Summary
In this genome-wide association study of colorectal cancer outcomes, multiple novel variants in the 6p12.1 region were identified as significantly associated with survival among individuals with distant-metastatic colorectal cancer.
Abstract
Genome-wide association studies have identified several germline single nucleotide polymorphisms (SNPs) significantly associated with colorectal cancer (CRC) incidence. Common germline genetic variation may also be related to CRC survival. We used a discovery-based approach to identify SNPs related to survival outcomes after CRC diagnosis. Genome-wide genotyping arrays were conducted for 3494 individuals with invasive CRC enrolled in six prospective cohort studies (median study-specific follow-up = 4.2–8.1 years). In pooled analyses, we used Cox regression to assess SNP-specific associations with CRC-specific and overall survival, with additional analyses stratified by stage at diagnosis. Top findings were followed-up in independent studies. A P value threshold of P < 5×10−8 in analyses combining discovery and follow-up studies was required for genome-wide significance. Among individuals with distant-metastatic CRC, several SNPs at 6p12.1, nearest the ELOVL5 gene, were statistically significantly associated with poorer survival, with the strongest associations noted for rs209489 [hazard ratio (HR) = 1.8, P = 7.6×10−10 and HR = 1.8, P = 3.7×10−9 for CRC-specific and overall survival, respectively). No SNPs were statistically significantly associated with survival among all cases combined or in cases without distant-metastases. SNPs in 6p12.1/ELOVL5 were associated with survival outcomes in individuals with distant-metastatic CRC, and merit further follow-up for functional significance. Findings from this genome-wide association study highlight the potential importance of genetic variation in CRC prognosis and provide clues to genomic regions of potential interest.
Introduction
Advances in colorectal cancer (CRC) early detection and treatment have led to considerable declines in CRC mortality rates (1). Nonetheless, 5-year relative survival for CRC is less than 65% in the United States (2). Although risk factors for incident CRC are relatively well-established, less is known about factors associated with CRC survival. At present, the strongest known predictor of CRC prognosis is stage (2); however, there is considerable heterogeneity in survival among individuals with the same stage at diagnosis (2). To extend our understanding of CRC pathogenesis and potentially direct treatment, there remains a need to identify markers of CRC prognosis. Information on the role of germline genetic factors in CRC prognosis represents an important gap in knowledge in this regard.
Genome-wide association studies (GWAS) for CRC susceptibility have identified several germline variants associated with CRC risk (3–12). Although these loci are only modestly associated with risk, they may provide important clues into the pathogenesis of CRC. The GWAS approach is similarly likely to provide valuable insights into CRC survival. To date, most studies evaluating genetic variation in relation to CRC survival have used candidate approaches, focusing on single nucleotide polymorphisms (SNPs) in genes involved in pathways of action for cancer therapeutics [e.g. the thymidylate synthase (TYMS) gene] (13,14). Other recent studies have explored the relationship between variation in CRC susceptibility SNPs, identified by GWAS for CRC incidence and survival after CRC diagnosis (e.g. rs4939827 in SMAD7) (5,15–18). Perhaps limited by small sample sizes or by the selection of unsuitable candidates, these studies have reported mostly null or only marginally significant associations, with little replication of findings.
Using a discovery-based approach with data from six prospective cohorts and follow-up in up to four independent studies, we evaluated the association between common genetic variation across the genome and CRC survival.
Materials and methods
Discovery study populations
Six cohort studies were included in primary discovery analyses: the Health Professionals Follow-up Study (HPFS) (19), the Nurses’ Health Study (NHS) (20–22), the Physicians’ Health Study (PHS) (23), the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO) (24,25), the VITamins And Lifestyle Study (VITAL) (26) and the Women’s Health Initiative (WHI) (27). These studies are included in the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) (3,4). All studies used a prospective design, with follow-up for incident cancer diagnoses and survival (19–27).
Discovery analyses were restricted to study participants with incident invasive CRC who self-reported European descent, and for whom genotype and survival data were available (N = 3494). Incident cancers were self-reported and confirmed by physician adjudication of medical records (HPFS, NHS, PHS, PLCO, WHI) and/or linkage to cancer registries (VITAL). Two subsets of cases were genotyped in the WHI: WHI1 included colon cancer patients from the WHI observational study diagnosed before September 2005 (4) and WHI2 included non-overlapping CRC patients diagnosed before August 2009. Similarly, two subsets of cases were genotyped in PLCO: PLCO1 included colon cancer patients, and PLCO2 included CRC cases not included in PLCO1. We excluded individuals for whom DNA was collected after CRC diagnosis. All participants provided informed consent for genetic testing. All studies were approved by their respective Institutional Review Boards.
Follow-up study populations
Four independent studies were used for follow-up of discovery-stage findings: the Cancer Prevention Study II Nutrition cohort (CPS-II) (28), the Diet, Activity and Lifestyle Study (DALS) (29), the Darmkrebs: Chancen der Verhütung durch Screening Study (DACHS) (30,31) and the UK Medical Research Council (MRC) combined COIN (32) and COIN-B trials (33). CPS-II, DALS and DACHS are included in GECCO. Study design details for these studies and COIN/COIN-B are published elsewhere (28–33). DALS and DACHS are population-based case–control studies for CRC incidence involving rapid case ascertainment and follow-up for survival; CPS-II is a prospective cohort study, with follow-up for incident cancers and survival; COIN/COIN-B are phase III treatment trials for advanced CRC. All studies were approved by their respective Institutional Review Boards.
Ascertainment of survival outcomes
Protocols for assessing survival in the included studies have been described previously (19,22,26,28–30,32–36). Most used active follow-up to ascertain vital status (HPFS, NHS, PHS, PLCO, WHI); dates and cause of death were confirmed via review of death certificates and/or medical records by trained adjudicators. Active follow-up was also used to ascertain survival outcomes in COIN/COIN-B, although information on cause of death was not available. For other studies (VITAL, CPS-II, DACHS, DALS), vital status was ascertained via linkage to the National Death Index, state cancer registries, state death records, or population registers with cause of death verified by death certificates. In all studies, patients alive at the most recent study follow-up or data linkage were censored on that date. In VITAL, individuals who moved outside Washington State were censored at their date of move.
Genotyping and quality control
Genotyping details for GECCO studies have been reported previously (3,4). Genomic DNA was extracted from blood or buccal samples using conventional methods. Genotyping was performed per manufacturer’s protocols for the Illumina HumanHap300 and HumanHap240S (PLCO1), 550K (WHI1, DALS1), 610K (PLCO1, WHI1, DALS1), HumanCytoSNP (PLCO2, VITAL, WHI2, DACHS1, DALS2) and HumanOmniExpress (HPFS, NHS, PHS, DACHS2) assays. CPS-II was genotyped on a custom Affymetrix Axiom array (1.3M SNPs). All genotyping underwent standard quality control (4), including concordance checks for blinded and unblinded duplicates, examination of sample and SNP call rates and testing for Hardy–Weinberg Equilibrium. The call rate was >97% for all samples and >98% for all SNPs.
Autosomal SNPs were imputed to the set of SNPs in HapMap II release 24 with MaCH (37), using Utah residents with Northern and Western European Ancestry from the Centre d’etude du polymorphisme humain (CEPH) collection (CEU) as the reference population. The present analysis included only those individuals who clustered with the CEU population. Imputed data were merged with genotyped data, giving preference to measured genotype when imputed and genotyped data were both available for a particular SNP. Evaluation was restricted to the ~2.7 million SNPs with a minor allele frequency ≥5% and an imputation accuracy R 2 > 0.3, excluding SNPs that were missing for >50% of included cases.
Two SNPs were evaluated in COIN/COIN-B follow-up analyses. Targeted genotyping of these SNPs was conducted using KASPar genotyping technology (LGC Genomics, London, UK).
Statistical analysis for discovery
Data were pooled across studies for discovery analyses. Survival time was calculated as the time from diagnosis to death or end of follow-up. We used Cox regression to calculate hazard ratios (HRs) and 95% confidence intervals (CIs) for SNP-specific associations. In analyses of CRC-specific survival, individuals who died from causes other than CRC were censored at the time of death. SNPs were modelled using a log-additive approach, relating genotype dose (i.e. number of copies of the minor allele) to survival outcomes. For imputed SNPs, ‘dosage’ was calculated on a scale from 0 to 2 based on imputation probabilities for each genotype (37).
We constructed separate models for overall and CRC-specific mortality. All models included age at diagnosis, sex, study and the first three principal components of genetic ancestry. We examined the Schoenfeld residuals to identify violations of the proportional hazards assumptions according to these covariates. We also conducted analyses stratified by stage at diagnosis. Because stage was classified according to Surveillance, Epidemiology and End Results (SEER) staging in some studies (i.e. local/regional/distant) and American Joint Committee on Cancer (AJCC) staging in others (i.e. I/II/III/IV), we stratified stage on harmonized groupings: non-distant (local/regional, stages I–III) and distant-metastatic (distant, stage IV). Genome-wide statistical significance was specified at P < 5×10–8 based on Wald P values in single-SNP models. We inspected Q–Q plots of −log10-transformed P values and assessed the influence of population stratification by calculating genomic control coefficients (38). Analyses were performed using R 2.15.3.
Statistical analysis for follow-up
Follow-up of top findings from discovery analyses (P < 5×10−6) was carried out in CPS-II, DALS1, DALS2, DACHS1 and DACHS2 (N = 3764), adjusting for age at diagnosis, sex, study sample and the first three principal components of genetic ancestry. Two findings from discovery analyses of overall survival in distant-metastatic cases were followed-up in COIN/COIN-B (N = 2234), with analyses adjusted for treatment arm, chemotherapy regimen, age at randomization, sex and time from diagnosis to randomization. Estimates were combined across discovery and follow-up sets using fixed effects meta-analysis. Among correlated SNPs with pairwise R 2 ≥ 0.8 in the HapMap CEU population, a representative SNP was selected for inclusion in Table 3.
Table 3.
Single nucleotide polymorphisms identified from discovery analyses as being associated with survival after colorectal cancer diagnosis at significance level P < 5×10−7
| SNP | Position/nearest gene | Minor allele | Minor allele frequency | Discovery | Follow-up | Combined Discovery + follow-up |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total N/N deaths | HR (95% CI)b | P value | Total N/N deaths | HR (95% CI)‡ | P value | HR (95% CI)b,c | P value | ||||
| All stages combined | |||||||||||
| Overall survival | |||||||||||
| rs11077289 | 16p13.2/TMEM114 | A | 0.26 | 3494/1223 | 0.8 (0.7–0.9) | 3.9×10−7 | 3764/1135 | 1.1 (1.0–1.2) | 0.380 | 0.9 (0.8–1.0) | 3.5×10−3 |
| Distant-metastatic disease casesa | |||||||||||
| Overall survival | |||||||||||
| rs17544464d | 6p12.1/ELOVL5 | C | 0.06 | 462/401 | 2.2 (1.6–2.9) | 1.7×10−7 | 2669/1975 | 1.1 (1.0–1.2) | 0.330 | 1.2 (1.1–1.4) | 1.5×10−3 |
| rs209489 | 6p12.1/ELOVL5 | C | 0.08 | 462/401 | 2.0 (1.5–2.5) | 2.2×10−7 | 435/363 | 1.6 (1.2–2.2) | 2.2×10−3 | 1.8 (1.5–2.1) | 3.7×10−9 |
| rs1442089§ | 18q21.2/DCC | C | 0.09 | 462/401 | 2.0 (1.5–2.6) | 4.8×10−7 | 2669/1975 | 1.0 (0.9–1.1) | 0.910 | 1.1 (1.0–1.3) | 0.045 |
| Disease-specific survival | |||||||||||
| rs17544464 | 6p12.1/ELOVL5 | C | 0.06 | 462/378 | 2.2 (1.7–3.0) | 7.5×10−8 | 435/339 | 1.5 (1.1–2.1) | 8.9×10−3 | 1.9 (1.5–2.3) | 9.1×10−9 |
| rs209489 | 6p12.1/ELOVL5 | C | 0.08 | 462/378 | 2.0 (1.6–2.6) | 9.7×10−8 | 435/339 | 1.6 (1.2–2.2) | 1.0×10−3 | 1.8 (1.5–2.2) | 7.6×10−10 |
aDistant-metastatic disease defined as distant stage per SEER staging or stage IV per AJCC stage classification.
bHazard ratios for discovery analyses adjusted for age at diagnosis, sex, study sample and first three principal components.
cHazard ratios for follow-up analyses adjusted for age at diagnosis, sex, study sample and first three principal components (DALS, DACHS, CPS II), or age at randomization, treatment arm, chemotherapy regimen, sex and time from diagnosis to randomization (COIN/COIN-B).
dFollow-up analyses for rs1442089 and rs17544464 for overall survival included COIN/COIN-B, DALS, DACHS and CPS II. COIN/COIN-B was not included in other follow-up analyses due to data availability.
Results
Characteristics of the discovery study populations are provided in Table 1. Median follow-up after diagnosis ranged from 4.2 to 8.1 years across studies. In total, 1223 (35%) CRC patients in discovery analyses died during follow-up; the proportion who died ranged from 22% (PLCO2) to 62% (PHS). Women accounted for 65% of the study population. Approximately 14% were diagnosed with distant-metastatic disease. Characteristics of follow-up study populations are provided in Table 2. Study population attributes, pooled across study phase, are also provided in Supplementary Table 1, available at Carcinogenesis Online.
Table 1.
Characteristics of colorectal cancer cases in study populations included in primary discovery analyses
| Health Professionals Follow-up Study | Nurses’ Health Study | Physicians’ Health Study | Prostate, Lung, Colon and Ovarian Cancer Screening Trial | VITamins and Lifestyle Study | Women’s Health Initiative | |||
|---|---|---|---|---|---|---|---|---|
| (Subset 1) | (Subset 2) | (Subset 1) | (Subset 2) | |||||
| Abbreviation | HPFS | NHS | PHS | PLCO1 | PLCO2 | VITAL | WHI1 | WHI2 |
| Genotyping platforma | 730K | 730K | 730K | 300/240S, 610K | 300K | 300K | 550K, 550Kduo, 610K | 300K |
| No. cases | 168 | 296 | 324 | 531 | 478 | 285 | 455 | 957 |
| No. deaths, total (% of cases) | 82 (49) | 118 (40) | 200 (62) | 180 (34) | 103 (22) | 117 (41) | 160 (35) | 263 (27) |
| No. deaths, CRC (% of deaths) | 47 (57) | 89 (75) | 131 (66) | 108 (60) | 77 (75) | 70 (60) | 115 (72) | 193 (73) |
| Median follow-up in years (SD) | 5.8 (3.7) | 6.7 (5.0) | 8.1 (7.2) | 6.6 (3.4) | 4.5 (3.6) | 4.9 (2.9) | 5.3 (3.5) | 4.2 (3.4) |
| % Female | 0 | 100 | 0 | 43 | 42 | 47 | 100 | 100 |
| Age at diagnosis, N (%) | ||||||||
| <65 years | 41 (24) | 101 (34) | 98 (30) | 125 (24) | 98 (21) | 61 (21) | 84 (18) | 160 (17) |
| 65–69 | 21 (13) | 66 (22) | 53 (16) | 145 (27) | 115 (24) | 57 (20) | 94 (21) | 205 (21) |
| 70–74 | 38 (23) | 63 (21) | 55 (17) | 161 (30) | 131 (27) | 96 (34) | 133 (29) | 242 (25) |
| 75–79 | 34 (20) | 46 (16) | 51 (16) | 88 (17) | 88 (18) | 58 (20) | 95 (21) | 196 (20) |
| ≥80 years | 34 (20) | 20 (7) | 67 (21) | 12 (2) | 46 (10) | 13 (5) | 49 (11) | 154 (16) |
| Stage at diagnosis, N (%) | ||||||||
| I/localized | 47 (36) | 61 (23) | 64 (28) | 193 (37) | 166 (35) | 135 (48) | 192 (43) | 427 (45) |
| II–III/regional | 61 (46) | 151 (58) | 121 (53) | 282 (54) | 246 (52) | 100 (36) | 197 (44) | 400 (42) |
| IV/distant | 24 (18) | 50 (19) | 44 (19) | 51 (10) | 65 (14) | 46 (16) | 61 (14) | 121 (13) |
| Unknown | 36 | 34 | 95 | 5 | 1 | 4 | 5 | 9 |
| Tumor site, N (%) | ||||||||
| Colon | 113 (75) | 228 (78) | 250 (78) | 514 (99) | 313 (66) | 215 (77) | 442 (98) | 701 (74) |
| Rectum | 38 (25) | 65 (22) | 70 (22) | 5 (1) | 160 (34) | 66 (23) | 10 (2) | 250 (26) |
| Unknown | 17 | 3 | 4 | 12 | 5 | 4 | 3 | 6 |
aAll platforms were Illumina assays.
Table 2.
Characteristics of colorectal cancer cases in study populations included in follow-up analyses
| MRC COIN and COIN B | Cancer Prevention Study II | Darmkrebs: Chancen der Verhütung durch Screening Study | Diet, Activity and Lifestyle Study | |||
|---|---|---|---|---|---|---|
| (Subset 1) | (Subset 2) | (Subset 1) | (Subset 2) | |||
| Abbreviation | COIN/COIN-B | CPS-II | DACHS1 | DACHS2 | DALS1 | DALS2 |
| Genotyping platforma | KASPar (targeted) | Custom Affymetrix Axiom array | 300K | 730K | 550K/610K | 300K |
| No. cases | 2234 | 523 | 1705 | 420 | 706 | 410 |
| No. deaths, total (% of cases) | 1612 (72) | 113 (22) | 573 (34) | 97 (23) | 241 (34) | 113 (28) |
| No. deaths, CRC (% of deaths) | Not available | 84 (74) | 414 (72) | 71 (73) | 133 (55) | 79 (70) |
| Median follow-up in years (SD) | 2.4 (2.2) | 2.8 (2.0) | 4.9 (1.7) | 2.9 (0.9) | 5.2 (2.5) | 4.6 (1.7) |
| % Female | 34 | 50 | 41 | 38 | 43 | 47 |
| Age at diagnosis, N (%) | ||||||
| <65 years | 1296 (58) | 12 (2) | 589 (34) | 149 (35) | 288 (41) | 161 (39) |
| 65–69 | 456 (20) | 79 (15) | 318 (19) | 66 (16) | 142 (20) | 85 (21) |
| 70–74 | 339 (15) | 138 (26) | 288 (17) | 78 (19) | 155 (22) | 96 (23) |
| 75–79 | 127 (6) | 172 (33) | 260 (15) | 61 (14) | 121 (17) | 68 (17) |
| ≥80 years | 14 (1) | 122 (23) | 250 (15) | 66 (16) | 0 (0) | 0 (0) |
| Stage at diagnosis, N (%) | ||||||
| I/localized | 0 (0) | 229 (46) | 412 (24) | 101 (24) | 260 (40) | 128 (35) |
| II–III/regional | 0 (0) | 223 (44) | 1051 (62) | 260 (63) | 331 (51) | 210 (58) |
| IV/distant | 2234 (100) | 51 (10) | 238 (14) | 55 (13) | 64 (10) | 27 (7) |
| Unknown | 0 | 20 | 4 | 4 | 51 | 45 |
| Tumor site, N (%) | ||||||
| Colon | 1017 (46) | 417 (81) | 1042 (61) | 234 (56) | 702 (100) | 410 (100) |
| Rectum | 1216 (54) | 101 (19) | 663 (39) | 186 (44) | 0 (0) | 0 (0) |
| Unknown | 1 | 5 | 0 | 0 | 4 | 0 |
aUnless otherwise stated, genotyping platforms were Illumina assays.
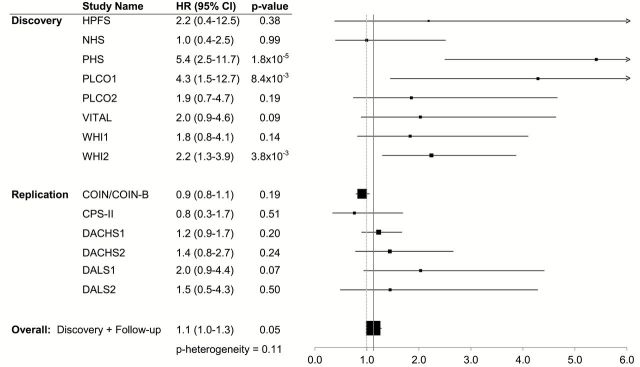
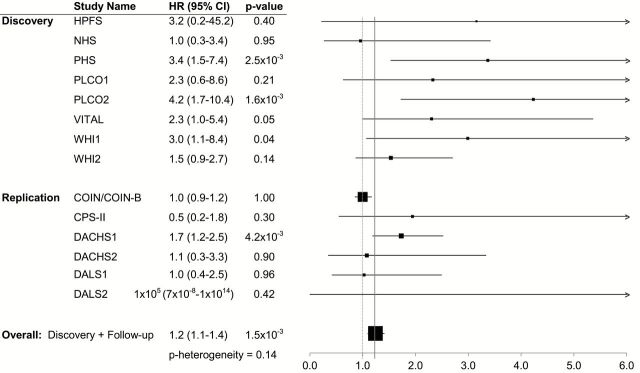
In discovery analyses of all cases combined (Supplementary Table 2; Supplementary Figures 1 and 2, available at Carcinogenesis Online), the minor allele at rs11077289 (16p13.2/TMEM114) was associated with more favorable overall survival (HR = 0.8, P = 3.9×10−7); however, this association was not evident in follow-up (Table 3). No SNPs emerged from analyses in non-distant CRC cases (Supplementary Table 3; Supplementary Figures 3 and 4, available at Carcinogenesis Online). In discovery analyses restricted to distant-metastatic CRC cases (Supplementary Figures 5–8, available at Carcinogenesis Online), the minor alleles at rs17544464 (6p12.1/ELOVL5), rs209489 (6p12.1/ELOVL5) and rs1442089 (18q21.2/DCC) were each associated with a 2.0- to 2.2-fold shorter overall survival (P = 1.7×10−7 to 4.8×10−7); P values were similar after adjusting for inflation factors (results not shown). This association with rs209489 persisted in follow-up (2.2×10−3) and was statistically significant in analyses of discovery and follow-up study populations combined (P = 3.7×10−9). Associations with rs209489 were similar and exceeded genome-wide significance in analyses of CRC-specific survival. Associations with overall survival for rs17544464 and rs1442089 were not evident in follow-up (P = 0.330 and P = 0.910, respectively), due largely to the contribution of COIN/COIN-B in the follow-up set (Figures 1 and 2). There was evidence of considerable heterogeneity across studies when including COIN/COIN-B in follow-up for these two SNPs (P heterogeneity = 1.3×10−4 and 3.7×10−5), but not when COIN/COIN-B was excluded from follow-up (P heterogeneity = 0.14 and 0.11, respectively). Other SNPs in linkage disequilibrium with or nearby rs17544464 or rs209489 were also strongly associated with survival among individuals with distant-metastatic CRC in analyses not including COIN/COIN-B (Supplementary Table 3, available at Carcinogenesis Online).
Figure 1.
Association between dose of rs1442089 minor allele and overall survival among those with distant-metastatic colorectal cancer by study population. Estimates for COIN/COIN-B are adjusted for treatment arm, chemotherapy regimen, age at randomization, sex and time from diagnosis to randomization. Estimates for all other studies are adjusted for age at diagnosis, sex and the first three principal components of genetic ancestry.
Figure 2.
Association between dose of rs17544464 minor allele and overall survival among those with distant-metastatic colorectal cancer by study population. Estimates for COIN/COIN-B are adjusted for treatment arm, chemotherapy regimen, age at randomization, sex and time from diagnosis to randomization. Estimates for all other studies are adjusted for age at diagnosis, sex and the first three principal components of genetic ancestry.
Discussion
In this discovery-based search for common genetic variants associated with CRC prognosis, multiple SNPs at 6p12.1 were identified as significantly associated with distant-metastatic CRC survival: the minor allele at rs209489 was associated with shorter overall and CRC-specific survival at a level of genome-wide significance, and the minor allele at rs17544464 was associated with significantly shorter CRC-specific survival. No SNPs were statistically significantly associated with survival among individuals with non-distant CRC or in analyses of all cases combined. To our knowledge, this is the first genome-wide examination of common genetic variation and CRC survival.
The loci that emerged from our combined analyses in those with distant-metastatic disease have not previously been described in relation to CRC survival or risk. Most SNPs that were identified as being associated with survival are located in or nearest to the ELOVL5 gene, which encodes a fatty acid elongase (ELOVL5). Knockout of ELOVL5 in mouse models appears to result in hepatic steatosis (39). Previous studies have found hepatic steatosis to be both an independent risk factor for distant-metastatic CRC (40) and a marker of lower risk of hepatic metastases of CRC (41). Nonetheless, associations between hepatic steatosis and CRC prognosis have been inconsistent (42,43). It is also plausible that noted associations with SNPs at 6p12.1 reflect activity of other nearby genes. The coding region for the intestinal cell (MAK-like) kinase (ICK) gene is located within 200kb downstream of the tagged region for rs209489. ICK encodes a protein kinase that localizes to the intestinal crypt and is thought to be important in epithelial cell proliferation and differentiation (44); knockdown of ICK in CRC cell lines has been shown to induce G1 cell cycle delay and slow cell growth (45). Other nearby genes include glutathione S-transferases alpha 1–5 (GSTA1, GSTA2, GSTA3, GSTA4, GSTA5). GST polymorphisms have been associated with CRC incidence and survival (46). Thus, although the functional significance of the SNPs at 6p12.1 found here to be associated with CRC survival has not been established, these findings merit further study.
Discovery analyses in cases with distant-metastatic CRC also suggested an association between the minor allele at rs1442089 (18q21.2/DCC) and shorter overall survival. DCC (i.e. Deleted in Colorectal Carcinoma) has been implicated in CRC etiology (47), and loss of DCC expression in CRC has been associated with a 2- to 4-fold poorer prognosis (48,49). However, results for rs1442089 were null in follow-up, suggesting our initial findings may have been spurious. Findings in the follow-up population were primarily driven by null results in the large COIN/COIN-B study. There are differences between the discovery study populations and COIN/COIN-B that may have contributed to discrepancies. In particular, the rigorous inclusion/exclusion criteria of the clinical trial setting may have resulted in a study population fundamentally and prognostically different from the population included in the observational studies that comprised the discovery set and the rest of the follow-up sample. Treatment differences may also have contributed. Differing methodologies, however, are unlikely to fully explain observed differences in results. Thus, although it remains possible that rs1442089 (18q21.2/DCC) is associated with prognosis in distant-metastatic CRC, the magnitude of such an association is likely not as strong as noted in our discovery analyses. Similarly, discovery analyses among all cases combined provided suggestive findings for a SNP in TMEM114 (rs11077289) that was not replicated. TMEM114 (transmembrane protein 114) has been implicated in cataract formation (50) but, to our knowledge, has not previously been associated with cancer risk or prognosis.
Previous analyses of genetic variation and CRC survival have taken a candidate approach, evaluating variation in specific pathways, genes, or SNPs based on a priori hypotheses. Several studies have focused on GWAS identified CRC susceptibility SNPs in relation to survival (5,15–18). Using this approach to interrogate 16 CRC susceptibility SNPs in a subset of the cases included in the present analysis, we previously reported a modest association between the minor allele in rs4939827 (SMAD7) and poorer CRC survival (P = 0.002) (15). Although results from our previous analysis and other candidate studies have generated suggestive findings, many such findings have not been replicated in subsequent analyses. The limited robustness of findings from prior studies may, in part, reflect the shortcomings of a candidate-based approach; i.e. the pathways, genes and SNPs most relevant to and most robustly associated with CRC survival may be ones without a previously understood role in CRC progression and prognosis.
In the present analysis, we used an agnostic discovery-based approach to search for variants associated with CRC survival. The GWAS approach has successfully identified several CRC susceptibility variants (3–12), most of which were not targets of earlier candidate studies. Based on our current findings, there is reason to suspect that the identified SNPs in the 6p12.1 region fit with this paradigm as loci important to CRC survival that would likely not have been considered through a candidate approach.
Our results should be interpreted in the context of study limitations. Treatment information was not available for studies in discovery analyses; therefore, we were unable to evaluate associations with response to specific treatments. Sample size limitations precluded extensive stratified analyses by other factors (e.g. tumor site). Lastly, one limitation inherent to the GWAS approach is the high likelihood of false-negative findings due to the stringent P-value threshold for genome-wide significance. This threshold is set to account for multiple testing and is designed to reduce the number of false-positive findings; however, a consequence of this stringency is that some important SNP-survival associations may have been missed.
The prospective nature of the studies included in discovery analyses constitutes an important strength; DNA specimens were collected prior to CRC diagnosis and, thus, inclusion in the analysis was not influenced by survival time. The included studies employed rigorous follow-up protocols to ensure the completeness of case ascertainment and vital status assessment. The large sample size and long duration of follow-up after diagnosis are also important strengths, as is the replication of findings in a large follow-up sample.
Just as GWAS for CRC risk have provided evidence for inherited susceptibility to CRC, findings from the present analysis support a role of common genetic variation in mediating CRC survival. SNPs at 6p12.1 were robustly associated with survival in individuals with distant-metastatic CRC in discovery and independent follow-up analyses, and merit further follow-up. The fact that the gene nearest to these SNPs, ELOVL5, has not previously been implicated in CRC etiology or progression highlights the utility of the agnostic GWAS approach, although it is also possible that the identified SNPs reflect the role of another nearby gene (e.g. ICK). Results also highlight the need for independent replication. Future well-powered GWAS with independent follow-up and consideration for stage at diagnosis may yield additional findings that further our understanding of the mechanisms underlying CRC progression.
Supplementary material
Supplementary Tables 1–4 and Supplementary Figures 1–8 can be found at http://carcin.oxfordjournals.org/.
Funding
National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services (U01CA137088 to U.P., R01CA059045 to U.P., R25CA094880, T32CA009168, K07CA172298 to A.I.P., K05CA152715 to P.A.N., R01 CA176272 to P.A.N. and A.T.C.). The COIN trial was funded by Cancer Research UK and the Medical Research Council, and its associated translational studies are supported by the Bobby Moore Fund from Cancer Research UK, Tenovus, the Kidani Trust and the National Institute for Social Care and Health Research Cancer Genetics Biomedical Research Unit. Additional funding support for individual studies is provided below:
CORECT: National Cancer Institute, National Institutes of Health under RFA # CA-09-002 (U19CA148107). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in CORECT, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or CORECT.
CPS-II: The American Cancer Society funds the creation, maintenance and updating of the Cancer Prevention Study-II (CPS-II) cohort. This study was conducted with Institutional Review Board approval.
DACHS: German Research Council (Deutsche Forschungsgemeinschaft, BR 1704/6-1, BR 1704/6-3, BR 1704/6-4 and CH 117/1-1) and the German Federal Ministry of Education and Research (01KH0404 and 01ER0814).
DALS: National Institutes of Health (R01 CA48998 to M.L.S).
HPFS: National Institutes of Health (P01CA055075, UM1CA167552, R01CA137178, P50CA127003, K24DK098311).
NHS: National Institutes of Health (R01CA137178, P01CA087969, P50CA127003, K24DK098311).
PHS: National Institutes of Health (R01CA42182, K24DK098311).
PLCO: Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH, DHHS.
VITAL: National Institutes of Health (K05CA154337 to E.W.).
WHI: National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, HHSN271201100004C. Supplementary Tables 1–4 and Figures 1–8 can be found at http://carcin.oxfordjournals.org/
Supplementary Material
Acknowledgements
GECCO: The authors would like to thank all those at the GECCO Coordinating Center for helping bring together the data and people that made this project possible. The authors acknowledge Dave Duggan and team members at TGEN (Translational Genomics Research Institute), the Broad Institute and the Génome Québec Innovation Center for genotyping DNA samples and for scientific input for GECCO.
CPS-II: The authors thank the CPS-II participants and Study Management Group for their invaluable contributions to this research. The authors would also like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention National Program of Cancer Registries and cancer registries supported by the National Cancer Institute Surveillance Epidemiology and End Results program.
DACHS: We thank all participants and cooperating clinicians, and Ute Handte-Daub, Renate Hettler-Jensen, Utz Benscheid, Muhabbet Celik and Ursula Eilber for excellent technical assistance.
HPFS, NHS and PHS: We would like to acknowledge Patrice Soule and Hardeep Ranu of the Dana Farber Harvard Cancer Center High-Throughput Polymorphism Core who assisted in the genotyping for NHS, HPFS and PHS under the supervision of Dr. Immaculata De Vivo and Dr. David Hunter, Qin (Carolyn) Guo and Lixue Zhu who assisted in programming for NHS and HPFS and Haiyan Zhang who assisted in programming for the PHS. We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-Up Study, for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
PLCO: The authors thank Drs. Christine Berg and Philip Prorok, Division of Cancer Prevention, National Cancer Institute, the Screening Center investigators and staff or the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial, Mr. Tom Riley and staff, Information Management Services, Inc., Ms. Barbara O’Brien and staff, Westat, Inc. and Drs. Bill Kopp, Wen Shao and staff, SAIC-Frederick. Most importantly, we acknowledge the study participants for their contributions to making this study possible.
WHI: The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Short%20List.pdf
Conflict of Interest Statement: Unrelated to the specific research detailed in this manuscript, several authors have received funding from Industry. The following authors have received funding in consulting or advisory roles: Dr. Andrew Chan (Bayer Healthcare, Pfizer), Dr. Charles Fuchs (Amgen, Roche, Genentech, Gilead, Merck, Bristol Myers Squibb, Bayer, Takeda, Eli Lilly, Acceleron, Vertex, Medimmune, Sanofi, Pfizer, Celgene). The following authors have received research funding from Industry: Dr. Hermann Brenner (Roche Diagnostics), Dr. Richard Kaplan (AstraZeneca, GSK), Robert Schoen (Shire). Additionally, Dr. Jeremy Cheadle has received patents or royalties (not related to the present research) from Myriad Genetics, and Dr. Richard Kaplan has received honoria from Celldex Therapeutics. Lastly, the following authors have stock or ownership in Industry: Keith Curtis (GenVec, Inc.), Manish Gala (New Amsterdam Genomics). In no instances were these Industry organizations directly involved in the research presented in this manuscript. Other authors reported no potential conflicts of interest.
Glossary
Abbreviations
- CRC
colorectal cancer
- CPS-II
Cancer Prevention Study II Nutrition cohort
- DALS
Diet, Activity and Lifestyle Study
- DACHS
Darmkrebs: Chancen der Verhütung durch Screening Study
- GWAS
Genome-wide association study
- SNP
single nucleotide polymorphism
References
- 1. Phipps A.I., et al. (2012) Temporal trends in incidence and mortality rates for colorectal cancer by tumor location: 1975-2007. Am. J. Public Health, 102, 1791–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O'Connell J.B., et al. (2004) Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J. Natl. Cancer Inst., 96, 1420–1425. [DOI] [PubMed] [Google Scholar]
- 3. Peters U., et al. (2013) Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology, 144, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peters U., et al. (2012) Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum. Genet., 131, 217–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tenesa A., et al. (2010) Ten common genetic variants associated with colorectal cancer risk are not associated with survival after diagnosis. Clin. Cancer Res., 16, 3754–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Houlston R.S., et al. (2008) Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet., 40, 1426–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Houlston R.S., et al. (2010) Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat. Genet., 42, 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tomlinson I.P., et al. (2008) A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat. Genet., 40, 623–630. [DOI] [PubMed] [Google Scholar]
- 9. Tomlinson I.P., et al. (2011) Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet., 7, e1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kocarnik J.D., et al. (2010) Characterization of 9p24 risk locus and colorectal adenoma and cancer: gene-environment interaction and meta-analysis. Cancer Epidemiol. Biomarkers Prev., 19, 3131–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hutter C.M., et al. (2010) Characterization of the association between 8q24 and colon cancer: gene-environment exploration and meta-analysis. BMC Cancer, 10, 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jia W.H., et al. (2013) Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat. Genet., 45, 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Afzal S., et al. (2011) The association of polymorphisms in 5-fluorouracil metabolism genes with outcome in adjuvant treatment of colorectal cancer. Pharmacogenomics, 12, 1257–1267. [DOI] [PubMed] [Google Scholar]
- 14. Curtin K., et al. (2007) Thymidylate synthase polymorphisms and colon cancer: associations with tumor stage, tumor characteristics and survival. Int. J. Cancer, 120, 2226–2232. [DOI] [PubMed] [Google Scholar]
- 15. Phipps A.I., et al. (2012) Association between colorectal cancer susceptibility loci and survival time after diagnosis with colorectal cancer. Gastroenterology, 143, 51–4.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xing J., et al. (2011) GWAS-identified colorectal cancer susceptibility locus associates with disease prognosis. Eur. J. Cancer, 47, 1699–1707. [DOI] [PubMed] [Google Scholar]
- 17. Cicek M.S., et al. (2009) Functional and clinical significance of variants localized to 8q24 in colon cancer. Cancer Epidemiol. Biomarkers Prev., 18, 2492–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Passarelli M.N., et al. (2011) Common colorectal cancer risk variants in SMAD7 are associated with survival among prediagnostic nonsteroidal anti-inflammatory drug users: a population-based study of postmenopausal women. Genes. Chromosomes Cancer, 50, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rimm E.B., et al. (1991) Prospective study of alcohol consumption and risk of coronary disease in men. Lancet, 338, 464–468. [DOI] [PubMed] [Google Scholar]
- 20. Belanger C.F., et al. (1978) The nurses' health study. Am. J. Nurs., 78, 1039–1040. [PubMed] [Google Scholar]
- 21. Belanger C., et al. (1980) The nurses' health study: current findings. Am. J. Nurs., 80, 1333. [DOI] [PubMed] [Google Scholar]
- 22. Colditz G.A., et al. (1997) The Nurses' Health Study: 20-year contribution to the understanding of health among women. J. Womens. Health, 6, 49–62. [DOI] [PubMed] [Google Scholar]
- 23. (1989) Final report on the aspirin component of the ongoing Physicians’ Health Study. N. Engl. J. Med., 321, 129–135. [DOI] [PubMed] [Google Scholar]
- 24. Prorok P.C., et al. (2000) Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening trial. Control Clin. Trials, 21, 273S–309S. [DOI] [PubMed] [Google Scholar]
- 25. Gohagan J.K., et al. (2000) The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial of the National Cancer Institute: history, organization, and status. Control Clin. Trials, 21, 251S–272S. [DOI] [PubMed] [Google Scholar]
- 26. White E., et al. (2004) VITamins And Lifestyle cohort study: study design and characteristics of supplement users. Am. J. Epidemiol., 159, 83–93. [DOI] [PubMed] [Google Scholar]
- 27. (1998) Design of the Women’s Health Initiative clinical trial and observational study. Control Clin. Trials, 19, 61–109. [DOI] [PubMed] [Google Scholar]
- 28. Calle E.E., et al. (2002) The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer, 94, 2490–2501. [DOI] [PubMed] [Google Scholar]
- 29. Slattery M.L., et al. (1997) Energy balance and colon cancer–beyond physical activity. Cancer Res., 57, 75–80. [PubMed] [Google Scholar]
- 30. Brenner H., et al. (2011) Protection from colorectal cancer after colonoscopy: a population-based, case-control study. Ann. Intern. Med., 154, 22–30. [DOI] [PubMed] [Google Scholar]
- 31. Lilla C., et al. (2006) Effect of NAT1 and NAT2 genetic polymorphisms on colorectal cancer risk associated with exposure to tobacco smoke and meat consumption. Cancer Epidemiol. Biomarkers Prev., 15, 99–107. [DOI] [PubMed] [Google Scholar]
- 32. Maughan T.S., et al. (2011) Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet, 377, 2103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wasan H., et al. (2014) Intermittent chemotherapy plus either intermittent or continuous cetuximab for first-line treatment of patients with KRAS wild-type advanced colorectal cancer (COIN-B): a randomised phase 2 trial. Lancet Oncol., 15, 631–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chan A.T., et al. (2009) Aspirin use and survival after diagnosis of colorectal cancer. JAMA, 302, 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Curb J.D., et al. (2003) Outcomes ascertainment and adjudication methods in the Women’s Health Initiative. Ann. Epidemiol., 13, S122–S128. [DOI] [PubMed] [Google Scholar]
- 36. Miller A.B., et al. (2000) Death review process in the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin. Trials, 21, 400S–406S. [DOI] [PubMed] [Google Scholar]
- 37. Li Y., et al. (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol., 34, 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Devlin B., et al. (1999) Genomic control for association studies. Biometrics, 55, 997–1004. [DOI] [PubMed] [Google Scholar]
- 39. Moon Y.A., et al. (2009) Deletion of ELOVL5 leads to fatty liver through activation of SREBP-1c in mice. J. Lipid Res., 50, 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin X.F., et al. (2014) Increased risk of colorectal malignant neoplasm in patients with nonalcoholic fatty liver disease: a large study. Mol. Biol. Rep., 41, 2989–2997. [DOI] [PubMed] [Google Scholar]
- 41. Augustin G., et al. (2013) Lower incidence of hepatic metastases of colorectal cancer in patients with chronic liver diseases: meta-analysis. Hepatogastroenterology, 60, 1164–1168. [DOI] [PubMed] [Google Scholar]
- 42. Lee Y.I., et al. (2012) Colorectal neoplasms in relation to non-alcoholic fatty liver disease in Korean women: a retrospective cohort study. J. Gastroenterol. Hepatol., 27, 91–95. [DOI] [PubMed] [Google Scholar]
- 43. Min Y.W., et al. (2012) Influence of non-alcoholic fatty liver disease on the prognosis in patients with colorectal cancer. Clin. Res. Hepatol. Gastroenterol., 36, 78–83. [DOI] [PubMed] [Google Scholar]
- 44. Togawa K., et al. (2000) Intestinal cell kinase (ICK) localizes to the crypt region and requires a dual phosphorylation site found in map kinases. J. Cell. Physiol., 183, 129–139. [DOI] [PubMed] [Google Scholar]
- 45. Fu Z., et al. (2009) Intestinal cell kinase, a MAP kinase-related kinase, regulates proliferation and G1 cell cycle progression of intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol., 297, G632–G640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McIlwain C.C., et al. (2006) Glutathione S-transferase polymorphisms: cancer incidence and therapy. Oncogene, 25, 1639–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Grady W.M. (2007) Making the case for DCC and UNC5C as tumor-suppressor genes in the colon. Gastroenterology, 133, 2045–2049. [DOI] [PubMed] [Google Scholar]
- 48. Aschele C., et al. (2004) Deleted in colon cancer protein expression in colorectal cancer metastases: a major predictor of survival in patients with unresectable metastatic disease receiving palliative fluorouracil-based chemotherapy. J. Clin. Oncol., 22, 3758–3765. [DOI] [PubMed] [Google Scholar]
- 49. Shibata D., et al. (1996) The DCC protein and prognosis in colorectal cancer. N. Engl. J. Med., 335, 1727–1732. [DOI] [PubMed] [Google Scholar]
- 50. Jamieson R.V., et al. (2007) Characterization of a familial t(16;22) balanced translocation associated with congenital cataract leads to identification of a novel gene, TMEM114, expressed in the lens and disrupted by the translocation. Hum. Mutat., 28, 968–977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.