Abstract
Recently, neuroimaging-based Alzheimer’s disease (AD) or mild cognitive impairment (MCI) diagnosis has attracted researchers in the field, due to the increasing prevalence of the diseases. Unfortunately, the unfavorable high-dimensional nature of neuroimaging data, but a limited small number of samples available, makes it challenging to build a robust computer-aided diagnosis system. Machine learning techniques have been considered as a useful tool in this respect and, among various methods, sparse regression has shown its validity in the literature. However, to our best knowledge, the existing sparse regression methods mostly try to select features based on the optimal regression coefficients in one step. We argue that since the training feature vectors are composed of both informative and uninformative or less informative features, the resulting optimal regression coefficients are inevidently affected by the uninformative or less informative features. To this end, we first propose a novel deep architecture to recursively discard uninformative features by performing sparse multi-task learning in a hierarchical fashion. We further hypothesize that the optimal regression coefficients reflect the relative importance of features in representing the target response variables. In this regard, we use the optimal regression co-efficients learned in one hierarchy as feature weighting factors in the following hierarchy, and formulate a weighted sparse multi-task learning method. Lastly, we also take into account the distributional characteristics of samples per class and use clustering-induced subclass label vectors as target response values in our sparse regression model. In our experiments on the ADNI cohort, we performed both binary and multi-class classification tasks in AD/MCI diagnosis and showed the superiority of the proposed method by comparing with the state-of-the-art methods.
Keywords: Alzheimer’s disease (AD), Mild cognitive impairment (MCI), Feature selection, Multi-task learning, Deep architecture, Sparse least squared regression, Magnetic resonance imaging (MRI), Positron emission topography (PET)
Introduction
As the population becomes older, the world is now facing an epidemic of dementia, the loss of mental functions such as memory, thinking, and reasoning, each of which is sufficient enough to interfere a person’s activities of daily life. Among various causes of dementia, Alzheimer’s disease (AD) is the most prevalent in elderly people, rising significantly every year in terms of the proportion of cause of death (Alzheimer’s Association 2012). Furthermore, it is reported that people with mild cognitive impairment (MCI), known as precursor to dementia in AD, progress to AD with an average conversion rate of 10 % per year (Busse et al. 2006; Alzheimer’s Association 2012). Although there is currently no pharmaceutical medicine to recover AD/MCI back to cognitive normal (CN), it is still important to detect the diseases for timely treatments that possibly delay the progress. Thus, it is of great interest for AD/MCI diagnosis or prognosis in the clinic.
With the advent of neuroimaging tools such as magnetic resonance imaging (MRI), positron emission tomography (PET), and functional MRI, many researchers have been devoting their efforts to investigate the underlying biological or neurological mechanisms and also to discover biomarkers for AD/MCI diagnosis or prognosis (Li et al. 2012; Zhang and Shen 2012). Recent studies have shown that information fusion of multiple modalities can help enhance the diagnostic performance (Perrin et al. 2009; Kohannim et al. 2010; Walhovd et al. 2010; Cui et al. 2011; Hinrichs et al. 2011; Zhang et al. 2011; Westman et al. 2012; Yuan et al. 2012; Zhang and Shen 2012; Suk et al. 2015). The main challenge in AD/MCI diagnosis or prognosis with neuroimaging arises from the fact that, while the data dimensionality is intrinsically high, in general, a small number of samples are available. In this regard, machine learning has been playing a pivotal role to overcome this so-called “large p, small n” problem (West 2003). Broadly, we can categorize the existing methods into a feature dimension-reduction approach and a feature selection approach. The feature dimension-reduction approach transforms the original features in an ambient space into a lower dimensional subspace, while the feature selection approach finds informative features in the original space. In neuroimaging data analysis, feature selection techniques have drawn much attention these days, due to its interpretational easiness of the results. In this work, we focus on the feature selection approach.
Among different feature selection techniques, sparse (least squares) regression methods, e.g., ℓ1-penalized linear regression (Tibshirani 1994), ℓ2,1-penalized group sparse regression (Yuan and Lin 2006; Nie et al. 2010), and their variants (Roth 2004; Wang et al. 2011; Wan et al. 2012; Zhu et al. 2014), have attracted researchers because of their theoretical strengths and effectiveness in various applications (Varoquaux et al. 2010; Fazli et al. 2011; de Brecht and Yamagishi 2012; Yuan et al. 2012; Zhang and Shen 2012; Suk et al. 2015).
For example, Wang et al. proposed a sparse multi-task1 regression and feature selection method to jointly analyze the neuroimaging and clinical data in prediction of the memory performance (Wang et al. 2011), where ℓ1- and ℓ2,1-norm regularizations were used for sparsity and facilitation of multi-task learning, respectively. Zhang and Shen exploited an ℓ2,1-norm based group sparse regression method to select features that could be used to jointly represent the clinical status, e.g., AD, MCI, or CN, and two clinical scores of Mini-Mental State Examination (MMSE) and Alzheimer’s Disease Assessment Scale-Cognitive (ADAS-Cog) (Zhang and Shen 2012). Varoquaux et al. (2010) formulated the subject-level functional connectivity estimation as multivariate Gaussian process and imposed a group constraint for a common structure on the graphical model in the population. Suk et al. (2013) proposed a supervised discriminative group sparse representation to estimate functional connectivity from fMRI by penalizing a large within-class variance and a small between-class variance of features. Recently, Yuan et al. (2012), Xiang et al. (2014), and Thung et al. (2014), independently, proposed a sparse regression-based feature selection method for AD/MCI diagnosis to maximally utilize features from multiple sources by focusing on a missing modality problem.
In the context of the data distribution, the previous sparse regression methods mostly assumed a unimodal distribution for a same group of subjects. However, due to the inter-subject variability in the same group (Fotenos et al. 2005; Noppeney et al. 2006; DiFrancesco et al. 2008), it is highly likely for neuroimaging data to have a complex data distribution, e.g., mixture of Gaussians. To this end, Suk et al. (2014) recently proposed a subclass-based sparse multi-task learning method, where they approximated the complex data distribution per class by means of clustering and defined subclasses to better encompass the distributional characteristics in feature selection.
Note that the above-mentioned sparse regression methods find the optimal regression coefficients for the respective objective function in one step, i.e., a single hierarchy, using the training feature vectors as regressors. Since the training feature vectors are composed of both informative and uninformative or less informative features, the resulting optimal regression coefficients are inevidently affected by uninformative or less informative features2. While the regularization terms drive the regression coefficients of the uninformative or less informative features to be zero or close to zero, and thus we can discard the corresponding features by thresholding, it is still problematic to find the optimal threshold for feature selection. As for the subclass-based feature selection method (Suk et al. 2014), the clustering is performed with the original full features. Therefore, the clustering results can be also affected by uninformative or less informative features, which sequentially can influence the sparse multi-task learning, feature selection, and classification accuracy.
In this paper, we propose a deep sparse multi-task learning method that can mitigate the effect of uninformative or less informative features in feature selection. Specifically, we iteratively perform subclass-based sparse multi-task learning by discarding uninformative features in a hierarchical fashion. That is, in each hierarchy, we first cluster the current feature samples for each original class. Based on the clustering results, we then assign new label vectors and perform sparse multi-task learning with an ℓ2,1-norm regularization. It should be noted that, unlike the conventional multi-task learning methods, which treat all features equally, we further propose to utilize the optimal regression coefficients learned in the lower hierarchy as context information to weight features adaptively. We validate the effectiveness of the proposed method on the ADNI cohort by comparing with the state-of-the-art methods.
Our main contributions can be threefold:
We propose a novel deep architecture to recursively discard uninformative features by performing sparse multi-task learning in a hierarchical fashion. The rationale of the proposed hierarchical feature selection is that, while the convex optimization algorithm finds optimal regression coefficients, it is still affected by the less informative features. Therefore, if we can discard uninformative features and perform the sparse multi-task learning iteratively, the optimal solution can be more robust to less informative features, and thus to select task-relevant features.
We also devise a weighted sparse multi-task learning using the optimal regression coefficients learned in one hierarchy as feature-adaptive weighting factors in the next deeper hierarchy. In this way, we can adaptively assign different weights for different features in each hierarchy and the features of small weights, which survived in the lower hierarchy, are less likely to be selected in the deeper hierarchy.
Motivated by Suk et al.’s work (2014), we also take into account the distributional characteristics of samples in each class and define clustering-induced label vectors. That is, in each hierarchy, we define subclasses by clustering the training samples but with only the selected feature set from the lower hierarchy, and then assign new label vectors. By taking this new label vectors as target response values, we perform the proposed weighted sparse multi-task learning.
Materials and image processing
Subjects
In this work, we use the ADNI cohort3, but consider only the baseline MRI, 18-fluoro-deoxyglucose PET, and cerebrospinal fluid (CSF) data acquired from 51 AD, 99 MCI, and 52 CN subjects4. For the MCI subjects, they were clinically further subdivided into 43 progressive MCI (pMCI), who progressed to AD in 18 months, and 56 stable MCI (sMCI), who did not progress to AD in 18 months. We summarize the demographics of the subjects in Table 1.
Table 1.
Demographic and clinical information of the subjects
| AD (N = 51) | Progressive MCI (N = 43) | Stable MCI (N = 56) | CN (N = 52) | |
|---|---|---|---|---|
| Female/male | 18/33 | 15/28 | 17/39 | 18/34 |
| Age (mean ± SD) | 75.2 ± 7.4 [59–88] | 75.7 ± 6.9 [58–88] | 75.0 ± 7.1 [55–89] | 75.3 ± 5.2 [62–85] |
| Education (mean ± SD) | 14.7 ± 3.6 [4–20] | 15.4 ± 2.7 [10–20] | 14.9 ± 3.3 [8–20] | 15.8 ± 3.2 [8–20] |
| MMSE (mean ± SD) | 23.8 ± 2.0 [20–26] | 26.9 ± 2.7 [20–30] | 27.0 ± 3.2 [18–30] | 29 ± 1.2 [25–30] |
| CDR (mean ± SD) | 0.7 ± 0.3 [0.5–1] | 0.5 ± 0 [0.5–0.5] | 0.5 ± 0 [0.5–0.5] | 0 ± 0 [0–0] |
MMSE mini-mental state examination, CDR clinical dementia rating, N number of subjects, SD standard deviation [min–max]
With regard to the general eligibility criteria in ADNI, subjects were in the age of between 55 and 90 with a study partner, who could provide an independent evaluation of functioning. General inclusion/exclusion criteria5 are as follows: (1) healthy subjects: Mini-Mental State Examination (MMSE) scores between 24 and 30 (inclusive), a Clinical Dementia Rating (CDR) of 0, non-depressed, non-MCI, and non-demented; (2) MCI subjects: MMSE scores between 24 and 30 (inclusive), a memory complaint, objective memory loss measured by education adjusted scores on Wechsler Memory Scale Logical Memory II, a CDR of 0.5, absence of significant levels of impairment in other cognitive domains, essentially preserved activities of daily living, and an absence of dementia; and (3) mild AD: MMSE scores between 20 and 26 (inclusive), CDR of 0.5 or 1.0, and meets the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS/ADRDA) criteria for probable AD.
Image processing and feature extraction
The MRI images were preprocessed by applying the typical procedures of Anterior Commissure (AC)–Posterior Commissure (PC) correction, skull stripping, and cerebellum removal. Specifically, we used MIPAV software6 for AC–PC correction, resampled images to 256 × 256 × 256, and applied N3 algorithm (Sled et al. 1998) to correct intensity inhomogeneity. An accurate and robust skull stripping (Wang 2014) was performed, followed by cerebellum removal. We further manually reviewed the skull-stripped images to ensure the clean and dura removal. Then, FAST in FSL package7 Zhang et al. (2001) was used for structural MRI image segmentation into three tissue types of gray matter (GM), white matter (WM) and CSF. We finally parcellated them into 93 regions of interest (ROIs) by warping Kabani et al.’s atlas (1998) to each subject’s space via HAMMER (Shen and Davatzikos 2002).
In this work, we considered only GM for classification, because of its relatively high relatedness to AD/MCI compared to WM and CSF (Liu et al. 2012). Regarding PET images, they were rigidly aligned to the corresponding MRI images, and then applied the parcellation propagated from the atlas by registration.
For each ROI, we used the GM tissue volume from MRI, and the mean intensity from PET as features, which are widely used in the field for AD/MCI diagnosis (Davatzikos et al. 2011; Hinrichs et al. 2011; Zhang and Shen 2012; Suk et al. 2015). Therefore, we have 93 features from an MRI image and the same dimensional features from a PET image. In addition, we have three CSF biomarkers of Aβ42, t-tau, and p-tau as features.
Method
Notations
In this paper, we denote matrices as boldface uppercase letters, vectors as boldface lowercase letters, and scalars as normal italic letters, respectively. For a matrix X = [xij], its i-th row and j-th column are denoted as xi and xj, respectively. We further denote a Frobenius norm and an ℓ2,1-norm of a matrix X as and , respectively. Let 1q and 0q denote q-dimensional row vectors whose elements are all 1 and 0, respectively, and be a cardinality of a set .
Preliminary
Let and denote, respectively, the D neuroimaging features and the corresponding class label vectors of N samples8 for C-class classification. In this work, without loss of generality, we represent a class label with a 0/1 encoding scheme. For example, in a binary classification problem, the class label of each training sample is represented by either o1 = [10] or o2 = [01]. Although it is more general to use scalar values of +1/ − 1 for a binary classification problem, in this work, for general applicability of the proposed method, we use a 0/1 encoding scheme, by which we can naturally apply our method to both binary and multi-class classification problems.
In the context of AD/MCI diagnosis, sparse (least squares) regression methods with different types of regularizers have been used for feature selection in neuroimaging data (Wang et al. 2011; Zhou et al. 2013; Suk et al. 2014; Zhu et al. 2014). The common assumption on these methods is that the target response values, which comprise the class labels in our work, can be predicted by a linear combination of the regressors, i.e., feature values in X, as follows:
| (1) |
where is a regression coefficient matrix and R(W) denotes a regularization function. Note that, since our main goal is to identify a clinical label based on the neuroimaging features, we constrain a common subset of features to be used in predicting the target values. In this regard, we can use an ℓ2,1-norm regularizer for R(W) in Eq. (1) and define a group sparse regression model (Zhou et al. 2013) as follows:
| (2) |
where λ denotes a group sparsity control parameter. By regarding the prediction of each target vector yi (i ∈ {1, …, C}) as a task, we designate this as sparse multi-task learning (SMTL). Due to the use of an ℓ2,1-norm regularizer in Eq. (2), the estimated optimal coefficient matrix will have some zero-valued row vectors, denoting that the corresponding features are not useful in prediction of the target response variables, i.e., class labels. Furthermore, the lower the ℓ2-norm of a row vector, the less informative the corresponding feature in X to represent the target response variables in Y.
In the meantime, while the neuroimaging is highly variable among subjects of a same group, the conventional sparse multi-task learning assumes a unimodal data distribution. That is, it overlooks the complicated distributional characteristics inherent in samples, and thus can fail to select task-relevant features. In this regard, Suk and Shen recently proposed a subclass-based sparse multi-task learning (S2MTL) method (Suk et al. 2014). Specifically, they used a clustering method to discover the complex distributional characteristics and defined subclasses based on the clustering results. Then, they encoded the respective subclasses, i.e., clusters, with their unique codes. Finally, by setting the codes as new label vectors of the training samples, they performed sparse multi-task learning as follows:
| (3) |
where denotes a new label matrix and C′ is the total number of response variables, i.e., the sum of the number of the original classes and the number of subclasses in each original class.
Deep weighted subclass-based sparse multi-task learning
The main limitation of the SMTL and S2MTL methods is that they find the optimal regression coefficients and then select task-relevant features based on the regression coefficients in one step, i.e., a single hierarchy. However, uninformative or less informative features, which are also included in regressors, can affect finding the optimal regression coefficients in both Eqs. (2) and (3). Thus, the features selected in a single hierarchy may not be optimal for classification. To mitigate the effects of uninformative or less informative features in optimizing coefficients and in selecting features, we propose a ‘deep weighted subclass-based sparse multi-task learning’ method. Specifically, rather than selecting features in one step, we iteratively discard uninformative features and perform sparse multi-task learning in a hierarchical fashion. In particular, we devise a novel sparse multi-task learning with a feature-adaptive weighting scheme under the hypothesis that the optimal regression coefficients reflect the relative importance of features in representing target response variables. Motivated by Suk and Shen’s work (2014), we also use the S2MTL framework combined with the proposed feature weighting scheme to reflect the distributional characteristics inherent in samples. Hereafter, we call the proposed method as deep weighted S2MTL (DW-S2MTL).
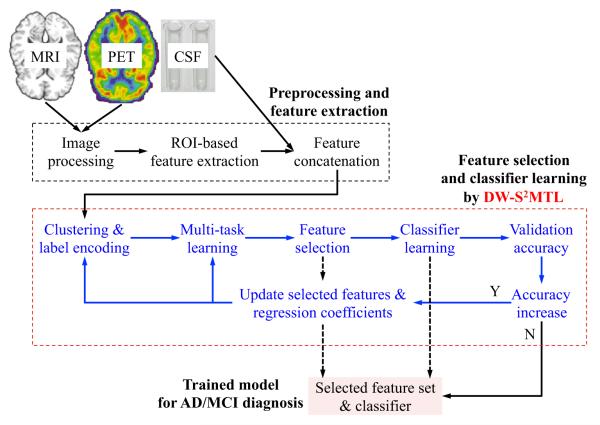
Figure 1 illustrates the overall framework of our method for AD/MCI diagnosis. Given multiple modalities of MRI, PET, and CSF, we extract features from MRI and PET, preceded by image preprocessing as described in “Image processing and feature extraction”, and then concatenate features of all modalities into a long vector for complementary information fusion. Using the concatenated features as regressors and the corresponding class labels as target response values, we perform the proposed DW-S2MTL for feature selection. In this step, we (1) perform S2MTL (clustering and label encoding and multi-task learning), (2) select features based on the learned optimal regression coefficients, (3) train a classifier using training samples but with only the selected features, and (4) compute validation accuracy. If the validation accuracy is higher than the previous one (initially, we set the previous validation accuracy as zero), we iterate the processes of (1) through (4) in a hierarchical manner. That is, in the following hierarchy, we consider only the selected features along with the corresponding regression coefficients learned from the current hierarchy. Once converged, i.e., there is no increase in the validation accuracy, we use the current feature set and the corresponding classifier to identify the clinical label of a testing sample.
Fig. 1.
A framework for AD/MCI diagnosis with the proposed deep weighted subclass-based sparse multi-task learning (DW-S2MTL) method
Now, let us describe the proposed method in detail. Assume that, at the h-th hierarchy, we have the dimension-reduced training samples , where denotes a set of features selected in the (h – 1)-th hierarchy9, along with the corresponding class labels Y. By regarding and Y as our current training samples, we perform clustering to find subclasses for each original class, by which we can facilitate the distributional characteristics in samples.
Earlier, Suk et al. (2014) used the K-means algorithm for this purpose due to its simplicity and computational efficiency. However, since it requires to predefine the number of clusters, i.e., K, for which a cross-validation technique is usually applied in the literature, it is limited to use the K-means algorithm in practical applications. To this end, in this work, we use affinity propagation (Frey and Dueck 2007), which can automatically select the optimal number of clusters and has been successfully applied to a variety of applications (Dueck and Frey 2007; Lu and Carreira-Perpinan 2008; Wang 2010; Shi et al. 2011; Alikhanian et al. 2013). For the details of affinity propagation, please refer to Appendix and Frey and Dueck (2007).
After clustering samples in via affinity propagation, we define subclasses and assign a new label to each sample. Let us consider a binary classification problem and assume that affinity propagation finds and numbers of clusters/exemplars for class 1 and class 2, respectively. Note that we regard the clusters as subclasses of the original class. Then, we define sparse codes for subclasses of the original class 1 and the original class 2 as follows:
where o1 = [10] and o2 = [01] denote the original class labels for class 1 and class 2, respectively, , , and and denote, respectively, subclass-indicator row vector in which only the l-th/m-th element is set to 1 and the others are 0. Thus, the full label set for binary classification becomes:
| (4) |
Now, without loss of generality, based on Eq. (4), we can extend the full label set for C-class classification as follows:
| (5) |
where and oc is a original class indicator row vector. Then, for the n-th training sample at the h-th hierarchy, if it belongs to the original class c and is assigned to a cluster m of the class, then its new label vector is set to .
By regarding the newly assigned label vectors as target response values, i.e., , we can learn the regression coefficients of an S2MTL model in Eq. (3). Here, it is noteworthy that the ℓ2-norm of a row vector in an optimal regression coefficient matrix quantifies the relevance of the corresponding feature in representing the target response variables. In our deep architecture, we use such context information to adaptively weight the selected features in the upper hierarchy. Specifically, we devise a novel weighted sparse multi-task learning method by exploiting the optimal regression coefficients learned in the lower hierarchy as feature weighting factors. We define an adaptive feature weighting vector at the h-th hierarchy as follows:
| (6) |
where is a normalizing constant. In our adaptive feature weighting scheme in Eq. (6), the higher the ℓ2-norm of the optimal regression coefficient vector , the smaller the weight for the i-th feature is assigned. By introducing this feature-adaptive weighting factor into a regularization term of a sparse regression model, we impose that in the upper hierarchy, the features of high ℓ2-norm values from the lower hierarchy have also high regression coefficients; meanwhile, those of low ℓ2-norm values from the lower hierarchy have low regression coefficients and ultimately become zero to be discarded. Thus, we formulate a weighted sparse multi-task learning method as follows:
| (7) |
where , , and ⊙ denotes an element-wise matrix multiplication. Note that the feature weights defined in Eq. (6) are used to guide the selection of informative features in the current hierarchy by adaptively adjusting the penalty levels of different features. That is, by giving small weights for the informative features in representing the target responses, we impose the corresponding regression coefficients to be larger, and thus to survive in feature selection. We should note that, since we use class labels as target responses, features corresponding to low regression coefficients would have low discriminative power for the classification of the respective classes. In this regard, the proposed method can be effective to remove such features by deep learning.
Based on the optimal regression coefficients , we select the features whose regression coefficient vector is non-zero, i.e., . With the selected features, we train a linear support vector machine (SVM), which has been successfully used in many applications (Zhang and Shen 2012; Suk and Lee 2013), and then compute the accuracy on the validation samples. If the validation accuracy is higher than the accuracy in the lower hierarchy10, we move to the next level of hierarchy, to further filter out uninformative features (if exist), and thus to reduce the dimensionality; otherwise, stop the deep learning. Algorithm 1 summarizes the overall procedures of the proposed DW-S2MTL method for feature selection.
Algorithm 1.
Pseudo algorithm of the proposed DW-S2MTL method.
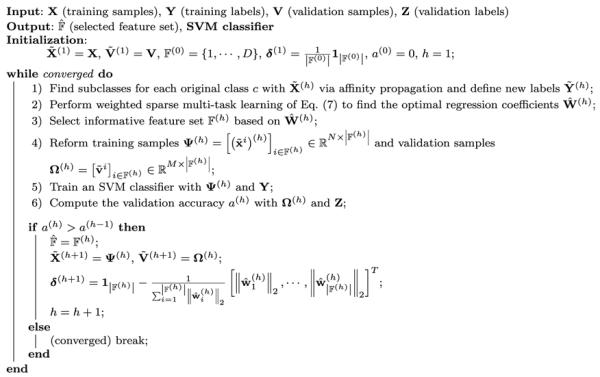
|
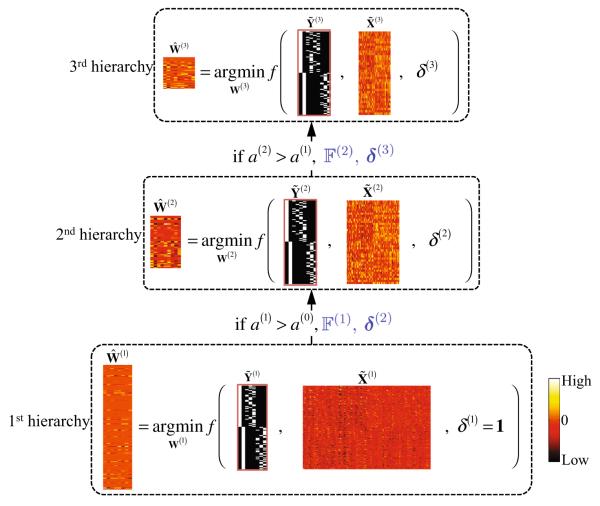
For better understanding, in Fig. 2, we present an example of applying the proposed DW-S2MTL for feature selection in binary classification. In the 1st hierarchy, we have the training feature samples and the new label vectors determined by clustering. In this hierarchy, since we have no prior weight information on the features, we treat all the features equally by setting . Note that the optimization problem in this hierarchy corresponds to S2MTL (Suk et al. 2014). Based on the learned optimal regression coefficients , we select a feature set and define δ(2) by Eq. (6). By taking account of the values of the selected features in and the original class labels Y, we train a linear SVM and compute the classification accuracy a(1) on a validation set. If a(1) is greater than a(0)(= 0), we set and the algorithm proceeds to the next hierarchy. For the 2nd hierarchy, we construct our feature samples from with only the selected features of and define new label vectors via clustering for each original class with feature samples in . We then learn the optimal regression coefficients by solving Eq. (7) with , , and δ(2) as inputs. Again, we select a feature set based on , and train a linear SVM with the feature samples of but only with features in and the original class labels Y. With the trained SVM, we compute the classification accuracy a(2) on a validation set. If the current validation accuracy a(2) is higher than a(1), we update our optimal feature set , compute the feature weights δ(3), and proceed to the 3rd hierarchy.
Fig. 2.
Schematic illustration of the proposed deep weighted subclass-based sparse multi-task learning for feature selection. denotes an objective function in Eq. (7), δ(h) is defined by Eq. (6), and a(h) (a(0) = 0) and denote, respectively, the validation accuracy and a set of the selected features at the h-th hierarchy
In a nutshell, in the h-th hierarchy, we sequentially perform the steps of (1) clustering samples to define subclasses and assigning a new label to the samples, (2) learning the optimal regression coefficients by taking into account the features selected in the (h – 1)-th hierarchy and the regression coefficients , (3) selecting informative feature set based on , (4) reorganizing training and validation samples by discarding the unselected features, and (5) training an SVM classifier and computing the validation accuracy a(h). If the current validation accuracy is higher than the previous one, i.e., a(h–1), which means that the current feature set is better suited for classification than the previous one, we repeat the steps from (1) to (5) until convergence, i.e., no improvement in the validation accuracy. Note that the number of features under consideration reduces gradually as advancing to the higher level in the hierarchy with the respective feature weights determined based on the optimal weight coefficients from the one level below.
Experimental results
In this section, we validate the effectiveness of the proposed deep weighted subclass-based sparse multi-task learning for feature selection in AD/MCI diagnosis. We conducted two sets of experiments, namely, binary and multi-class classification problems. For the binary classification, we considered three tasks: (1) AD vs. CN, (2) MCI vs. CN, and (3) progressive MCI (pMCI), who converted to AD in 18 months, vs. stable MCI (sMCI), who did not converted to AD in 18 months. Meanwhile, for the multi-class classification, we performed two tasks of (1) AD vs. MCI vs. CN (3-class) and (2) AD vs. pMCI vs. sMCI vs. CN (4-class). In the classifications of MCI vs. CN (binary) and AD vs. MCI vs. CN (3-class), we labeled both pMCI and sMCI as MCI.
Experimental setting
For performance comparison, we consider five competing methods as follows:
Sparse multi-task learning (SMTL) (Zhou et al. 2013) that assumes a unimodal data distribution and selects features in a single hierarchy.
Subclass-based SMTL (S2MTL) (Suk et al. 2014) that takes into account a complex data distribution and selects features in a single hierarchy.
Deep weighted SMTL (DW-SMTL) that assumes a unimodal data distribution and selects features in a hierarchical fashion using the proposed deep sparse multi-task learning with a feature weighting scheme.
Deep S2MTL (D-S2MTL) that takes into account a complex data distribution and also selects features in a hierarchical fashion using the proposed deep sparse multi-task learning but without a feature weighting scheme.
Deep weighted S2MTL (DW-S2MTL) that takes account a complex data distribution and also features in a hierarchical fashion using the deep sparse multi-task learning with a feature weighting scheme.
For the S2MTL method, unlike the original work in Suk et al. (2014), we used affinity propagation to define classes in order for fair comparison with D-S2MTL DW-S2MTL. It should be noted that the main difference among the competing methods lies in the characteristics such as the use of data distribution (unimodal or complex), the number of hierarchies (single or multiple), and the use of context information, i.e., feature weights. We compare their characteristics in Table 2.
Table 2.
Characteristics of the competing methods considered in our experiments
| SMTL | S2MTL | DW-SMTL | D-S2MTL | DW-S2MTL | |
|---|---|---|---|---|---|
| Distribution | Unimodal | Complex | Unimodal | Complex | Complex |
| Hierarchy | Single | Single | Multiple | Multiple | Multiple |
| Use of context information | No | No | Yes | No | Yes |
SMTL sparse multi-task learning, S2 MTL subclass-based SMTL, DW-SMTL deep weighted SMTL, D-S2MTL deep S2MTL, DW-S2MTL deep weighted S2MTL
Due to the limited number of samples, we evaluated the performance of all the competing methods by applying a tenfold cross-validation technique in each classification problem and taking the average of the results. Specifically, we randomly partitioned the samples of each class into 10 subsets with approximately equal size without replacement. We then used 9 out of 10 subsets for training and the remaining one for testing. We repeated this process 10 times. It is noteworthy that for fair comparison among the competing methods, we used the same training and testing samples in our cross-validation.
Regarding model selection of the sparsity control parameter λ in sparse regression models and the soft margin parameter C in SVM (Burges 1998), we defined the parameter spaces as λ ∈ {0.001,0.005,0.01,0.05,0.1,0.3,0.5} and C ∈ {2−10, …,25}, and performed a grid search. The parameters that achieved the best classification accuracy in the inner cross-validation were finally used in testing. In our implementation, we used a SLEP toolbox11 for optimization of the respective objective function and an LIBSVM toolbox12 for SVM classifier learning. As for the multi-class classification, we applied a one-versus-all strategy (Milgram et al. 2006) and chose the class which classified the test sample with the greatest margin.
We used 93 MRI features, 93 PET features, and/or 3 CSF features as regressors in all the competing methods. Regarding the multimodality neuroimaging fusion, e.g., MRI + PET (MP for short) and MRI + PET + CSF (MPC for short), we constructed a long feature vector by concatenating features of the modalities.
Performance comparison
Let TP, TN, FP, and FN denote, respectively, true positive, true negative, false positive, and false negative. We considered the following metrics to measure the performance of the methods:
ACCuracy (ACC) = (TP + TN)/(TP + TN + FP + FN)
SENsitivity (SEN) = TP/(TP + FN)
SPECificity (SPEC) = TN/(TN + FP)
Balanced ACcuracy (BAC) = (SEN + SPEC)/2
Positive Predictive Value (PPV) = TP/(TP+FP)
Negative Predictive Value (NPV) = TN/(TN+FN)
The accuracy that counts the number of correctly classified samples in a test set is the most direct metric for comparison among methods. Regarding the sensitivity and specificity, the higher the values of these metrics, the lower the chance of misdiagnosing to the respective clinical label.
Note that in our dataset, since the number of samples available for each class is imbalanced, it is likely to have an inflated performance estimates for two binary classification tasks, i.e., MCI (99) vs. CN (52) and pMCI (43) vs. sMCI (56), and one multi-class classification task, i.e., AD (51) vs. MCI (99) vs. CN (52). For this reason, we also considered a balanced accuracy and positive/negative predictive values (Wei and Dunbrack 2013).
Binary classification results
We summarized the performances of the competing methods with various modalities in Tables 3, 4, 5. In discrimination between AD and CN (Table 3), SMTL achieved the ACCs of 86.55 % (MRI), 80.45 % (PET), 87.64 % (MP), and 92.45 % (MPC), while S2MTL achieved the ACCs of 86.55 % (MRI), 85.36 % (PET), 93.18 % (MP), and 92.36 % (MPC). When applying the proposed deep and feature-adaptive weighting scheme to these methods, we obtained the ACCs of 88.36 % (MRI), 82.45 % (PET), 90.45 % (MP), and 92.45 % (MPC) by DW-SMTL and the ACCs of 90.36 % (MRI), 89.27 % (PET), 93.18 % (MP), and 95.09 % (MPC) by DW-S2MTL. Note that thanks to the proposed deep and feature-adaptive weighting scheme, we could improve the ACCs by 1.85 % (MRI), 2 % (PET), and 2.81 % (MP) in comparison between SMTL and DW-SMTL and by 3.91 % (MRI), 3.91 % (PET), and 2.73 % (MPC) in comparison between S2MTL and DW-S2MTL. Regarding the proposed feature weighting scheme, we could also verify its effectiveness by comparison between D-S2MTL and DW-S2MTL. Overall, the proposed DW-S2MTL outperformed the other four competing methods. It is worth noting that since the discrimination between AD and NC is relatively easier than the other classification tasks described below, all the competing methods achieved good performance, i.e., higher than 90 % in accuracy. Thus, there is no substantial difference among the competing methods.
Table 3.
A summary of the performances for AD vs. CN classification
| Method | Modality | ACC (%) | SEN (%) | SPEC (%) | BAC (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|
| SMTL | MRI | 86.55 ± 3.02 | 88.33 | 84.33 | 86.33 | 87.55 | 85.28 |
| PET | 80.45 ± 3.26 | 80.33 | 80.67 | 80.50 | 88.49 | 69.11 | |
| MP | 87.64 ± 3.31 | 90.33 | 85.00 | 87.67 | 85.53 | 89.96 | |
| MPC | 92.45 ± 2.69 | 94.00 | 90.67 | 92.33 | 92.04 | 92.94 | |
| DW-SMTL | MRI | 88.36 ± 3.20 | 84.33 | 92.33 | 88.33 | 91.55 | 85.68 |
| PET | 82.45 ± 3.22 | 82.33 | 82.67 | 82.50 | 89.70 | 71.85 | |
| MP | 90.45 ± 3.13 | 90.33 | 90.33 | 90.33 | 94.67 | 83.65 | |
| MPC | 92.45 ± 2.69 | 94.00 | 90.67 | 92.33 | 92.05 | 92.94 | |
| S2MTL | MRI | 86.55 ± 3.02 | 78.33 | 94.33 | 86.33 | 92.90 | 82.14 |
| PET | 85.36 ± 3.29 | 84.00 | 86.67 | 85.33 | 85.86 | 84.90 | |
| MP | 93.18 ± 2.52 | 90.00 | 96.33 | 93.17 | 96.05 | 90.68 | |
| MPC | 92.36 ± 2.70 | 94.00 | 90.33 | 92.17 | 92.33 | 92.40 | |
| D-S2MTL | MRI | 83.64 ± 3.70 | 84.33 | 83.00 | 83.67 | 89.78 | 74.48 |
| PET | 89.36 ± 2.68 | 90.00 | 88.67 | 89.33 | 89.54 | 89.16 | |
| MP | 88.27 ± 2.78 | 82.00 | 94.33 | 88.17 | 93.32 | 84.43 | |
| MPC | 90.36 ± 2.77 | 90.00 | 90.33 | 90.17 | 89.25 | 91.33 | |
| DW-S2MTL | MRI | 90.36 ± 2.53 | 82.33 | 98.33 | 90.33 | 98.00 | 84.86 |
| PET | 89.27 ± 2.97 | 82.33 | 96.33 | 89.33 | 95.80 | 84.27 | |
| MP | 93.18 ± 2.82 | 90.00 | 96.33 | 93.17 | 96.05 | 90.68 | |
| MPC | 95.09 ± 2.28 | 92.00 | 98.00 | 95.00 | 97.74 | 92.86 |
Boldface denotes the best performance and the maximum performance in each metric
SMTL sparse multi-task learning, S2 MTL subclass-based SMTL, DW-SMTL deep weighted SMTL, D-S2MTL deep S2MTL, DW-S2MTL deep weighted S2MTL
Table 4.
A summary of the performances for MCI vs. CN classification
| Method | Modality | ACC (%) | SEN (%) | SPEC (%) | BAC (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|
| SMTL | MRI | 70.90 ± 3.04 | 80.78 | 52.00 | 66.39 | 76.30 | 58.58 |
| PET | 64.98 ± 2.68 | 75.89 | 44.00 | 59.94 | 72.27 | 48.69 | |
| MP | 66.76 ± 3.29 | 73.56 | 53.67 | 63.61 | 75.35 | 51.33 | |
| MPC | 68.32 ± 3.48 | 74.89 | 56.00 | 65.44 | 76.14 | 54.32 | |
| DW-SMTL | MRI | 68.89 ± 2.85 | 76.67 | 54.00 | 65.33 | 76.13 | 54.74 |
| PET | 64.31 ± 2.82 | 73.89 | 46.00 | 59.94 | 72.34 | 47.96 | |
| MP | 70.94 ± 3.04 | 80.89 | 52.00 | 66.44 | 76.23 | 58.84 | |
| MPC | 72.77 ± 3.40 | 79.78 | 59.33 | 69.56 | 79.00 | 60.48 | |
| S2MTL | MRI | 70.32 ± 3.18 | 82.78 | 46.67 | 64.72 | 74.66 | 58.81 |
| PET | 67.60 ± 3.22 | 78.89 | 46.33 | 62.61 | 73.47 | 53.81 | |
| MP | 69.65 ± 2.56 | 76.78 | 56.33 | 66.56 | 76.66 | 56.49 | |
| MPC | 67.02 ± 2.95 | 78.78 | 44.67 | 61.72 | 73.07 | 52.55 | |
| D-S2MTL | MRI | 68.85 ± 3.15 | 76.56 | 54.33 | 65.44 | 75.94 | 55.17 |
| PET | 68.89 ± 2.96 | 76.89 | 54.00 | 65.44 | 75.68 | 55.66 | |
| MP | 70.98 ± 2.91 | 77.78 | 58.33 | 68.06 | 77.64 | 58.53 | |
| MPC | 68.98 ± 3.30 | 76.78 | 54.33 | 65.56 | 75.95 | 55.47 | |
| DW-S2MTL | MRI | 77.57 ± 2.92 | 90.89 | 52.00 | 71.44 | 78.42 | 74.83 |
| PET | 74.90 ± 2.55 | 96.00 | 34.67 | 65.33 | 73.69 | 81.97 | |
| MP | 80.11 ± 2.64 | 93.89 | 53.67 | 73.78 | 79.54 | 82.07 | |
| MPC | 78.77 ± 2.47 | 90.78 | 56.00 | 73.39 | 79.64 | 76.21 |
Boldface denotes the best performance and the maximum performance in each metric
SMTL sparse multi-task learning, S 2MTL subclass-based SMTL, DW-SMTL deep weighted SMTL, D-S 2MTL deep S2MTL, DW-S 2MTL deep weighted S2MTL
Table 5.
A summary of the performances for pMCI vs. sMCI classification
| Method | Modality | ACC (%) | SEN (%) | SPEC (%) | BAC (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|
| SMTL | MRI | 51.44 ± 3.68 | 39.50 | 61.00 | 50.25 | 44.78 | 55.74 |
| PET | 50.92 ± 3.97 | 40.50 | 59.33 | 49.92 | 44.56 | 55.27 | |
| MP | 54.48 ± 3.88 | 41.50 | 65.00 | 53.25 | 49.01 | 57.82 | |
| MPC | 60.69 ± 4.06 | 46.50 | 72.00 | 59.25 | 56.96 | 62.80 | |
| DW-SMTL | MRI | 55.46 ± 3.65 | 29.00 | 76.33 | 52.67 | 49.14 | 57.68 |
| PET | 54.12 ± 4.28 | 38.00 | 66.33 | 52.17 | 46.09 | 58.55 | |
| MP | 56.71 ± 4.28 | 44.00 | 67.00 | 55.50 | 51.91 | 59.64 | |
| MPC | 58.56 ± 4.09 | 36.50 | 75.67 | 56.08 | 53.78 | 60.57 | |
| S2MTL | MRI | 61.00 ± 3.47 | 53.00 | 66.33 | 59.67 | 51.19 | 67.93 |
| PET | 57.87 ± 4.13 | 44.50 | 68.33 | 56.42 | 52.36 | 61.15 | |
| MP | 65.33 ± 3.84 | 59.50 | 70.67 | 65.08 | 65.01 | 65.58 | |
| MPC | 67.04 ± 4.38 | 67.00 | 66.67 | 66.83 | 61.48 | 69.89 | |
| D-S2MTL | MRI | 63.71 ± 2.43 | 25.50 | 92.00 | 58.75 | 70.24 | 62.52 |
| PET | 55.25 ± 4.60 | 50.00 | 59.67 | 54.83 | 51.07 | 58.64 | |
| MP | 67.82 ± 3.41 | 37.00 | 93.33 | 65.17 | 82.12 | 64.16 | |
| MPC | 70.73 ± 3.41 | 42.50 | 93.00 | 67.75 | 82.73 | 67.22 | |
| DW-S2MTL | MRI | 69.84 ± 2.68 | 44.00 | 89.00 | 66.50 | 74.79 | 68.19 |
| PET | 65.71 ± 3.64 | 29.50 | 95.00 | 62.25 | 82.68 | 62.49 | |
| MP | 74.15 ± 3.35 | 50.50 | 92.67 | 71.58 | 84.36 | 70.51 | |
| MPC | 73.04 ± 3.51 | 53.00 | 89.00 | 71.00 | 79.33 | 70.39 |
Boldface denotes the best performance and the maximum performance in each metric
SMTL sparse multi-task learning, S 2MTL subclass-based SMTL, DW-SMTL deep weighted SMTL, D-S 2MTL deep S2MTL, DW-S 2MTL deep weighted S2MTL
For the task of MCI vs. CN classification (Table 4), the proposed DW-S2MTL achieved the best ACCs of 77.57 % (MRI), 74.90 % (PET), 80.11 % (MP), and 78.77 % (MPC), while D-S2MTL/DW-SMTL achieved the ACCs of 68.85/68.89 % (MRI), 68.89/64.31 % (PET), and 70.98/70.94 % (MP), and 68.98/72.77 % (MPC). In the meantime, SMTL/S2MTL achieved the ACCs of 70.90/70.32 % (MRI), 64.98/67.90 % (PET), 66.76/69.65 % (MP), and 68.32/67.02 % (MPC), respectively. By applying the proposed deep and feature-adaptive weighting scheme, DW-SMTL improved the ACCs by 4.18 % (MP) and 4.45 % (MPC) compared to SMTL. It is remarkable that compared to S2MTL, DW-S2MTL improved by 7.25 % (MRI), 7.30 % (PET), 10.46 % (MP), and 11.75 % (MPC).
Lastly, in the classification of pMCI and sMCI (Table 5), which is clinically the most important because the timely symptomatic treatment can potentially delay the progression (Francis et al. 2010), DW-S2MTL outperformed the other competing methods again, and the proposed deep and feature-adaptive weighting scheme helped improve the accuracies for both SMTL and S2MTL. Concretely, we obtained the ACCs of 69.84 % (MRI), 65.71 % (PET), 74.15 % (MP), and 73.04 % (MPC) by DW-S2MTL and the ACCs of 63.71/55.46 % (MRI), 55.25/54.12 % (PET), 67.82/56.71 % (MP), 70.73/58.56 % (MPC) by D-S2MTL/DW-SMTL. In comparison between S2MTL and DW-S2MTL, the improvements were 8.84 % (MRI), 7.84 % (PET), 8.82 % (MP), and 6 % (MPC). It is also noteworthy that the subclass-based methods, i.e., S2MTL and DW-S2MTL, that encompass the characteristics of a complex distribution were superior to both SMTL and DW-SMTL that assumed a unimodal data distribution.
Multi-class classification results
From a clinical standpoint, while there exist multiple stages in the spectrum of AD and CN, the previous work mostly focused on binary classification problems. By taking account of more practical applications, we also performed experiments of multi-class classifications. Note that no change in our framework is required for multi-class classification, except for the class labels.
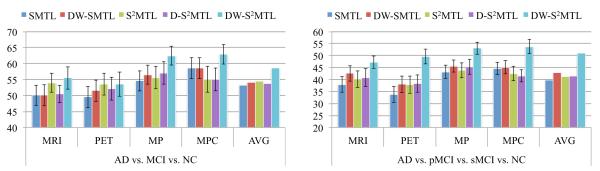
Figure 3 summarizes the performances on two multi-class classification tasks. Same as the binary classification results, we observed that the proposed DW-S2MTL method outperformed the competing methods for both three-class and four-class classification tasks. Concretely, in three-class classification, SMTL achieved the ACCs of 50.10 % (MRI), 49.52 % (PET), 54.57 % (MP), and 58.55 % (MPC), and DW-SMTL achieved the ACCs of 50.10 % (MRI), 51.50 % (PET), 56.52 % (MP), and 58.55 % (MPC). Meanwhile, DW-S2MTL achieved 55.50 % (MRI), 53.50 % (PET), 62.43 % (MP), and 62.93 % (MPC). In four-class classification, the maximal ACC of 53.72 % was produced by the proposed DW-S2MTL method with MPC data, improving the ACC by 9.08 % (vs. SMTL), 8.63 % (vs. DW-SMTL), 11.22 % (vs. S2MTL), and 12.21 % (vs. D-S2MTL), respectively.
Fig. 3.
Performance comparison on two multi-class classification problems. (AVG average of accuracies over different modalities, SMTL sparse multi-task learning, S 2MTL subclass-based SMTL, DW-SMTL deep weighted SMTL, D-S 2MTL deep S2MTL, DW-S 2MTL deep weighted S2MTL)
Classification results on a large MRI dataset
Since the focus on AD/MCI diagnosis or prognosis appears to be mostly on MRI, we further performed experiments with a large number of MRI data. Specifically, we considered 805 subjects of 198 (AD), 167 (pMCI), 236 (sMCI), and 229 (NC). With this large dataset, we conducted experiments for the same tasks as considered above. The classification accuracies and the respective standard deviations are presented in Fig. 4. In all classification tasks, the proposed DW-S2MTL clearly surpassed the other four competing methods, by achieving the ACCs of 90.27 % (AD vs. NC), 70.86 % (MCI vs. NC), 73.93 % (pMCI vs. sMCI), 57.74 % (AD vs. MCI vs. NC), and 47.83 % (AD vs. pMCI vs. sMCI vs. NC), respectively.
Fig. 4.
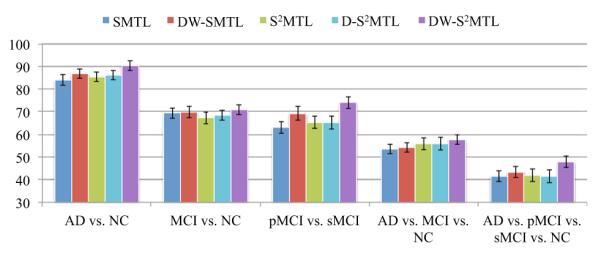
Performance comparison on a large MRI dataset from ADNI. (SMTL sparse multi-task learning, S 2MTL subclass-based SMTL, DW-SMTL deep weighted SMTL, D-S 2MTL deep S2MTL, DW-S 2MTL deep weighted S2MTL)
Discussions
Based on our experiments of binary and multi-class classifications, we observed two interesting results: (1) when comparing SMTL with S2MTL and also DW-SMTL with DW-S2MTL, the subclass-based approaches, i.e., S2MTL and DW-S2MTL, outperformed the respective competing methods, i.e., SMTL and DW-SMTL; (2) the proposed deep sparse multi-task learning method with a feature-adaptive weighting scheme helped enhance the diagnostic accuracies, i.e., DW-SMTL and DW-S2MTL showed better performance than SMTL, and S2MTL and D-S2MTL, respectively. In this section, we further discuss the results in various perspectives.
Data distributions
In our experiments, the subclass-based methods, i.e., S2MTL and DW-S2MTL, were superior to the respective competing methods, i.e., SMTL and DW-SMTL. To justify the results, we performed Henze–Zirkler’s multivariate normality test (Henze and Zirkler 1990) that statistically determines how well samples can be modeled by a multivariate normal distribution, and summarized the results in Table 6. In our test, the null hypothesis was that the samples could come from a multivariate normal distribution. Regarding MRI, the null hypothesis was rejected for both AD and MCI. With respect to PET, the test rejected the hypothesis for MCI. In the meantime, it turned out that the CSF samples of all the disease labels did not follow a multivariate Gaussian distribution. Based on these statistical evaluations, we can confirm the complex data distributions and also justify the necessity of using the subclass-based approach, which can efficiently handle such a complex distribution problem.
Table 6.
A summary of Henze–Zirkler’s multivariate normality test on our dataset
| Modality | AD | MCI | NC |
|---|---|---|---|
| MRI | 0.0005 (R) | 0.0004 (R) | 0.6967 (A) |
| PET | 0.4273 (A) | 0.0239 (R) | 0.3150 (A) |
| CSF | 0.0049 (R) | <0.0001 (R) | <0.0001 (R) |
‘R’ or ‘A’ in parentheses denotes whether the null hypothesis (that the samples could come from a multivariate normal distribution) is rejected or accepted at the 5 % significance level
Effect of deep architecture in feature selection
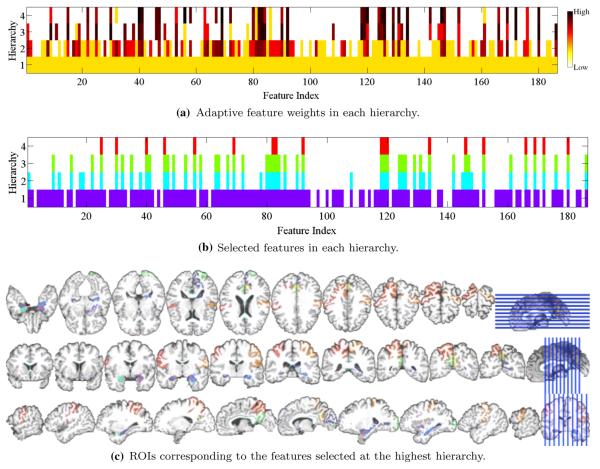
To see the effect of the proposed deep learning scheme in a sparse regression framework, in Fig. 5a and b, respectively, we illustrate the change of the weights for each feature and the selected features over hierarchies by DW-S2MTL from one of the tenfolds in three-class classification with MP data. From the figure, it is clear that in the 1st hierarchy that corresponds to S2MTL, the weights for the features are equal and more than 80 % of the total features were selected. But, as the algorithm forwarded to the higher hierarchy, it gradually discarded uninformative or less informative features, whose weights from the optimal regression coefficients in the lower hierarchy were relatively low, and after the 4-th hierarchy, it finally selected only 19 features (approximately 10 % of the total features). The ROIs corresponding to the finally selected features, i.e., weighted high for classification, included hippocampal formation left/right, amygdala left/right (in a medial temporal lobe that involves a system of anatomically related structures that are vital for declarative or long-term memory) (Braak and Braak 1991; Visser et al. 2002; Mosconi 2005; Lee et al. 2006; Devanand et al. 2007; Frisoni et al. 2008; Burton et al. 2009; Desikan et al. 2009; Ewers et al. 2012; Walhovd et al. 2010), precuneus left/right (Karas et al. 2007), cuneus left (Bokde et al. 2006; Singh et al. 2006; Davatzikos et al. 2011), uncus left, anterior cingulate gyrus left, occipital pole left, subthalamic nucleus left, postcentral gyrus left/right, superior parietal lobule right, anterior limb of internal capsule right, and angular gyrus left (Schroeter et al. 2009; Nobili et al. 2010; Yao et al. 2012). From a biological perspective, we could understand that some of the ROIs such as hippocampal formation, amygdala, and precuneous selected from our MRI features were related to the volume atrophy in medial temporal cortex, while precuneous, cingulate gyrus, and parietal lobule selected from our PET features could be concerned with hypometabolism (Joie et al. 2012). For reference, we also summarized the statistics of the number of hierarchies built with the proposed DW-S2MTL in the tasks of binary and multi-class classification with different modalities in Table 7.
Fig. 5.
An example of the change of the selected features over hierarchies with MP in AD vs. MCI vs. CN
Table 7.
A summary of the statistics (mean ± std [min–max]) of the number of hierarchies with the proposed DW-S2MTL in the tasks of binary and multi-class classification with modalities
| Task | MRI | PET | MP | MPC |
|---|---|---|---|---|
| AD/CN | 1.1 ± 0.3 [1–2] | 1.4 ± 0.7 [1–3] | 1.5 ± 0.7 [1–3] | 1.6 ± 1.0 [1–4] |
| MCI/CN | 1.5 ± 0.8 [1–3] | 1.8 ± 0.8 [1–3] | 1.4 ± 0.5 [1–2] | 2.0 ± 1.1 [1–4] |
| pMCI/sMCI | 1.1 ± 0.3 [1–2] | 1.2 ± 0.4 [1–2] | 1.3 ± 0.5 [1–2] | 1.4 ± 0.7 [1–3] |
| AD/MCI/CN | 1.4 ± 0.5 [1–2] | 1.7 ± 0.8 [1–3] | 1.6 ± 1.0 [1–4] | 1.5 ± 0.7 [1–3] |
| AD/pMCI/sMCI/CN | 1.4 ± 1.0 [1–4] | 1.8 ± 0.8 [1–3] | 1.4 ± 0.8 [1–3] | 1.5 ± 0.5 [1–2] |
Performance interpretation
In “Binary classification results” and “Multi-class classification results”, we showed the superiority of the proposed DW-S2MTL method compared to the competing methods in the context of classification accuracy. For the binary classifications of MCI vs. CN and pMCI vs. sMCI, the proposed DW-S2MTL method with MP data showed better performance than with MPC data, even though the later provided additional information from CSF. Note that in this work, we treated different modalities equally, i.e., uniform weight across modalities. However, should we apply a modality-adaptive weighting scheme similar to Zhang et al. (2011), we then expect to obtain enhanced performances with MPC data.
Regarding sensitivity and specificity, the higher the sensitivity, the lower the chance of misdiagnosing AD/MCI patients; also the higher the specificity, the lower the chance of misdiagnosing CN to AD/MCI. In our three binary classification tasks, although the proposed DW-S2MTL method achieved the best accuracies, it did not necessarily obtain the best sensitivity or specificity (but still reported high sensitivity and specificity). It is noteworthy that due to the imbalanced samples between classes, we obtained low sensitivity in pMCI vs. sMCI and low specificity in MCI vs. CN. In this regard, we also computed the balanced accuracy that avoids inflated performance estimates on imbalanced datasets by taking the average of sensitivity and specificity. Based on this metric, we clearly see that the proposed DW-S2MTL method outperformed the competing methods by achieving the maximal BACs of 95 % (MPC) in AD vs. CN, 73.78 % (MP) in MCI vs. CN, and 71.58 % (MP) in pMCI vs. sMCI.
The metrics of sensitivity and specificity have been widely considered in the fields of the computer-aided AD diagnosis. However, note that since both sensitivity and specificity are defined on the basis of people with or without a disease, there is no practical use to estimate the probability of disease in an individual patient (Akobeng 2007). We rather need to know the positive/negative predictive values (PPV/NPV for short), which describe a patient’s probability of having disease once the classification results are known. Furthermore, PPV and NPV are highly related to the prevalence of disease. That is, the higher the disease prevalence, the higher the PPV, i.e., the more likely a positive diagnostic result; the lower disease prevalence, the lower the PPV, i.e., the less likely a positive diagnostic result. NPV would show exactly the opposite trends. In our experiments, the proposed DW-S2MTL method achieved the maximal PPVs/NPVs of 97.74 % (MPC)/92.86 % (MPC) in AD vs. CN, 79.64 % (MPC)/82.07 % (MP) in MCI vs. CN, and 84.36 % (MP)/70.51 % (MP) in pMCI vs. sMCI. It is remarkable that in pMCI vs. sMCI classification, which is clinically the most important, the proposed DW-S2MTL showed PPV improvements by 28.4 % (vs. SMTL with MPC), 30.58 % (vs. DW-SMTL with MPC), 22.88 % (vs. S2MTL with MPC), and 1.63 % (vs. D-S2MTL) and NPV improvements by 7.71 % (vs. SMTL with MPC), 9.94 % (vs. DW-SMTL with MP), 0.62 % (vs. S2MTL with MPC), and 3.29 % (vs. D-S2MTL with MPC).
Comparison with the state-of-the-art methods
In Table 8, we also compared the classification accuracies of the proposed DW-S2MTL method with those of the state-of-the-art methods that fused multiple modalities for the classifications of AD vs. NC and MCI vs. NC. Note that, due to different datasets and different approaches for extracting features and building classifiers, it is not fair to directly compare the performances among the methods. Nevertheless, the proposed method showed the highest accuracies among the methods in both binary classification problems. In particular, it is noteworthy that compared to Zhang and Shen’s work (2011) in which they used the same dataset as ours, the proposed method enhanced the accuracies by 1.89 and 3.71 % for the classifications of AD/CN and MCI/CN, respectively. Furthermore, in comparison with Liu et al.’s work (2013), where they also used both the same types of features from MRI and PET and the same number of subjects with ours, our method improved the accuracies by 0.72 % (AD/CN) and 1.31 % (MCI/CN), respectively. We also performed statistical significance tests to compare with Liu et al.’s and Zhang et al.’s methods. In summary, the null hypothesis was rejected beyond the 99 % of the confidence level based on the p-values of 0.00024 (vs. Liu et al.’s method) and 0.00012 (vs. Zhang et al.’s method).
Table 8.
Comparison of classification accuracies (%) with the state-of-the-art methods that used multimodal neuroimaging for AD/CN and MCI/CN. The boldface denotes the maximum performance in each classification problem. (MP: MRI+PET, MPC: MRI+PET+CSF)
| Methods | Subjects (AD/MCI/NC) | Modalities | AD/CN | MCI/CN |
|---|---|---|---|---|
| Kohannim et al. (2010) | 40/83/43 | MPC | 90.7 | 75.8 |
| Hinrichs et al. (2011) | 48/119/66 | MP | 92.4 | n/a |
| Zhang et al. (2011) | 51/99/52 | MPC | 93.2 | 76.4 |
| Liu et al. (2013) | 51/99/52 | MP | 94.37 | 78.80 |
| Proposed DW-S2MTL | 51/99/52 | MPC | 95.09 | 80.11 |
Conclusions
In neuroimaging-based AD/MCI diagnosis, the ‘high-dimension and small sample’ problem has been one of the major issues. To tackle this problem, sparse regression methods have been widely exploited for feature selection, thus reducing the dimensionality. To our best knowledge, most of the existing methods select informative features in a single hierarchy. However, during the optimization of the regression coefficients, the weights of informative features are inevitably affected by non-informative or noisy features, and thus there is a high possibility of having the informative features underestimated or the uninformative features overestimated. In this regard, we proposed a deep sparse multi-task learning method along with a feature-adaptive weighting scheme for feature selection in AD/MCI diagnosis. The main contributions of this work can be threefold: (1) Rather than selecting informative features in a single hierarchy, the proposed method iteratively filters out uninformative features in a hierarchical fashion. (2) Furthermore, at different hierarchies, our method utilizes the regression coefficients optimized in the lower hierarchy as context information to better determine informative features for classification. (3) Last but not least, our method reflects the complex distributional characteristics in each class via a subclass labeling scheme.
In our experimental results on the ADNI cohort, we validated the effectiveness of the proposed method in both binary classification and multi-class classification tasks, outperforming the competing methods in various metrics.
It is noteworthy that in this work, we regarded the importance of features from different modality equally. However, as demonstrated by Zhang et al. (2011), different modalities may have different impacts on making a clinical decision. If a multi-kernel SVM (Gönen and Alpaydin 2011) is used to replace the linear SVM in our framework, then it would be possible to learn modality-adaptive weights and thus can obtain the relative importance of different modalities.
According to a recent broad spectrum of studies, there are increasing evidences that subjective cognitive complaint is one of the important genetic risk factors, which increases the risk of progression to MCI or AD (Loewenstein et al. 2012; Mark and Sitskoorn 2013). That is, among the cognitively normal elderly individuals who have subjective cognitive impairment, there exists a high possibility for some of them to be in the stage of ‘pre-MCI’. However, this issue has been underestimated in the field. Thus, we believe that it is important to design and develop diagnostic methods by taking into account such information as well. In addition, to our best knowledge, most of the existing computational methods have focused on improving diagnostic accuracy or finding the potential biomarkers. However, for practical application of those computational tools as an expert system, it is required to present the grounds for the clinical decision. For example, when a diagnostic system makes a decision to MCI, then it would be beneficial for doctors to know which parts of the brain regions are distinct or abnormal compared to those of the normal healthy controls.
Acknowledgments
This work was supported in part by NIH grants EB006733, EB008374, EB009634, AG041721, MH100217, and AG042599, and also supported by ICT R&D program of MSIP/IITP. [B0101-15-0307, Basic Software Research in Human-level Lifelong Machine Learning (Machine Learning Center)].
Appendix. Affinity propagation
Here, we briefly review the affinity propagation (Frey and Dueck 2007), by which we find subclasses in each original class. Let denote the pairwise similarities13 between each pair of N samples in . The affinity propagation algorithm works on the similarity matrix and attempts to find ‘exemplars’ that maximize the overall sum of similarities between all exemplars and their member samples. Methodologically, the algorithm defines two types of messages, namely, responsibility and availability, exchanged among samples: Responsibility represents the accumulated evidence for how well-suited sample j is to serve as the exemplar for sample i; Availability reflects the accumulated evidence for how appropriate it would be for sample i to choose sample j as its exemplar. Using these messages, the exemplar of sample i is determined by the one that maximizes the following objective function:
| (8) |
In Algorithm 1, both and are initially set to zero matrices, and then their values are iteratively updated as below until converged:
Footnotes
In a least squares regression framework, one task corresponds to find optimal regression coefficients to represent the values of a target response variable. So, when we consider multiple target response variables simultaneously, it is regarded as multi-task learning (Argyriou et al. 2008).
In this work, we define the uninformative and less informative features based on their optimal regression coefficients. Specifically, the features whose regression coefficients are zero or close to zero, are regarded, respectively, as uninformative or less informative in representing the target response variables.
Available at ‘http://www.loni.ucla.edu/ADNI’.
Although there exist in total more than 800 subjects in ADNI database, only 202 subjects have the baseline data including all the modalities of MRI, PET, and CSF.
Refer to ‘http://www.adniinfo.org’ for more details.
Available at ‘http://mipav.cit.nih.gov/clickwrap.php’.
Available at ‘http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/’.
In our experiments on the ADNI cohort, we have one sample per subject.
denotes the original full feature set.
Initially, we set the current best accuracy zero.
Available at ‘http://www.public.asu.edu/~jye02/Software/SLEP/index.htm’.
Available at ‘http://www.csie.ntu.edu.tw/~cjlin/libsvm/’.
In this work, we use a negative Euclidian distance for similarity computation.
Conflict of interest The authors declare that they have no conflict of interest.
Ethical standard This article does not contain any studies with human participants performed by any of the authors.
Informed consent Informed consent was obtained from all individual participants included in the study.
References
- Akobeng AK. Understanding diagnostic tests 1: sensitivity, specificity and predictive values. Acta Pediatr. 2007;96(3):338–341. doi: 10.1111/j.1651-2227.2006.00180.x. [DOI] [PubMed] [Google Scholar]
- Alikhanian H, Crawford JD, DeSouza JFX, Cheyne D, Blohm G. Adaptive cluster analysis approach for functional localization using magnetoencephalography. Front Neurosci. 2013;7(73) doi: 10.3389/fnins.2013.00073. doi: 10.3389/fnins.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Association Alzheimer’s 2012 Alzheimer’s disease facts and figures. Alzheimer’s Dementia. 2012;8(2):131–168. doi: 10.1016/j.jalz.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Argyriou A, Evgeniou T, Pontil M. Convex multi-task feature learning. Mach Learn. 2008;73(3):243–272. [Google Scholar]
- Bokde ALW, Lopez-Bayo P, Meindl T, Pechler S, Born C, Faltraco F, Teipel SJ, Möller HJ, Hampel H. Functional connectivity of the fusiform gyrus during a face-matching task in subjects with mild cognitive impairment. Brain. 2006;129(5):1113–1124. doi: 10.1093/brain/awl051. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- de Brecht M, Yamagishi N. Combining sparseness and smoothness improves classification accuracy and interpretability. NeuroImage. 2012;60(2):1550–1561. doi: 10.1016/j.neuroimage.2011.12.085. [DOI] [PubMed] [Google Scholar]
- Burges CJC. A tutorial on support vector machines for pattern recognition. Data Mining Knowl Discov. 1998;2(2):121–167. [Google Scholar]
- Burton EJ, Barber R, Mukaetova-Ladinska EB, Robson J, Perry RH, Jaros E, Kalaria RN, O’Brien JT. Medial temporal lobe atrophy on MRI differentiates Alzheimer’s disease from dementia with lewy bodies and vascular cognitive impairment: a prospective study with pathological verification of diagnosis. Brain. 2009;132(1):195–203. doi: 10.1093/brain/awn298. [DOI] [PubMed] [Google Scholar]
- Busse A, Angermeyer MC, Riedel-Heller SG. Progression of mild cognitive impairment to dementia: a challenge to current thinking. Br J Psychiatry. 2006;189:399–404. doi: 10.1192/bjp.bp.105.014779. [DOI] [PubMed] [Google Scholar]
- Cui Y, Liu B, Luo S, Zhen X, Fan M, Liu T, Zhu W, Park M, Jiang T, Jin JS, the Alzheimer’s Disease Neuroimaging Initiative Identification of conversion from mild cognitive impairment to Alzheimer’s disease using multivariate predictors. PLoS ONE. 2011;6(7):e21896. doi: 10.1371/journal.pone.0021896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol Aging. 2011;32(12):2322.e19–2322.e27. doi: 10.1016/j.neurobiolaging.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Cabral H, Hess C, Dillon W, Salat D, Buckner R, Fischl B, Initiative ADN. Automated MRI measures identify individuals with mild cognitive impairment and Alzheimer’s disease. Brain. 2009;132:2048–2057. doi: 10.1093/brain/awp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanand DP, Pradhaban G, Liu X, Khandji A, De Santi S, Segal S, Rusinek H, Pelton GH, Hoing LS, Mayeux R, Stern Y, Tabert MH, de Leon JJ. Hippocampal and entorhinal atrophy in mild cognitive impairment. Neurology. 2007;68:828–836. doi: 10.1212/01.wnl.0000256697.20968.d7. [DOI] [PubMed] [Google Scholar]
- DiFrancesco M, Hollandm S, Szaflarski J. Simultaneous EEG/functional magnetic resonance imaging at 4 tesla: correlates of brain activity to spontaneous alpha rhythm during relaxation. J Clin Neurophysiol. 2008;25(5):255–264. doi: 10.1097/WNP.0b013e3181879d56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueck D, Frey B. Non-metric affinity propagation for unsupervised image categorization. 2007 IEEE international conference on computer vision (ICCV).2007. pp. 1–8. [Google Scholar]
- Ewers M, Walsh C, Trojanowski JQ, Shaw LM, Petersen RC, Jr., Feldman HH, Bokde AL, Alexander GE, Scheltens P, Vellas B, Dubois B, Weiner M, Hampel H. Prediction of conversion from mild cognitive impairment to Alzheimer’s disease dementia based upon biomarkers and neuropsychological test performance. Neurobiol Aging. 2012;33(7):1203–1214.e2. doi: 10.1016/j.neurobiolaging.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazli S, Danczy M, Schelldorfer J, Mller KR. ℓ1-penalized linear mixed-effects models for high dimensional data with application to BCI. NeuroImage. 2011;56(4):2100–2108. doi: 10.1016/j.neuroimage.2011.03.061. [DOI] [PubMed] [Google Scholar]
- Fotenos A, Snyder A, Girton L, Morris J, Buckner R. Normative estimates of cross-sectional and longitudinal brain volume decline in aging and AD. Neurology. 2005:1032–1039. doi: 10.1212/01.WNL.0000154530.72969.11. 2005. [DOI] [PubMed] [Google Scholar]
- Francis PT, Ramírez MJ, Lai MK. Neurochemical basis for symptomatic treatment of Alzheimer’s disease. Neuropharmacology. 2010;59(4–5):221–229. doi: 10.1016/j.neuropharm.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315(5814):972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Ganzola R, Canu E, Rüb U, Pizzini FB, Alessandrini F, Zoccatelli G, Beltramello A, Caltagirone C, Thompson PM. Mapping local hippocampal changes in Alzheimer’s disease and normal ageing with MRI at 3 Tesla. Brain. 2008;131(12):3266–3276. doi: 10.1093/brain/awn280. [DOI] [PubMed] [Google Scholar]
- Gönen M, Alpaydin E. Multiple kernel learning algorithms. J Mach Learn Res. 2011;12:2211–2268. [Google Scholar]
- Henze N, Zirkler B. A class of invariant consistent tests for multivariate normality. Commun Stat Theory Methods. 1990;19(10):3595–3617. [Google Scholar]
- Hinrichs C, Singh V, Xu G, Johnson SC. Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. NeuroImage. 2011;55(2):574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joie RL, Perrotin A, Barré L, Hommet C, Mézenge F, Ibazizene M, Camus V, Abbas A, Landeau B, Guilloteau D, de La Sayette V, Eustache F, Desgranges B, Chételat G. Region-specific hierarchy between atrophy, hypometabolism, and beta-amyloid (Aβ) load in Alzheimer’s disease dementia. J Neurosci. 2012;32:16265–16273. doi: 10.1523/JNEUROSCI.2170-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani N, MacDonald D, Holmes C, Evans A. A 3D atlas of the human brain. NeuroImage. 1998;7(4):S717. [Google Scholar]
- Karas G, Scheltens P, Rombouts S, van Schijndel R, Klein M, Jones B, van der Flier W, Vrenken H, Barkhof F. Precuneus atrophy in early-onset Alzheimer’s disease: a morphometric structural MRI study. Neuroradiology. 2007;49(12):967–976. doi: 10.1007/s00234-007-0269-2. [DOI] [PubMed] [Google Scholar]
- Kohannim O, Hua X, Hibar DP, Lee S, Chou YY, Toga AW, Jr, Jack CR, Weiner MW, Thompson PM. Boosting power for clinical trials using classifiers based on multiple biomarkers. Neurobiol Aging. 2010;31(8):1429–1442. doi: 10.1016/j.neurobiolaging.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ACH, Buckley MJ, Gaffan D, Emery T, Hodges JR, Graham KS. Differentiating the roles of the hippocampus and perirhinal cortex in processes beyond long-term declarative memory: a double dissociation in dementia. J Neurosci. 2006;26(19):5198–5203. doi: 10.1523/JNEUROSCI.3157-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang Y, Wu G, Shi F, Zhou L, Lin W, Shen D. Discriminant analysis of longitudinal cortical thickness changes in Alzheimer’s disease using dynamic and network features. Neurobiol Aging. 2012;33(2):427.e15–427.e30. doi: 10.1016/j.neurobiolaging.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wee CY, Chen H, Shen D. Inter-modality relationship constrained multi-task feature selection for AD/MCI classification. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Medical image computing and computer-assisted intervention (MICCAI) Vol. 8149. Springer; Berlin: 2013. pp. 308–315. Lecture Notes in Computer Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang D, Shen D. Ensemble sparse classification of Alzheimer’s disease. NeuroImage. 2012;60(2):1106–1116. doi: 10.1016/j.neuroimage.2012.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewenstein DA, Greig MT, Schinka JA, Barker W, Shen Q, Potter E, Raj A, Brooks L, Varon D, Schoenberg M, Banko J, Potter H, Duara R. An investigation of PreMCI: subtypes and longitudinal outcomes. Alzheimer’s Dementia. 2012;8(3):172–179. doi: 10.1016/j.jalz.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Carreira-Perpinan M. Constrained spectral clustering through affinity propagation. 2008 IEEE conference on computer vision and pattern recognition (CVPR).2008. pp. 1–8. [Google Scholar]
- Mark RE, Sitskoorn MM. Are subjective cognitive complaints relevant in preclinical Alzheimer’s disease? A review and guidelines for healthcare professionals. Rev Clin Gerontol. 2013;23:61–74. [Google Scholar]
- Milgram J, Cheriet M, Sabourin R. “One against one” or “one against all”: which one is better for handwriting recognition with SVMs?. In: Lorette G, editor. Tenth international workshop on frontiers in handwriting recognition; Suvisoft; 2006. [Google Scholar]
- Mosconi L. Brain glucose metabolism in the early and specific diagnosis of Alzheimer’s disease. Eur J Nucl Med Mol Imaging. 2005;32(4):486–510. doi: 10.1007/s00259-005-1762-7. [DOI] [PubMed] [Google Scholar]
- Nie F, Huang H, Cai X, Ding CH. Efficient and robust feature selection via joint ℓ2,1-norms minimization. In: Lafferty J, Williams C, Shawe-Taylor J, Zemel R, Culotta A, editors. Advances in neural information processing systems. Vol. 23. 2010. pp. 1813–1821. [Google Scholar]
- Nobili F, Mazzei D, Dessi B, Morbelli S, Brugnolo A, Barbieri P, Girtler N, Sambuceti G, Rodriguez G, Pagani M. Unawareness of memory deficit in amnestic MCI: FDG-PET findings. J Alzheimer’s Dis. 2010;22(3):993–1003. doi: 10.3233/JAD-2010-100423. [DOI] [PubMed] [Google Scholar]
- Noppeney U, Penny WD, Price CJ, Flandin G, Friston KJ. Identification of degenerate neuronal systems based on inter-subject variability. NeuroImage. 2006;30(3):885–890. doi: 10.1016/j.neuroimage.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Perrin RJ, Fagan AM, Holtzman DM. Multimodal techniques for diagnosis and prognosis of Alzheimer’s disease. Nature. 2009;461:916–922. doi: 10.1038/nature08538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth V. The generalized LASSO. IEEE Trans Neural Netw. 2004;15(1):16–28. doi: 10.1109/TNN.2003.809398. [DOI] [PubMed] [Google Scholar]
- Schroeter ML, Stein T, Maslowski N, Neumann J. Neural correlates of Alzheimer’s disease and mild cognitive impairment: a systematic and quantitative meta-analysis involving 1351 patients. NeuroImage. 2009;47(4):1196–1206. doi: 10.1016/j.neuroimage.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imaging. 2002;21(11):1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Shi F, Wang L, Gilmore J, Lin W, Shen D. Learning-based meta-algorithm for MRI brain extraction. In: Fichtinger G, Martel A, Peters T, editors. Medical image computing and computer-assisted intervention (MICCAI), Lecture Notes in Computer Science. Vol. 6893. 2011. pp. 313–321. [DOI] [PubMed] [Google Scholar]
- Singh V, Chertkow H, Lerch JP, Evans AC, Dorr AE, Kabani NJ. Spatial patterns of cortical thinning in mild cognitive impairment and Alzheimer’s disease. Brain. 2006;129(11):2885–2893. doi: 10.1093/brain/awl256. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17(1):87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Suk HI, Lee SW. A novel Bayesian framework for discriminative feature extraction in brain–computer interfaces. IEEE Trans Pattern Anal Mach Intell. 2013;35(2):286–299. doi: 10.1109/TPAMI.2012.69. [DOI] [PubMed] [Google Scholar]
- Suk HI, Lee SW, Shen D. Subclass-based multi-task learning for Alzheimer’s disease diagnosis. Front Aging Neurosci. 2014;6(168) doi: 10.3389/fnagi.2014.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk HI, Lee SW, Shen D. Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain Struct Funct. 2015;220(2):841–859. doi: 10.1007/s00429-013-0687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk HI, Wee CY, Shen D. Discriminative group sparse representation for mild cognitive impairment classification. Mach Learn Med Imaging Lect Notes Comput Sci. 2013;8184:131–138. [Google Scholar]
- Thung KH, Wee CY, Yap PT, Shen D. Neurodegenerative disease diagnosis using incomplete multi-modality data via matrix shrinkage and completion. NeuroImage. 2014;91:386–400. doi: 10.1016/j.neuroimage.2014.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the LASSO. J R Stat Soc Ser B. 1994;58:267–288. [Google Scholar]
- Varoquaux G, Gramfort A, Poline JB, Thirion B. Brain covariance selection: better individual functional connectivity models using population prior. In: Lafferty JD, Williams CKI, Shawe-Taylor J, Zemel RS, Culotta A, editors. Advances in neural information processing systesms. Vol. 23. 2010. pp. 2334–2342. [Google Scholar]
- Visser PJ, Verhey FRJ, Hofman PAM, Scheltens P, Jolles J. Medial temporal lobe atrophy predicts Alzheimer’s disease in patients with minor cognitive impairment. J Neurol Neurosurg Psychiatry. 2002;72(4):491–497. doi: 10.1136/jnnp.72.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd K, Fjell A, Brewer J, McEvoy L, Fennema-Notestine C, Jr, Hagler DJ, Jennings R, Karow D, Dale A, the Alzheimer’s Disease Neuroimaging Initiative Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. Am J Neuroradiol. 2010;31:347–354. doi: 10.3174/ajnr.A1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang Z, Yan J, Li T, Rao B, Fang S, Kim S, Risacher S, Saykin A, Shen L. Sparse Bayesian multi-task learning for predicting cognitive outcomes from neuroimaging measures in Alzheimer’s disease. 2012 IEEE conference on computer vision and pattern recognition (CVPR).2012. pp. 940–947. [Google Scholar]
- Wang H, Nie F, Huang H, Risacher S, Ding C, Saykin A, Shen L. Sparse multi-task regression and feature selection to identify brain imaging predictors for memory performance. 2011 IEEE international conference on computer vision (ICCV); 2011. pp. 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Chen L, Yap PT, Wu G, Shen D. Groupwise registration based on hierarchical image clustering and atlas synthesis. Human Brain Mapp. 2010;31(8):1128–1140. doi: 10.1002/hbm.20923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nie J, Yap PT, Li G, Shi F, Geng X, Guo L, Shen D, for the Alzheimer’s Disease Neuroimaging Initiative Knowledge-guided robust MRI brain extraction for diverse large-scale neuroimaging studies on humans and non-human primates. PLoS ONE. 2014;9(1):e77810. doi: 10.1371/journal.pone.0077810. doi: 10.1371/journal.pone.0077810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Dunbrack, Lehmann RL. The role of balanced training and testing data sets for binary classifiers in bioinformatics. PLoS ONE. 2013;8(7):e67863. doi: 10.1371/journal.pone.0067863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M. Bayesian factor regression models in the “large p, small n” paradigm. Bayesian statistics. 2003:723–732. [Google Scholar]
- Westman E, Muehlboeck JS, Simmons A. Combining MRI and CSF measures for classification of Alzheimer’s disease and prediction of mild cognitive impairment conversion. NeuroImage. 2012;62(1):229–238. doi: 10.1016/j.neuroimage.2012.04.056. [DOI] [PubMed] [Google Scholar]
- Xiang S, Yuan L, Fan W, Wang Y, Thompson PM, Ye J. Bi-level multi-source learning for heterogeneous block-wise missing data. NeuroImage. 2014;102(1):192–206. doi: 10.1016/j.neuroimage.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Hu B, Liang C, Zhao L, Jackson M, the Alzheimer’s Disease Neuroimaging Initiative A longitudinal study of atrophy in amnestic mild cognitive impairment and normal aging revealed by cortical thickness. PLoS ONE. 2012;7(11):e48973. doi: 10.1371/journal.pone.0048973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Wang Y, Thompson PM, Narayan VA, Ye J. Multi-source feature learning for joint analysis of incomplete multiple heterogeneous neuroimaging data. NeuroImage. 2012;61(3):622–632. doi: 10.1016/j.neuroimage.2012.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. J R Stat Soc Ser B. 2006;68(1):49–67. [Google Scholar]
- Zhang D, Shen D. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. NeuroImage. 2012;59(2):895–907. doi: 10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shen D. Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PLoS One. 2012;7(3):e33182. doi: 10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage. 2011;55(3):856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- Zhou J, Liu J, Narayan VA, Ye J. Modeling disease progression via multi-task learning. NeuroImage. 2013;78:233–248. doi: 10.1016/j.neuroimage.2013.03.073. [DOI] [PubMed] [Google Scholar]
- Zhu X, Suk HI, Shen D. Matrix-similarity based loss function and feature selection for Alzheimer’s disease diagnosis. 2014 IEEE conference on computer vision and pattern recognition (CVPR); 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]