SUMMARY
Plants use cell surface-resident receptor-like kinases (RLKs) to sense diverse extrinsic and intrinsic cues and elicit distinct biological responses. In Arabidopsis, the ERECTA family RLKs recognize EPIDERMAL PATTERNING FACTORS (EPFs) to specify stomatal patterning. However, little is known about the molecular link between ERECTA activation and intracellular signaling. We report here that the SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) family RLKs regulate stomatal patterning downstream of EPF ligands and upstream of a MAP kinase cascade. EPF ligands induce the heteromerization of ERECTA and SERK family RLKs. SERKs and ERECTA family RLKs transphosphorylate each other. In addition, SERKs associate with the receptor-like protein (RLP) TMM, a signal modulator of stomata development, in a ligand-independent manner, suggesting that ERECTA, SERKs and TMM form a multi-protein receptorsome consisting of different RLKs and RLP perceiving peptide ligands in regulating stomatal patterning. In contrast to the differential requirement of individual SERK members in plant immunity, cell death control and BR signaling, all four functional SERKs are essential but with unequal genetic contributions to stomatal patterning with descending order of importance from SERK3/BAK1, SERK2, SERK1 to SERK4. Although BR signaling connects stomatal development via multiple components, the function of SERKs in stomatal patterning is uncoupled from their involvement in BR signaling. Our results reveal that the SERK family is a shared key module in diverse Arabidopsis signaling receptorsomes and different combinatorial codes of individual SERK members regulate distinct functions.
INTRODUCTION
Plants possess a largely expanded number of receptor-like kinases (RLKs) that are potentially involved in sensing intrinsic and extrinsic cues and lead to complex cellular networks with distinct signaling outputs [1, 2]. RLKs regulate a wide range of biological processes including plant growth, development, symbiosis and immunity via perception of diverse signals likely through different extracellular domains. The Arabidopsis genome contains more than 200 RLKs with extracellular leucine-rich repeat (LRR) domains [1]. An LRR-RLK typically contains an extracellular domain with different number of LRRs, a single transmembrane domain and an intracellular kinase domain. Some well-known examples of LRR-RLKs include the BRI1 receptor for brassinosteroids (BRs), a class of plant hormones with essential roles in growth and development [3]; FLS2 which recognizes bacterial flagellin or flg22 (the 22-amino-acid peptide of flagellin) and initiates plant immune signaling [4]; and the ERECTA (ER) family LRR-RLKs that recognize the endogenous peptides EPIDERMAL PATTERNING FACTOR 1 (EPF1) and EPF2 to control stomatal patterning [5, 6].
Stomata are epidermal pores that control water vapor and gas exchange between plants and the atmosphere and consist of two highly specialized guard cells (GCs) that surround each stomatal pore. In Arabidopsis, the stomatal lineage is initiated from a subset of protodermal cells that undergo a cellular transition to become meristemoid mother cells (MMCs) [7, 8]. An asymmetric entry division of the MMC generates a smaller, triangular cell called meristemoid and a larger cell called stomatal lineage ground cell (SLGC). The meristemoid either differentiates into a round-shaped guard mother cell (GMC) that further divides once into two GCs, or undergoes several amplifying divisions to produce more SLGCs. The SLGC either directly expands and differentiates into a pavement cell, or undergoes an asymmetric cell division to produce a satellite meristemoid that is oriented away from existing meristemoids or stomata [7, 8]. The “spacing” division of SLGCs ensures that stomata are always separated by at least one pavement cell, the so-called “one-cell-spacing rule”.
The signaling pathway controlling stomatal patterning is initiated by the secreted peptide ligands EPF1 and EPF2 that act as negative regulators with distinct functions. EPF1 functions mainly in the orientation of the cell spacing division, whereas EPF2 primarily controls asymmetric entry cell division [5, 9, 10]. The ER family LRR-RLKs, ER, ER-LIKE1 (ERL1) and ERL2 possess overlapping and distinct functions in the control of stomatal patterning [6]. EPF2-ER and EPF1-ERL1 function as ligand-receptor pairs to specify asymmetric entry division and spacing division respectively [5]. TOO MANY MOUTHS (TMM), an LRR-receptor-like protein (LRR-RLP), associates with ER family RLKs and differentially modulates stomatal development in different organs with a negative role in cotyledons and positive role in hypocotyls and stems [5, 11]. A MAP kinase (MAPK) cascade composed of YDA (MAPKKK), MKK4/MKK5 (MAPKKs) and MPK3/MPK6 (MAPKs), functions downstream of ER family RLKs and negatively regulates stomatal development [12–14]. Potential targets of the MAPK cascade include the transcription factors SPEECHLESS (SPCH), MUTE, FAMA, SCRM1 and SCRM2 [15–18]. SPCH directly targets key regulators of cell lineage specification and asymmetric cell division [19]. However, little is known about the molecular link between ER family receptor activation and intracellular signaling in stomatal development.
Receptor dimerization often constitutes the first step in the activation of downstream intracellular modules in RLK signaling [1]. BAK1, originally identified as a BRI1-associated receptor kinase mediating BR signaling [20, 21], is an important player in plant immunity via association with FLS2 and other immune sensors [22–25]. BAK1 is also known as SOMATIC EMBRYOGENESIS RECEPTOR KINASE 3 (SERK3), belonging to a subfamily of LRR-RLKs with 5 members [26]. Except for SERK5, which is likely a nonfunctional kinase [27], SERK1 to SERK4 possess diverse functions in male gametophyte development, BR-mediated growth, plant defense and cell death control [28, 29]. In this study, we report that the SERK family RLKs regulate stomatal development and patterning through ligand-induced heteromerization and transphosphorylation with ER and ERL1. Successive mutation of four SERK genes causes excessive stomatal clustering, reminiscent of the loss-of-function mutant for the entire ER family. Importantly, each SERK member makes an unequal contribution to stomatal patterning with descending order of importance from SERK3/BAK1, SERK2, SERK1 to SERK4. Our study indicates that the SERK family RLKs act as co-receptors for the ER family RLKs in regulating stomatal patterning and suggests that the combinatorial codes of individual SERK members control distinct cellular functions in cell fate determination, growth and immunity.
RESULTS
Ectopic expression of bacterial effector AvrPto or AvrPtoB impairs stomatal patterning upstream of YDA
Pathogenic bacteria inject a repertoire of effector proteins into host cells to modulate diverse host cellular activities and physiology [30, 31]. Interestingly, ectopic expression of the bacteria Pseudomonas syringae pv tomato (Pst) effector AvrPto in Arabidopsis transgenic plants under the control of a dexamethasone (Dex)-inducible promoter led to excessively clustered stomata in the cotyledon epidermis, which violated the one-cell-spacing rule in stomatal development (Figures 1A and S1A). The stomatal density indicated by the stomatal index was also much higher in the Dex∷AvrPto transgenic plants after Dex treatment than that without Dex treatment (Figure S1B). Similarly, expression of AvrPtoB, another Pst effector sharing certain overlapping host targets with AvrPto [32], also caused a strong stomatal clustering phenotype (Figure 1A). However, transgenic plants expressing AvrRpt2 or AvrRpm1, which has distinct virulence mechanisms from AvrPto and AvrPtoB [33], exhibited a similar stomatal patterning as wild-type (WT) Col-0 plants (Figure 1A). The MAPK cascade YDA-MKK4/MKK5-MPK3/MPK6 functions downstream of ER family RLKs in regulating stomatal development [12, 13]. AvrPto-mediated interference on stomatal development likely occurs upstream of YDA since expression of a constitutively active form of YDA (YDAac) rescued the AvrPto-induced stomatal patterning defects (Figure 1B). In addition, overexpression of AvrPto in Arabidopsis protoplasts did not interfere with the YDAac-mediated activation of MPK3 and MPK6 (Figure 1C), which is consistent with its suppression function in plant immune signaling [34]. These results suggest that AvrPto and AvrPtoB target a common signaling component(s) upstream of YDA to interfere with stomatal development in Arabidopsis. Since stomatal pore is a natural entry point for pathogen invasion [31], specific bacterial effectors may modulate stomatal density and patterning to promote pathogenicity.
Figure 1. Ectopic expression of effector protein AvrPto or AvrPtoB impairs stomatal patterning.
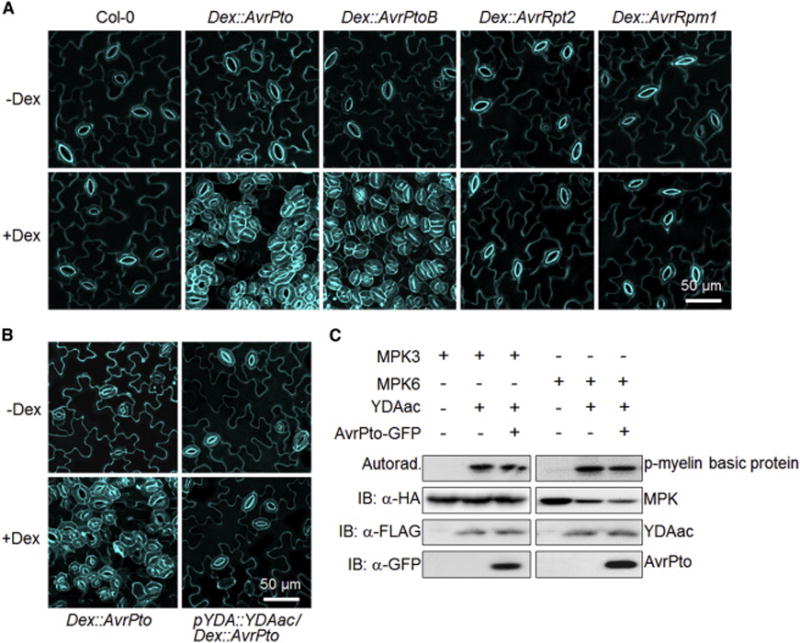
(A) Dex-induced expression of AvrPto or AvrPtoB but not AvrRpt2 or AvrRpm1 in Arabidopsis transgenic plants leads to severe stomatal clustering phenotypes. (B) Expression of YDAac rescues the AvrPto-induced stomatal patterning defects. Confocal images were taken on the abaxial cotyledon epidermis of 10-day-old seedlings grown on ½ MS medium with (A and B, bottom panels) or without (A and B, top panels) 100 μM Dex. Cell outlines were visualized with propidium iodide staining. The representative images in A and B were selected from at least five replicates. (C) Expression of AvrPto does not affect YDAac-mediated activation of MPK3 and MPK6 in Arabidopsis protoplasts. The HA-tagged MPK3/MPK6 and FLAG-tagged YDAac were co-expressed with or without AvrPto in protoplasts. The MPK3/MPK6 proteins were immunoprecipitated with α-HA agarose beads for an in vitro kinase assay using myelin basic protein as the substrate. The phosphorylation of myelin basic protein by MPK3/MPK6 is shown with autoradiograph (top panel), and the protein expression is shown with immunoblotting (bottom three panels). The experiments were repeated three times with similar results. (see also Figure S1).
The SERK family RLKs redundantly regulate stomatal patterning
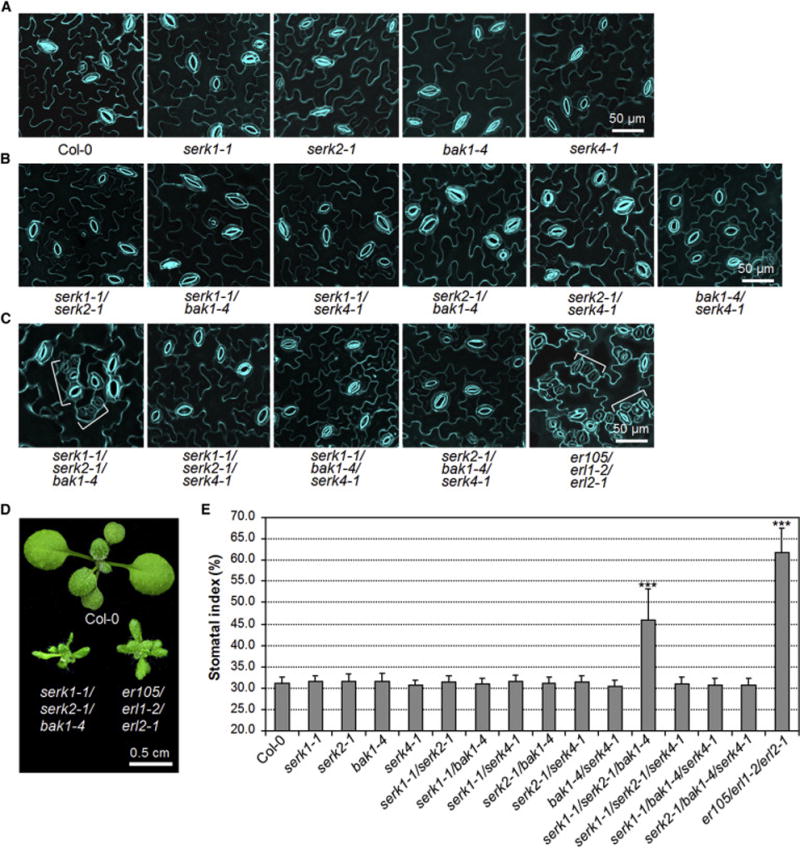
BAK1 is one of the physiological targets of AvrPto and AvrPtoB as supported by structural analysis of the BAK1-AvrPtoB complex and reduced virulence function of AvrPto/AvrPtoB in the bak1 mutant [32, 35, 36]. In addition, AvrPto and AvrPtoB also interact with other SERKs including SERK1, SERK2 and SERK4 (Figures S1C and S1D) [32]. Therefore, we tested whether the stomatal patterning defects in the AvrPto and AvrPtoB transgenic plants were caused by the dysfunction of BAK1 and other SERKs. However, neither the serk1-1, serk2-1, bak1-4 nor serk4-1 single null mutants displayed abnormal stomatal patterning compared to WT Col-0 plants (Figure 2A). To reveal the potential functional redundancy, we systemically generated different combinations of serk higher-order mutants. The stomatal patterning is normal in the cotyledon of all double mutants, including serk1-1/serk2-1, serk1-1/bak1-4, serk1-1/serk4-1, serk2-1/bak1-4, serk2-1/serk4-1 and bak1-4/serk4-1 (Figure 2B). Remarkably, clustered stomata were observed in the cotyledon epidermis of the serk1-1/serk2-1/bak1-4 triple mutant, but not in the other triple mutants including serk1-1/serk2-1/serk4-1, serk1-1/bak1-4/serk4-1 or serk2-1/bak1-4/serk4-1 (Figure 2C). The stomatal clusters in the serk1-1/serk2-1/bak1-4 mutant often consist of more than two stomata, similar to that of the er105/erl1-2/erl2-1 triple mutant, which harbors loss-of-function mutations in all three ER family genes, ER, ERL1 and ERL2 [6] (Figure 2C). In addition, the serk1-1/serk2-1/bak1-4 mutant, but not other mutants, exhibited similar growth morphology as the er105/erl1-2/erl2-1 mutant (Figures 2D and S2). Consistently, the stomatal index is also much higher in the cotyledon of the serk1-1/serk2-1/bak1-4 mutant than that in WT and other mutant plants (Figure 2E). The clustered stomata were also observed in the true leaves of serk1-1/serk2-1/bak1-4 triple mutant, but not in any other single, double or triple mutants (Figure S3). These results indicate that BAK1, SERK1 and SERK2 redundantly regulate stomatal development. The data are consistent with that both AvrPto and AvrPtoB target multiple SERK family members in Arabidopsis (Figures S1C and S1D) [32]. Notably, the extent of stomatal clustering in the serk1-1/serk2-1/bak1-4 mutant is weaker than that in AvrPto transgenic plants or the er105/erl1-2/erl2-1 mutant (Figures 1A, 2C and 2E). It is possible that SERK4 may also play certain roles in this process. However, the serk1-1/serk2-1/bak1-4/serk4-1 quadruple null mutant is embryolethal [27], which precludes the possibility to examine its stomatal development.
Figure 2. Redundant function of SERK family RLKs in stomatal patterning.
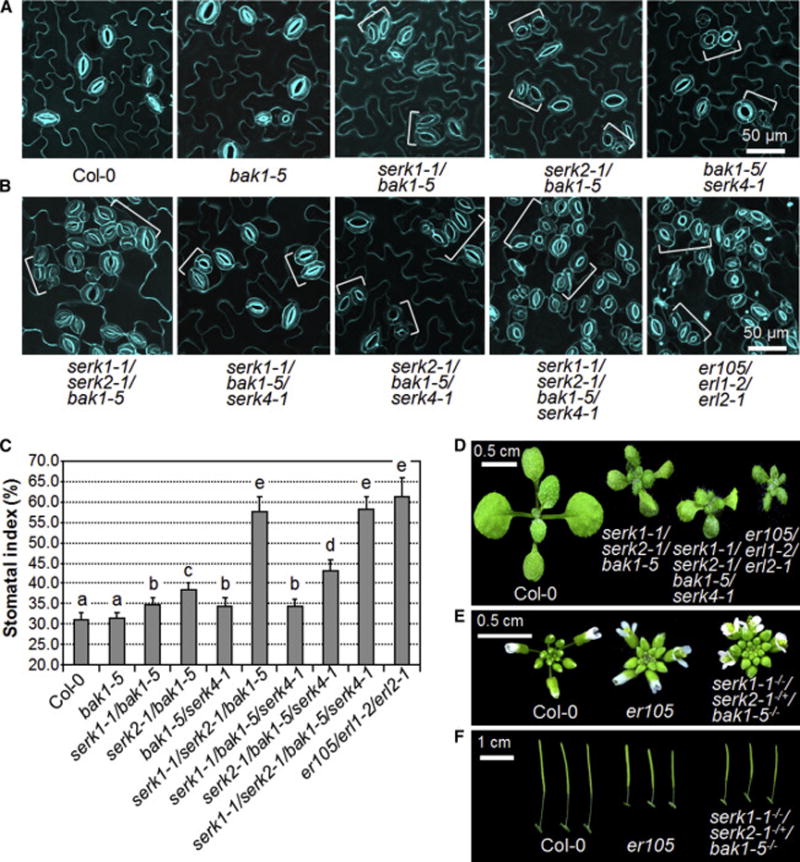
(A–C) The serk1-1/serk2-1/bak1-4 mutant but not other serk mutants shows stomatal patterning defects. Confocal images of indicated genotypes were taken on the abaxial cotyledon epidermis of 10-day-old seedlings grown on ½ MS plates. The representative images were selected from at least five replicates. Brackets indicate clustered stomata (C). (D) The seedling phenotypes of two-week-old serk1-1/serk2-1/bak1-4 and er105/erl1-2/erl2-1 mutants grown on soil. (E) Abaxial cotyledon stomatal index of 10-day-old seedlings, expressed as percentage of the number of stomata to the total number of epidermal cells. The data are shown as mean + SD (n=8). Asterisks above the columns indicate significant difference compared with the data from WT plants (*** P<0.0001, Student’s t-test). The experiments were repeated three times with similar results. (see also Figures S2 and S3).
Unequal redundancy of individual SERK members in stomatal patterning
In contrast to the null mutant bak1-4, the bak1-5 mutant, a semi-dominant allele with a mis-sense mutation in the kinase domain, is not impaired in cell death control or BR signaling, yet is severely compromised in immune responses [37]. To circumvent the embryonic lethality and further explore the roles of different SERK members in stomatal development, we generated higher-order serk mutants in the bak1-5 background. Although the bak1-5 single mutant exhibited normal stomatal patterning, the serk1-1/bak1-5, serk2-1/bak1-5 and bak1-5/serk4-1 double mutants displayed moderate stomatal clustering in the cotyledon compared to WT plants (Figure 3A). BAK1 is likely the most important SERK member in stomatal development since stomatal patterning defects were only observed in the cotyledon of serk double and triple mutants harboring the bak1 mutation but not in any other combinations (Figures 2B, 2C, 3A and 3B). Apparently, the stomatal clustering was more pronounced in the cotyledon of serk2-1/bak1-5 than those in the serk1-1/bak1-5 and bak1-5/serk4-1 mutants (Figures 3A and 3C), suggesting that SERK2 plays a more prominent role than SERK1 and SERK4 in stomatal patterning. The stomatal clustering and index of serk1-1/bak1-5/serk4-1 were similar with those of serk1-1/bak1-5 and bak1-5/serk4-1 (Figures 3A–3C), reinforcing the importance of SERK2 in stomatal patterning. Introduction of the serk4 mutation in bak1-5 or serk2-1/bak1-5 slightly but significantly increased stomatal clustering and index (Figures 3A–3C), indicating that SERK4 also plays an important but relatively minor role in stomatal development compared to BAK1 and SERK2. The stomatal clustering was much more severe in the cotyledon of serk1-1/serk2-1/bak1-5 than that in serk2-1/bak1-5/serk4-1 (Figures 3B and 3C), suggesting that SERK1 likely contributes more than SERK4 in stomatal development. Notably, the stomatal clustering and index in the cotyledon of serk1-1/serk2-1/bak1-5 and serk1-1/serk2-1/bak1-5/serk4-1 mutants were comparable to that in the er105/erl1-2/erl2-1 mutant (Figures 3B and 3C). Similarly, the stomatal clustering in descending order of severity was observed in the true leaves of serk1-1/serk2-1/bak1-5, serk2-1/bak1-5/serk4-1 and serk2-1/bak1-5 (Figure S4). The extent of stomatal clustering in the true leaves of serk1-1/serk2-1/bak1-5 was also comparable to that in er105/erl1-2/erl2-1 (Figure S4). However, we did not observe the stomatal clustering in the true leaves of serk1-1/bak1-5, bak1-5/serk4-1 and serk1-1/bak1-5/serk4-1 plants (Figure S4). Taken together, based on the extent of stomatal clustering in different serk double, triple and quadruple mutants, it appears that each SERK member contributes differentially to stomatal development with descending order of importance from BAK1, SERK2, SERK1 to SERK4. In contrast to the stomatal lineage cell-specific genes such as EPF1 and EPF2 [9, 10], the expression of BAK1-GFP under the control of BAK1 native promoter was observed ubiquitously in the epidermal cells including stomatal lineage cells in the pBAK1-BAK1-GFP transgenic plants (Figure S5A), which is consistent with the multifunctionality of SERK family RLKs in diverse signaling pathways [28, 29].
Figure 3. Differential contributions of SERK family RLKs in stomatal patterning.
(A, B) The stomatal clustering phenotypes of serk higher-order mutants in the bak1-5 background. Confocal images were taken on the abaxial cotyledon epidermis at 10 days after germination on ½ MS medium. Brackets indicate clustered stomata. (C) Abaxial cotyledon stomatal indexes of indicated genotypes. The data are shown as mean + SD (n=8). The mean values marked with different letters are significantly different from each other (P<0.05, Student’s t-test). The experiments were repeated three times with similar results. (D) The phenotypes of two-week-old seedlings grown on soil. (E, F) The serk1-1−/−/serk2-1−/+/bak1-5−/− plants phenocopy the er105 mutant in inflorescence architecture (E) and pedicel length (F). (see also Figures S4 and S5).
In addition, the serk1-1/serk2-1/bak1-5 and serk1-1/serk2-1/bak1-5/serk4-1 mutants morphologically mimic the er105/erl1-2/erl2-1 mutant in seedling stage (Figure 3D), whereas the morphologies of serk1-1/bak1-5/serk4-1 and serk2-1/bak1-5/serk4-1 are relatively normal compared to WT plants (Figure S5B). Moreover, the serk1-1−/−/serk2-1−/+/bak1-5−/− mutant also phenocopies the er105 mutant in inflorescence architecture (Figure 3E) and pedicel length (Figure 3F) [38]. Compared with WT plants, both the serk1-1−/−/serk2-1−/+/bak1-5−/− and er105 mutants exhibited clustered inflorescences (Figure 3E), which were associated with the shortened pedicels of these mutants (Figure 3F). The serk1-1−/−/serk2-1−/+/bak1-5−/− mutant was used here is because serk1-1/serk2-1 homozygous mutant is male sterile and does not produce any seeds [39, 40]. The similar stomatal clustering and growth phenotypes in serk and er mutants suggest genetic interaction between SERK and ER family RLKs.
Uncoupled functions of SERKs in stomatal patterning and BR signaling
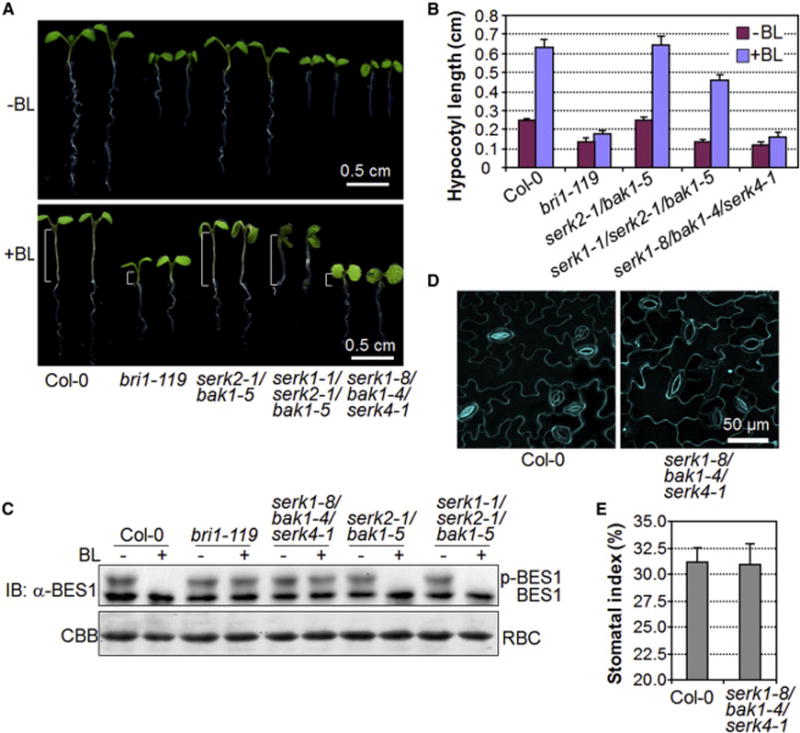
Members of the SERK family are also essential regulators of BR perception and signaling via complexing with the BR receptor BRI1 [20, 21, 27, 41]. It has been shown that BR regulates stomatal development through phosphorylation of YDA, MKK4/MKK5 and/or SPCH by the GSK3-like kinase BIN2 downstream of the BRI1-BAK1 complex [42–44]. To address whether the stomatal patterning defects in the serk mutants are caused by altered BR signaling, we examined the BR responses of the serk2-1/bak1-5 and serk1-1/serk2-1/bak1-5 mutants that displayed moderate and severe stomatal clustering, respectively. In contrast to the bri1-119 mutant, which no longer exhibited hypocotyl elongation in response to exogenous brassinolide (BL) treatment, both serk2-1/bak1-5 and serk1-1/serk2-1/bak1-5 mutants showed elongated hypocotyls upon BL treatment, similar to that observed in WT plants (Figures 4A and 4B). In addition, exogenous BL treatment induced the dephosphorylation of BES1 in both serk2-1/bak1-5 and serk1-1/serk2-1/bak1-5 mutants, comparable to that in WT plants (Figure 4C). Apparently, the BR sensitivity of the serk2-1/bak1-5 and serk1-1/serk2-1/bak1-5 mutants is similar to that of WT plants. These data support that the stomatal patterning defects in serk2-1/bak1-5 and serk1-1/serk2-1/bak1-5 mutants are not due to impaired BR signaling. In addition, the serk1-8/bak1-4/serk4-1 triple null mutant, in which BR signaling is completely abolished (Figures 4A–4C) [27], exhibited normal stomatal patterning and index (Figures 4D and 4E), reinforcing the uncoupled functions of SERK family RLKs in BR signaling and stomatal patterning. Notably, SERK2 is not required for BR signaling [27], whereas SERK2 is essential in stomatal development (Figures 2 and 3), suggesting the functional specificity of individual SERK family members.
Figure 4. Uncoupled functions of SERK family RLKs in stomatal patterning and BR signaling.
(A, B) The serk2-1/bak1-5 and serk1-1/serk2-1/bak1-5 mutants show normal hypocotyl elongation in response to brassinolide (BL) treatment. The seedlings were grown under the light for 10 days on ½ MS plates with or without 100 nM BL (A), and hypocotyl lengths were quantified (B). Brackets indicate hypocotyl (A). The data are shown as mean + SD (n=15) (B). (C) BL treatment induces the dephosphorylation of BES1 in serk2-1/bak1-5 and serk1-1/serk2-1/bak1-5 mutants. Ten-day-old seedlings grown in liquid ½ MS medium were treated with 0 or 1 μM BL for 2 hr, and the total proteins were analyzed by immunoblotting with α-BES1 antibody (Top panel). The protein loading is shown by Coomassie Brilliant Blue (CBB) staining for RuBisCO (RBC) (bottom panel). (D, E) The serk1-8/bak1-4/serk4-1 mutant exhibits normal stomatal patterning and index. Confocal images were taken on the abaxial cotyledon epidermis of 10-day-old seedlings (D), and the stomatal indexes were quantified (E). The experiments were repeated twice with similar results.
Interaction and transphosphorylation between SERK and ER family RLKs
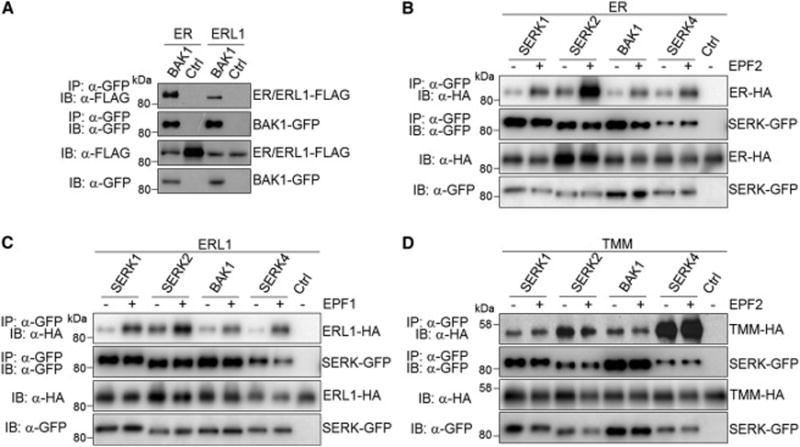
We next tested whether BAK1 and other SERKs associate with ER or ERL1 for regulating stomatal development. A co-immunoprecipitation (Co-IP) assay using co-expressed FLAG-tagged SERKs and HA-tagged ER or ERL1 in Arabidopsis protoplasts indicates that SERK1, SERK2, BAK1 and SERK4 were able to co-immunoprecipitate both ER and ERL1 (Figure S6A). We further crossed pBAK1∷BAK1-GFP transgenic Arabidopsis plants with pER∷ER-FLAG or pERL1∷ERL1-FLAG transgenic plants for the Co-IP assay. BAK1 could co-immunoprecipitate both ER and ERL1 when expressed under the control of their native promoters in transgenic Arabidopsis plants, indicating that they associate in vivo (Figure 5A). We further examined whether the EPF1 or EPF2 ligand could regulate the ER/ERL1-BAK1/SERK association dynamics. EPF1-ERL1 and EPF2-ER have been shown to function as ligand-receptor pairs specifying different steps of stomatal development [5]. Thus, we tested the ER-BAK1/SERK association in the presence of bioactive EPF2 peptide and the ERL1-BAK1/SERK association in the presence of EPF1 peptide. Importantly, EPF2 induced the association of ER with SERK1, SERK2, BAK1 and SERK4 (Figure 5B), and EPF1 induced the association of ERL1 with different SERKs (Figure 5C). The LRR-RLP TMM associates with ER and ERL1 and functions as a signal modulator in regulating stomatal patterning [5, 11]. TMM also shows binding ability to EPF2 but not EPF1 [5]. Interestingly, TMM also associated with SERK1, SERK2, BAK1 and SERK4 in Co-IP assays (Figure 5D). Apparently, the ligand EPF2 did not affect the association dynamics of TMM-BAK1/SERKs (Figure 5D). Taken together, these results suggest that ER/ERL1, BAK1/SERKs and TMM form a multi-protein receptor complex consisting of different RLKs and RLP to perceive and transduce EPF peptide signals and regulate stomatal development.
Figure 5. Interactions between SERK and ER family RLKs.
(A) BAK1 associates with ER and ERL1 in pBAK1∷BAK1-GFP/pER∷ER-FLAG and pBAK1∷BAK1-GFP/pERL1∷ERL1-FLAG transgenic plants. Protein extracts from transgenic plants were immunoprecipitated with α-GFP antibody (IP: α-GFP), and immunoblotted with α-FLAG (IB: α-FLAG) or α-GFP antibody (IB: α-GFP) (top two panels). The protein inputs are shown with immunoblotting before immunoprecipitation (bottom two panels). The pER∷ER-FLAG and pERL1∷ERL1-FLAG plants were used as controls here. (B) EPF2 induces the association of ER with SERKs in Arabidopsis protoplasts. SERK-GFP and ER-HA were transiently co-expressed in Arabidopsis protoplasts. After protoplasts were treated with or without 1 μM EPF2 for 5 min, protein extracts were immunoprecipitated with α-GFP antibody (IP: α-GFP), and immunoblotted with α-HA (IB: α-HA) or α-GFP antibody (IB: α-GFP) (top two panels). The protein inputs are shown with immunoblotting before immunoprecipitation (bottom two panels). (C) EPF1 induces the association of ERL1 with SERKs in Arabidopsis protoplasts. (D) SERKs associate with TMM in Arabidopsis protoplasts. Protoplasts were co-transfected with SERK-GFP and TMM-HA, and then treated with or without 1 μM EPF2 for 5 min. The experiments were repeated three times with similar results. (see also Figure S6A).
To test whether BAK1 directly interacts with ER through their cytosolic kinase domains (CD), we performed an in vitro pull-down assay. The maltose-binding protein (MBP)-tagged BAK1CD (MBP-BAK1CD) could be pulled down by the glutathione S-transferase (GST)-tagged ERCD (GST-ERCD), but not by GST itself (Figure 6A). Moreover, in vitro kinase assays show that MBP-BAK1CD phosphorylated GST-ERCD (Figure 6B), and GST-ERCD phosphorylated a kinase-inactive mutant of BAK1CD (MBP-BAK1CDKm) (Figure 6C), indicating the transphosphorylation of the ER-BAK1 receptor complex. Notably, although both BAK1 and ER are RD-type RLKs (Figure S6B), the kinase activity of ER is very weak compared with that of BAK1. This allowed us to demonstrate the in vitro phosphorylation of ER by BAK1 using WT ERCD (Figure 6B). Taken together, these data support that the SERK family RLKs transduce stomatal development signaling through transphosphorylation with the ER family RLKs.
Figure 6. Transphosphorylation between the cytosolic kinase domains of BAK1 and ER.
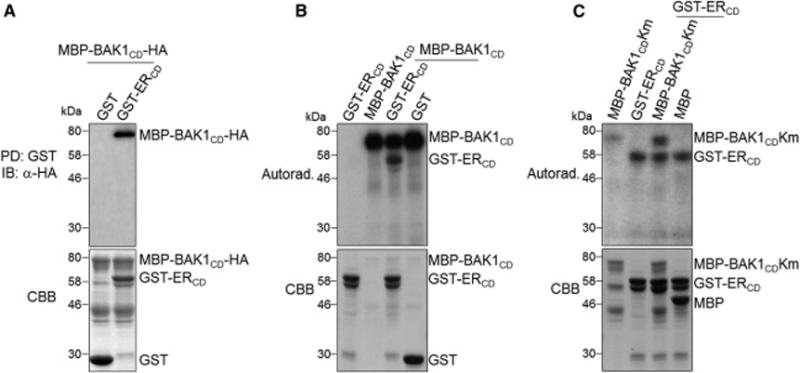
(A) BAK1CD interacts with ERCD in vitro. MBP-BAK1CD-HA proteins were incubated with GST or GST-ERCD glutathione beads, and the pull-down (PD) proteins were immunoblotted with α-HA antibody (top panel). The CBB staining of input proteins is shown on the bottom panel. (B) The phosphorylation of ERCD by BAK1CD (top panel). (C) The phosphorylation of BAK1CD by ERCD (top panel). The kinase assays were performed using ERCD and BAK1CD kinase mutant (BAK1CDKm) proteins as substrates in (B) and (C) respectively. The CBB staining of input proteins is shown on the bottom panels. The experiments were repeated three times with similar results. (see also Figure S6B).
SERKs function downstream of EPFs and upstream of YDA in regulating stomatal development
To examine whether SERKs are required for EPF1- and EPF2-mediated stomatal development, we treated the serk1-1/serk2-1/bak1-5 seedlings with bioactive EPF1 or EPF2 peptides, and introduced the estradiol (Est)-inducible EPF1 or EPF2 transgene into the serk1-1/serk2-1/bak1-5 mutant (Figure S7A). Similar with the previous report [5], application of EPF1 peptide or Est-induced overexpression of EPF1 in WT seedlings rendered the epidermis devoid of stomata with arrested meristemoids (Figures 7A and 7B). In contrast, seedlings of serk1-1/serk2-1/bak1-5 still exhibited excessively clustered stomata upon exogenous EPF1 treatment (Figure 7A) or induction of EPF1 overexpression (Figure 7B). In addition, application of EPF2 peptide or overexpression of EPF2 resulted in the epidermis with only pavement cells in WT seedlings, whereas the serk1-1/serk2-1/bak1-5 seedlings were insensitive to EPF2 application or overexpression and still exhibited severe stomatal clustering (Figures 7A and 7B). These demonstrate that EPF1- and EPF2-mediated stomatal development requires SERK family RLKs, and provide genetic evidence that SERKs function together with ER and ERL1 in regulating EPF2- and EPF1-mediated stomatal patterning.
Figure 7. SERKs function downstream of EPFs and upstream of YDA in regulating stomatal development.
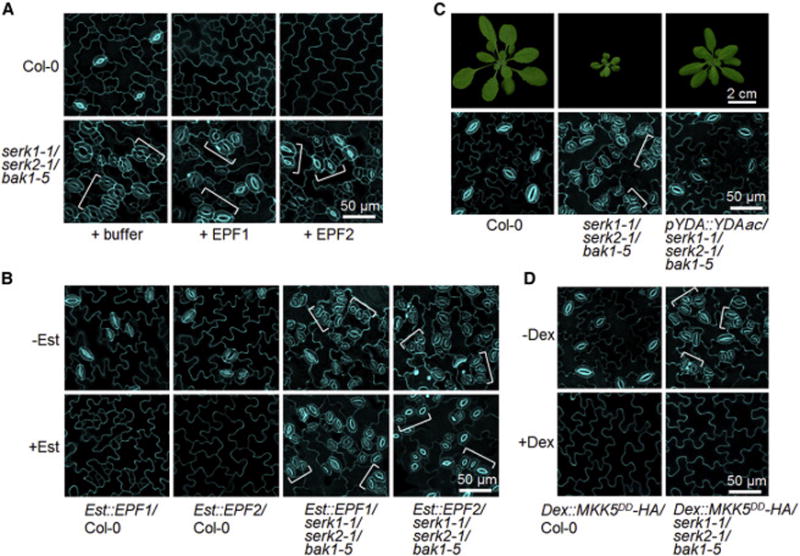
(A, B) SERKs are required for EPF1- and EPF2-mediated stomatal development. Confocal images were taken on the abaxial cotyledon epidermis of 6-day-old Col-0 and serk1-1/serk2-1/bak1-5 seedlings grown in ½ MS liquid medium containing 2.5 μM EPF1 or EPF2 (A) and 10-day-old transgenic seedlings of Est∷EPF1 or Est∷EPF2 grown on ½ MS plates with or without 10 μM estradiol (B). (C) Expression of YDAac driven by its native promoter rescues the growth and stomatal patterning defects of serk1-1/serk2-1/bak1-5. The images were taken on 4-week-old plants (top panels) or 10-day-old cotyledon epidermis (bottom panels). (D) Ectopic expression of MKK5DD eliminates stomata in serk1-1/serk2-1/bak1-5 mutant. Confocal images were taken on the abaxial cotyledon epidermis of 10-day-old transgenic seedlings of Dex∷MKK5DD with or without 0.02 μM Dex treatment. Brackets indicate clustered stomata. At least two transgenic lines for each construct in B-D were used, and the similar results were obtained. The representative images were selected from at least five replicates. (see also Figure S7).
To determine the genetic relationship between SERK family RLKs and the YDA-MKK4/MKK5-MPK3/MPK6 cascade, we transformed a constitutively active form of YDA (YDAac) driven by its native promoter into the serk1-1/serk2-1/bak1-5 mutant. As shown in Figure 7C, heterozygous YDAac was capable of fully rescuing the stomatal clustering defects in the serk1-1/serk2-1/bak1-5 mutant. Notably, heterozygous YDAac was also able to rescue the growth defects of serk1-1/serk2-1/bak1-5 plants (Figure 7C). Furthermore, a constitutively active MKK5 variant (MKK5DD) under the control of a Dex-inducible promoter was able to completely reverse the stomatal clustering phenotype in the serk1-1/serk2-1/bak1-5 mutant and resulted in the epidermis solely composed of pavement cells (Figures 7D and S7B). Collectively, these data further demonstrate that SERKs function in the same pathway with ER/ERL receptors upstream of the YDA-MKK4/MKK5-MPK3/MPK6 cascade in regulating stomatal development.
DISCUSSION
The SERK family RLKs connect complex signaling networks via association with various RLK receptors and modulate distinct cellular responses [26, 29]. From the observation that ectopic expression of pathogen effectors targeting the SERK family members led to clustered stomata in Arabidopsis, our study provides novel insights into the host cellular signaling that BAK1, SERK1, SERK2 and SERK4 negatively regulate stomatal development via ligand-induced heteromerization and transphosphorylation with the ER and ERL1 receptors downstream of the EPF1 and EPF2 ligands and upstream of the YDA-MKK4/MKK5-MPK3/MPK6 cascade. Our study elucidates that the SERK family RLKs function as a shared signaling node that modulates the interconnected architecture of complex cellular signaling networks yet disseminates diverse biological outcomes, including cell differentiation, growth and immunity. Identification of the SERK family RLKs as important regulators in stomatal development via association with the ER family receptors substantiates the similarity of signaling pathways downstream of multiple RLK receptors.
Apparently, a diverse combinatorial code of individual SERK family RLKs contributes to their functional specificity. BAK1 and SERK4, but not SERK1 or SERK2, are important regulators in plant innate immunity and cell death control [25, 45, 46]. In contrast, SERK1 and SERK2, but not BAK1 or SERK4, have a crucial and redundant function in anther development [39, 40]. BAK1, SERK1 and SERK4, but not SERK2, play an essential role in BR signaling [27]. We show here that all four functional SERKs (SERK1, SERK2, SERK3/BAK1 and SERK4) are involved in stomatal patterning (Figures 2 and 3). By comparison of stomatal clustering phenotypes in different combinations of serk higher-order mutants in the bak1-5 background (Figures 3A–3C and S4), we reveal the differential contributions of the SERK family RLKs in stomatal patterning, with descending order of importance from BAK1, SERK2, SERK1 to SERK4. This unequal functional redundancy of different SERKs was also observed in plant immunity and BR signaling pathways [25, 27].
Accumulating evidence indicates that the diverse functions of SERK family RLKs are uncoupled. For instance, the involvement of BAK1 and SERK4 in cell death control is independent of their function in BR signaling [45, 46]. The function of BAK1 in innate immunity can be separated from its involvement in cell death control and BR signaling [37]. Similarly, several lines of evidence suggest that SERKs regulate stomatal patterning independently of BR signaling: (1) Despite showing normal BR responses, the serk1-1/serk2-1/bak1-5 mutant displayed severe defects in stomatal patterning (Figure 3B, 4A–4C); (2) The serk1-8/bak1-4/serk4-1 mutant, in which the BR signaling is completely abolished [27], exhibited normal stomatal patterning (Figures 4D and 4E); (3) The serk2 mutation in either serk single or higher-order mutants had an undetectable effect on BR signaling [27], whereas the introduction of serk2 mutation in the serk1/bak1 double mutant dramatically exacerbated the stomatal clustering phenotype (Figure 3); (4) The BR receptor mutant bri1 and biosynthesis mutant det2 showed much weaker stomatal clustering phenotypes than the serk1/serk2/bak1 mutants [42, 44] (Figures 2 and 3). Thus, it appears that the SERK family RLKs function independently in different signaling pathways.
With a relatively short extracellular LRR domain, the SERK family RLKs appear not to be directly involved in the binding of ligands such as BL or flg22. Recent crystal structure analyses of the BL-BRI1-BAK1 and flg22-FLS2-BAK1 complexes indicate that BAK1 is involved in ligand sensing through contacting the BL-BRI1- or flg22-FLS2- binding interface [47–49]. Thus, although BAK1 itself does not confer BL- or flg22-binding activity, these structural studies support that the SERK family RLKs function as co-receptors to interact directly with the ligand-receptor complexes. In consistent with this model, BL and flg22 induce the heterodimerization of SERKs with BRI1 and FLS2 respectively [22, 23, 25, 50]. Similarly, we observed that EPF2 and EPF1 peptides induce the heterodimerization of SERKs with their corresponding receptors ER and ERL1 respectively (Figures 5B and 5C). Therefore, it is likely that the SERK family RLKs also serve as the co-receptors for ER and ERL in sensing EPF peptide signals. However, unlike FLS2 that does not oligomerize [49], both ER and ERL1 form receptor homomers and associate with the LRR-RLP TMM [5]. Lacking an obvious intracellular domain, TMM may not be directly involved in signal transduction. Genetic and biochemical studies indicate complex interactions with both antagonistic and cooperative roles between the ER family receptors and TMM in regulating stomatal patterning [5, 6, 11]. Interestingly, SERK family RLKs also associate with TMM (Figure 5D), suggesting that ER/ERL1, SERKs and TMM form a multi-protein receptor complex that perceives and transduces EPF peptide signals to regulate stomatal patterning. EPF ligands induce associations of the cognate receptors ER and ERL1 but not the signal modulator TMM with SERKs (Figures 5B, 5C and 5D), indicating the signaling role of ER/ERL1-SERK heterodimerization. It is likely that TMM may be involved in modulating the dimerization and/or activation of the ER/ERL1-SERK complexes. Future structural study of the EPF receptorsome consisting of multiple LRR-RLKs (ER/ERL and SERKs) and LRR-RLP (TMM) will provide insights into the activation mechanism of the ligand-receptor-co-receptor complex.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
Arabidopsis thaliana Columbia (Col-0) accession was used as wild-type (WT). The mutants bri1-119, bak1-4, er105/erl1-2/erl2-1, and the transgenic plants of pBAK1∷BAK1-GFP, Dex∷AvrPto, Est∷EPF1 and Est∷EPF2 in the Col-0 background, pER∷ER-FLAG in er105 and pERL1∷ERL1-FLAG in erl1-2 were reported previously [5, 6, 32, 34]. The other serk T-DNA insertional mutants were obtained from the Arabidopsis Biological Resource Center (serk1-1, SALK_044330; serk2-1, SALK_058020; serk4-1, SALK_057955). The Dex∷AvrRpt2 (in the rps2-101C mutant background) and the Dex∷AvrRpm1 (in the rpm1 mutant background) transgenic plants were obtained from Dr. Frederic Ausubel. The Dex∷AvrPtoB transgenic plants in Nd-0 background were obtained from Drs. John Mansfield and Murray Grant [51]. The bak1-5 mutant was obtained from Dr. Cyril Zipfel [37] and the serk1-8/bak1-4/serk4-1 mutant from Dr. Jia Li [27]. The serk double, triple and quadruple mutants, and pBAK1∷BAK1-GFP/pER∷ER-FLAG and pBAK1∷BAK1-GFP/pERL1∷ERL1-FLAG transgenic plants were generated by genetic crosses. Arabidopsis seeds were surface sterilized with 50% bleach and grown on half-strength Murashige and Skoog (½ MS) medium or on soil in a growth room at 23°C, 45% humidity and 75 μE m−2s−1 light with a 12 hr light/12 hr dark photoperiod.
Plasmid construction, protoplast transient assay and generation of transgenic plants
The Est∷EPF1 and Est∷EPF2 constructs were reported previously [5]. The pYDA∷YDAac construct was obtained from Dr. Wolfgang Lukowitz [52]. To make the Dex∷MKK5DD construct, the PCR product of a MKK5 variant containing constitutively active Ser-to-Asp mutations (MKK5DD) was introduced into a modified pTA7002 vector and fused with an HA epitope-tag at its C-terminus. ER, ERL1, SERK1, SERK2 and SERK4 genes were amplified by PCR from Col-0 cDNA, and cloned into the plant expression vector for transient protein expression in protoplasts. The ER cytosolic domain was subcloned into a modified pGEX4T-1 vector (Pharmacia) for GST fusion protein expression in E. coli, and the MBP fusion constructs of BAK1CD and BAK1CDKm were generated previously [53]. Protoplast transient assay was carried out as described previously [54]. Arabidopsis transgenic plants were generated using Agrobacterium-mediated transformation by the floral-dip method. For all transgenic plants, >20 T1 plants per construct were screened for transgene expression using reverse transcription-polymerase chain reaction (RT-PCR) or immunoblotting, and two to three T2 lines with a single insertion and similar transgene expression levels were subjected to phenotypic characterization. The primers are listed in Supplementary Table S1.
Histochemical analysis and microscopy
To visualize epidermal cell outlines, seedlings were stained with 0.2 mg/ml propidium iodide (PI) for 5 min, and then washed twice with water. Confocal images were taken using a Zeiss LSM700 microscope with a 20× objective lens. Histochemical staining of epidermis using toluidine blue O (TBO) (Sigma) was performed as described previously [5]. Stomatal index was quantified as the percentage of the number of stomata to the total number of epidermal cells using TBO-stained epidermal samples.
Chemical and peptide treatments
To characterize the BL-induced hypocotyl elongation, seeds were germinated on ½ MS plates containing 100 nM BL, and the hypocotyl length was measured 10 days after germination. To examine the BL-induced BES1 dephosphorylation, 10-day-old seedlings grown in ½ MS liquid medium were treated with 1 μM BL for 2 hr, and total proteins were analyzed by immunoblotting with an α-BES1 antibody (a generous gift from Dr. Yanhai Yin). Expression, purification and refolding of recombinant bioactive EPF1 and EPF2 peptides were performed as described previously [5]. For peptide treatment, either buffer alone (50 mM Tris-HCl, pH 8.0) or with 2.5 μM EPF peptides were applied to 1-day-old Arabidopsis plants germinated on ½ MS medium. After 5 days of further incubation in ½ MS liquid medium containing each peptide, stomatal phenotypes of abaxial cotyledons were analyzed with a confocal microscope. For chemical induction of transgenes, Est∷EPF1, Est∷EPF2 and Dex∷MKK5DD transgenic seeds were germinated on ½ MS plates containing 10 μM estradiol or 0.02 μM Dex, and stomatal phenotypes were examined 10 days after germination.
Coimmunoprecipitation, GST pull-down and in vitro phosphorylation assays
For the Co-IP assay, transfected protoplasts or leave tissues of 4-week-old soil-grown transgenic plants were lysed with 0.5–1 ml extraction buffer (10 mM HEPEs, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% Glycerol, 0.5% Triton X-100 and 1:200 complete protease inhibitor cocktail from Sigma). After vortexing vigorously for 30 sec, the samples were centrifuged at 16, 000×g for 10 min at 4°C, and the supernatant was then incubated with α-FLAG (Sigma) or α-GFP agarose beads (ChromoTek) for 2 hr at 4°C with gentle shaking. The immunoprecipitated proteins were analyzed by immunoblotting with α-HA (Roche) or α-FLAG (Sigma) antibody. Expression and purification of the GST and MBP fusion proteins were performed using standard protocols. For the GST pull-down assay, 10 μg of MBP-BAK1CD-HA proteins were incubated with prewashed GST or GST-ERCD glutathione beads in 0.5 ml pull-down buffer (10 mM HEPEs, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% Glycerol, 1% Triton X-100) for 2 hr at 4°C with gentle shaking. The pull-down proteins were analyzed by immunoblotting with α-HA antibody. For in vitro kinase assay, reactions were performed in 30 μl kinase buffer (20 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 5 mM EGTA, 100 mM NaCl, and 1 mM DTT) containing 10 μg of fusion proteins with 0.1 mM cold ATP and 5 μCi of [32P]-γ-ATP at room temperature for 2 hr with gentle shaking. The reactions were stopped by adding 4× SDS loading buffer, and the phosphorylation of fusion proteins was analyzed by autoradiography after separation with SDS/PAGE.
Supplementary Material
Highlights.
The SERK family receptor-like kinases redundantly regulate stomatal patterning
SERKs act downstream of the EPF ligands and upstream of the YDA MAPKKK
SERKs associate with the ERECTA family receptors in a ligand-induced manner
SERKs regulate stomatal patterning independently of brassinosteroid signaling
Acknowledgments
We thank Drs. Frederick Ausubel, Murray Grant, John Mansfield, Jia Li, Jianmin Li, Cyril Zipfel and the Arabidopsis Biological Resource Center for various Arabidopsis mutant and transgenic seeds, Dr. Wolfgang Lukowitz for the YDAac construct, Dr. Yanhai Yin for α-BES1 antibody, Dr. Julian Avila for assistance in purification of EPF peptides, Drs. Jen Sheen and Tim Devarenne for the critical reading of the manuscript, and the members of the laboratories of L.S., P.H., and K.U.T. for discussions and comments of the experiments. The work was supported by National Institutes of Health (NIH) (R01GM092893) and National Science Foundation (NSF) (IOS-1252539) to P.H., NIH (R01GM097247) and the Robert A. Welch foundation (A-1795) to L.S., and the Gordon and Betty Moore Foundation (GBMF3035) to K.U.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and one table.
AUTHOR CONTRIBUTIONS
X.M., X.C., K.U.T., P.H., and L.S. designed experiments, X.M., X.C., H.M., C.L., X.Y., and X.G. performed experiments and analyzed data, X.M., P.H., and L.S. wrote the manuscript with input from all co-authors.
The authors have declared no conflict of interests.
References
- 1.Belkhadir Y, Yang L, Hetzel J, Dangl JL, Chory J. The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors. Trends Biochem Sci. 2014;39:447–456. doi: 10.1016/j.tibs.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shiu SH, Bleecker AB. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 4.Gomez-Gomez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 5.Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, McAbee JM, Sarikaya M, Tamerler C, Torii KU. Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 2012;26:126–136. doi: 10.1101/gad.179895.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shpak ED, McAbee JM, Pillitteri LJ, Torii KU. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science. 2005;309:290–293. doi: 10.1126/science.1109710. [DOI] [PubMed] [Google Scholar]
- 7.Dong J, Bergmann DC. Stomatal patterning and development. Curr Top Dev Biol. 2010;91:267–297. doi: 10.1016/S0070-2153(10)91009-0. [DOI] [PubMed] [Google Scholar]
- 8.Pillitteri LJ, Torii KU. Mechanisms of stomatal development. Annu Rev Plant Biol. 2012;63:591–614. doi: 10.1146/annurev-arplant-042811-105451. [DOI] [PubMed] [Google Scholar]
- 9.Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007;21:1720–1725. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt L, Gray JE. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr Biol. 2009;19:864–869. doi: 10.1016/j.cub.2009.03.069. [DOI] [PubMed] [Google Scholar]
- 11.Nadeau JA, Sack FD. Control of stomatal distribution on the Arabidopsis leaf surface. Science. 2002;296:1697–1700. doi: 10.1126/science.1069596. [DOI] [PubMed] [Google Scholar]
- 12.Bergmann DC, Lukowitz W, Somerville CR. Stomatal development and pattern controlled by a MAPKK kinase. Science. 2004;304:1494–1497. doi: 10.1126/science.1096014. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell. 2007;19:63–73. doi: 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Wang P, Shao W, Zhu JK, Dong J. The BASL Polarity Protein Controls a MAPK Signaling Feedback Loop in Asymmetric Cell Division. Developmental cell. 2015;33:136–149. doi: 10.1016/j.devcel.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell. 2008;20:1775–1785. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lampard GR, MacAlister CA, Bergmann DC. Arabidopsis Stomatal Initiation Is Controlled by MAPK-Mediated Regulation of the bHLH SPEECHLESS. Science. 2008;322:1113–1116. doi: 10.1126/science.1162263. [DOI] [PubMed] [Google Scholar]
- 17.MacAlister CA, Ohashi-Ito K, Bergmann DC. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature. 2007;445:537–540. doi: 10.1038/nature05491. [DOI] [PubMed] [Google Scholar]
- 18.Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007;445:501–505. doi: 10.1038/nature05467. [DOI] [PubMed] [Google Scholar]
- 19.Lau OS, Davies KA, Chang J, Adrian J, Rowe MH, Ballenger CE, Bergmann DC. Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science. 2014;345:1605–1609. doi: 10.1126/science.1256888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Wen JQ, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 21.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 22.Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JDG, Felix G, Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–U412. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 23.Heese A, Hann DR, Gimenez-Ibanez S, Jones AME, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. P Natl Acad Sci USA. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Postel S, Kufner I, Beuter C, Mazzotta S, Schwedt A, Borlotti A, Halter T, Kemmerling B, Nurnberger T. The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur J Cell Biol. 2010;89:169–174. doi: 10.1016/j.ejcb.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tor M, de Vries S, Zipfel C. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell. 2011;23:2440–2455. doi: 10.1105/tpc.111.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aan den Toorn M, Albrecht C, de Vries S. On the Origin of SERKs: Bioinformatics Analysis of the Somatic Embryogenesis Receptor Kinases. Mol Plant. 2015;8:762–782. doi: 10.1016/j.molp.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 27.Gou X, Yin H, He K, Du J, Yi J, Xu S, Lin H, Clouse SD, Li J. Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 2012;8:e1002452. doi: 10.1371/journal.pgen.1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J. Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr Opin Plant Biol. 2010;13:509–514. doi: 10.1016/j.pbi.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Liebrand TW, van den Burg HA, Joosten MH. Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 2014;19:123–132. doi: 10.1016/j.tplants.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Macho AP, Zipfel C. Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr Opin Microbiol. 2015;23:14–22. doi: 10.1016/j.mib.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 31.Xin XF, He SY. Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol. 2013;51:473–498. doi: 10.1146/annurev-phyto-082712-102321. [DOI] [PubMed] [Google Scholar]
- 32.Shan L, He P, Li J, Heese A, Peck SC, Nurnberger T, Martin GB, Sheen J. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S, Dangl JL, Mackey D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 34.He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nurnberger T, Sheen J. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell. 2006;125:563–575. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 35.Cheng W, Munkvold KR, Gao H, Mathieu J, Schwizer S, Wang S, Yan YB, Wang J, Martin GB, Chai J. Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase-interacting domains in a type III Effector. Cell Host Microbe. 2011;10:616–626. doi: 10.1016/j.chom.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou J, Wu S, Chen X, Liu C, Sheen J, Shan L, He P. The Pseudomonas syringae effector HopF2 suppresses Arabidopsis immunity by targeting BAK1. Plant J. 2014;77:235–245. doi: 10.1111/tpj.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 2011;7:e1002046. doi: 10.1371/journal.pgen.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shpak ED, Berthiaume CT, Hill EJ, Torii KU. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development. 2004;131:1491–1501. doi: 10.1242/dev.01028. [DOI] [PubMed] [Google Scholar]
- 39.Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S. The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell. 2005;17:3337–3349. doi: 10.1105/tpc.105.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell. 2005;17:3350–3361. doi: 10.1105/tpc.105.036731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karlova R, Boeren S, Russinova E, Aker J, Vervoort J, de Vries S. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell. 2006;18:626–638. doi: 10.1105/tpc.105.039412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gudesblat GE, Schneider-Pizon J, Betti C, Mayerhofer J, Vanhoutte I, van Dongen W, Boeren S, Zhiponova M, de Vries S, Jonak C, et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nature cell biology. 2012;14:548–554. doi: 10.1038/ncb2471. [DOI] [PubMed] [Google Scholar]
- 43.Khan M, Rozhon W, Bigeard J, Pflieger D, Husar S, Pitzschke A, Teige M, Jonak C, Hirt H, Poppenberger B. Brassinosteroid-regulated GSK3/Shaggy-like kinases phosphorylate mitogen-activated protein (MAP) kinase kinases, which control stomata development in Arabidopsis thaliana. The Journal of biological chemistry. 2013;288:7519–7527. doi: 10.1074/jbc.M112.384453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim TW, Michniewicz M, Bergmann DC, Wang ZY. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature. 2012;482:419–422. doi: 10.1038/nature10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He K, Gou X, Yuan T, Lin H, Asami T, Yoshida S, Russell SD, Li J. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr Biol. 2007;17:1109–1115. doi: 10.1016/j.cub.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 46.Kemmerling B, Schwedt A, Rodriguez P, Mazzotta S, Frank M, Qamar SA, Mengiste T, Betsuyaku S, Parker JE, Mussig C, et al. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol. 2007;17:1116–1122. doi: 10.1016/j.cub.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 47.Santiago J, Henzler C, Hothorn M. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science. 2013;341:889–892. doi: 10.1126/science.1242468. [DOI] [PubMed] [Google Scholar]
- 48.Sun Y, Han Z, Tang J, Hu Z, Chai C, Zhou B, Chai J. Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 2013;23:1326–1329. doi: 10.1038/cr.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science. 2013;342:624–628. doi: 10.1126/science.1243825. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 51.de Torres M, Mansfield JW, Grabov N, Brown IR, Ammouneh H, Tsiamis G, Forsyth A, Robatzek S, Grant M, Boch J. Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. Plant J. 2006;47:368–382. doi: 10.1111/j.1365-313X.2006.02798.x. [DOI] [PubMed] [Google Scholar]
- 52.Lukowitz W, Roeder A, Parmenter D, Somerville C. A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell. 2004;116:109–119. doi: 10.1016/s0092-8674(03)01067-5. [DOI] [PubMed] [Google Scholar]
- 53.Lin W, Li B, Lu D, Chen S, Zhu N, He P, Shan L. Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc Natl Acad Sci U S A. 2014;111:3632–3637. doi: 10.1073/pnas.1318817111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He P, Shan L, Sheen J. The use of protoplasts to study innate immune responses. Methods Mol Biol. 2007;354:1–9. doi: 10.1385/1-59259-966-4:1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


