Abstract
The host immune system plays a significant role in tumor control, although most cancers escape immune surveillance through a variety of mechanisms. The aim of the present study was to evaluate the clinicopathological significance of a novel co‐inhibitory receptor, B and T lymphocyte attenuator (BTLA), the anergy cell marker Casitas–B‐lineage lymphoma protein‐b (Cbl‐b), and clinical implications of tumor‐infiltrating immune cells in gallbladder cancer (GBC) tissues. We investigated 211 cases of GBC, 21 cases of chronic cholecystitis (CC), and 11 cases of xanthogranulomatous cholecystitis (XGC) using immunohistochemistry to detect tissue‐infiltrating immune cells and their expression of BTLA and Cbl‐b, and carried out correlation and survival analyses. The density of infiltrating T cells was significantly higher in CC and XGC than in GBC. The density ratio of BTLA + cells to CD8+ T cells (BTLA/CD8) and that of Cbl‐b+ cells to CD8+T cells (Cbl‐b/CD8) were significantly higher in GBC than in CC and XGC. The FOXP3/CD4, BTLA/CD8, and Cbl‐b/CD8 ratios were significantly correlated with each other, and also with malignant phenotypes. Survival analyses revealed that a lower density of tumor‐infiltrating CD8+ cells, and higher Foxp3/CD4, BTLA/CD8, and Cbl‐b/CD8 ratios were significantly associated with shorter overall survival and disease‐free survival in GBC patients. Multivariate analyses showed that M factor, perineural invasion, BTLA/CD8, and Cbl‐b/CD8 were closely associated with shorter overall survival. These findings suggest that higher ratios of BTLA/CD8 and Cbl‐b/CD8 are independent indicators of unfavorable outcome in GBC patients, and that upregulation of BTLA in cancer tissues is involved in inhibition of antitumor immunity.
Keywords: Anergy, BTLA, gallbladder cancer, immune checkpoint, immune microenvironment
Gallbladder cancer (GBC) is the most common malignant biliary neoplasm and the seventh most common gastrointestinal cancer.1 Complete surgical resection is the standard treatment for patients with localized disease, and the only potentially definitive curative therapy.2, 3 However, because most cases of GBC are diagnosed at an advanced stage, when the disease is not amenable to surgical resection, this malignancy is highly lethal, with a 5‐year survival rate of <5% for such patients.4, 5 Therefore, a more thorough understanding of GBC is essential for the development of new therapeutic strategies.
Based on a growing body of evidence from both animal and human models, it is generally accepted that naturally occurring immunity can play a significant role in the control of tumor development and progression.6 Most clinically evident cancers exhibit immune escape, where the cancer microenvironment can induce immune tolerance through a variety of mechanisms, such as the production of soluble immunosuppressive factors and the recruitment of suppressor immune cells.7, 8 Elucidation of the molecular and cellular events responsible for tumor immune escape is essential in order to make anticancer therapy truly effective in a clinical setting.9 Tumor‐infiltrating immune cells are one of the representative cellular components of host antitumor immune responses and tumor immune escape. Tumor‐infiltrating immune cells are composed of different cell subsets, which determine the overall protumor or antitumor characteristics. For example, a high proportion of CD8+ T cells infiltrating the cancer tissue can be a favorable prognostic indicator in colorectal,10, 11 ovarian,12 esophageal,13 liver,14 and pancreatic15, 16 cancers. In contrast, patients whose cancers show marked infiltration of regulatory T cells tend to have a poorer prognosis in several types of cancer.17, 18, 19 Two groups have studied the tumor‐infiltrating immune cells in GBC using small numbers of cases (45 cases20 and 69 cases21), and showed that tumor‐infiltrating CD8+ T cells and CD4+ T cells were indicators of favorable prognosis, whereas tumor‐infiltrating FOXP3+ cells or CD20+ B cells had no significant impact on outcome.
In addition to immunosuppressive cells, tumor cells deploy distinct mechanisms to evade immune attack, including expression of ligands for co‐inhibitory receptors on T cells.22 Recently, the distinct role of co‐inhibitory receptors, such as CTL‐associated protein 4 (CTLA‐4) and programmed cell death 1 (PD‐1), in this process has been extensively investigated.23 The function of antigen‐specific CD8+ T cells, which may protect against both infectious and malignant diseases, can be impaired by ligation of these inhibitory receptors. Expression of these receptors has been linked to functional impairment of T cells in cancer, and therapeutic blockade of these two pathways has shown clinical promise. Antagonist antibodies have been developed in order to overcome immune evasion, and anti‐CTLA‐4 and anti‐PD1 antibodies have been tested in clinical trials with encouraging results.24, 25, 26, 27 Anti‐CTLA4 antibody was the first agent shown to confer a survival benefit on patients with advanced melanoma, and was approved by the US FDA in 2010.
B and T lymphocyte attenuator (BTLA) has been identified as a novel co‐inhibitory receptor, expressed by a majority of lymphocytes, and shows structural and functional similarities to CTLA‐4 and PD‐1.28 Because tumor‐infiltrating immune cells express multiple co‐inhibitory receptors, it is assumed that dual or triple blockade of co‐inhibitory receptors will enhance antitumor immunity. Indeed, combined blockade of the PD‐1/programmed death‐ligand 1 (PD‐L1) and CTLA‐4 pathways, and that of the PD‐1/PD‐L1 and lymphocyte activation gene 3 (LAG3) pathways, has been shown to enhance antitumor effects in a human clinical study and animal model studies.29, 30, 31 Thus, BTLA is expected to be a new target for interventions aimed at reversal of immune evasion and boosting of antitumor immunity in cancer patients. Expression of BTLA has been reported in B‐cell small lymphocytic lymphoma/chronic lymphocytic leukemia cells and gastric cancer cells.32, 33 However, there are currently no data on the immunohistochemical expression of BTLA in the microenvironment of human tumors, and the clinical significance of BTLA expression in tumor‐infiltrating immune cells remains to be identified. It have also been reported that BTLA plays a critical role in the induction of peripheral T cell anergy in vivo.34 T cell anergy is also widely accepted as an important mechanism of tumor immune escape,35 although no previous studies have shown anergy cells in tissues. Casitas–B‐lineage lymphoma protein‐b (Cbl‐b), an E3 ubiquitin ligase, is critical for establishing the threshold for T cell activation and for induction of T cell anergy.36, 37
The aim of this study was to evaluate the clinicopathological impact of BTLA as well as the clinical implications of tumor‐infiltrating immune cells in patients with GBC to assess their potential as new targets for cancer immunotherapy.
Materials and Methods
Study population
Clinical and pathologic data and the specimens used for immunohistochemical analysis were obtained through a detailed retrospective review of the medical records of all 211 patients with GBC who had undergone surgical resection between 1985 and 2012 at the National Cancer Center Hospital (Tokyo, Japan). All of the patients included in this study underwent macroscopic curative resection, and all had primary carcinomas of the gallbladder. Neuroendocrine neoplasms were excluded. The median follow‐up period was 28 months (range, 1–289 months) for the patients overall: 89 patients (42.2%) were alive, 84 (39.8%) had died because of GBC, and 38 (18.0%) had died of other causes. During the study period, adjuvant chemotherapy was not carried out. In addition, we analyzed cases of chronic cholecystitis (CC) (n = 21) and xanthogranulomatous cholecystitis (XGC) (n = 11) as controls for evaluating the significance of tumor‐infiltrating immune cells.
This study was approved by the Institutional Review Board of the National Cancer Center, Japan. Informed consent was obtained from all participants involved in the study, and all clinical investigations were carried out in line with the principles of the Declaration of Helsinki.
Pathological examination
All of the carcinomas were examined pathologically and classified according to the World Health Organization classification,38 Union for International Cancer Control TNM classification,39 and the Japanese Society of Biliary Surgery classification of biliary tract carcinoma.40 Tumors were staged and the histopathologic variables (histopathological grading, lymphatic, venous, and perineural invasion) were evaluated and described in accordance with their classifications.39, 40
Immunohistochemistry
Immunohistochemistry was carried out on formalin‐fixed, paraffin‐embedded tissue sections using the avidin–biotin complex method as described previously.41 We used 4‐μm‐thick serial sections of representative blocks with antibodies against the following: CD3 (PS1; 1:100) from Santa Cruz Biotechnology (Santa Cruz, CA, USA), CD4 (368; 1:100) and CD8 (4B11; 1:200) from Leica Microsystems (Newcastle‐upon‐Tyne, UK), FOXP3 (42; 1:100) produced in house,18 BTLA (HPA047211; 1:500) from Atlas Antibodies (Stockholm, Sweden), and Cbl‐b (246C5A; 1:50) from Abcam (Cambridge, UK). Immunohistochemistry without the primary antibody was used as a negative control.
Double immunostaining
We carried out double staining on formalin‐fixed paraffin‐embedded sections. First, the 4‐μm‐thick sections were immunostained using anti‐BTLA antibody or anti‐Cbl‐b antibody as the primary antibody, and visualized with 3,3′‐diaminobenzidine. After the tissue sections had been treated with glycine–HCl (pH 2.5), they were subjected to immunofluorescence staining using antibodies against each of the following antigens: CD1a (O10, Lab Vision, Fremont, CA, USA), CD3, CD4, CD8, CD14 (7, Leica Microsystems), CD20 (L26, DAKO), CD56 (1B6, Leica Microsystems), CD68 (KP1, DAKO), CD207 (12D6, Leica Microsystems), CD208 (104.G4, Immunotech, Fullerton, CA, USA), FOXP3, BTLA, and Cbl‐b. Immunostained tissue sections were analyzed with a confocal microscope (LSM5 Pascal; Carl Zeiss, Jena, Germany) equipped with a 15‐mW Kr/Ar laser.
Quantitative evaluation of tumor‐infiltrating T cell subsets, BTLA‐positive cells, and Cbl‐b‐positive cells
After immunohistochemistry, the microscopic images were imported as digital photo files using a NanoZoomer Digital Pathology system (Hamamatsu Photonics, Hamamatsu, Japan), and the density of the immunolabeled cells was analyzed using the image analysis software, Tissue Studio (Definiens, Munich, Germany). We manually selected one area as region of interest (ROI), in which the CD3‐labeled T cells had infiltrated into the tumor most densely in the specimen, when we checked it in low‐power view. In each individual case, the same ROI was applied to all the other immunostained images. The immunolabeled cells inside the ROI were automatically counted on the basis of staining intensity. In each analysis we confirmed that the immunohistochemically positive lymphocytes were appropriately detected. The density of positive cells was calculated by dividing their number by the ROI area (cells/μm2). Also, we calculated the density ratio of FOXP3 to CD4 (FOXP3/CD4), that of BTLA to CD3 or CD8 (BTLA/CD3, BTLA/CD8), and that of Cbl‐b to CD3 or CD8 (Cbl‐b/CD3, Cbl‐b/CD8). For survival and correlation analyses, patients were divided into two groups showing high and low cell infiltration, using the median value as a cut‐off.
Statistical analysis
We expressed continuous data as median and range and compared them using the Mann–Whitney U‐test. We compared categorical data by Pearson's X2 or Fisher's exact test, as appropriate. We constructed survival curves by the Kaplan–Meier method and compared them using the log–rank test. We calculated the length of overall survival (OS) from the date of surgical resection to the date of death from any cause, calculated the length of disease‐free survival (DFS) from the date of surgical resection to the date of the first radiologic findings for recurrence, and censored those on the date of the last follow‐up. To evaluate the prognostic significance of tumor‐infiltrating lymphocytes and cells expressing inhibitory molecules in patients with GBC, univariate and multivariate Cox analyses were applied. The factors found to be significant by univariate analysis were subjected to multivariate analysis. P < 0.05 was considered to denote statistical significance for all analyses. Statistical analyses were carried out using spss version 20.0 (SPSS, Chicago, IL, USA).
Results
Immunophenotype of BTLA+ cells and Cbl‐b+ cells
To examine the immunophenotype of BTLA+ cells and Cbl‐b+ cells, double immunostaining was carried out (Fig. 1). BTLA was present in a proportion of CD3+ T cells, CD4+ T cells, CD8+ T cells, CD20+ B cells, CD14+ monocytes, CD68+ macrophages, CD1a+ dendritic cells (DCs), CD207+ DCs, or CD208+ DCs, although the majority of BTLA+ cells were CD4+. In contrast, no expression of BTLA was found in FOXP3+ cells, CD56+ natural killer (NK) cells, or Cbl‐b+ cells. Cbl‐b was expressed in a small proportion of CD3+ T cells, CD4+ T cells, CD8+ T cells, Foxp3+ cells, and CD20+ B cells, and was not expressed in CD56+ NK cells, CD14+ monocytes, CD68+ macrophages, or DCs. There were no BTLA+Cbl‐b+ cells in the cancer tissues, although T cells that had infiltrated into cancer cell nests and become attached to cancer cells were often positive for Cbl‐b, and sometimes for BTLA. No tumor cells expressed BTLA or Cbl‐b in our series.
Figure 1.
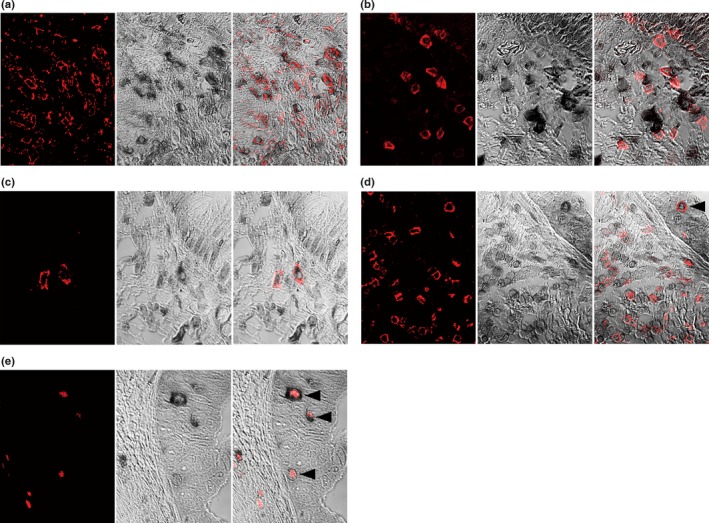
Double immunostaining features in gallbladder cancer samples. (a) Most B and T lymphocyte attenuator (BTLA)+ cells (black) are CD4+ T cells (red). (b, c) BTLA (black) staining is sometimes present in CD8+ T cells (red) (b) or CD1a+ (red) dendritic cells infiltrated in the cancer stroma (c). (d) Tumor‐infiltrating CD8+ T cells (red) are often Casitas–B‐lineage lymphoma protein‐b (Cbl‐b)+ (black). CD8+ T cells that have infiltrated and become attached to cancer cells (arrowhead) are Cbl‐b+ anergic cells. (e) FOXP3+ regulatory T cells (red) often express Cbl‐b (black). Arrowheads indicate regulatory T cells that have infiltrated and become attached to cancer cells. (a–e) Left column shows immunofluorescence staining for CD4 (a), CD8 (b, d), CD1a (c), or FOXP3 (e). Center column shows immunohistochemical staining for BTLA (a–c) or Cbl‐b (d, e). Right column shows merged images of left and center photographs.
Infiltration of T cell subsets, BTLA+ cells, and Cbl‐b+ cells
The median area of the ROI was 10 129 950 μm2 among the 212 cases of GBC, 1 336 266 μm2 among the 21 cases of CC, and 4 922 487 μm2 among the 11 cases of XGC. Representative photos of each type of positive cell are shown in Figure 2. Although there were too few samples of normal gallbladder tissue to compare with cancer tissue statistically, we found several CD3+ cells, CD4+ cells, and CD8+ cells in lamina propria. And a few FOXP3+ cells or Cbl‐b+ cells but no BTLA+ cells were found. Figure 3(a) shows a comparison of T cell subsets, BTLA‐positive cells, and Cbl‐b‐positive cells infiltrating into CC (n = 21), XGC (n = 11), and GBC (n = 211). The density of CD3+ cells, CD4+ cells, and CD8+ cells in CC and XGC was significantly higher than that in GBC, respectively (P < 0.01). In contrast, the density of FOXP3+ cells, BTLA+ cells, and Cbl‐b+ cells did not differ significantly among the diseases, except for the density of BTLA+ cells between CC and GBC. The number of suppressor cells such as regulatory T cells (Tregs) is often affected by the entire T cell infiltration.16, 18 It is often better to estimate the immune microenvironment from the ratio of the number of tumor‐infiltrating suppressor cells to that of the immune responsive cell population, rather than the absolute number of tumor‐infiltrating suppressor cells.16, 18, 42 We used the ratio of FOXP3 density to CD4 density (FOXP3/CD4), the ratio of BTLA density to CD8 density (BTLA/CD8), and the ratio of Cbl‐b density to CD8 density (Cbl‐b/CD8); comparisons of these ratios between the diseases are depicted in Figure 3(b). FOXP3/CD4, BTLA/CD8, and Cbl‐b/CD8 in GBC were significantly higher than those in CC and XGC, respectively (P < 0.01). Comparisons of the BTLA/CD3 and Cbl‐b/CD3 ratios among GBC, CC, and XGC showed similar profiles to those of BTLA/CD8 and Cbl‐b/CD8, respectively (Fig. S1).
Figure 2.
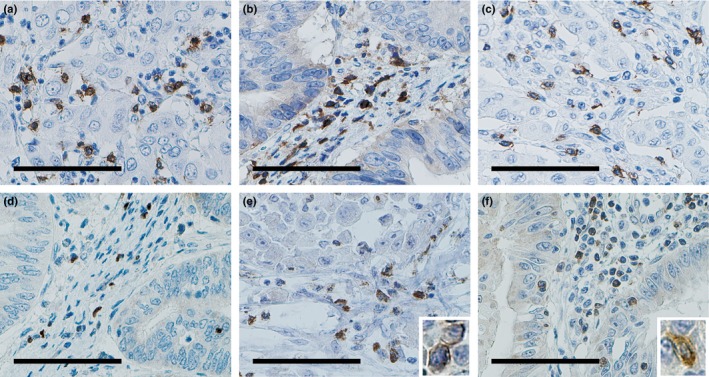
Immunohistochemical features of tumor‐infiltrating cells. These representative photos show: (a) CD3+ T cells, (b) CD4+ T cells, (c) CD8+ T cells, (d) FOXP3+ T cells, (e) B and T lymphocyte attenuator (BTLA)+ cells, and (f) Casitas–B‐lineage lymphoma protein‐b (Cbl‐b+) cells. Scale bar = 100 μm. High power views are shown in insets of (e) and (f).
Figure 3.
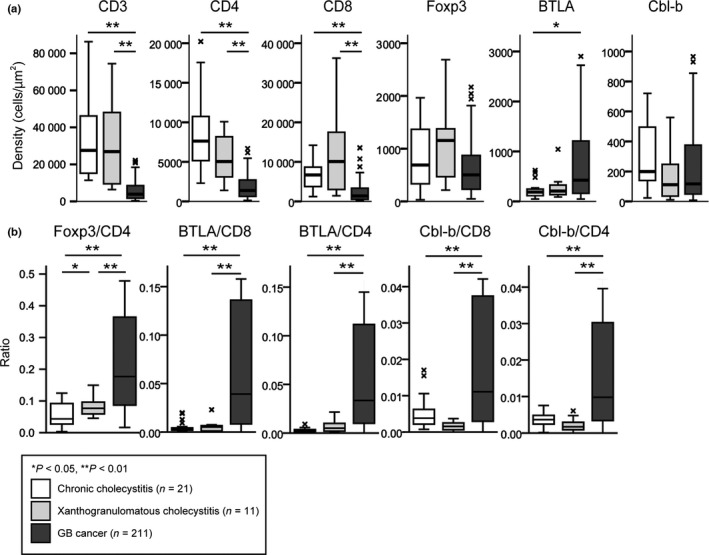
Comparison of cells infiltrating into chronic cholecystitis (CC, n = 21), xanthogranulomatous cholecystitis (XGC, n = 11), and gallbladder cancer (GBC, n = 211). (a) Density of immunolabeled cells (cells/μm2). (b) Density ratio of FOXP3+ cells to CD4+ T cells (FOXP3/CD4), that of B and T lymphocyte attenuator (BTLA)+ cells to CD8+ T cells (BTLA/CD8), that of BLTA + cells to CD4+ T cells (BTLA/CD4), that of Casitas–B‐lineage lymphoma protein‐b (Cbl‐b)+ cells to CD8+ T cells (Cbl‐b/CD8), and that of Cbl‐b+ cells to CD4+ T cells (Cbl‐b/CD4). The line in the middle of the boxes shows the median value. The bottom and top of the box indicates the 25th and 75th percentiles, respectively. The T‐bars that extend from the boxes show inner fences and “x” indicates outliers. Significant values were *P < 0.05 and **P < 0.01.
Interrelationships between clinicopathological variables and tumor‐infiltrating cells
We analyzed the interrelationships between clinicopathological variables of GBC and tumor‐infiltrating T cell subsets, BTLA+ cells, and Cbl‐b+ cells (Table 1). FOXP3/CD4, BTLA/CD8, and Cbl‐b/CD8 showed significant and positive correlations with each other. FOXP3/CD4 was significantly correlated with most of the conventional clinicopathological variables: tumor main location, T factor, N factor, M factor, histopathological grading, venous invasion, and perineural invasion. Likewise, BTLA/CD8 was significantly correlated with T factor, M factor, histopathological grading, venous invasion, and perineural invasion, and Cbl‐b/CD8 was significantly correlated with T factor and M factor. The interrelationships of BTLA/CD3 and Cbl‐b/CD3 with various factors were similar to those of BTLA/CD8 and Cbl‐b/CD8, respectively (data not shown).
Table 1.
Interrelationships between clinicopathological variables and tumor‐infiltrating cells
| Variables | CD3 | CD4 | CD8 | FOXP3 | FOXP3/CD4 | BTLA | BTLA/CD8 | BTLA/CD4 | Cbl‐b | Cbl‐b/CD8 | Cbl‐b/CD4 | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | P‐value | Low | High | P‐value | Low | High | P‐value | Low | High | P‐value | Low | High | P‐value | Low | High | P‐value | Low | High | P‐value | Low | High | P‐value | Low | High | P‐value | Low | High | P‐value | Low | High | P‐value | |
| Age, years | 0.302 | 0.033 | 0.063 | 0.016 | 0.837 | 0.046 | 0.242 | 0.046 | 0.630 | 0.302 | 0.535 | ||||||||||||||||||||||
| <68 (n = 105) | 56 | 49 | 60 | 45 | 59 | 46 | 61 | 44 | 53 | 52 | 45 | 60 | 48 | 57 | 45 | 60 | 54 | 51 | 49 | 56 | 50 | 55 | |||||||||||
| ≥68 (n = 106) | 49 | 57 | 45 | 61 | 46 | 60 | 44 | 62 | 52 | 54 | 60 | 46 | 57 | 49 | 60 | 46 | 51 | 55 | 57 | 49 | 55 | 51 | |||||||||||
| Gender | 0.046 | 0.948 | 0.833 | 0.002 | 0.537 | 0.948 | 0.627 | 0.733 | 0.046 | 0.627 | 0.085 | ||||||||||||||||||||||
| Female (n = 100) | 57 | 43 | 50 | 50 | 49 | 51 | 61 | 39 | 52 | 48 | 50 | 50 | 48 | 52 | 51 | 49 | 57 | 43 | 52 | 48 | 56 | 44 | |||||||||||
| Male (n = 111) | 48 | 63 | 55 | 56 | 56 | 55 | 44 | 67 | 53 | 58 | 55 | 56 | 57 | 54 | 54 | 57 | 48 | 63 | 54 | 57 | 49 | 62 | |||||||||||
| Tumor main location | 0.308 | 0.009 | 0.422 | 0.681 | 0.025 | 0.158 | 0.617 | 0.845 | 0.422 | 0.158 | 0.025 | ||||||||||||||||||||||
| Gf, Gb (n = 104, 46) | 78 | 72 | 66 | 84 | 72 | 78 | 76 | 74 | 82 | 68 | 70 | 80 | 73 | 77 | 74 | 76 | 72 | 78 | 80 | 70 | 82 | 68 | |||||||||||
| Gn, C (n = 40, 21) | 27 | 34 | 39 | 22 | 33 | 28 | 29 | 32 | 23 | 38 | 35 | 26 | 32 | 29 | 31 | 30 | 33 | 28 | 26 | 35 | 23 | 38 | |||||||||||
| T | 0.538 | 0.295 | 0.060 | 0.951 | 0.023 | 0.023 | <0.001 | 0.005 | 0.372 | 0.003 | 0.011 | ||||||||||||||||||||||
| 1, 2 (n = 28, 66) | 49 | 45 | 43 | 51 | 40 | 54 | 47 | 47 | 55 | 39 | 55 | 39 | 62 | 32 | 57 | 37 | 50 | 44 | 58 | 36 | 56 | 38 | |||||||||||
| 3, 4 (n = 74, 43) | 56 | 61 | 62 | 55 | 65 | 52 | 58 | 59 | 50 | 67 | 50 | 67 | 43 | 74 | 48 | 69 | 55 | 62 | 48 | 69 | 49 | 68 | |||||||||||
| N | 0.836 | 0.448 | 0.448 | 0.063 | 0.033 | 0.007 | 0.113 | 0.301 | 0.113 | 0.946 | 0.190 | ||||||||||||||||||||||
| 0 (n = 107) | 54 | 53 | 56 | 51 | 56 | 51 | 60 | 47 | 61 | 46 | 63 | 44 | 59 | 48 | 57 | 50 | 59 | 48 | 54 | 53 | 58 | 49 | |||||||||||
| 1 (n = 104) | 51 | 53 | 49 | 55 | 49 | 55 | 45 | 59 | 44 | 60 | 42 | 62 | 46 | 58 | 48 | 56 | 46 | 58 | 52 | 52 | 47 | 57 | |||||||||||
| M | 0.963 | 0.522 | 0.126 | 0.398 | 0.009 | 0.083 | 0.003 | 0.020 | 0.802 | 0.034 | 0.009 | ||||||||||||||||||||||
| 0 (n = 145) | 72 | 73 | 70 | 75 | 67 | 78 | 75 | 70 | 81 | 64 | 78 | 67 | 82 | 63 | 80 | 65 | 73 | 72 | 80 | 65 | 81 | 64 | |||||||||||
| 1 (n = 66) | 33 | 33 | 35 | 31 | 38 | 28 | 30 | 36 | 24 | 42 | 27 | 39 | 23 | 43 | 25 | 41 | 32 | 34 | 26 | 40 | 24 | 42 | |||||||||||
| Histopathological grading | 0.303 | 0.239 | 0.727 | 0.063 | 0.003 | 0.063 | 0.033 | 0.033 | 0.632 | 0.146 | 0.191 | ||||||||||||||||||||||
| 1 (n = 99) | 53 | 46 | 45 | 54 | 48 | 51 | 56 | 43 | 60 | 39 | 56 | 43 | 57 | 42 | 57 | 42 | 51 | 48 | 55 | 44 | 54 | 45 | |||||||||||
| 2, 3, 4 (n = 62, 45, 5) | 52 | 60 | 60 | 52 | 57 | 55 | 49 | 63 | 45 | 67 | 49 | 63 | 48 | 64 | 48 | 64 | 54 | 58 | 51 | 61 | 51 | 61 | |||||||||||
| Lymphatic invasion | 0.016 | 0.352 | 0.962 | 0.069 | 0.128 | 0.035 | 0.069 | 0.128 | 0.128 | 0.403 | 0.128 | ||||||||||||||||||||||
| 0 (n = 68) | 42 | 26 | 37 | 31 | 34 | 34 | 40 | 28 | 39 | 29 | 41 | 27 | 40 | 28 | 39 | 29 | 39 | 29 | 37 | 31 | 39 | 29 | |||||||||||
| 1 (n = 143) | 63 | 80 | 68 | 75 | 71 | 72 | 65 | 78 | 66 | 77 | 64 | 79 | 65 | 78 | 66 | 77 | 66 | 77 | 69 | 74 | 66 | 77 | |||||||||||
| Venous invasion | 0.010 | 0.537 | 0.436 | 0.083 | 0.022 | 0.083 | 0.022 | 0.146 | 0.146 | 0.058 | 0.022 | ||||||||||||||||||||||
| 0 (n = 88) | 53 | 35 | 46 | 42 | 41 | 47 | 50 | 38 | 52 | 36 | 50 | 38 | 52 | 36 | 49 | 39 | 49 | 39 | 51 | 37 | 52 | 36 | |||||||||||
| 1 (n = 123) | 52 | 71 | 59 | 64 | 64 | 59 | 55 | 68 | 53 | 70 | 55 | 68 | 53 | 70 | 56 | 67 | 56 | 67 | 55 | 68 | 53 | 70 | |||||||||||
| Perineural invasion | 0.148 | 0.442 | 0.295 | 0.372 | 0.002 | 0.735 | 0.045 | 0.372 | 0.951 | 0.110 | 0.023 | ||||||||||||||||||||||
| 0 (n = 94) | 52 | 42 | 44 | 50 | 43 | 51 | 50 | 44 | 58 | 36 | 48 | 46 | 54 | 40 | 50 | 44 | 47 | 47 | 53 | 41 | 55 | 39 | |||||||||||
| 1 (n = 117 | 53 | 64 | 61 | 56 | 62 | 55 | 55 | 62 | 47 | 70 | 57 | 60 | 51 | 66 | 55 | 62 | 58 | 59 | 53 | 64 | 50 | 67 | |||||||||||
| CD3 | <0.001 | <0.001 | <0.001 | 0.005 | 0.063 | 0.046 | 0.002 | 0.063 | 0.302 | 0.046 | |||||||||||||||||||||||
| Low (n = 105) | 77 | 28 | 74 | 31 | 71 | 34 | 42 | 63 | 59 | 46 | 45 | 60 | 41 | 64 | 59 | 46 | 49 | 56 | 45 | 60 | |||||||||||||
| High (n = 106) | 28 | 78 | 31 | 75 | 34 | 72 | 63 | 43 | 46 | 60 | 60 | 45 | 64 | 42 | 46 | 60 | 57 | 49 | 60 | 46 | |||||||||||||
| CD4 | <0.001 | <0.001 | <0.001 | 0.007 | 0.011 | <0.001 | <0.001 | 0.003 | <0.001 | ||||||||||||||||||||||||
| Low (n = 105) | 81 | 24 | 70 | 35 | 29 | 76 | 62 | 43 | 43 | 62 | 35 | 70 | 67 | 38 | 42 | 63 | 37 | 68 | |||||||||||||||
| High (n = 106) | 24 | 82 | 35 | 71 | 76 | 30 | 43 | 63 | 62 | 44 | 70 | 36 | 38 | 68 | 64 | 42 | 68 | 38 | |||||||||||||||
| CD8 | <0.001 | <0.001 | 0.630 | <0.001 | <0.001 | 0.016 | <0.001 | 0.001 | |||||||||||||||||||||||||
| Low (n = 105) | 71 | 34 | 37 | 68 | 54 | 51 | 29 | 76 | 37 | 68 | 61 | 44 | 32 | 73 | 40 | 65 | |||||||||||||||||
| High (n = 106) | 34 | 72 | 68 | 38 | 51 | 55 | 76 | 30 | 68 | 38 | 44 | 62 | 74 | 32 | 65 | 41 | |||||||||||||||||
| FOXP3 | 0.001 | <0.001 | 0.945 | 0.837 | 0.001 | 0.449 | 0.837 | ||||||||||||||||||||||||||
| Low (n = 105) | 64 | 41 | 68 | 37 | 52 | 53 | 53 | 52 | 64 | 41 | 50 | 55 | 53 | 52 | |||||||||||||||||||
| High (n = 106) | 41 | 65 | 37 | 69 | 53 | 53 | 52 | 54 | 41 | 65 | 56 | 50 | 52 | 54 | |||||||||||||||||||
| FOXP3/CD4 | 0.063 | <0.001 | <0.001 | 0.837 | <0.001 | <0.001 | |||||||||||||||||||||||||||
| Low (n = 105) | 59 | 46 | 69 | 36 | 75 | 30 | 53 | 52 | 68 | 37 | 74 | 31 | |||||||||||||||||||||
| High (n = 106) | 46 | 60 | 36 | 70 | 30 | 76 | 52 | 54 | 39 | 67 | 31 | 75 | |||||||||||||||||||||
| BTLA | <0.001 | <0.001 | <0.001 | <0.001 | 0.016 | ||||||||||||||||||||||||||||
| Low (n = 105) | 80 | 25 | 78 | 27 | 69 | 36 | 67 | 38 | 61 | 44 | |||||||||||||||||||||||
| High (n = 106) | 25 | 81 | 27 | 79 | 36 | 70 | 39 | 67 | 44 | 62 | |||||||||||||||||||||||
| BTLA/CD8 | <0.001 | 0.007 | <0.001 | <0.001 | |||||||||||||||||||||||||||||
| Low (n = 105) | 88 | 17 | 62 | 43 | 80 | 25 | 70 | 35 | |||||||||||||||||||||||||
| High (n = 106) | 17 | 89 | 43 | 63 | 26 | 80 | 35 | 71 | |||||||||||||||||||||||||
| BTLA/CD4 | 0.033 | <0.001 | <0.001 | ||||||||||||||||||||||||||||||
| Low (n = 105) | 60 | 45 | 73 | 32 | 73 | 32 | |||||||||||||||||||||||||||
| High (n = 106) | 45 | 61 | 33 | 73 | 32 | 74 | |||||||||||||||||||||||||||
| Cbl‐b | <0.001 | <0.001 | |||||||||||||||||||||||||||||||
| Low (n = 105) | 76 | 29 | 75 | 30 | |||||||||||||||||||||||||||||
| High (n = 106) | 30 | 76 | 30 | 76 | |||||||||||||||||||||||||||||
| Cbl‐b/CD8 | <0.001 | ||||||||||||||||||||||||||||||||
| Low (n = 105) | 90 | 16 | |||||||||||||||||||||||||||||||
| High (n = 106) | 15 | 90 | |||||||||||||||||||||||||||||||
Density ratios are shown of FOXP3 to CD4 (FOXP3/CD4), B and T lymphocyte attenuator (BTLA) to CD8 (BTLA/CD8), BTLA to CD4 (BTLA/CD4), Casitas–B‐lineage lymphoma protein‐b (Cbl‐b) to CD8 (Cbl‐b/CD8), and Cbl‐b to CD4 (Cbl‐b/CD4). Bold values, P < 0.05; underlined values, positively correlated. C, cystic duct; Gb, gallbladder body; Gf, gallbladder fundus; Gn, gallbladder neck.
Prognostic significance of tumor‐infiltrating T cell subsets, BTLA+ cells, and Cbl‐b+ cells
Kaplan–Meier survival analyses revealed that a lower density of tumor‐infiltrating CD8+ T cells, and higher FOXP3/CD4, BTLA/CD8, and Cbl‐b/CD8 ratios were significantly associated with both shorter OS and DFS in GBC patients (Fig. 4). Higher BTLA/CD3 and Cbl‐b/CD3 ratios were closely associated with both shorter OS and DFS (Fig. S2). Five‐year survival rate, median survival time, and Cox analyses in the groups categorized by each of the parameters of tumor‐infiltrating cells and the conventional clinicopathological variables are summarized in Tables 2 and S1. When the variables that had been found to be significant by univariate analysis were subjected to multivariate analysis, M factor, perineural invasion, BTLA/CD8, and Cbl‐b/CD8 were closely associated with shorter OS. In addition, T factor, M factor, perineural invasion, and Cbl‐b/CD8 were significantly associated with shorter DFS.
Figure 4.
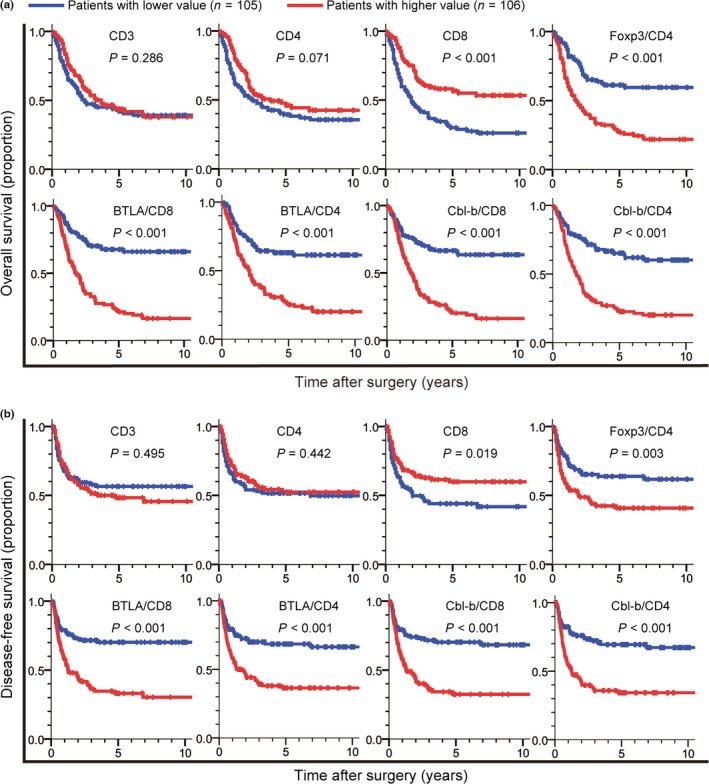
Kaplan–Meier survival curves comparing (a) overall survival and (b) disease‐free survival in gallbladder cancer patients between the high (red) and low (blue) value groups with regard to the density of tumor‐infiltrating CD3+ T cells, CD4+ T cells, CD8+ T cells and the density ratio of the FOXP3+ T cells to CD4+ T cells (Foxp3/CD4), that of B and T lymphocyte attenuator (BTLA)+ cells to CD8+ T cells (BTLA/CD8), that of BTLA + cells to CD4+ T cells (BTLA/CD4), that of Casitas–B‐lineage lymphoma protein‐b (Cbl‐b)+ cells to CD8+ T cells (Cbl‐b/CD8), and that of Cbl‐b+ cells to CD4+ T cells (Cbl‐b/CD4). P‐values were obtained by log–rank test.
Table 2.
Univariate and multivariate analyses for variables associated with overall survival (OS) and disease‐free survival (DFS) in patients with gallbladder cancer
| Variables | Category | n | 5‐year survival, % | Median survival time, months | OS | DFS | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||||||||||||||||
| HR | 95%CI | P‐value | HR | 95%CI | P‐value | HR | 95%CI | P‐value | HR | 95%CI | P‐value | |||||||||
| Age ≥68 years | No/yes | 105/106 | 40.8/44.5 | 34.1/38.2 | 0.958 | 0.669 | 1.371 | 0.813 | 1.024 | 0.679 | 1.545 | 0.910 | ||||||||
| Gender | F/M | 100/111 | 47.6/38.1 | 56.1/27.8 | 1.308 | 0.911 | 1.876 | 0.145 | 1.395 | 0.915 | 2.127 | 0.122 | ||||||||
| Location | Gf,Gb/Gn,C | 104,46/40,21 | 48.4/29.6 | 54.6/27.1 | 1.422 | 0.973 | 2.078 | 0.069 | 1.667 | 1.092 | 2.543 | 0.018 | ||||||||
| T | 1,2/3,4 | 28,66/74,43 | 71.5/19.2 | 215.2/17.8 | 4.587 | 3.013 | 6.986 | <0.001 | 7.453 | 4.255 | 13.055 | <0.001 | 2.906 | 1.431 | 5.898 | 0.003 | ||||
| N | 0/1 | 107/104 | 58.3/25.8 | 215.2/19.5 | 2.713 | 1.870 | 3.937 | <0.001 | 3.868 | 2.467 | 6.064 | <0.001 | ||||||||
| M | 0/1 | 145/66 | 54.7/15.1 | 138.8/12.9 | 3.772 | 2.607 | 5.457 | <0.001 | 2.156 | 1.462 | 3.179 | <0.001 | 3.922 | 2.562 | 6.004 | <0.001 | 2.318 | 1.493 | 3.598 | <0.001 |
| Grading | 1/2,3,4 | 99/62,45,5 | 61.5/26.1 | 215.2/19.5 | 3.071 | 2.049 | 4.535 | <0.001 | 3.659 | 2.303 | 5.814 | <0.001 | ||||||||
| L | 0/1 | 68/143 | 71.7/27.9 | 215.2/22.9 | 3.500 | 2.195 | 5.581 | <0.001 | 5.737 | 3.042 | 10.818 | <0.001 | ||||||||
| V | 0/1 | 88/123 | 69.4/22.3 | 215.2/19.8 | 3.861 | 2.505 | 5.814 | <0.001 | 4.984 | 2.956 | 8.404 | <0.001 | ||||||||
| Pn | 0/1 | 94/117 | 71.7/18.7 | 215.2/17.2 | 4.715 | 3.093 | 7.187 | <0.001 | 3.833 | 2.453 | 5.989 | <0.001 | 7.758 | 4.424 | 13.605 | <0.001 | 3.325 | 1.631 | 6.779 | 0.001 |
| CD3 | Low/high | 105/106 | 41.5/43.7 | 29.4/43.6 | 0.824 | 0.576 | 1.179 | 0.291 | 1.152 | 0.762 | 1.742 | 0.501 | ||||||||
| CD4 | Low/high | 105/106 | 39.3/46.0 | 29.7/46.4 | 0.721 | 0.503 | 1.033 | 0.075 | 0.853 | 0.565 | 1.287 | 0.448 | ||||||||
| CD8 | Low/high | 105/106 | 30.0/56.6 | 22.9/191.0 | 0.478 | 0.331 | 0.691 | <0.001 | 0.616 | 0.406 | 0.933 | 0.022 | ||||||||
| Foxp3/CD4 | Low/high | 105/106 | 61.1/26.8 | NR/20.8 | 2.783 | 1.894 | 4.088 | <0.001 | 1.855 | 1.217 | 2.826 | 0.004 | ||||||||
| BTLA/CD8 | Low/high | 105/106 | 65.8/20.0 | 191.0/20.8 | 3.577 | 2.387 | 5.359 | <0.001 | 2.154 | 1.400 | 3.313 | <0.001 | 2.757 | 1.766 | 4.302 | <0.001 | ||||
| BTLA/CD4 | Low/high | 105/106 | 63.0/25.2 | NR/24.6 | 2.892 | 1.957 | 4.274 | <0.001 | 2.362 | 1.530 | 3.648 | <0.001 | ||||||||
| Cbl‐b/CD8 | Low/high | 105/106 | 65.6/18.8 | 191.2/21.6 | 3.282 | 2.213 | 4.868 | <0.001 | 2.374 | 1.565 | 3.602 | <0.001 | 2.759 | 1.766 | 4.309 | <0.001 | 2.445 | 1.555 | 3.847 | <0.001 |
| Cbl‐b/CD4 | Low/high | 105/106 | 65.2/22.5 | NR/21.2 | 3.080 | 2.088 | 4.544 | <0.001 | 2.675 | 1.723 | 4.153 | <0.001 | ||||||||
| Number, OS, and DFS in subgroups categorized by the status of BTLA/CD8 and Cbl‐b/CD8 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Combined variables | n | 5‐year survival, % | Median survival time, months | Median disease‐free survival time, months | ||||||||||||||
| BTLA/CD8high and Cbl‐b/CD8high | 80 | 17.1 | 20.8 | 15.2 | ||||||||||||||
| BTLA/CD8high and Cbl‐b/CD8low | 26 | 31.3 | 18.7 | 14.2 | ||||||||||||||
| BTLA/CD8low and Cbl‐b/CD8high | 25 | 31.2 | 27.1 | 19.9 | ||||||||||||||
| BTLA/CD8low and Cbl‐b/CD8low | 80 | 78.3 | NR | NR | ||||||||||||||
Density ratios are shown of FOXP3 to CD4 (FOXP3/CD4), B and T lymphocyte attenuator (BTLA) to CD8 (BTLA/CD8), BTLA to CD4 (BTLA/CD4), Casitas–B‐lineage lymphoma protein‐b (Cbl‐b) to CD8 (Cbl‐b/CD8), and Cbl‐b to CD4 (Cbl‐b/CD4). C, cystic duct; CI, confidence interval; F, female; Gb, gallbladder body; Gf, gallbladder fundus; Gn, gallbladder neck; HR, hazard ratio; L, lymphatic invasion; M, male; NR, not reached; Pn, perineural invasion; V, venous invasion. Bold values, P < 0.05.
Discussion
Here we evaluated for the first time the clinical impact of BTLA in cancer tissues. First, we assessed whether tumor‐infiltrating immune cells reflected the character of the tumor immune microenvironment, as it has been reported in various other cancers. The present study, including 211 cases of GBC, revealed that greater infiltration of CD8+ T cells and CD4+ T cells in the cancer tissue was a favorable prognostic indicator, as has been reported for many other cancers and for GBC.20, 21 It also showed that a higher prevalence of Tregs (FOXP3/CD4) was significantly correlated with malignant phenotypes and an unfavorable outcome, suggesting that Tregs play a role in controlling the immune response to GBC. Furthermore, a higher ratio of Cbl‐b+ anergic lymphocytes to CD8+ T cells (or to total T cells) was significantly correlated with advanced T factor, distant metastasis, and poorer prognosis. Therefore, the importance of antitumor immunity, represented by tumor‐infiltrating immune cells, in the outcome and control of cancer was confirmed for GBC.
Tumor cells are able to evade immune recognition and destruction through a variety of mechanisms. One such mechanism is the immunosuppressive action of co‐inhibitory molecules.23 BTLA is a co‐inhibitory molecule whose expression has been reported in mice and human blood cells. However, no previous study has investigated the expression of BTLA in the microenvironment of human tumors. Here, we found that BTLA was expressed on a proportion of several different types of tumor‐infiltrating immune cells, including CD3+ T cells, CD4+ T cells, CD8+ T cells, CD20+ B cells, CD14+ monocytes, CD68+ macrophages, CD1a+ DCs, CD207+ DCs, and CD208+ DCs. Our clinicopathological investigation revealed that BTLA/CD8 was increased in malignant disease (GBC) relative to benign disease (CC and XGC) and that the BTLA/CD8 ratio in tumor‐infiltrating immune cells was enhanced in patients with more advanced cancer. Furthermore, a higher BTLA/CD8 ratio was significantly associated with shorter DFS and OS. These findings suggest that BTLA is involved in the formation of a protumor microenvironment, supporting the contention that BTLA is an important co‐inhibitory molecule in the orchestration of immunosuppressive networks in cancer, and therefore a potential new target for interventions aimed at reversal of immune evasion and boosting of antitumor immunity in cancer patients.
It has been shown that BLTA binds to the herpes virus entry mediator (HVEM),43 inhibits T‐cell proliferation and cytokine production in vitro, and mediates the negative regulation of CD8+ T‐cell homeostasis and memory cell generation in vivo.44 In the context of cancer immunology, BTLA–HVEM is known to be another inhibitory pathway used by cancer cells to impair the antitumor immune response.45, 46, 47 HVEM is constitutively expressed on naive T cells and downregulated following T cell activation, only to be re‐expressed later on effector and memory T cells.48 It is also broadly expressed on cells of the immune system such as Tregs, B cells, monocytes, neutrophils, NK cells, and DCs, in addition to epithelial cells.49, 50 Although it would have been informative to examine the distribution of HVEM immunohistochemically, we were unable to do so, because of the lack of reliable antibody specific for HVEM. In contrast to the CTLA‐4 and PD‐1 pathways, HVEM binds to several receptor molecules, generating immune‐positive and ‐negative signals according to the receptors involved. Therefore, the total expression of BTLA in the tumor microenvironment might be a simpler and more accurate indicator of the protumor microenvironment, rather than expression of HVEM. It remains to be investigated whether a specific cell type with high BTLA expression exerts a predominantly inhibitory action or whether various types of immune cells function in a coordinated manner.
Maintenance of tolerance and induction of T cell anergy is critical for prevention of autoimmunity. However, in cases of malignancy, tumor‐induced T cell anergy and/or tolerance induces cancer‐associated immune paralysis, which contributes, at least in part, to uncontrolled tumor growth and metastasis. No previous studies have, in fact, demonstrated anergy cells in tissues. In the present study, we observed that T cells that had infiltrated and become attached to cancer cells were often positive for Cbl‐b, and sometimes positive for BTLA. Although no BTLA+Cbl‐b+ cells were found, T cells that had infiltrated within cancer nests were suggested to have become anergic under the influence of BTLA signals. Our clinicopathological analyses revealed that Cbl‐b/CD8 was increased in GBC relative to CC and XGC, and that Cbl‐b/CD8 in tumor‐infiltrating immune cells was higher in patients with more advanced cancer, as was the case for BTLA expression. Moreover, Cbl‐b/CD8 was significantly and positively correlated with BTLA/CD8 and FOXP3/CD4, and a higher Cbl‐b/CD8 ratio was an independent indicator of poor prognosis in terms of DFS and OS for patients with GBC. Accordingly, it is suggested that T cell anergy in the tumor microenvironment, reflected by the expression of Cbl‐b, represents promotion of a protumor microenvironment in GBC, in concert with BTLA. In addition, Cbl‐b expression may be a useful marker for evaluating T cell anergy in the tumor microenvironment. Here we have shown for the first time the usefulness of Cbl‐b for evaluation of the tumor microenvironment.
Therapeutic blockade of the immune checkpoint pathways, CTLA‐4 and PD‐1/PD‐L1, has shown promise in a variety of malignancies,23, 51 although the effectiveness of the reagents used, especially anti‐CTLA4 antibody, has been rather limited for major epithelial cancers in comparison with melanoma, and even in melanoma cases showing basic or acquired resistance have often been observed. In addition, a wide range of immune‐related adverse events have also been observed following treatment.23, 51 To increase the antitumor effect and to reduce the incidence of immune‐related adverse events, optimization of the dose and schedule of reagents, and characterization of biomarkers associated with disease outcome, have been extensively investigated. In addition, various combination approaches have been considered, including blockade of immune checkpoints together with the use of other anticancer treatments such as chemotherapy, radiotherapy, targeted therapy, and other forms of immunotherapy. Some combination approaches have achieved enhanced antitumor effects in animal models and human clinical studies,29, 30, 31 in which monotherapy has exerted only modest effects. Novel immune checkpoints are currently being investigated based on experience with CTLA‐4 and PD‐1/PD‐L1. Various combination approaches using these novel checkpoints may yield a therapeutic edge in the battle against a range of cancers. Therefore, it is suggested that our data for BTLA in GBC will be useful for future studies aimed at developing this new approach.
In conclusion, our findings suggest that BTLA in the tumor microenvironment plays an inhibitory role in the immune reaction against GBC, in concert with other factors. Higher expression of BTLA and Cbl‐b in tumor‐infiltrating immune cells appears to be an indicator of poor prognosis, whereas a higher ratio of non‐anergic CD8+ T cells is an independent favorable prognostic factor in GBC patients. Targeting BTLA for reversal of immune evasion may represent a promising new therapeutic approach.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. Comparison of infiltrating cells in gallbladder lesion.
Fig. S2. Kaplan–Meier survival curves.
Table S1. Univariate and multivariate survival analyses.
Acknowledgments
This work was supported by a Grant‐in‐Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (N.H.), and the National Cancer Center Research and Development Fund (N.H.). The authors thank Ms. Keiko Gomisawa for excellent technical assistance.
Cancer Sci 106 (2015) 1750–1760
Funding Information
Ministry of Education, Culture, Sports, Science and Technology of Japan, Japan Society for the Promotion of Science (‘KAKENHI 25460486’), National Cancer Center Research and Development Fund.
References
- 1. Reid KM, Ramos‐De la Medina A, Donohue JH. Diagnosis and surgical management of gallbladder cancer: a review. J Gastrointest Surg 2007; 11: 671–81. [DOI] [PubMed] [Google Scholar]
- 2. D'Angelica M, Dalal KM, DeMatteo RP, Fong Y, Blumgart LH, Jarnagin WR. Analysis of the extent of resection for adenocarcinoma of the gallbladder. Ann Surg Oncol 2009; 16: 806–16. [DOI] [PubMed] [Google Scholar]
- 3. Hueman MT, Vollmer CM Jr, Pawlik TM. Evolving treatment strategies for gallbladder cancer. Ann Surg Oncol 2009; 16: 2101–15. [DOI] [PubMed] [Google Scholar]
- 4. Gourgiotis S, Kocher HM, Solaini L, Yarollahi A, Tsiambas E, Salemis NS. Gallbladder cancer. Am J Surg 2008; 196: 252–64. [DOI] [PubMed] [Google Scholar]
- 5. Shukla PJ, Neve R, Barreto SG et al A new scoring system for gallbladder cancer (aiding treatment algorithm): an analysis of 335 patients. Ann Surg Oncol 2008; 15: 3132–7. [DOI] [PubMed] [Google Scholar]
- 6. Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu Rev Immunol 2011; 29: 235–71. [DOI] [PubMed] [Google Scholar]
- 7. Poschke I, Mougiakakos D, Kiessling R. Camouflage and sabotage: tumor escape from the immune system. Cancer Immunol Immunother 2011; 60: 1161–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rabinovich GA, Gabrilovich D, Sotomayor EM. Immunosuppressive strategies that are mediated by tumor cells. Annu Rev Immunol 2007; 25: 267–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer 2005; 5: 263–74. [DOI] [PubMed] [Google Scholar]
- 10. Naito Y, Saito K, Shiiba K et al CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res 1998; 58: 3491–4. [PubMed] [Google Scholar]
- 11. Pages F, Berger A, Camus M et al Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med 2005; 353: 2654–66. [DOI] [PubMed] [Google Scholar]
- 12. Zhang L, Conejo‐Garcia JR, Katsaros D et al Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med 2003; 348: 203–13. [DOI] [PubMed] [Google Scholar]
- 13. Schumacher K, Haensch W, Roefzaad C, Schlag PM. Prognostic significance of activated CD8(+) T cell infiltrations within esophageal carcinomas. Cancer Res 2001; 61: 3932–6. [PubMed] [Google Scholar]
- 14. Wada Y, Nakashima O, Kutami R, Yamamoto O, Kojiro M. Clinicopathological study on hepatocellular carcinoma with lymphocytic infiltration. Hepatology 1998; 27: 407–14. [DOI] [PubMed] [Google Scholar]
- 15. Hiraoka N, Ino Y, Yamazaki‐Itoh R, Kanai Y, Kosuge T, Shimada K. Intratumoral tertiary lymphoid organ is a favourable prognosticator in patients with pancreatic cancer. Br J Cancer 2015; 112: 1782–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ino Y, Yamazaki‐Itoh R, Shimada K et al Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br J Cancer 2013; 108: 914–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Curiel TJ, Coukos G, Zou L et al Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 2004; 10: 942–9. [DOI] [PubMed] [Google Scholar]
- 18. Hiraoka N, Onozato K, Kosuge T, Hirohashi S. Prevalence of FOXP3+ regulatory T cells increases during the progression of pancreatic ductal adenocarcinoma and its premalignant lesions. Clin Cancer Res 2006; 12: 5423–34. [DOI] [PubMed] [Google Scholar]
- 19. Kobayashi N, Hiraoka N, Yamagami W et al FOXP3+ regulatory T cells affect the development and progression of hepatocarcinogenesis. Clin Cancer Res 2007; 13: 902–11. [DOI] [PubMed] [Google Scholar]
- 20. Nakakubo Y, Miyamoto M, Cho Y et al Clinical significance of immune cell infiltration within gallbladder cancer. Br J Cancer 2003; 89: 1736–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goeppert B, Frauenschuh L, Zucknick M et al Prognostic impact of tumour‐infiltrating immune cells on biliary tract cancer. Br J Cancer 2013; 109: 2665–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen L, Flies DB. Molecular mechanisms of T cell co‐stimulation and co‐inhibition. Nat Rev Immunol 2013; 13: 227–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012; 12: 252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brahmer JR, Tykodi SS, Chow LQ et al Safety and activity of anti‐PD‐L1 antibody in patients with advanced cancer. N Engl J Med 2012; 366: 2455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hodi FS, O'Day SJ, McDermott DF et al Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010; 363: 711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robert C, Thomas L, Bondarenko I et al Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med 2011; 364: 2517–26. [DOI] [PubMed] [Google Scholar]
- 27. Topalian SL, Hodi FS, Brahmer JR et al Safety, activity, and immune correlates of anti‐PD‐1 antibody in cancer. N Engl J Med 2012; 366: 2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Watanabe N, Gavrieli M, Sedy JR et al BTLA is a lymphocyte inhibitory receptor with similarities to CTLA‐4 and PD‐1. Nat Immunol 2003; 4: 670–9. [DOI] [PubMed] [Google Scholar]
- 29. Curran MA, Montalvo W, Yagita H, Allison JP. PD‐1 and CTLA‐4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci USA 2010; 107: 4275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsuzaki J, Gnjatic S, Mhawech‐Fauceglia P et al Tumor‐infiltrating NY‐ESO‐1‐specific CD8+ T cells are negatively regulated by LAG‐3 and PD‐1 in human ovarian cancer. Proc Natl Acad Sci USA 2010; 107: 7875–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wolchok JD, Kluger H, Callahan MK et al Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med 2013; 369: 122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. M'Hidi H, Thibult ML, Chetaille B et al High expression of the inhibitory receptor BTLA in T‐follicular helper cells and in B‐cell small lymphocytic lymphoma/chronic lymphocytic leukemia. Am J Clin Pathol 2009; 132: 589–96. [DOI] [PubMed] [Google Scholar]
- 33. Feng XY, Wen XZ, Tan XJ et al Ectopic expression of B and T lymphocyte attenuator in gastric cancer: a potential independent prognostic factor in patients with gastric cancer. Mol Med Rep 2015; 11: 658–64. [DOI] [PubMed] [Google Scholar]
- 34. Hurchla MA, Sedy JR, Gavrieli M, Drake CG, Murphy TL, Murphy KM. B and T lymphocyte attenuator exhibits structural and expression polymorphisms and is highly induced in anergic CD4+ T cells. J Immunol 2005; 174: 3377–85. [DOI] [PubMed] [Google Scholar]
- 35. Loeser S, Penninger JM. The ubiquitin E3 ligase Cbl‐b in T cells tolerance and tumor immunity. Cell Cycle 2007; 6: 2478–85. [DOI] [PubMed] [Google Scholar]
- 36. Heissmeyer V, Macian F, Im SH et al Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat Immunol 2004; 5: 255–65. [DOI] [PubMed] [Google Scholar]
- 37. Jeon MS, Atfield A, Venuprasad K et al Essential role of the E3 ubiquitin ligase Cbl‐b in T cell anergy induction. Immunity 2004; 21: 167–77. [DOI] [PubMed] [Google Scholar]
- 38. Albores‐Saavedra J, Adsay NV, Crawford JM et al Carcinoma of the gallbladder and extrahepatic ducts In: Bosman FT, Carneiro F, Hruban RH, Theise ND, eds. WHO Classification of Tumours of the Digestive System, 4th edn Lyon, France: IARC, 2010; 266–73. [Google Scholar]
- 39. Sobin LH, Gospodarowicz MK, Wittekind C. TNM Classification of Malignant Tumors, 7th edn New York: Wiley‐Blackwell, 2009. [Google Scholar]
- 40. Japanese Society of Biliary Surgery . Classification of Biliary Tract Carcinoma, 2nd edn Tokyo, Japan: Kanehara, 2004. [Google Scholar]
- 41. Takahashi Y, Hiraoka N, Onozato K et al Solid‐pseudopapillary neoplasms of the pancreas in men and women: do they differ? Virchows Arch 2006; 448: 561–9. [DOI] [PubMed] [Google Scholar]
- 42. Sato E, Olson SH, Ahn J et al Intraepithelial CD8+ tumor‐infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA 2005; 102: 18538–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sedy JR, Gavrieli M, Potter KG et al B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat Immunol 2005; 6: 90–8. [DOI] [PubMed] [Google Scholar]
- 44. Krieg C, Boyman O, Fu YX, Kaye J. B and T lymphocyte attenuator regulates CD8+ T cell‐intrinsic homeostasis and memory cell generation. Nat Immunol 2007; 8: 162–71. [DOI] [PubMed] [Google Scholar]
- 45. Derre L, Rivals JP, Jandus C et al BTLA mediates inhibition of human tumor‐specific CD8+ T cells that can be partially reversed by vaccination. J Clin Invest 2010; 120: 157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fourcade J, Sun Z, Pagliano O et al CD8(+) T cells specific for tumor antigens can be rendered dysfunctional by the tumor microenvironment through upregulation of the inhibitory receptors BTLA and PD‐1. Cancer Res 2012; 72: 887–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hobo W, Norde WJ, Schaap N et al B and T lymphocyte attenuator mediates inhibition of tumor‐reactive CD8+ T cells in patients after allogeneic stem cell transplantation. J Immunol 2012; 189: 39–49. [DOI] [PubMed] [Google Scholar]
- 48. del Rio ML, Lucas CL, Buhler L, Rayat G, Rodriguez‐Barbosa JI. HVEM/LIGHT/BTLA/CD160 cosignaling pathways as targets for immune regulation. J Leukoc Biol 2010; 87: 223–35. [DOI] [PubMed] [Google Scholar]
- 49. Morel Y, Truneh A, Sweet RW, Olive D, Costello RT. The TNF superfamily members LIGHT and CD154 (CD40 ligand) costimulate induction of dendritic cell maturation and elicit specific CTL activity. J Immunol 2001; 167: 2479–86. [DOI] [PubMed] [Google Scholar]
- 50. Xu H, Cao D, Guo G, Ruan Z, Wu Y, Chen Y. The intrahepatic expression and distribution of BTLA and its ligand HVEM in patients with HBV‐related acute‐on‐chronic liver failure. Diagn Pathol 2012; 7: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol 2015; 33: 1974–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Comparison of infiltrating cells in gallbladder lesion.
Fig. S2. Kaplan–Meier survival curves.
Table S1. Univariate and multivariate survival analyses.


