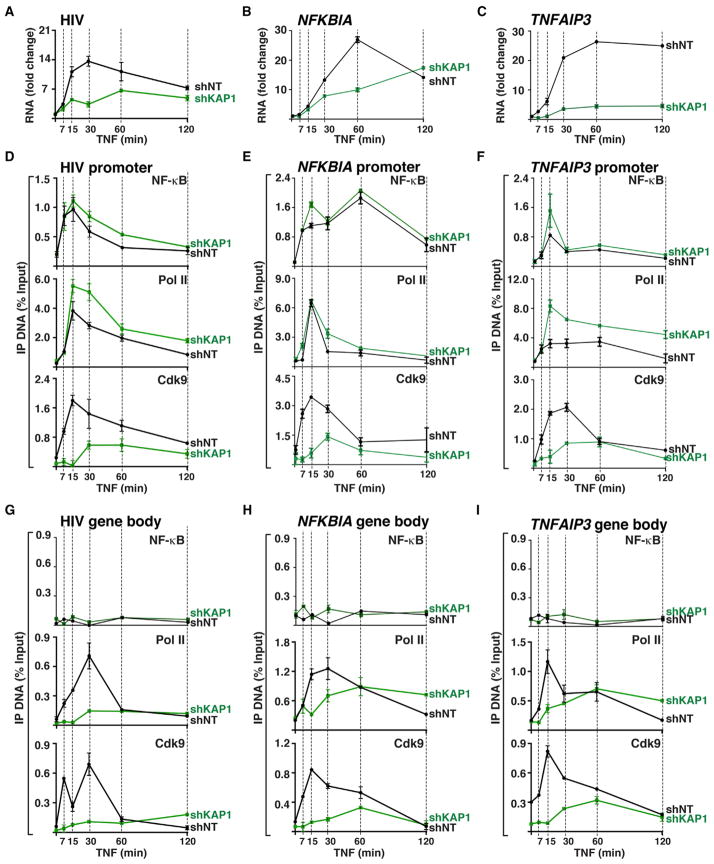
Given that KAP1 recruits the 7SK snRNP complex to the promoter-proximal regions of PRGs regulated by NF-κB, we reasoned that loss of 7SK snRNP from these sites in response to KAP1 KD might alter gene induction in response to stimulation. To examine this in detail, we monitored transcriptional kinetics of three NF-κB-regulated PRGs (HIV, NFKBIA, and TNFAIP3) in response to TNF in the shNT and shKAP1 cell lines (Figure 4). Given that in response to TNF, HIV is sequentially activated by NF-κB and the viral activator Tat, we used the HIV Tat null cell line 2B2D (Pearson et al., 2008) (here referred to as HIV for simplicity) to solely monitor NF-κB activation. Interestingly, we observed a sharp decrease in the magnitude of RNA synthesis in response to TNF in the shKAP1 cell line compared to the shNT control (Figure 4A; Figure S3A). Strikingly, this decrease in magnitude was also observed, albeit with gene-specific differences, in NFKBIA and TNFAIP3 (Figures 4B and 4C; Figures S3B and S3C). The reduced magnitude of RNA synthesis demonstrates that KAP1-mediated 7SK snRNP recruitment to the promoter-proximal regions of PRGs is required for proper gene induction upon stimulation.
Figure 4. Loss of KAP1 Reduces P-TEFb Recruitment to Promoters, Thereby Decreasing Elongation and Gene Induction in Response to Stimulation.
(A) The 2B2D shNT and shKAP1 cell lines were used to monitor HIV (+2,627 amplicons) induction after TNF treatment using qRT-PCR. Graphs show fold RNA change ± TNF normalized to ACTB (mean ± SEM; n = 3).
(B) Same as in (A) for NFKBIA.
(C) Same as in (A) for TNFAIP3.
(D) ChIP assays at the HIV promoter (mean ± SEM; n = 3).
(E) Same as in (D) for the NFKBIA promoter.
(F) Same as in (D) for the TNFAIP3 promoter.
(G) ChIP assays at the HIV gene body (mean ± SEM; n = 3).
(H) Same as in (G) for the NFKBIA gene body.
(I) Same as in (G) for the TNFAIP3 gene body.
See also Figure S3.
To test whether reduced gene activation was due to the inability of NF-κB and Pol II to be properly recruited to the promoter in response to stimulation, we treated both cell lines with TNF (time points preceding and following the kinetics of RNA synthesis) and monitored NF-κB and Pol II occupancy at the three target promoters using ChIP. In response to TNF, NF-κB is recruited rapidly and similarly to the three promoters examined in both cell lines, albeit with gene-specific differences in kinetics (Figures 4D–4F), but not to their gene bodies (Figures 4G–4I).
Whereas the kinetics of NF-κB and Pol II recruitment to the HIV promoter in response to TNF was not impaired, Cdk9 recruitment was reduced throughout the time course (>3-fold less density in the shKAP1 cell line compared to the shNT control) (Figure 4D). We then asked whether this delayed recruitment of Cdk9 to the promoter was also observed in the NFKBIA and TNFAIP3 promoters. Consistently, neither NF-κB nor Pol II was affected in their recruitment to the two cellular gene promoters in response to TNF (Figures 4E and 4F). However, Cdk9 recruitment to these two gene promoters was strongly antagonized in the shKAP1 cell line (Figures 4E and 4F). This finding demonstrates that KAP1-mediated 7SK snRNP recruitment to promoter-proximal regions (before and after stimulation) facilitates the delivery of the P-TEFb kinase to promote gene activation.
To test whether decreased RNA levels were a result of a non-processive Pol II as a consequence of delayed Cdk9 recruitment, we again performed ChIP assays after a temporal TNF treatment to monitor levels of Cdk9 and Pol II in the gene body of the three PRGs. Consistent with the ChIP results at the promoter, Cdk9 and Pol II occupancy in the gene body of all three PRGs was decreased in the shKAP1 cell line in response to TNF (Figures 4G–4I). Notably, reduced Cdk9 and Pol II occupancy in the gene bodies is consistent with decreased RNA levels in response to stimulation, indicative of a restriction in productive elongation. In the case of HIV and TNFAIP3, more Pol II was recruited to the promoter in response to TNF (Figures 4D and 4F). However, this Pol II was not elongation competent, because gene body levels were lower in the shKAP1 cell line compared to the shNT control for both genes (Figures 4G and 4I), in agreement with the requirement of KAP1 for pause release and the low levels of Cdk9 observed in the gene body.
Together, we have established a new role for KAP1 in transcription elongation control through deposition of localized, but catalytically inactive, P-TEFb at promoters.
KAP1 Is Dynamically Recruited to Gene Promoters to Enable 7SK snRNP Loading and P-TEFb Delivery in Response to Stimulation
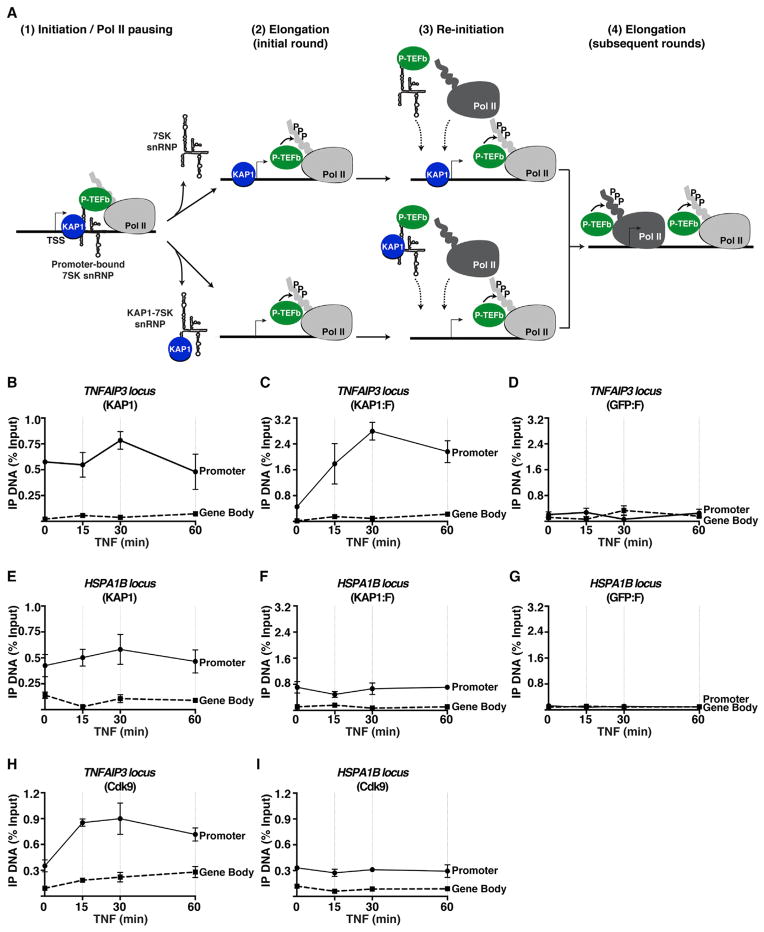
The finding that KAP1 mediates 7SK snRNP recruitment to promoter-proximal regions in basal conditions does not completely explain why more P-TEFb kinase is recruited in response to stimulation (to facilitate NF-κB-mediated Pol II pause release). Given that the KAP1-7SK snRNP complex is present both on and off promoters, we hypothesized that additional P-TEFb molecules are recruited to act on paused Pol II to facilitate rapid elongation upon stimulation. Because KAP1 indirectly interacts with P-TEFb through the 7SK snRNP (Figure 1), we reasoned that in addition to 7SK snRNP recruitment to the promoter-proximal region (which is required for the initial round of elongation), KAP1 might facilitate recruitment of additional 7SK snRNP molecules during stimulation to continuously supply more P-TEFb to facilitate subsequent rounds of elongation (Figure 5A).
Figure 5. KAP1 and P-TEFb Are Continuously Recruited to Promoter-Proximal Regions in Response to Stimulation.
(A) Two potential models of KAP1-7SK snRNP recruitment to promoters and gene activation.
(B) HEK293 T-REx cells were treated with TNF and ChIP (KAP1) was performed at the TNFAIP3 locus. Error bars represent mean ± SEM (n = 3) throughout the figure.
(C) HEK293 T-REx KAP1:F cells were treated with TNF and ChIP (FLAG) was performed at the TNFAIP3 locus.
(D) HEK293 T-REx GFP:F cells were treated with TNF and ChIP (FLAG) was performed at the TNFAIP3 locus.
(E) HEK293 T-REx cells were treated with TNF and ChIP (KAP1) was performed at the HSPA1B locus.
(F) HEK293 T-REx KAP1:F cells were treated with TNF and ChIP (FLAG) was performed at the HSPA1B locus.
(G) HEK293 T-REx GFP:F cells were treated with TNF and ChIP (FLAG) was performed at the HSPA1B locus.
(H) HEK293 T-REx KAP1:F cells were treated with TNF and ChIP (Cdk9) was performed at the TNFAIP3 locus.
(I) HEK293 T-REx KAP1:F cells were treated with TNF and ChIP (Cdk9) was performed at the HSPA1B locus.
We envisioned two potential mechanisms that take place once the promoter-bound 7SK snRNP is disassembled upon stimulation and P-TEFb is released to facilitate the initial round of elongation: (1) KAP1 remains bound to the promoter to promote the recruitment of subsequent 7SK snRNP molecules (Figure 5A, top) or (2) the entire KAP1-7SK snRNP complex is continuously resupplied to promoters upon stimulation (Figure 5A, bottom). To discern between these two possible mechanisms, we took advantage of our inducible cell lines, where KAP1:F (expressed at similar levels to endogenous KAP1; Figure S1A) or GFP:F is only expressed in the presence of doxycycline (Figure S1A), and performed competition ChIP assays. After a short time induction of KAP1:F and GFP:F, we monitored whether there is any KAP1 turnover at promoters after a TNF time-course treatment through competition of endogenous KAP1 with induced KAP1:F. In the event that new KAP1-7SK snRNP is recruited upon stimulation, the FLAG ChIP signal would increase temporally, thereby showing newly synthesized KAP1:F competition with endogenous KAP1 for promoter binding. Conversely, if KAP1 remains stably bound at the promoter and more 7SK snRNP is recruited upon stimulation, then KAP1:F would not replace promoter-bound endogenous KAP1.
We observed that upon stimulation with TNF, KAP1 occupancy at the TNFAIP3 promoter remains virtually constant and low to undetectable in the gene body (Figure 5B). However, a rapid recruitment of KAP1:F, but not GFP:F, to the promoter is observed, signifying that the endogenous KAP1 is being competed off with KAP1:F (Figures 5C and 5D). This phenomenon was exclusive to the TNFAIP3 promoter, as there was no difference between GFP:F and KAP1:F recruitment to the gene body.
Importantly, as a control, we also examined factor occupancy at the HSPA1B locus, which also shows evidence of KAP1 recruitment to the promoter but not to the gene body (Figure 5E), alongside the 7SK snRNP and paused Pol II (Figure S2B). Because HSPA1B is nonresponsive to TNF, we reasoned that no turnover of KAP1 should occur at the promoter after stimulation, and therefore KAP1:F should not compete off endogenous KAP1 assembled at the promoter. Whereas low levels of KAP1:F, but not GFP:F, were detected at the promoter before stimulation, like at the TNFAIP3 locus, no additional recruitment of KAP1:F was observed upon stimulation (Figures 5F and 5G), indicating that at nonresponsive, highly paused genes, KAP1 does not undergo any turnover at the promoter in response to stimulation. Notably, Cdk9 levels in the TNFAIP3 promoter and gene body (but not at the HSPA1B locus) increase with similar kinetics to KAP1, indicating that they are corecruited to the promoter (Figures 5H and 5I).
Collectively, the data demonstrate that KAP1 is dynamically recruited to promoters in response to stimulation to continuously recruit the 7SK snRNP to facilitate gene activation.
KAP1 and the 7SK snRNP Co-occupy Most Gene Promoter-Proximal Regions with Paused Pol II
Our results support a model in which KAP1 mediates placement of the 7SK snRNP complex to promoter-proximal regions. To obtain genome-wide information, we performed ChIP-seq (sequencing) of KAP1, 7SK snRNP components, and Pol II. Given our proposed link between active and paused genes and placement of KAP1-7SK snRNP, we used the HCT116 cell line to compare levels of Pol II and KAP1-7SK snRNP at promoters with transcriptional activity by using the nascent RNA-sequencing data (GRO-seq) generated by the Espinosa laboratory (Galbraith et al., 2013).
After ranking all RefSeq genes in the human genome (30,180 unique TSSs) based on decreasing promoter Pol II levels, we observed that KAP1 and components of the 7SK snRNP co-occupy promoter-proximal regions and that their distribution is very similar to Pol II (Figure 6A). Expectedly, the distribution of KAP1-7SK snRNPs at promoter-proximal regions also mirrors that of the active chromatin signature (H3K4me3), as well as H3K27ac, but not the enhancer signature (H3K4me1) (Figure 6A), which is absent from promoter-proximal regions (Calo and Wysocka, 2013). Additionally, analysis of the GRO-seq dataset sorted based on ranked Pol II occupancy indicates that KAP1 and the 7SK snRNP co-occupy genes containing transcriptionally engaged Pol II (Figure 6A), including transcripts derived from both DNA strands (Figure S4A).
Figure 6. KAP1 and the 7SK snRNP Co-occupy Most Promoter-Proximal Regions Containing Paused Pol II.
(A) Heatmap representation of factor distribution at all RefSeq genes relative to the TSS and GRO-seq in the HCT116 cell line. All heatmaps are clustered to Pol II.
(B) Metagene analysis showing the distribution of each marker relative to the TSS.
(C) The number of genes containing Hexim1, Larp7, and Cdk9 (7SK snRNP target genes) at promoter-proximal regions was calculated.
(D) The number of genes containing the 7SK snRNP, KAP1, and paused Pol II at promoter-proximal regions was calculated. Genes containing all three markers are referred to as KEC target genes.
(E) Genome browser view of ChIP-seq at the KEC target gene TNFAIP3 along with GRO-seq data showing transcripts from the Watson (blue) and Crick (red) DNA strands.
See also Figure S4.
To further define the relative proximity of KAP1 and the 7SK snRNP respective to the TSS, we generated metagene profiles. We observed that KAP1 and all 7SK snRNP components peak at +200 to +250 bp from the TSS (Figure S4B), slightly downstream of the metagene Pol II paused peak located at about +100 bp, in agreement with previous reports (Adelman and Lis, 2012; Muse et al., 2007). The location of paused Pol II and KAP1-7SK snRNP also matches the expected distribution of the active markers (H3K4me3 and H3K27ac) at promoter-proximal regions, which are devoid of H3K4me1 (Figure 6B).
To identify genes regulated by KAP1-7SK snRNP, we first defined 7SK snRNP target genes as those marked by three components of the 7SK snRNP (Hexim1, Larp7, and Cdk9) at promoter-proximal regions. We used a −250 to +1,000 bp window respective to the TSS because we noted that 6,184 genes (20.5% of all annotated genes) contain paused Pol II and H3K4me3 peaks in the +251 to +1,000 bp region, possibly indicating that at these genes, Pol II can pause further downstream than expected or that their TSS annotations are not accurate (see Supplemental Experimental Procedures for complete details). Using these parameters, we identified 15,850 7SK snRNP target genes (52.5% of all annotated genes) (Figure 6C). Although most of the genes are marked by all three 7SK snRNP components, we are probably underestimating this number, due to the absence of one of the three markers from some genes based on the high confidence threshold used for peak calling.
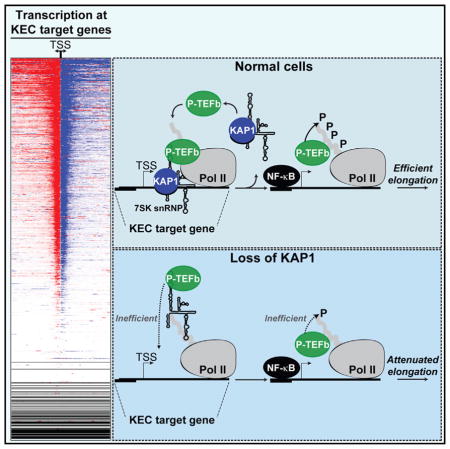
Using the previously identified 7SK snRNP target genes, we then determined genes that are also marked by KAP1 and paused Pol II in the −250 to +1,000 bp region surrounding the TSS, and where both KAP1 and the 7SK snRNP are within −250 to +250 bp of the Pol II peak summit. This filter guarantees that the factors are within a window where Pol II may realistically be considered paused, and that they are all within a distance to interact with one another. Notably, we found that 12,211 genes (40.5% of all annotated genes and 70.5% of genes containing paused Pol II) contained KAP1, the 7SK snRNP, and transcriptionally engaged Pol II (Figure 6D), suggesting a significant enrichment of KAP1-7SK snRNP and Pol II at promoter-proximal regions (p value < 2.2204−16, machine zero, hypergeometric test). We thus refer to these genes as KEC (KAP1-7SK snRNP early elongation complex) target genes.
In contrast to KEC target genes, only 551 genes (3.2% of all genes containing paused Pol II) completely lack KAP1-7SK snRNP at promoter-proximal regions as defined above, indicating significant de-enrichment of genes having paused Pol II but no KAP1-7SK snRNP at these sites (p value < 2.2204−16, machine zero, hypergeometric test). Thus, most of the times we observed paused Pol II at promoter-proximal regions, we also detected the KAP1-7SK snRNP complex.
Inspection of genome browser views reveals examples of KEC target genes such as TNFAIP3 (Figure 6E), in agreement with the ChIP-qPCR data generated in Jurkat cells (Figure 3), as well as NFKBIA and HSPA1B (Figures S4C and S4D). Moreover, active genes such as GAPDH and RPL19 are KEC targets (Figures S4E and S4F), but Pol II and KAP1-7SK snRNP distribution extends beyond the promoter-proximal region. Compared to transcriptionally paused (TNFAIP3, NFKBIA, and HSPA1B) and active (GAPDH and RPL19) genes, transcriptionally inactive genes (IL2 and IL8) did not show evidence of KAP1-7SK snRNP assembly or paused Pol II (Figures S4G and S4H), thus supporting the previous enrichment analysis.
Our findings that KAP1 occupies most genes containing promoter-proximal Pol II pausing prompted us to test the effect of KAP1’s loss on gene expression under basal conditions. For this, we created HCT116 shNT and shKAP1 cell lines (showing an ~85% KD) (Figure S4I) and used them for qRT-PCR assays to measure steady-state RNA levels. We found that the majority of the transcripts examined (GAPDH, NFKBIA, TNFAIP3, HSPA1B, and HSP90) have decreased RNA levels in the shKAP1 cell line (Figure S4J). However, MYC, GREB1, RPL19, and ACTB did not show any noticeable changes in RNA levels. Although GRO-seq experiments will be needed to obtain genome-wide transcription information, the data support that KAP1 indeed has an effect in facilitating basal transcription in addition to playing a critical role in the activation of PRGs in response to stimulation.
A Model for KAP1-Mediated Recruitment of the 7SK snRNP Complex to Promoter-Proximal Regions and Transcription Elongation
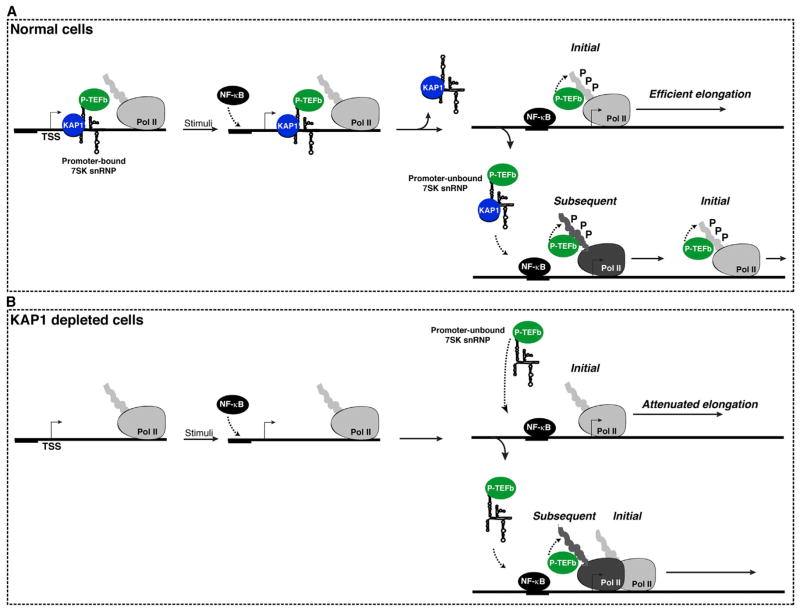
Collectively, KAP1-mediated 7SK snRNP recruitment to promoter-proximal regions plays a critical role in gene activation. We therefore propose a model in which early transcription elongation primes a gene for rapid induction through assembly of the KAP1-7SK snRNP complex at these sites. In normal cells, TFs (such as NF-κB) locally capture P-TEFb for rapid gene activation in response to induction stimuli through KAP1-mediated placement of the 7SK snRNP. Importantly, recruitment of KAP1-7SK snRNP to promoters does not affect the kinetics of TF-promoter DNA interaction in response to stimuli; rather, it facilitates the capture of P-TEFb by the TF to promote transcription elongation (initial round). From there, additional molecules of promoter-unbound KAP1-7SK snRNP are further recruited to promote subsequent rounds of elongation and robust gene induction in response to stimulation (Figure 7A). Once P-TEFb is delivered to the promoter, KAP1 might be evicted from chromatin or travel with elongating Pol II (to fulfill other downstream functions). This step may allow the further recruitment of additional KAP1-7SK snRNP molecules (“cycling”) to continuously facilitate elongation.
Figure 7. Model for the Role of the KAP1-7SK snRNP Complex in Transcriptional Pause Release.
(A) The KEC is assembled at paused genes. In response to stimulation, NF-κB recognizes target promoters and captures P-TEFb released from the KAP1-7SK snRNP. KAP1 and inhibitory snRNP components are then evicted from the promoter. These molecular events allow for the initial round of elongation through P-TEFb-mediated phosphorylation (P) of Pol II (light gray). The promoter-bound KEC facilitates elongation (initial round). Incoming pools of KEC provide a continuous source of P-TEFb that phosphorylates newly incorporated Pol II (dark gray) to promote productive elongation (subsequent rounds).
(B) Loss of KAP1 results in the elimination of the 7SK snRNP from promoter-proximal regions, thereby antagonizing P-TEFb recruitment and attenuating Pol II elongation.
In KAP1-depleted cells, NF-κB and Pol II are recruited to target gene promoters in response to stimuli as well as in normal cells. By contrast, the loss of KAP1 drastically affects P-TEFb recruitment to promoter-proximal regions, delaying Pol II phosphorylation and attenuating both the initial and subsequent rounds of transcription elongation (Figure 7B).
The genome-wide distribution of KAP1-7SK snRNP along with paused Pol II at most promoter-proximal regions underscores the importance of this complex in gene activation in basal conditions. However, due to the nature of transcriptional activation of constitutive and inducible genes, the last set of genes appears to be much more sensitive to the loss of KAP1, probably due to their critical requirement of elongation factors for rapid and robust gene activation.
DISCUSSION
Several studies have documented that the 7SK snRNP complex is recruited to promoter-proximal regions (D’Orso and Frankel, 2010; Ji et al., 2013; McNamara et al., 2013). However, the molecular basis of this recruitment mechanism and its precise role in the transcriptional cycle have yet to be reconciled. In this study, we have established that the KAP1 protein physically recruits the 7SK snRNP to the integrated HIV 5′-LTR and most cellular gene promoters containing transcriptionally engaged Pol II. Mechanistically, KAP1 recruits the 7SK snRNP onto chromatin by directly contacting the Larp7 subunit. Recruitment of the 7SK snRNP facilitates delivery of P-TEFb to promoters to induce Pol II phosphorylation, pause release, and rapid activation of PRGs in response to stimulation.
Besides controlling the rapid induction of PRGs, we showed that KAP1-7SK snRNP is deposited at most genes containing promoter-proximal paused Pol II, thus providing a critical role in transcriptional homeostasis. We propose that KAP1-mediated localized placement of inactive but primed P-TEFb kinase in the vicinity of paused Pol II allows TFs to locally capture P-TEFb to couple transcription initiation with elongation. TFs such as NF-κB bind P-TEFb to stimulate transcriptional elongation by Pol II (Barboric et al., 2001). Although TNF stimulation triggers the recruitment of P-TEFb to NF-κB-regulated genes, it remains poorly understood whether P-TEFb is tethered to DNA through interaction with the TF or through another mechanism. Our findings suggest that P-TEFb is already assembled at promoter-proximal regions (even in the absence of the inducible TF) and that additional P-TEFb is recruited through the KAP1-7SK snRNP interaction in response to stimulation.
Notably, KAP1-7SK snRNP is recruited to genes containing paused Pol II but not to inactive genes. In agreement with this model, chemical inhibition of initiation with triptolide, which interferes with formation of the open complex, antagonizes assembly of the KAP1-7SK snRNP, PIC assembly at the promoter, and Pol II recruitment and pausing (data not shown). This is further exemplified through the use of multiple clonal cell lines containing a single HIV integrant in which only viral genomes with transcriptionally engaged Pol II (active paused genomes), but not transcriptionally inactive HIV genomes, show evidence of KAP1-7SK snRNP assembly. It is noteworthy that this recruitment requires a nucleosome-free region in the promoter-proximal region and ongoing transcription initiation, because proviruses integrated in silent genomic regions (heterochromatin) or transcriptionally inactive genes in the human genome do not show evidence of KAP1-7SK snRNP and promoter-proximal Pol II pausing.
Whereas our work has focused on the assembly of KAP1-7SK snRNP at promoter-proximal regions, one previous study reported that the 7SK snRNP complex occupies distal enhancers and plays an important role in transcriptional pause release through a specialized set of chromatin-modifying enzymes (Liu et al., 2013). It is thus possible that KAP1-7SK snRNP functions from multiple genomic domains (enhancers and promoters) to properly control gene activation.
Historically, KAP1 has been functionally linked to epigenetic silencing in embryonic stem cells through the deposition of repressive chromatin marks (H3K9me3) and the formation of a heterochromatic environment in developmental genes and retro-elements (Rowe et al., 2010; Wolf and Goff, 2007). However, our findings indicate an additional function of KAP1 in facilitating the rapid induction of PRGs through recruitment of the 7SK snRNP complex to promoter-proximal regions and probably controlling transcription elongation genome-wide. In the case of the transcriptional silencing function, KAP1 is recruited through interaction with the tetrapod-restricted family of KRAB-containing zinc finger proteins (Iyengar and Farnham, 2011). Therefore, it would be informative for the field to further define the mode of KAP1 recruitment to promoter-proximal regions of all KEC target genes. Recently, Bunch et al. reported that KAP1 appears to have single-stranded DNA-binding activity (Bunch et al., 2014). Additionally, KAP1 contains a bromodomain that can contact acetylated lysine residues on histones, indicating that KAP1 might be recruited by contacting DNA and/or chromatin directly. Further examination of these properties is needed to precisely ascertain the recruitment mechanism.
Recent work has indicated that the loss of KAP1 triggers a subtle increase in RNA levels in basal conditions for a subset of genes (Bunch et al., 2014). Based on these data, the authors hypothesized a role for KAP1 in the control of Pol II pausing. Our data (both in basal and stimulated conditions) indicate that KAP1 is needed for transcription elongation at most Pol II-regulated genes, with the most dramatic effect observed in the attenuation of PRG induction in response to stimulation. Although our findings focused on the stimulus-dependent transcriptional regulation by KAP1, we defined that KEC target genes comprise over 40.5% of all annotated genes (70.5% of genes containing paused Pol II). Therefore, further work to define how the KEC maintains transcriptional homeostasis would be needed.
Interestingly, KAP1 belongs to a family of transcriptional regulators known as transcription intermediary factor 1 composed of three members: TIF1α/TRIM24, TIF1β/TRIM28, and TIF1γ/TRIM33. The TIF1γ member controls erythroid cell fate by regulating transcription elongation of a selective small class of genes through P-TEFb (Bai et al., 2010; Rowe et al., 2010; Tsai et al., 2010). Although it is unknown whether TIF1γ interacts with the 7SK snRNP to indirectly recruit P-TEFb to promoters (such as KAP1), TIF1γ’s strikingly high homology to KAP1 would support a similar mechanism of interaction (Iyengar and Farnham, 2011). Therefore, further work in this arena will enhance our understanding of the TIF1/TRIM family in transcription elongation control through the 7SK snRNP.
In conclusion, we have discovered and characterized an unprecedented transcriptional signaling network that facilitates the rapid induction of genes upon stimulation. Through the KAP1-mediated recruitment of the 7SK snRNP to promoter-proximal regions, we propose that the P-TEFb kinase can be activated on site, allowing for proper Pol II pause release. This localized placement of transcriptional regulators allows the cell to efficiently respond to activating stimuli, a mechanism that HIV has hijacked for rapid viral activation and spread.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Standard cell-culture techniques were employed. The primers, plasmids, and antibodies used in this study are listed in Tables S3–S5, respectively.
shRNA-Mediated Knockdown, RNA Extraction, and qRT-PCR
RNA isolation and qRT-PCR were performed and analyzed as previously described (McNamara et al., 2013).
Immunoprecipitation of Endogenous Proteins
Endogenous proteins were immunopurified with antibodies conjugated to protein G Dynabeads (Life Technologies) using routine protocols.
Preparation of Nuclear Extracts for Affinity Purification and Mass Spectrometry
Nuclei from HEK293 T-REx cells inducibly expressing Larp7:S and GFP:S were prepared using the Dignam method (Dignam et al., 1983).
LC-MS/MS, Protein Identification, and Quantitation
Samples were analyzed using an in-house data analysis pipeline from the UT Southwestern Proteomics service. Table S1 lists the top ten high-confidence Larp7 interactors alongside the peptide and coverage calculations. Table S2 lists the proteins identified across all samples and provides spectral counts and spectral index values/ratios for all samples/ratios.
Indirect Immunofluorescence
Standard protocols were followed as previously described (McNamara et al., 2013).
ChIP-qPCR Assays and ChIP-Seq
ChIP assays were performed as previously described (McNamara et al., 2013).
Computational Analysis
All scripting was performed using Python 2.7.6 (http://www.python.org). All ChIP-seq binding events were loaded into a custom MySQL database (http://www.mysql.com) to allow for efficient comparisons of multiple factors’ binding loci.
Supplementary Material
Highlights.
KAP1 recruits 7SK snRNP to most genes containing promoter-proximal paused Pol II
KAP1 delivers inactive P-TEFb kinase to gene promoters for on-site activation
Inducible pathways rely on KAP1-7SK snRNP for P-TEFb delivery and Pol II elongation
HIV exploits KAP1-7SK snRNP to transcribe its genome in response to stimulation
Acknowledgments
We thank T. Kim and N. Conrad for critical reading of this manuscript; the UT Southwestern Proteomics and Live Cell Imaging cores for their assistance with protein identification and high-resolution microscopy, respectively; J. Espinosa for sharing and providing advice on GRO-seq; and C. Guzman for computational support. We are grateful to J. Karn for generously sharing the Jurkat HIV E4, 2B2D, and 2B5 clones. The 6.3, 8.4, and 9.2 Jurkat HIV clones were obtained from the NIH AIDS Reagent Program kindly deposited by E. Verdin. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the NIH under award numbers R56AI106514 and R01AI114362, and Welch Foundation grant I-1782 (to I.D.). R.P.M. was supported by NIH training grant 2T32AI007520-16. C.W.B. and E.A.M. were supported by NIH training grant 5T32GM8203-27.
Footnotes
ACCESSION NUMBERS
The accession number for the ChIP-seq raw sequence tags and bigwig files reported in this paper is NCBI GEO: GSE72622.
Supplemental Information includes Supplemental Experimental Procedures, four figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2015.11.004.
References
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Kim J, Yang Z, Jurynec MJ, Akie TE, Lee J, LeBlanc J, Sessa A, Jiang H, DiBiase A, et al. TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell. 2010;142:133–143. doi: 10.1016/j.cell.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- Bunch H, Zheng X, Burkholder A, Dillon ST, Motola S, Birrane G, Ebmeier CC, Levine S, Fargo D, Hu G, et al. TRIM28 regulates RNA polymerase II promoter-proximal pausing and pause release. Nat Struct Mol Biol. 2014;21:876–883. doi: 10.1038/nsmb.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Flynn RA, Martin L, Spitale RC, Chang HY, Wysocka J. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature. 2015;518:249–253. doi: 10.1038/nature13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrier T, Le Douce V, Eilebrecht S, Riclet R, Marban C, Dequiedt F, Goumon Y, Paillart JC, Mericskay M, Parlakian A, et al. CTIP2 is a negative regulator of P-TEFb. Proc Natl Acad Sci USA. 2013;110:12655–12660. doi: 10.1073/pnas.1220136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamant G, Dikstein R. Transcriptional control by NF-κB: elongation in focus. Biochim Biophys Acta. 2013;1829:937–945. doi: 10.1016/j.bbagrm.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- D’Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol. 2010;17:815–821. doi: 10.1038/nsmb.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler T, Sen R, Roy AL. Regulation of primary response genes. Mol Cell. 2011;44:348–360. doi: 10.1016/j.molcel.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, Allen MA, Bensard CL, Wang X, Schwinn MK, Qin B, Long HW, Daniels DL, Hahn WC, Dowell RD, Espinosa JM. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013;153:1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell. 2008;29:588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem. 2011;286:26267–26276. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadlowsky JK, Wong JY, Graham AC, Dobrowolski C, Devor RL, Adams MD, Fujinaga K, Karn J. Negative elongation factor is required for the maintenance of proviral latency but does not induce promoter-proximal pausing of RNA polymerase II on the HIV long terminal repeat. Mol Cell Biol. 2014;34:1911–1928. doi: 10.1128/MCB.01013-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Thérien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153:855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Defechereux P, Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 2001;20:1726–1738. doi: 10.1093/emboj/20.7.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem. 2005;280:28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal AK, Rosenfeld MG. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Xiang Y, Fujinaga K, Bartholomeeusen K, Nilson KA, Price DH, Peterlin BM. Release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein (snRNP) activates hexamethylene bisacetamide-inducible protein (HEXIM1) transcription. J Biol Chem. 2014;289:9918–9925. doi: 10.1074/jbc.M113.539015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara RP, McCann JL, Gudipaty SA, D’Orso I. Transcription factors mediate the enzymatic disassembly of promoter-bound 7SK snRNP to locally recruit P-TEFb for transcription elongation. Cell Rep. 2013;5:1256–1268. doi: 10.1016/j.celrep.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, Bensaude O. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- Pearson R, Kim YK, Hokello J, Lassen K, Friedman J, Tyagi M, Karn J. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J Virol. 2008;82:12291–12303. doi: 10.1128/JVI.01383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Smale ST. Selective transcription in response to an inflammatory stimulus. Cell. 2010;140:833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.