Abstract
Waterborne pathogens pose significant threat to the global population and early detection plays an important role both in making drinking water safe, as well as in diagnostics and treatment of water-borne diseases. We present an innovative centrifugal sedimentation immunoassay platform for detection of bacterial pathogens in water. Our approach is based on binding of pathogens to antibody-functionalized capture particles followed by sedimentation of the particles through a density-media in a microfluidic disk. Beads at the distal end of the disk are imaged to quantify the fluorescence and determine the bacterial concentration. Our platform is fast (20 min), can detect as few as ∼10 bacteria with minimal sample preparation, and can detect multiple pathogens simultaneously. The platform was used to detect a panel of enteric bacteria (Escherichia coli, Salmonella typhimurium, Shigella, Listeria, and Campylobacter) spiked in tap and ground water samples.
INTRODUCTION
Safety and quality of drinking water is necessary for the well-being of the human population worldwide. Bacterial contamination of water sources is responsible for diarrheal diseases with possible outcomes such as fever, nausea, dehydration, and, in extreme cases, death. Fresh, uncontaminated water is quickly becoming a scarce commodity and it is estimated that two and a half billion people in developing countries do not yet have acceptable water sources and sanitation.1 It is estimated that by the year 2025, more than half of the world population still will not have enough water necessary to support a healthy lifestyle.2 For people living in third world countries, the risk of infection is much higher due to unsanitary conditions, large population increases, and the inability to improve water treatment and distribution processes. In addition to this, out of 4 × 109 occurrences of waterborne infection worldwide, there is an estimated 2 × 106 deaths, most of them in the third world countries among children and adults with weakened immune systems.3
The ultimate solution for eliminating or minimizing infections with waterborne pathogens is to develop cost-effective and scalable water purification systems. However, a large part of the world does not have the proper resources and is not likely to have them for years to come. The next best solution then is to develop rapid, inexpensive tests for detecting the deadly pathogens in water so that the contaminated water sources are quickly identified and eliminated or treated. However, there is a dearth of cost-effective, sensitive detection systems. For example, the infectious dose for Salmonellosis and Listeriosis is approximately 103 CFU/ml, while the current level of detection using conventional enzyme-linked immunosorbent assay (ELISA) is approximately 105 and 104 CFU/ml, respectively,4–6 additionally, these tests require multiple time-consuming preparation and incubation steps. The infectious dose of Campylobacter is 10–100 CFU, but it can only be detected at concentrations of 104–105 CFU/ml with traditional methods.7,8 There exist numerous portable methods and devices for pathogen identification, but they suffer from critical disadvantages. The fecal indicator test is a widely available indirect method for checking for pathogens based on assumption that if there are any traceable amounts of fecal indicators in water, there must be a presence of bacteria too. Although this test is inexpensive and simple, the correlation between presence of feces and pathogen has not been consistent.9,10 Pathogen identification using polymeraze chain reaction (PCR) is a popular method due to its ability to detect low levels of pathogens within a few hours.11,12 However, PCR assays remain laboratory-based, requiring dedicated personnel and instrumentation. Furthermore, multiplexed PCR assays become increasingly costly and labor-intensive. The DNA microarray technology, with two-step PCR, has been shown to effectively identify the pathogen and the species, but the prerequisite of having a PCR reaction prior to hybridization implies longer assay time and significant cost.13,14 Some of the EPA-approved methods using ELISAs show a detection limit of approximately 105 CFU/ml with total hands-on time of 2 h.15 However, EPA Method 1200 for drinking and surface water analysis requires 24 to 42 h incubation period, following the 24-h enrichment step in tryptic soy broth.16 There are also many home-use rapid kits for bacterial detection in house water or swimming pools. However, most of these suffer from poor sensitivity, specificity, long assay time (up to 38 h) and can only detect one pathogen at a time.
Microfluidic platforms, because of their ability to analyze minute amounts of sample rapidly, have attracted considerable attention for sensing applications including pathogen detection and medical diagnostics.17,18 A few innovative microfluidic platforms have been described recently including surface plasmon resonance detection of Escherichia coli and Staphylococcus aureus with the limit of detection (LOD) 1000 CFU/ml,19 a microfluidic impedance-based biosensor with the limit of detection 3000 CFU/ml,20 detection of E. coli by microflow cytometer,21 and detection of Streptococcus agalactiae using magnetoresistive sensor.22
Centrifugal microfluidics, a method that relies on centrifugal force to actuate flow, has proven to be a versatile platform because of its simplicity and ability to integrate many assay steps into a single device.23 Many centrifugal immunoassay platforms have been described, including adaptations of data compact discs, adaptation of conventional immunoassays, and protein microarray based immunoassays.23,24 Researchers at Samsung have developed a fully automated centrifugal platform for performing immunoassays.24 While these methods are innovative, they are fairly complex requiring several valves and on-disk reagent reservoirs. The need for multiple wash steps to remove non-specifically bound reagents necessitates incorporation of multiple wash steps. These result in complex disk architecture that reduces the potential for multiplexing and workflow requiring longer assay durations.
In this paper, we present an innovative approach to detect bacterial targets in ground water using a sedimentation-based immunoassay, which does not require any wash steps or reservoirs. Antibody-derivatized silica beads are incubated with water sample and fluorescently labeled antibodies. The mixture is loaded on top of a density media in the disc. When the disc is spun, bead-bound bacterial target and fluorophore-labeled antibody sediment through the density media, leaving behind the unbound analyte and/or fluorophore-labeled antibody. The beads are concentrated at the distal end of the disk and fluorescence is measured to quantify the analyte. Multiple channels in one disk allow parallel assays leading to detection of multiple pathogens in a single run. This general experimental set-up can be modified to detect any pathogen of interest by substituting its corresponding capture and detection antibodies. In addition, the set-up uses low-cost equipment with minimal hands-on approach, no enrichment steps requiring addition of media to promote growth of bacteria, with overall experimental time (incubation steps, spin through density media, and result read-out) of approximately 20 min and limit of detection of ∼10 bacteria/sample. The approach does require pre-concentration of water sample using a commercial filtration system to concentrate bacteria from 20 ml to ∼100 μl volume.
MATERIALS AND METHODS
Unless otherwise specified, reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Antibodies: Antibodies for Shigella (PA1-7245), Listeria (PA1-7230), Salmonella (PA1-7244), and E. coli (PA1-7213) were obtained from Pierce (Rockford, IL). Bac-Trace Campylobacter (01-92-93) antibody was obtained from KPL Gaithersburg, MD.
Bacterial Pathogens: Salmonella typhimurium (50-74-01), Shigella (50-90-01), Listeria (50–90-90), Campylobacter (50-92-93), and E. coli O157:H7 (50-95-90) were obtained from KPL (Gaithersburg, MD).
Microspheres: Alexa 488 Fluorescent Microspheres (T-8864), Alexa 647 Fluorescent Microspheres (A-20186) were obtained from Invitrogen (Eugene, OR). Carboxylic acid functionalized silica microspheres were obtained from, Bangs Laboratories, Inc. (SC06N/11491; Fishers, IN).
Wash buffer contained 10 mg bovine serum albumin (BSA) (0.1% w/v), 5 mg NaN3 (0.05% w/v), 10 mg Pluronic F-127 (0.1% w/v) in 10 ml PBS. Other buffers used were phosphate buffer saline (PBS) (138 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), 2-(N-morpholino)ethanesulfonic acid (MES) (0.1 M), and sodium bicarbonate (1 M). Density media used was Histopaque 1119 from Sigma Aldrich (11191-100 ml).
Silica beads functionalization: 20 mg of 5 μm carboxylated silica beads were washed with 500 μl of 0.1 M MES buffer twice, sonicated 20 min, washed twice with MES, and resuspended in 800 μl MES. 10 mg each of 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and N-Hydroxysuccinimide (NHS) were dissolved in 200 μl MES, added to the washed silica beads and allowed to incubate for 30 min at 25 °C with continuous rotation. The beads were washed twice with 1 ml MES, twice with 1 ml PBS, and resuspended in 900 ml PBS. 75 mg of poly(ethylene glycol) bis(amine), MW 6000 and 100 μl of 1 M of sodium bicarbonate (NaHCO3) were added to the reaction and allowed to incubate for 2 h at 25 °C with continuous rotation. After two washes with PBS, the beads were resuspended in 500 μl PBS and 20 mg of succinic anhydride was slowly added to the reaction not to allow pH drop below 8.0, adding NaHCO3 as needed. The reaction was incubated for 2 h at 25 °C with continuous rotation, washed twice in PBS, twice in MES, and resuspended in 800 μl MES. 10 mg each of EDC and NHS were dissolved in 200 μl MES, added to the washed silica beads and allowed to incubate for 30 min at 25 °C with continuous rotation. After two washes with MES and PBS, the beads were resuspended in 900 μl PBS, 100 μl NaHCO3, and 20 mg of desalted antibody was added to reaction and allowed to incubate overnight at 4 °C with continuous rotation. After two washes with PBS, the beads were blocked with 1% BSA at 25 °C with continuous rotation for 30 min, washed with wash buffer and resuspended in 500 μl wash buffer. The beads can be stored in wash buffer at 4 °C up to six months.
Antibody labeling with fluorophore: Using 100 kDa filter, the antibody was concentrated to at least 10 mg/ml, and brought to 45 μl final volume with PBS and 5μl NaHCO3 (0.1 M). 10 μl of stock Alexa 488 Fluorescent microspheres (9.51E + 12 spheres/ml) were mixed with 10 μl DMSO, 8 μl of mix were added to detection antibody solution and incubated 1 h at 25 °C with continuous rotation. The reaction was eluted with Zeba 7 kDa column based on manufacturer's protocol and the eluent was measured at A280 and A488 to determine the degree of labeling (approximately 5–6 fluors per antibody).The concentrated fluorophore-labeled antibody could be stored in 1 mg/ml BSA at 4 °C up to six months and antibody diluted to a working concentration can be stored at 4 °C up to a week.
Microfluidic disk fabrication: Microfluidic disks were designed in AutoCAD (Autodesk) and fabricated by sandwiching a double-sided pressure-sensitive adhesive (9474-12 × 12, 9474LE 300 LSE, 3 M, St. Paul, MN) between two acrylic sheets (PD-15401710, 0.060 × 12 × 24, McMaster Carr, Atlanta, GA). Channel patterns were cut in the pressure-sensitive adhesive layer before assembly by using a computer-controlled laser-cutter (Universal Laser Systems 60 W VLS 6.60). The top acrylic sheet contained fluid access and vent openings also cut by the laser-cutter. The three layers were assembled by alignment and sealing under applied pressure using Jack Richeson & Co. “Baby Press” (Kimberly, WI). The total final volume of each of the 21 wells on the disk was 15μl.
Water Filtration and Sample recovery: Ground water was collected and filtered through a Whatman filter paper (WHA1001325) to remove debris as a pre-filtration step. The typical volume used for assay was 5–10 μl, and hence, a preconcentration step was used to reduce the volume from 20 ml to 100μl using Vivaspin 20 centrifugal concentrators (Cole-Parmer, WU-36224–81). 20 mm batches of tap or ground water were spiked with bacteria in serial dilution from 104 to 102 bacteria/ml. The 20 ml aliquot of ground or tap water was added to the concentrator, which was then loaded in a bench-top centrifuge. After centrifugation for 10 min at 3000 g, the volume retained above the filter, approximately 100 μl, was recovered. Ten out of 100μl of the concentrated bacteria was mixed with corresponding capture antibody on silica beads and detection antibody with fluorophore, as explained in the “Centrifugal immunoassay” section. The flow-through (containing no bacteria) from the concentrator was used as negative control.
Centrifugal immunoassay
Two percent of silica beads (w/v) functionalized with antibody were added to 10 μl of bacterial sample in 10-fold dilution series from 104 cells/ml to 3 × 102 cells/ml in wash buffer, tap, or ground water and allowed to incubate for 5 min with each dilution in an individual microcentrifuge tube. After addition of 3 μl of fluorophore-modified antibody (10 nM), the reaction was allowed to proceed for another 15 min. Meanwhile, 3 μl of Histopaque 1119 is loaded in each well in the microfluidic disk and the disk is spun at 8000 RPM for 1 min to allow the density gradient to settle. After the incubation is done, the 5 μl of reaction volume is transferred from microcentrifuge tube to the disk. The disk is spun at 8000 RPM for 1 min to allow the beads with bound pathogens and their corresponding detection antibody to pellet at the distant tip of the well. The major difference of microfluidics approach from conventional centrifugation in individual tubes is that the density gradient acts as a washing medium, separating the unbound analyte and fluorescently labeled antibodies from the pathogen/fluorescent antibody complex bound to the beads. Furthermore, the disk, unlike standard microcentrifuge tubes, can be directly analyzed via microscopy following centrifugation. The “free” analyte and antibodies by themselves, when not bound to beads, do not have sufficient density to pass through the density gradient. After centrifugation, the signal from the beads is quantitated by fluorescent microscopy (Nikon Inverted Microscope Eclipse Ti, Melville, NY). The microscope lens with magnification 10× and exposure time of 40 ms were used for all experiments. Standard exposure, gain, and excitation intensity were used for image acquisition. No significant photobleaching was observed throughout our experiments due to relatively photostable probes, large working distance (7.5 mm), and illumination of samples through 10× (low-power) objective.25–28 The exposure of sample to the light was minimized to 1–2 s during focusing. An area of 400 000 pixels (0.33 mm2, approximately 1/3 of the total bead area) was designated as “reading area” to record the mean intensity of each individual sample. The mean intensities of three replicas of each sample were collected and averaged to determine the average values and standard deviation of samples, as explained in the “Statistical analysis” section.
Statistical analysis
The average background fluorescence signal (n = 3, mean intensity of silica beads only, without analyte and detection antibody) was subtracted from the collected individual raw data points; then, the average value of the signal intensity for every bacterial concentration was calculated. To normalize the data, all the averaged values were divided by the averaged fluorescence intensity of the highest bacterial concentration. The standard deviation was determined by calculating the square root of the sum of squared standard deviations of background and averaged data points. The error bars represent ±1 standard deviation. Kaleidagraph software was used for curve fitting and calculations of limit of detection and chi-squared values, allowing 0.1% error and first-degree parameter partial derivatives. All curves are represented as sigmoidal fit and their R2 values are higher than 0.97. The LOD and the limit of quantification (LOQ) were set as three and ten standard deviations, respectively, higher than the signal for negative control sample, which included functionalized silica beads incubated with corresponding fluorescently labeled antibody but without the analyte of interest.
RESULTS AND DISCUSSION
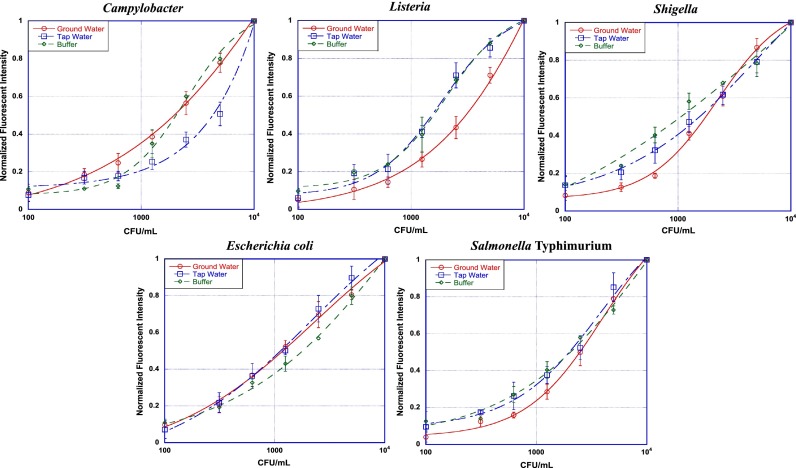
Contamination of drinking water with enteric bacteria is a growing problem and leads to high mortality and morbidity in many parts of the world. Access to inexpensive, portable technology to test water for presence of bacteria will be of considerable value to stop spread of diseases by treating/quarantining of contaminated water sources. Towards this goal, we have developed multiplexed immunoassays that can be implemented in a portable microfluidic platform developed by our group previously.29,30 The sensing method used was sandwich immunoassays performed using silica beads (Figure 1). High specificity antibodies (capture antibodies) were covalently attached to the silica beads using a PEG linker. Based on A280 and A488 measurements, there were 5–6 fluorophores attached to each antibody. Based on the concentration of antibodies provided by the supplier, there were ∼1000 antibodies per bead. The antibody-bearing beads were incubated with water sample containing bacterial cells and fluorescently labeled antibodies (detection antibodies). This mixture was added to the disc (Fig. 1(J)) containing a density medium and the disc was spun to centrifuge the beads to the distal end of a channel (periphery of the disc). The signal was measured using a fluorescent microscope. The results are shown in Figure 2 for the 5 bacteria tested. The limits of detection in buffer were ∼103 CFU/ml as determined by sigmoidal fit (R2 values were greater than 0.99) and are shown in Table I. The limit of detection in terms of number of bacteria detected per assays was Campylobacter: 12, Listeria: 19, Shigella: 3, E. coli: 8, and Salmonella: 22. The entire assay was completed in less than 30 min, as compared to hours for commercial ELISAs and >1 day for culture-based assays. Moreover, our approach requires minimal sample preparation other than concentration of bacteria from ∼20 ml volume to ∼100 μl using EPA-approved method.
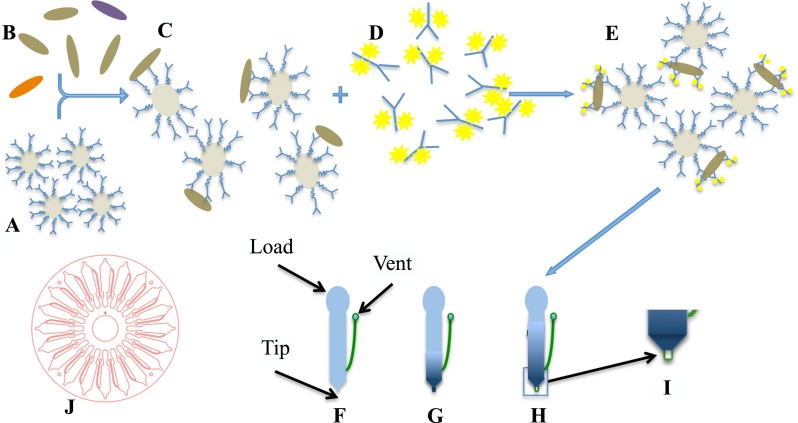
FIG. 1.
Carboxylated silica beads with anti-bacteria antibodies attached to a PEG-linker (A) are mixed with bacteria of interest (B) and allowed to incubate (C). Anti-bacteria antibodies labeled with Alexa 488 fluorophore (D) are added and incubated (E). To an empty well on the disk (F), density gradient is added (G), after which 6 μl of reaction from (E) are added to the well and centrifuged to separate beads with specifically bound bacteria and fluorophore-labeled antibody from unbound reagents and sample (H). The pelleted beads at the distal end are imaged with a fluorescence microscope (I). (J) Schematic of a microfluidic disk.
FIG. 2.
Bacterial detection in assay buffer (n = 3), tap water (n = 9), and ground water (n = 9). For detection in buffer, each bacteria in ten-fold serial dilution was incubated with its corresponding capture (5 min) and detection (15 min) antibody and spun at 8000 RPM for 1 min through density gradient. For detection in tap and ground water, 20 ml of tap or ground water were spiked with bacteria and concentrated using VivaSpin columns. Corresponding buffers were added for each bacteria and the assay were performed as explained earlier. The fluorescent signal was quantified using fluorescent microscopy. The normalized fluorescence for negative control sample is represented by concentration 100 CFU/ml.
TABLE I.
Limits of detection (LOD), Limit of Quantification (LOQ), chi-squared, and R2 values of bacterial detection (CFU/ml).
| LOD (CFU/ml) | LOD (bacteria/assay) | LOQ (CFU/ml) | LOQ (bacteria/assay) | chi2 | R2 | ||
|---|---|---|---|---|---|---|---|
| Buffer | Campylobacter | 3.58 × 103 | 12 | 1.54 × 104 | 51 | 1.37 × 10−3 | 0.9993 |
| Listeria | 5.80 × 103 | 19 | 6.65 × 104 | 222 | 1.89 × 10−3 | 0.99892 | |
| Shigella | 9.09 × 102 | 3 | 5.28 × 104 | 176 | 2.20 × 10−3 | 0.99834 | |
| E. coli | 2.33 × 103 | 8 | 4.01 × 104 | 134 | 7.30 × 10−3 | 0.99167 | |
| Salmonella | 6.72 × 103 | 22 | 1.34 × 105 | 447 | 2.55 × 10−3 | 0.99681 | |
| Ground water | Campylobacter | 5.63 × 103 | 19 | 2.18 × 104 | 73 | 3.70 × 10−3 | 0.99638 |
| Listeria | 9.19 × 103 | 31 | 2.70 × 104 | 90 | 5.11 × 10−3 | 0.99571 | |
| Shigella | 4.67 × 103 | 16 | 1.16 × 104 | 39 | 2.18 × 10−3 | 0.99812 | |
| E. coli | 1.14 × 103 | 4 | 3.69 × 103 | 12 | 4.90 × 10−3 | 0.99473 | |
| Salmonella | 4.58 × 103 | 15 | 1.40 × 104 | 47 | 2.70 × 10−3 | 0.99757 | |
| Tap water | Campylobacter | 9.55 × 103 | 32 | 3.51 × 104 | 117 | 2.88 × 10−2 | 0.97294 |
| Listeria | 4.51 × 103 | 15 | 1.52 × 104 | 51 | 5.61 × 10−3 | 0.99499 | |
| Shigella | 4.97 × 103 | 17 | 2.13 × 104 | 71 | 4.35 × 10−3 | 0.9952 | |
| E. coli | 3.57 × 103 | 12 | 1.39 × 104 | 46 | 2.73 × 10−3 | 0.99728 | |
| Salmonella | 9.43 × 102 | 3 | 3.8 × 103 | 13 | 5.01 × 10−3 | 0.99292 | |
| Multiplexed | Campylobacter | 2.41 × 103 | 8 | 8.09 × 103 | 27 | 3.04 × 10−3 | 0.99588 |
| Listeria | 5.13 × 103 | 17 | 2.06 × 104 | 69 | 1.36 × 10−3 | 0.9985 | |
| Shigella | 4.69 × 103 | 16 | 1.37 × 104 | 46 | 1.48 × 10−2 | 0.98556 | |
| Salmonella | 5.61 × 103 | 19 | 1.40 × 104 | 47 | 1.54 × 10−3 | 0.99834 |
The first step in developing our bead-based sandwich immunoassays was screening of commercial antibodies to find high-sensitivity pairs for each bacteria. Criteria used for selection of the optimum pair include LOD and LOQ in relevant sample matrix, non-specific binding, and cross-reactivity with other bacterial species.
The immunoassays we developed have higher detection sensitivity than most commercial ELISAs. The enhanced sensitivity of our platform is attributable to (a) isolation of the beads from the sample during sedimentation removes background samples species improving signal/noise and (b) pelleting the beads at the end of the channel allows fluorescence to be integrated over hundreds of beads.
Detection of bacteria in tap water
While tap water is typically considered safe from microbiological contamination because it is delivered after treatment (filtration and chlorination), in most parts of the world it gets contaminated due to leaks or improper storage. Hence, it is desirable to have portable detectors to test tap water where chances of microbiological contamination are high. We successfully developed immunoassays for detection of water-borne bacteria spiked in tap water as shown in Figure 2. Bacteria were spiked in 20 ml of tap water and the water was filtered to concentrate the bacteria in 100 μl volume. The recovery rate was found to be approximately 95% (data not shown) after filtration for each of the 5 bacteria. As tap water has very low ionic strength and variable pH, we mixed it 1:1 with buffer (PBS) to achieve optimal conditions for binding of antibodies. We also evaluate different salt concentrations to improve signal and reduce background and found that E. coli and Shigella assays had the best performance at 100 mM NaCl, while Campylobacter, Salmonella, and Listeria require 50 mM MgSO4 and 50 mM (NH3)2SO4. The incubation times were optimized to yield the largest dynamic range. Even after optimization, the limits of detection in tap water were slightly higher than those observed in buffer and are listed in Table I. This could be because of the presence of chlorine in tap water. However, the detection limit of our assays is better than most commercially available ELISAs, including those that adhere to EPA guidance.
Detection of bacteria in ground water
Ground water remains a significant source of drinking water in rural parts of the USA and many parts of the third world. With ever-increasing contamination from open sewage systems and leaky septic tanks, many sources of groundwater around the world are at risk for microbiological contamination and it is recommended to test them periodically to ensure their safety. Our centrifugal platform was able to detect bacteria spiked in ground water with high sensitivity. Since ground water contains particulates, the water was pre-filtered through a Whatman filter (11 μm pore size). The assumption was made that any bacteria present in water will pass through the filter and will only be collected at the concentration step. The 20 ml aliquots were spiked with bacteria and assays were performed using the same buffers and salts as with the tap water. We were able to detect bacteria with high sensitivity as shown in Figure 1 and Table I.
Multiplexed assay
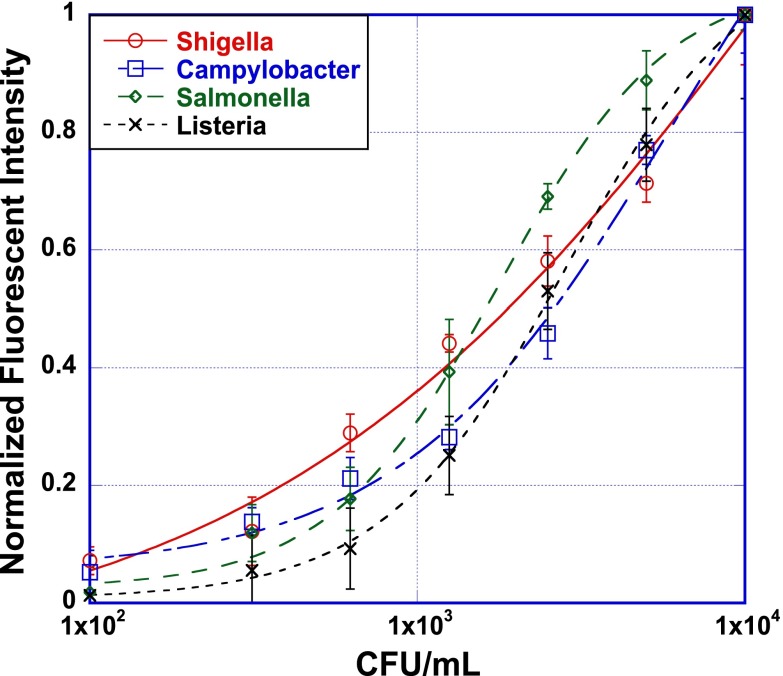
The aforementioned immunoassays were done with single type of bacteria spiked in water. However, for a portable detector to have broad applicability, it is desirable to have the ability to detect multiple pathogens. This is fairly easily accomplished in our platform because each disk can have as many as 21 channels, with each channel capable of detecting one pathogen. We demonstrated the multiplexing capability by spiking ground water with 4 species of bacteria and then detecting one each in individual channels. The results are presented in Figure 3. The twenty-milliliter ground water sample spiked with four bacteria (Campylobacter, Listeria, Salmonella, and Shigella) at the initial concentration of 104 each bacteria per milliliter and concentrated to 100 μl with VivaSpin columns as earlier described. The negative control (“zero” sample) for each bacteria contained 104 CFU/ml of bacteria that did not correspond to the capture and detection antibody. For example, the negative control for Salmonella contained 104 CFU of each Shigella, Listeria, and Campylobacter.
FIG. 3.
Multiplexing assay in ground water (n = 3). Bacteria at concentration 104 CFU/ml each was spiked into the pre-filtered 20 ml ground water sample. The concentration and assay steps were performed as described in sections above. The normalized fluorescence for negative control sample is represented by concentration 100 CFU/ml.
Since E. coli and Shigella are the two species that are very closely related,24,31 we were unable to find antibodies that can differentiate the two. Hence, E. coli was omitted from the multiplexing experiments. It is possible to differentiate between the two but that is unlikely with a pair of polyclonal antibodies (such as those used in this study); species-specific monoclonal antibodies might have a higher probability but they are not commercially available. Some PCR assays can distinguish between E. coli and Shigella, but only if the amplicons are of very different sizes,32 and require a larger number of bacteria per reaction in a relatively clean background (i.e., low amounts of other background microflora).33 The limits of detection for each bacteria were: Campylobacter: 2.41 × 103 CFU/ml or, 8 bacteria/assay, Listeria: 5.13 × 103 CFU/ml or 17 bacteria/assay, Salmonella: 5.61 × 103 CFU/ml or 16 bacteria/assay, and Shigella: 4.69 × 103 CFU/ml or 19 bacteria/assay. These values are marginally higher than the LODs obtained in groundwater using individual bacteria suggesting low levels of cross-reactivity and non-specific binding.
CONCLUSIONS
Our overarching goal is to develop a rapid, reliable, cost-effective device for testing of water for presence of pathogenic bacteria. We envision a simple device and assay workflow in which manual intervention is limited to introducing the water sample into a disk, loading the disk into a reader, and hitting the start button. The centrifugal platform we have developed is well suited for this as it is fast (<30 min assay time), multiplexed, sensitive, and requires minimal sample preparation. Low detection limits of enteric bacteria panel (E. coli, Listeria, S. typhimurium, Shigella, and Campylobacter) were achieved using our centrifugal platform for single bacteria in buffer, tap water, and ground water, as well as for multiple bacteria in ground water. Comparing the sensitivity of our assay to ELISA, where the detection limit for most bacteria ranges from 104 to 106 CFU/ml, we detect 10- to 1000-fold lower concentration of bacteria at levels of approximately 103 CFU/ml (or, as low as 10 bacteria per reaction).
Finally, this technique is highly adaptable for rapid development of new assays. The disk architecture is generic and disconnected from the assay reagents (beads). Hence, we can develop immunoassays for other pathogens fairly easily and rapidly (∼2 weeks) by preparing beads with antibodies against new analytes.
ACKNOWLEDGMENTS
This research was funded by NIAID (Grant No. R01AI0988530). Sandia National Laboratories is a multi-program laboratory managed and operated by Sandia Corporation, a wholly owned subsidiary of Lockheed Martin Corporation, for the U.S. Department of Energy's National Nuclear Security Administration under contract DE-AC04-94AL85000. SAND2015-1581J.
References
- 1.World World Health Organization, in UNICEF, United States: WHO/UNICEF Joint Monitoring Programme for Water Supply and Sanitation, 2012. [Google Scholar]
- 2. Robertson L. J., Epidemiol. Infect. 142, 1118 (2014). 10.1017/S0950268814000132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization, Diarrhoea: Why Children are still dying and what can be done ( UNICEF, 2009). [Google Scholar]
- 4. Chattopadhyay S., Kaur A., Jain S., and Singh H., Biosens. Bioelectron. 45, 274 (2013). 10.1016/j.bios.2013.01.032 [DOI] [PubMed] [Google Scholar]
- 5. Chunglok W., Wuragil D. K., Oaew S., Somasundrum M., and Surareungchai W., Biosens. Bioelectron. 26, 3584 (2011). 10.1016/j.bios.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 6. Ueda S. and Kuwabara Y., Biocontrol Sci. 15, 91 (2010). 10.4265/bio.15.91 [DOI] [PubMed] [Google Scholar]
- 7. Oyarzabal O. A. and Battie C., in Trends in Immunolabelled and Related Techniques, edited by Abuelzein E. ( INTECH Open Access Publisher, 2012). [Google Scholar]
- 8. Nyati K. K. and Prasad K. N., Med. Microbiol. Immunol. 199, 109 (2009). [DOI] [PubMed] [Google Scholar]
- 9. Savichtcheva O. and Okabe S., Water Res. 40, 2463 (2006). 10.1016/j.watres.2006.04.040 [DOI] [PubMed] [Google Scholar]
- 10. Field K. G. and Samadpour M., Water Res. 41, 3517 (2007). 10.1016/j.watres.2007.06.056 [DOI] [PubMed] [Google Scholar]
- 11. Lee D. Y., Seto P., and Korczak R., J. Microbiol. Methods 80, 129 (2009). [DOI] [PubMed] [Google Scholar]
- 12. Cai D., Xiao M., Xu P., Xu Y. C., and Du W., Lab Chip 14(20), 3917 (2014). 10.1039/C4LC00669K [DOI] [PubMed] [Google Scholar]
- 13. Brinkman N. E. and Fout G. S., J. Virol. Methods 156, 8 (2009). 10.1016/j.jviromet.2008.03.010 [DOI] [PubMed] [Google Scholar]
- 14. Leski T. A., Lin B., Malanoski A. P., Wang Z., Long N. C., Meador C. E., Barrows B., Ibrahim S. et al. , PLOS ONE 4, e.6569 (2009). 10.1371/journal.pone.0006569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.See http://www.epa.gov/ncer/publications/workshop/microorganisms_dw_092109.pdf, Philadelphia, PA, 2009.
- 16.EPA, Office of Water, see http://www.epa.gov/safewater for Method 1200: Analytical Protocol for Non-Typhoidal Salmonella in Drinking Water and Surface Water, 2012.
- 17. Meagher R. J., Light Y. K., and Singh A. K., Lab Chip 8(4), 527 (2008). 10.1039/b716462a [DOI] [PubMed] [Google Scholar]
- 18. Curtis T. L., Chin D., Cheung Y. K., Steinmiller D., Linder V., and Sia S. K. et al. , Nat. Med. 17, 1015 (2011). [DOI] [PubMed] [Google Scholar]
- 19. Chen L., Mungroo N., Daikuara L., and Neethirajan S., J. NanoBioTechnol. 13, 45 (2015). 10.1186/s12951-015-0106-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dastider S. G., Barizuddin S., Yuksek N. S., Dweik M., and Almasri M. F., J. Sens. 2015, 293461 (2015). 10.1155/2015/293461 [DOI] [Google Scholar]
- 21. Guo T., Wei Y., Xu C., Watts B. R., Zhang Z., Fang Q., Zhang H., Selvaganapathy P. R., and Deen M. J., Electrophoresis 36, 298–304 (2015). 10.1002/elps.201400211 [DOI] [PubMed] [Google Scholar]
- 22. Fernandes A. C., Duarte C. M., Cardoso F. A., Bexiga R., Cardoso S., and Freitas P. P., Sensors 14(8), 15496 (2014). 10.3390/s140815496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hugo S., Land K., Madou M., and Kido H., S. Afr. J. Sci. 110, 1 (2014). 10.1590/sajs.2014/20130091 [DOI] [Google Scholar]
- 24. Rahman M. Z., Akter S., Azmuda N., Sultana M., Weill F.-X., Khan S. I., Grimont P. A. D., and Birkeland N.-K., Curr. Microbiol. 67(5), 590 (2013). 10.1007/s00284-013-0405-7 [DOI] [PubMed] [Google Scholar]
- 25. Model M. A., Reese J. L., and Fraizer G. C., Cytometry, Part A 75(10), 874 (2009). 10.1002/cyto.a.20787 [DOI] [PubMed] [Google Scholar]
- 26. Waters J. C., J. Cell Biol. 185(7), 1135 (2009). 10.1083/jcb.200903097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Regev T., Meyers N., Zarivach R., and Fishov I., PLoS ONE 7(5), e36441 (2012). 10.1371/journal.pone.0036441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ohkura M., Sasaki T., Sadakari J., Gengyo-Ando K., Kagawa-Nagamura Y., Kobayashi C. et al. , PLoS One 7(12), e51286 (2012). 10.1371/journal.pone.0051286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schaff U. Y. and Sommer G. J., Clin. Chem. 57, 753 (2011). 10.1373/clinchem.2011.162206 [DOI] [PubMed] [Google Scholar]
- 30. Koh C. Y., Schaff U. Y., Piccini M. E., Stanker L. H., Cheng L. W., Ravichandran E., Singh B. R., Sommer G. J., and Singh A. K., Anal. Chem. 87, 922 (2015). 10.1021/ac504054u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chart H., Daniel R. M. A., and Cheasty T., FEMS Microbiol. Lett. 292, 21 (2008). [DOI] [PubMed] [Google Scholar]
- 32. Li Y. and Mustapha A., J. Food Prot. 67, 27 (2004). [DOI] [PubMed] [Google Scholar]
- 33. Li Y., Zhuang S., and Mustapha A., Meat Sci. 71, 402 (2005). 10.1016/j.meatsci.2005.04.013 [DOI] [PubMed] [Google Scholar]