Abstract
The GINS complex associates with cell division cycle (Cdc) protein 45 and mini‐chromosome maintenance (Mcm) proteins 2–7 to form the Cdc45–Mcm–GINS (CMG) complex, which is essential for DNA duplication. One member of the GINS complex is Psf3. We previously found that increased Psf3 expression was strongly associated with poor survival in lung adenocarcinoma. Here, we investigated the role of Psf3 expression in non‐small‐cell lung cancer (NSCLC). We verified Psf3 expression in human NSCLC tissues (180 patients) and cell lines. Immunohistochemical analysis revealed that the overexpression of Psf3 was significantly associated with vessel invasion (P = 0.016), lymphatic invasion (P = 0.002), and pleural invasion (P = 0.036). The overall survival rate in patients with Psf3 overexpression was significantly lower than that in patients without Psf3 overexpression (P = 0.006). Multivariate survival analysis revealed Psf3 expression to be an independent risk factor for an unfavorable outcome (P = 0.049). A proximal ligation assay showed interactions between Psf3 and other CMG components (such as Mcm2 and Cdc45) in both NSCLC specimens and cell lines, indicating that Psf3 acted as the CMG complex, which could lead to excessive proliferation. Knockdown of Psf3 inhibited the proliferation of both cell lines by delaying the S phase, which revealed that Psf3 played an important role in cancer proliferation. Thus, Psf3 acted as the CMG complex, promoting excessive proliferation. These results suggest that Psf3 inhibition might be a therapeutic target for NSCLC with Psf3 overexpression.
Keywords: Cell line, GINS3 human protein, non‐small‐cell lung carcinoma, pathology, RNA interference
Psf3 is a member of the GINS (Go‐Ichi‐Ni‐San) complex, a heterotetrameric assembly of SLD5, Psf1, Psf2, and Psf3. In eukaryotic cells, the GINS complex associates with cell division cycle (Cdc) protein 45 and mini‐chromosome maintenance (Mcm) proteins 2–7 to form the CMG (Cdc45–Mcm–GINS) complex, which constitutes the DNA replicative helicase, and is essential for both the initiation and progression of DNA duplication.1, 2, 3 The CMG complex unwinds double‐stranded DNA at replication forks, and also appears to interact with and stimulate polymerase activities.4, 5, 6
In addition to their role in DNA replication, several reports have suggested that GINS components are required for the acute proliferation of cells, especially immature cells such as stem cells and progenitor cells.7, 8, 9 For instance, Psf1 expression is essential for early embryogenesis, maintenance of immature hematopoietic cell pool size, and acute bone marrow regeneration in mice. It was observed that loss of Psf1 lead to embryonic lethality around the implantation stage, which was caused by the inability of cells in the inner cell mass to proliferate.7 Moreover, GINS components not only play a role in immature cells, but also play a role in cancer cells. Previous studies have suggested that GINS components are unregulated in cancer cells (such as in colon carcinoma, breast carcinoma, intrahepatic cholangiocarcinoma, and melanoma), and some GINS components may be useful for detecting cancer stem cells.10, 11, 12, 13, 14, 15
In our previous report, immunohistochemical observation revealed that increased Psf3 expression was associated with poor prognosis in primary lung adenocarcinoma.16 Psf3 expression was significantly correlated with the Ki67 (MIB‐1) expression index, which was used as an indicator of cell proliferation. Importantly, however, staining for Psf3 showed a clustered pattern, which differed from the pattern associated with Ki67 in the same serial section. These findings suggested that the aberrant accumulation of Psf3 was related to the malignant behavior of tumors in a cell cycle‐independent manner, at least in the case of lung adenocarcinoma. However, the significance of abnormal Psf3 overexpression in cancer cells remains unclear.
In this study, we showed that Psf3 expression was upregulated in human non‐small‐cell lung cancer (NSCLC) tissue and cell lines. Additionally, we examined the interactions between Psf3 and the other CMG components in lung cancer specimens and cell lines, and assessed whether Psf3 expression in cancer cells functioned as the CMG complex. Moreover, we used RNAi methodology to knock down the expression of Psf3 and thereby determine the significance of abnormal Psf3 expression in NSCLC.
Materials and Methods
Materials
All resected specimens were obtained from patients who underwent surgery for NSCLC at Hyogo Cancer Center (Akashi, Japan) in 2005. The samples were fixed in 10% (v/v) neutral buffered formalin and then embedded in paraffin. The Hyogo Cancer Center institutional review boards approved the study and each participant provided informed consent.
Immunohistochemistry
Formalin‐fixed, paraffin‐embedded specimens were cut into 5‐mm‐thick slices at the maximal area of tumor mass. The sections were deparaffinized with xylene and rehydrated with ethanol. Mouse anti‐human Psf3 mAbs (1:500; GeneStem, Osaka, Japan), rabbit anti‐human Mcm2 (1:2000; Universal Biological, Cambridge, UK), and rabbit anti‐human Cdc45 (1:100; Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used as the primary antibodies for the detection of Psf3, Mcm2, and Cdc45, respectively. The Histofine Simple Stain MAX PO kit (Nichirei Biosciences, Tokyo, Japan) was used for endogenous peroxidase blocking, treatment with a secondary antibody against anti‐mouse and anti‐rabbit immunoglobulin antibodies, and the visualization of HRP. Hematoxylin staining was used as the counterstain. Photographs of the stained sections were obtained using a camera mounted on a Keyence BZ‐8000 digital microscope (Keyence, Osaka, Japan).
A single representative tissue section from each tumor was surveyed microscopically at 200× magnification in at least two or three fields having high Psf3 intensity in positive tumor cells. The percentage of nuclei stained for Psf3 in cancer cells was evaluated by two investigators (Y.S. and D.H.) who were blinded to clinical data. The percentage of positively stained tumor cells were categorized as 0–24%, 25–49%, 50–74%, or 75–100% of cells stained.
Cell culture and RNA interference
The A549 and EBC1 cell lines were obtained from the Cell Resource Center for Biomedical Research, Institute of Development, Aging and Cancer, Tohoku University (Sendai, Japan). The PC14 and RFRF‐LCSQ1 cell lines were obtained from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan). These cell lines were maintained in RPMI‐1640 medium supplemented with penicillin (100 U/mL), streptomycin (100 U/mL), and 10% bovine calf serum (at 37°C, 5% CO2). RNA interference was carried out with commercially available siRNA for Psf3 (#HSS127764; Invitrogen, Carlsbad, CA, USA) and a non‐silencing control siRNA (Invitrogen) according to the manufacturer's instructions. Briefly, 6 pmol transfection reagent was suspended in 1 μL Lipofectamine RNAiMAX (Invitrogen) and 100 μL Opti‐MEM (Invitrogen), finally containing 10 μM siRNA. After a 15‐min incubation at room temperature, the mixture was added to cell lines (60‐mm round dish with 4 mL culture medium containing 10% FBS and antibiotics mentioned above) and grown to 80% confluence; the final concentration of the siRNA was 10 nM. After 48 h (at 37°C, 5% CO2), these cells were examined to assay for gene knockdown.
In situ proximal ligation assay analyses
Proximal ligation assay (PLA) is a novel method that can enable the detection of dual protein interactions.17, 18 Paraffin‐embedded resected specimens were deparaffinized according to the same method described under “Immunohistochemistry”. In cases with cultured cells, cells were grown on the chamber slide as described as above and fixed with 4% paraformaldehyde in PBS for 20 min. Fixed cells were permeabilized with 0.1% Triton X‐100 in PBS for 3 min. Proximal ligation assay was carried out using the Duolink in situ Starter kit (Olink Bioscience, Uppsala, Sweden) following the manufacturer's protocol. Samples were blocked in a humidified chamber. Mouse anti‐Psf3 (1:500; GeneStem) and rabbit Mcm2 (1:2000; Universal Biological) primary antibodies were diluted and co‐incubated overnight at 4°C. The antibodies used for another analysis included mouse anti‐Psf3 (1:500; GeneStem) and rabbit Cdc45 (1:100; Santa Cruz Biotechnology). Next, PLA MINUS and PLA PLUS secondary probes (Olink Bioscience) conjugated with oligonucleotides were diluted and co‐incubated in a humidified chamber. The ligation solution consisted of two oligonucleotides and ligase, which hybridized to the two PLA probes and joined to a closed circle if they were in close proximity. The ligation solution and the amplification solution, which consisted of nucleotides and fluorescently labeled oligonucleotides, was added together with DNA polymerase and incubated in a humidified chamber. The oligonucleotide arm of one of the PLA probes acted as a primer for a rolling circle amplification reaction, hybridizing to fluorescently red‐labeled oligonucleotides. The PLA probe dilution/incubation time and rolling circle amplification times were all optimized for this specific application. Samples were mounted with coverslips using Duolink in situ Mounting Medium (Olink Bioscience) with DAPI during the detection step of the PLA reaction and examined with a Keyence BZ‐8000 digital microscope (Keyence) under a 20× objective.
Western blot analysis
Cultured cells were washed with PBS and then lysed with 100 μL Laemmli sample buffer, after which 10 μL samples were separated using SDS PAGE. The separated proteins were then transferred to nitrocellulose membranes (GE Healthcare, Little Chalfont, UK), which were washed with PBS–Tween‐20 (PBS‐T) and then blocked for 30 min with a PBS‐T solution containing 5% skim milk. Blocked membranes were then rinsed twice with PBS‐T and incubated overnight at 4°C with Psf3 antibody (GeneStem), which was diluted 1:100 with 5% BSA/PBS‐T. After the membranes were washed with PBS‐T, membranes were incubated (30 min, room temperature) with the secondary peroxidase‐labeled donkey anti‐rabbit Ig whole antibody (GE Healthcare), which was diluted 1:5000 with PBS‐T. Membranes were washed with PBS‐T and then treated with a chemiluminescent detection kit (GE Healthcare) before they were visualized using a Luminoimage analyzer (LAS‐3000; Fujifilm, Tokyo, Japan). As a control assay, immunoblotting was carried out on the same membranes with a primary antibody directed against β‐actin (#4967; Cell Signaling Technology, Beverly, MA, USA), followed by a peroxidase‐labeled secondary antibody (GE Healthcare).
Co‐immunoprecipitation assay
Co‐immunoprecipitation was carried out using the Thermo Scientific (Thermo Fisher Scientific Waltham, MA, USA) Pierceco‐IP kit following the manufacturer's protocol. Briefly, each antibody (rabbit polyclonal anti‐Mcm2 antibody [1:100; Universal Biological] and rabbit anti‐Cdc45 antibody [1:100; Santa Cruz Biotechnology]) was first immobilized for 2 h using AminoLink Plus coupling resin (Thermo Fisher Scientific Waltham, MA, USA). The resin was then washed and incubated with cell lysate overnight. After incubation, the resin was again washed and protein eluted using elution buffer. Lysate supernatant was used as an input control. A negative control received the same treatment except that the samples did not include lysate, and the same samples except that the rabbit IgG was immobilized in the resin. Samples were analyzed by Western blotting using rabbit anti‐Mcm2 (1:1000; Universal Biological), rabbit anti‐Cdc45 (1:100; Santa Cruz Biotechnology), and mouse anti‐Psf3 (1:500; GeneStem) primary antibodies, and HRP‐labeled secondary antibodies (GE Healthcare).
Bromodeoxyuridine fluorescence‐activated cell sorting
Using this method, the cell cycle positions and DNA synthetic activities of cells can be determined by analyzing the correlated expression of total DNA and incorporated BrdU levels. At first, each cell was incubated with 10 μM BrdU (Becton Dickinson, Franklin Lakes, NJ, USA) for 30 min. Cells were then treated with DNase and incubated with FITC for 20 min at room temperature. Next, we added 7‐aminoactinomycin D, which can bind to total DNA. Using flow cytometric analysis with the combination of FITC‐labeled BrdU and 7‐aminoactinomycin D, we evaluated the enumeration and characterization of cells that were actively synthesizing DNA in terms of their cell cycle position.
Statistical analysis
Associations between Psf3 overexpression in cancer cells and clinicopathological features were determined using the χ2‐test. Survival was examined using the Kaplan–Meier method, and the significance of difference was evaluated by a log–rank tests. Variable effects on survival time were investigated using Cox's regression model. Statistical significance was determined as P < 0.05. All statistical analyses were carried out using JMP software, version 8 (SAS Institute, Cary, NC, USA).
Results
Expression of Psf3 in NSCLC
We previously reported the expression status of Psf3 in lung adenocarcinoma, determining that increased Psf3 expression enhanced cancer progression and proliferation, resulting in poor prognosis.16 In some adenocarcinoma tissues, the nuclei of cancer cells were stained in a scattered pattern (Fig. 1a). In contrast, other tissues showed stained nuclei clustered in some areas of adenocarcinoma tissues (Fig. 1b). In the present study, we investigated whether Psf3 might also be involved in squamous cell carcinoma. In well‐differentiated tumors, Psf3 expression was localized to proliferating cells directly above a defined epithelial basal layer (Fig. 1c). In contrast, in poorly differentiated tumors, in which such topological restriction was abolished, Psf3 expression showed a diffusely scattered pattern (Fig. 1d). Interestingly, we found that Psf3 expression was basically stained in the areas surrounding round nodules with concentric, laminated layers; this is the so‐called “keratinous pearl,” in which tumor cells transform into keratinized squames (Fig. 1e). In other words, Psf3 expression was especially present in the proliferating cells, suggesting that Psf3 might play an important role in the proliferation of squamous cell carcinomas.
Figure 1.
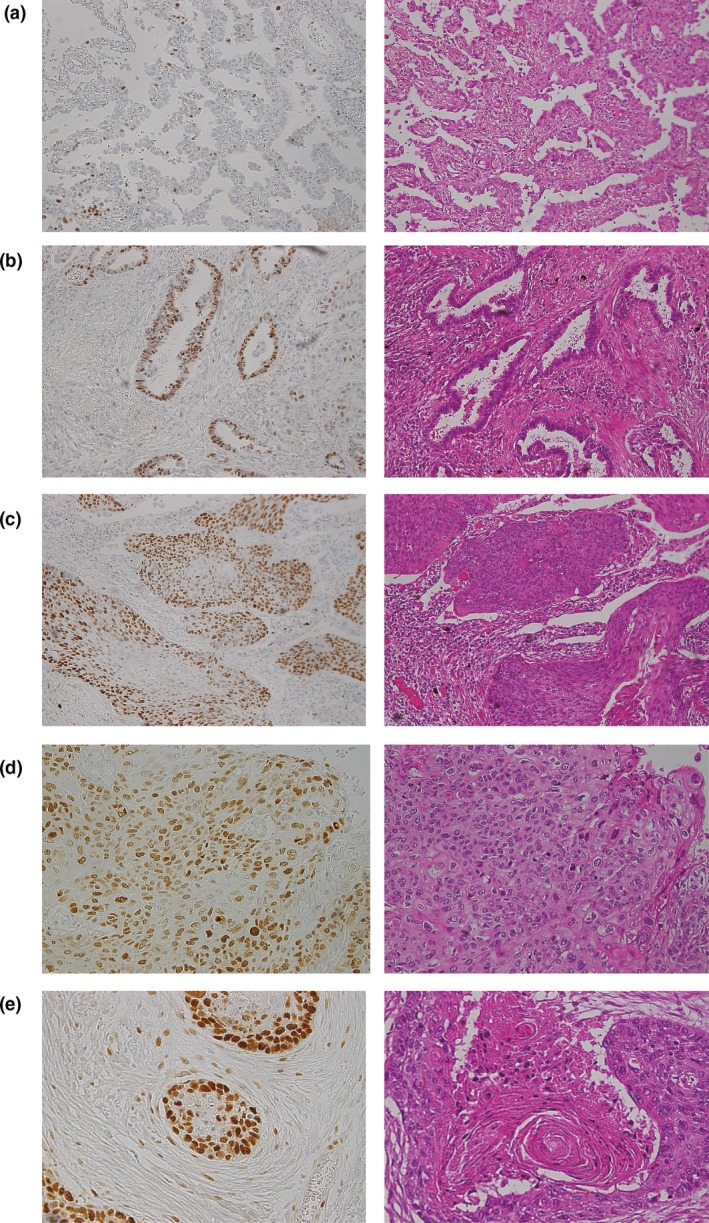
Nuclear Psf3 expression was examined by immunohistochemistry in lung adenocarcinoma and squamous cell carcinoma. (a) In some tissues, the nuclei of cancer cells were stained in a scattered pattern. (b) In contrast, other tissues showed stained nuclei clustered in certain areas of adenocarcinoma tissues. (c,d) Psf3 expression was restricted to proliferating cells directly above a defined epithelial basal layer in well‐differentiated squamous cell carcinoma but showed a diffusely scattered pattern in poorly differentiated tumors, where such topological restriction was abolished. (e) Interestingly, Psf3‐positive cells were localized in the outer layer of the cancer pearl, where proliferating cells were abundant.
Expression of Psf3 and its relationship to clinicopathological characteristics and prognosis
A total of 180 surgically resected NSCLC tissues were investigated. The distribution of cases by staining category for Psf3 is listed in Table 1. Notably, the rate of Psf3‐positive cells was higher in squamous cell carcinoma than in adenocarcinoma. This trend reflected the observation that Psf3 expression was present in the basal layer in all squamous cell carcinoma tissues.
Table 1.
Distribution of Psf3 expression in 180 surgically resected non‐small‐call lung cancer tissues by histological subtype
| Tumor cell staining category (%) | Ad (%) | Sq (%) | Others (%) |
|---|---|---|---|
| 0–24 | 35 (32.1) | 4 (8.5) | 2 (8.3) |
| 25–49 | 15 (13.8) | 11 (23.4) | 7 (29.2) |
| 50–74 | 38 (34.8) | 15 (31.9) | 8 (33.3) |
| 75–100 | 21 (19.3) | 17 (36.2) | 7 (29.2) |
Ad, adenocarcinoma; Sq, squamous cell carcinoma.
Next, we determined the status of Psf3 expression. In squamous cell carcinoma, more than 75% staining was considered positive for Psf3, whereas in adenocarcinoma and other types, more than 50% staining was considered positive for Psf3.
Of all the specimens examined, 91 were positive for Psf3 expression and 89 were negative. Table 2 shows the association between Psf3 findings and clinicopathological factors. The results of the analysis revealed that Psf3 expression was significantly associated with vessel invasion (P = 0.016), lymphatic invasion (P = 0.002), and pleural invasion (P = 0.036). The association between Psf3 expression and clinicopathological factors was assessed separately in adenocarcinoma and squamous cell carcinoma (Tables 3, 4). Psf3 expression was significantly related to gender (P = 0.004), high Brinkman index (P = 0.005), vessel invasion (P < 0.001), lymphatic invasion (P < 0.001), pleural invasion (P = 0.007), and the type of surgery (P = 0.019) in adenocarcinoma, but not in squamous cell carcinoma.
Table 2.
Association between Psf3 expression and clinicopathological characteristics in 180 patients with non‐small‐cell lung cancer
| Factor | Total | Psf3 | P‐value | |
|---|---|---|---|---|
| Positive | Negative | |||
| No. of patients (%) | 180 | 91 (50.5) | 89 (49.5) | |
| Age, years (mean ± SD) | 66.3 ± 10.3 | 65.1 ± 11.1 | 67.4 ± 9.2 | 0.130 |
| Gender (male/female) | 123/57 | 67/24 | 56/33 | 0.122 |
| Brinkman index (≥400/<400) | 104/76 | 56/35 | 48/41 | 0.301 |
| Pathologic stage (I/II/III/IV) | 84/53/38/5 | 37/32/20/2 | 47/21/18/3 | 0.287 |
| T factor (T1/T2/T3/T4) | 68/74/32/6 | 28/43/17/3 | 40/31/15/3 | 0.242 |
| N factor (N0/N1/N2) | 109/44/27 | 51/27/13 | 58/17/14 | 0.252 |
| M factor (M0/M1) | 175/5 | 89/2 | 86/3 | 0.631 |
| Vessel invasion (negative/positive) | 95/85 | 40/51 | 55/34 | 0.016a |
| Lymphatic invasion (negative/positive) | 86/94 | 33/58 | 53/36 | 0.002a |
| Pleural invasion (negative/positive) | 118/62 | 53/38 | 65/24 | 0.036a |
| Histology | 0.053 | |||
| Adenocarcinoma | 109 | 59 | 50 | |
| Squamous cell carcinoma | 47 | 17 | 30 | |
| Others | 24 | 15 | 9 | |
| Surgery | 0.176 | |||
| Lobectomy | 143 | 77 | 66 | |
| Segmentectomy/wedge | 31 | 11 | 20 | |
| Pneumonectomy | 6 | 3 | 3 | |
Statistically significant.
Table 3.
Association between Psf3 expression and clinicopathological characteristics in 109 patients with adenocarcinoma
| Factor | Total | Psf3 | P‐value | |
|---|---|---|---|---|
| Positive | Negative | |||
| No. of patients (%) | 109 | 59 (53.2) | 50 (46.8) | |
| Age, years (mean ± SD) | 64.5 ± 10.8 | 63.5 ± 11.8 | 65.7 ± 9.7 | 0.283 |
| Gender (male/female) | 62/47 | 41/18 | 21/29 | 0.004a |
| Brinkman index (≥400/<400) | 46/63 | 32/27 | 14/36 | 0.005a |
| Pathologic stage (I/II/III/IV) | 62/24/19/4 | 28/17/12/2 | 34/7/7/2 | 0.141 |
| T factor (T1/T2/T3/T4) | 51/42/11/5 | 21/27/8/3 | 30/15/3/2 | 0.075 |
| N factor (N0/N1/N2) | 74/21/14 | 37/13/9 | 37/8/5 | 0.445 |
| M factor (M0/M1) | 105/4 | 57/2 | 48/2 | 0.866 |
| Vessel invasion (negative/positive) | 66/43 | 27/32 | 39/11 | <0.001a |
| Lymphatic invasion (negative/positive) | 53/56 | 19/40 | 34/16 | <0.001a |
| Pleural invasion (negative/positive) | 73/36 | 33/26 | 40/10 | 0.007a |
| Surgery | 0.019a | |||
| Lobectomy | 83 | 51 | 32 | |
| Segmentectomy/wedge | 24 | 7 | 17 | |
| Pneumonectomy | 2 | 1 | 1 | |
Statistically significant.
Table 4.
Association between Psf3 expression and clinicopathological characteristics in 47 patients with squamous cell carcinoma
| Factor | Total | Psf3 | P‐value | |
|---|---|---|---|---|
| Positive | Negative | |||
| No. of patients (%) | 47 | 17 (36.2) | 30 (63.8) | |
| Age, years (mean ± SD) | 70.2 ± 7.4 | 71.5 ± 6.7 | 69.5 ± 7.7 | 0.370 |
| Gender (male/female) | 45/2 | 16/1 | 29/1 | 0.683 |
| Brinkman index (≥400/<400) | 43/4 | 14/3 | 29/1 | 0.097 |
| Pathologic stage (I/II/III/IV) | 14/20/12/1 | 4/8/5/0 | 10/12/7/1 | 0.673 |
| T factor (T1/T2/T3/T4) | 13/18/15/1 | 5/6/6/0 | 8/12/9/1 | 0.781 |
| N factor (N0/N1/N2) | 23/17/7 | 6/10/1 | 17/7/6 | 0.042a |
| M factor (M0/M1) | 46/1 | 17/0 | 29/1 | 0.340 |
| Vessel invasion (negative/positive) | 22/25 | 8/9 | 14/16 | 0.979 |
| Lymphatic invasion (negative/positive) | 25/22 | 10/7 | 15/15 | 0.559 |
| Pleural invasion (negative/positive) | 30/17 | 12/5 | 18/12 | 0.465 |
| Surgery | 0.228 | |||
| Lobectomy | 39 | 12 | 27 | |
| Segmentectomy/wedge | 6 | 4 | 2 | |
| Pneumonectomy | 2 | 1 | 1 | |
Statistically significant.
Next, using the data collected from 180 patients, we evaluated their prognosis and its relationship to the expression of Psf3. The median follow‐up period was 37.0 ± 10.2 months. We examined the overall survival of Psf3‐positive and ‐negative groups and found that overall survival in patients in the Psf3‐positive group was lower than that for patients in the Psf3‐low‐positive group (P = 0.006; Fig. 2a). Furthermore, in the Psf3‐positive group, the same trend toward lower overall survival was observed in subgroups of the patients with adenocarcinoma (P = 0.021) and squamous cell carcinoma (P = 0.037; Fig. 2b,c). Additionally, we used univariate and multivariate analyses to detect prognostic factors among the examined clinicopathological factors (Table 5). Multivariate analysis indicated that Psf3 expression was an independent prognostic factor in NSCLC (P = 0.049).
Figure 2.
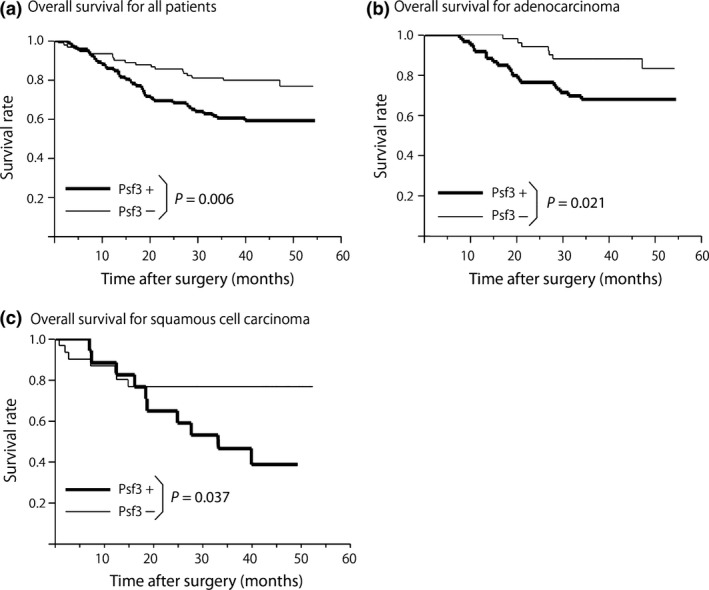
Postoperative overall survival curve among patients with positive and negative Psf3 expressions. (a) Strong Psf3 expression was significantly associated with poor survival (P = 0.006). Within the Psf3‐positive group, the same trends were observed in subgroups of adenocarcinoma (P = 0.021) (b) and squamous cell carcinoma (P = 0.037) (c).
Table 5.
Univariate and multivariate analyses of the association between overall survival and prognostic factors in 180 patients with non‐small‐cell lung cancer
| Variable | HR (95%CI) | P‐value |
|---|---|---|
| Univariate | ||
| Age | 1.01 (0.97–1.04) | 0.017* |
| Gender (male vs female) | 2.51 (1.32–5.27) | 0.004* |
| Smoking status (Brinkman index ≥400) | 1.71 (0.99–3.08) | 0.052 |
| Pathologic stage (I vs II–IV) | 2.27 (1.72–3.00) | <0.001* |
| T factor (T1/T2/T3/T4) | 2.65 (1.98–3.53) | <0.001* |
| N factor (N0/N1/N2) | 2.01 (1.46–2.75) | <0.001* |
| M factor (M0/M1) | 3.68 (1.11–9.01) | 0.035* |
| Vessel invasion (positive vs negative) | 2.49 (1.45–4.38) | <0.001* |
| Lymphatic invasion (positive vs negative) | 4.90 (2.63–10.0) | <0.001* |
| Pleural invasion (positive vs negative) | 3.91 (2.30–6.80) | <0.001* |
| Type of surgery (lobar vs sublobar) | 2.44 (1.07–7.03) | 0.031* |
| Psf3 (positive vs negative) | 2.13 (1.24–3.79) | 0.006* |
| Multivariate | ||
| Age | 1.04 (1.01–1.08) | 0.003* |
| Gender (male vs female) | 2.30 (1.17–4.99) | 0.014* |
| Pathologic stage (I vs II–IV) | 1.02 (0.46–2.19) | 0.951 |
| T factor (T1/T2/T3/T4) | 2.58 (1.54–4.41) | <0.001* |
| N factor (N0/N1/N2) | 1.57 (0.82–3.07) | 0.174 |
| M factor (M0/M1) | 1.14 (0.24–6.33) | 0.871 |
| Vessel invasion (positive vs negative) | 1.19 (0.43–1.65) | 0.604 |
| Lymphatic invasion (positive vs negative) | 2.52 (1.15–5.91) | 0.021* |
| Pleural invasion (positive vs negative) | 1.36 (0.69–2.72) | 0.369 |
| Type of surgery (lobar vs sublobar) | 1.53 (0.47–4.35) | 0.456 |
| Psf3 (positive vs negative) | 1.76 (1.01–3.22) | 0.049* |
CI, confidence interval; HR, hazard ratio. *Statistically significant.
Expression of Psf3, Cdc45, and Mcm2 and their interaction in NSCLC specimens
The expressions of Cdc45 and Mcm2, which contact with the GINS complex to form the CMG complex, were analyzed by immunohistochemistry in NSCLC specimens. Immunohistochemical stainings for Cdc45 and Mcm2 were carried out on the same serial specimens in which Psf3‐positive nuclei of cancer cells were clustered. Both Cdc45‐ and Mcm2‐positive cancer cells showed a similar pattern to Psf3‐positive cells (Fig. 3). These findings showed that Psf3 expression might occur in correspondence with Cdc45 and Mcm2 expressions.
Figure 3.
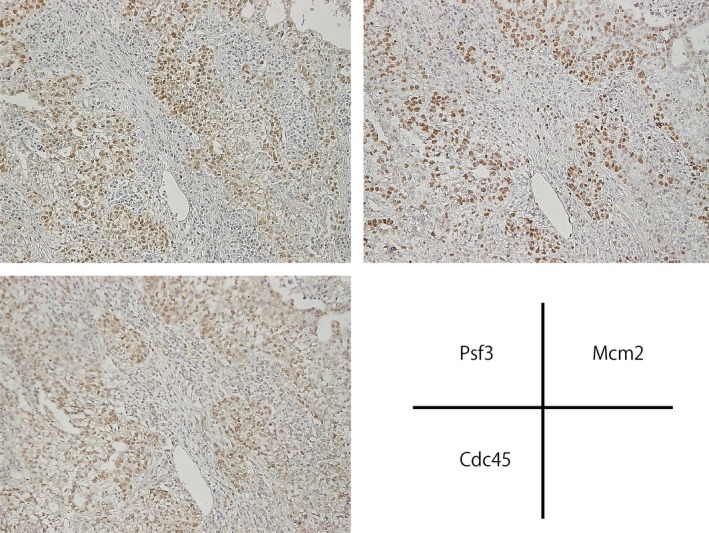
Immunohistochemical stainings for Psf3, Cdc45, and Mcm2 on the same serial specimens, in which Psf3‐positive nuclei of cancer cells were clustered. Both Cdc45‐ and Mcm2‐positive cancer cells showed a similar pattern to Psf3‐positive cells.
To assess whether Psf3 acts as the CMG complex in NSCLC, we used PLA methodology to examine the interaction between Psf3 and other CMG components, such as Cdc45 and Mcm2. The PLA revealed that interactions between Psf3 and Mcm2 proteins were observed in approximately 10% of cancer cells, in which both Psf3 and Mcm2 were strongly expressed in a clustered pattern (Fig. 4a‐1). However, these interactions were not observed in areas that did not include the expressions of these proteins. We also investigated interactions between Psf3 and Cdc45 proteins, obtaining results that resembled our findings for Psf3–Mcm2 protein interactions (Fig. 4a‐2). These findings suggested that some part of the abnormal Psf3 expression interacted with Cdc45 and Mcm2, and functioned as the CMG complex in NSCLC.
Figure 4.
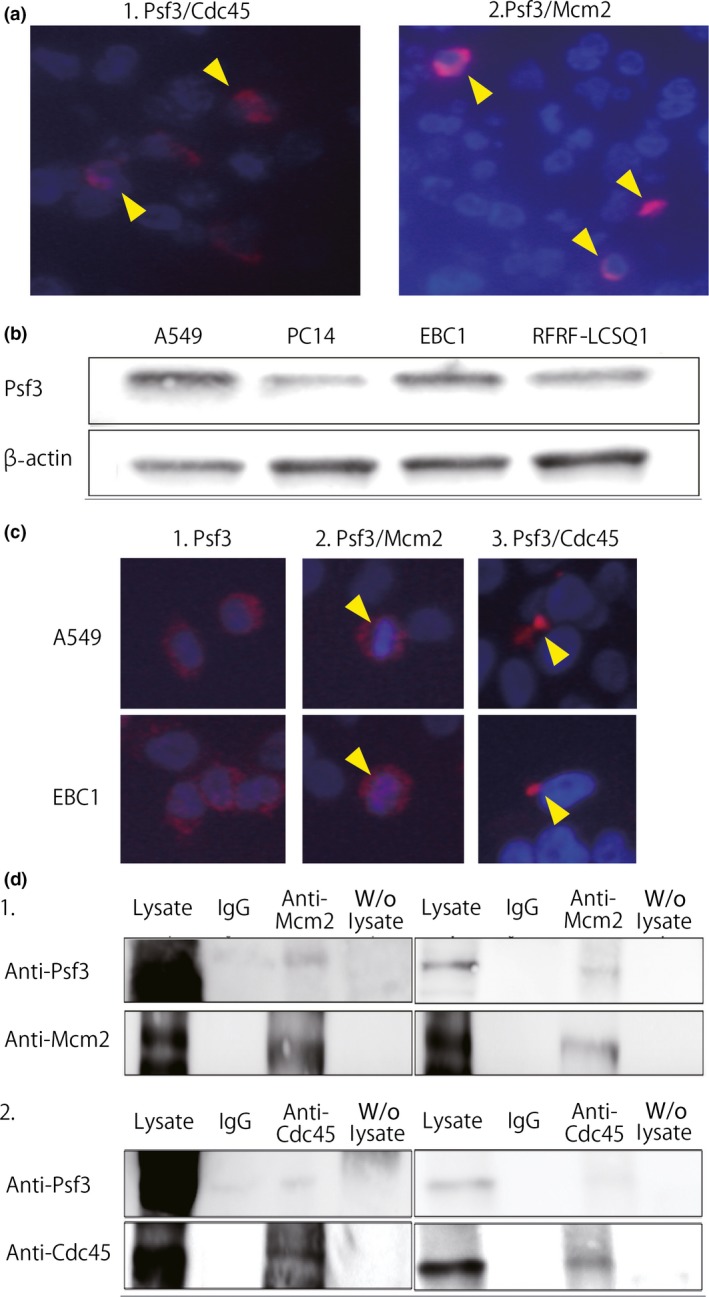
(a) Proximal ligation assay showed interactions between Psf3 and other Cdc45–Mcm–GINS (CMG) components, such as Cdc45 (a‐1) and Mcm2 (a‐2), in non‐small‐cell lung cancer specimens (arrowheads). (b) Western blotting for Psf3 and β‐actin in lung adenocarcinoma cell lines A549 and PC14 and lung squamous cell carcinoma cell lines RFRF‐LCSQ1 and EBC1. We detected strong expression of Psf3 in A549 and EBC1. (c) Immunofluorescent staining for Psf3 expression in A549 and EBC1 cells. DNA was counterstained with DAPI (blue). Nuclear accumulations of Psf3 were observed with a diffuse pattern in both cell lines (red). Proximal ligation assay analysis of the interaction between Psf3 and Mcm2. In both A549 and EBC1, dual protein interactions were observed in a portion of the Psf3‐positive cells, located around the nuclei. Interactions between Psf3 and Cdc45 were also observed in both cell lines. (d) Lysates from A549 and EBC1 cells were subjected to immunoprecipitation with anti‐Mcm2 and anti‐Cdc45 antibodies. The precipitates were separated by SDS‐PAGE followed by immunoblotting with anti‐Psf3, anti‐Mcm2, and anti‐Cdc45 antibodies. In both cell lines, Psf3‐positive signals were clearly detected in anti‐Mcm2 and anti‐Cdc45 precipitates.
Expression and interactions of Psf3 in lung cancer cell lines
We examined the same expressions and interactions in human lung cancer cell lines. Western blot analysis was carried out to determine whether Psf3 was expressed in human cell lines of adenocarcinoma (A549, PC14) and squamous cell carcinoma (RFRF‐LCSQ1, EBC1). The Psf3 expressions in the A549 and EBC1 cell lines were stronger than those in the PC14 and RFRF‐LCSQ1 cell lines (Fig. 4b). Hence, we used A549 and EBC1 in our evaluation of Psf3 expression and the interactions with Cdc45 and Mcm2. First, we used immunocytochemistry to evaluate Psf3 expression in A549 and EBC1 cells. Nuclear accumulations of Psf3 were observed with a diffuse pattern in both cell lines (Fig. 4c‐1). Next, we used PLA to evaluate the interaction between Psf3 and CMG complex components, such as Cdc45 and Mcm2. In both A549 and EBC1 cell lines, interactions between Psf3 and Cdc45 were observed in a portion of the cells, located around the nuclei (Fig. 4c‐2). Likewise, interactions between Psf3 and Mcm2 were observed in both cell lines (Fig. 4c‐3).
Moreover, we carried out co‐immunoprecipitation to examine complex formation between Psf3 and CMG complex components. In both cell lines, a Psf3‐positive signal was clearly detected in anti‐Mcm2 and anti‐Cdc45 precipitates, but not in precipitates incubated with control rabbit IgG (Fig. 4d). Thus, these results indicated that cancer cell lines also show CMG complex activity in Psf3‐positive cancer cells.
Effect of RNAi‐mediated knockdown of Psf3 on cell proliferation
We used RNAi methodology and knocked down the expression of Psf3 to assess the proliferative role of Psf3 in lung cancer cells. We carried out Western blots to check if we were able to inhibit Psf3 in A549 and EBC1 cell lines. Both A549 and EBC1 cells transfected with Psf3 siRNAs showed reductions in Psf3 expression of approximately 80% and 50%, respectively, as compared with cells transfected with non‐silencing control siRNAs (Fig. 5a). Subsequently, we assessed the proliferative effects of Psf3 on each cell line on normal tissue culture plates (60‐mm round dish with 4 ml culture medium containing 10% FBS and antibiotics, as mentioned above). These plates contained 20 × 104 A549 and 5 × 104 EBC1 cells transfected with either Psf3 RNAi or control non‐silencing siRNA. The number of cells in each transfected cell line was counted every 24 h from culture initiation for 72 h. The cells transfected with Psf3 RNAi grew more slowly than other cell lines, which revealed that the knockdown of Psf3 inhibits the proliferation of A549 and EBC1 (Fig. 5b). Moreover, we used BrdU–FACS to evaluate whether the knockdown of Psf3 led to delayed S phase. The percentages in the S‐phase gating zone were decreased by Psf3 gene knockdown in both cell lines (Fig. 6). Thus, Psf3 gene knockdown in human lung carcinoma cell lines resulted in growth inhibition characterized by delayed S‐phase progression.
Figure 5.
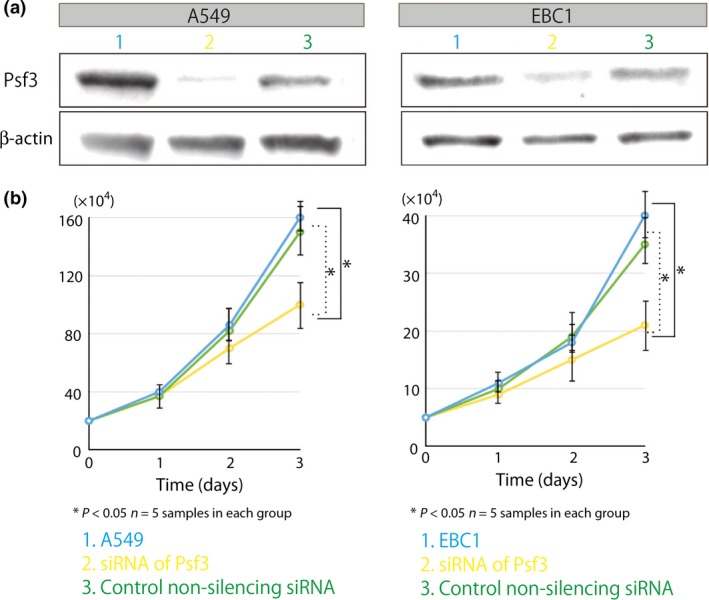
Comparison of proliferative activities in Psf3‐transfected lung adenocarcinoma A549 and lung squamous cell carcinoma EBC1 cell lines. In both A549 and EBC1, cells transfected with Psf3 RNAi grew slower than normal cells and control‐treated cells (n = 5 samples for each group).
Figure 6.
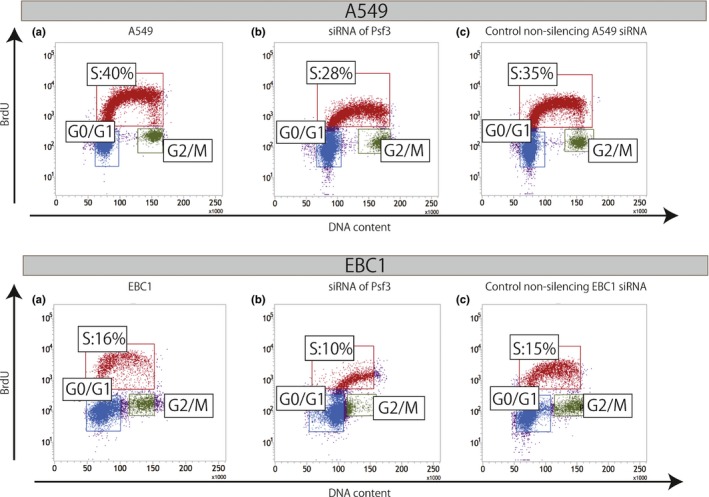
Cellular BrdU incorporation assay for the evaluation of the DNA duplicative activities of Psf3‐transfected lung adenocarcinoma A549 and lung squamous cell carcinoma EBC1 cell lines. In both A549 and EBC1, the percentage of the S‐phase gating zone was decreased by gene knockdown of Psf3.
Discussion
Highly efficient unwinding of double‐stranded DNA is an essential prerequisite for the duplication of genetic material. In eukaryotic cells, unwinding is catalyzed by the CMG complex, a macromolecular assembly of three replication factors: the Cdc45 protein, the Mcm complex, and the GINS complex.1, 2, 3 Psf3 is a member of the GINS complex, as are Sld5, Psf1, and Psf2. In our previous study, the expression status of Psf3 was immunohistochemically examined in 125 lung adenocarcinomas. We found that increased Psf3 expression was strongly predictive of poor prognosis. Furthermore, we compared the staining patterns of Psf3 and Ki67 in the same serial sections and found that their staining patterns differed. Ki67 expression showed a scattered staining pattern in the nuclei of cancer cells because it increased during the middle of cell division and was detected in a cell cycle‐dependent manner.19 However, the Psf3 expression showed a clustered staining pattern, indicating that the aberrant accumulation of Psf3 might play a role in cancer cells, irrespective of the cell cycle. Therefore, we hypothesized that the abnormal role of persistent Psf3 expression outside of the cell cycle might have contributed to malignant behavior in lung cancer.
We found that the “upregulation” of Psf3 resulted in an abnormal spatial distribution, in addition to the abnormal temporal distribution. We immunohistochemically examined squamous cell carcinoma specimens and found that Psf3‐positive cells were especially localized in the outer layer of the cancer pearl, where proliferating cells were abundant. These results indicated that abnormal Psf3 expression could lead to excessive proliferation in cancer cells. In addition, Psf3‐positive cells showed strong expression in keratinized areas, frequently shown in well‐differentiated squamous cell carcinoma, whereas they showed a clustered excessive staining in invasive adenocarcinoma. Thus, it is quite a matter of interest that the staining pattern of Psf3 differs according to the histological subtype.
The proper orchestration of DNA replicative proteins and pre‐replicative complex proteins leads to the correct initiation of DNA replication. Insufficient activation or overactivation of these proteins can result in abnormal DNA replication, which itself can promote genomic instability and subsequent tumorigenesis.20, 21, 22, 23 For instance, Hermand and Nurse suggested that pre‐replicative complex proteins are essential for accurate DNA replication/S‐phase progression, effective S‐phase checkpoint signaling, and genomic stability.23 Thus, it is not surprising that several of these proteins are deregulated in cancer. Indeed, it has been reported that several DNA replication proteins are over‐expressed in cancer cells and may serve as good tumor makers.24, 25, 26 For example, Yang et al.25 showed that higher expression of Mcm2 is significantly associated with poor prognosis in cases of NSCLC, which suggests that higher tumor proliferation and motility may be important to the prognosis of NSCLC. In the present study, we carried out immunohistochemical analyses of DNA replicative proteins Cdc45 and Mcm2 on the same serial sections in which Psf3‐positive nuclei of cancer cells were clustered. We found that Cdc45 and Mcm2 had staining patterns that resembled the stainings for Psf3. Together with the results of previous studies, our findings suggested that these DNA replication proteins were also upregulated in NSCLC. We also investigated the interaction between Psf3 and other CMG components using PLA methodology. We found that Psf3 interacted with Cdc45 and Mcm2 in a portion of Psf3‐positive cancer cells. These findings suggested that upregulated Psf3 assembles with Cdc45 and Mcm2 to form the CMG complex in lung cancer. To best of our knowledge, this is the first study to determine whether CMG complexes are detected in tumor specimens. However, these CMG complexes were only detected in a small proportion of the Psf3‐positive cancer cells. It is not clear why overexpressed Psf3 did not always function as the CMG complex. However, we assume that it was only possible to detect a portion of the Psf3 that functioned as CMG complexes, because of the short durations for which these proteins form such complexes.
In this study, Psf3 gene knockdown resulted in growth inhibition in human lung carcinoma cell lines. In addition, BrdU–FACS analysis showed that the S‐phase proportion decreased significantly in Psf3‐silenced cancer cell lines, leading to the inhibition of proliferative activity. Consequently, we expect that Psf3 should be a good candidate for molecular‐targeting cancer therapy in NSCLC. However, we also pose several questions that have not been settled by the present study: Is the Psf3 expression in cancer cells oncogenic? If it is oncogenic, how does it contribute to the DNA replication process? Further studies of Psf3 should be performed to assess its gene profile, to examine its potential as a biomarker for cancer stem cells, and to investigate its possible value as a therapeutic target for lung cancers with Psf3 overexpression.
In conclusion, Psf3 expression was strongly correlated with tumor progression in NSCLC. Furthermore, overexpressed Psf3, which acts as a CMG complex, is crucial for cancer cell proliferation in human lung cancer.
Disclosure Statement
The authors have no conflict of interest.
Cancer Sci 106 (2015) 1625–1634
Funding Information No sources of funding were declared for this study.
References
- 1. Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2‐7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A 2006; 103: 10236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bauerschmidt C, Pollok S, Kremmer E, Nasheuer HP, Grosse F. Interactions of human Cdc45 with the Mcm2‐7 complex, the GINS complex, and DNA polymerases delta and epsilon during S phase. Genes Cells 2007; 12: 745–58. [DOI] [PubMed] [Google Scholar]
- 3. Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2‐7 helicase by association with Cdc45 and GINS proteins. Mol Cell 2010; 37: 247–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bermudez VP, Farina A, Raghavan V, Tappin I, Hurwitz J. Studies on human DNA polymerase epsilon and GINS complex and their role in DNA replication. J Biol Chem 2011; 286: 28963–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kang YH, Galal WC, Farina A, Tappin I, Hurwitz J. Properties of the human Cdc45/Mcm2‐7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc Natl Acad Sci U S A 2012; 109: 6042–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kang YH, Farina A, Bermudez VP et al Interaction between human Ctf4 and the Cdc45/Mcm2‐7/GINS (CMG) replicative helicase. Proc Natl Acad Sci U S A 2013; 110: 19760–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ueno M, Itoh M, Kong L, Sugihara K, Asano M, Takakura N. PSF1 is essential for early embryogenesis in mice. Mol Cell Biol 2005; 25: 10528–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Han Y, Ueno M, Nagahama Y, Takakura N. Identification and characterization of stem cell‐specific transcription of PSF1 in spermatogenesis. Biochem Biophys Res Commun 2009; 380: 609–13. [DOI] [PubMed] [Google Scholar]
- 9. Ueno M, Itoh M, Sugihara K, Asano M, Takakura N. Both alleles of PSF1 are required for maintenance of pool size of immature hematopoietic cells and acute bone marrow regeneration. Blood 2009; 113: 555–62. [DOI] [PubMed] [Google Scholar]
- 10. Obama K, Ura K, Satoh S, Nakamura Y, Furukawa Y. Up‐regulation of PSF2, a member of the GINS multiprotein complex, in intrahepatic cholangiocarcinoma. Oncol Rep 2005; 14: 701–6. [PubMed] [Google Scholar]
- 11. Hayashi R, Arauchi T, Tategu M, Goto Y, Yoshida K. A combined computational and experimental study on the structure‐regulation relationships of putative mammalian DNA replication initiator GINS. Genomics Proteomics Bioinformatics 2006; 4: 156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ryu B, Kim DS, Deluca AM, Alani RM. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS ONE 2007; 2: e594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nagahama Y, Ueno M, Haraguchi N, Mori M, Takakura N. PSF3 marks malignant colon cancer and has a role in cancer cell proliferation. Biochem Biophys Res Commun 2010; 392: 150–4. [DOI] [PubMed] [Google Scholar]
- 14. Nagahama Y, Ueno M, Miyamoto S et al PSF1, a DNA replication factor expressed widely in stem and progenitor cells, drives tumorigenic and metastatic properties. Cancer Res 2010; 70: 1215–24. [DOI] [PubMed] [Google Scholar]
- 15. Nakahara I, Miyamoto M, Shibata T et al Up‐regulation of PSF1 promotes the growth of breast cancer cells. Genes Cells 2010; 15: 1015–24. [DOI] [PubMed] [Google Scholar]
- 16. Hokka D, Maniwa Y, Tane S et al Psf3 is a prognostic biomarker in lung adenocarcinoma. Lung Cancer 2013; 79: 77–82. [DOI] [PubMed] [Google Scholar]
- 17. Jarvius M, Paulsson J, Weibrecht I et al In situ detection of phosphorylated platelet‐derived growth factor receptor beta using a generalized proximity ligation method. Mol Cell Proteomics 2007; 6: 1500–9. [DOI] [PubMed] [Google Scholar]
- 18. Soderberg O, Leuchowius KJ, Kamali‐Moghaddam M et al Proximity ligation: a specific and versatile tool for the proteomic era. Genet Eng 2007; 28: 85–93. [DOI] [PubMed] [Google Scholar]
- 19. Scholzen T, Gerdes J. The Ki‐67 protein: from the known and the unknown. J Cell Physiol 2000; 182: 311–22. [DOI] [PubMed] [Google Scholar]
- 20. Oehlmann M, Score AJ, Blow JJ. The role of Cdc6 in ensuring complete genome licensing and S phase checkpoint activation. J Cell Biol 2004; 165: 181–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol 2006; 18: 658–67. [DOI] [PubMed] [Google Scholar]
- 22. Dutta A. Chaotic license for genetic instability and cancer. Nat Genet 2007; 39: 10–1. [DOI] [PubMed] [Google Scholar]
- 23. Hermand D, Nurse P. Cdc18 enforces long‐term maintenance of the S phase checkpoint by anchoring the Rad3–Rad26 complex to chromatin. Mol Cell 2007; 26: 553–63. [DOI] [PubMed] [Google Scholar]
- 24. Ramnath N, Hernandez FJ, Tan DF et al MCM2 is an independent predictor of survival in patients with non‐small‐cell lung cancer. J Clin Oncol 2001; 19: 4259–66. [DOI] [PubMed] [Google Scholar]
- 25. Yang J, Ramnath N, Moysich KB et al Prognostic significance of MCM2, Ki‐67 and gelsolin in non‐small cell lung cancer. BMC Cancer 2006; 6: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pollok S, Bauerschmidt C, Sanger J, Nasheuer HP, Grosse F. Human Cdc45 is a proliferation‐associated antigen. FEBS J 2007; 274: 3669–84. [DOI] [PubMed] [Google Scholar]


