Abstract
Differentiating malignant pleural mesothelioma (MPM) cells morphologically from reactive mesothelial hyperplasia cells is problematic. Homozygous deletion (HD) of p16 (CDKN2A), detected by FISH, is a good marker of malignancy and is useful to differentiate between these cells. However, the correlation between the p16 status of effusion smears and that of the underlying MPM tissues has not been investigated. We used p16‐specific FISH to investigate 20 cases of MPM from which both effusion cytologic smears and histologic specimens were available. In five cases, histologic specimens included both an invasive component and surface mesothelial proliferation. In 14 cases (70%), MPM cells in both tissue sections and effusion smears were p16 HD‐positive. Conversely, MPM cells in the remaining six tumors (30%) were p16 HD‐negative in both tissue sections and effusion smears. For all five MPM cases with surface mesothelial proliferations and invasive components, the effusion smears, surface mesothelial proliferations, and invasive MPM components all displayed p16 deletion. Moreover, the extent to which p16 was deleted in smears highly correlated with the extent of p16 deletion in tissues. The p16 deletion percentages were also similar among smears, tissue surface proliferations, and invasive components. In cases with clinical and radiologic evidence of a diffuse pleural tumor, detection of p16 deletion in cytologic smear samples may permit MPM diagnosis without additional tissue examination. However, the absence of p16 deletion in cytologic smear samples does not preclude MPM.
Keywords: Cytology, fluorescence in situ hybridization, malignant pleural mesothelioma, p16, reactive mesothelial hyperplasia
Malignant pleural mesothelioma (MPM) is relatively rare but is the most common primary tumor of the pleura. The widespread use of asbestos fibers, regarded as the principal carcinogen that causes MPM, has led to a rising incidence of MPM worldwide; unfortunately, this trend is expected to continue for the next one to two decades.1 In Japan, deaths from mesothelioma rose from 500 in 1995 to 1400 in 2012, and experts anticipate 103 000 deaths in the 40‐year period following 2005.1, 2, 3
Malignant pleural mesothelioma is an aggressive malignancy, and prognosis remains very poor. Overall survival rarely exceeds 12 months.2, 4, 5, 6 This is partly because of the difficulty of diagnosing MPM early; patients are often diagnosed when they become symptomatic and show advanced pleural lesions.7 Because prompt treatment of MPM early in the progression of the disease would improve outcomes,2 early diagnosis of MPM is a significant clinical priority.
The first clinical sign of MPM is often pleural effusion, which is reportedly present in 54–89% of MPM patients.8, 9, 10, 11, 12, 13 Effusion cytology is often the first diagnostic approach, and detecting mesothelioma cells in pleural effusions is critical to the early diagnosis of MPM.8, 10, 13 However, even though the cytologic features of mesothelioma cells have been described and refined for more than 50 years, it is still unknown whether cytologic analysis alone could establish a definitive diagnosis.2 The published sensitivity of cytologic diagnosis of MPM ranges between 32% and 76%.2, 13 At present, the International Mesothelioma Interest Group and the European Respiratory Society/European Society of Thoracic Surgeons discourage basing a diagnosis on cytologic findings alone, but instead recommend MPM diagnosis with tissue confirmation.8, 9
In routine practice, immunohistochemistry (IHC) and immunocytochemistry (ICC) have been proposed for the definitive diagnosis of MPM. By using an appropriate panel of IHC/ICC markers, cells of mesothelial origin can be readily distinguished from pleural metastases of other primary carcinomas in most cases. However, there is still no reliable marker separating benign from malignant mesothelial proliferations with sufficient sensitivity and specificity.14, 15, 16
Recent studies have revealed cytogenetic and molecular changes in MPM. One of the most common genetic alterations is the homozygous deletion (HD) of the 9p21 locus, which includes a cluster of genes such as cyclin‐dependent kinase inhibitor 2A (CDKN2A, also known as p16), CDKN2B (also known as p15), p14 ARF, and methylthioadenosine phosphorylase (MTAP).9, 17, 18, 19, 20 Previous studies reported that homozygous deletion of p16 can be detected by FISH in up to 70–80% of MPM cases. The overall sensitivity of p16 FISH has been reported to be between 56% and 79%, and, notably, the positive predictive value and specificity are both 100%.9, 21, 22, 23 Detection of homozygous p16 deletion by FISH seems to be feasible and helpful in confirming a diagnosis of mesothelioma in cytologic and surgical specimens.9, 14
Biopsy specimens are sometimes too small to detect invasion for diagnostic purposes. However, Hwang et al.14 showed that an MPM diagnosis may be made with only small surface tissue by demonstrating the correlation of p16 FISH status between the surface mesothelial proliferation and the underlying invasive mesothelioma. When a biopsy shows only surface mesothelial proliferation, the finding of homozygous p16 deletion in such a biopsy allows a diagnosis of mesothelioma without an additional biopsy.14 To the best of our knowledge, there is no previous study to evaluate the correlation of p16 FISH status in mesothelioma cells between cytologic and histologic specimens. Effusion cytology may reflect the status of whole MPM lesions more precisely than a small biopsy, especially in the early lesions that show no remarkable pleural changes on radiological observation. If p16 FISH status of cytologic preparations can predict that of the underlying MPM, it may be possible to diagnose MPM in cytologic preparations. Therefore, detecting the p16 FISH status by effusion smears may facilitate early treatment and thus lead to consequent improvement of survival for patients with MPM.
The aim of our study was to establish the correlation between the p16 deletion status of effusion cytology and that of underlying MPM tissues by FISH analysis for MPM cases in which both materials were available. We also examined the deletion status among effusion smears, surface mesothelial proliferations, and underlying invasive components in cases where all of these materials were available. The results indicate the diagnostic utility of p16 FISH for MPM.
Materials and Methods
Case selection
This study included 20 cases of MPM for which both effusion cytology and histologic specimens were available. In five cases, tissue samples included not only deeper areas of tumor invading the adipose tissue (invasive components) but also proliferations of mesothelial cells along the pleural surface (surface proliferations). Additionally, cytologic preparations of 28 non‐mesothelioma cases with reactive mesothelial cells (RMC) and histologic specimens of 25 cases with reactive mesothelial hyperplasia (RMH) were evaluated to determine the cut‐off value for p16 FISH. The cytologic preparations were obtained from pleural effusions associated with tuberculosis, pneumonia, cardiovascular disease, or lung cancer. The histologic specimens with RMH were all obtained from patients with bullae. All cases were derived from the pleural lesion files of the Department of Pathology, Fukuoka University Hospital (Fukuoka, Japan), including consultation cases, between 2007 and 2013. Anonymous use of redundant tissues and cells is part of the standard treatment agreement with patients in our hospital when no objection is expressed. Furthermore, the study protocol was approved by the Ethics Committee of Fukuoka University. The diagnosis and classification of mesothelioma was carried out according to the WHO classification 2003 guidelines.24 The mesothelial nature of the tumor was confirmed by IHC/ICC, using a combination of calretinin, podoplanin (D2‐40), cytokeratin 5/6, and Wilms’ tumor‐1 staining as positive mesothelial markers and thyroid transcription factor‐1, Ber‐EP4, and carcinoembryonic antigen as negative markers.
Fluorescence in situ hybridization assay
The FISH studies were carried out on histologic specimens with formalin‐fixed, paraffin‐embedded, 4‐μm‐thick tissue sections and on cytologic specimens fixed in 95% ethanol using dual‐color probes: a spectrum green‐labeled chromosome 9 centromeric probe and a spectrum orange‐labeled, locus‐specific p16 probe (Vysis LSI p16/CEP 9 probe; Abbott Japan, Tokyo, Japan), as previously described.10
Briefly, cytologic specimens were first treated in xylene overnight to remove the mounting medium, rehydrated with descending alcohol dilutions, and refixed in Carnoy's solution at room temperature (RT) for 15 min. Tissue sections were deparaffinized in xylene, followed by rehydration as above. Both cytologic and tissue preparations were then incubated in 2× SSC buffer containing 0.3% Tween‐20 (Sigma, St. Louis, MO, USA) at 37°C for 1–12 h and then in pretreatment solution (Histology FISH Accessory Kit; Dako, Carpinteria, CA, USA) (1:20 dilution) at 95°C for 10 min. These specimens were then digested with pepsin solution (Dako) at 37°C for 1–2 min for cytologic and 5 min for tissue preparations. Hereafter, both cytologic and tissue preparations were treated similarly. After refixation in 10% buffered formalin at RT for 5 min, the preparations were treated in 2× SSC buffer containing 0.3% Tween‐20 at 45°C for 30 min, dehydrated in ethanol, and dried, followed by addition of the two probes. Both the probes and preparations were denatured at 80°C for 5 min in the probe solution included with the kit (Abbott Japan) followed by hybridization at 37°C for 48 h in the ThermoBrite unit (Abbott Japan). The preparations were washed in 2× SSC containing 0.3% Tween‐20 at 72°C for 3–5 min and in 2× SSC containing 0.1% Tween‐20 at RT for 3–5 min. Nuclei were counterstained with DAPI in the antifade reagent (Vector Laboratories, Burlingame, CA, USA). Analyses were carried out using a fluorescence microscope (Axio Imager Z1; Carl Zeiss Microimaging, Jena, Germany) and the Isis analysis system (MetaSystems, Altlussheim, Germany) equipped with filter sets with single‐ and dual‐band excitors for spectrum green, spectrum orange, and DAPI (UV, 360 nm).
Homozygous deletion was defined as lack of both 9p21 signals. Heterozygous deletion was assumed when only one 9p21 signal was present or when the total number of 9p21 signals did not exceed half the total number of the centromeric signals. At least 100 cells were scored in each case, and if homozygous deletion of 9p21 was identified in >10% of mesothelial cells, p16 homozygous deletion was considered positive. Lymphocytes in each preparation served as internal negative controls and showed two signals per FISH probe. This confirmed that loss of p16 FISH signals was not due to preanalytic factors such as fixation or processing.
Statistical analysis
To analyze the relationship between the cytologic and histologic specimen p16 FISH results, simple linear regression was carried out, and the coefficient of determination (R 2) was calculated accordingly. Spearman's rank correlation was used to assess the relationship among cytologic preparations, surface mesothelial proliferations, and underlying invasive components. All statistical analyses were carried out using JMP 11.0 software (SAS Institute, Cary, NC, USA), and P‐values of less than 0.05 were considered statistically significant.
Results
Determination of the cut‐off values for p16 FISH
The cut‐off values were calculated as the mean percentage ± three standard deviations of nuclei showing loss of both 9p21 signals for homozygous deletion, as previously described.10 Among cases with cytologic preparations with RMC and histologic specimens of RMH, a homozygous deletion pattern was observed in 0–3.6% (mean, 1.3%) and 0.8–4.7% (mean, 2.6%), respectively (Fig. 1a,b). Based on these results, a cut‐off value of >10% for nuclei with a homozygous deletion pattern was set for homozygous deletion. Although the actual values were 5.5% for cytologic preparations with RMC and 6.1% for histologic specimens of RMH, the value of >10% was used to exclude the possibility of pseudopositives.
Figure 1.
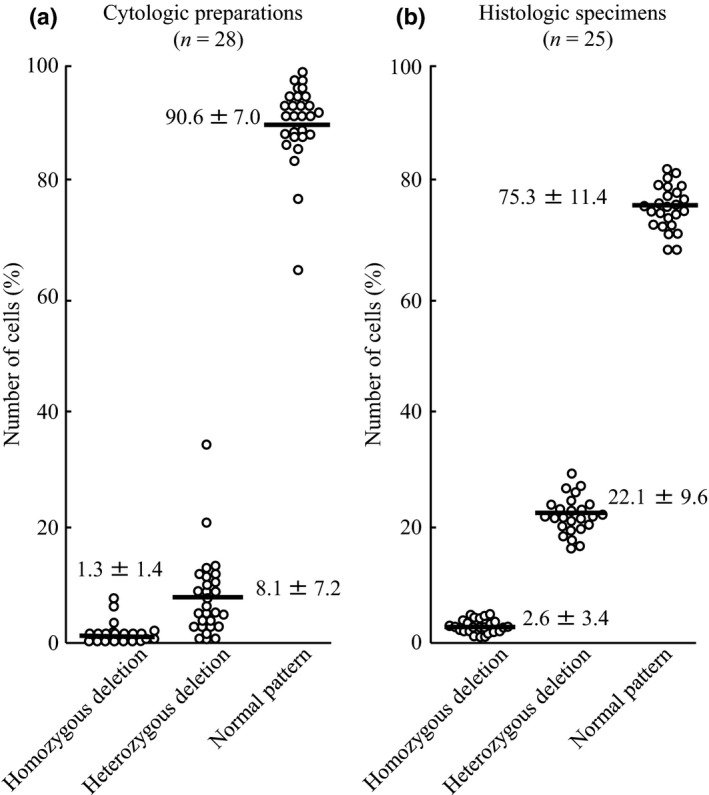
Number of cells showing each p16 FISH pattern in benign mesothelial proliferations. (a) Reactive mesothelial cells in cytologic preparations (n = 28). (b) Reactive mesothelial hyperplasia in histologic specimens (n = 25). The bold lines indicate mean numbers of cells. Data are given as mean ± SD.
Correlation between cytology and histology
The deletion status of p16 determined by FISH in cytologic and histologic specimens in each case is summarized in Table 1. Of the 20 patients included in this study, 17 were male and three were female (M:F ratio, 5.7:1). The mean age at diagnosis was 66.6 years (range, 55–81 years). All cases were of the epithelioid histologic subtype.
Table 1.
Clinical data and p16 FISH results in 20 cases of malignant pleural mesothelioma with cytology and histology
| Case no. | Age, decade | Sex | HD‐positive cells, % | Result | |
|---|---|---|---|---|---|
| Cytology | Histology | ||||
| 1 | 6th | M | 4 | 1 | − |
| 2 | 7th | M | 0 | 2 | − |
| 3 | 6th | M | 91 | 59 | + |
| 4 | 6th | M | 25 | 30 | + |
| 5 | 8th | M | 87 | 80 | + |
| 6 | 8th | M | 11 | 18 | + |
| 7 | 6th | M | 98 | 96 | + |
| 8 | 9th | M | 97 | 90 | + |
| 9 | 7th | F | 92 | 98 | + |
| 10 | 8th | M | 92 | 98 | + |
| 11 | 7th | M | 98 | 91 | + |
| 12 | 7th | M | 90 | 94 | + |
| 13 | 7th | M | 99 | 94 | + |
| 14 | 7th | M | 2 | 4 | − |
| 15 | 8th | M | 0 | 2 | − |
| 16 | 7th | M | 96 | 74 | + |
| 17 | 7th | M | 1 | 3 | − |
| 18 | 8th | M | 85 | 73 | + |
| 19 | 8th | F | 92 | 94 | + |
| 20 | 7th | F | 1 | 4 | − |
| Mean ± SE | 58.1 ± 9.9 | 54.5 ± 9.1 | |||
| Median (range) | 88 (0–99) | 73 (1–98) | |||
F, female; HD, homozygous deletion; M, male.
Fourteen of the 20 patients (70%) showed homozygous deletion of p16 in both MPM tissue and atypical mesothelial cells in effusion smears (Fig. 2, Table 2). Although the atypical mesothelial cells in smears may have contained MPM cells and also some RMC, no significant difference was detected in the proportions of p16 homozygous deletion between effusion smears and MPM tissues. The remaining six patients (30%) were p16 deletion negative in both tissues and smears (Fig. 3, Table 2). There were no p16 FISH result discrepancies between the preparations for any of the cases (Fig. 4a, Table 2). The average percentages of homozygous deletion across all cases (mean ± SE) were 54.5 ± 9.1 (tissues) vs 58.1 ± 9.9 (smears) (Table 1).
Figure 2.
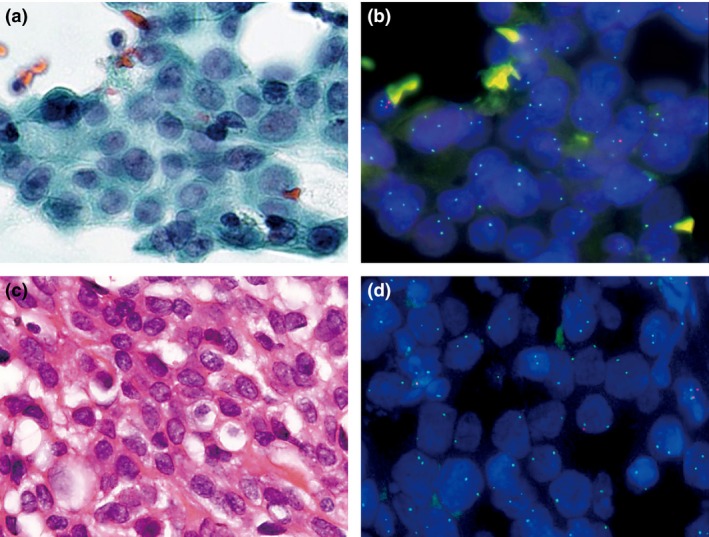
Example of a p16 deletion‐positive malignant pleural mesothelioma. Atypical mesothelial cells in pleural effusion (a) and corresponding p16 FISH image (b). Invasive mesothelioma (c) and corresponding p16 FISH image (d).
Table 2.
Comparison of malignant pleural mesothelioma cytology and histology statistics based on p16 deletion status using FISH
| Case type | n (%) | Mean % deletion (±SE) Median (range) | |
|---|---|---|---|
| Cytology | Histology | ||
|
Cytology (+) Histology (+) |
14/20 (70) |
82.4 ± 7.4 92 (11–99) |
76.8 ± 6.8 91 (18–98) |
|
Cytology (−) Histology (−) |
6/20 (30) |
1.5 ± 0.6 1 (0–4) |
2.6 ± 0.5 2 (1–4) |
|
Cytology (−) Histology (+) |
0/20 (0) |
NA | NA |
|
Cytology (+) Histology (−) |
0/20 (0) |
NA | NA |
NA, not applicable; (+), homozygous deletion‐positive; (−), homozygous deletion‐negative.
Figure 3.
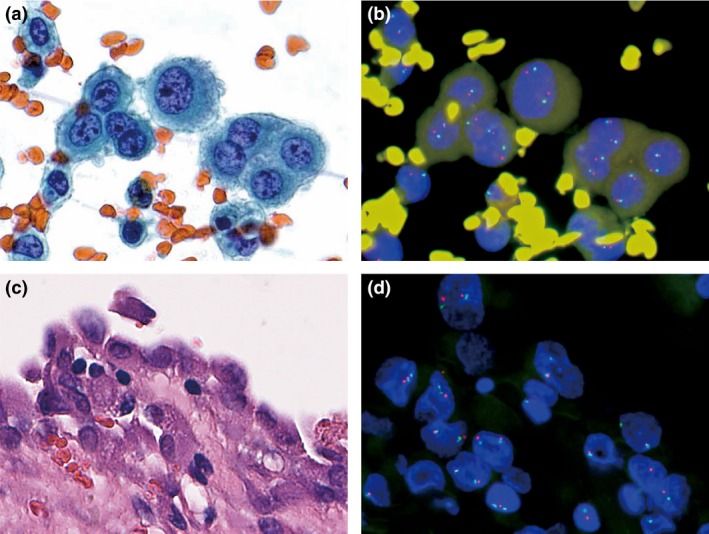
Example of a p16 deletion‐negative malignant pleural mesothelioma. Atypical mesothelial cells in pleural effusion (a) and corresponding p16 FISH image (b). Invasive mesothelioma (c) and corresponding p16 FISH image (d).
Figure 4.
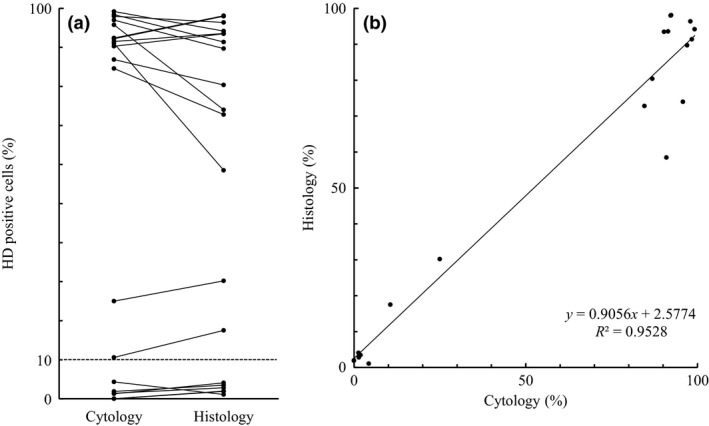
Statistical analysis of the cytologic and histologic findings from 20 malignant pleural mesothelioma cases. (a) Correspondence of each proportion of p16 homozygous deletion (HD)‐positive cells. The threshold value is set at 10% (dashed line). (b) Simple linear regression. The regression line equation and coefficient of determination (R 2) are shown.
More than 90% of the tumor cells showed homozygous deletion of p16 in 12 of the 14 deletion‐positive cases, whereas only 10–30% showed homozygous deletion in both preparations of the remaining two cases. However, the proportions of p16 homozygous deletion‐positive tumor cells in the cytologic preparations tended to be similar to those of the histologic specimens in each case (Fig. 4a, Table 2). There was excellent concordance in the results of the regression analysis of positivity of homozygous deletion between cytologic and histologic specimens (Fig. 4b, R 2 = 0.9528).
Correlation among effusion smears, surface mesothelial proliferations and invasive MPM components
As shown in Table 3, all five positive cases that included both surface mesothelial proliferations and invasive components in histologic specimens showed similar p16 homozygous deletion rates in all three specimens: effusion smears, surface mesothelial proliferations, and underlying invasive MPM components (%HD‐positive cells, 43.5 ± 21.4 [smears], 66.3 ± 16.5 [surface tissues], 68.0 ± 14.7 [invasive tissues]) (Table 3, Fig. 5). Three of these cases showed homozygous deletion in more than 80% of the tumor cells, and the remaining two cases showed low positivity in all three preparations, although all cases were considered homozygous deletion‐positive. There was a trend toward positive correlation in homozygous p16 deletion MPM cells among the three preparations, although the number of cases was not enough (smears vs surface tissues, Spearman's ρ = 0.8000, P = 0.1041; smears vs invasive tissues, ρ = 0.8000, P = 0.1041; surface vs invasive tissues, ρ = 1.0000, P < 0.0001).
Table 3.
Results of p16 FISH in five malignant pleural mesothelioma cases with effusion cytology, surface mesothelial proliferations, and underlying invasive components
| Case no. | HD‐positive mesothelial cells, % | ||
|---|---|---|---|
| Cytologic smear | Surface proliferation | Invasive component | |
| 4 | 25 | 23 | 32 |
| 6 | 11 | 29 | 33 |
| 11 | 98 | 97 | 97 |
| 12 | 90 | 90 | 83 |
| 13 | 99 | 93 | 96 |
| Mean ± SE | 43.5 ± 21.4 | 66.3 ± 16.5 | 68.0 ± 14.7 |
| Median (range) | 90 (11–99) | 90 (23–97) | 83 (32–97) |
HD, homozygous deletion.
Figure 5.
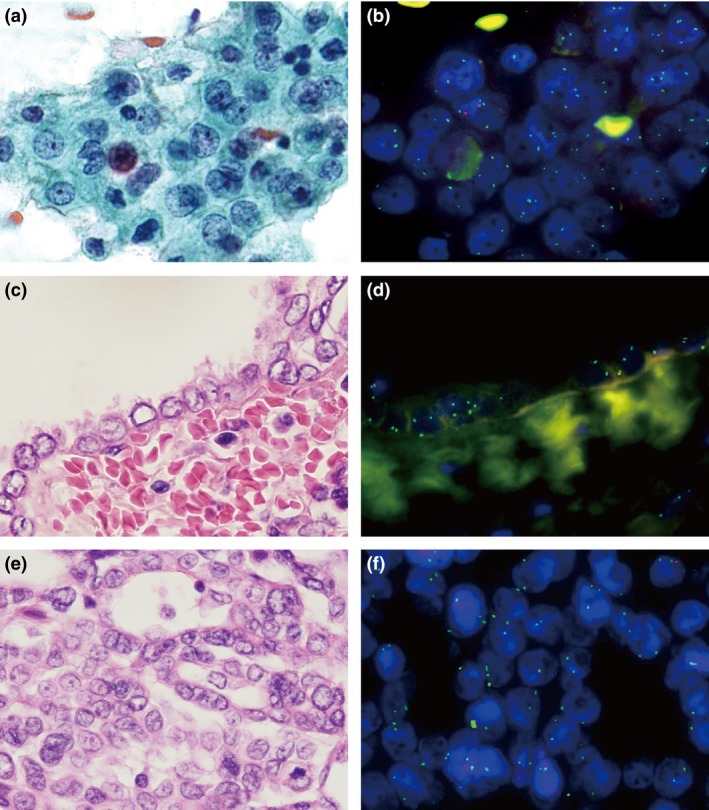
Example of malignant pleural mesothelioma (MPM) with effusion cytology, surface mesothelial proliferation, and underlying invasive component. Atypical mesothelial cells in pleural effusion (a) and corresponding p16 FISH image (b). Surface mesothelial proliferation (c) and corresponding p16 FISH image (d). Invasive MPM component (e) and corresponding p16 FISH image (f).
Discussion
This report is the first to establish the correlation between the p16 deletion status of effusion cytology and that of the underlying MPM. The tumor cells in these two preparations showed good concordance of positivities of p16 homozygous deletion. Accordingly, we may be able to predict the p16 deletion status of underlying MPM by analyzing the status of tumor cells in pleural effusion. This may contribute to the earlier detection and diagnosis of MPM using effusion smear cytology specimens, although the current guidelines for diagnosis of MPM require the detection of invasion in histologic specimens and do not permit the diagnosis to be based solely on effusion cytology.8, 9
Diagnosis of MPM involves first determining the mesothelial origin of the lesion by morphology and IHC/ICC and then differentiating between benign and malignant conditions. An MPM diagnosis based on effusion cytology alone has previously not been reliable due to the difficulty in differentiating MPM from other conditions on the basis of morphology. There are numerous cytomorphologic mimics of MPM, such as reactive mesothelial proliferations caused by infections, collagen vascular diseases, pulmonary infarction, drug reactions, reactions to pneumothorax, subpleural lung carcinomas, surgery, trauma, and non‐specific inflammation in the pleural space.9 These conditions often show complex growth, increased cellularity, cytologic atypia, mitoses, architectural atypia such as papillary excrescences and luminal formation, and pseudoinvasion (entrapment).14, 25 Thus, the term atypical mesothelial proliferation is more commonly used on effusion cytology than surgical specimens.
The use of various molecules in IHC/ICC to diagnose MPM has been challenged. Recently, new biomarkers such as glucose transporter‐1 (GLUT‐1), CD146, and oncofetal protein IMP3 have been described for confirmed diagnosis in cytologic and histologic specimens.26 Although IHC/ICC is a powerful tool to identify cells of mesothelial origin and differentiate MPM from pleural metastases of other primary carcinomas, the ability of this technique to distinguish between benign and malignant mesothelial proliferations remains controversial.14, 15, 16
Recent cytogenetic and molecular studies have identified several frequent genetic alterations in mesothelioma, the most common of which is homozygous deletion of the 9p21 locus containing a cluster of genes, including CDKN2A, CDKN2B, p14 ARF, and MTAP.18, 22, 23, 27, 28, 29, 30 The p16 protein, encoded by CDKN2A, is a CDK inhibitor and a member of the INK4 (inhibitor of CDK4) family of proteins that bind and generally inhibit D‐type CDKs. The p16 protein is present in all normal cells, acts through CDK4/CDK6, and blocks the phosphorylation of the retinoblastoma (RB) protein.9, 14, 22, 31 Deletions of p16 have been reported in up to 80% of primary pleural mesotheliomas, depending on the histologic subtype (90–100% of the sarcomatoid subtype, 70% of epithelioid and mixed subtypes).9 Point mutations and DNA methylation occur less frequently at this locus and may occur in benign mesothelial cells as well.28, 32 The detection of p16 homozygous deletion by FISH is considered helpful in discriminating MPM cells from RMCs.23 Moreover, the presence of p16 homozygous deletion correlates with shorter survival in patients with MPM.9
The analysis of p16 deletion status by FISH may provide another diagnostic confirmation, along with the presence of tissue invasion. Hwang et al.14 revealed that it is possible to diagnose MPM using the detection of p16 deletion in surface mesothelial proliferations, even if tumor invasion was not evident, because the p16 deletion status of mesothelial surface proliferations and that of underlying mesotheliomas are correlated. Additionally, in the current study, we showed a good correlation between the p16 deletion status of MPM cells in pleural effusions and that of underlying invasive MPM tissues. Moreover, we also revealed the correlation of p16 deletion status among effusion cytologic smears, surface mesothelial proliferations, and underlying invasive MPM components in five cases. Each case showed similar cellular proportions of p16 deletion in all of the preparations. These lines of evidence suggest that it is possible to diagnose MPM based on cytologic preparations or small tissue sections without definitive evidence of tumor invasion in deeper tissues through the use of p16 FISH.
In the present study, there were two groups of p16 homozygous deletion‐positive cases, those with high‐level homozygous deletion (>90%) and those with low‐level homozygous deletion (11–30%). Two cases belonged to the latter group and showed relatively high proportions of p16 heterozygous deletion in both smears and tissues (smears/tissues, 63%/59% and 62%/66%, respectively). Recent studies have suggested the utility of p16 heterozygous deletion for the diagnosis of MPM using appropriate cut‐off values.10, 15 The results presented in this study may indicate the possible correlation in the heterozygous deletion status between smears and tissues of MPM. However, the biological difference between the high‐ and low‐level homozygous deletion groups and the clinical significance of p16 heterozygous deletion dominancy remain unclear. Further studies with more cases should clarify these issues.
Histology, when it detects tumor invasion, is powerful and conclusive in the diagnosis of MPM. However, sampling errors sometimes occur, especially in the early stages of the disease; the lesion might not be continuous or not evident on thoracoscopic examination. In these cases, effusion smear cytology may be more effective because desquamated MPM cells can accumulate in effusions. These limitations of current diagnostic techniques support the usefulness of applying p16 FISH to smear cytology.
Our study had a few limitations. First, the number of cases analyzed was relatively small. In particular, there was a dearth of cases for which all three preparations (effusion smears, surface mesothelial proliferations, and underlying invasive MPM) were available to obtain statistical significance. Although similar proportions of p16 deletion were observed among these three preparations, a larger‐scale study is necessary to determine statistical correlation. Second, the absence of p16 deletion in the tumor cells in pleural effusion does not rule out underlying MPM because some MPM cases do not show p16 deletion. Furthermore, we also cannot differentiate MPM from mesothelioma‐like metastatic lesions from other malignancies, including carcinomas of the lung, bladder, breast, and prostate, by p16 deletion status.14 We should carefully diagnose MPM in such cases by using IHC/ICC together with H&E‐stained sections and clinical information. Third, our study includes only epithelioid MPMs, which often present with pleural effusions. Finally, we should recognize that there are also some cases of MPM without pleural effusions, such as those of the sarcomatoid subtype, which cannot be diagnosed by effusion cytology.9
In conclusion, p16 FISH analysis is a powerful tool to differentiate MPM from benign mesothelial proliferations. Detection of p16 deletion in smear cytologic samples may permit diagnosis of MPM without additional tissue examination in cases with clinical and radiologic evidence of a diffuse pleural tumor, although the absence of p16 deletion in cytologic smear samples does not preclude MPM.
Disclosure Statement
The authors declare no conflict of interest.
Acknowledgments
The authors thank Ms. K. Yano, Department of Pathology, Fukuoka University School of Medicine and Hospital, for her skillful help in p16 FISH. This work was supported in part by a grant from the Research Center for Advanced Molecular Medicine, Fukuoka University, the Izumo City Supporting Cancer Research Project, and the Ministry of the Environment, Japan.
Cancer Sci (2015)
Funding Information
This work was supported in part by a grant from the Research Center for Advanced Molecular Medicine, Fukuoka University, Izumo City Supporting Cancer Research Project (ICSCRP), and the Ministry of the Environment.
References
- 1. Robinson BW, Lake RA. Advances in malignant mesothelioma. N Engl J Med 2005; 353: 1591–603. [DOI] [PubMed] [Google Scholar]
- 2. Tsao AS, Wistuba I, Roth JA, Kindler HL. Malignant pleural mesothelioma. J Clin Oncol 2009; 27: 2081–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Statistics and Information Department, Minister's Secretariat, Ministry of Health, Labor and Welfare. Vital Statistics of Japan, 2012. Tokyo, Japan: Health and Welfare Statistics Association, 2014. [Google Scholar]
- 4. Dacic S, Kothmaier H, Land S et al Prognostic significance of p16/cdkn2a loss in pleural malignant mesotheliomas. Virchows Arch 2008; 453: 627–35. [DOI] [PubMed] [Google Scholar]
- 5. Neragi‐Miandoab S. Multimodality approach in management of malignant pleural mesothelioma. Eur J Cardiothorac Surg 2006; 29: 14–9. [DOI] [PubMed] [Google Scholar]
- 6. Robinson BW, Musk AW, Lake RA. Malignant mesothelioma. Lancet 2005; 366: 397–408. [DOI] [PubMed] [Google Scholar]
- 7. Milano MT, Zhang H. Malignant pleural mesothelioma: a population‐based study of survival. J Thorac Oncol 2010; 5: 1841–8. [DOI] [PubMed] [Google Scholar]
- 8. Scherpereel A, Astoul P, Baas P et al Guidelines of the European Respiratory Society and the European Society of Thoracic Surgeons for the management of malignant pleural mesothelioma. Eur Respir J 2010; 35: 479–95. [DOI] [PubMed] [Google Scholar]
- 9. Husain AN, Colby T, Ordonez N et al Guidelines for pathologic diagnosis of malignant mesothelioma: 2012 update of the consensus statement from the International Mesothelioma Interest Group. Arch Pathol Lab Med 2013; 137: 647–67. [DOI] [PubMed] [Google Scholar]
- 10. Matsumoto S, Nabeshima K, Kamei T et al Morphology of 9p21 homozygous deletion‐positive pleural mesothelioma cells analyzed using fluorescence in situ hybridization and virtual microscope system in effusion cytology. Cancer Cytopathol 2013; 121: 415–22. [DOI] [PubMed] [Google Scholar]
- 11. Senyiğit A, Bayram H, Babayiğit C et al Malignant pleural mesothelioma caused by environmental exposure to asbestos in the Southeast of Turkey: CT findings in 117 patients. Respiration 2000; 67: 615–22. [DOI] [PubMed] [Google Scholar]
- 12. Chapman A, Mulrennan S, Ladd B, Muers MF. Population based epidemiology and prognosis of mesothelioma in Leeds, UK. Thorax 2008; 63: 435–9. [DOI] [PubMed] [Google Scholar]
- 13. Savic S, Franco N, Grilli B et al Fluorescence in situ hybridization in the definitive diagnosis of malignant mesothelioma in effusion cytology. Chest 2010; 138: 137–44. [DOI] [PubMed] [Google Scholar]
- 14. Hwang H, Tse C, Rodriguez S, Grown A, Churg A. p16 FISH deletion in surface epithelial mesothelial proliferations is predictive of underlying invasive mesothelioma. Am J Surg Pathol 2014; 38: 681–8. [DOI] [PubMed] [Google Scholar]
- 15. Chung CT, Santos Gda C, Hwang DM et al FISH assay development for the detection of p16/CDKN2A deletion in malignant pleural mesothelioma. J Clin Pathol 2010; 63: 630–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kato Y, Tsuta K, Seki K et al Immunohistochemical detection of GLUT‐1 can discriminate between reactive mesothelium and malignant mesothelioma. Mod Pathol 2007; 20: 215–20. [DOI] [PubMed] [Google Scholar]
- 17. Prins JB, Williamson KA, Kamp MM et al The gene for the cyclin‐dependent‐kinase‐4 inhibitor, CDKN2A, is preferentially deleted in malignant mesothelioma. Int J Cancer 1998; 75: 649–53. [DOI] [PubMed] [Google Scholar]
- 18. Cheng JQ, Jhanwar SC, Klein WM et al P16 alterations and deletion mapping of 9p21‐P22 in malignant mesothelioma. Cancer Res 1994; 54: 5547–51. [PubMed] [Google Scholar]
- 19. Singhal S, Wiewrodt R, Malden LD et al Gene expression profiling of malignant mesothelioma. Clin Cancer Res 2003; 9: 3080–97. [PubMed] [Google Scholar]
- 20. Musti M, Kettunen E, Dragonieri S et al Cytogenetic and molecular genetic changes in malignant mesothelioma. Cancer Genet Cytogenet 2006; 170: 9–15. [DOI] [PubMed] [Google Scholar]
- 21. Illei PB, Rusch VW, Zakowski MF, Ladanyi M. Homozygous deletion of CDKN2A and codeletion of the methylthioadenosine phosphorylase gene in the majority of pleural mesotheliomas. Clin Cancer Res 2003; 9: 2108–13. [PubMed] [Google Scholar]
- 22. Illei PB, Ladanyi M, Rusch VW, Zakowski MF. The use of CDKN2A deletion as a diagnostic marker for malignant mesothelioma in body cavity effusions. Cancer 2003; 99: 51–6. [DOI] [PubMed] [Google Scholar]
- 23. Monaco SE, Shuai Y, Bansal M, Krasinskas AM, Dacic S. The diagnostic utility of p16 FISH and GLUT‐1 immunohistochemical analysis in mesothelial proliferations. Am J Clin Pathol 2011; 135: 619–27. [DOI] [PubMed] [Google Scholar]
- 24. Travis WD, Brambilla E, Müller‐Hermelink HK, Harris CC. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. Lyon, France: International Agency for Research on Cancer Press, 2004. [Google Scholar]
- 25. Chiosea S, Krasinskas A, Cagle PT, Mitchell KA, Zander DS, Dacic S. Diagnostic importance of 9p21 homozygous deletion in malignant mesothelioma. Mod Pathol 2008; 21: 742–7. [DOI] [PubMed] [Google Scholar]
- 26. Minato H, Kurose N, Fukushima M et al Comparative immunohistochemical analysis of IMP3, GLUT1, EMA, CD146, and desmin for distinguishing malignant mesothelioma from reactive mesothelial cells. Am J Clin Pathol 2014; 141: 85–93. [DOI] [PubMed] [Google Scholar]
- 27. Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors. Mesothelioma. Cancer Genet Cytogenet 2001; 127: 93–110. [DOI] [PubMed] [Google Scholar]
- 28. Hirao T, Bueno R, Chen CJ, Gordon GJ, Heilig E, Kelsey KT. Alterations of the p16(INK4) locus in human malignant mesothelial tumors. Carcinogenesis 2002; 23: 1127–30. [DOI] [PubMed] [Google Scholar]
- 29. Björkqvist AM, Tammilehto L, Anttila S, Mattson K, Knuutila S. Recurrent DNA copy number changes in 1q, 4q, 6q, 9p, 13q, 14q and 22q detected by comparative genomic hybridization in malignant mesothelioma. Br J Cancer 1997; 75: 523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xio S, Li D, Vijg J, Sugarbaker DJ, Corson JM, Fletcher JA. Codeletion of p15 and p16 in primary malignant mesothelioma. Oncogene 1995; 11: 511–5. [PubMed] [Google Scholar]
- 31. Stahel RA. Malignant pleural mesothelioma: a new standard of care. Lung Cancer 2006; 54: S9–14. [DOI] [PubMed] [Google Scholar]
- 32. Pu RT, Sheng ZM, Michael CW, Rhode MG, Clark DP, O'Leary TJ. Methylation profiling of mesothelioma using real‐time methylation‐specific PCR: a pilot study. Diagn Cytopathol 2007; 35: 498–502. [DOI] [PubMed] [Google Scholar]


