Abstract
The regions of the genome that interact frequently with the nucleolus have been termed Nucleolar Associated Domains (NADs). Deep-sequencing and DNA-FISH experiments have revealed that these domains are enriched for repetitive elements, regions of the inactive X chromosome (Xi), and several RNA polymerase III-transcribed genes. NADs are often marked by chromatin modifications characteristic of heterochromatin, including H3K27me3, H3K9me3, and H4K20me3, and artificial targeting of genes to this area is correlated with reduced expression. It has therefore been hypothesized that NAD localization to the nucleolar periphery contributes to the establishment and/or maintenance of heterochromatic silencing. Recently published studies from several multicellular eukaryotes have begun to reveal the trans-acting factors involved in NAD localization, including the insulator protein CTCF, chromatin assembly factor CAF-1 subunit p150, several nucleolar proteins, and two long non-coding RNAs (lncRNAs). The mechanisms by which these factors coordinate with one another in regulating NAD localization and/or silencing are still unknown. This review will summarize recently published studies, discuss where additional research is required, and speculate about the mechanistic and functional implications of genome organization around the nucleolus.
Keywords: Nucleolus, perinucleolar region, NADs, genome organization, lncRNAs, heterochromatin
1. Introduction: The Nucleolus
The nucleolus was first described by Wagner (1835) and Valentin (1836) through light microscopy observations, highlighting the prominence of the nucleolus as a sub-nuclear body visible under crude light microscopy conditions (Wagner 1835; Valentin 1836; Valentin 1839). In the early 1930s, Heitz and McClintock independently discovered that the nucleoli are organized around specific genomic loci, which were later termed nucleolus organizer regions (NORs) (Heitz 1931; McClintock 1934). An explosion of discoveries were made in the mid 1960’s, culminating in the landmark discovery that nucleoli are the sites of ribosomal biogenesis (reviewed in (Pederson 2011)). In addition to its primary role as the site of ribosomal transcription and maturation, the nucleolus hosts many other biological processes, including replication of various viruses (Li 1997; Boyne and Whitehouse 2006; Sonntag et al. 2010), signal recognition particle biosynthesis (Jacobson and Pederson 1998; Pederson and Politz 2000; Politz et al. 2000; Ciufo and Brown 2000; Grosshans et al. 2001), sequestration of cell cycle regulators such as p53 and mdm2 (Weber et al. 1999), and sequestration of the transcription factor Hand1 prior to stem cell differentiation (Martindill et al. 2007). This review will focus on how the periphery of the nucleolus contacts particular regions of the genome and will outline what is known about the functionality of these interactions.
2. Organization of the Genome via Association with Specific Sub-Nuclear Regions
2A. Lamina-Associated Domains (LADs)
With the advent of high-throughput sequencing, scientists have devised several genome-scale methods to test whether nuclear structures associate with the genome in a random or non-random manner. One important method, termed Dam-ID, was developed by Bas van Steensel and Steven Henikoff (van Steensel and Henikoff 2000). Dam-ID involves the fusion of selected proteins with E. coli DNA adenine methyltransferase (Dam), followed by isolation and deep sequencing-based identification of DNA containing methylated adenine. Eukaryotes lack adenine methylation; therefore genome-scale mapping of this orthologous mark reveals genomic loci that were in close proximity to the fused protein of interest. Studies in a D. melanogaster embryonic cell line (Pickersgill et al. 2006) and human fibroblasts (Guelen et al. 2008) fused B-type lamins with Dam to detect peripherally-localized genomic regions, which were termed lamina-associated domains (LADs). LADs tend to be gene-poor and enriched for heterochromatic silencing marks such as H3K9me2 (Kind et al. 2013). Mouse and human genomes contain up to 1,400 LADs encompassing approximately 40% of the genome, ranging in size from 40 kilobases to over 30 megabases (Peric-Hupkes et al. 2010; Kind and van Steensel 2010).
The mechanisms that govern tethering of LADs to the nuclear periphery are still largely unclear, but recent studies suggest this tethering may be crucial in regulating the transcriptional status of the LADs. This was tested by using a LacO array proximal to a reporter gene and expressing a LacI fused to a protein which directly interacts with the inner nuclear membrane, such as EMD or Lap2β (Finlan et al. 2008; Reddy et al. 2008; Dialynas et al. 2010). In these experiments, targeting various reporter genes to the nuclear lamina (NL) resulted in decreased reporter expression. Likewise, in a comparison of LADs in mouse embryonic stem cells (ESCs) and neural precursor cells (NPCs), an increase in NL association was correlated with a decrease in expression level. Conversely, gene ontology (GO) analysis revealed that ~20% of genes that featured decreased association with the NL during ESC→NPC differentiation were required for neural physiology. These neural physiology genes generally displayed increased expression during neural differentiation, suggesting that release from the NL is an important step during the induction of lineage-specific gene expression (Peric-Hupkes et al. 2010). In summation, these studies suggest that positioning of LADs at the NL is an important method for physically and functionally compartmentalizing eukaryotic genomes.
2B. Nucleolar Associated Domains (NADs)
In 2010, two independent studies isolated and sequenced the genomic DNA associated with purified nucleoli (van Koningsbruggen et al. 2010; Németh et al. 2010). Both studies found that these nucleolar-associated domains (NADs) are relatively gene-poor compared to the rest of the genome and are highly enriched for satellite DNA repeats. Additionally, both studies also found enrichment for specific types of genes including those coding for the 5S rRNA, immunoglobulins, olfactory receptors, and zinc-finger proteins. These gene classes often exist as multigene arrays, however it remains unknown to what extent primary sequences contribute to perinucleolar localization (see concluding remarks below).
Along with these similarities in the two NAD datasets, there are several notable differences that may be attributable to the different cell types or experimental procedures used (notably, the use of crosslinking). The Längst group isolated nucleoli from formaldehyde-crosslinked HeLa cells and observed that NADs were significantly enriched for tRNA genes, which are transcribed by RNA polymerase III (Németh et al. 2010). This finding is compatible with previous observations that RNA polymerase III is especially active around the nucleolar periphery (Matera et al. 1995; Thompson et al. 2003; Haeusler and Engelke 2006). The Lamond group analyzed NADs in non-crosslinked HT-1080 fibrosarcoma cells and emphasized that the majority of the NAD peaks overlap with previously published LADs (van Koningsbruggen et al. 2010). To explore this overlap, the Lamond group photoactivated a GFP-tagged histone around the periphery of the nucleolus and then tracked the localization of that chromatin through the cell cycle. It was found that after mitosis, the photoactivated chromatin could localize to either the perinucleolar (PN) region or the NL, indicating that the PN and the NL may be interchangeable addresses for some loci. These data are consistent with other studies which found that LADs often re-localize to the PN region after mitosis (Kind et al. 2013). LADs can also redistribute to either the PN or pericentromeric (PC) heterochromatin regions upon a short treatment with actinomycin D at a dose that selectively inhibits RNA polymerase I (Ragoczy et al. 2015). However, the transcriptional activity of the relocalized loci were not altered during this treatment, suggesting that the PN and NL serve as dynamic, functionally overlapping regions for genome organization and silencing (reviewed in (Padeken and Heun 2014)).
2C. The Perinculeolar Compartment (PNC) of Cancer Cells
The HeLa and HT-1080 cells used by the Lamond and Längst laboraties (van Koningsbruggen et al. 2010; Németh et al. 2010) both contain a cancer-specific sub-nuclear structure known as the perinucleolar compartment (PNC) (Norton et al. 2008). The PNC is localized on a portion of the nucleolar surface (Huang et al. 1997) and thus may copurify with nucleoli. This suggests that some of the NADs described in the previous section may be cancer cell-specific. The presence of the PNC is correlated with metastasis and inversely correlated with patient survival and relapse (Kamath et al. 2005). The PNC is enriched in proteins that regulate the splicing and polyadenylation of RNA polymerase II transcripts (Ghetti et al. 1992; Matera et al. 1995; Timchenko et al. 1996; Hall et al. 2004). This compartment also contains specific RNA polymerase III transcripts, including RNases P, MRP, Y RNAs (Matera et al. 1995), Alu RNAs, and signal recognition particle RNA (Wang et al. 2003). These RNA species are not actively transcribed within the PNC (Matera et al. 1995; Wang et al. 2003) and the compartment is devoid of most RNA polymerase III transcripts, notably tRNAs (Matera et al. 1995). The function of the PNC is still unknown, but is of great interest to the field of cancer biology (reviewed in (Pollock and Huang 2010)). In the interest of space, the remainder of this review will focus on NAD associations which occur in both primary and cancer cells.
3. NAD Function: PN Localization Linked to Heterochromatin Silencing
3A. The Inactive X Chromosome
In 1949 Barr and Bertram described a “nucleolar satellite” which protruded from the nucleolus of female cat motor neurons (Barr and Bertram 1949). This structure was later identified as the inactive X chromosome. During embryonic development of female mammalian cells, one of the X chromosomes is silenced in order to provide dosage compensation, ensuring that female cells with two X chromosomes do not overexpress X-linked genes (Lyon 1961). During gastrulation and after most of the DNA methylation imprints from the parents are erased, the two X-chromosomes pair and each chromosome is randomly assigned to become active (Xa) or inactive (Xi). The designated Xi then transcribes a long non-coding RNA (lncRNA) known as Xist (Brown et al. 1991), which spreads across the Xi in cis (Clemson et al. 1996). Xist recruits Polycomb Repressive Complexes 1 and 2 (PRC1 and PRC2), which induce methylation of histone H3 lysine 27 and silence the transcriptional activity of most of the genes on the Xi ((Plath et al. 2003; Plath et al. 2004); reviewed in (Lucchesi et al. 2005; Thorvaldsen et al. 2006; Lee 2012)). Thus, X inactivation is a prominent example of gene regulation via alteration of chromatin state.
Several studies have shown that the Xi can associate with either the NL or PN regions of the nucleus (Barton et al. 1965; Bourgeois et al. 1985). A study by the Lee laboratory found that Xi association with nucleoli is most prevalent during mid-late S-phase of the cell cycle (Zhang et al. 2007). This study also showed that the interaction is dependent upon the X inactivation center (Xic), the region encoding the Xist locus, because autosomes bearing Xic translocations also preferentially associate with nucleoli. Conversely, deletion of Xist reduces nucleolar association, H3K27 methylation, and derepression of Xic-proximal genes in some cell lines analyzed. This PN localization occurs during mid-late S-phase when heterochromatin is replicated, suggesting that replication in the PN region helps to maintain the silent state of the Xi. Additional tools to perturb Xi-nucleolar associations without having to delete central silencing factors such as Xist will be important to further test this hypothesis. However, it seems likely that the Xi-PN association by itself is not essential for maintaining the bulk of Xi silencing, consistent with previous observations of a large degree of functional overlap between Xist and other factors (Csankovszki et al. 2001).
3B. The 5S rDNA
Eukaryotic ribosomes are comprised of large (60S) and small (40S) subunits. The major RNA species found in mature ribosomes (28S, 18S, and 5.8S rRNA) are encoded within the 47S rRNA primary transcripts that are produced from repeated templates on the short arms of the five acrocentric chromosomes in humans (reviewed in (Boisvert et al. 2007; Sirri et al. 2008; Németh and Längst 2011)). An additional rRNA species is encoded by an array of approximately 100 RNA polymerase III-transcribed 5S rDNA genes located on Chromosome 1 (Steffensen et al. 1974; Stults et al. 2008). As noted above, several studies have found that RNA polymerase III transcribed genes, including the 5S rDNA, are enriched in the PN region in a variety of different species and cell types (Matera et al. 1995; Thompson et al. 2003; van Koningsbruggen et al. 2010; Németh et al. 2010). The 5S nucleolar association in humans was described in HeLa cells, but its localization was noted as being outside of the PNC region (Matera et al. 1995). One possible rationale for the close proximity of the 5S array to nucleoli would be to increase the efficiency of ribosome assembly (Haeusler and Engelke 2006). However, a study by the Magnuson lab suggests instead that nucleolar localization of 5S rDNA repeats may facilitate transcriptional silencing. In this study, the 119-bp 5S rDNA and a reporter gene was randomly inserted into the genome of mouse ES cells, and these transgenes frequently associated with nucleoli. This association was correlated with reduced reporter gene expression and increased H3K9me3 enrichment. Furthermore, endogenous mouse 5S pseudoegenes, which maintain internal RNA polymerase III transcription factor binding sites but do not produce a functional transcript, also frequently associate with the PN region. ChIP-qPCR analysis demonstrated that many of these pseudogenes feature low RNA polymerase III transcription factor occupancy, suggesting that the 5S rDNA sequence and not the RNA Polymerase III transcription machinery is responsible for PN localization (Fedoriw et al. 2012b). The cis-acting sequences and trans-acting factors required for these types of higher-order genome interactions are of major interest, and specific examples related to nucleoli are discussed below (Table 1).
Table 1.
Summary of Known Perinucleolar Region Regulators Protein or RNA
| Protein or RNA | Known Interphase Nuclear Localization |
Regulation Target (Species) |
|---|---|---|
| CTCF | Insulator Elements (Kim et al. 2007) | Insulators (H. sapiens)(Yusufzai et al. 2004) Clustered Centromeres (D. melanogaster) (Padeken et al. 2013) Xi (M. musculus) (Yang et al. 2015) |
| NLP (NCL) | Nucleolar Fibrillar Centers(Lischwe et al. 1981; Spector et al. 1984) Nucleolar Dense Fibrillar Component (Spector et al. 1984; Escande et al. 1985) Nucleolar Granular Component (Escande et al. 1985) |
Clustered Centromeres (D. melanogaster)(Padeken et al. 2013) |
| Modulo (NPM1) | Nucleolar Granular Component (Spector et al. 1984) | Clustered Centromeres (D. melanogaster) (Padeken et al. 2013) |
| CAF-1 p150 | DNA Damage Foci (Martini et al. 1998; Green and Almouzni 2003) Heterochromatin (Murzina et al. 1999) Perinucleolar Region (Smith et al. 2014) Replication Foci (Krude 1995) |
5S rDNA (H. sapiens) (Smith et al. 2014) Alpha Satellite DNA (H. sapiens) (Smith et al. 2014) D4Z4 (Chromosome 10q) (H. sapiens) (Smith et al. 2014) |
| Ki-67 | Satellite DNA (Bridger et al. 1998) Nucleolar Dense Fibrillar Component (Kill 1996) |
47S rDNA (H. sapiens) (Chromosome 13p) (Booth et al. 2014) |
| Kcnq1ot1 | Kcnq1 Locus (Pandey et al. 2008) | Kcnq1 locus (H. sapiens, M. musculus) (Pandey et al. 2008; Mohammad et al. 2008) |
| Firre | 6 gene-containing loci (Firre, Slc25a12, Ypel4, Eef1a1, Atf, Ppp1r10), and 34 non-gene loci (Hacisuleyman et al. 2014) | Xi (M. musculus) (Yang et al. 2015) |
4. Protein Regulators of PN Structure and Function
Tethering of NADs to the PN region is now known to require several trans-acting factors, which presumably directly or indirectly bind to NAD DNA. These NAD-bound factors must either interact with the ribosomal DNA of the nucleolus or with nucleolar proteins in order to facilitate nucleolar localization. The following sections will briefly introduce the protein and RNA factors thus far discovered to be involved in NAD localization.
4A. CTCF
The CCCTC-Binding Factor (CTCF) is a DNA-binding protein with between 55,000–65,000 sites of enrichment throughout the human genome (Chen et al. 2012). CTCF is particularly enriched on insulator elements (Kim et al. 2007; Song et al. 2011; DeMare et al. 2013), which block interaction between promoters and enhancers. Importantly, CTCF regulates the three-dimensional interaction of these regulatory elements with distal promoters, thereby regulating transcription (Sanyal et al. 2012; Shen et al. 2012). Insulator occupancy by CTCF is cell-type specific (Barski et al. 2007; Kim et al. 2007; Chen et al. 2008; Cuddapah et al. 2009; Shen et al. 2012), suggesting that CTCF contributes to regulatory networks responsible for changes in cell-lineage specific nuclear architecture. One study in human leukemia cells showed that CTCF binding sites within an exogenous insulator conferred a higher propensity to associate with nucleoli. This study further showed that CTCF interacts with the nucleolar protein nucleophosmin (NPM1/B23), and that NPM1 enrichment on the exogenous insulator was dependent on the presence of CTCF binding sites (Yusufzai et al. 2004). One plausible model to explain this tethering would be that CTCF links insulator elements to the nucleoli via interaction with the nucleolar protein NPM1. Later sections in this review will discuss the role of CTCF in regulating the nucleolar localization of clustered centromeres and the Xi.
4B. The Nucleophosmin Homolog NLP and the Nucleolin Homolog Modulo
Nucleolin (NCL/C23) is a multifunctional protein that primarily localizes to the nucleolus (Bugler et al. 1982), interacts with snoRNAs (Sáez-Vasquez et al. 2004), regulates the folding and assembly of ribosomes (Ginisty et al. 1998), and regulates 47S rDNA transcription (Roger et al. 2002; Rickards et al. 2007). NPM1 is also a multifunctional nucleolar protein (Michalik et al. 1981) that interacts with several ribosomal proteins (Yu et al. 2006; Lindström and Zhang 2008) and has multiple roles in nucleolar biology including acting as a pre-rRNA endoribonuclease (Herrera et al. 1995; Savkur and Olson 1998) and facilitating 40S and 60S nuclear export (Maggi et al. 2008). NPM1 interact with a wide variety of proteins, including the centromere-specific histone variant CENP-A (Foltz et al. 2006). Centromeres often cluster together and localize to the nucleolus in many species and cell types (Weierich et al. 2003). The Heun laboratory showed that depletion of NLP, a Drosophila homolog of NPM1, or the Drosophila CTCF insulator protein resulted in de-clustering of centromeres and a decrease in centromere association with nucleoli. Depletion of Modulo, the Drosophila homolog of NCL, also resulted in de-clustering of centromeres and disruption of nucleolar structure, making it difficult to determine whether Modulo is required for PN positioning. NLP or Modulo depletions also resulted in de-repression of several classes of transposable elements and cells showed signs of genomic instability, including increased double stranded breaks and lagging chromosomes during mitosis (Padeken et al. 2013). These data are consistent with the idea that the frequent nucleolar localization of centromeres is functionally important for maintaining a chromatin structure optimal for proper chromosome segregation.
4C. Chromatin Assembly Factor-1 p150
Chromatin Assembly Factor 1 (CAF-1) is an evolutionarily conserved complex comprised of three subunits known as p150, p60, and p48 in humans (Kaufman et al. 1995). CAF-1 was first identified as a factor required for chromatin assembly during in vitro replication of SV40 DNA (Smith and Stillman 1989). CAF-1 deposits histone (H3/H4)2 heterotetramers onto replicating DNA, thereby colocalizing with foci of BrdU incorporation during S-phase (Krude 1995). This histone deposition activity is also important for restoring chromatin structure after several types of DNA damage repair (Gaillard et al. 1996; Green and Almouzni 2003; Polo et al. 2006) and for delivery of suitably modified histones for maintenance of heterochromatic silencing (Murzina et al. 1999; Reese et al. 2003; Quivy et al. 2008; Huang et al. 2010).
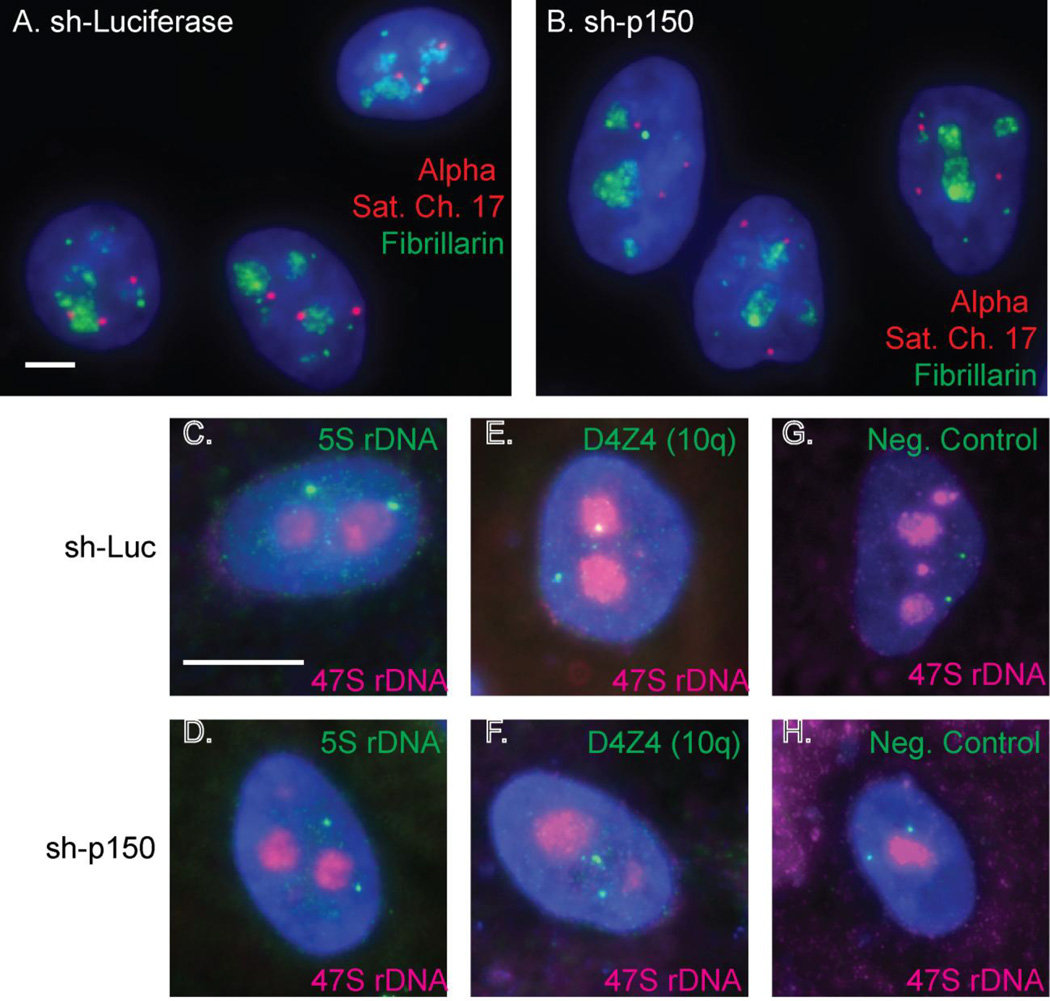
A recent study from the Kaufman lab found an additional function for CAF-1 that appears unrelated to histone deposition. In this study, mass spectrometry identified several new p150-interacting proteins including nucleolar proteins Ki-67, NCL and NPM1. Upon depletion of p150 in HeLa cells, NCL, NPM, Ki-67, and several other nucleolar proteins lost their nucleolar localization. In addition, repetitive DNA elements in MCF-10A mammary epithelial cells, including the 5S rDNA, alpha satellite DNA from chromosome 17, and the macrosatellite D4Z4, showed decreased association with nucleoli upon p150 depletion (Figure 1). p150 depletion in primary human fibroblasts also reduced the nucleolar association of alpha satellite DNA, suggesting that this phenomenon is not specific to the cancer cell-specific PNC structure. Structure-function studies demonstrated that the N-terminus of p150 is sufficient to maintain both the nucleolar protein and repetitive DNA associations. These results indicate that this nuclolear activity is distinct and separable from the histone deposition activity of the CAF-1 complex, which requires the p60 and p48 binding sites elsewhere in the protein. This study additionally showed that a sumoylation interacting motif (SIM) within p150 is required for the association of these repetitive elements (Smith et al. 2014). Of note, the p150 SIM is also required for the nucleolar localization of Ki-67 (Smith et al. 2014), another protein which was recently shown to regulate NAD localization (Booth et al. 2014).
Figure 1. CAF-1 p150 is required for normal localization of several repetitive elements to the perinucleolar region.
Panels A–B: DNA-FISH analysis of HeLa cells, showing proximity of chromosome 17 alpha satellite DNA (red) to nucleoli (fibrillarin, green) in cells expressing a shRNA directed against luciferase (A) or CAF-1 p150 (B). Panels C–H: DNA-FISH analysis of MCF-10A cells displaying proximity of nucleoli (rDNA, red) to the 5S rDNA array (C and D), the D4Z4 array on chromosome 10q (E and F), or a non-NAD negative control BAC from 10q (Németh et al. 2010) (G and H). Cells expressed an shRNA directed against luciferase (panels C, E, G) or CAF-1 p150 (panels D, F, H). The scale bar is 5 µm in panel A and 10 µm in panel C. Methods, probes, and antibodies described in (Smith et al. 2014).
4D. Ki-67
Ki-67 was first identified as an epitope recognized by a monoclonal antibody raised against nuclei from a Hodgkin lymphoma cell line (Gerdes et al. 1983). Since its initial discovery, Ki-67 has been used as a cell proliferation marker in thousands of clinical studies examining growth rates of various types of cancers. Despite this, relatively little is known about the molecular functions of Ki-67. In interphase cells Ki-67 primarily localizes to the nucleolus (Kill 1996; Cheutin et al. 2003), is enriched on the 47S rDNA gene (Bullwinkel et al. 2006), and is required for normal levels of 47S rDNA transcription (Rahmanzadeh et al. 2007; Booth et al. 2014). In early G1, Ki-67 localizes to distinct foci which co-localize with several different classes of repeats enriched within the NADs, including centromeric alpha satellite, telomeric repeats, and Sat III (Bridger et al. 1998). A recent study by the Vagnarelli and Earnshaw laboratories (Booth et al. 2014) found that Ki-67 is required for the formation of the human perichromosomal layer, a proteinaceous sheath that coats condensed chromosomes during mitosis (reviewed in (Van Hooser et al. 2005)). Ki-67 is also required for normal nucleolar association of an rDNA-proximal NAD sequence containing a LacO reporter array (Booth et al. 2014). Future studies should examine whether the NAD and perichromosomal regulation activities of Ki67 are interrelated.
5. lncRNAs as Regulators of PN Structure and Function
5A. Kcnq1ot1
The Kcnq1 locus in mouse and human cells is regulated through maternal imprinting. While genes within this locus are expressed on the maternal chromosome, the paternal chromosome expresses an antisense lncRNA known as Kcnq1ot1 in order to facilitate silencing of the paternal genes (Pandey et al. 2004; Thakur et al. 2004; Mancini-DiNardo et al. 2006; Pandey et al. 2008). In contrast to the paternally-derived chromosome, Kcnq1ot1 expression on the maternal chromosome is inhibited due to imprinted methylation of the Kcnq1ot1 promoter (Fitzpatrick et al. 2002). The Kanduri laboratory discovered that the Kcnq1ot1 locus is often enriched for heterochromatic histone silencing marks in mouse placenta cells, but not in fetal liver cells. In placenta cells, the Kcnq1ot1 locus is also associated with nucleoli twice as frequently as observed in fetal liver cells (Pandey et al. 2008), supporting the correlation between nucleolar localization and the establishment and/or maintenance of heterochromatin. Another study found that an 890-bp region near the 5’-end of the human Kcnq1ot1 transcript is required for silencing of the other Kcnq1 locus genes. Furthermore, when this silencing domain was inserted into an episomal vector, the vector localized to nucleoli during mid S-phase and a flanking reporter gene was silenced. When the silencing domain was inserted in reverse orientation, the vector failed to localize to nucleoli and the reporter gene was expressed (Mohammad et al. 2008). These results support the hypothesis that the transcribed Kcnq1ot1 lncRNA and not the DNA sequence encoding it is required for localization to the PN region and silencing of the vector reporter genes.
5B. Firre
One of the X-linked genes which escapes silencing during X-inactivation encodes the lncRNA Firre (Yang et al. 2010). Firre is important for long-range chromosomal interactions, interacting with the RNA-binding protein hnRNPU to facilitate localization of the Xi to regions from five different chromosomes (Hacisuleyman et al. 2014). Firre is also required for normal association of the mouse Xi with nucleoli, as the frequency of PN localization of the X-linked Firre and DXZ4 macrosatellite loci decrease upon depletion of the Firre lncRNA (Yang et al. 2015). Firre depletion also decreases the enrichment of the heterochromatic silencing mark H3K27me3 on the Xi without decreasing the expression levels of Xist. However, depletion of Firre did not result in significant transcriptional changes on the Xi. Future experiments will be required to distinguish whether these data result from functional redundancy among repressive factors which govern Xi silencing, or because transcriptional regulation is not a major functional consequence of NAD localization.
The Firre and DXZ4 loci also feature enrichment of the insulator protein CTCF (Hacisuleyman et al. 2014; Yang et al. 2015). CTCF depletion decreased PN association of both the Firre and DXZ4 loci, reduced expression of the Firre lncRNA, and diminished levels of H3K27me3 on the Xi (Yang et al. 2015). Therefore, CTCF may regulate PN localization and silencing of the Xi as both a direct interaction factor and as a transcriptional regulator of Firre expression. Additional studies should explore whether the trans-chromosomal interactions regulated by Firre also localize to the periphery of the nucleolus and are regulated by CTCF.
Concluding Remarks: What defines the NADs, and what is their functional significance?
What defines the NADs?
One possible determinant of NAD localization is genomic DNA sequence. Primary DNA sequence is likely critical for some classes of NADs, as is the case for 5S pseudogenes, which can associate with the nucleoli independently of RNA Polymerase III machinery (Fedoriw et al. 2012b). However, primary DNA sequence cannot be the only determinant of all nucleolar localization. For example, in the case of Firre-dependent PN localization of the inactive Xi chromosome (Figure 2)(Yang et al. 2015), the primary sequences of the Xi must be insufficient for nucleolar association because the Xa chromosome does not localize to the PN region. The discovery that lncRNAs are required for some NAD associations is an exciting development, as over 100,000 lncRNAs have been annotated in the human genome (Volders et al. 2013), and the functions of most of these molecules remain unknown. Additional research is required to elucidate how proteins and lncRNAs coordinate NAD-PN interactions. One likely possibility is that the lncRNAs directly interact with nucleolar targeting proteins, and recent advances in high throughput RNA-binding protein identification will be instrumental in determining the binding partners for Kcnq1ot1 and Firre (Chu et al. 2015; McHugh et al. 2015). In the case of Firre, a particularly relevant interacting protein is the RNA binding protein hnRNPU. hnRNPU binds to Firre and is required for the inter-chromosomal interactions mediated by Firre (Hacisuleyman et al. 2014). Another protein of interest, CTCF, binds to both the Firre RNA and DNA (Hacisuleyman et al. 2014; Yang et al. 2015) and this interaction may be mediated by hnRNPU. Many more contributing factors are likely to be discovered as investigations of these higher-order interactions continue.
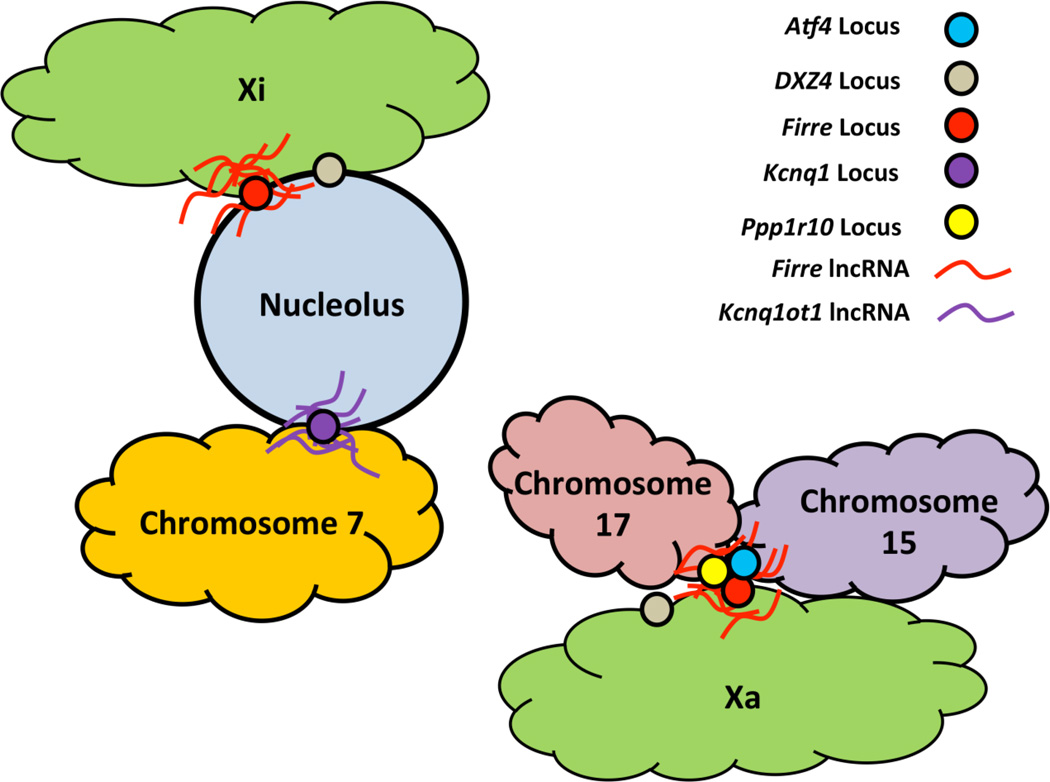
Figure 2. Schematic of lncRNAs Facilitating Trans-Chromosomal Interactions in Mouse.
The Kcnq1ot1 lncRNA tethers the Kcnq1 locus to the nucleolus (Mohammad et al. 2008), while the lncRNA Firre mediates nucleolar association of the Firre and DXZ4 loci (Yang et al. 2015). Firre also mediates transchromosomal interactions of the Firre locus with 5 other loci, including the Atf4 locus on chromosome 15 and the Ppp1r10 locus on chromosome 17 (Hacisuleyman et al. 2014).
What is the functional significance of NADs?
The major question to be answered is whether nucleolar localization affects the biological activities of NAD sequences. Multiple studies have correlated localization of NADs to the PN region with heterochromatin formation and transcriptional silencing (Zhang et al. 2007; Pandey et al. 2008; Mohammad et al. 2008; Fedoriw et al. 2012a; Fedoriw et al. 2012b; Yang et al. 2015). In the example of the Xi, failure to localize to the PN region during S-phase results in decreased heterochromatic silencing marks (Zhang et al. 2007; Yang et al. 2015) without changes in gene expression at most Xi loci (Zhang et al. 2007). Likewise, loci which failed to associate with the nucleolus often re-localize to the NL or PC regions and transcriptional silencing is maintained (van Koningsbruggen et al. 2010; Ragoczy et al. 2015). Together, these data are consistent with the idea that PN localization is one of several functionally overlapping mechanisms for transcriptional repression, although there is a lack of data for a unique role of NAD localization in regulating gene expression. It should be considered that transcriptional regulation may not be the only or even a major outcome of NAD localization to the PN region. In contrast, the data from the Heun laboratory in the Drosophila system points to genome stability as a critical outcome of proper NLP (NPM1) and Modulo (NCL) function (Padeken et al. 2013). There is precedent for deleterious hyper-recombination phenotypes within the rDNA upon chromatin perturbation in budding yeast (Lindstrom et al. 2011; Cesarini et al. 2012; Ide et al. 2013), but these types of events have not been examined in conjunction with NAD analysis in metazoans.
We do not presently understand how NADs migrate among different repressive nuclear regions, nor is it likely that we know all of the proteins and/or RNA factors involved in these transitions. Further study is needed to determine whether and how association of NADs with the nucleolus affects nucleolar structure and function. These questions will be especially challenging to explore because many of the proteins required for NAD localization also regulate rDNA transcription and/or nucleolar structure. For example, CTCF (van de Nobelen et al. 2010; Huang et al. 2013), NCL (Roger et al. 2002; Rickards et al. 2007; Cong et al. 2012), NPM1 (Murano et al. 2008), and Ki-67 (Rahmanzadeh et al. 2007; Booth et al. 2014) all regulate rDNA transcription. Likewise, depletion of Modulo (NCL) in flies disrupts nucleolar structure as demonstrated by immunofluorescence (Padeken et al. 2013), and Ki-67 depletion in human cells results in fewer and smaller nucleoli (Booth et al. 2014). However, not all perturbations necessarily affect all structural aspects of the nucleolus. For example, depletion of CAF-1 p150 causes mislocalization of multiple nucleolar proteins (Smith et al. 2014) but does not appear to alter the macrostructure of the rDNA (videos 1 and 2). Therefore, it remains to be determined whether the structure or function phenotypes observed in the depletion of these proteins is caused by mislocalization of NADs or merely correlated with it, and mutations that separate these functions will be required to assess this.
Understanding the structure of the genome is critical for understanding human disease, as many diseases are dependent upon genomic three-dimensional structure (reviewed in (Misteli 2010)). For example, translocations are associated with a variety of different cancers, and these translocation events occur most frequently between genomic elements in close proximity to one another (Zhang et al. 2012). This has been particularly well documented in the case of ABL-BCR translocations that drive chronic myeloid leukemia, because the BCR gene on chromosome 9 is often found in close proximity to the ABL gene on chromosome 22 in hematopoietic cells (Lukášová et al. 1997; Neves et al. 1999). Although the periphery of the nucleolus is a small fraction of nuclear volume, many higher order chromosomal interactions occur there. Discovering the extent to which this organizational hub is coordinated with other nuclear elements will be critical in comprehending the three-dimensional structures of metazoan genomes, and how these structures are perturbed in disease states.
Supplementary Material
Video 1 shows a HeLa S3 nucleus after 72 hours of expression of a control shRNA directed against luciferase. DAPI staining is in blue (A), the nucleolar and Cajal body protein Nopp140 in green (B) and the ribosomal rDNA in red (C). Video 2 shows a HeLa S3 nucleus after 72 hours of expression of an shRNA directed against CAF-1 p150 with the same channels as in Video 1. Note that Nopp140 does not localize to the nucleolus in video 2C, but does maintain localization to two different Cajal bodies. In contrast, the morphology of the nucleolus visualized by rDNA hybridization is not significantly different. (Methods, probes, and antibodies described in (Smith et al. 2014). The Nopp140 antibody (RS8) was a generous gift of U. Thomas Meier, Albert Einstein College of Medicine, New York, NY (Kittur et al. 2007). Z-stack images were taken on a Leica TCS SP5 II Laser Scanning Confocal Microscope and videos were generated using the Leica Application Suite AF version 2.5.1.6757)
Acknowledgements
Funding
This study was funded by NIH R01 GM055712. We thank Dr. Thoru Pederson for helpful review of this manuscript.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by either of the authors.
References
- Barr ML, Bertram EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, et al. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Barton DE, David FN, Merrington M. The relative positions of the chromosomes in the human cell in mitosis. Ann Hum Genet. 1965;29:139–146. doi: 10.1111/j.1469-1809.1965.tb00509.x. [DOI] [PubMed] [Google Scholar]
- Boisvert F, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Booth DG, Takagi M, Sanchez-Pulido L, et al. Ki-67 is a PP1-interacting protein that organises the mitotic chromosome periphery. Elife. 2014;2014:1–22. doi: 10.7554/eLife.01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois CA, Laquerriere F, Hemon D, et al. New data on the in situ position of the inactive X chromosome in the interphase nucleus of human fibroblasts. Hum Genet. 1985;69:122–129. doi: 10.1007/BF00293281. [DOI] [PubMed] [Google Scholar]
- Boyne JR, Whitehouse A. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc Natl Acad Sci U S A. 2006;103:15190–15195. doi: 10.1073/pnas.0604890103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger JM, Kill IR, Lichter P. Association of pKi-67 with satellite DNA of the human genome in early G1 cells. Chromosom Res. 1998;6:13–24. doi: 10.1023/a:1009210206855. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio a, Rupert JL, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Bugler B, Caizergues-Ferrer M, Bouche G, et al. Detection and localization of a class of proteins immunologically related to a 100-kDa nucleolar protein. Eur J Biochem. 1982;128:475–480. doi: 10.1111/j.1432-1033.1982.tb06989.x. [DOI] [PubMed] [Google Scholar]
- Bullwinkel J, Baron-Lühr B, Lüdemann A, et al. Ki-67 protein is associated with ribosomal RNA transcription in quiescent and proliferating cells. J Cell Physiol. 2006;206:624–635. doi: 10.1002/jcp.20494. [DOI] [PubMed] [Google Scholar]
- Cesarini E, D’Alfonso A, Camilloni G. H4K16 acetylation affects recombination and ncRNA transcription at rDNA in Saccharomyces cerevisiae. Mol Biol Cell. 2012;23:2770–2781. doi: 10.1091/mbc.E12-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Tian Y, Shu W, et al. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS One. 2012 doi: 10.1371/journal.pone.0041374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, et al. Integration of External Signaling Pathways with the Core Transcriptional Network in Embryonic Stem Cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cheutin T, O’Donohue MF, Beorchia A, et al. Three-dimensional organization of pKi-67: a comparative fluorescence and electron tomography study using FluoroNanogold. J Histochem Cytochem. 2003;51:1411–1423. doi: 10.1177/002215540305101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, et al. Systematic discovery of xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciufo LF, Brown JD. Nuclear export of yeast signal recognition particle lacking Srp54p by the Xpo1p/Crm1p NES-dependent pathway. Curr Biol. 2000;10:1256–1264. doi: 10.1016/s0960-9822(00)00743-0. [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong R, Das S, Ugrinova I, et al. Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase i transcription. Nucleic Acids Res. 2012;40:9441–9454. doi: 10.1093/nar/gks720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in Maintaining X Chromosome Inactivation. J Cell Biol. 2001;153:773–783. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMare LE, Leng J, Cotney J, et al. The genomic landscape of cohesin-Associated chromatin interactions. Genome Res. 2013;23:1224–1234. doi: 10.1101/gr.156570.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dialynas G, Speese S, Budnik V, et al. The role of Drosophila Lamin C in muscle function and gene expression. Development. 2010;137:3067–3077. doi: 10.1242/dev.048231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escande ML, Gas N, Stevens BJ. Immunolocalization of the 100 K nucleolar protein in CHO cells. Biol Cell. 1985;53:99–109. doi: 10.1111/j.1768-322x.1985.tb00359.x. [DOI] [PubMed] [Google Scholar]
- Fedoriw AM, Calabrese JM, Mu W, et al. Differentiation-driven nucleolar association of the mouse imprinted Kcnq1 locus. G3 (Bethesda) 2012a;2:1521–1528. doi: 10.1534/g3.112.004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoriw AM, Starmer J, Yee D, Magnuson T. Nucleolar association and transcriptional inhibition through 5S rDNA in mammals. PLoS Genet. 2012b doi: 10.1371/journal.pgen.1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, Higgins MJ. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet. 2002;32:426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Black BE, et al. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Gaillard PH, Martini EM, Kaufman PD, et al. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- Gerdes J, Schwab U, Lemke H, Stein H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;31:13–20. doi: 10.1002/ijc.2910310104. [DOI] [PubMed] [Google Scholar]
- Ghetti A, Piñol-Roma S, Michael WM, et al. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CM, Almouzni G. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 2003;22:5163–5174. doi: 10.1093/emboj/cdg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Deinert K, Hurt E, Simos G. Biogenesis of the Signal Recognition Particle (Srp) Involves Import of Srp Proteins into the Nucleolus, Assembly with the Srp-Rna, and Xpo1p-Mediated Export. J Cell Biol. 2001;153:745–762. doi: 10.1083/jcb.153.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, Engelke DR. Spatial organization of transcription by RNA polymerase III. Nucleic Acids Res. 2006;34:4826–4836. doi: 10.1093/nar/gkl656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MP, Huang S, Black DL. Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol Biol Cell. 2004;15:774–786. doi: 10.1091/mbc.E03-09-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz E. Nukleolar und chromosomen in der gattung. Vicia Planta. 1931;15:495–505. [Google Scholar]
- Herrera JE, Savkur R, Olson MO. The ribonuclease activity of nucleolar protein B23. Nucleic Acids Res. 1995;23:3974–3979. doi: 10.1093/nar/23.19.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Yu Z, Zhang S, et al. Drosophila CAF-1 regulates HP1-mediated epigenetic silencing and pericentric heterochromatin stability. J Cell Sci. 2010;123:2853–2861. doi: 10.1242/jcs.063610. [DOI] [PubMed] [Google Scholar]
- Huang K, Jia J, Wu C, et al. Ribosomal RNA gene transcription mediated by the master genome regulator protein CCCTC-binding factor (CTCF) is negatively regulated by the condensin complex. J Biol Chem. 2013;288:26067–26077. doi: 10.1074/jbc.M113.486175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Deerinck TJ, Ellisman MH, Spector DL. The dynamic organization of the perinucleolar compartment in the cell nucleus. J Cell Biol. 1997;137:965–974. doi: 10.1083/jcb.137.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide S, Saka K, Kobayashi T. Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast. PLoS Genet. 2013 doi: 10.1371/journal.pgen.1003410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Pederson T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc Natl Acad Sci U S A. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RV, Thor AD, Wang C, et al. Perinucleolar compartment prevalence has an independent prognostic value for breast cancer. Cancer Res. 2005;65:246–253. [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. doi: S0092-8674(05)80015-7 [pii] [DOI] [PubMed] [Google Scholar]
- Kill IR. Localisation of the Ki-67 antigen within the nucleolus. Evidence for a fibrillarin-deficient region of the dense fibrillar component. J Cell Sci. 1996;109(Pt 6):1253–1263. doi: 10.1242/jcs.109.6.1253. [DOI] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, et al. Analysis of the Vertebrate Insulator Protein CTCF-Binding Sites in the Human Genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- Kind J, van Steensel B. Genome-nuclear lamina interactions and gene regulation. Curr Opin Cell Biol. 2010;22:320–325. doi: 10.1016/j.ceb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Kittur N, Zapantis G, Aubuchon M, et al. The nucleolar channel system of human endometrium is related to endoplasmic reticulum and R-rings. Mol Biol Cell. 2007;18:2296–2304. doi: 10.1091/mbc.E07-02-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- Lee JT. Epigenetic Regulation by Long Noncoding RNAs. Sci (Washington, DC, U S) 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Li YP. Protein B23 is an important human factor for the nucleolar localization of the human immunodeficiency virus protein Tat. J Virol. 1997;71:4098–4102. doi: 10.1128/jvi.71.5.4098-4102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom DL, Leverich CK, Henderson KA, Gottschling DE. Replicative age induces mitotic recombination in the ribosomal RNA gene cluster of Saccharomyces cerevisiae. PLoS Genet. 2011 doi: 10.1371/journal.pgen.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström MS, Zhang Y. Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J Biol Chem. 2008;283:15568–15576. doi: 10.1074/jbc.M801151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischwe MA, Richards RL, Busch RK, Busch H. Localization of phosphoprotein C23 to nucleolar structures and to the nucleolus organizer regions. Exp Cell Res. 1981;136:101–109. doi: 10.1016/0014-4827(81)90041-0. [DOI] [PubMed] [Google Scholar]
- Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- Lukášová E, Kozubek S, Kozubek M, et al. Localisation and distance between ABL and BCR genes in interphase nuclei of bone marrow cells of control donors and patients with chronic myeloid leukaemia. Hum Genet. 1997;100:525–535. doi: 10.1007/s004390050547. [DOI] [PubMed] [Google Scholar]
- Lyon M. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Maggi LB, Kuchenruether M, Dadey DYa, et al. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the Mammalian ribosome. Mol Cell Biol. 2008;28:7050–7065. doi: 10.1128/MCB.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini-DiNardo D, Steele SJS, Levorse JM, et al. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindill DMJ, Risebro CA, Smart N, et al. Nucleolar release of Hand1 acts as a molecular switch to determine cell fate. Nat Cell Biol. 2007;9:1131–1141. doi: 10.1038/ncb1633. [DOI] [PubMed] [Google Scholar]
- Martini E, Roche DMJ, Marheineke K, et al. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J Cell Biol. 1998;143:563–575. doi: 10.1083/jcb.143.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Frey MR, Margelot K, Wolin SL. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The relationship of a particular chro-mosomal element to the development of the nucleoli in Zea mays. Z ZellforschMikrosk. 1934;21:294–398. [Google Scholar]
- McHugh CA, Chen CK, Chow A, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015 doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalik J, Yeoman LC, Busch H. Nucleolar localization of protein B23 (37/5.1) by immunocytochemical techniques. Life Sci. 1981;28:1371–1379. doi: 10.1016/0024-3205(81)90411-2. [DOI] [PubMed] [Google Scholar]
- Misteli T. Higher-order genome organization in human disease. Cold Spring Harb. Perspect. Biol. 2010;2:85–92. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Pandey RR, Nagano T, et al. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol Cell Biol. 2008;28:3713–3728. doi: 10.1128/MCB.02263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murano K, Okuwaki M, Hisaoka M, Nagata K. Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity. Mol Cell Biol. 2008;28:3114–3126. doi: 10.1128/MCB.02078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- Németh A, Conesa A, Santoyo-Lopez J, et al. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh A, Längst G. Genome organization in and around the nucleolus. Trends Genet. 2011;27:149–156. doi: 10.1016/j.tig.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Neves H, Ramos C, da Silva MG, et al. The nuclear topography of ABL, BCR, PML, and RARalpha genes: evidence for gene proximity in specific phases of the cell cycle and stages of hematopoietic differentiation. Blood. 1999;93:1197–1207. [PubMed] [Google Scholar]
- Norton JT, Pollock CB, Wang C, et al. Perinucleolar compartment prevalence is a phenotypic pancancer marker of malignancy. Cancer. 2008;113:861–869. doi: 10.1002/cncr.23632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padeken J, Heun P. Nucleolus and nuclear periphery: Velcro for heterochromatin. Curr Opin Cell Biol. 2014;28C:54–60. doi: 10.1016/j.ceb.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Padeken J, Mendiburo MJ, Chlamydas S, et al. The nucleoplasmin homolog NLP mediates centromere clustering and anchoring to the nucleolus. Mol Cell. 2013;50:236–249. doi: 10.1016/j.molcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Ceribelli M, Singh PB, et al. NF-Y regulates the antisense promoter, bidirectional silencing, and differential epigenetic marks of the Kcnq1 imprinting control region. J Biol Chem. 2004;279:52685–52693. doi: 10.1074/jbc.M408084200. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, et al. Kcnq1ot1 Antisense Noncoding RNA Mediates Lineage-Specific Transcriptional Silencing through Chromatin-Level Regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pederson T. The nucleolus. Cold Spring Harb Perspect Biol. 2011;3:1–15. doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T, Politz JC. The nucleolus and the four ribonucleoproteins of translation. J. Cell Biol. 2000;148:1091–1095. doi: 10.1083/jcb.148.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, et al. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Plath K, Talbot D, Hamer KM, et al. Developmentally regulated alterations in polycomb repressive complex 1 proteins on the inactive X chromosome. J Cell Biol. 2004;167:1025–1035. doi: 10.1083/jcb.200409026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Yarovoi S, Kilroy SM, et al. Signal recognition particle components in the nucleolus. Proc Natl Acad Sci. 2000;97:55–60. doi: 10.1073/pnas.97.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock C, Huang S. The perinucleolar compartment. J Cell Biochem. 2010;107:189–193. doi: 10.1002/jcb.22107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Quivy JP, Gérard A, Cook AJL, et al. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol. 2008;15:972–979. doi: 10.1038/nsmb.1470. [DOI] [PubMed] [Google Scholar]
- Ragoczy T, Telling A, Scalzo D, et al. Functional redundancy in the nuclear compartmentalization of the late-replicating genome. Nucleus. 2015;5:626–635. doi: 10.4161/19491034.2014.990863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmanzadeh R, Hüttmann G, Gerdes J, Scholzen T. Chromophore-assisted light inactivation of pKi-67 leads to inhibition of ribosomal RNA synthesis. Cell Prolif. 2007;40:422–430. doi: 10.1111/j.1365-2184.2007.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Reese BE, Bachman KE, Baylin SB, Rountree MR. The methyl-CpG binding protein MBD1 interacts with the p150 subunit of chromatin assembly factor 1. Mol Cell Biol. 2003;23:3226–3236. doi: 10.1128/MCB.23.9.3226-3236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickards B, Flint SJ, Cole MD, LeRoy G. Nucleolin is required for RNA polymerase I transcription in vivo. Mol Cell Biol. 2007;27:937–948. doi: 10.1128/MCB.01584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger B, Moisand A, Amalric F, Bouvet P. Repression of RNA polymerase I transcription by nucleolin is independent of the RNA sequence that is transcribed. J Biol Chem. 2002;277:10209–10219. doi: 10.1074/jbc.M106412200. [DOI] [PubMed] [Google Scholar]
- Sáez-Vasquez J, Caparros-Ruiz D, Barneche F, Echeverría M. A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol Cell Biol. 2004;24:7284–7297. doi: 10.1128/MCB.24.16.7284-7297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur RS, Olson MOJ. Preferential cleavage in pre-ribosomal RNA by protein B23 endoribonuclease. Nucleic Acids Res. 1998;26:4508–4515. doi: 10.1093/nar/26.19.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: The fascinating nuclear body. Histochem Cell Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Matheson TD, Trombly DJ, et al. A separable domain of the p150 subunit of human chromatin assembly factor-1 promotes protein and chromosome associations with nucleoli. Mol Biol Cell. 2014;25:2866–2881. doi: 10.1091/mbc.E14-05-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag F, Schmidt K, Kleinschmidt JA. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci U S A. 2010;107:10220–10225. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Ochs RL, Busch H. Silver staining, immunofluorescence, and immunoelectron microscopic localization of nucleolar phosphoproteins B23 and C23. Chromosoma. 1984;90:139–148. doi: 10.1007/BF00292451. [DOI] [PubMed] [Google Scholar]
- Steffensen DM, Duffey P, Prensky W. Localisation of 5S ribosomal RNA genes on human chromosome 1. Nature. 1974;252:741–743. doi: 10.1038/252741a0. [DOI] [PubMed] [Google Scholar]
- Stults DM, Killen MW, Pierce HH, Pierce AJ. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008;18:13–18. doi: 10.1101/gr.6858507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur N, Tiwari VK, Thomassin H, et al. An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol Cell Biol. 2004;24:7855–7862. doi: 10.1128/MCB.24.18.7855-7862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen JL, Verona RI, Bartolomei MS. X-tra! X-tra! News from the Mouse X Chromosome. Dev Biol. 2006;298:344–353. doi: 10.1016/j.ydbio.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, et al. Identification of a (CUG)(n) triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin G. Repertorium fur Anatomie und Physiologie. Verlag von Veit und Comp Berlin. 1836;1:1–293. [Google Scholar]
- Valentin G. Repertorium fur Anatomie und Physiologie. Verlag von Veit und Comp Berlin. 1839;4:1–275. [Google Scholar]
- Van de Nobelen S, Rosa-Garrido M, Leers J, et al. CTCF regulates the local epigenetic state of ribosomal DNA repeats. Epigenetics Chromatin. 2010;3:19. doi: 10.1186/1756-8935-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser AA, Yuh P, Heald R. The perichromosomal layer. Chromosoma. 2005;114:377–388. doi: 10.1007/s00412-005-0021-9. [DOI] [PubMed] [Google Scholar]
- Van Koningsbruggen S, Gierlinski M, Schofield P, et al. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21:3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- Volders PJ, Helsens K, Wang X, et al. LNCipedia: A database for annotated human IncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:246–251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R. Einige Bemerkungen und Fragen uber das Keimblaschen (vesicular germinativa) Muller’s Arch Anat Physiol Wiss Med. 1835:373–377. [Google Scholar]
- Wang C, Politz JC, Pederson T, Huang S. RNA polymerase III transcripts and the PTB protein are essential for the integrity of the perinucleolar compartment. Mol Biol Cell. 2003;14:2425–2435. doi: 10.1091/mbc.E02-12-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, et al. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Weierich C, Brero A, Stein S, et al. Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosom Res. 2003;11:485–502. doi: 10.1023/a:1025016828544. [DOI] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Deng X, Ma W, et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015 doi: 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Maggi LB, Brady SN, et al. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol Cell Biol. 2006;26:3798–3809. doi: 10.1128/MCB.26.10.3798-3809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF Tethers an Insulator to Subnuclear Sites, Suggesting Shared Insulator Mechanisms across Species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- Zhang L-F, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video 1 shows a HeLa S3 nucleus after 72 hours of expression of a control shRNA directed against luciferase. DAPI staining is in blue (A), the nucleolar and Cajal body protein Nopp140 in green (B) and the ribosomal rDNA in red (C). Video 2 shows a HeLa S3 nucleus after 72 hours of expression of an shRNA directed against CAF-1 p150 with the same channels as in Video 1. Note that Nopp140 does not localize to the nucleolus in video 2C, but does maintain localization to two different Cajal bodies. In contrast, the morphology of the nucleolus visualized by rDNA hybridization is not significantly different. (Methods, probes, and antibodies described in (Smith et al. 2014). The Nopp140 antibody (RS8) was a generous gift of U. Thomas Meier, Albert Einstein College of Medicine, New York, NY (Kittur et al. 2007). Z-stack images were taken on a Leica TCS SP5 II Laser Scanning Confocal Microscope and videos were generated using the Leica Application Suite AF version 2.5.1.6757)