Abstract
Thalassemia is among the most common genetic diseases worldwide. α-Thalassemia is usually caused by deletion of one or more of the duplicated HBA genes on chromosome 16. In contrast, most β-thalassemia results from point mutations that decrease or eliminate expression of the HBB gene on chromosome 11. Deletions within the HBB locus result in thalassemia or hereditary persistence of fetal Hb. Although routine diagnostic testing cannot distinguish thalassemia deletions from point mutations, deletional hereditary persistence of fetal Hb is notable for having an elevated HbF level with a normal mean corpuscular volume. A small number of deletions accounts for most α-thalassemias; in contrast, there are no predominant HBB deletions causing β-thalassemia. To facilitate the identification and characterization of deletions of the HBA and HBB globin loci, we performed array-based comparative genomic hybridization using a custom oligonucleotide microarray. We accurately mapped the breakpoints of known and previously uncharacterized HBB deletions defining previously uncharacterized deletion breakpoints by PCR amplification and sequencing. The array also successfully identified the common HBA deletions --SEA and --FIL. In summary, comparative genomic hybridization can be used to characterize deletions of the HBA and HBB loci, allowing high-resolution characterization of novel deletions that are not readily detected by PCR-based methods.
Thalassemia is one of the most common genetic diseases and is responsible for significant morbidity worldwide. α-Thalassemia is most commonly caused by deletion of one or more of the duplicated HBA genes on chromosome 16. In contrast, most β-thalassemia results from point mutations that cripple or completely inactivate expression from the HBB gene on chromosome 11. These mutations commonly result in a decrease in HBB and a relative increase in HBD protein expression, resulting in an elevated HbA2 level. There is frequently a relative increase in HBG1/2 expression as well, with a corresponding increase in HbF. Deletions within the HBB locus occur and can eliminate HBB expression. If the HBD and HBG genes are deleted along with the HBB gene, the characteristic increases in HbA2 and HbF will not be seen, making it impossible to distinguish from α-thalassemia on the basis of HbA2 quantification alone. Deletions resulting in levels of HBD and HBG polypeptide production insufficient to balance that from the HBA locus result in δβ- or γδβ-thalassemia with a decreased mean corpuscular volume (MCV). In contrast, some deletions result in elevated levels of HBG sufficient to balance that from the HBA locus, resulting in hereditary persistence of fetal Hb (HPFH), the hallmark of which is a normal MCV. In both of these cases, an HBB locus deletion is assumed. HBB deletions causing thalassemia may be more common than generally thought because they may not be recognized during routine thalassemia diagnostic procedures, especially in the rare deletions that do not result in HbF elevation.
Several well-characterized deletions account for most α-thalassemia; thus, gap-PCR–based methods are routinely used to identify HBA gene deletions.1 In contrast, HBB deletions, when they occur, are diverse, so routine PCR-based methods are not efficient or routinely available. This is especially true in the United States, where no set of mutations predominates because of the ethnic diversity of this population. To provide a rapid inexpensive and efficient platform to identify common, previously described, and unique deletions of either the HBA or HBB loci, we developed a microarray-based comparative genomic hybridization (CGH) assay. This method can be used to genotype patients with deletional forms of α- or β-thalassemia, HPFH, or complex mixed HBA and HBB globin deletions, whose genotype is not apparent from protein-based studies, such as electrophoresis and high-performance liquid chromatography (HPLC). We used this array to characterize both previously characterized and novel deletions of the HBB locus. This array has been used in patients with apparent thalassemia trait but no evidence of α-thalassemia and no increase in HbA2, providing diagnostic information that allows accurate genetic counseling.
Materials and Methods
Patient Specimens and Diagnostic Studies
Patient specimens were sent to the University of Washington Red Cell Disorders laboratory for routine clinical testing. Clinical testing at the University of Washington included qualitative Hb analysis by isoelectric focusing (Perkin-Elmer Multiphor II; Perkin-Elmer, Waltham MA) and HPLC (Bio-Rad Variant II; Bio-Rad, Hercules, CA). HbF and HbA2 were quantified by HPLC. Detection of HbH inclusions was performed by brilliant cresyl blue staining as previously described.2 Iron deficiency was assessed by determining the zinc protoporphyrin/heme ratio3; serum iron, ferritin, total iron binding capacity, and transferrin saturation were determined in a subset of cases. For samples collected at the University of Washington, DNA was isolated using a Corbett automated DNA extractor (Qiagen, Valencia, CA). PCR to detect HBA deletions was performed as previously described.4 Additional specimens were obtained from Boston Children's Hospital. All studies were approved by the University of Washington Human Subjects Division.
Microarray Design
Custom arrays were designed essentially as described, including the software used for probe design and repetitive sequence masking.5 Genome coordinates used throughout this article are from the human hg38 assembly (December 2013). For chromosome 11, probes were tiled at a median density of 60 bp across an approximately 450-kbp region surrounding the HBB locus (coordinates chr11:5,022,271-5,472,133), and at a median density of 57 to 62 kbp on either side of this region for the remainder of chromosome 11 (coordinates chr11:218,364-5,022,271 and chr11:5,472,133-134,998,453). For chromosome 16, probes were tiled at a median density of 64 bp across an approximately 400-kbp region surrounding the HBA loci (coordinates chr16:11,177-414,936) and a median density of 56,261 bp for the remainder of chromosome 16 (chr16:429,413-90,081,925). An additional 883 probes representing the other chromosomes (median spacing of 1.8 Mbp) were included for the purpose of signal normalization. The array design is accessible in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GPL20136).
Array Hybridization, Scanning, and Data Analysis
Subject genomic DNA was labeled with Cy5-dUTP, and normal human genomic DNA (Promega, Madison, WI) was labeled with Cy3-dUTP using the Agilent Genomic DNA Labeling kit PLUS (Agilent Technologies, Santa Clara, CA). Array hybridization, scanning, and data analysis were performed as described previously.5 The raw data are accessible at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE68587).
Confirmation of Deletion Breakpoints
PCR primers flanking breakpoints identified by CGH were designed using Primer 3 software version 1.1.0 (http://bioinfo.ut.ee/primer3, last accessed June 1, 2009). DNA sequencing was performed using standard BigDye Terminator reagents (Applied Biosystems, Foster City, CA) using the same PCR primers (Table 1).
Table 1.
PCR and Sequencing Primers Used in This Study
| Subject No. | Forward PCR primer | Reverse PCR primer | Additional sequencing primers, if used |
|---|---|---|---|
| 4 | 5′-TAGATCCCTTTGCCATTATG-3′ | 5′-TTGGGTTTCTGATAGGCACTG-3′ | |
| 5 | 5′-AGCCTCATGGTAGCAGAATC-3′ | 5′-TGGTATCTGCAGCAGTTGCC-3′ | |
| 7∗ | 5′-CCTTTTTCTTGTGGTAAATGCTT-3′ | 5′-TTTCCTTGTGTTTGAAAGTGCT-3′ | F: 5′-ACCCTTTGAGTAATAGTTTCCTGA-3′ R: 5′-GCAAATAAGCACACATATATTCCAA-3′ |
| 5′-ATGCCAGTGCTCTCCACAAT-3′ | 5′-ATAATAAGCCTGCGCCCTTC-3′ | R: 5′-AGTTTCTTGTTTACTCTGGA-3′ R: 5′-GGCCATATAGGGTAACTTTCTGAC-3′ R: 5′-AAAAGTGTGCCATGGTTTTAATG-3′ |
|
| 8 | 5′-CATCCACCACTTTCTGATAGG-3′ | 5′-TAGCATGCATGAGCAAATTAAGA-3′ | |
| 9 | 5′-GCCACATGGTATGGGAGGTA-3′ | 5′-TGTTACTTGTCTGGTGTGGCTAA-3′ |
F, forward; R, reverse.
Upper line shows sequences for telomeric breakpoint; lower line shows sequences for centromeric breakpoint.
Results
Deletions within the HBB Locus
Subjects at the University of Washington were suspected of having a deletional form of β-thalassemia if they were microcytic without evidence of iron deficiency, had normal levels of HbA2, and had no evidence of α-thalassemia trait (eg, negative HbH inclusion body assay). Other subjects were identified because of unexplained elevations of HbF or unusually high HbA2. Hematologic findings are detailed in Table 2.
Table 2.
Clinical Data for Patients with HBB Deletions
| Subject no. | Age | Sex | MCV, fL | HbA2, % | HbF, % | Hb, g/dL | Hct, % | Electrophoresis | Ethnicity |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 months | F | 93 | 0.9 | 91.1 | 11.3 | HbF, HbS | African | |
| 2 | 20 months | F | 76 | 3 | 34.1 | 11.6 | HbS, HbF | African | |
| 3 | 26 years | F | 82 | 16.5 | 11.7 | 13.1 | HbA, Hb Kenya | African | |
| 4 | 25 years | F | 69 | ND | 44.2 | 35 | HbE, HbF | Cambodian | |
| 5 | 25 years | F | 79 | 4.3 | 20.6 | 35 | HbA, HbF, HbA2 | Vietnamese | |
| 6 | 36 years | M | 83 | 1.9 | 40.8 | 16.6 | 50.6 | HbA, HbF, HbA2 | Sri Lankan |
| 7 | 30 years | F | 65 | 8.2 | 1.2 | 34 | Kurdish | ||
| 8 | 29 years | F | 72 | 7 | 9.5 | 10.2 | HbA, HbF, HbA2 | Irish/Romanian | |
| 9 | 12 years | M | 60.5 | 3 | ND | 11.6 | HbA, HbA2 | Belgian | |
| 10 | 4.5 years | F | 69.1 | 2.6 | 16.8 | 12 | 36.8 | HbA, HbF, HbA2 | Sri Lankan |
| 11 | 2 months | F | 66 | 2.1 | 26.3 | 7.4 | HbA, HbF, HbA2 | White |
F, female; M, male; Hct, hematocrit; MCV, mean corpuscular volume; ND, not determined.
For array CGH, Cy5-labeled subject DNA was combined with Cy3-labeled reference DNA, and the labeled DNA was hybridized to a microarray containing oligonucleotide probes spanning the HBA and HBB loci. The density of probes varied (see Materials and Methods for genomic coordinates), with a high density of probes spanning structural genes of the loci and their upstream regulatory elements (approximately one probe per 60 to 64 bp) and a low density of probes in the flanking regions of the loci (approximately one probe per 56 to 57 kbp). Additional probes from the other chromosomes were included as normalization controls.
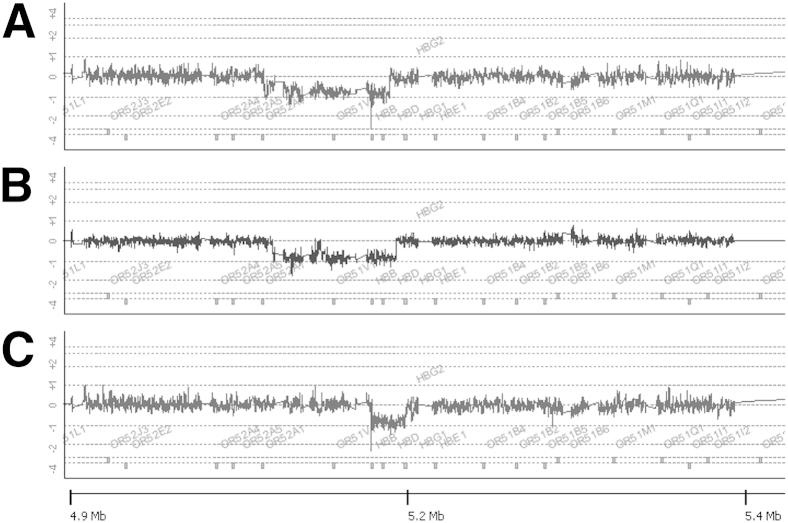
Figure 1 shows the results of array CGH for four DNA specimens from patients (subjects 1 to 4) with previously characterized HBB locus deletions (a generous gift of Dr. David H.K. Chui, Boston University, Boston, MA), including HPFH-1 (subject 1) (Figure 1A), HPFH-2 (subject 2) (Figure 1B), and Hb Kenya (subject 3) (Figure 1C). The HPFH-1 deletion is shown as a region of Cy5:Cy3 ratio of log2 −1 extending from chromosome 11 coordinate chr11:5,153,221 within a cluster of olfactory receptor genes at the telomeric end of the locus to coordinate chr11:5,238,138 between the HBD and HBG1 genes (Figure 1A), resulting in deletion of the HBB and HBD genes. This deletion is most commonly found in populations of African ancestry and results in very high HbF levels (25% to 30%) in heterozygotes.6 The HPFH-2 deletion is very similar to the HFPH-1 deletion, with breakpoints just centromeric to the HPFH-1 breakpoints at both ends (telomeric breakpoint at chr11:5,158,438, centromeric breakpoint at chr11:5,242,749). The HPFH-2 deletion is also common in individuals of African ancestry, with HbF levels similar to those seen with HPFH-1.6 Hb Kenya (Figure 1C) is a deletion that excises sequences between the HBB and HBG2 genes (telomeric breakpoint chr11:5,226,631 to centromeric breakpoint at chr11:5,249,422), resulting in a HBG1-HBB fusion gene and protein.7 The abnormal fusion protein co-elutes with HbA2 on HPLC, explaining the elevated level of HbA2 in subject 3; the intact HBG2 gene in cis is expressed at an elevated level as well.
Figure 1.
Examples of array comparative genomic hybridization showing previously characterized HBB deletions. Deletions identified include hereditary persistence of fetal Hb (HPFH)-1 (subject 1, A), HPFH-2 (subject 2, B), and Hb Kenya (subject 3, C). Numbers at the bottom of the figure represent distance from the chromosome 11p telomere. The ratio of Cy5 to Cy3 is 1 in the absence of copy number change, reflected in a log2 of 0. Heterozygous deletions are identified by a 50% loss (log2 change of −1) of Cy5 signal in a group of contiguous probes. Homozygous deletions result in a log2 change of −∞.
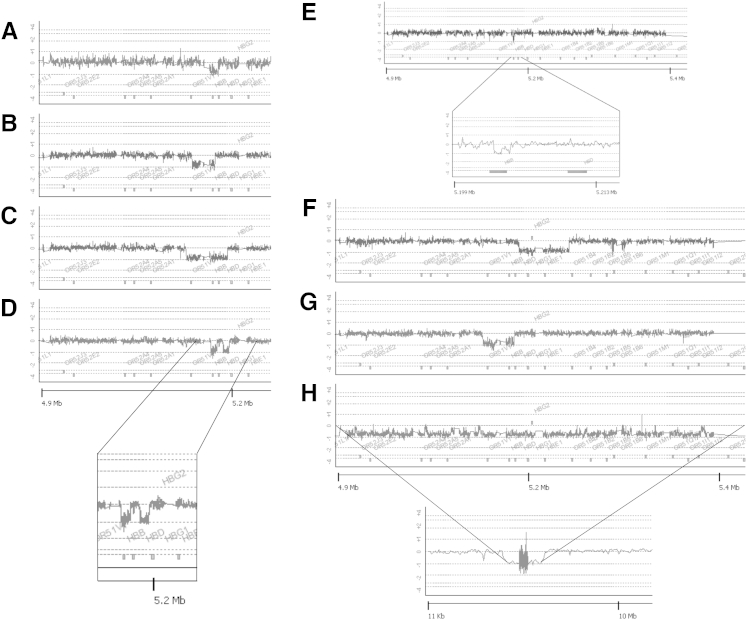
Figure 2 shows the results obtained from subjects with HPFH or deletional β-thalassemia whose HBB locus deletions had not been characterized molecularly (subjects 4 to 11). All these deletion breakpoints were sequenced by designing PCR primers that flanked the breakpoints and determining the sequence of the resulting PCR products using Sanger sequencing other than subject 11, whose breakpoints occurred in sequences tiled at low density. Breakpoint sequence data for deletions not shown elsewhere can be found in Supplemental Figure S1 and Supplemental Table S1. A diagram showing the extent of the deletions with respect to the globin genes on chromosome 11 is shown in Figure 3. Figure 2A is a DNA specimen from an adult Cambodian patient with HbE, markedly elevated HbF, and no HbA, suggesting compound heterozygosity for HbE and a HBB gene deletion (subject 4). The lack of anemia, very high HbF level, and MCV consistent with HbE trait suggested a deletional HPFH rather than β0-thalassemia trait on the non-E allele. This patient was found to have a 12.6-kbp deletion from telomeric breakpoint chr11:5,220,623 to centromeric breakpoint chr11:5,233,234, resulting in deletion of the HBB gene and a small segment of the 3′ end of the HBD gene. This deletion has been termed the Thai/Vietnamese (δβ)0 deletion.8 Figure 2B shows a 27.4-kbp deletion (telomeric breakpoint chr11:5,201,647 to centromeric breakpoint chr11:5,229,059) in a Vietnamese patient with an elevated HbF level, slightly elevated HbA2 level, and mild microcytosis (subject 5) having the SEA HPFH deletion.9 The HBB gene is deleted, but the HBD and HBG genes are intact, resulting in a relative increase in both HBD and HBG expression. Figure 2C shows a 49.8-kbp HBB-HBD deletion (telomeric breakpoint chr11:5,194,453 to centromeric breakpoint chr11:5,244,225) consistent with HPFH-310, 11 in a patient from Sri Lanka (subject 6). Figure 2D shows a complex deletion in a Kurdish family that was previously described but only partially characterized12 (subject 7). This is a discontinuous deletion that removes the HBB (deleted nucleotides chr11:5,223,960-5,231,570) and HBBP1 (deleted nucleotides chr11:5,238,759-5,246,317) genes but spares the HBD gene and results in microcytosis consistent with β-thalassemia trait but with more elevated HbA2 and normal HbF levels. Figure 2E shows the results from a patient of Irish/Romanian ancestry (subject 8) with a previously unreported 1518-bp deletion of the HBB promoter region (telomeric breakpoint chr11:5,225,894 to centromeric breakpoint chr11:5,227,412), resulting in a β-thalassemic phenotype with relatively high levels of both HBG and HBD. Figure 2F shows a 59.9-kbp deletion (telomeric breakpoint chr11:5,238,245 to centromeric breakpoint chr11:5,298,126) in a Belgian male with microcytosis and normal HbF level (subject 9). This deletion spares the HBB gene but removes the remaining globin genes and the locus control region. Figure 2G shows a 38.2-kbp deletion (telomeric breakpoint chr11:5,194,461 to centromeric breakpoint chr11:5,232,686) in a Sri Lankan patient, causing δβ-thalassemia with increased HbF (subject 10). Of note, this deletion shares a common breakpoint downstream of the locus with the HPFH-3 deletion but differs in the breakpoint within the locus, located immediately telomeric to the HBD gene and has been termed Sri Lankan (δβ)0 thalassemia.10 Finally, Figure 2H shows a deletion of >2 Mbp in a 2-month-old girl with microcytosis (subject 11). Because the resolution of the array is lower at large distances from the structural HBB genes, we were not able to precisely map the breakpoints of this deletion. However, the telomeric breakpoint is between chr11:4,109,643-4,188,151, and the centromeric breakpoint is between chr11:6,384,408-6,422,730. Of interest, this patient had early neurologic developmental abnormalities and white matter changes, raising the possibility that this large deletion is responsible for more than the hematologic changes observed.
Figure 2.
Array comparative genomic hybridization used to characterize deletions in patient samples that had not been defined molecularly. These samples include a deletional hereditary persistence of fetal Hb co-inherited with HbE trait in a Cambodian subject (subject 4, A), an HBB deletion in a Vietnamese subject (subject 5, B), a deletion causing HPFH in a Sri Lankan individual that is identical to HPFH-3 (subject 6, C), a noncontiguous deletion removing the HBBP1 and HBB genes while sparing the intervening HBD gene in a Kurdish individual (subject 7, D, expanded in inset), a novel deletion causing δβ-thalassemia in an individual of Irish/Romanian ancestry (subject 8, E, expanded in inset), a deletion causing β-thalassemia that includes the β-globin locus control region and the HBE1, HBG1/2, and HBD genes that spares the linked HBB gene (subject 9, F), a deletion causing δβ-thalassemia in a Sri Lankan individual (subject 10, G), and a very large (>2 Mbp) deletion causing β0-thalassemia with an inset showing an overview of the 11-Mb region containing the deletion (subject 11, H). Numbers at the bottom of the figure represent distance from the chromosome 11p telomere. Note that the inset for H shows a sequence span of approximately 10 Mb.
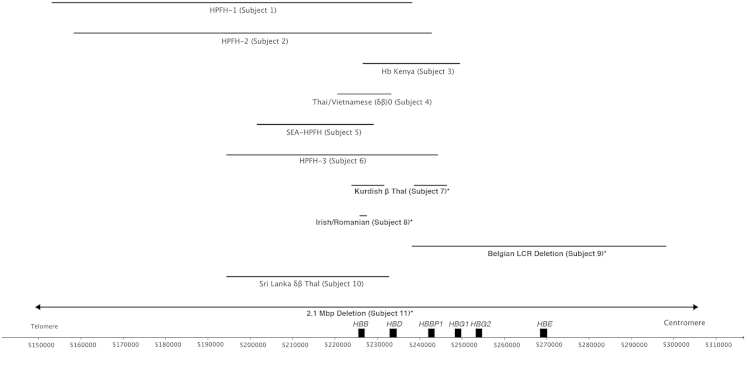
Figure 3.
Comparison of deletions for subjects with HBB locus deletions. Coordinates of chromosome 11 are shown at the bottom. Deletions not previously described in the literature are indicated by asterisks.
Deletions Causing α-Thalassemia
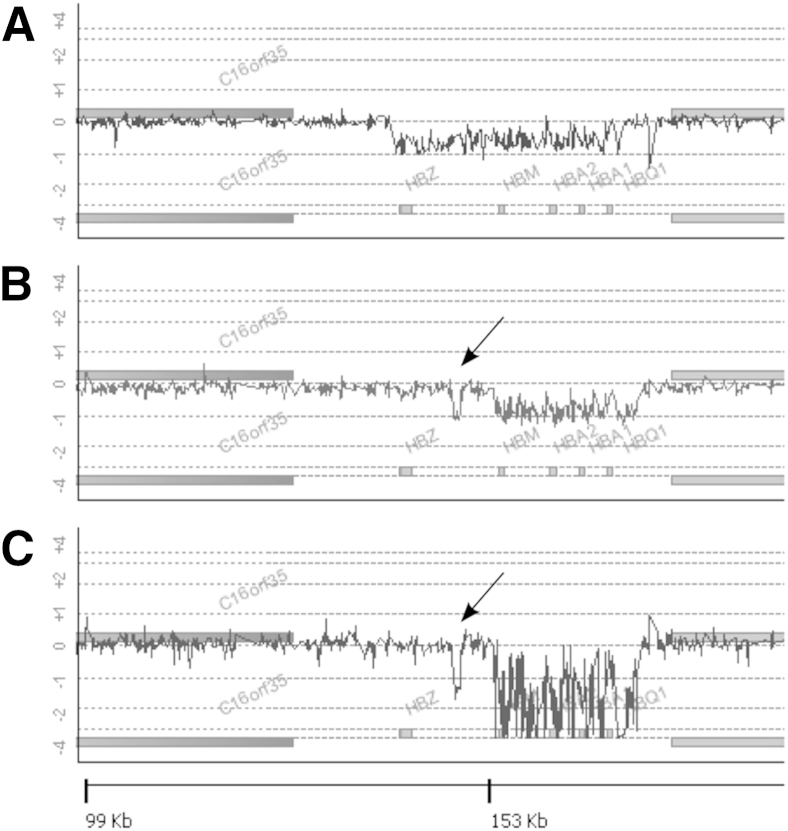
To determine the ability to characterize HBA deletions by array CGH, DNA samples that had been characterized previously by gap-PCR were studied using the globin array. Figure 4 shows results from patients with cis (two-gene, α-thalassemia-1) deletions. A sample heterozygous for the Filipino deletion (αα/--FIL) yielded a 50% decrease in signal spanning nucleotides chr16:150,821-182,671, including the HBZ, the two HBA, and the HBQ1 genes (Figure 4A), consistent with accurate mapping of the mutation. A Southeast Asian deletion (αα/--SEA) sample similarly resulted in the expected 50% decrease in signal from nucleotides chr16:165,401-184,701, sparing the HBZ gene, and removing the HBM, the two HBA, and the HBQ genes (Figure 4B). Finally, a case of a homozygous --SEA deletion, associated with Hb Bart hydrops fetalis, reveals a complete loss of signal in the deleted region (Figure 4C). Of interest, the --SEA specimens (heterozygous and homozygous) had a shorter α-globin inter-ζ hypervariable region (HVR)13 when compared with the reference DNA, telomeric to the large deletion. This finding appears to be in linkage disequilibrium with the --SEA deletion because it is present in all --SEA specimens examined for this study. Similarly, the --FIL specimen appears to have a shorter α-globin 3′ HVR14 than the reference DNA, seen as a decrease in signal just centromeric to the deleted region (Figure 4A) and the --SEA homozygous specimen apparently has a longer 3′ HVR as evidenced by increased signal (Figure 4C).
Figure 4.
Array comparative genomic hybridization characterization of HBA two-gene deletions in cis-causing α-thalassemia. These deletions include heterozygosity for the common Filipino αα/--FIL deletion (A), heterozygosity for the common Southeast Asian deletion αα/--SEA (B), and homozygosity for the --SEA deletion (C). The arrows in B and C indicate the inter-ζ hypervariable region, which is shorter in individuals with the --SEA deletion than in individuals without the deletion. Numbers at the bottom of the figure represent distance from the chromosome 16p telomere.
We also investigated the utility of this array to detect the common single HBA gene deletions. Interpretation of the plots is difficult due to the sequence homology between the two HBA genes because DNA from the remaining (nondeleted) HBA gene in cis hybridizes to the probes spanning the deleted HBA gene, as well as its own probes. Although it is not possible to determine the extent of these deletions from array CGH a priori, the patterns of signal loss for each deletion are reproducible. Examples are shown in Supplemental Figure S2.
Discussion
Molecular diagnosis of thalassemia is a useful adjunct to the more common techniques of Hb electrophoresis and HPLC when genetic counseling or prenatal diagnosis is a concern. It is especially important when standard methods cannot be used for a definitive diagnosis, for example, in the setting of β-thalassemia or HbE traits, where α-thalassemia can be masked, or in the setting of deletional β-thalassemia, where the characteristic increase in HbA2 is often not present. In this study, we found that array CGH can be accurately used to diagnose patients as having deletions of the HBA and HBB loci.
We designed a custom oligonucleotide array representing 400- to 450-kbp genomic segments surrounding the HBA and HBB loci with a median probe spacing of 60 to 64 bp, enabling high-resolution mapping of deletions involving these loci. As a demonstration of the ability of this technique to map known deletions, we accurately map the 5′ and 3′ breakpoints of the previously characterized HPFH-1, HPFH-2, and Hb Kenya deletions (Figure 1). We then reveal that deletions that were mapped previously using lower-resolution DNA blots can be mapped at high resolution using the CGH array, allowing a definitive molecular characterization after the design of a single set of PCR primers and a single Sanger sequencing run. This technique allows the rapid identification of novel mutations and facilitates their rapid sequencing. Finally, we reveal that CGH produces reproducible patterns for deletional α-thalassemia. Two other groups have reported using similar technology to identify HBA and HBB deletions,15, 16 but sequencing of breakpoints was not reliable. In contrast, the higher resolution of our array has facilitated one-step PCR and sequencing when breakpoints occur in the high-density region of the array.
HBB Deletions
Diagnosis of deletional β-thalassemia can be challenging. Most β-thalassemias are due to point mutations of the HBB structural or regulatory sequences and are routinely characterized by DNA sequencing.17 In populations where specific mutations are very common, methods are available to detect these mutations using allele-specific reagents.18, 19, 20, 21 Similarly, gap-PCR methods, in which primers are used that flank specific deletion breakpoints, may be used to confirm specific mutations when suspected from less specific studies or family history.22 To detect unknown deletions, DNA blot analysis has been the standard approach but is not commonly used in clinical laboratories because it is labor intensive, low resolution, and slow. Quantitative PCR using primer sets spaced throughout the HBB locus followed by capillary electrophoresis has been reported for characterizing HBB deletions.23 More recently, multiplex ligation-dependent probe amplification has been used to characterize deletions of both the HBA and HBB loci.24, 25, 26, 27 These latter techniques, although useful to reveal the presence of deletions, do not have the resolution available with the CGH method presented in this study.
Our CGH approach was able to map previously uncharacterized HBB locus deletions with high enough resolution to design PCR primers that could amplify the DNA sequences flanking the deletion breakpoints and obtain definitive sequence analysis in a single run. This finding resulted in the identification and characterization of novel HPFH and (γ) (δ)β0-thalassemia deletions as well as definitive sequencing of prior mutations that were only imprecisely mapped. Subject 4, an adult Cambodian woman, was a compound heterozygote for HbE trait and β-thalassemia with 44.2% HbF. CGH array analysis revealed chromosome 11 breakpoints that correspond to previously described Vietnamese/Laotian/Thai HPFH deletions.8, 28, 29 In a previously reported case,28 the HbF level was 21.3%. In our case, the HbF level is roughly twice that in previous reports. This can be accounted for by the fact that expression of βE is approximately half that of βA.
Both subjects 6 and 10, of Sri Lankan ethnicity, had elevated HbF levels but with quite different phenotypes. Subject 6, a 36-year-old man, has a very high HbF level of 40.8% with low-normal MCV. CGH revealed the 49,772-bp HPFH-3 deletion.10 Subject 10, a 4.5-year-old girl, was microcytic with a moderately elevated HbF of 16.8% and a novel 38,225-bp deletion we define as Sri Lankan (δβ)0-thalassemia.10 The telomeric breakpoints were almost identical for these two patients (chr11:5,194,453 for subject 6 versus chr11:5,194,461 for subject 10), whereas the centromeric breakpoints were 11,539 bp apart (chr11:5,244,225 for subject 6 versus chr11:5,232,686 for subject 10). The region between chr11:5,244,225 and chr11:5,232,686, which is immediately centromeric to the HBD gene, contains sequences that repress HBG expression based on the large difference in HbF levels between these two subjects, and chromatin immunoprecipitation has reveasled a binding site for the key HbF silencing factor BCL11A in this interval,30 along with binding sites for corepressors and repressive histone and chromatin marks.10 This site has also been associated with regulation of HBG expression in genome-wide association studies.31, 32 Subject 7, a 30-year-old Kurdish woman, has β-thalassemia with typical microcytosis and a significantly elevated HbA2 level but no significant increase in HbF. CGH analysis revealed two noncontiguous deletions removing the HBB and HBBP1 genes but sparing the HBD gene. The retained sequences include this BCL11A binding site.10 Other mechanisms must exist for elevated HBG expression, however, because the deletions in subjects 4 and 5 do not remove the BCL11A binding site centromeric to the HBD gene, yet these subjects have elevated HbF levels of 44.2% and 20.6%, respectively. Perhaps the increased HbF in these subjects is due to a telomeric enhancer brought into proximity of the HBG genes.
One other interesting feature of subject 10's Sri Lankan (δβ)0 thalassemia deletion is that the HBD gene is intact, yet there is no increase in HbA2. The breakpoint occurs approximately 200 bp 3′ (telomeric) to the HBD structural sequences and 3′ untranslated region. This result suggests the presence of an HBD-positive regulatory element located somewhere telomeric to this breakpoint.
HBA Deletions
We also report the utility of array CGH for detecting HBA gene deletions. For subjects heterozygous for large deletions of the HBA locus, such as the --SEA and --FIL deletions, the expected 50% decrease in signal intensity is observed throughout the deleted region. CGH is also able to characterize some of the polymorphic repeat regions in the HBA locus, including the inter-ζ HVR, a polymorphic region on chromosome 16 that lies between the HBZ and HBZP1 genes13 and the 3′ HVR centromeric to the HBQ gene.14 It appears that the inter-ζ HVR is smaller and the 3′ HVR is larger for the --SEA allele than for the corresponding regions of the reference DNA used for the CGH study (Figure 4, A and B). The --FIL allele appears to have a shorter 3′ HVR (Figure 4A). Identification of alleles lacking one of two HBA genes, such as the -α3.7 and -α4.2 deletions, is more difficult because the two HBA genes are highly homologous, and when only one is deleted, labeled DNA fragments from the nondeleted HBA genes (and their homologous pseudogenes) cross-hybridize to probes covering the deleted regions, resulting in a signal ratio of subject to reference DNA of >0.5 (Supplemental Figure S2). Standard gap-PCR is adequate for detecting the common HBA deletions leading to α-thalassemia,1 but the array CGH method still has potential utility to identify unusual deletions not tested for by gap-PCR and novel deletions of the HBA locus. Although we did not have samples to test, array CGH should also be able to identify and characterize triplicated HBA genes.
Conclusions
We found that CGH using a high-density custom oligonucleotide array can be used to rapidly and efficiently define both previously described and unique deletions of the HBA and HBB loci. This method will be a useful adjunct to current diagnostic methods for patients suspected of having thalassemia where standard diagnostic techniques are inconclusive. The typical patient would be microcytic with normal iron studies and have a normal level of HbA2. If gap-PCR to detect the common HBA deletions is negative, an HBB or uncommon HBA deletion is likely. The lack of HbA2 elevation makes an HBB point mutation unlikely, so Sanger sequencing of HBB would not likely be diagnostic, and thus proceeding directly to array CGH would be the most efficient diagnostic pathway. The other clinical scenario where HBB deletion analysis would be useful is HPFH, where there is a large variety of deletions that are not amenable to gap-PCR, particularly in an ethnically diverse population. Next-generation sequencing should eventually replace CGH and most other genetic technologies for clinical testing, although currently CGH represents a less labor- and data-intensive way to characterize deletions, such as those in the globin loci, and thus could remain more accessible to laboratories with limited resources.
Acknowledgment
HBB locus deletions were a generous gift of Dr. David H.K. Chui (Boston University, Boston, MA).
Footnotes
Supported by NIH grant DK44746 (M.A.B.) and funds from the Department of Laboratory Medicine, University of Washington.
Disclosures: None declared.
Current address of H.A.G.: LabCorp, Seattle. WA.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2015.07.011.
Contributor Information
Daniel E. Sabath, Email: dsabath@uw.edu.
Harvey A. Greisman, Email: hagreisman@gmail.com.
Supplemental Data
Sanger sequencing chromatograms of the breakpoints of β-globin deletions characterized by array comparative genomic hybridization (CGH) that had not been finely mapped previously. PCR primers were designed based on the breakpoints as determined by array CGH. The resulting PCR products were sequenced by Sanger sequencing. Vertical black lines indicate where the breakpoints occur. Orange bars indicate sequences that are not present in the hg38 reference human germline sequence. Tracings are for subject 4 (A, Thai/Vietnamese (δβ)0 thalassemia), subject 5 (B, SEA-HPFH), subject 7 (C, Kurdish deletion; note that there are two breakpoints for this deletion shown), subject 8 (D, Irish/Romanian), and subject 9 (E, Belgian). Sequence data for subjects 6 and 10 can be found in the supplemental material for reference 5. Because the breakpoints for the 2.1-Mb deletion (subject 11) occur in low-density tiled regions of chromosome 11, it was not possible to design PCR primers and sequence the breakpoints.
Array comparative genomic hybridization (CGH) characterization of HBA single-gene deletions causing α-thalassemia. These include heterozygosity for the –α3.7 deletion (A), heterozygosity for the –α4.7 deletion (B), homozygosity for the –α3.7 deletion (C), and compound heterozygosity for the –α3.7 and –α4.7 deletions (D). Numbers at the bottom of the figure represent distance from the chromosome 16p telomere. In DNA samples heterozygous for either the –α3.7 or the –α4.2 deletion, a loss of signal of <50% can be seen in the region of the HBA genes (A and B, respectively). With DNA containing a homozygous –α3.7 deletion, the loss in signal is closer to the expected 50% in the stretch spanning nucleotides chr16:173,301-177,104, including the 3′ end of the HBA2 gene to the 5′ end of the HBA1 gene (C). Again, although there is loss of genetic material from both alleles in this region, there is only effectively a 50% loss because there are two remaining α-globin genes, which hybridize to the probes for the deleted region. A similar result is obtained with DNA from a compound heterozygote for –α3.7 and –α4.2 (D).
References
- 1.Chong S.S., Boehm C.D., Higgs D.R., Cutting G.R. Single-tube multiplex-PCR screen for common deletional determinants of α-thalassemia. Blood. 2000;95:360–362. [PubMed] [Google Scholar]
- 2.Sabath D.E., Cross S.T., Mamiya L.Y. An improved method for detecting red cells with hemoglobin H inclusions that does not require glass capillary tubes. Clin Lab Haematol. 2003;25:87–91. doi: 10.1046/j.1365-2257.2003.00502.x. [DOI] [PubMed] [Google Scholar]
- 3.Labbe R.F., Rettmer R.L. Zinc protoporphyrin: a product of iron-deficient erythropoiesis. Semin Hematol. 1989;26:40–46. [PubMed] [Google Scholar]
- 4.Chong S.S., Boehm C.D., Cutting G.R., Higgs D.R. Simplified multiplex-PCR diagnosis of common Southeast Asian deletional determinants of α-thalassemia. Clin Chem. 2000;46:1692–1695. [PubMed] [Google Scholar]
- 5.Greisman H.A., Hoffman N.G., Yi H.S. Rapid high-resolution mapping of balanced chromosomal rearrangements on tiling CGH arrays. J Mol Diagn. 2011;13:621–633. doi: 10.1016/j.jmoldx.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuan D., Feingold E., Newman M., Weissman S.M., Forget B.G. Different 3′ end points of deletions causing δβ-thalassemia and hereditary persistence of fetal hemoglobin: implications for the control of γ-globin gene expression in man. Proc Natl Acad Sci U S A. 1983;80:6937–6941. doi: 10.1073/pnas.80.22.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huisman T.H., Wrightstone R.N., Wilson J.B., Schroeder W.A., Kendall A.G. Hemoglobin Kenya, the product of fusion of γ and β polypeptide chains. Arch Biochem Biophys. 1972;153:850–853. doi: 10.1016/0003-9861(72)90408-0. [DOI] [PubMed] [Google Scholar]
- 8.Chalaow N., Thein S.L., Viprakasit V. The 12.6 kb-deletion in the β-globin gene cluster is the known Thai/Vietnamese (δβ)0-thalassemia commonly found in Southeast Asia. Haematologica. 2013;98:e117–e118. doi: 10.3324/haematol.2013.090613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X.M., Li Z.Q., Liu Z.Y., Zhong X.L., Zhao Y.Z., Mo Q.H. Molecular characterization and PCR detection of a deletional HPFH: application to rapid prenatal diagnosis for compound heterozygotes of this defect with β-thalassemia in a Chinese family. Am J Hematol. 2000;65:183–188. doi: 10.1002/1096-8652(200011)65:3<183::aid-ajh1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 10.Sankaran V.G., Xu J., Byron R., Greisman H.A., Fisher C., Weatherall D.J., Sabath D.E., Groudine M., Orkin S.H., Premawardhena A., Bender M.A. A functional element necessary for fetal hemoglobin silencing. N Engl J Med. 2011;365:807–814. doi: 10.1056/NEJMoa1103070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henthorn P.S., Mager D.L., Huisman T.H., Smithies O. A gene deletion ending within a complex array of repeated sequences 3′ to the human β-globin gene cluster. Proc Natl Acad Sci U S A. 1986;83:5194–5198. doi: 10.1073/pnas.83.14.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oner C., Oner R., Gurgey A., Altay C. A new Turkish type of β-thalassaemia major with homozygosity for two non-consecutive 7.6 kb deletions of the ψβ and β genes and an intact δ gene. Br J Haematol. 1995;89:306–312. doi: 10.1111/j.1365-2141.1995.tb03305.x. [DOI] [PubMed] [Google Scholar]
- 13.Goodbourn S.E., Higgs D.R., Clegg J.B., Weatherall D.J. Molecular basis of length polymorphism in the human ζ-globin gene complex. Proc Natl Acad Sci U S A. 1983;80:5022–5026. doi: 10.1073/pnas.80.16.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarman A.P., Nicholls R.D., Weatherall D.J., Clegg J.B., Higgs D.R. Molecular characterisation of a hypervariable region downstream of the human α-globin gene cluster. EMBO J. 1986;5:1857–1863. doi: 10.1002/j.1460-2075.1986.tb04437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blattner A., Brunner-Agten S., Ludin K., Hergersberg M., Herklotz R., Huber A.R., Röthlisberger B. Detection of germline rearrangements in patients with α- and β-thalassemia using high resolution array CGH. Blood Cells Mol Dis. 2013;51:39–47. doi: 10.1016/j.bcmd.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Phylipsen M., Chaibunruang A., Vogelaar I.P., Balak J.R., Schaap R.A., Ariyurek Y., Fucharoen S., den Dunnen J.T., Giordano P.C., Bakker E., Harteveld C.L. Fine-tiling array CGH to improve diagnostics for α- and β-thalassemia rearrangements. Hum Mutat. 2012;33:272–280. doi: 10.1002/humu.21612. [DOI] [PubMed] [Google Scholar]
- 17.Chan O.T., Westover K.D., Dietz L., Zehnder J.L., Schrijver I. Comprehensive and efficient HBB mutation analysis for detection of β-hemoglobinopathies in a pan-ethnic population. Am J Clin Pathol. 2010;133:700–707. doi: 10.1309/AJCP7HQ2KWGHECIO. [DOI] [PubMed] [Google Scholar]
- 18.Shaji R.V., Edison E.S., Poonkuzhali B., Srivastava A., Chandy M. Rapid detection of β-globin gene mutations and polymorphisms by temporal temperature gradient gel electrophoresis. Clin Chem. 2003;49:777–781. doi: 10.1373/49.5.777. [DOI] [PubMed] [Google Scholar]
- 19.Vrettou C., Traeger-Synodinos J., Tzetis M., Malamis G., Kanavakis E. Rapid screening of multiple β-globin gene mutations by real-time PCR on the LightCycler: application to carrier screening and prenatal diagnosis of thalassemia syndromes. Clin Chem. 2003;49:769–776. doi: 10.1373/49.5.769. [DOI] [PubMed] [Google Scholar]
- 20.Wang W., Kham S.K., Yeo G.H., Quah T.C., Chong S.S. Multiplex minisequencing screen for common Southeast Asian and Indian β-thalassemia mutations. Clin Chem. 2003;49:209–218. doi: 10.1373/49.2.209. [DOI] [PubMed] [Google Scholar]
- 21.Xiong F., Huang Q., Chen X., Zhou Y., Zhang X., Cai R., Chen Y., Xie J., Feng S., Wei X., Xiao Q., Zhang T., Luo S., Yang X., Hao Y., Qu Y., Li Q., Xu X. A melting curve analysis–based PCR assay for one-step genotyping of β-thalassemia mutations: a multicenter validation. J Mol Diagn. 2011;13:427–435. doi: 10.1016/j.jmoldx.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Craig J.E., Barnetson R.A., Prior J., Raven J.L., Thein S.L. Rapid detection of deletions causing δβ thalassemia and hereditary persistence of fetal hemoglobin by enzymatic amplification. Blood. 1994;83:1673–1682. [PubMed] [Google Scholar]
- 23.De Andrade T.G., Saad S.T.O., Sonati M.F., Costa F.F. Simple fluorescent PCR method for detection of large deletions in the β-globin gene cluster. Am J Hematol. 2003;72:225–227. doi: 10.1002/ajh.10291. [DOI] [PubMed] [Google Scholar]
- 24.Colosimo A., Gatta V., Guida V., Leodori E., Foglietta E., Rinaldi S., Cappabianca M.P., Amato A., Stuppia L., Dallapiccola B. Application of MLPA assay to characterize unsolved α-globin gene rearrangements. Blood Cells Mol Dis. 2011;46:139–144. doi: 10.1016/j.bcmd.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Harteveld C.L., Voskamp A., Phylipsen M., Akkermans N., den Dunnen J.T., White S.J., Giordano P.C. Nine unknown rearrangements in 16p13.3 and 11p15.4 causing α- and β-thalassaemia characterised by high resolution multiplex ligation-dependent probe amplification. J Med Genet. 2005;42:922–931. doi: 10.1136/jmg.2005.033597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kipp B.R., Roellinger S.E., Lundquist P.A., Highsmith W.E., Dawson D.B. Development and clinical implementation of a combination deletion PCR and multiplex ligation-dependent probe amplification assay for detecting deletions involving the human α-globin gene cluster. J Mol Diagn. 2011;13:549–557. doi: 10.1016/j.jmoldx.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phylipsen M., Vogelaar I.P., Schaap R.A., Arkesteijn S.G., Boxma G.L., van Helden W.C., Wildschut I.C., de Bruin-Roest A.C., Giordano P.C., Harteveld C.L. A new α0-thalassemia deletion found in a Dutch family (–AW) Blood Cells Mol Dis. 2010;45:133–135. doi: 10.1016/j.bcmd.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Ghedira E.S., Lecerf L., Faubert E., Costes B., Moradkhani K., Bachir D., Galacteros F., Pissard S. Estimation of the difference in HbF expression due to loss of the 5′ δ-globin BCL11A binding region. Haematologica. 2013;98:305–308. doi: 10.3324/haematol.2012.061994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trent R.J., Svirklys L., Jones P. Thai (δβ)0-thalassemia and its interaction with γ-thalassemia. Hemoglobin. 1988;12:101–114. doi: 10.3109/03630268808998017. [DOI] [PubMed] [Google Scholar]
- 30.Sankaran V.G., Menne T.F., Xu J., Akie T.E., Lettre G., Van Handel B., Mikkola H.K., Hirschhorn J.N., Cantor A.B., Orkin S.H. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 31.Galarneau G., Palmer C.D., Sankaran V.G., Orkin S.H., Hirschhorn J.N., Lettre G. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat Genet. 2010;42:1049–1051. doi: 10.1038/ng.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farrell J.J., Sherva R.M., Chen Z.Y., Luo H.Y., Chu B.F., Ha S.Y., Li C.K., Lee A.C., Li R.C., Li C.K., Yuen H.L., So J.C., Ma E.S., Chan L.C., Chan V., Sebastiani P., Farrer L.A., Baldwin C.T., Steinberg M.H., Chui D.H. A 3-bp deletion in the HBS1L-MYB intergenic region on chromosome 6q23 is associated with HbF expression. Blood. 2011;117:4935–4945. doi: 10.1182/blood-2010-11-317081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sanger sequencing chromatograms of the breakpoints of β-globin deletions characterized by array comparative genomic hybridization (CGH) that had not been finely mapped previously. PCR primers were designed based on the breakpoints as determined by array CGH. The resulting PCR products were sequenced by Sanger sequencing. Vertical black lines indicate where the breakpoints occur. Orange bars indicate sequences that are not present in the hg38 reference human germline sequence. Tracings are for subject 4 (A, Thai/Vietnamese (δβ)0 thalassemia), subject 5 (B, SEA-HPFH), subject 7 (C, Kurdish deletion; note that there are two breakpoints for this deletion shown), subject 8 (D, Irish/Romanian), and subject 9 (E, Belgian). Sequence data for subjects 6 and 10 can be found in the supplemental material for reference 5. Because the breakpoints for the 2.1-Mb deletion (subject 11) occur in low-density tiled regions of chromosome 11, it was not possible to design PCR primers and sequence the breakpoints.
Array comparative genomic hybridization (CGH) characterization of HBA single-gene deletions causing α-thalassemia. These include heterozygosity for the –α3.7 deletion (A), heterozygosity for the –α4.7 deletion (B), homozygosity for the –α3.7 deletion (C), and compound heterozygosity for the –α3.7 and –α4.7 deletions (D). Numbers at the bottom of the figure represent distance from the chromosome 16p telomere. In DNA samples heterozygous for either the –α3.7 or the –α4.2 deletion, a loss of signal of <50% can be seen in the region of the HBA genes (A and B, respectively). With DNA containing a homozygous –α3.7 deletion, the loss in signal is closer to the expected 50% in the stretch spanning nucleotides chr16:173,301-177,104, including the 3′ end of the HBA2 gene to the 5′ end of the HBA1 gene (C). Again, although there is loss of genetic material from both alleles in this region, there is only effectively a 50% loss because there are two remaining α-globin genes, which hybridize to the probes for the deleted region. A similar result is obtained with DNA from a compound heterozygote for –α3.7 and –α4.2 (D).