Abstract
BACKGROUND
The renin–angiotensin–aldosterone system (RAAS) plays an important role in blood pressure (BP) regulation. The current study uses single-marker and gene-based analyses to examine the association between RAAS genes and longitudinal BP phenotypes in a Han Chinese population.
METHODS
A total of 1,768 participants from the Genetic Epidemiology Network of Salt Sensitivity (GenSalt) follow-up study were included in the current study. Twenty-seven BP measurements were taken using random-zero sphygmomanometers at baseline and 2 follow-up visits. Mixed-effect models were used to assess the additive associations of 106 single-nucleotide polymorphisms (SNPs) in 10 RAAS genes with longitudinal BP changes and hypertension incidence. Gene-based analyses were conducted using the truncated product method. Attempts were made to replicate significant findings among Asian participants of the Multi-ethnic Study of Atherosclerosis (MESA). False discovery rate procedures were used to adjust for multiple testing.
RESULTS
During an average of 7.2 years of follow-up, average systolic and diastolic BP increased, and 32.1% (512) of participants free from hypertension at baseline developed hypertension. NR3C2 SNPs rs7694064 and rs6856803 were significantly associated with longitudinal changes in systolic BP (P interaction = 6.9×10−5 and 8.2×10−4, respectively). Through gene-based analysis, NR3C2 was found to be significantly associated with longitudinal systolic BP change (P value of 1.00×10−7), even after removal of significant markers rs7694064 and rs6856803 from the analysis. The association between NR3C2 and longitudinal systolic BP change was replicated in Asian MESA participants (P value of 1.00×10−4).
CONCLUSIONS
These findings indicate that NR3C2 may play an important role in BP progression and development of hypertension.
Keywords: blood pressure, blood pressure changes, genetics, hypertension, rennin–angiotensin–aldosterone system (RAAS).
Hypertension is a major, worldwide public health challenge due to its high prevalence and associated risk of cardiovascular disease and all-cause mortality.1–3 Blood pressure (BP) is a complex trait, as it is influenced by multiple factors. One factor that contributes to interindividual variability in BP is genetics, with heritability estimates ranging 31%–68%.4 However, the exact genomic mechanisms influencing BP are largely unknown.
The renin–angiotensin–aldosterone system (RAAS) plays a central role in BP regulation by maintaining sodium and water homeostasis and vascular tone.5,6 Several genetic variants in the RAAS, for example, the I/D polymorphism of angiotensin-converting enzyme (ACE), M235T of angiotensinogen (AGT), A1166C of angiotensin II type-1 receptor (AGTR1), and C344T of aldosterone synthase (CYP11B2), have been associated with high BP and renal sodium excretion in cross-sectional studies.7–12 RAAS genetic variants were also associated with salt sensitivity of BP.13,14 However, there have been a limited number of investigations on the association between RAAS single-nucleotide polymorphism (SNPs) with longitudinal BP phenotypes and none examining the aggregate associations of RAAS genes with these phenotypes in a cohort study. Since time may modify the effects of genetic variants on BP, examination of longitudinal BP measures could provide important information regarding the role of RAAS in BP regulation.15 Furthermore, gene-based analyses may provide increased statistical power to detect the modest effects of common variants on BP changes and hypertension incidence.16
In this study, we conducted single-marker and gene-based analyses to examine the associations of 10 RAAS candidate genes (renin (REN); hydroxysteroid (11-beta) dehydrogenase 1 (HSD11B1); angiotensinogen (AGT) angiotensin II type 1 receptor (AGTR1); nuclear receptor subfamily 3, group C, member 2 (NR3C2); cytochrome P450, family 11, subfamily B, polypeptide 1 (CYP11B1); cytochrome P450, family 11, subfamily B, polypeptide 2 (CYP11B2); angiotensin-converting enzyme (ACE); angiotensin II type 1 receptor 2 (AGTR2); and renin-binding protein (RENBP)) with longitudinal BP changes and hypertension incidence among 1,768 Han Chinese individuals who participated in the Genetic Epidemiology Network of Salt Sensitivity (GenSalt) follow-up study.
METHODS
Study Population
The GenSalt study is a family-based feeding study that was created to examine the interaction of genes and dietary sodium and potassium intake on BP.17 Participants of the study were recruited from 6 northern, rural Chinese populations with high habitual sodium intake. The design and methods of the GenSalt study has been described in detail in past papers.18 Briefly, among persons aged 18–60 years, community-based BP screenings were performed in the 6 study villages to identify potential probands. Those with mean systolic BP of 130–160mm Hg and/or diastolic BP of 85–100mm Hg and no use of antihypertensive medication were recruited, as well as with their parents, siblings, spouses, and offspring. Individuals with stage 2 hypertension, secondary hypertension, a history of clinical cardiovascular disease or diabetes, or were pregnant, heavy alcohol drinkers, or currently on a low-sodium diet were excluded from the study.
Written informed consents were obtained from all GenSalt participants after detailed explanation of the study. The study has been approved by the Institutional Review Boards at all participating institutions.
Baseline Data Collection
The baseline examination and subsequent dietary intervention occurred from 2003 to 2005. In this examination, a standard questionnaire was given by trained staff to collect information regarding family pedigrees, demographic characteristics, personal and family medical history, and lifestyle risk factors. Three BP measurements were obtained according to a standard protocol on each morning of the 3-day baseline observation. BP readings were taken by trained and certified technicians using a random-zero sphygmomanometer after participants rested for 5 minutes in the sitting position. Also, participants were advised to avoid alcohol, cigarette smoking, coffee/tea, and exercise for at least 30 minutes before their BP measurements were taken. Mean BP at baseline was calculated as the average of the 9 measurements taken from the baseline observation. Body weight and height were measured twice in light indoor clothing without shoes. Body mass index was calculated as kilograms per square meter (kg/m2).
Follow-Up Data Collection
As part of the GenSalt follow-up study, participants took part in 2 follow-up examinations which were conducted in 2008–2009 and again in 2011–2012. During each follow-up visit, a clinical examination lasting 3 days was conducted using the same protocol as that of the baseline examination. Mean BP was calculated as the average of 9 BP measurements which were taking during each of the two 3-day follow-up visits. Hypertension was defined as having a systolic BP ≥140mm Hg and/or diastolic BP ≥90mm Hg or use of antihypertensive medication.
Among 1,906 eligible individuals from the 633 families who participated in the GenSalt baseline examination, 117 individuals were missing BP data at both follow-up visits and another 21 individuals were missing genotype data. In total, 1,768 participants (92.3%) were included in the analysis.
Genotype Data and Quality Control
Ten genes in the RAAS (REN, HSD11B1, AGT, AGTR1, NR3C2, CYP11B1, CYP11B2, ACE, AGTR2, and RENBP) were selected based on their potential biological effect on BP regulation. Within the 10 candidate genes, 265 SNPs were genotyped on the Affymetrix 6.0 platform (Affymetrix, Santa Clara, CA). Quality control excluded 55 SNPs based on low minor allele frequency <1%, low genotyping call rate (<95%), or deviation from the Hardy–Weinberg equilibrium after using the false discovery rate procedure to correct for multiple testing. Among the remaining 210 SNPs, we used empirical patterns of linkage disequilibrium structure within the GenSalt sample and Tagger software (Dr Paul de Bakker, Broad Institute, Cambridge, MA) to select 106 tag-SNPs with r 2 <0.8 for inclusion in the statistical analysis.19 Detailed information on these SNPS, including their genomic locations, major/minor alleles, minor allele frequency, call rates, and Hardy–Weinberg equilibrium-P values can be found in the Supplementary Table.
Statistical Analysis
The phenotypic and genotypic data of GenSalt follow-up study participants were presented as mean ± SD for continuous variables and as percentages for categorical variables. To account for the longitudinal, family-based design of the GenSalt study, mixed-effect linear regression models were used to examine the associations between each SNP and longitudinal BP phenotypes.20,21 For the assessment of BP change over time, a genotype by follow-up time interaction term and the main effects of these variables were included in the models. Autoregressive and compound symmetry covariance matrices were used to accommodate the correlations of repeated measurements within individuals and of individuals within families, respectively. Models were additionally adjusted for the fixed effects of age, gender, and body mass index using the PROC MIXED procedure in SAS (version 9.2; SAS Institute, Cary, NC). For participants taking antihypertensive medication (N = 159 at baseline examination), BP was imputed by adding 10 and 5mm Hg to systolic BP and diastolic BP values, respectively.22
After excluding 173 participants with hypertension at baseline, the additive association of SNPs with hypertension incidence was assessed using a multilevel logistic regression model.23 Autoregressive and compound symmetry covariance matrices were once again used to account for the correlations of repeated measurements within individuals and of individuals within families, respectively. After accommodating for the longitudinal family design, the fixed effects of age, gender, body mass index, and follow-up time were adjusted in multivariable analyses using the PROC GLIMMIX procedure in SAS.
The truncated product method was used to determine the overall association of each RAAS gene (with at least 2 genotyped SNPs) with longitudinal BP changes and hypertension incidence.24,25 This method is particularly suitable for combining P values across genes or genomic loci due to its ability to accommodate correlations among SNPs and its increased power to detect gene-based associations compared to other meta-analysis techniques.24 When determining a gene’s association with longitudinal BP changes, the P value for the genotype by follow-up time interaction term was used. When determining a gene’s association with hypertension incidence, the P value of the genotype term was used. The truncation point was set as τ = 0.10, and the P value for truncated product method was estimated through simulation (10,000 replications). Gene-based analysis was performed using R software (Version 3.0.1; http://www.r-project.org). Furthermore, using publicly available data from the NCBI Database of Genotypes and Phenotypes (dbGaP), attempts were made to replicate significant SNPs and genes among 775 Chinese-Americans who took part in the longitudinal Multi-Ethnic Study of Atherosclerosis (MESA) (dbGaP accession phs000209v11.p3.c2 and phs00420v4.p3.c2).26,27 False discovery rate procedures were used to adjust for multiple testing in all analyses.
RESULTS
Table 1 shows the baseline characteristics of the 1,768 GenSalt participants who were eligible for analysis. At baseline, the average participant had an age of 39.0 years, a body mass index of 23.4kg/m2, a systolic BP of 116.9mm Hg, and a diastolic BP of 73.8mm Hg. Of the 1,768 participants, 924 (52.3%) were male, 173 (9.8%) had hypertension, and 159 (9.0%) were taking antihypertensive medications. After excluding the individuals with hypertension at baseline, 512 (32.1%) participants developed hypertension during an average of 7.2 years of follow-up.
Table 1.
Baseline characteristics of GenSalt follow-up study participants (N = 1,768)
| Variable | Mean ± SD or % |
|---|---|
| Age, years | 39.0±9.2 |
| Male | 52.3 |
| Body mass index, kg/m2 | 23.4±3.2 |
| Systolic blood pressure, mm Hg | 116.9±14.1 |
| Diastolic blood pressure, mm Hg | 73.8±10.2 |
| Hypertension | 9.8 |
| Antihypertension medication | 9.0 |
| Incidence of hypertension during follow-upa | 32.1 |
aExcludes 173 participants with hypertension at baseline.
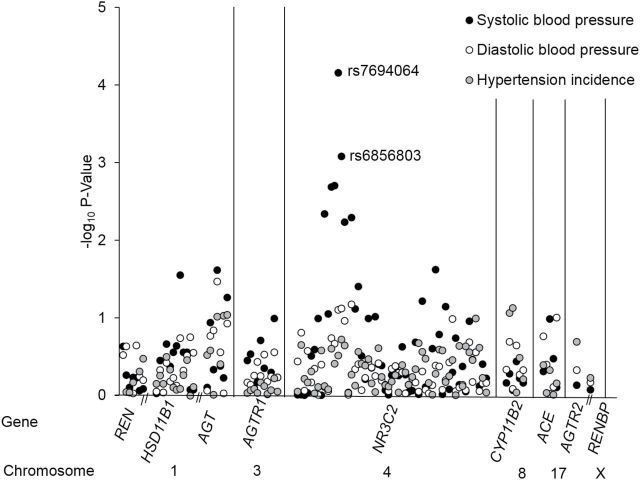
Figure 1 displays the association of 106 SNPs in 10 RAAS genes with BP changes and hypertension incidence. NR3C2 SNPs rs7694064 and rs6856803 were both significantly associated with longitudinal changes in systolic BP after correcting for multiple testing (P interaction = 6.9×10−5 and 8.2×10−4 respectively) (Table 2). However, these findings could not be replicated among a smaller sample of 775 Asian participants of the MESA study (P = 0.3 and 0.5 for rs7684064 and rs6856803, respectively). No SNPs had significant associations with longitudinal changes in diastolic BP or hypertension incidence after adjustment for multiple testing.
Figure 1.
Log10 P values for the association of 106 single nucleotide polymorphisms (SNPs) in REN, HSD11B1, AGT, AGTR1, NR3C2, CYP11B2, ACE, AGTR2, and RENBP with longitudinal changes in systolic and diastolic blood pressure, as well as hypertension incidence. The black and white circles show P values for the testing of genotype by follow-up time interactions for systolic blood pressure and diastolic blood pressure, respectively, while the gray circles show P values for the testing of the effect of SNPs on hypertension incidence. Two labeled SNPs were significantly associated with longitudinal changes in systolic BP after using false discovery rate procedures for multiple testing.
Table 2.
Association of significant RAAS SNPs with BP changes and hypertension incidence among GenSalt study participants
| HGNC gene symbol | SNP | Chr. | Position | Genotype | Systolic BP change | Diastolic BP change | HTN incidence | |||
|---|---|---|---|---|---|---|---|---|---|---|
| β (SE) | P valuea | β (SE) | P valuea | OR (95% CI) | P valueb | |||||
| NR3C2 | rs7694064 | 4 | 149302668 | A/A (195) | 2.26 (0.16) | 6.9×10–5c | 1.33 (0.11) | 0.08 | 1.33 (0.93, 1.89) | 0.30 |
| G/A (828) | 1.76 (0.07) | 1.23 (0.05) | 1.00 (0.78, 1.28) | |||||||
| G/G (856) | 1.56 (0.07) | 1.15 (0.05) | Ref. | |||||||
| rs6856803 | 4 | 149306577 | C/C (415) | 1.51 (0.10) | 8.2×10–4c | 1.08 (0.07) | 0.08 | 0.83 (0.60, 1.14) | 0.19 | |
| T/C (918) | 1.66 (0.07) | 1.21 (0.05) | 0.75 (0.57, 0.99) | |||||||
| T/T (548) | 1.97 (0.10) | 1.28 (0.07) | Ref. | |||||||
Abbreviations: BP, blood pressure; Chr. = chromosome; HGNC = HUGO gene nomenclature committee; HTN = hypertension; OR = odds ratio; RAAS = renin–angiotensin–aldosterone system; SNP = single nucleotide polymorphism.
a P value for genotype × time interaction term.
b P value for additive genetic association.
cSignificant after adjustment for multiple testing.
Systolic and diastolic BP changes as well as odds ratios for hypertension incidence according to genotype for the 2 significant NR3C2 SNPs are shown in Table 2. For rs7694064, the minor A allele was associated with dose-dependent increases in longitudinal systolic BP, while the minor C allele of rs6856803 was associated with dose-dependent decreases in systolic BP over time. Similar but nonsignificant trends were noted for diastolic BP changes and odds ratios for hypertension incidence.
Table 3 presents the results of gene-based analyses. Two genes, AGTR2 and RENBP, could not be examined since genotype data were available on only one SNP within each of these genes (Supplementary Table). Among the remaining genes, NR3C2 was found to be significantly associated with longitudinal systolic BP change (P value of 1.00×10−7). NR3C2 was still significant after adjustment for multiple testing and the simultaneous removal of significant SNPs (rs7694064 and rs6856803) from gene-based analyses. Furthermore, the significant association between NR3C2 and systolic BP change was replicated in MESA (P value of 1.00×10−4). No genes were significantly associated with longitudinal diastolic BP changes or hypertension incidence.
Table 3.
Associations of 8 RAAS genes containing at least 2 SNPs with longitudinal BP phenotypes in the GenSalt study
| HGNC symbol | k | Systolic BP change | Diastolic BP change | HTN incidence |
|---|---|---|---|---|
| REN | 7 | 0.31 | 0.46 | 0.36 |
| HSD11B1 | 12 | 0.48 | 0.47 | 0.79 |
| AGT | 7 | 0.07 | 0.28 | 0.01 |
| AGTR1 | 10 | 0.53 | 0.25 | 0.42 |
| NR3C2 | 57 | 1.00×10−7a,b,c | 0.61 | 1.00 |
| CYP11B1 | 3 | 0.18 | 0.25 | 0.05 |
| CYP11B2 | 3 | 0.17 | 0.17 | 0.26 |
| ACE | 5 | 0.31 | 0.45 | 0.46 |
Abbreviations: BP = blood pressure; HGNC = HUGO gene nomenclature committee; HTN = hypertension; k = number of SNPs included in gene-based analyses; RAAS = renin–angiotensin–aldosterone system; SNP = single nucleotide polymorphism.
aSignificant after adjustment for multiple testing.
bSignificant after simultaneous removal of significant SNPs from gene-based analyses.
cEvidence of replication in the Multi-ethnic Study of Atherosclerosis (P = 1.00×10−4 for NR3C2 – systolic BP change association).
DISCUSSION
This is the first study to investigate the association between RAAS genes with longitudinal BP changes and hypertension incidence in a Han Chinese population. Two variants of the NR3C2 gene, rs7694064 and rs6856803, were found to be significantly associated with systolic BP changes over an extended period of time. The minor allele of rs7694064 was related to an increased systolic BP change over time compared to its more frequent counterpart, while the minor allele for rs6856803 was significantly associated with a decrease in longitudinal systolic BP change compared to its more frequent counterpart. Consistent with these findings, gene-based analyses also revealed an association of the NR3C2 gene with systolic BP changes over time. In aggregate, these findings contribute strong genomic evidence for a role of the mineralocorticoid receptor, encoded by NR3C2, in BP regulation.
Similar to the current genomic findings, physiological studies have long demonstrated an influence of the mineralocorticoid receptor on BP. Its effect on BP is achieved through the control of renal salt handling. Specifically, the interaction of aldosterone with the mineralocorticoid receptor in renal tubular cells increases activity of the epithelial sodium channel, leading to net salt and water reabsorption. The increased salt and water retention expands extracellular fluid volume and increases cardiac output, raising BP.28,29 Studies of monogenic disorders have further confirmed the physiological relevance of the mineralocorticoid receptor on BP. An NR3C2 gain-of-function mutation results in an early onset form of severe salt-sensitive hypertension that is exacerbated by pregnancy,30 while a loss-of-function mutation to the NR3C2 gene is responsible for a severe salt-wasting hypotension disorder known as pseudohypoaldosteronism, type I.31
Of the 106 SNPs analyzed in the present study, 2 SNPs in NR3C2 (rs7694064 and rs6856803) were found to be significantly associated with systolic BP change over time. Both rs7694064 and rs6856803 are located in intronic regions of the NR3C2, unlikely representing functional variants but rather the synthetic associations of unobserved causal variants. While these single-marker findings are promising, their associations with longitudinal systolic BP could not be replicated in MESA. However, given MESA’s relatively small sample of Asian participants, such an analysis was likely underpowered to detect the modest effects of these variants. In contrast, others have identified SNPs in NR3C2 that influence BP phenotypes.10,31,32 Of particular interest, NR3C2 rs6810951, which is in strong linkage disequilibrium with rs6856803 (r 2 = 0.906), was found to be significantly associated with both systolic BP and diastolic BP in a population of 7551 individuals among the Ansung and Ansan communities in South Korea.10 Similar to the effect directions noted in the present study, Song and colleagues demonstrated higher systolic BP and diastolic BP associated with the major compared to the minor allele.10 In populations of European ancestry, NR3C2 rs5522 was significantly associated with risk of hypertension among 1,502 Spanish adults32 while NR3C2 rs2070951 was associated with systolic BP among 1,754 Dutch men.33 Although the present study did not replicate rs5522 (potentially due to differences in linkage disequilibrium structure) and genotype data were not available for rs2070951, these findings support a role of the NR3C2 gene in BP regulation.
In addition to the association of single NR3C2 markers with the longitudinal systolic BP phenotype, NR3C2 aggregately associated with longitudinal systolic BP change in gene-based analysis. These findings were significant even after removal of rs7694064 and rs6856803, indicating that gene-based analyses were not solely driven by the observed single-marker associations. This suggests that there may be additional variants in NR3C2 influencing systolic BP change that were not discovered in this study. Such findings highlight the potential relevance of gene-based analysis for increasing statistical power to detect meaningful genomic mechanisms underlying BP progression. Furthermore, with strong evidence for replication among MESA participants, these results implicate NR3C2 as an important factor in BP progression.
The current study has several strengths. It is one of few studies to examine the associations between RAAS gene variants and longitudinal BP phenotypes and the first to conduct gene-based analyses in a cohort study. Study attributes, such as the recruitment of all Han Chinese participants, should make the analysis robust to population stratification.34 Also, the study had a very high follow-up rate (92.3%). Stringent quality control procedures were employed for genotyping and data collection. The ability to detect association was further enhanced by using the average of 9 separate BP measures that were collected at baseline and each follow-up examination, which should have greatly reduced measurement error. In addition, false discovery rate procedures were performed in order to account for multiple testing. However, this study also does have its limitations. The single-marker findings from this paper need to be replicated in a larger study population before any definitive conclusions can be made. While the genotyping platform used generally provides good coverage of common SNPs across the genome,35 some RAAS SNPs may not be tagged by this array. Any associations of such SNPs with longitudinal BP phenotypes would be missed by the current study. Less frequent genetic variants may have also been left out of the current study. Furthermore, due to the limited number of SNPS in AGTR2 and RENBP, gene-based analyses could not be conducted for these RAAS genes. Future research will be needed to explore their gene-based associations with longitudinal BP phenotypes.
Based on single-marker and gene-based analyses, the present study provides strong evidence for a role of the NR3C2 gene in BP progression in a Han Chinese population. Although single-marker results still require confirmation by an independent sample, the gene-based findings were successfully replicated among 775 Asian participants of the MESA study. Findings from the current study emphasize the potential for using longitudinal BP measures and gene-based analyses to help elucidate the biological pathways underlying hypertension susceptibility. Furthermore, this works contributes to the cumulative understanding of the genomic mechanisms that regulate BP.
SUPPLEMENTARY MATERIAL
Supplementary material is available at American Journal of Hypertension (http://ajh.oxfordjournals.org).
DISCLOSURE
There is no conflict of interest.
Supplementary Material
ACKNOWLEDGMENTS
The Genetic Epidemiology Network of Salt Sensitivity is supported by research grants (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD. The Multi-Ethnic Study of Atherosclerosis (MESA) and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC- 95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, and CTSA UL1-RR-024156. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, CA) and the Broad Institute of Harvard and MIT (Boston, MA) using the Affymetrix Genome-Wide Human SNP Array 6.0.
REFERENCES
- 1. Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: analysis of worldwide data. Lancet 2005; 365:217–223. [DOI] [PubMed] [Google Scholar]
- 2. Lawes CM, Vander Hoorn S, Rodgers A. Global burden of blood-pressure-related disease, 2001. Lancet 2008; 371:1513–1518. [DOI] [PubMed] [Google Scholar]
- 3. Bromfield S, Muntner P. High blood pressure: the leading global burden of disease risk factor and the need for worldwide prevention programs. Curr Hypertens Rep 2013; 15:134–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ehret GB. Genome-wide association studies: contribution of genomics to understanding blood pressure and essential hypertension. Curr Hypertens Rep 2010; 12:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ames RP. The role of the renin–angiotensin–aldosterone system in blood pressure regulation. Am J Hypertens 2002; 15(7 Pt 1):653–654. [DOI] [PubMed] [Google Scholar]
- 6. Weir MR, Dzau VJ. The renin-angiotensin-aldosterone system: a specific target for hypertension management. Am J Hypertens 1999; 12:205S–213S. [DOI] [PubMed] [Google Scholar]
- 7. Paillard F, Chansel D, Brand E, Benetos A, Thomas F, Czekalski S, Ardaillou R, Soubrier F. Genotype-phenotype relationships for the renin-angiotensin-aldosterone system in a normal population. Hypertension 1999; 34:423–429. [DOI] [PubMed] [Google Scholar]
- 8. Lynch AI, Arnett DK, Pankow JS, Miller MB, North KE, Eckfeldt JH, Hunt SC, Rao DC, Djoussé L. Sex-specific effects of ACE I/D and AGT-M235T on pulse pressure: the HyperGEN Study. Hum Genet 2007; 122:33–40. [DOI] [PubMed] [Google Scholar]
- 9. Siani A, Russo P, Paolo Cappuccio F, Iacone R, Venezia A, Russo O, Barba G, Iacoviello L, Strazzullo P. Combination of renin-angiotensin system polymorphisms is associated with altered renal sodium handling and hypertension. Hypertension 2004; 43:598–602. [DOI] [PubMed] [Google Scholar]
- 10. Song SB, Jin HS, Hong KW, Lim JE, Moon JY, Jeong KH, Ihm CG, Lee TW, Oh B, Lee SH. Association between renin-angiotensin-aldosterone system-related genes and blood pressure in a Korean population. Blood Press 2011; 20:204–210. [DOI] [PubMed] [Google Scholar]
- 11. Zhu X, Chang YP, Yan D, Weder A, Cooper R, Luke A, Kan D, Chakravarti A. Associations between hypertension and genes in the renin-angiotensin system. Hypertension 2003; 41:1027–1034. [DOI] [PubMed] [Google Scholar]
- 12. Li X, Ling Y, Lu D, Lu Z, Liu Y, Chen H, Gao X. Common polymorphism rs11191548 near the CYP17A1 gene is associated with hypertension and systolic blood pressure in the Han Chinese population. Am J Hypertens 2013; 26:465–472. [DOI] [PubMed] [Google Scholar]
- 13. Kelly TN, He J. Genomic epidemiology of blood pressure salt sensitivity. J Hypertens 2012; 30:861–873. [DOI] [PubMed] [Google Scholar]
- 14. Poch E, González D, Giner V, Bragulat E, Coca A, de La Sierra A. Molecular basis of salt sensitivity in human hypertension. Evaluation of renin-angiotensin-aldosterone system gene polymorphisms. Hypertension 2001; 38:1204–1209. [DOI] [PubMed] [Google Scholar]
- 15. Smith EN, Chen W, Kahonen M, Kettunen J, Lehtimaki T, Peltonen L, Raitakari OT, Salem RM, Schork NJ, Shaw M, Srinivasan SR, Topol EJ, Viikari JS, Berenson GS, Murray SS. Longitudinal genome-wide association of cardiovascular disease risk factors in the Bogalusa Heart Study. PLOS Genet 2010; 6: e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ma L, Clark AG, Keinan A. Gene-based testing of interactions in association studies of quantitative traits. PLoS Genet 2013; 9:e1003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He J, Kelly TN, Zhao Q, Li H, Huang J, Wang L, Jaquish CE, Sung YJ, Shimmin LC, Lu F, Mu J, Hu D, Ji X, Shen C, Guo D, Ma J, Wang R, Shen J, Li S, Chen J, Mei H, Chen CS, Chen S, Chen J, Li J, Cao J, Su X, Wu X, Rice TK, Gu CC, Schwander K, Hamm LL, Liu D, Rao DC, Hixson JE, Gu D. Genome-wide association study identifies 8 novel loci associated with blood pressure responses to interventions in Han Chinese. Circ Cardiovasc Genet 2013; 6:598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. GenSalt Collaborative Research Group. GenSalt: rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens 2007; 21:639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet 2005; 37:1217–1223. [DOI] [PubMed] [Google Scholar]
- 20. Shi G, Rice TK, Gu CC, Rao DC. Application of three-level linear mixed-effects model incorporating gene-age interactions for association analysis of longitudinal family data. BMC Proc 2009; 3(Suppl 7):S89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singer JD. Using SAS PROC MIXED to fit multilevel models, hierarchical models, and individual growth models. J Educ Behav Stat 1998; 23:323–355. [Google Scholar]
- 22. Law MR, Morris JK, Wald NJ. Use of blood pressure lowering drugs in the prevention of cardiovascular disease: meta-analysis of 147 randomised trials in the context of expectations from prospective epidemiological studies. BMJ 2009; 338:b1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schabenberger O. Introducing the GLIMMIX procedure for generalized linear mixed models. SUGI 2008; 30:196. [Google Scholar]
- 24. Zaykin DV, Zhivotovsky LA, Westfall PH, Weir BS. Truncated product method for combining P-values. Genet Epidemiol 2002; 22:170–185. [DOI] [PubMed] [Google Scholar]
- 25. Sheng X, Yang J. Panel unit root test by combining dependent p-values: a comparative study. J Prob Stat 2011; 2011:1–17. [Google Scholar]
- 26. Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Khodol M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet 2007; 39:1181–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tryka KA, Hao L, Sturcke A, Jin Y, Wang ZY, Ziyabari L, Lee M, Popova N, Sharopova N, Kimura M, Feolo M. NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res 2014; 42:D975–D979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fuller PJ, Young MJ. Mechanisms of mineralocorticoid action. Hypertension 2005; 46:1227–1235. [DOI] [PubMed] [Google Scholar]
- 29. Lifton RP. Molecular genetics of human blood pressure variation. Science 1996; 272:676–680. [DOI] [PubMed] [Google Scholar]
- 30. Geller DS, Farhi A, Pinkerton N, Fradley M, Moritz M, Spitzer A, Meinke G, Tsai FT, Sigler PB, Lifton RP. Activating mineralocorticoid receptor mutation in hypertension exacerbated by pregnancy. Science 2000; 289:119–123. [DOI] [PubMed] [Google Scholar]
- 31. Geller DS, Rodriguez-Soriano J, Vallo Boado A, Schifter A, Bayer S, Chang M, Lifton RP . Mutations in the mineralocorticoid receptor gene cause autosomal dominant pseudohypoaldosteronism, type I. Nat Genet 1998; 19:279–281. [DOI] [PubMed] [Google Scholar]
- 32. Martinez F, Mansego ML, Escudero JC, Redon J, Chaves FJ. Association of a mineralocorticoid receptor gene polymorphism with hypertension in a Spanish population. Am J Hypertens 2009; 22:649–655. [DOI] [PubMed] [Google Scholar]
- 33. van Leeuwen N, Caprio M, Blaya C, Fumeron F, Sartorato P, Ronconi V, Giacchetti G, Mantero F, Fernandes-Rosa FL, Simian C, Peyrard S, Zitman FG, Penninx BW, de Kloet ER, Azizi M, Jeunemaitre X, Derijk RH, Zennaro MC. The functional c.-2G>C variant of the mineralocorticoid receptor modulates blood pressure, renin, and aldosterone levels. Hypertension 2010; 56:995–1002. [DOI] [PubMed] [Google Scholar]
- 34. Gu D, Kelly TN, Hixson JE, Chen J, Liu D, Chen JC, Rao DC, Mu J, Ma J, Jaquish CE, Rice TK, Gu C, Hamm LL, Whelton PK, He J. Genetic variants in the renin-angiotensin-aldosterone system and salt sensitivity of blood pressure. J Hypertens 2010; 28:1210–1220. [PMC free article] [PubMed] [Google Scholar]
- 35. Nishida N, Koike A, Tajima A, Ogasawara Y, Ishibashi Y, Uehara Y, Inoue I, Tokunaga K. Evaluating the performance of Affymetrix SNP Array 6.0 platform with 400 Japanese individuals. BMC Genomics 2008; 9:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.