Abstract
Metastatic breast cancer remains a highly lethal disease, and it is very important to evaluate the biomarkers associated with the distant metastasis. MicroRNA (miRNA) are small non‐protein coding RNA that regulate various cellular functions. Recent investigations have demonstrated the importance of some miRNA in breast cancer, but the significance of the great majority of miRNA remains largely unclear in breast cancer metastasis. Therefore, in this study, we first examined expression profiles of miRNA in stage IV breast carcinoma tissues, comparing stage I–III cases by miRNA PCR array, and identified miR‐1 as the miRNA which was the most associated with the distant metastasis. However, miR‐1 has not yet been examined in breast carcinoma tissue, and its significance remains unknown. Therefore, we further examined miR‐1 expression in breast carcinoma using in situ hybridization (ISH). miR‐1 was localized in carcinoma cells in 20% of breast carcinoma cases, but it was negligible in non‐neoplastic mammary glands or stroma. miR‐1 ISH status was significantly associated with stage, pathological T factor, lymph node metastasis, distant metastasis, histological grade, estrogen receptor, progesterone receptor and Ki‐67 in breast carcinoma. Moreover, the miR‐1 status was demonstrated using multivariate analysis as an independent worse prognostic factor for both disease‐free and breast cancer‐specific survival. These findings suggest that abnormal miR‐1 expression is associated with an aggressive phenotype of breast carcinoma and that miR‐1 status is a potent prognostic factor in human breast cancer patients.
Keywords: Breast cancer, in situ hybridization, microRNA, PCR array, prognosis
Breast cancer is the most common malignancy among women throughout the world. Despite the recent advances in early detection and treatment,1 6–7% of breast cancer presents distant metastasis at diagnosis (stage IV)2 and approximately 30% will develop metastasis during the evolution of their disease.3 Metastatic breast cancer remains a highly lethal disease, and the 5‐year overall survival ranges from 4 to 28%.4 Therefore, it is very important to evaluate the clinical and/or biological markers associated with the distant metastasis and to clarify molecular mechanisms of distant metastasis to improve the prognosis of breast cancer patients.
MicroRNA (miRNA) are small (18–24 nucleotides) non‐protein coding RNA that post‐transcriptionally negatively regulate target mRNA by binding to their 3′ untranslated regions.5, 6 Single miRNA binds to multiple target mRNA, and regulates various cellular functions including proliferation, differentiation and metastasis.7 Altered expression levels of miRNA have been reported in several types of human cancer, and some of them are suggested to contribute to tumor progression or suppression.8 miRNA have been also investigated in breast cancer,9, 10, 11 and some miRNA (e.g. miR‐10b and miR‐21) have been reported to be associated with metastasis.12, 13 However, the significance and function of the great majority of miRNA in breast cancer metastasis remain unclear. Therefore, in the present study, we first studied the expression profiles of miRNA in stage IV breast carcinoma tissues based on miRNA PCR array, and newly demonstrated that miR‐1 is the most closely associated with the distant metastasis of breast cancer.
In humans, miR‐1 is processed from two different precursors: miR‐1‐1 and miR‐1‐2.14, 15 miR1‐1 and miR1‐2 are located in an intron of C20orf166 and MIB1 (mindbomb E3 ubiquitin protein ligase 1) genes, respectively.14, 16 MIB1 is essential for activation of Notch signaling17 which regulates various cellular functions,18 and is also involved in oncogenesis in many human carcinomas.19 This evidence suggests an important role for miR‐1 in breast cancer, but to the best of our knowledge, miR‐1 has not been studied in breast carcinoma tissues. Therefore, in this study, we examined miR‐1 localization in human breast carcinoma tissues by in situ hybridization (ISH) to clarify its clinicopathological significance.
Materials and Methods
Patients and tissues
In the present study, 163 specimens of invasive ductal carcinoma (IDC) of the breast were evaluated. All specimens had been fixed in 10% formalin and embedded in paraffin wax. Among these, 22 specimens were stage IV IDC obtained from women who underwent surgical treatment from 1995 to 2013 in the Department of Surgery, Tohoku University Hospital, Sendai, Japan. The metastatic sites of breast cancer at diagnosis were bone (n = 12), lung (n = 11) and liver (n = 3) in these patients. In addition, 141 specimens of Stages I–III IDC were obtained from women who underwent surgical treatment in two different periods, 1995–1999 (n = 42) and 2007–2008 (n = 99), in the Department of Surgery, Tohoku University Hospital, Sendai, Japan. Among these, 111 patients received adjuvant endocrine therapy after the surgery, and tamoxifen and aromatase inhibitors were mainly used in the former and later periods, respectively. In contrast, 77 patients received adjuvant chemotherapy. The clinical outcome was evaluated by disease‐free and breast cancer‐specific survival of the stage I–III patients according to a previous report,20 and the mean follow‐up time was 72 months (range, 2–168 months). Breast cancer‐specific survival was defined as the time from surgery to death from the breast cancer.
MicroRNA PCR array was performed in 11 estrogen receptor (ER)‐positive cases (six stage IV cases and five stage I–III cases) among these samples. Snap‐frozen specimens were also available for five cases of stage IV breast carcinoma, and these specimens were used for microarray analysis.
Research protocols for the present study were approved by the Ethics Committee at the Tohoku University School of Medicine.
MicroRNA PCR array
Formalin‐fixed paraffin‐embedded breast carcinoma tissues were cut into 10‐μm sections and five serial sections were collected. After dissection of the area where the tumor cells were contained more than 80%, miRNA was extracted using a miRNeasy FFPE Kit (QIAGEN, Hilden, Germany).
MicroRNA PCR array was performed according to a previous report.21 Briefly, cDNA for miRNA PCR array was synthesized using a miScript II RT kit (QIAGEN), then cDNA was preamplicated using a miScript PreAMP PCR kit (QIAGEN). Specimens were analyzed for the expression of a panel of 88‐cancer related miRNA using miScript miRNA PCR Arrays (QIAGEN). PCR was performed in the ABI7500 Real‐Time PCR System (Applied Biosystems, Foster city, CA, USA) at the Biomedical Research Core of Tohoku University (Sendai, Japan). Data analyses were performed using the miScript miRNA PCR Array Web‐based software (http://pcrdataanalysis.sabiosciences.com/mirna/arrayanalysis.php).
Microarray analysis
Gene expression profiles of breast carcinoma cells were examined using microarray analysis. Briefly, total RNA was extracted from five breast carcinoma tissues using a miRNeasy Mini kit (QIAGEN). A SurePrint G3 Human GE 8 × 60K v2 Microarray Kit (G4851B, ID 039494 [Agilent Technologies, Waldbronn, Germany]) was used, and sample preparation and processing were performed according to the manufacturer's protocol. The putative miR‐1 target genes were predicted by four different prediction tools (i.e. TargetScan [http://www.targetscan.org/], PicTar [http://pictar.mdc-berlin.de/], miDB [http://mirdb.org/miRDB/] and microRNA.org [http://www.microrna.org/microrna/home.do]) in this study.
in situ hybridization
MicroRNA ISH Buffer and Controls kit (Exiqon, Vedbaek, Denmark) were used for ISH in the present study according to the manufacturer's protocol. Briefly, formalin‐fixed paraffin‐embedded breast carcinoma tissues were cut into 4‐μm sections and deparaffinized. After treatment with proteinase K and post‐fixation with 4% paraformaldehyde, hybridization mixture containing 5 nM double‐digoxigenin labeled miRCURY LNA for miR‐1 was applied and hybridized for 1 h at 50°C. The probe for miR‐1 used in this study is complementary to human mature miR‐1 and the sequence was 5′‐ATACATACTTCTTTACATTCCA‐3′. For signal detection, Anti‐digoxigenin‐AP Fab fragments (1:1000; Roche Applied Science, Mannheim, Germany) were used as primary antibody, and the slides were incubated with NBT/BCIP solution (Roche). Counterstaining was performed by Nuclear Fast Red (Chroma, Stuttgart, Germany).
As a negative control, scrambled negative control (5′‐TTCACAATGCGTTATCGGATGT‐3′; Exiqon) was applied instead of the miR‐1 specific probe. We used skeletal muscle tissue as a positive control.22 miR‐1 signal was detected in the cytoplasm of breast carcinoma cells, and the cases that had more than 10% of the positive carcinoma cells were considered positive for miR‐1 ISH status in this study.
Immunohistochemistry
Immunohistochemistry for ER (CONFIRM anti‐ER [SP1]) and progesterone receptor (PR; CONFIRM anti‐PR [1E2]; Roche Diagnostics Japan, Tokyo, Japan) was performed with Ventana Benchmark XT (Roche Diagnostics Japan), and that for HER2 was performed by HercepTest (DAKO). Ki‐67 (MIB1) was purchased from DAKO (Carpinteria, CA, USA), and a Histofine kit (Nichirei Bioscience, Tokyo, Japan) was used for the immunohistochemistry.
Immunoreactivity for ER, PR and Ki‐67 was detected in the nuclei and was evaluated in more than 1000 carcinoma cells for each case. Subsequently, the percentage of immunoreactivity (labeling index [LI]) was determined, and cases with ER LI or PR LI of more than 1% were considered ER‐positive or PR‐positive according to a previous report.23 HER2 status was evaluated according to the grading system proposed in the HercepTest (DAKO), and strongly circumscribed membrane immunoreactivity of HER2 present in more than 10% carcinoma cells (score 3+) was considered positive. In addition, HER2 gene amplification was investigated by FISH in intermediate scoring (score 2+) cases, and the score 2+ cases that were positive were considered positive for HER2 status.
Statistical analysis
To evaluate miR‐1 ISH status and clinicopathological factors, Student's t‐test or a cross‐table using the χ2‐test were used. Disease‐free and breast cancer‐specific survival curves were generated using the Kaplan–Meier method, and statistical significance was calculated using the log‐rank test. Univariate and multivariate analyses were evaluated by a proportional hazard model (Cox). In the present study, P < 0.05 and 0.05 ≤ P < 0.10 were considered significant and borderline significant, respectively.24 The statistical analyses were performed using JMP Pro version 9.02 (SAS Institute Inc., Cary, NC, USA).
Results
MicroRNA expression profile in stage IV breast carcinoma
We first compared expression profiles of 88‐cancer related miRNA between ER‐positive stage IV and stages I–III breast carcinoma tissues (n = 6 and n = 5, respectively) by miRNA PCR array to evaluate the characteristics of miRNA expression in stage IV breast carcinoma. When the expression ratio of a particular miRNA in the stage IV group compared to that in the stages I–III group was > 2.0 or < 0.5, we tentatively determined that the miRNA was predominantly expressed in either the stage IV or stages I–III group in this study.20 As shown in Figure 1a, two miRNA (2.2%), that is, miR‐1 (8.5‐fold) and miR‐200a (2.2‐fold), were predominantly expressed in the stage IV group, while one miRNA (1.1%; miR‐155 [0.47‐fold]) was predominantly expressed in the stages I–III group, among 88 miRNA examined. A great majority of miRNA (85 miRNA [96.6%]) had a similar expression level between the stage IV and stages I–III groups (ratio, 2.0–0.5).
Figure 1.
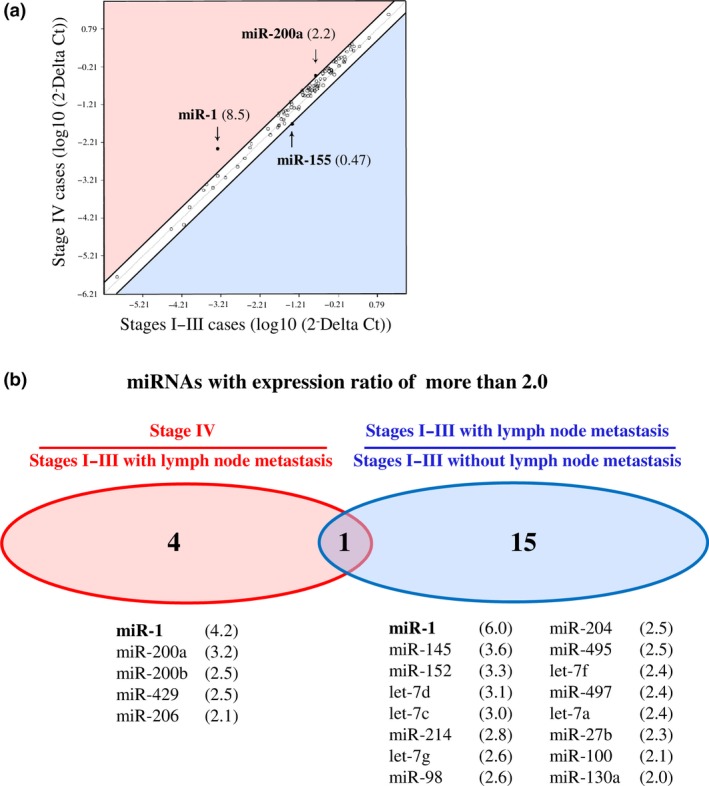
miRNA PCR array data containing 88‐cancer related miRNA in breast carcinoma. (a) Scatter plot analysis in ER‐positive stage IV breast carcinoma tissues (n = 6) comparing ER‐positive stages I–III cases (n = 5). miRNA with the expression ratio of more than 2.0 and < 0.5 were located outside diagonal lines, and indicated by arrows with their fold change in parenthesis. (b) Venn diagrams representing number of miRNA identified with expression ratio of more than in stage IV cases (n = 6) comparing stages I–III cases with lymph node metastasis (n = 3) and that in stages I–III with lymph node metastasis (n = 3) comparing stages I–III without lymph node metastasis (n = 2). All the stage IV cases represented lymph node metastasis in this study. The lower panels summarized their miRNA lists with the fold change in parenthesis. Only miR‐1 listed in both panels and described in bold.
All six stage IV cases examined showed lymph node metastasis, while three out of five stages I–III cases were positive for lymph node metastasis. When we classified the stages I–III group into two groups according to the lymph node status and further analyzed the miRNA expression profiles, 5 miRNA (miR‐1, miR‐200a, miR‐200b, miR‐429 and miR‐206) were predominantly expressed in the stage IV group compared to the stages I–III with lymph node metastasis group, and 16 miRNA were predominantly expressed in the stages I–III with lymph node metastasis group comparing those without lymph node metastasis (Fig. 1b). Interestingly, miR‐1 showed the highest ratio in all the analyses examined, suggesting possible involvement of miR‐1 in the distant and lymph node metastasis of breast carcinoma.
Gene expression profile of miR‐1‐positive stage IV breast carcinoma
To explore the functional significance of miR‐1 in the breast carcinoma, we next compared gene expression profiles of stage IV breast carcinoma according the miR‐1 status by microarray analysis. As shown in Figure 2a, a scatter plot revealed that 456 genes (1.1%) were predominantly expressed in the miR‐1‐positive group (Group A; miR‐1‐positive group/miR‐1‐negative group ratio > 2.0) and 889 genes (2.1%) were mainly expressed in the miR‐1‐negative group (Group B; ratio < 0.5), while a great majority of genes (42 470 genes [96.8%]) had a similar expression level in both groups (Group C; ratio 0.5–2.0).
Figure 2.
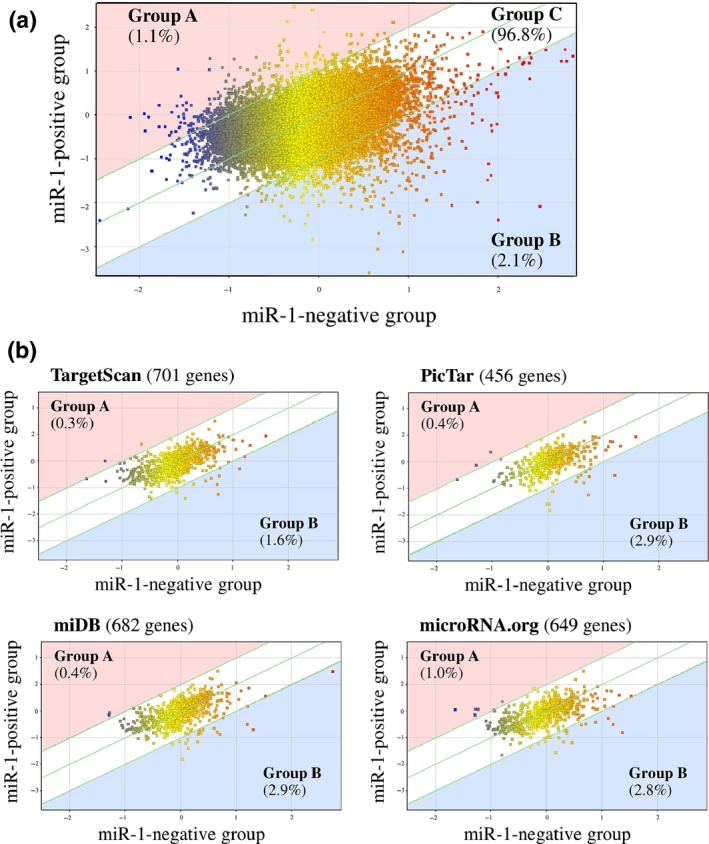
Scatter plot analysis of microarray data of stage IV breast carcinoma according to the miR‐1 status. (a) whole genes (n = 42 545) and (b) putative miR‐1 target genes predicted by using four target prediction tools. Genes are plotted on the logarithmic graph. Genes those were more than 2.0‐fold higher in the miR‐1‐positive group (n = 3) or miR‐1‐negative group (n = 2) are located outside of the diagonal line, and classified as group A or group B respectively. Genes ≦ 2.0‐fold changes are classified as group C.
Because miRNA is primarily involved in the negative regulation of the target gene expression, we then focused on the putative miR‐1 target genes using 4 different prediction tools (n = 1716 in total). As shown in Figure 2b, the percentage of the gene number tended to be decreased in group A (0.3–0.9%) and increased in group B (1.6–2.9%) compared to that in Figure 2a (1.1 and 2.1%, respectively). The gene list of group B in the putative miR‐1 target genes is summarized in Table S1.
We next performed gene ontology (GO) analysis in the group A (n = 456) and Group B (n = 889) genes. As shown in Table 1, we detected 16 GO terms that were significantly enriched in the Group A genes (Table 1a), and 40 GO terms significantly enriched in the Group B genes (Table 1b). Interestingly, 10 out of 16 (63%) GO terms identified in Group A included the words “extracellular” or “collagen,” and 25 out of 40 (63%) GO terms in group B were associated with “morphogenesis,” “development” or “differentiation” in this study.
Table 1.
List of GO terms significantly enriched in group (a) A gene and (b) B gene
| GO acession | GO term | P‐value |
|---|---|---|
| (a) | ||
| GO:0032963 | Collagen metabolic process | 2.3 × 10−5 |
| GO:0044259 | Multicellular organismal macromolecule | 2.5 × 10−5 |
| metabolic process | ||
| GO:0030574 | Collagen catabolic process | 2.5 × 10−5 |
| GO:0044236 | Multicellular organismal metabolic process | 2.5 × 10−5 |
| GO:0005615 | Extracellular space | 2.5 × 10−5 |
| GO:0044243 | Multicellular organismal catabolic process | 3.5 × 10−5 |
| GO:0031012 | Extracellular matrix | 8.2 × 10−4 |
| GO:0030198 | Extracellular matrix organization | 8.2 × 10−4 |
| GO:0043062 | Extracellular structure organization | 8.2 × 10−4 |
| GO:0005578 | Proteinaceous extracellular matri | 8.3 × 10−4 |
| GO:0022617 | Extracellular matrix disassembly | 8.6 × 10−4 |
| GO:0005576 | Extracellular region | 0.0021 |
| GO:0006955 | Immune response | 0.0021 |
| GO:0002376 | Immune system process | 0.0042 |
| GO:0030199 | Collagen fibril organization | 0.029 |
| GO:0006952 | Defense response | 0.035 |
| (b) | ||
| GO:0005576 | Extracellular region | 4.9 × 10−8 |
| GO:0060429 | Epithelium development | 1.7 × 10−5 |
| GO:0044421 | Extracellular region part | 5.1 × 10−5 |
| GO:0009888 | Tissue development | 6.7 × 10−5 |
| GO:0048856 | Anatomical structure development | 3.2 × 10−4 |
| GO:0098590 | Plasma membrane region | 3.2 × 10−4 |
| GO:0048731 | System development | 5.3 × 10−4 |
| GO:0005615 | Extracellular space | 0.0018 |
| GO:0009653 | Anatomical structure morphogenesis | 0.0019 |
| GO:0030855 | Epithelial cell differentiation | 0.005 |
| GO:0048468 | Cell development | 0.0052 |
| GO:0016323 | Basolateral plasma membrane | 0.0057 |
| GO:0030879 | Mammary gland development | 0.0057 |
| GO:0007399 | Nervous system development | 0.0057 |
| GO:0071466 | Cellular response to xenobiotic stimulus | 0.0057 |
| GO:0009410 | Response to xenobiotic stimulus | 0.0062 |
| GO:0044767 | Single‐organism developmental process | 0.0062 |
| GO:0048762 | Mesenchymal cell differentiation | 0.0062 |
| GO:0014033 | Neural crest cell differentiation | 0.0072 |
| GO:0032502 | Developmental process | 0.0072 |
| GO:0048732 | Gland development | 0.0072 |
| GO:0016324 | Apical plasma membrane | 0.0081 |
| GO:0014031 | Mesenchymal cell development | 0.0084 |
| GO:0007275 | Multicellular organismal development | 0.010 |
| GO:0022612 | Gland morphogenesis | 0.012 |
| GO:0000902 | Cell morphogenesis | 0.012 |
| GO:0006805 | Xenobiotic metabolic process | 0.012 |
| GO:0014032 | Neural crest cell development | 0.012 |
| GO:0045177 | Apical part of cell | 0.018 |
| GO:0021675 | Nerve development | 0.018 |
| GO:0048513 | Organ development | 0.025 |
| GO:0060444 | Branching involved in mammary gland duct morphogenesis | 0.026 |
| GO:0001763 | Morphogenesis of a branching structure | 0.045 |
| GO:0009887 | Organ morphogenesis | 0.045 |
| GO:0043230 | Extracellular organelle | 0.045 |
| GO:0060485 | Mesenchyme development | 0.045 |
| GO:0065010 | Extracellular membrane‐bounded organelle | 0.045 |
| GO:0070062 | Extracellular vesicular exosome | 0.045 |
| GO:1903561 | Extracellular vesicle | 0.045 |
| GO:0002009 | Morphogenesis of an epithelium | 0.048 |
(a) Bold indicates GO term including “extracellular” and “collagen.”
(b) Bold indicates GO term related to “morphogenesis” and “development” and “differentiation.”
miR‐1 localization in breast carcinoma
When we next performed ISH for miR‐1 in breast carcinoma, it was localized in the cytoplasm of carcinoma cells (Fig. 3a). The number of miR‐1 positive breast carcinoma was 32 out of the 163 (20%) cases examined (Fig. 3b). miR‐1 signal was weakly observed in some non‐neoplastic mammary glands, but was negative in stroma (Fig. 3c). The miR‐1 signal was strongly detected in the skeletal muscle tissue as a positive control (Fig. 3d, left panel), but not when we used a scrambled negative control probe instead of the miR‐1 specific probe (Fig. 3d, right panel).
Figure 3.
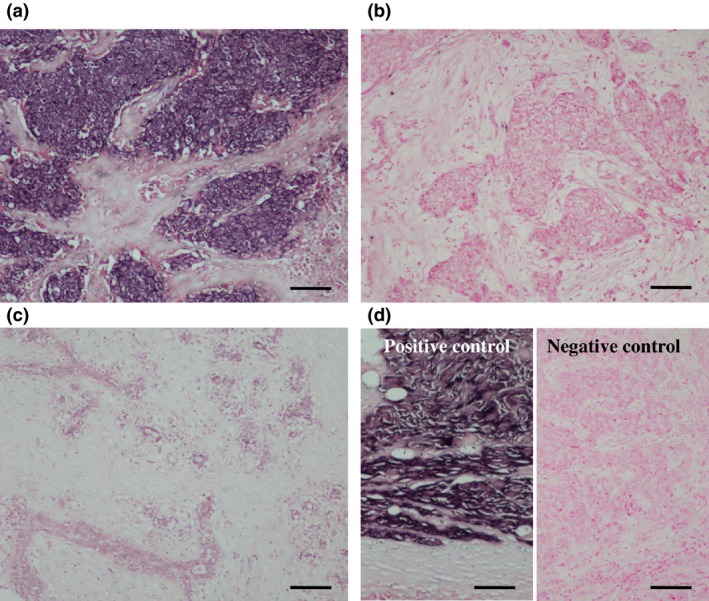
In situ hybridization (ISH) for miR‐1 in breast carcinoma. (a) miR‐1 was localized in the cytoplasm of carcinoma cells. (b) miR‐1‐negative breast carcinoma. (c) Hybridization signal for miR‐1 was focally and weakly detected in the morphologically normal mammary epithelium. (d) Positive control (skeletal muscle tissue; left panel) and negative control (scrambled negative control probe in breast carcinoma; right panel) for miR‐1 ISH. Bar = 100 μm, respectively.
Association between miR‐1 ISH status and various clinicopathological parameters in breast carcinoma is summarized in Table 2. miR‐1 status was significantly associated with stage (P < 0.0001), pathological T factor (pT) (P < 0.0001), lymph node metastasis (P = 0.0001), distant metastasis (P < 0.0001), histological grade (P < 0.0001) and Ki‐67 LI (P < 0.0001), and inversely correlated with ER status (P = 0.0098) and PR status (P = 0.0049). In contrast, no significant association was detected between miR‐1 and patients' age, menopausal status and HER2 status. The positive association between miR‐1 status and stage, pT or distant metastasis was significant regardless of the ER status of these cases (Table S2). miR‐1 status was also significantly associated with stage, pT, lymph node metastasis, histological grade, ER status, PR status and Ki‐67 LI in the stages I–III cases (Table S3).
Table 2.
Association between miR‐1 ISH status and clinicopathological parameters in 163 breast carcinomas
| Variable | miR‐1 status | P‐value | |
|---|---|---|---|
| + (n = 32) | − (n = 131) | ||
| Agea(years) | 56.5 ± 2.1 | 56.3 ± 1.1 | 0.93 |
| Menopausal status | |||
| Premenopausal | 12 | 49 | |
| Postmenopausal | 20 | 82 | 0.76 |
| Stage | |||
| I | 3 | 65 | |
| II | 7 | 41 | |
| III | 8 | 17 | |
| IV | 14 | 8 | <0.0001 |
| Pathological T factor (pT) | |||
| pT1 | 5 | 80 | |
| pT2‐4 | 27 | 51 | <0.0001 |
| Lymph node metastasis | |||
| Positive | 24 | 49 | |
| Negative | 8 | 82 | 0.0001 |
| Distant metastasis | |||
| Positive | 14 | 8 | |
| Negative | 18 | 123 | <0.0001 |
| Histological grade | |||
| 1 (well) | 5 | 42 | |
| 2 (moderate) | 10 | 60 | |
| 3 (poor) | 17 | 29 | <0.0001 |
| ER status | |||
| Positive | 19 | 106 | |
| Negative | 13 | 25 | 0.0098 |
| PR status | |||
| Positive | 14 | 92 | |
| Negative | 18 | 39 | 0.0049 |
| HER2 status | |||
| Positive | 6 | 21 | |
| Negative | 26 | 110 | 0.71 |
| Ki‐67 LIa (%) | 26.8 ± 4.0 | 14.4 ± 1.1 | <0.0001 |
Data are presented as mean ± SEM. All other values represent the number of cases P‐values < 0.05 were considered significant, and are presented in bold.
Association between miR‐1 status and metastatic sites in stage IV cases is shown in Table 3. miR‐1‐positive breast carcinoma was marginally (P = 0.076) associated with the lung metastasis in this study.
Table 3.
miR‐1 status of breast carcinoma tissue according to metastatic sites in stage IV cases (n = 22)
| Metastatic site | miR‐1 status of breast carcinoma tissue | P‐value | |
|---|---|---|---|
| + (n = 14) | − (n = 8) | ||
| Lung | |||
| + | 9 | 2 | |
| − | 5 | 6 | 0.076 |
| Bone | |||
| + | 6 | 6 | |
| − | 8 | 2 | 0.14 |
| Liver | |||
| + | 2 | 1 | |
| − | 12 | 7 | 0.91 |
Data represent the number of cases. 0.05 ≤ P < 0.10 was considered borderline significant and is listed in italic type.
Association between miR‐1 status and clinical outcome of breast cancer patients
As demonstrated in Figure 4a, miR‐1 status was significantly associated with an increased incidence of recurrence in stages I–III breast cancer patients (n = 141; P < 0.0001 by log‐rank test). A significant association was also detected between miR‐1 status and adverse clinical outcome of these patients (P < 0.0001 by log‐rank test [Fig. 4b]). Similar tendency was detected regardless of the sample‐collection periods (1995–1999 [n = 42] and 2007–2008 [n = 99]; Fig. S1a–d). Association between miR‐1 status and worse clinical outcome of the patients was also detected in the cases with lymph node metastasis (n = 56; P < 0.0001 for disease‐free survival [Fig. 4c] and P = 0.0007 for breast cancer‐specific survival), pT2‐4 cases (n = 57; P < 0.0001 [Fig. 4d] and P = 0.0003, respectively), patients who received adjuvant endocrine therapy (n = 111; P = 0.0029 [Fig. 4e] and P < 0.0001, respectively) and patients who received adjuvant chemotherapy (n = 77; P < 0.0001 [Fig. 4f] and P = 0.0009, respectively).
Figure 4.
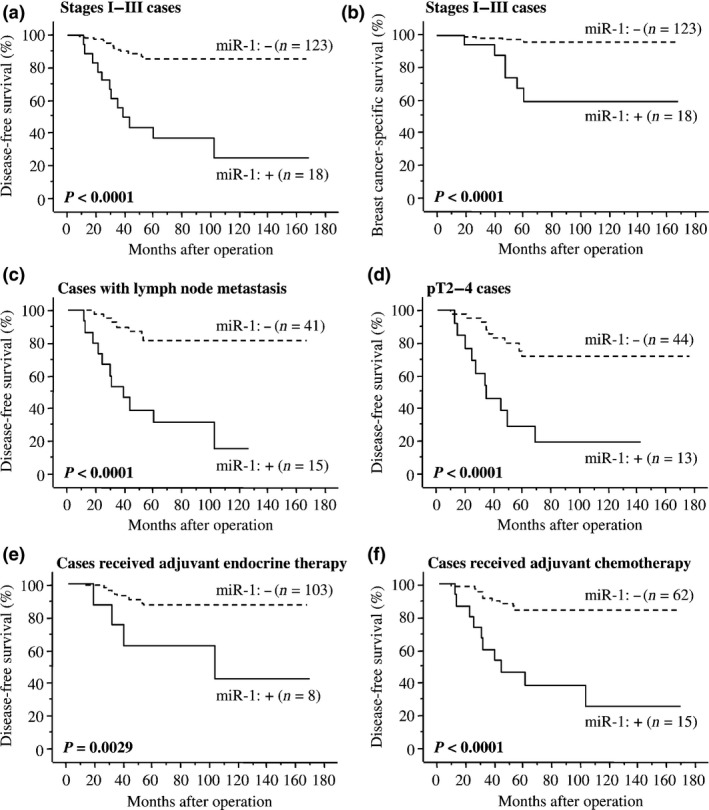
Disease‐free (a,c–f) and breast cancer‐specific survival (b) of stages I–III breast cancer patients according to miR‐1 ISH status using the Kaplan–Meier method. The solid line shows miR‐1‐positive cases and the dashed line shows miR‐1‐negative cases. Statistical analysis was performed using the log‐rank test. P‐values < 0.05 were considered significant and are shown in bold.
Univariate analysis of disease‐free survival by Cox (Table 4), miR‐1 ISH status, Ki‐67 LI, pT, adjuvant endocrine therapy, PR status and lymph node metastasis were revealed significant prognostic parameters for disease‐free survival in the 141 stages I–III breast cancer patients, and ER status and histological grade were also detected as the borderline significance. Subsequent multivariate analysis demonstrated that pT (P = 0.0098) and miR‐1 status (P = 0.017) were independent prognostic factors. As shown in Table 5, univariate analyses for breast cancer‐specific survival revealed miR‐1 status, Ki67‐LI, PR status, histological grade, pT, ER status, adjuvant endocrine therapy and lymph node metastasis as significant prognostic variables in these patients, and following multivariate analysis it turned out that only miR‐1 (P = 0.032) was an independent parameter of these patients in this study.
Table 4.
Univariate and multivariate analyses of disease‐free survival in stages I–III breast cancer patients (n = 141)
| Variable | Univariate | Mutivariate | ||
|---|---|---|---|---|
| P‐value | Relative risk (95% CI) | P‐value | Relative risk (95% CI) | |
| miR‐1 status (positive/negative) | <0.0001 * | 6.90 (3.25–14.64) | 0.017 | 3.50 (1.26–9.78) |
| Ki‐67 LI ** (1–98%) | <0.0001 * | 1.05 (1.01–1.09) | 0.12 | 1.03 (0.99–1.06) |
| pT (pT1/pT2‐4) | 0.0001 * | 0.19 (0.08–0.44) | 0.0098 | 0.22 (0.07–0.70) |
| Adjvant endocrine therapy (received/not received) | 0.0009 * | 0.27 (0.12–0.59) | 0.43 | 0.63 (0.20–1.98) |
| PR status (positive/negative) | 0.0065 * | 0.35 (0.17–0.75) | 0.64 | 0.34 (0.11–1.07) |
| Lymph node metastasis (positive/negative) | 0.0066 * | 2.92 (1.35–6.33) | 0.73 | 0.83 (0.28–2.46) |
| ER status (positive/negative) | 0.079 * | 0.50 (0.23–1.08) | 0.37 | 1.84 (0.49–6.86) |
| Histological grade (1,2/3) | 0.091 * | 0.52 (0.24–1.11) | 0.26 | 2.04 (0.59–7.06) |
| Ajuvant chemotherapy (received/not received) | 0.15 | 1.84 (0.80–4.02) | ||
| HER2 status (positive/negative) | 0.33 | 1.81 (0.55–6.00) | ||
Statistical analysis was evaluated by a proportional hazard model (Cox). P < 0.05 and 0.05 ≤ P < 0.10 were considered significant and borderline significant, and are listed in bold and italic, respectively. *Significant (P < 0.05) and borderline‐significant (0.05 ≤ P < 0.10) values were examined in the multivariate analyses in this study. **Data were evaluated as continuous variables, and all other data were evaluated as dichotomized variables. 95% CI, 95% confidence interval.
Table 5.
Univariate and multivariate analyses of breast cancer‐specific survival in stages I–III breast cancer patients (n = 141)
| Variable | Univariate | Mutivariate | ||
|---|---|---|---|---|
| P‐value | Relative risk (95% CI) | P‐value | Relative risk (95% CI) | |
| miR‐1 status (positive/negative) | 0.0001 * | 11.74 (3.31–41.69) | 0.032 | 6.72 (1.18–38.37) |
| Ki‐67 LI** (1–98%) | 0.0002 * | 1.07 (1.04–1.11) | 0.43 | 1.02 (0.97–1.01) |
| PR status (positive/negative) | 0.0069 * | 0.58 (0.01–0.46) | 0.055 | 0.08 (0.01–1.06) |
| Histological grade (1,2/3) | 0.0083 * | 0.16 (0.04–0.63) | 0.47 | 2.24 (0.25–20.01) |
| pT (pT1/pT2‐4) | 0.011 * | 0.07 (0.01–0.54) | 0.13 | 0.13 (0.01–1.86) |
| ER status (positive/negative) | 0.013 * | 0.2 (0.06–0.071) | 0.98 | 1.02 (0.13–7.97) |
| Adjuvant endcrine therapy (received/not received) | 0.014 * | 0.21 (0.06–0.073) | 0.88 | 1.16 (0.18–7.49) |
| Lymph node metastasis (positive/negative) | 0.022 * | 6.12 (1.30–28.84) | 0.79 | 0.75 (0.09–6.21) |
| Ajuvant chemotherapy (received/not received) | 0.15 | 3.11 (0.66–14.63) | ||
| HER2 status (positive/negative) | 0.52 | 0.86 (0.18–4.04) | ||
Statistical analysis was evaluated by a proportional hazard model (Cox). P < 0.05 and 0.05 ≤ P < 0.10 were considered significant and borderline significant, and are listed in bold and italic respectively. *Significant (P < 0.05) and borderline‐significant (0.05 ≤ P < 0.10) values were examined in the multivariate analyses in this study. **Data were evaluated as continuous variables, and all other data were evaluated as dichotomized variables. 95% CI, 95% confidence interval.
Discussion
To the best of our knowledge, this is the first report to demonstrate expression profiles of miRNA in stage IV breast carcinoma tissues. The PCR array data revealed five miRNA that are potentially associated with distant metastasis in ER‐positive breast cancer patients (Fig. 1b). Among these, Dykxhoorn et al.25 and Le et al.26 report that miR‐200a, miR‐200b and miR‐429, belonging to the miR‐200 family, regulate mesenchymal‐to‐epithelial transition (MET) and promote breast cancer cell metastasis. In the present study, miR‐1 showed the highest expression ratio in stage IV cases compared to stages I–III cases (8.5‐fold). Moreover, it was also the highest expression ratio both in stage IV cases in comparison with stages I–III cases with lymph node metastasis (4.2‐fold) and in stages I–III cases with lymph node metastasis compared to those without lymph node metastasis (6.0‐fold). These findings suggest that miR‐1 is the most pronouncedly linked to distant and lymph node metastasis in breast carcinoma. However, to the best of our knowledge, miR‐1 expression has not been examined in breast carcinoma tissues, and its clinicopathological significance has remained unknown.
In the present ISH analysis, miR‐1 expression was detected in 20% of breast carcinoma cases. Previous studies have demonstrated that miR‐1 expression is downregulated in thyroid carcinomas,27 and head and neck squamous cell carcinomas,28 and it is considered to be associated with tumor suppression in some cancers. In contrast, miR‐1 is specifically overexpressed in the multiple myeloma in comparison with normal plasma cells,29 and Liu et al.30 show that serum miR‐1 was markedly upregulated in gastric cancer patients compared to controls. Moreover, Chan et al.31 demonstrate that serum miR‐1 level was significantly higher in breast cancer patients than that in healthy controls. Considering that miR‐1 expression was negligible in morphologically normal mammary glands in the present ISH analysis, it is suggested that miR‐1 is abnormally overexpressed and plays important roles in a subset of breast carcinomas. The mechanism of miR‐1 overexpression is unclear in breast carcinoma. However, considering that miR‐1 was specifically overexpressed in the multiple myeloma with t(14;16),29 it may be partly caused by chromosomal aberration. Further investigations are required.
In this ISH study, miR‐1 expression was significantly associated with distant metastasis in the breast carcinomas regardless of the ER status, which is in good agreement with our present miRNA PCR array data. In addition, our results showed that miR‐1 expression tended to be associated with lung metastasis in the stage IV cases. Lung metastasis is frequently detected in the triple negative breast carcinoma compared to other subtypes,32 and some cascades selectively involved in the lung metastasis have been reported by Knowles et al.33 Moreover, in this study, miR‐1 expression was significantly associated with stage, pT, lymph node metastasis, histological grade and Ki‐67 LI in the breast carcinomas. Biological function of miR‐1 remains unclear in breast carcinoma. However, serum miR‐1 level was correlated to stage and marginally associated with liver metastasis in gastric carcinoma patients,30, 34 and miR‐1 was associated with cell proliferation of acute myeloid leukemia.35 Therefore, it is suggested that miR‐1 is overexpressed in an aggressive phenotype of breast carcinoma and is involved in a variety of functions, such as the growth and metastatic processes.
In the present study, miR‐1 status was significantly associated with recurrence and worse prognosis in breast cancer patients, and a similar tendency was also detected in the patients who received endocrine therapy and/or chemotherapy. Moreover, results of multivariate analyses demonstrated that miR‐1 ISH status turned out to be an independent prognostic factor for both disease‐free and breast cancer‐specific survival. Very recently, Huang et al.34 reported that the higher level of serum miR‐1 was significantly correlated with a worse response rate to the first‐line chemotherapy in the gastric carcinoma, which is consistent with the results of our present study. No information is available about the effects of endocrine therapy on miR‐1. However, because Masuda et al.21 did not find significant change in miR‐1 expression with estrogen treatment in MCF‐7 breast carcinoma cells using the same miRNA PCR array as ours, miR‐1 functions might not be influenced by estrogen actions or endocrine therapy in breast carcinoma.
The results of our microarray analysis revealed that GO terms associated with “morphogenesis,” “development” and “differentiation” were frequently decreased in the miR‐1‐positive breast carcinoma. miR‐1 is a muscle‐specific miRNA, and plays a role in myogenesis and muscle regeneration.36, 37 miR‐1 regulates embryonic stem cells differentiation to cardiac lineage, and Huang et al.38 demonstrated that miR‐1 overexpression in mesenchymal stem cells promoted various cardiomyocyte markers, including a lineage selector gene GATA4, which has been reported as a worse prognostic factor in breast cancer.39 Therefore, abnormal expression of miR‐1 might cause alternative lineages and/or dedifferentiation in the breast carcinoma. Our present results also showed that GO terms associated with “extracellular” and “collagen” were frequently enriched in the miR‐1‐positive breast carcinoma, and Liu et al.40 reported that miR‐1 has roles in regulating epithelial–mesenchymal transition and mesenchymal differentiation. Because miR‐1 expression is associated with a variety of biological functions as described in this section through regulating the expression of a multitude of target genes, residual carcinoma cells following surgical treatment in miR‐1‐positive breast carcinomas could still have the potential to rapidly recur despite adjuvant therapy. However, the number of cases examined was limited (n = 163) and the mean follow‐up period was 72 months in this study; replication studies with a larger sample set with a longer follow up period are needed to confirm the clinical significance of miR‐1 in breast carcinoma. In addition, further examinations are required to clarify the molecular functions of miR‐1 in human breast carcinoma.
In summary, we examined the expression profile of miRNA in ER‐positive stage IV breast carcinoma tissues by miRNA PCR array, and demonstrated that miR‐1 expression was most closely associated with the distant metastasis of breast carcinoma. A subsequent ISH analysis revealed that miR‐1 was localized in 20% of breast cancer cases, and miR‐1 status was significantly associated with stage, pT, lymph node metastasis, distant metastasis, histological grade, ER status, PR status and Ki‐67 LI. Moreover, multivariate analysis demonstrated that the miR‐1 status was an independent worse prognostic factor for both disease‐free and breast cancer‐specific survival. These results suggest that miR‐1 plays important roles in the progression of breast carcinoma, and miR‐1 status is a potent prognostic factor in breast cancer patients.
Disclosure Statement
The authors have no conflict of interest to declare.
Supporting information
Fig. S1. Association between miR‐1 status and disease‐free (a,c) and breast cancer‐specific survival (b,d) of the breast cancer patients according to the sample collection periods (a,b: 1995–1999, and c,d: 2007–2008). The survival curve was generated using the Kaplan–Meier method. The solid line shows miR‐1‐positive cases and the dashed line shows miR‐1‐negative cases. Statistical analysis was performed using the log‐rank test. P‐values <0.05 were considered significant and are shown in bold. The P‐value is not estimated (NE) in Figure S1b, because no patients had died in the miR‐1‐negative group.
Table S1. List of miR‐1 target genes classified into group B.
Table S2. Association between miR‐1 ISH status and clinicopathological parameters according to the ER status in 163 breast carcinomas.
Table S3. Association between miR‐1 ISH status and clinicopathological parameters according to the distant metastasis in 163 breast carcinomas.
Acknowledgments
This work was partly supported by a Grant‐in‐Aid for Scientific Research (25460410 and 26860229) from the Japanese Ministry of Education, Culture, Sports, Science and Technology.
Cancer Sci (2015)
Funding Information
JSPS KAKENHI Grant number 25460410 and 26860229.
References
- 1. Giordano SH, Buzdar AU, Smith TL, Kau SW, Yang Y, Hortobagyi GN. Is breast cancer survival improving? Cancer 2004; 100: 44–52. [DOI] [PubMed] [Google Scholar]
- 2. Khodari W, Sedrati A, Naisse I, Bosc R, Belkacemi Y, AROME . Impact of loco‐regional treatment on metastatic breast cancer outcome: a review. Crit Rev Oncol Hematol 2013; 87: 69–79. [DOI] [PubMed] [Google Scholar]
- 3. Gerratana L, Fanotto V, Bonotto M et al Pattern of metastasis and outcome in patients with breast cancer. Clin Exp Metastasis 2015; 32: 125–33. [DOI] [PubMed] [Google Scholar]
- 4. Khanfir A, Lahiani F, Bouzguenda R, Ayedi I, Daoud J, Frikha M. Prognostic factors and survival in metastatic breast cancer: a single institution experience. Rep Pract Oncol Radiother 2013; 18: 127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ambros V. microRNAs: tiny regulators with great potential. Cell 2001; 107: 823–6. [DOI] [PubMed] [Google Scholar]
- 6. Leung AK, Sharp PA. MicroRNAs: a safeguard against turmoil? Cell 2007; 130: 581–5. [DOI] [PubMed] [Google Scholar]
- 7. Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 2006; 94: 776–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell 2009; 136: 586–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iorio MV, Ferracin M, Liu CG et al MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65: 7065–70. [DOI] [PubMed] [Google Scholar]
- 10. Yan LX, Huang XF, Shao Q et al MicroRNA miR‐21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 2008; 14: 2348–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo L, Zhao Y, Yang S, Cai M, Wu Q, Chen F. Genome‐wide screen for aberrantly expressed miRNAs reveals miRNA profile signature in breast cancer. Mol Biol Rep 2013; 40: 2175–86. [DOI] [PubMed] [Google Scholar]
- 12. Ma L, Teruya‐Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA‐10b in breast cancer. Nature 2007; 449: 682–8. [DOI] [PubMed] [Google Scholar]
- 13. Huang TH, Wu F, Loeb GB et al Up‐regulation of miR‐21 by HER2/neu signaling promotes cell invasion. J Biol Chem 2009; 284: 18515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nohata N, Hanazawa T, Enokida H, Seki N. microRNA‐1/133a and microRNA‐206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget 2012; 3: 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han C, Yu Z, Duan Z, Kan Q. Role of microRNA‐1 in human cancer and its therapeutic potentials. Biomed Res Int 2014; 2014: 428371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu CC, Gan P, Zhang RY, Xue JX, Ran LK. Identification of prostate cancer LncRNAs by RNA‐Seq. Asian Pac J Cancer Prev 2014; 15: 9439–44. [DOI] [PubMed] [Google Scholar]
- 17. Itoh M, Kim CH, Palardy G et al Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell 2003; 4: 67–82. [DOI] [PubMed] [Google Scholar]
- 18. Miele L, Osborne B. Arbiter of differentiation and death: notch signaling meets apoptosis. J Cell Physiol 1999; 181: 393–409. [DOI] [PubMed] [Google Scholar]
- 19. Wang Z, Li Y, Sarkar FH. Notch signaling proteins: legitimate targets for cancer therapy. Curr Protein Pept Sci 2010; 11: 398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suzuki S, Takagi K, Miki Y et al Nucleobindin 2 in human breast carcinoma as a potent prognostic factor. Cancer Sci 2012; 103: 136–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Masuda M, Miki Y, Hata S et al An induction of microRNA, miR‐7 through estrogen treatment in breast carcinoma. J Transl Med 2012; 10: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schneider M, Andersen DC, Silahtaroglu A et al Cell‐specific detection of microRNA expression during cardiomyogenesis by combined in situ hybridization and immunohistochemistry. J Mol Histol 2011; 42: 289–99. [DOI] [PubMed] [Google Scholar]
- 23. Hammond ME, Hayes DF, Dowsett M et al American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch Pathol Lab Med 2010; 134: e48–72. [DOI] [PubMed] [Google Scholar]
- 24. Sato‐Tadano A, Suzuki T, Amari M et al Hexokinase II in breast carcinoma: a potent prognostic factor associated with hypoxia‐inducible f factor‐1α and Ki‐67. Cancer Sci 2013; 104: 1380–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dykxhoorn DM, Wu Y, Xie H et al miR‐200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS ONE 2009; 4: e7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Le MT, Hamar P, Guo C et al miR‐200‐containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest 2014; 124: 5109–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leone V, D'Angelo D, Rubio I et al miR‐1 is a tumor suppressor in thyroid carcinogenesis targeting CCND2, CXCR4, and SDF‐1alpha. J Clin Endocrinol Metab 2011; 96: E1388–98. [DOI] [PubMed] [Google Scholar]
- 28. Nohata N, Sone Y, Hanazawa T et al miR‐1 as a tumor suppressive microRNA targeting TAGLN2 in head and neck squamous cell carcinoma. Oncotarget 2011; 2: 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gutiérrez NC, Sarasquete ME, Misiewicz‐Krzeminska I et al Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling. Leukemia 2010; 24: 629–37. [DOI] [PubMed] [Google Scholar]
- 30. Liu R, Zhang C, Hu Z et al A five‐microRNA signature identified from genome‐wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer 2011; 47: 784–91. [DOI] [PubMed] [Google Scholar]
- 31. Chan M, Liaw CS, Ji SM et al Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res 2014; 19: 4477–87. [DOI] [PubMed] [Google Scholar]
- 32. Chikarmane SA, Tirumani SH, Howard SA, Jagannathan JP, DiPiro PJ. Metastatic patterns of breast cancer subtypes: what radiologists should know in the era of personalized cancer medicine. Clin Radiol 2015; 70: 1–10. [DOI] [PubMed] [Google Scholar]
- 33. Knowles LM, Gurski LA, Engel C, Gnarra JR, Maranchie JK, Pilch J. Integrin αvβ3 and fibronectin upregulate Slug in cancer cells to promote clot invasion and metastasis. Cancer Res 2013; 73: 6175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang D, Wang H, Liu R et al miRNA27a is a biomarker for predicting chemosensitivity and prognosis in metastatic or recurrent gastric cancer. J Cell Biochem 2014; 115: 549–56. [DOI] [PubMed] [Google Scholar]
- 35. Gómez‐Benito M, Conchillo A, García MA et al EVI1 controls proliferation in acute myeloid leukaemia through modulation of miR‐1‐2. Br J Cancer 2010; 103: 1292–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen JF, Mandel EM, Thomson JM et al The role of microRNA‐1 and microRNA‐133 in skeletal muscle proliferation and differentiation. Nat Genet 2006; 38: 228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kukreti H, Amuthavalli K, Harikumar A et al Muscle‐specific microRNA1 (miR1) targets heat shock protein 70 (HSP70) during dexamethasone‐mediated atrophy. J Biol Ch 2013; 288: 6663–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang F, Tang L, Fang ZF, Hu XQ, Pan JY, Zhou SH. miR‐1‐mediated induction of cardiogenesis in mesenchymal stem cells via downregulation of Hes‐1. Biomed Res Int 2013; 2013: 216286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takagi K, Moriguchi T, Miki Y et al GATA4 immunolocalization in breast carcinoma as a potent prognostic predictor. Cancer Sci 2014; 105: 600–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu YN, Yin JJ, Abou‐Kheir W et al MiR‐1 and miR‐200 inhibit EMT via Slug‐dependent and tumorigenesis via Slug‐independent mechanisms. Oncogene 2013; 32: 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Association between miR‐1 status and disease‐free (a,c) and breast cancer‐specific survival (b,d) of the breast cancer patients according to the sample collection periods (a,b: 1995–1999, and c,d: 2007–2008). The survival curve was generated using the Kaplan–Meier method. The solid line shows miR‐1‐positive cases and the dashed line shows miR‐1‐negative cases. Statistical analysis was performed using the log‐rank test. P‐values <0.05 were considered significant and are shown in bold. The P‐value is not estimated (NE) in Figure S1b, because no patients had died in the miR‐1‐negative group.
Table S1. List of miR‐1 target genes classified into group B.
Table S2. Association between miR‐1 ISH status and clinicopathological parameters according to the ER status in 163 breast carcinomas.
Table S3. Association between miR‐1 ISH status and clinicopathological parameters according to the distant metastasis in 163 breast carcinomas.


