Abstract
The monitoring of viral load is critical for proper management of antiretroviral therapy for HIV-positive patients. Unfortunately, in the developing world, significant economic and geographical barriers exist, limiting access to this test. The complexity of current viral load assays makes them expensive and their access limited to advanced facilities. We attempted to address these limitations by replacing conventional RNA extraction, one of the essential processes in viral load quantitation, with a simplified technique known as immiscible filtration assisted by surface tension (IFAST). Furthermore, these devices were produced via the embossing of wax, enabling local populations to produce and dispose of their own devices with minimal training or infrastructure, potentially reducing the total assay cost. In addition, IFAST can be used to reduce cold chain dependence during transportation. Viral RNA extracted from raw samples stored at 37°C for 1 week exhibited nearly complete degradation. However, IFAST-purified RNA could be stored at 37°C for 1 week without significant loss. These data suggest that RNA isolated at the point of care (eg, in a rural clinic) via IFAST could be shipped to a central laboratory for quantitative RT-PCR without a cold chain. Using this technology, we have demonstrated accurate and repeatable measurements of viral load on samples with as low as 50 copies per milliliter of sample.
Diagnostic technology in the developing world lags behind the developed world as a result of several factors, including economics, lack of reliable laboratory infrastructure, and limited access to personnel with technology-specific training.1 Although moderate-to advanced laboratories exist in urban clinics and large hospitals in the developing world, the facilities are often inaccessible to a large proportion of the population. Samples cannot be reliably transported to these facilities because of a lack of safe and continuous cold storage in remote areas.1 Without reliable cold storage, clinically important biomarkers (eg, nucleic acids, proteins, whole cells) may be partially or completely degraded by endogenous proteases and nucleases, resulting in a false-negative result. Thus, there exists a need for technologies that can isolate and stabilize these biomarkers while also rendering them noninfectious for safer handling. These technologies should have few requirements for laboratory infrastructure or trained personnel.
Currently, the predominant technology for preserving blood-based biomarkers is the dried blood spot (DBS). Although DBS enables biomarker analyses in remote settings, it possesses limitations, especially when quantitative results are required. Use of DBS adds extra steps to the diagnostic workflow, including spotting of the blood, contamination-free drying of the spot, and elution of the specimen from the DBS medium. Each of these DBS-specific steps requires extra labor and introduces additional and significant opportunity for error, particularly when performed in the field.2 As an alternative to DBS, sample preparation (ie, the extraction of the biomarker from the bulk sample) can be performed at the site of sample acquisition to isolate the biomarker into a nuclease/protease-free buffer or other preservative solution. Unfortunately, conventional sample preparation techniques (eg, phenol/chloroform extraction, spin columns) are quite involved, requiring an advanced laboratory setting and a skilled technician or robotic platform.
More recently, advanced technologies have emerged that enable simple nucleic acid extraction in a low-resource setting with minimal infrastructure requirements. One set of such technologies applies the lab-on-a-chip concept to miniaturize and automate the entire sample preparation process—and occasionally additional processes—on a single chip. These platforms capture nucleic acids on functionalized surfaces3, 4, 5 or immobilized paramagnetic particles (PMPs),6, 7 and then use microfluidic networks to wash and elute the nucleic acids. Another set of advanced sample preparation technologies leverage immiscible phase filtration, where a PMP-bound analyte is drawn from the sample through a different fluid (eg, oil, air) and into an elution buffer, greatly simplifying the sample preparation workflow.8, 9, 10, 11, 12
An important consideration in field-compatible sample preparation is proper disposal of medical waste generated as a result of sample acquisition and preparation. In low-resource settings, most medical waste is disposed via landfill or low-temperature incinerators without environmental controls (eg, a wood fire contained in a barrel).13 Most field-compatible sample preparation devices are intended for single use to limit cross-contamination. These devices are typically fabricated from a variety of materials that include plastic, elastomer, silicon, glass, and in some cases, electronic components. Low-temperature incineration (approximately 400°C) of these materials can prove problematic, particularly for some plastic devices, which can emit harmful chemicals, including dioxins, furans, and coplanar PCBs, on incineration.14 Exposure to these chemicals may lead to the impairment of the immune system and the impairment of the development of the nervous system, endocrine system, and reproductive functions.14
In addition to infrastructural limitations, viral load quantitation remains cost prohibitive for much of the developing world. Although accurate monitoring of viral load is essential to proper management of antiretroviral therapy, the high cost of viral load quantitation prevents many HIV-positive patients from obtaining routine measurements. Because of its complexity, sample preparation is a major contributor to the total cost of a viral load test because multiple PMP captures and liquid transfer steps must be performed to obtain suitable purity for efficient quantitative RT-PCR (RT-qPCR). Although complete sample-to-answer solutions remain highly appealing, we anticipate that significant reduction in sample preparation complexity and cost will substantially reduce the overall cost of the viral load assay.
Here, we designed and tested a sample preparation platform manufactured entirely from wax. In the microfabrication field, wax has been used extensively as a soft lithography mold,15 an adhesive,16 or to pattern channels on paper-based devices.17 Here, we fabricated the entire device from wax using a simple stamping process. Following operation, this device can be incinerated at a low temperature without producing the emissions linked to low-temperature incineration of some plastics. The sample preparation device is a modified version of immiscible filtration assisted by surface tension (IFAST), an immiscible phase filtration technique previously developed by our lab and manufactured in a plastic/elastomeric form.10 IFAST uses surface tension to position aqueous and oil phases side by side, such that nucleic acid (or another analyte) can be bound to a PMP in a sample and drawn through an oil phase into an elution buffer, thus purifying the nucleic acid in a single step. More recently, IFAST has been demonstrated to be highly amenable to parallel processing and automation,18, 19 such that high-throughput operation could be implemented when required. The IFAST device is simple enough for field operation, potentially eliminating the need for a cold chain; however, it is sufficiently inexpensive to substantially reduce the overall cost of a viral load assay.
Materials and Methods
Device Design and Fabrication
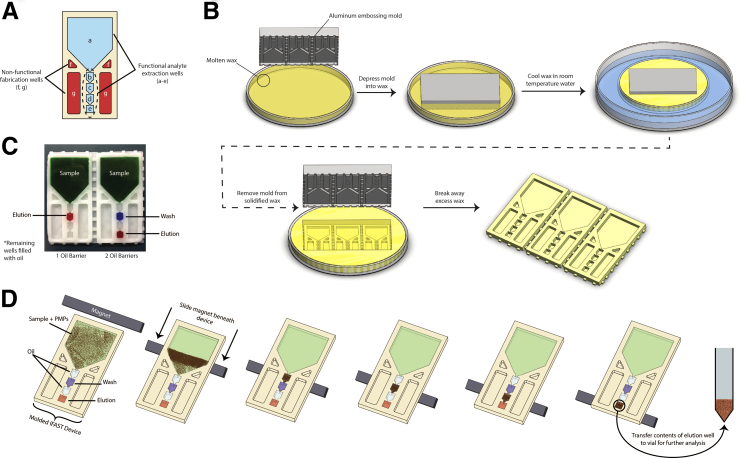
The wax IFAST uses adjacent immiscible liquids to purify target analytes by pinning subsequent droplets in small wells. The basic geometry consists of a large sample input well followed by several smaller wash/elution wells downstream (Figure 1A). Adjacent to the smaller wells, two pairs of pockets were molded into the wax to prevent heat concentrations from forming during the molding process. To form the devices, a negative mold containing all of the desired features was first machined from a plate of aluminum (alloy 6061; MetalsDepot, Winchester, KY). Glass Petri dishes were then thoroughly cleaned and filled with shavings of wax (B7347; Sasol Wax, Hayward, CA) before being placed on a 115°C hot plate. Once the wax had completely melted, the aluminum mold was depressed into the molten wax. The Petri dish was then removed from the hotplate and placed into a larger dish partially filled with room temperature water. When fully cooled, the wax disk was removed from the Petri dish. Excess wax around the mold was broken away and remelted, and the imprinted device removed from the mold. A schematic of this process can be seen in Figure 1B. Devices were qualitatively assessed for well-surface smoothness, with only those deemed sufficiently smooth used for analyte extraction. Those deemed insufficiently smooth or wrinkled were remelted to form new devices. Further information on well-surface quality is presented in the Discussion section.
Figure 1.
Diagram of the IFAST design. Connected wells in the center are used for analyte extraction, whereas wells on either side are for fabrication purposes only (A); schematic of wax IFAST fabrication (B); image illustrating the difference between using a single oil barrier (left) and two oil barriers (right) (C); schematic of IFAST sample extraction procedure (D).
Device Operation
Once fabricated, devices were placed on a flat surface and loaded such that the aqueous phases were separated by an immiscible oil phase. Sample, containing PMPs, was added to the input well (Figure 1A) of the device at a volume ranging from 100 to 500 μL depending on which size device was used. See VLP Detection in Varying Sample Volumes within the Materials and Methods and Results sections for further discussion of various sizes. As illustrated in Figure 1C, if a one-oil-barrier configuration was used, 20 μL of elution buffer was added to the middle well (Figure 1A). If a two-oil-barrier configuration was used, an aqueous wash buffer was added to the middle well, and elution buffer was added to the final well, both at a volume of 20 μL (Figure 1A). Once the aqueous buffers were in position, 20 μL of oil was added to the remaining wells to complete device filling. All necessary aqueous phases must be dispensed before addition of any oil. In this study, complete loading of each device was performed in <1 minute. Figure 1D depicts the basic IFAST extraction procedure. To operate the loaded wax IFASTs, magnets were slid underneath the devices to draw the PMPs from the input well into the output well. For these experiments, magnets were operated by hand at a velocity of approximately 1 to 2 mm/second. Small magnetic cubes (BX333-N52; K&J Magnetics, Pipersville, PA) were used for these experiments; however, a wide variety of magnets may be used for analyte extraction. For example, although these small cube magnets were used for single-device isolations in this study, longer magnetic bars (BX041-N52; K&J Magnetics) have been used for simultaneous operation of several devices in parallel.
Preparation of the HIV Model System (Viral-Like Particles)
As previously described, diagnostics in developing countries are often limited by labile biomarkers. Lack of effective HIV viral load monitoring in the developing world illustrates how diagnostic limitations affect patient outcomes. With that in mind, noninfectious HIV viral-like particles (VLPs) were created to serve as an initial model system for preliminary wax IFAST testing. These particles have been used extensively in virology studies, and additional characterization data have been previously reported.20 HEK293T cells (a generous gift from Dr. Elaine Alarid, University of Wisconsin–Madison) were plated onto 100-mm-diameter cell culture dishes at a density of 4.0 × 106 cells per dish. Cells were permitted to adhere overnight at 37°C in Dulbecco’s modified Eagle’s medium containing 4.5g/L d-glucose, 0.6 g/L l-glutamine (11968; Gibco, Grand Island, NY), and 10% fetal bovine serum (Gibco). Cells were transfected with 7.5 μg of p24 Gag-Pol-Vif expression plasmid (1 μg/μL) and 2.5 μg of pRev expression plasmid (1 μg/μL) (both expression plasmids were a generous gift from Dr. Nathan Sherer, University of Wisconsin–Madison), and 30 μL of FuGENE 6 (E2691; Promega, Madison, WI) in serum containing medium. The transfection medium was replaced with fresh medium 24 hours post-transfection. At 48 and 72 hours post-transfection, the medium was removed from the cell culture dishes and filtered through a 0.2-μm filter system (564-0020 Nalgene; Fisher Scientific, Waltham, MA) and then fresh medium was added to dishes at the 48-hour time point. The filtrate was centrifuged through a 20% sucrose gradient (S0389; Sigma-Aldrich, St. Louis, MO) for 2 hours at 21,000 relative centrifugal force at 4°C. The supernatant was discarded, and the pellet containing the viral-like particles was resuspended in 100 μL of PBS (MP Biomedicals 2810305; Fisher Scientific) supplemented with 10% fetal bovine serum. These particles were then stored at −80°C until use. Viral particles were spiked into fetal bovine serum (Gibco) to create mock patient samples. Spiked samples were mixed with an equal volume of lysis/binding buffer (Buffer MFL; Qiagen, Valencia, CA) that was preloaded with PMPs (MagAttract Suspension F, 1 μL per sample; Qiagen) and incubated at room temperature for 5 minutes to allow for lysis and binding. IFAST devices were then loaded with this lysed VLP solution and operated as previously described.
RT-qPCR Detection of VLPs
To quantify viral RNA extraction, RT-qPCR was performed on the IFAST-extracted RNA. After IFAST operation, the eluent containing the PMP-bound RNA was extracted via pipette and mixed with an equal volume of 2× reverse transcription master mix (High Capacity cDNA Master Mix; Life Technologies, Carlsbad, CA). RT was performed in a thermal cycler (Techne TC-412; Burlington, NJ) at 37°C for 1 hour followed by 85°C for 5 minutes. Following reverse transcription, cDNA was mixed with qPCR master mix (TaqMan Gene Expression Master Mix; Life Technologies) and primers and a probe specific to the long terminal repeat region of the HIV genome (forward primer: 5′-GCCTCAATAAAGCTTGCC-3′; reverse primer: 5′-GGCGCCACTGCTAGAGATTTT-3′; probe: 5′-AAGTAGTGTGTGCCC-3′; taken from Avettand-Fènoël et al21 and synthesized by Life Technologies). Data by other groups have shown that this PCR assay compares well with established commercial HIV assays.22 The PCR mixture was amplified for 40 cycles of 95°C for 15 seconds followed by 60°C for 1 minute using a real-time thermal cycler (LightCycler 480; Roche Applied Science, Indianapolis, IN). Serial dilutions of plasmid with known long terminal repeat copy numbers were used to establish a standard curve linking RT-qPCR threshold cycle (CT) to copy number. Briefly, plasmid DNA concentration was measured with a spectrophotometer (NanoDrop 1000; NanoDrop Technologies, Wilmington, DE), and this measurement was used to calculate the long terminal repeat copy number per microliter of plasmid stock solution. The standard curve was constructed by running qPCR on plasmid dilutions ranging from 50 to 5 × 1010 copies per reaction. Using this standard curve, viral loads were calculated from CT values for the IFAST-purified samples.
VLP Detection in Varying Sample Volumes
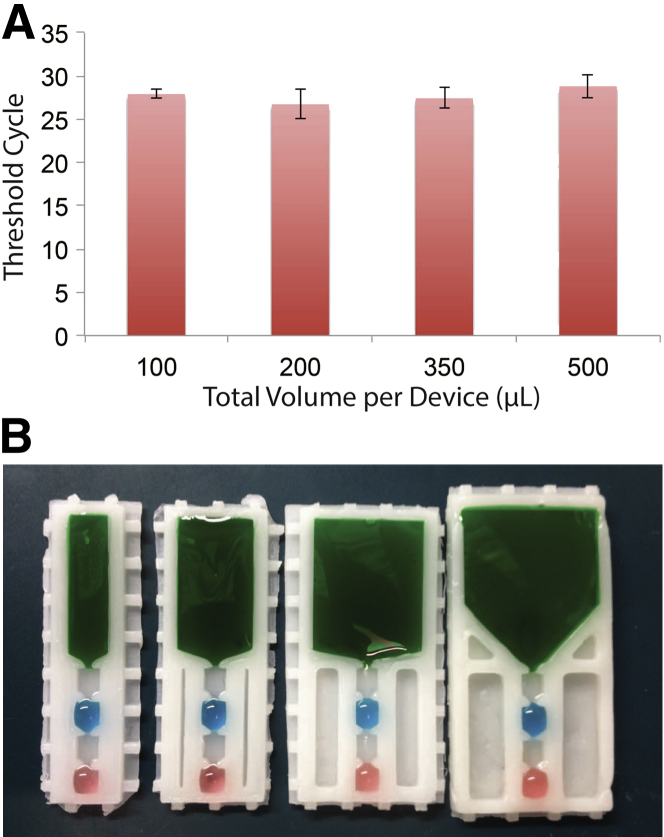
IFAST has already proven effective for extraction of various analytes at volumes between 5 to 10 μL.10 These volumes, however, are not conducive to sensitive detection of analytes in patient samples, which typically require several hundred microliters of input as a result of the low biomarker abundance. As such, we investigated the effects of scaling up input volumes on the quality of output as well as general device operation. A particular benefit of the wax IFAST device is its ability to be rapidly tailored to the specific requirements of individual assays. By changing the geometry of the mold, custom devices may be manufactured with any number of wells at varying volumes. Devices were fabricated with input well volumes of 100 μL, 200 μL, 350 μL, and 500 μL (Figure 2; see Supplemental Figure S1 for a dimensioned drawing of the mold). These were filled as previously described, and all sample wells were spiked with approximately 1.4 × 103 VLPs. One microliter of PMPs were then pulled through the device, and the eluent was collected, after which RT-qPCR was performed to determine mean CT as previously described.
Figure 2.
A: Correlation between sample input volume and RT-qPCR threshold cycle. Approximately 1400 VLPs were spiked into serum and loaded into each IFAST device. RNA was extracted and the level of recovered HIV RNA was measured via RT-qPCR. Error bars represent ±1 SD. B: Examples of each device size (100, 200, 350, and 500 μL).
Quantification of Carryover
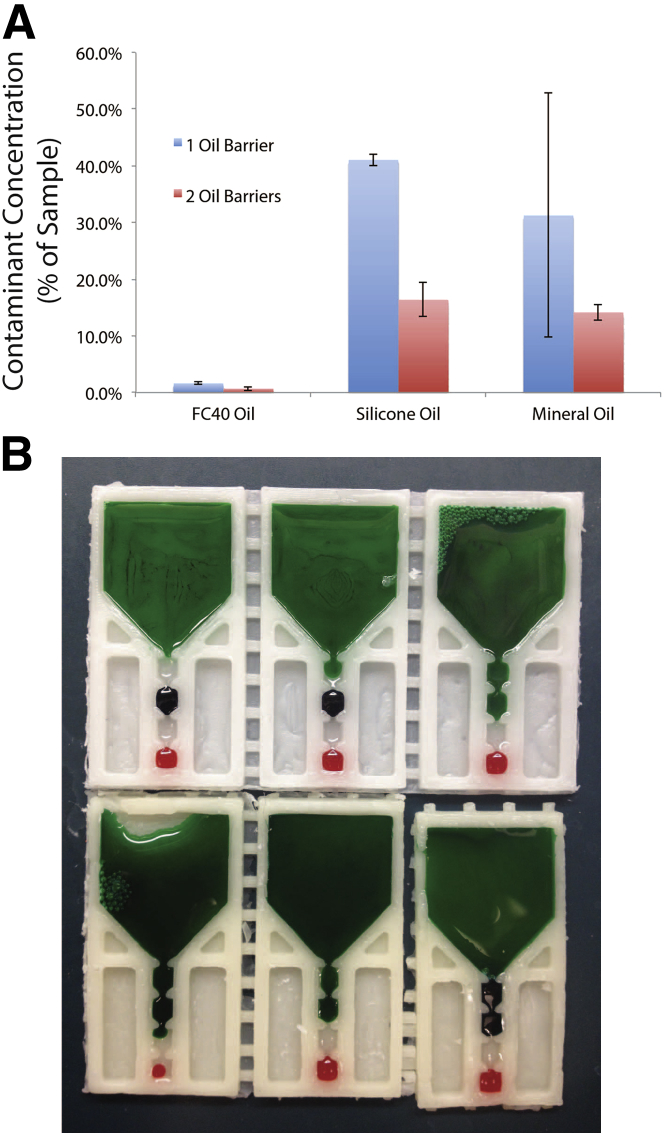
In previous studies with IFAST,23 we observed that a small volume (typically 0% to 2%) of input sample is nonspecifically transferred through the oil barrier during operation. However, these devices were of limited use for highly sensitive and/or dilute biomarkers because they only accommodated small sample volumes (5 to 10 μL) Therefore, we pursued studies investigating larger-volume devices. Of particular interest were performance differences between one and two oil barriers and performance differences between Fluorinert FC-40 oil (3M Company, St. Paul, MN), silicone oil (Fisher Scientific #S159-500), and mineral oil (Acros Organics #415080010; Geel, Belgium). To quantify the effect of oil on nonspecific carryover during wax IFAST operation, acridine orange (10 mg/mL stock solution) was added to a representative fetal bovine serum/lysis buffer (Buffer MFL; Qiagen) sample at a dilution of 1 μL of acridine orange stock solution to 100 μL of sample. PMPs were also added at a concentration of 1 μL of PMP stock solution per 500 μL of sample. For one-oil-barrier testing, wells were loaded in the following manner in accordance with Figure 1A: elution buffer (Buffer MFE; Qiagen) in well c, sample in a, and oil in b, d, and e. Oil added to d and e functions to maintain hydrostatic pressure in the system and prevent spillage into upstream wells. For two-oil-barrier testing, wells were filled in the following order: elution buffer in e, wash buffer (Buffer MFW2; Qiagen) in c, sample in a, and oil in b and d. In both schemes, 20 μL was added to wells b through e, whereas 500 μL was added to sample well a. After loading, devices were operated as previously described. Carryover was quantified by measuring acridine orange fluorescence with an Invitrogen Qubit 2.0 Fluorimeter using blue light excitation. Fifteen microliters was removed from the elution well (c for one barrier, e for two) and added to 185 μL of nuclease-free H2O in a Qubit Assay Tube (Q32866; Life Technologies). Sample fluorescence readings were compared to a previously constructed standard curve, and percent carryover was determined.
Sample Degradation
Viral RNA is susceptible to degradation via ribonucleases; therefore, methods to reduce degradation (eg, continuous cold chain, DBS) are typically required when transporting HIV plasma samples from the point of collection to the testing laboratory. To simulate these real-world conditions, we stored VLP samples at 37°C for 1 day or 1 week, and then purified RNA via IFAST and detected viral RNA via RT-qPCR as previously described. We used the wax IFAST device to purify RNA within 1 hour of preparing the mock sample, stored the eluent at 37°C for 1 week, and then detected viral RNA using RT-qPCR. Finally, we extracted RNA immediately after preparing the sample and stored it at −80°C for 1 week. The purpose of this experiment was to determine the effects of unreliable/unpredictable shipping practices (where the sample is exposed to warm temperatures for extended periods of time) on the quality of the measurement and compare it to an IFAST-extracted sample exposed to the worst-case shipping condition tested (37°C for 1 week).
Testing of IFAST Devices with the HIV Model System
To demonstrate the sensitivity of the optimized wax IFAST device, samples were prepared with low viral loads. Specifically, samples were prepared by loading serum with VLP at concentrations ranging from 100 to 10,000 copies per milliliter of serum. Additional samples were prepared by loading unmodified HIV virus (subtype B reference strain, a gift from Dr. David O’Connor) into serum at concentrations of 50 to 5000 copies per milliliter. Samples were loaded into IFAST devices with 500-μL input wells (to accommodate the maximum volume of sample) and two oil barriers in series (Figure 1C). IFAST purification and RT-qPCR were performed as previously described. The previously established VLP standard curve was then used to calculate measured viral load from the qPCR CT values.
Results
VLP Detection in Varying Input Sample Volumes
PCR mean CT was determined for all input volumes and is presented in Figure 2. Mean CT values within groups were the following: for 100 μL, mean CT = 27.926 ± 0.271; for 200 μL, mean CT = 26.742 ± 2.791; for 350 μL, mean CT = 27.456 ± 1.324; and for 500 μL, mean CT = 28.833 ± 1.799.
In short, mean CT values between groups were all within approximately two cycles. However, for reasons presented in the Discussion, the 500-μL device was selected for further analysis.
Quantification of Carryover
Using FC-40 oil resulted in the lowest percent carryover at 1.7% ± 0.2% and 0.7% ± 0.3% for one and two oil barriers, respectively (Figure 3). Loading devices with mineral oil resulted in 31.3% ± 21.5% carryover for one barrier and 14.2% ± 1.4% for two, whereas silicone oil yielded 41.1% ± 1.0% and 16.4% ± 3.0% accordingly. Qualitatively, it was observed that the aqueous phases would often connect on addition of mineral or silicone oil (Figure 3). Specifically, the aqueous sample wells were observed to creep underneath the adjacent oil phase, eventually connecting to the next aqueous well, resulting in mixing of the well contents. This failure mode was especially evident when devices were picked up for operation, likely because of the slight tilt imparted to the device. FC-40 oil demonstrated superior pinning stability, preventing excessive carryover of sample into the output elution well, and thus was selected for subsequent trials.
Figure 3.
Correlation between oil used and contaminant carryover. A: Error bars in the graph represent ±1 SD. B: All devices depicted in this figure were filled using silicone oil, illustrating its inability to adequately pin and separate the aqueous phases as demonstrated by the sample in the input well flooding into downstream extraction wells.
Sample Degradation
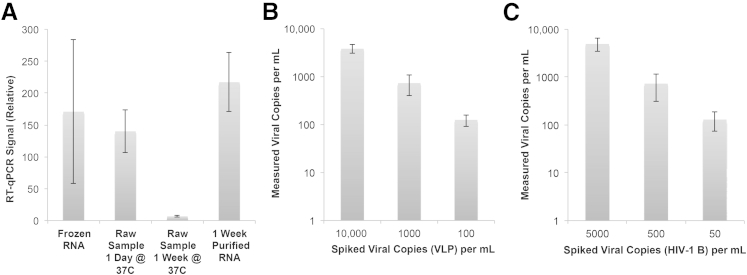
Raw samples stored at 37°C experienced substantial degradation, particularly after 1 week. After 1 day, the qPCR signal had decreased to 80% of the value obtained from frozen samples, and after 1 week, the signal had decayed to 4% of its original value. Surprisingly, the frozen sample exhibited a high degree of variation between the runs, potentially suggesting variable degradation associated with freezing and thawing samples. For those samples in which RNA was extracted via IFAST and stored at 37°C for 1 week, minimal degradation was observed. Although there were no significant differences between the IFAST-extracted samples and those stored at −80°C and 37°C for 1 day, there was significant difference between IFAST-extracted samples and those stored at 37°C for 1 week (P = 0.02, two-tailed Student's t-test) (Figure 4A). Importantly, both of these samples were stored at 37°C for 1 week, where one sample was prepared via IFAST before freezing, and one sample was left in raw form, demonstrating the utility of sample preparation at the point of sample acquisition.
Figure 4.
A: RT-qPCR measurements of RNA isolated from VLP-spiked serum samples that was prepared using IFAST and frozen immediately, raw sample stored at 37°C for 1 day, 1 week, or prepared via IFAST and then stored at 37°C for 1 week. B: Viral loads were quantified from VLP-spiked serum samples using IFAST as the RNA extraction method, allowing comparison of actual (spiked) values with the measurements. C: Viral loads were quantified from serum spiked with unmodified subtype B HIV, again using IFAST as the extraction method. Error bars represent ±1 SD.
Testing with the HIV Model System
Viral RNA purified from wax IFAST devices could be detected via RT-qPCR at low viral load concentrations. Using both noninfectious VLPs and unmodified, subtype B HIV virus, we demonstrated that wax IFAST devices are a viable sample preparation process. On average, viral load measurements obtained with spiked VLP samples were within 80% of the expected values with a coefficient of variation (defined as the SD divided by the mean) of 31% (Figure 4B). This level of performance with samples that have a relatively low viral load enables quantification of viral load in patients near the low end of the measurable viral load range. As a comparison, modern gold standard tests report limits of detection between 20 and 50 copies per milliliter [eg, the Roche COBAS TaqMan HIV-1 version 2.0 test (Roche Molecular Diagnostics, Pleasanton, CA) reported a limit of detection = 20 copies/mL, and the Abbott RealTime HIV-1 assay (Abbott Laboratories. Abbott Park, IL) reported a limit of detection = 40 copies/mL]. Similarly, viral load measurements obtained with spiked HIV samples were within 51% of the expected values, with a coefficient of variation of 44% (Figure 4C).
Discussion
We have demonstrated the efficacy of the wax IFAST platform for isolation of labile analytes. Of note, however, is the slight difference observed according to varying device input volume. This discrepancy may be due to a number of factors such as ease of PMP reclamation, VLP aggregation, and/or dilution of VLPs in higher volumes when combined with PMPs. One challenge encountered when fabricating the devices was controlling well-surface smoothness. A few of the completed devices exhibited a rough, almost wrinkled or wavy surface, examples of which are illustrated in Supplemental Figure S2. In some cases, this roughness had a tendency to trap some PMPs, preventing them from exiting the input well of the device, resulting in decreased detection levels. The effects of this phenomenon were more evident as the size of the input well increased, because the larger well surface provided more surface area for wrinkles. The primary explanation for the nonuniform surfaces likely stems from cooling kinetics between the mold and the wax. Additional studies from our group indicate that careful control of temperature throughout the molding process has a tremendous effect on the final surface quality.19 We were able to achieve consistently smooth devices with an automated, Peltier-heated embossing system; however, here, we wanted to demonstrate that highly functional devices could be manufactured from low-tech processes and methodology.
Similarly, any amount of VLP aggregation may have potentially thrown off the actual levels detected. VLPs not adequately and evenly resuspended in the sample solution may not have adhered to the PMPs, resulting in lowered detection levels. The third possible explanation for the detection variability could lie in the changing VLP–PMP binding kinetics at increasing input volumes. As total volume increases, effective VLP concentration decreases. Thus, at larger volumes, it is conceivable that fewer VLPs will bind to PMPs, yielding lower detection.
Despite variation among device designs, this slight variability in CT may be permissible, as increased sample volume should result in an improved limit-of-detection. For example, although the 500-μL device generated a higher CT value than the smaller-volume devices, we anticipate that this device may ultimately give the best overall sensitivity, given its ability to accommodate the largest amount of sample. For this reason, the 500-μL device was used for all other experiments.
Although IFAST, in principle, could be performed with any immiscible liquid, our trials clearly indicated that FC-40 outperformed the alternatives. Observed differences between oils may be due to the relative densities of each. Both silicone and mineral oils are less dense (0.913 g/mL and 0.84 g/mL at 25°C, respectively) than water, whereas FC-40 is substantially denser (1.85 g/mL at 25°C). Given that they do not pin in the wax wells, the oils easily flow among wells within the device. When the less dense oils make contact with aqueous phases, a combination of density imbalance and greater aqueous phase hydrostatic pressure may explain the inability of the silicone and mineral oils to sufficiently contain the other buffers. In addition, it is possible that certain components of the aqueous buffers, especially those containing detergents, may cause the oil phases to delaminate from the device surface, as reported in our previous work.10
Overall, although there are other methods to stabilize RNA at the point of sample acquisitions [eg, DBS, RNAlater (Life Technologies) treatments], these methods add steps (and associated costs) to the workflow. By contrast, performing sample preparation using IFAST at the point of acquisition does not add any steps to the workflow, but rather moves processes typically performed in the laboratory to the point of acquisition. Therefore, we believe these performance metrics demonstrate proof-of-concept feasibility for using a relatively low-tech sample preparation process that can be manufactured and operated in remote point-of-care settings.
Conclusions
We have demonstrated that HIV viral RNA extraction can be performed in wax IFAST devices and that the extracted RNA is suitable for high-sensitivity quantitation via RT-qPCR. In the developing world, viral load monitoring is constrained by both cost and laboratory resource availability. Adoption of the wax IFAST device for HIV viral load analysis overcomes both of these constraints without sacrificing accuracy or precision. Importantly, IFAST isolation of RNA at the point of acquisition results in RNA stabilization, enabling prepared RNA samples to remain stable during transport to a central laboratory for RT-qPCR quantitation without requiring a cold chain over the course of a week at 37°C. Furthermore, wax IFAST devices can be produced, operated, and disposed of in low-resource settings at minimal cost, thus substantially reducing the total cost of an HIV viral load assay. Taken together, we expect that these advantages can improve access to viral load testing, enabling proper HIV antiretroviral therapy management, particularly in the developing world.
Acknowledgments
We thank Drs. Elaine Alarid, Nathan Sherer, and David O’Connor (University of Wisconsin–Madison) for providing HEK293T cells; p24 Gag-Pol-Vif and pRev expression plasmids; and the unmodified HIV virus (subtype B reference strain), respectively.
Footnotes
Supported by the Bill and Melinda Gates Foundation through the Grand Challenges in Global Health Initiative and by NIH grant R33 CA160344 (D.J.B.).
Disclosures: S.M.B. and D.J.B. are co-founders of Salus Discovery, which has licensed some of the technology reported in this paper.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2014.01.004.
Supplemental Data
Dimensioned drawing of the mold used to produce the 500-μL input volume wax IFAST devices (all dimensions in millimeters).
Examples of a very smooth device deemed suitable for analyte extraction (left) and of a wrinkled device deemed unsuitable for analyte extraction (right).
References
- 1.Urdea M., Penny L.A., Olmsted S.S., Giovanni M.Y., Kaspar P., Shepherd A., Wilson P., Dahl C.A., Buchsbaum S., Moeller G., Burgess D.C.H. Requirements for high impact diagnostics in the developing world. Nature. 2006;444:73–79. doi: 10.1038/nature05448. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization: Technical Brief on HIV Viral Load Technologies. June 2010. Geneva, Switzerland, WHO Press, 2010
- 3.Cady N.C., Stelick S., Kunnavakkam M.V., Batt C.A. Real-time PCR detection of Listeria monocytogenes using an integrated microfluidics platform. Sens Actuators B Chem. 2005:332–341. [Google Scholar]
- 4.Anderson R.C., Su X., Bogdan G.J., Fenton J. A miniature integrated device for automated multistep genetic assays. Nucleic Acids Res. 2000;28:e60. doi: 10.1093/nar/28.12.e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witek M.A., Hupert M.L., Park D.S.-W., Fears K., Murphy M.C., Soper S.A. 96-well polycarbonate-based microfluidic titer plate for high-throughput purification of DNA and RNA. Anal Chem. 2008;80:3483–3491. doi: 10.1021/ac8002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu R.H., Yang J., Lenigk R., Bonanno J., Grodzinski P. Self-contained, fully integrated biochip for sample preparation, polymerase chain reaction amplification, and DNA microarray detection. Anal Chem. 2004;76:1824–1831. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- 7.Marcus J.S., Anderson W.F., Quake S.R. Microfluidic single-cell mRNA isolation and analysis. Anal Chem. 2006;78:3084–3089. doi: 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- 8.Okochi M., Tsuchiya H., Kumazawa F., Shikida M., Honda H. Droplet-based gene expression analysis using a device with magnetic force-based-droplet-handling system. J Biosci Bioeng. 2010;109:193–197. doi: 10.1016/j.jbiosc.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Sur K., McFall S.M., Yeh E.T., Jangam S.R., Hayden M.A., Stroupe S.D., Kelso D.M. Immiscible phase nucleic acid purification eliminates PCR inhibitors with a single pass of paramagnetic particles through a hydrophobic liquid. J Mol Diagn. 2010;12:620–628. doi: 10.2353/jmoldx.2010.090190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berry S.M., Alarid E.T., Beebe D.J. One-step purification of nucleic acid for gene expression analysis via Immiscible Filtration Assisted by Surface Tension (IFAST) Lab Chip. 2011;11:1747–1753. doi: 10.1039/c1lc00004g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bordelon H., Adams N.M., Klemm A.S., Russ P.K., Williams J.V., Talbot H.K., Wright D.W., Haselton F.R. Development of a low-resource RNA extraction cassette based on surface tension valves. ACS Appl Mater Interfaces. 2011;3:2161–2168. doi: 10.1021/am2004009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuchiya H., Okochi M., Nagao N., Shikida M., Honda H. On-chip polymerase chain reaction microdevice employing a magnetic droplet-manipulation system. Sens Actuators B Chem. 2008;130:583–588. [Google Scholar]
- 13.Diaz L.F., Savage G.M., Eggerth L.L. Alternatives for the treatment and disposal of healthcare wastes in developing countries. Waste Manag. 2005;25:626–637. doi: 10.1016/j.wasman.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization. Health-care waste management. Fact sheet N°281. October 2011. Geneva, Switzerland, WHO Press, 2011
- 15.Díaz-González M., Baldi A. Fabrication of biofunctionalized microfluidic structures by low-temperature wax bonding. Anal Chem. 2012;84:7838–7844. doi: 10.1021/ac301512f. [DOI] [PubMed] [Google Scholar]
- 16.Gong X., Yi X., Xiao K., Li S., Kodzius R., Qin J., Wen W. Wax-bonding 3D microfluidic chips. Lab Chip. 2010;10:2622–2627. doi: 10.1039/c004744a. [DOI] [PubMed] [Google Scholar]
- 17.Carrilho E., Martinez A.W., Whitesides G.M. Understanding wax printing: a simple micropatterning process for paper-based microfluidics. Anal Chem. 2009;81:7091–7095. doi: 10.1021/ac901071p. [DOI] [PubMed] [Google Scholar]
- 18.Berry S.M., Regehr K.J., Casavant B.P., Beebe D.J. Automated operation of immiscible filtration assisted by surface tension (IFAST) arrays for streamlined analyte isolation. J Lab Autom. 2013;18:206–211. doi: 10.1177/2211068212462023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guckenberger D., Thomas P., Rothbauer J., LaVanway A., Anderson M., Gilson D., Fawcett K., Berto T., Barrett K., Beebe D., Berry S.M. A combined fabrication and instrumentation platform for sample preparation. J Lab Autom. 2014 doi: 10.1177/2211068213518312. http://dx.doi.org/10.1177/2211068213518312 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 20.Sherer N.M., Swanson C.M., Papaioannou S., Malim M.H. Matrix mediates the functional link between human immunodeficiency virus type 1 RNA nuclear export elements and the assembly competency of Gag in murine cells. J Virol. 2009;83:8525–8535. doi: 10.1128/JVI.00699-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avettand-Fènoël V., Chaix M.L., Blanche S., Burgard M., Floch C., Toure K., Allemon M.C., Warszawski J., Rouzioux C., French Pediatric Cohort Study ANRS-CO 01 Group LTR real-time PCR for HIV-1 DNA quantitation in blood cells for early diagnosis in infants born to seropositive mothers treated in HAART area (ANRS CO 01) J Med Virol. 2009;81:217–223. doi: 10.1002/jmv.21390. [DOI] [PubMed] [Google Scholar]
- 22.Rouet F., Chaix M.-L., Nerrienet E., Ngo-Giang-Huong N., Plantier J.-C., Burgard M., Peeters M., Damond F., Ekouevi D.K., Msellati P., Ferradini L., Rukobo S., Maréchal V., Schvachsa N., Wakrim L., Rafalimanana C., Rakotoambinina B., Viard J.-P., Seigneurin J.-M., Rouzioux C. Impact of HIV-1 genetic diversity on plasma HIV-1 RNA quantification: usefulness of the Agence Nationale de Recherches sur le SIDA second-generation long terminal repeat-based real-time reverse transcriptase polymerase chain reaction test. J Acquir Immune Defic Syndr. 2007;45:380–388. doi: 10.1097/QAI.0b013e3180640cf5. [DOI] [PubMed] [Google Scholar]
- 23.Berry S.M., Strotman L.N., Kueck J.D., Alarid E.T., Beebe D.J. Purification of cell subpopulations via immiscible filtration assisted by surface tension (IFAST) Biomed Microdevices. 2011;13:1033–1042. doi: 10.1007/s10544-011-9573-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dimensioned drawing of the mold used to produce the 500-μL input volume wax IFAST devices (all dimensions in millimeters).
Examples of a very smooth device deemed suitable for analyte extraction (left) and of a wrinkled device deemed unsuitable for analyte extraction (right).