Summary
Nucleosome positioning varies between cell types. By deep sequencing cell-free DNA (cfDNA) isolated from circulating blood plasma, we generated maps of genome-wide in vivo nucleosome occupancy, and found that short cfDNA fragments harbor footprints of transcription factors. The cfDNA nucleosome occupancies correlate well with the nuclear architecture, gene structure and expression observed in cells, suggesting that they could indicate the cell-type of origin. Nucleosome spacing inferred from cfDNA in healthy individuals correlates most strongly with epigenetic features of lymphoid and myeloid cells, consistent with hematopoietic cell death as the normal source of cfDNA. We build on this observation to show how nucleosome footprints can be used to infer cell types contributing to cfDNA in pathological states such as cancer. Since this strategy does not rely on differing cfDNA and cellular DNA genotypes, it may enable the noninvasive monitoring of a much broader set of clinical conditions than currently possible.
Introduction
Cell-free DNA (cfDNA) is present in the circulating plasma, urine, and other bodily fluids of humans (Chan et al., 2003). The cfDNA comprises double-stranded DNA fragments that are overwhelmingly short (<200 base-pairs (bp)) and normally at a low concentration (Fleischhacker and Schmidt, 2007). In healthy individuals, plasma cfDNA is believed to derive primarily from apoptosis of normal cells of the hematopoietic lineage, with minimal contributions from other tissues (Lui et al., 2002). The short half-life of cfDNA in the circulation (Lo et al., 1999) suggests a model of ongoing release from apoptotic cells and rapid degradation or filtration. The size distribution of cfDNA fragments bears correspondence with these origins – specifically, peaks corresponding to nucleosomes (~147 bp) and chromatosomes (nucleosome + linker histone; ~167 bp) have been noted (Fan et al., 2008; Lo et al., 2010) – and some proportion of cfDNA may circulate as nucleosomes or chromatosomes, rather than as free DNA (Holdenrieder et al., 2005; Wimberger et al., 2010).
In the context of specific physiological conditions or disease processes, a substantial proportion of cfDNA may be derived from a different distribution of tissues than during the typical, healthy state. This fact has been exploited in recent years to achieve noninvasive diagnostics based on cfDNA composition. In pregnant women, ~10–15% of cfDNA originates from placental trophoblasts, and cfDNA-based screening for fetal genetic abnormalities is now common in high-risk pregnancies (Chiu et al., 2008; Fan et al., 2008). In oncology, the monitoring of advanced cancers by quantifying mutations or aneuploidy in tumor-shed cfDNA is gaining traction (Diaz and Bardelli, 2014). In transplant medicine, allograft rejection events can be correlated with abnormally high levels of donor-derived cfDNA fragments contributed by the transplanted solid organ (Snyder et al., 2011).
Despite these advances, a common limitation is the requirement for genetic differences to distinguish between contributing tissues, e.g. fetus vs. mother, tumor vs. normal, or donor vs. recipient. Conditions such as myocardial infarction (Chang et al., 2003), stroke (Rainer et al., 2003) and autoimmune disorders (Galeazzi et al., 2003) are associated with elevations in cfDNA levels, possibly consequent to tissue damage, but cannot be specifically monitored via cfDNA because of the lack of such genetic differences. Furthermore, even as mutations enable monitoring of tumor-derived cfDNA, they only weakly inform a tumor’s tissue-of-origin.
We hypothesized that if cfDNA is the detritus of cell death, and if the boundaries of cfDNA fragments are biased by their association with nucleosomes, then the fragmentation patterns observed in an individual’s cfDNA might contain evidence of the epigenetic landscape(s) of the cells giving rise to these fragments – and thus, of their tissue(s)-of-origin – i.e., a strategy that does not rely on genotypic differences between contributing cell types.
To evaluate this hypothesis, we first set out to deeply sequence cfDNA to better understand the processes that give rise to it. We use the resulting data to build a map of nucleosome occupancy that approaches saturation of the mappable human genome. By optimizing protocols to recover short fragments, we discover that the in vivo occupancies of transcription factors (TFs) such as CTCF are also directly footprinted by cfDNA. Finally, we show that nucleosome spacing in regulatory elements and gene bodies, as revealed by cfDNA sequencing in healthy individuals, correlates most strongly with DNase I hypersensitivity (DHS) and gene expression in lymphoid and myeloid cell lines. To test whether we can infer additional contributing tissues in non-healthy states, we sequenced cfDNA samples from five late-stage cancer patients. The patterns of nucleosome spacing in these samples reveal additional contributions to cfDNA that correlate most strongly with non-hematopoietic tissues or cell lines, often matching the anatomical origin of the patient’s cancer.
Results
cfDNA fragments correspond to chromatosomes and contain substantial DNA damage
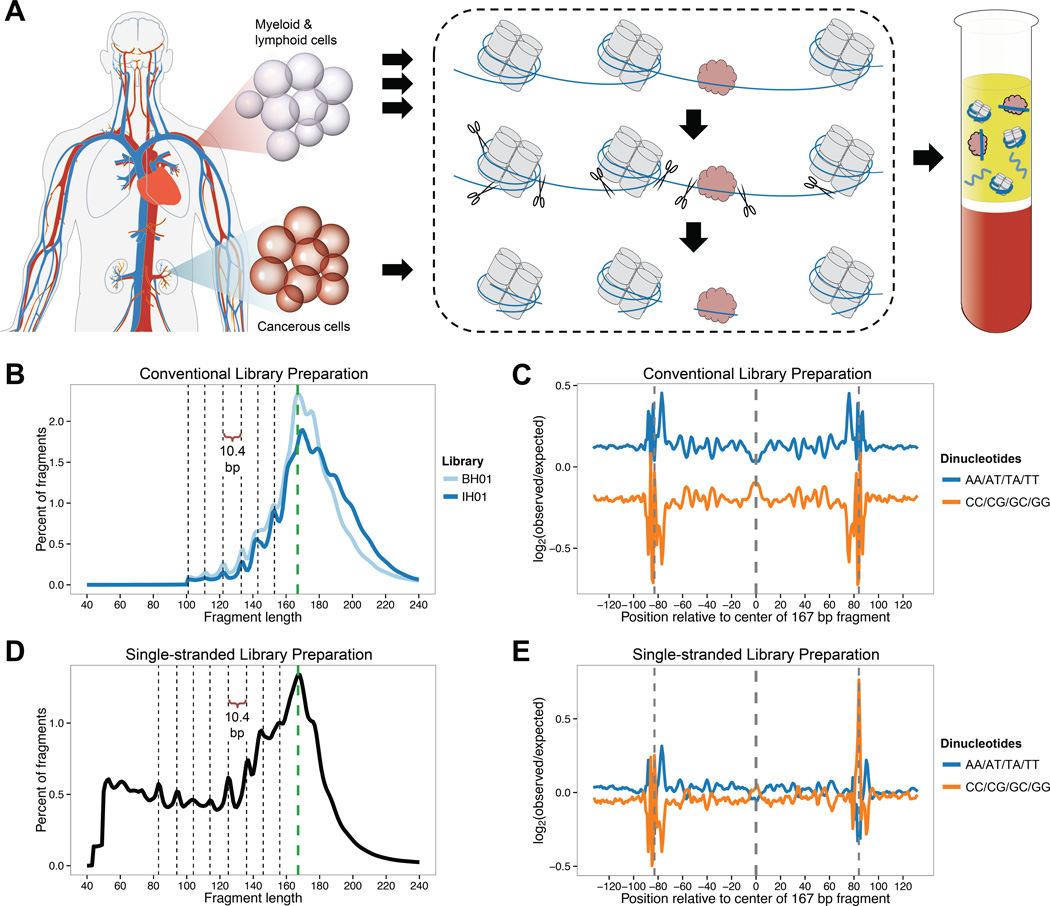
We prepared conventional sequencing libraries by end-repair and adaptor ligation to cfDNA fragments purified from plasma pooled from an unknown number of healthy individuals (‘BH01’) or a single individual (‘IH01’) (Figure 1A; Table S1). We sequenced these libraries to 96- and 105-fold coverage (1.5 billion (G) and 1.6G fragments). The fragment length distributions have a dominant peak at ~167 bp (coincident with the length of DNA associated with a chromatosome), and ~10.4 bp periodicity in the 100–160 bp range (Figure 1B). These distributions support a model in which cfDNA fragments are preferentially protected from nuclease cleavage by association with proteins – in this case, by the nucleosome core particle (NCP) and linker histone – but where some degree of additional nicking or cleavage occurs in relation to the helical pitch of nucleosome-bound DNA (Fan et al., 2008; Lo et al., 2010). Further supporting this model is the dinucleotide composition of these fragments, which recapitulates key features of earlier studies of MNase-derived, nucleosome-associated fragments (e.g. bias against A/T dinucleotides at the dyad) (Gaffney et al., 2012) and supports the notion that the NCP is symmetrically positioned with respect to the chromatosome (Harshman et al., 2013) (Figure 1C).
Figure 1. Origins and characteristics of cfDNA fragments in human plasma.
(A) Schematic overview of cfDNA fragmentation. Apoptotic or necrotic cell death results in near-complete digestion of native chromatin. Protein-bound DNA fragments, typically associated with histones or TFs, preferentially survive digestion and are released into the circulation, while naked DNA is lost. Fragments can be recovered from peripheral blood plasma following proteinase treatment. In healthy individuals, cfDNA is primarily derived from myeloid and lymphoid cell lineages, but contributions from one or more additional tissues may be present in certain medical conditions. (B) Fragment length of cfDNA observed with conventional sequencing library preparation, inferred from alignment of paired-end reads. A reproducible peak in fragment length at 167 bp (green dashed line) is consistent with association with chromatosomes. Additional peaks evidence ~10.4 bp periodicity, corresponding to the helical pitch of DNA on the nucleosome core. Enzymatic end-repair during library preparation removes 5’ and 3’ overhangs and may obscure true cleavage sites. (C) Dinucleotide composition of 167 bp fragments and flanking genomic sequence in conventional libraries. Observed dinucleotide frequencies in the BH01 library were compared to expected frequencies from simulated fragments. (D) Fragment length of cfDNA in single-stranded sequencing library preparation. No enzymatic end-repair is performed to template molecules during library preparation. Short fragments of 50–120 bp are highly enriched compared to conventional libraries. While ~10.4 bp periodicity remains, its phase is shifted by ~3 bp. (E) Dinucleotide composition of 167 bp fragments and flanking genomic sequence in single-stranded library IH02, calculated as in (C). The apparent difference in the background level of bias between BH01 and IH02 relate to differences between the simulations, rather than the real libraries (data not shown). See also Figure S1 and Table S1.
A prediction of this model is widespread DNA damage, e.g. single-strand nicks as well as 5’ and 3’ overhangs. During conventional library preparation, damaged as well as short dsDNA molecules (Mouliere et al., 2014) may be poorly recovered. To address this, we prepared a single-stranded cfDNA library from an additional healthy individual (‘IH02’) using a protocol adapted from studies of ancient DNA (Figure S1A; Table S2) (Gansauge and Meyer, 2013), and sequenced it to 30-fold coverage (779M fragments). The fragment length distribution again exhibited a dominant peak at ~167 bp, but was considerably enriched for shorter fragments relative to conventional library preparation (Figures 1D, 1E, and S1B–D). Although all libraries exhibit ~10.4 bp periodicity, the fragment sizes are offset by 3 bp for the two methods, consistent with damaged or non-flush input molecules whose true endpoints are more faithfully represented in single-stranded libraries.
A genome-wide map of in vivo nucleosome protection based on deep cfDNA sequencing
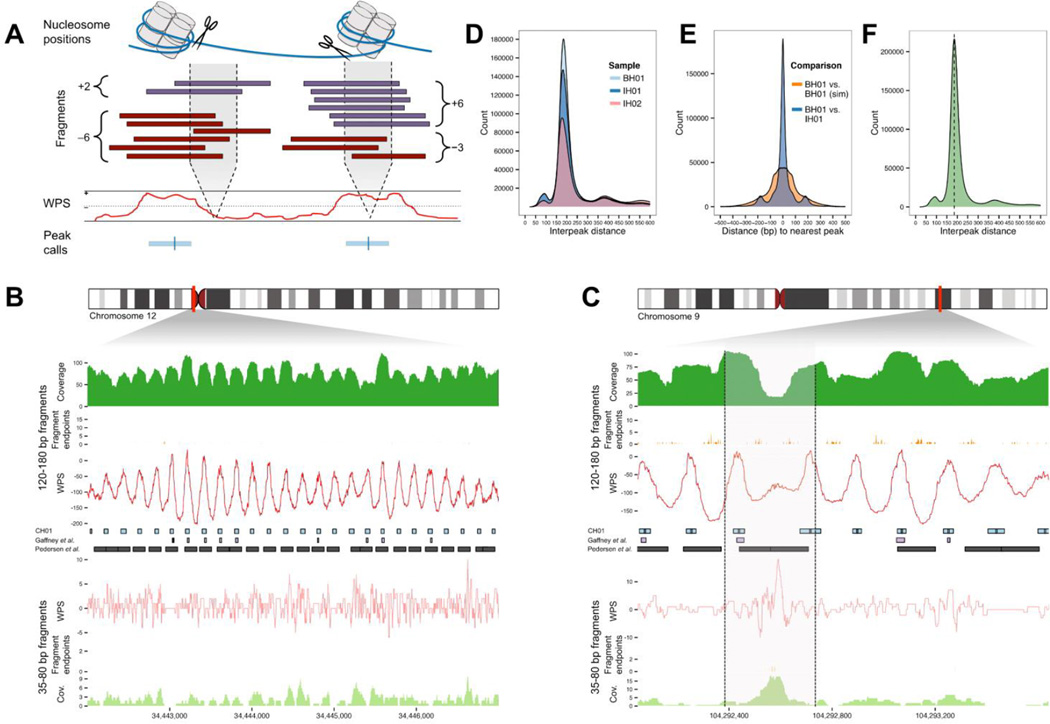
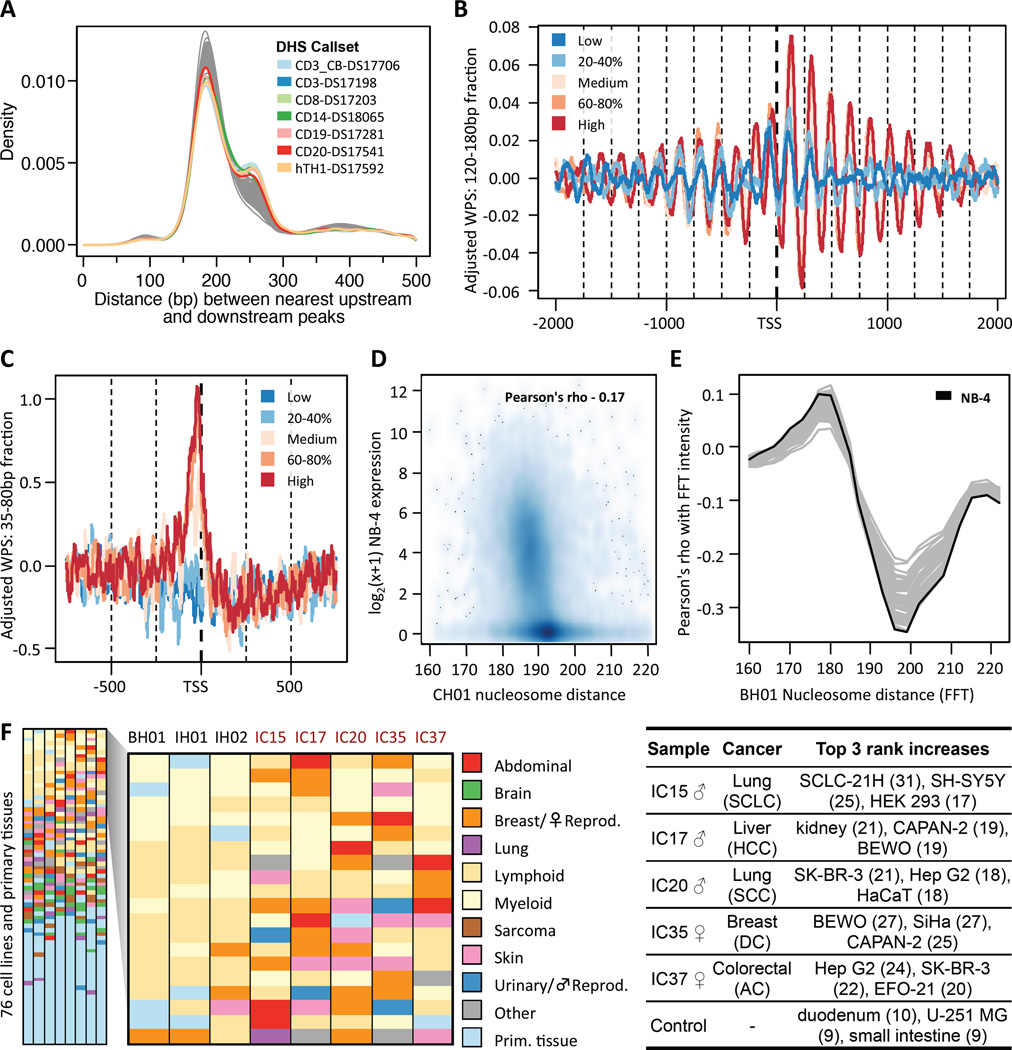
We next asked whether the predominant local positions of nucleosomes in tissue(s) contributing to cfDNA could be inferred from the distribution of aligned fragment endpoints. Specifically, we expect that cfDNA fragment endpoints should cluster adjacent to NCP boundaries, while also being depleted on the NCP itself. To quantify this, we developed a Windowed Protection Score (WPS), which is the number of DNA fragments completely spanning a 120 bp window centered at a given genomic coordinate, minus the number of fragments with an endpoint within that same window (Figure 2A). As expected, the WPS correlates with the locations of nucleosomes within strongly positioned arrays, as mapped by other groups with in vitro methods (Gaffney et al., 2012; Valouev et al., 2012) or ancient DNA (Pedersen et al., 2014) (Figure 2B). At other sites, the WPS correlates with genomic features such as DHS sites, e.g. consistent with the repositioning of nucleosomes flanking a distal regulatory element (Figure 2C).
Figure 2. Genome-wide determination of nucleosome positions from cfDNA fragmentation patterns.
(A) Schematic of inference of nucleosome positioning. A per-base windowed protection score (WPS) is calculated by subtracting the number of fragment endpoints within a 120 bp window from the number of fragments completely spanning the window. High WPS values indicate increased protection of DNA from digestion; low values indicate that DNA is unprotected. Peak calls identify contiguous regions of elevated WPS. (B) Strongly positioned nucleosomes at a well-studied alpha-satellite array. Coverage, fragment endpoints, and WPS values from sample CH01 are shown for long fragment (120 bp window; 120–180 bp fragments) or short fragment (16 bp window; 35–80 bp fragments) bins at a pericentromeric locus on chromosome 12. Nucleosome calls from CH01 (middle, blue boxes) are regularly spaced across the locus. Nucleosome calls from two published callsets (Gaffney et al., 2012; Pedersen et al., 2014) (middle, purple and black boxes) are also displayed. (C) Inferred nucleosome positioning around a DHS site. Coverage, fragment endpoints, WPS values, and nucleosome calls are shown as in (B). The hypersensitive region (gray shading), is marked by reduced coverage in the long fragment bin. Nucleosome calls adjacent to the DHS site are spaced more widely than typical adjacent pairs, consistent with accessibility of the intervening sequence to regulatory proteins including TFs. Coverage of short fragments, which may be associated with such proteins, is increased at the DHS site, which overlaps with several annotated TFBSs (not shown). (D) Distances between adjacent peaks by sample. Distances are measured between adjacent peak centers. (E) Comparison of peak calls between samples. For each pair of samples, the distances between each peak call in the sample with fewer peaks and the nearest peak call in the other sample are shown. Negative and positive numbers indicate the nearest peak is upstream or downstream, respectively. (F) Distances between adjacent peaks, sample CH01. The dotted black line indicates the mode of the distribution (185 bp). See also Figure S2.
We applied a heuristic peak-calling algorithm to the genome-wide WPS of the BH01, IH01 and IH02 datasets to identify and score 12.6M, 11.9M, and 9.7M local maxima of nucleosome protection (Figures 2 and S2A). In each sample, the mode distance between adjacent peaks is 185 bp with low variance (Figure 2D), consistent with previous analyses of the nucleosome repeat length in mammalian cells (Teif et al., 2012; Valouev et al., 2012). The positions of peak calls are concordant between samples (Figures 2E and S2B), e.g. the median (absolute) distance from a BH01 peak call to a nearest-neighbor IH01 peak call is 23 bp overall, but <10 bp for the most highly scored peaks (Figure S2C). Furthermore, the density of peak calls is constant across the spectrum of GC content (Figure S2D).
As biases introduced by nuclease specificity or library preparation might artifactually contribute to the signal of nucleosome protection, we also simulated fragment endpoints, matching for the depth, size distribution and terminal dinucleotide frequencies (Figures S1C–D). We then calculated genome-wide WPS and called 10.3M, 10.2M, and 8.0M local maxima by the same heuristic, for simulated datasets matched to BH01, IH01 and IH02, respecitvely. Peaks from simulated datasets are associated with lower scores than peaks from real datasets (Figure S2E) and do not align well with the locations of peaks called from real datasets (Figures 2E and S2F).
We next pooled and reanalyzed data from BH01, IH01, and IH02 (‘CH01’; 231-fold coverage; 3.8G fragments; Table S1). The resulting map of in vivo nucleosome occupancy comprises 12.9M peaks, with higher scores and approaching saturation (Figure S2E). Considering all peak-to-peak distances below 500 bp (Figure 2F), the CH01 peaks span 2.53 gigabases.
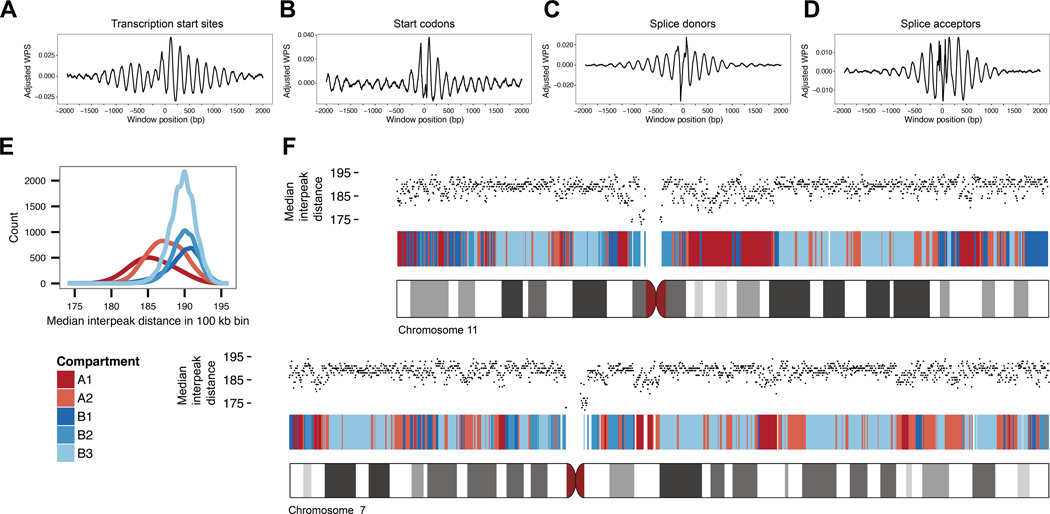
Nucleosomes are known to be well-positioned in relation to landmarks of gene regulation, e.g. transcriptional start sites (Pedersen et al., 2014) and exon-intron boundaries (Andersson et al., 2009; Chodavarapu et al., 2010). We observe such positioning in our data as well, in relation to landmarks of transcription, translation and splicing (Figures 3A–D and S3). We further examined the median peak-to-peak spacing within 100 kilobase (kb) windows that had been assigned to compartment A (enriched for open chromatin) or compartment B (enriched for closed chromatin) on the basis of chromatin contact maps in a lymphoblastoid cell line (Rao et al., 2014). Nucleosomes in compartment A exhibit tighter spacing than nucleosomes in compartment B (median 187 bp (A) vs. 190 bp (B)), with further differences between subcompartments (Figure 3E). Along the length of chromosomes, no general pattern is seen, except that median nucleosome spacing drops sharply in pericentromeric regions, presumably driven by strong positioning across arrays of alpha satellites (Figures 2B and 3F).
Figure 3. Nucleosome positioning and spacing correlates with genomic features.
(A) Aggregate, adjusted windowed protection scores (WPS; 120 bp window) around 22,626 transcription start sites (TSS). TSS are aligned at the 0 position after adjusting for strand and direction of transcription. Aggregate WPS is tabulated for both real data and simulated data by summing per-TSS WPS at each position relative to the centered TSS. The values plotted represent the difference between the real and simulated aggregate WPS (see Experimental Procedures for details). (B) Aggregate, adjusted WPS around 22,626 start codons. (C and D) Aggregate, adjusted WPS around 224,910 splice donor (C) and 224,910 splice acceptor (D) sites. (E) Nucleosome spacing in A/B compartments. Median nucleosome spacing in non-overlapping 100 kb bins, each containing ~500 nucleosome calls, is calculated genome-wide. A/B compartment predictions, also with 100 kb resolution, are shown for GM12878. Compartments A and B are associated with open and closed chromatin, respectively. (F) Nucleosome spacing and A/B compartments on chromosomes 7 and 11. A/B segmentation (red and blue bars) largely recapitulates chromosomal G-banding (ideograms, gray bars). Median nucleosome spacing (black dots) is calculated in 100 kb bins. See also Figure S3.
Short cfDNA fragments directly footprint CTCF and other TFs
Previous studies of DNase I cleavage patterns identified two dominant classes of fragments: longer fragments associated with cleavage between nucleosomes, and shorter fragments associated with cleavage adjacent to transcription factor binding sites (TFBS) (Vierstra et al., 2013). To ask whether in vivo-derived cfDNA fragments also result from two classes of sensitivity to nuclease cleavage, we partitioned sequence reads (CH01) on the basis of inferred fragment length and recalculated the WPS using long fragments (120–180 bp; 120 bp window; the same as the WPS described for nucleosome calling) or short fragments (35–80 bp; 16 bp window) separately (Figures 2B and 2C). To obtain a set of well-defined TFBSs enriched for actively bound sites in our data, we intersected clustered FIMO predictions (Grant et al., 2011; Maurano et al., 2012) with a unified set of ChIP-seq peaks from ENCODE for each TF.
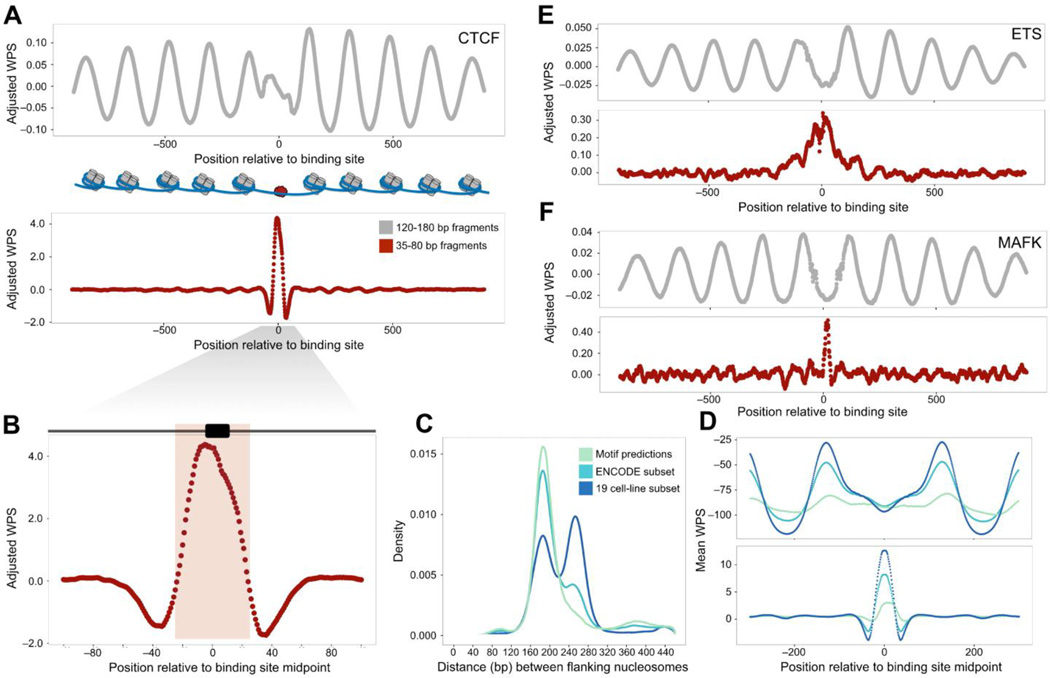
Consistent with observations by others (Fu et al., 2008; Pedersen et al., 2014; Teif et al., 2012), the long fraction WPS (L-WPS) supports strong organization of nucleosomes near CTCF binding sites (Figure 4A). However, we also observe a strong signal in the short fraction WPS (S-WPS) coincident with the CTCF site itself (Ong and Corces, 2014) (Figures 4A and 4B). We stratified CTCF sites based on our confidence that they are bound in vivo. Experimentally well-supported CTCF sites exhibit substantially broader spacing between the flanking −1 and + 1 nucleosomes based on the L-WPS, consistent with their repositioning upon CTCF binding (~190 bp → ~260 bp; Figures 4C and S4A). Experimentally well-supported CTCF sites also exhibit a much stronger S-WPS signal over the CTCF binding site itself (Figures 4D and S4B).
Figure 4. Short cfDNA fragments footprint CTCF and other TF binding sites.
Clustered FIMO predictions were intersected with ChIP-seq data to obtain confident sets of binding site predictions for various TFs. Aggregate, adjusted WPS were calculated for both the long (120–180 bp) and short (35–80 bp) fractions of cfDNA fragments. Higher WPS values indicate greater nucleosome or TF protection, respectively. (A) Aggregate, adjusted WPS for 93,530 predicted CTCF binding sites for the long (top) and short (bottom) cfDNA fractions. Binding of CTCF results in strong positioning of neighboring nucleosomes. (B) Aggregate, adjusted WPS, calculated for 93,530 predicted CTCF sites as in (A) and magnified for detail, for 35–80 bp cfDNA fragments. The pink shading indicates the larger 52 bp CTCF binding motif, and the black box shows the location of the 17 bp motif used for FIMO predictions. (C) Density of −1 to +1 nucleosome spacing around CTCF sites derived from clustered FIMO predictions (purely motif-based: 518,632 sites), a subset of these predictions overlapping with ENCODE ChIP-seq peaks (93,530 sites), and a further subset active across 19 cell lines (23,723 sites). Flanking nucleosome spacing at the least stringent set of sites (motif-based) mirrors the genome-wide average (~185 bp), while spacing at the most stringent set of sites is highly enriched for greater distances (~260 bp), consistent with active CTCF binding and repositioning of adjacent nucleosomes. (D) Mean WPS calculated for the long (top) and short (bottom) cfDNA fractions at the sets of CTCF sites in (C). (E and F) Aggregate, adjusted WPS calculated for both long (top) and short (bottom) cfDNA fractions at predicted binding sites for ETS (210,798 sites) (E) and MAFK (32,159 sites) (F). For both factors, short fraction WPS is consistent with TF-conferred protection of the binding site, whereas long fraction WPS evidences regular, local positioning of surrounding nucleosomes. See also Figure S4.
We performed similar analyses for additional TFs for which both FIMO predictions and ENCODE ChIP-seq data were available (Figure S4C). For many of these, e.g. ETS and MAFK (Figures 4E and 4F), we observe a short fraction footprint that is accompanied by periodic signal in the L-WPS, consistent with strong positioning of nucleosomes surrounding bound TFBS. Overall, these data support the view that short cfDNA fragments, which are much better recovered by the single-stranded protocol (Figures 1B and 1D), directly footprint the in vivo occupancy of DNA-bound TFs including CTCF and others.
Nucleosome spacing patterns inform cfDNA tissues-of-origin
We next asked whether in vivo nucleosome protection, as measured through cfDNA, could be used to infer the cell types contributing to cfDNA in healthy individuals. We examined the peak-to-peak spacing of nucleosome calls within DHS sites defined in 116 diverse biological samples (Maurano et al., 2012). Similar to bound CTCF sites (Figure 4C), we observe substantially broader spacing for nucleosome pairs within a subset of DHS sites, plausibly corresponding to sites at which the nucleosomes are repositioned by intervening TF binding in the cell type(s) giving rise to cfDNA (~190 bp → ~260 bp; Figure 5A). Indeed, the proportion of widened nucleosome spacing (~260 bp) varies considerably depending on which cell type’s DHS sites are used. However, all of the cell types for which this proportion is highest are lymphoid or myeloid in origin (e.g. CD3_CB-DS17706, etc. in Figure 5A), consistent with hematopoietic cell death as the dominant source of cfDNA in healthy individuals (Lui et al., 2002).
Figure 5. Inference of mixtures of cell types contributing to cell-free DNA in healthy states and cancer.
(A) The distribution of nucleosome spacing for peaks flanking DHS sites in 116 callsets is bimodal, plausibly corresponding to widened nucleosome spacing at active DHS sites due to intervening TF binding (~190 bp → 260 bp). Lymphoid or myeloid callsets have the largest proportions of DHS sites with widened nucleosome spacing, consistent with hematopoietic cell death as the dominant source of cfDNA in healthy individuals. (B and C) Partitioning adjusted WPS scores around TSS into five gene expression bins (quintiles) defined for NB-4 (an acute promyelocytic leukemia cell line) reveals differential nucleosome spacing and positioning. (B) Highly expressed genes show strong nucleosome phasing within the transcript body. Upstream of the TSS, −1 nucleosomes are well-positioned across expression bins, but −2 and −3 nucleosomes are well-positioned only for medium to highly expressed genes. (C) For medium to highly expressed genes, a short fragment WPS peak is observed between the TSS and the −1 nucleosome, plausibly footprinting some or all of the transcription preinitiation complex at transcriptionally active genes. (D) Median nucleosome spacing in the transcript body is negatively correlated with gene expression in NB-4 (ρ = −0.17, n = 19,677 genes). Spacing in genes with low or no expression is 193 bp, while spacing in expressed genes ranges from 186 to 193 bp. (E) To deconvolve multiple contributions, intensities from fast Fourier transformation (FFT) quantified the specific frequency contributions in the long fragment WPS for 10 kb windows downstream of each TSS. Shown are correlation trajectories for RNA expression in 76 cell lines and primary tissues at different frequencies. Correlations are strongest for intensities in the 193–199 bp frequency range. (F) The ranks of correlation for 76 RNA expression datasets with average intensity in the 193–199 bp frequency range for various cfDNA libraries are shown, categorized by type and listed from highest (top row) to lowest rank (bottom row). Correlation values and full cell line or tissue names are provided in Table S3. All of the strongest correlations for all three healthy samples (BH01, IH01 and IH02; first three columns) are with lymphoid and myeloid cell lines or with bone marrow. In contrast, cfDNA samples obtained from stage IV cancer patients (IC15, IC17, IC20, IC35, IC37; last five columns) show top correlations with various cancer cell lines, e.g. IC17 (hepatocellular carcinoma, HCC) showing highest correlations with HepG2 (HCC cell line), and IC35 (breast ductal carcinoma, DC) with MCF7 (metastatic breast adenocarcinoma cell line). When comparing cell line/tissue ranks observed for the cancer samples to each of the three healthy samples and averaging the rank changes, maximum rank changes are over two-fold higher than those observed from comparing the three healthy samples with each other and averaging rank changes (‘Control’). For example, for IC15 (small cell lung carcinoma, SCLC) the rank of SCLC-21H (SCLC cell line) increased by an average of 31 positions, for IC20 (squamous cell lung carcinoma, SCC) SK-BR-3 (metastatic breast adenocarcinoma cell line) increased by an average rank of 21, and for IC37 (colorectal adenocarcinoma, AC) HepG2 increased by 24 ranks. See also Figure S5 and Table S4.
We next re-examined the signal of nucleosome protection in the vicinity of transcriptional start sites (TSS) (Figure 3A). If we stratify based on gene expression in a lymphoid cell line, NB-4, we observe strong differences in the patterns of nucleosome protection in relation to the TSS, in highly vs. lowly expressed genes (Figure 5B). Furthermore, if we examine the S-WPS, we observe a clear footprint immediately upstream of the TSS whose intensity also strongly correlates with expression (Figure 5C). This plausibly reflects footprinting of the transcription pre-initiation complex, or some component thereof, at transcriptionally active genes.
These observations support our thesis that cfDNA fragmentation patterns indeed contain signal that might be used to infer the tissue(s) or cell-type(s) giving rise to cfDNA. However, a challenge is that relatively few reads in a genome-wide cfDNA library directly overlap DHS sites and TSSs.
It was previously observed that nucleosome spacing varies between cell types as a function of chromatin state and gene expression (Teif et al., 2012; Valouev et al., 2012). In general, open chromatin and transcription are associated with a shorter nucleosome repeat length, consistent with our analyses of compartment A vs. B (Figure 3E). In our peak calls, we also observe a correlation between nucleosome spacing across gene bodies and their expression levels, with tighter spacing associated with higher expression (Figure 5D; ρ = −0.17; n = 19,677 genes). The correlation is highest for the gene body itself, relative to adjacent regions (upstream 10 kb ρ = −0.08; downstream 10 kb ρ = −0.01). If we limit this analysis to gene bodies that span at least 60 nucleosome calls, the correlation is much stronger (ρ = −0.50; n = 12,344 genes).
An advantage of exploiting signals such as nucleosome spacing across gene bodies or other domains is that a much larger proportion of cfDNA fragments will be informative, and moreover we might be able to detect mixtures of signals resulting from multiple cell types contributing to cfDNA. To test this, we performed fast Fourier transformation (FFT) on the L-WPS across the first 10 kb of gene bodies and on a gene-by-gene basis. The intensity of the FFT signal is correlated with gene expression at specific frequency ranges, with a maximum at 177–180 bp for positive correlation and a minimum at ~199 bp for negative correlation (Figure 5E). In performing this analysis against a dataset of 76 expression datasets for human cell lines and primary tissues (Uhlén et al., 2015), we observe that the strongest correlations are with hematopoietic lineages (Figure 5E). For example, the most highly ranked negative correlations with average intensity in the 193–199 bp frequency range for each of three healthy samples (BH01, IH01, IH02) are all to lymphoid cell lines, myeloid cell lines, or bone marrow tissue (Figure 5F; Table S3). These top correlation ranks are robust to downsampling (Figure S5A).
Nucleosome spacing in cancer patients’ cfDNA identifies non-hematopoietic contributions
We next sought to ask whether we could detect signatures of non-hematopoietic cell types contributing to circulating cfDNA in non-healthy states. We first screened 44 plasma samples from individuals with clinical diagnoses of a variety of Stage IV cancers with light sequencing of single-stranded libraries prepared from cfDNA (Table S4; median 2.2-fold coverage; of note, with same protocol and many in the same batch as IH02). Because matched tumor genotypes were not available, we scored each sample on two metrics of aneuploidy to identify a subset likely to contain a high proportion of tumor-derived cfDNA: first, the deviation from the expected proportion of reads derived from each chromosome (Leary et al., 2012); and second, the per-chromosome allele balance profile for a panel of common single nucleotide polymorphisms. Based on these metrics, we sequenced single-stranded libraries derived from five individuals (with a small cell lung cancer, a squamous cell lung cancer, a colorectal adenocarcinoma, a hepatocellular carcinoma, and a ductal carcinoma in situ breast cancer) to a depth similar to that of IH02 (Table S5).
We again performed FFT on the L-WPS across gene bodies and correlated the average intensity in the 193–199 bp frequency range against the same 76 expression datasets for human cell lines and primary tissues (Uhlén et al., 2015). In contrast with the three samples from healthy individuals (where all of the top 10, and nearly all of the top 20, correlations were to lymphoid or myeloid lineages), we observe that many of the most highly ranked cell lines or tissues represent non-hematopoietic lineages, in some cases aligning with the cancer type (Figure 5F). For example, for IC17, where the patient had a hepatocellular carcinoma, the top-ranked correlation is with HepG2, a hepatocellular carcinoma cell line. For IC35, where the patient had a ductal carcinoma in situ breast cancer, the top-ranked correlation is with MCF7, a metastatic breast adenocarcinoma cell line. In other cases, the cell lines or primary tissues that exhibit the greatest change in correlation rank align with the cancer type. For example, for IC15, where the patient had small-cell lung cancer, the largest change in correlation rank (−31) is for a small-cell lung cancer cell line (SCLC-21H), and the second largest change (−25) is for a neuroblastoma cancer cell line (SH.SY5Y). For IC20 (a lung squamous cell carcinoma) and IC35 (a colorectal adenocarcinoma), there are many non-hematopoietic cancer cell lines displacing the lymphoid/myeloid cell lines in terms of correlation rank, but the alignment of these to the specific cancer type is less clear. It is possible that the molecular profile of these patients’ cancers is not well-represented amongst our 76 expression datasets (none are lung squamous cell carcinomas; CACO-2 is a cell line derived from a colorectal adenocarcinoma, but is highly heterogeneous (Sambuy et al., 2005)). As with samples from healthy individuals, the top correlation ranks associated with the samples from cancer patients are robust to downsampling (Figure S5A). However, in silico “dilution” of samples from cancer patients with samples from healthy patients results in proportionally lower ranks for non-hematopoietic cell lines, consistent with expectation (Figure S5B).
Discussion
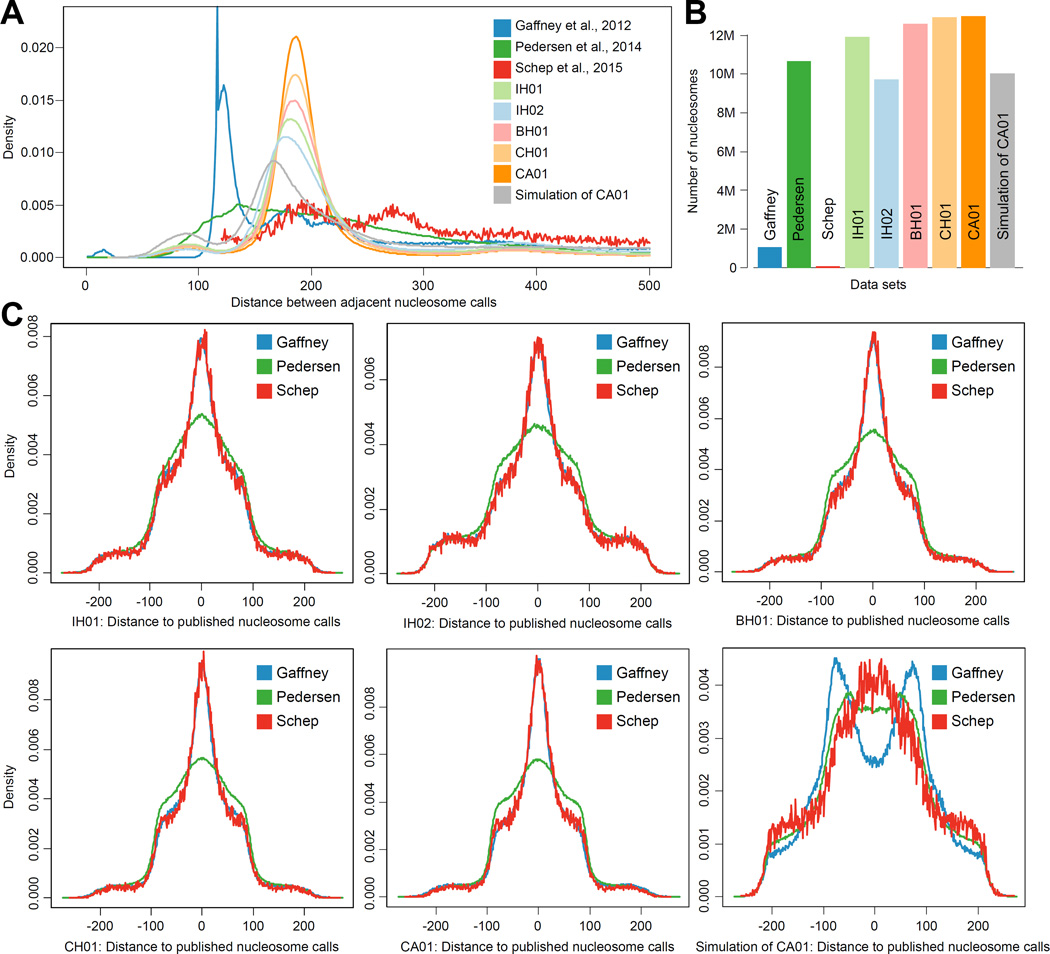
We present a dense, genome-wide map of in vivo nucleosome protection inferred from plasma-borne cfDNA fragments. Although the number of peaks is essentially saturated in CH01, other metrics of quality continued to be a function of sequencing depth (Figure S2C). We therefore constructed an additional genome-wide nucleosome map based on all of the cfDNA sequencing that we have performed to date (‘CA01’, 14.5G fragments; 700-fold coverage; 13.0M peaks). Although this map exhibits even more uniform spacing (Figure 6A) and more highly supported peak calls (not shown), we caution that it is based on cfDNA from both healthy and non-healthy individuals (Tables S1 and S5).
Figure 6. Comparison of nucleosome callsets.
(A) Distance between nucleosome peak calls across three published data sets (Gaffney et al. 2012, Pedersen et al. 2014, Schep et al. 2015) and calls produced in this study. Previously published callsets lack one defined mode at the canonical ~185 bp nucleosome spacing, possibly due to sparse sampling or wide call ranges. In contrast, all the nucleosome calls from cfDNA show one well-defined mode, the magnitude of which increases with the number of fragments examined. The callset produced from simulation has a lower mode (166 bp) and a wider distribution. (B) Number of calls in each set. The densest cfDNA-derived callset contains nearly 13M nucleosome calls. (C–G) Comparison of peak locations between samples. For each pair of samples, the distribution of distances between each peak call in the sample with fewer peaks and the nearest peak call in the other sample is shown. Negative numbers indicate the nearest peak is upstream; positive numbers indicate the nearest peak is downstream. Concordance between callsets increases with the number of cfNDA fragments examined. (H), As in (G), comparing other callsets to the matched simulation of CA01. See also Table S5.
Our work builds directly on previous efforts to map nucleosome occupancy in human cells genome-wide (Gaffney et al., 2012; Pedersen et al., 2014; Schones et al., 2008; Teif et al., 2012; Valouev et al., 2012), but our callset is substantially more complete and uniform (Figure 6). The fragments that we observe are generated by endogenous physiological processes, avoiding the technical variation associated with in vitro MNase digestion. A limitation of our map is that the cell types that give rise to cfDNA are inevitably heterogeneous (e.g. a mixture of lymphoid and myeloid cell types in healthy individuals). Nonetheless, the map’s relative completeness may facilitate a deeper understanding of the interplay of nucleosome positioning and spacing with primary sequence, epigenetic regulation, transcriptional output, and nuclear architecture.
A second goal of this study was to explore whether the nucleosome footprints contained in cfDNA fragments can be used to infer contributing cell types. Through comparisons with gene expression and regulatory site profiles, we identify the epigenetic signature of hematopoietic lineages contributing to cfDNA in healthy individuals, with plausible additional contributions from one or more non-hematopoietic tissues in a small panel of individuals with advanced cancers. For this proof-of-concept, we stacked the odds in our favor by focusing individuals that appeared to have large burdens of tumor-derived DNA. However, it should be emphasized that in the context of cancer, our goal is not necessarily to outperform the sensitivity of mutation-based monitoring of circulating tumor DNA. Rather, we envision that a unique application of this approach may be to non-invasively classify cancers at time-of-diagnosis by matching the epigenetic signature of cfDNA fragmentation patterns against reference datasets corresponding to diverse cancer types. For example, this may have value for non-invasively and molecularly classifying “cancers of unknown primary”, which comprise 4–5% of all invasive cancers (Greco and Hainsworth, 2009), as well as cancers where invasive biopsies are currently required for definitive diagnosis and/or for subtyping (e.g. lung cancer).
In addition, there are a range of non-malignant conditions for which it may be valuable to explore the nucleosome and TF footprints contained in cfDNA as markers for acute or chronic tissue damage, e.g. myocardial infarction (Chang et al., 2003), stroke (Rainer et al., 2003) and autoimmune disorders (Galeazzi et al., 2003). Contributions from these tissues to cfDNA cannot be readily detected under the current paradigm of analyzing genotypic differences, which are effectively non-existent in these conditions. By contrast, the approach presented here should generalize to detecting contributions to cfDNA from any non-hematopoietic cell lineage (and, possibly, grossly aberrant contributions from hematopoietic cell lineages).
Alternative “genotype-independent” approaches for using circulating nucleic acids as markers for disease include cell-free RNA (Koh et al., 2014) and DNA methylation. Although these merit exploration, tumor-derived cell-free RNA appears to be much less stable than nucleosome-bound cfDNA (García-Olmo et al., 2013), while bisulfite sequencing libraries are challenging to robustly construct from small amounts of starting material.
A limitation of this study is the small number of samples studied (n = 8) and the relatively small size of the reference dataset of cell lines and tissues against which these samples were compared (n = 76). We anticipate that increasing the number of samples studied, as well as the range of physiological states and diseases with which these samples are associated, is necessary to fully evaluate the potential and limitations of this approach. Furthermore, expanding the breadth and quality of the reference datasets against which these samples are compared (e.g., directly comparing to cell-type-specific nucleosome maps, rather than to expression profiles), is likely to improve the ability to robustly assign and quantify contributing cell types.
Cell-free DNA has tremendous potential as a “liquid biopsy”, and indeed its use in non-invasive prenatal screening for fetal trisomies has vastly outpaced all other applications of DNA sequencing in terms of clinical uptake. In contrast with current paradigms for analyzing cfDNA, we show how the information contained in cfDNA fragmentation patterns, effectively the footprints of protein-DNA interactions, can be used to infer contributing cell types without relying on genotypic differences. To the extent that cfDNA composition is impacted by cell death consequent to malignancy, acute or chronic tissue damage, or other conditions, this method may substantially expand the range of clinical scenarios in which cfDNA sequences comprise a clinically useful biomarker.
Experimental Procedures
Plasma Samples
Bulk human peripheral blood plasma, containing contributions from an unknown number of healthy individuals, was obtained from STEMCELL Technologies (Vancouver, British Columbia, Canada). Anonymous, individual human peripheral blood plasma from healthy donors, donors with clinical diagnosis of Stage IV cancer, and donors with clinical diagnosis with autoimmune disease (Tables S1, S4 and S5) was obtained from Conversant Bio (Huntsville, Alabama, USA) or PlasmaLab International (Everett, Washington, USA). Plasma was stored at −80°C and thawed on the bench-top immediately before use. Cell-free DNA was purified from each sample with the QiaAMP Circulating Nucleic Acids kit (Qiagen) as per the manufacturer’s protocol. DNA was quantified with a Qubit fluorometer (Invitrogen).
Preparation of sequencing libraries
Conventional, double-stranded sequencing libraries were prepared with the ThruPLEX-FD or ThruPLEX DNA-seq kits (Rubicon Genomics), comprising a proprietary series of end-repair, ligation, and amplification reactions. Libraries were prepared with 0.5–30.0 ng of cfDNA input and individually barcoded. Library amplification was monitored by real-time PCR and was typically terminated after 4–6 cycles. Single-stranded sequencing libraries were prepared with a protocol adapted from Gansauge and Meyer (2013). A detailed description can be found in Supplemental Experimental Procedures.
Sequencing and primary data processing
All libraries were sequenced on HiSeq 2000 or NextSeq 500 instruments (Illumina). Details of sequencing are provided in Tables S1 and S5. Barcoded paired-end (PE) sequencing data was split allowing up to one substitution in the barcode sequence. Fragments shorter than or equal to the read length were consensus-called and adapter-trimmed. Remaining consensus single-end reads (SR) and the individual PE reads were aligned to the human reference genome (GRCh37, 1000 Genomes phase 2 technical reference, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/phase2_reference_assembly_sequence/) using the ALN algorithm in BWA v0.7.10 (Li and Durbin, 2010). PE reads were further processed with BWA SAMPE to resolve ambiguous placement of read pairs or to rescue missing alignments by a more sensitive alignment step around the location of one placed read end. Aligned SR and PE data were stored in BAM format using the samtools API (Li et al., 2009). BAM files for each sample were merged across lanes and sequencing runs.
Simulations
Sequencing data was simulated procedurally to mimic observed cleavage and ligation biases and length distributions. See Supplemental Experimental Procedures for details.
Coverage, fragment endpoints, and windowed protection scores
Fragment endpoint coordinates were extracted from BAM files with the SAMtools API. Both outer alignment coordinates of PE data were extracted for properly paired reads. Both end coordinates of SR alignments were extracted when PE data was collapsed to SR data by adapter trimming. A fragment’s coverage is defined as all positions between the two (inferred) fragment ends, inclusive of endpoints. We define the Windowed Protection Score (WPS) of a window of size k as the number of molecules spanning the window minus those with an endpoint within the window. We assign the determined WPS to the center of the window. For 35–80 bp fragments (short fraction, S-WPS), k=16; for 120–180 bp fragments (long fraction, L-WPS), k=120.
Nucleosome peak calling
L-WPS is locally adjusted to a running median of zero in 1 kb windows and smoothed using a Savitzky-Golay filter (Savitzky and Golay, 1964) (window size 21, 2nd order polynomial). The L-WPS track is then segmented into above-zero regions (allowing up to 5 consecutive positions below zero). If the resulting region is 50–150 bp, we identify the median L-WPS value of that region and search for the maximum-sum contiguous window above the median. We report the start, end and center coordinates of this window as the “peak,” or local maximum of nucleosome protection. All calculations involving distances between peaks are based on these center coordinates. A score for each peak is determined as the distance between maximum value in the window and the average of the two adjacent L-WPS minima neighboring the region. If the identified region is 150–450 bp, we apply the same above median contiguous window approach, but only report those windows that are 50–150 bp. For score calculation of multiple windows derived from 150–450 bp regions, we set the neighboring minima to zero. We discard regions <50 bp or >450 bp.
Analysis of TFBS, DHS sites, and genic features
Features were aggregated and aligned at starting coordinates while adjusting for strand and direction of transcription. TFBS sets were obtained by filtering motif predictions with ChIP-seq peaks. For most features, L-WPS values were adjusted to account for signal observed in matched simulations. See Supplemental Experimental Procedures for details.
Gene expression analysis
FPKM gene expression (GE) values measured for 20,344 Ensembl gene identifiers in 44 human cell lines and 32 primary tissues by the Human Protein Atlas (Uhlén et al., 2015) was downloaded from http://www.proteinatlas.org/download/rna.csv.zip. Genes with 3 or more non-zero expression values were retained (n=19,378 genes). The GE data set is provided with one decimal precision for the FPKM values. Thus, a zero GE value (0.0) indicates expression in the interval [0, 0.05) Unless otherwise noted, we set the minimum GE value to 0.04 FPKM before log2-transformation.
Fourier transformation and correlation with expression
L-WPS was used to calculate periodograms of genomic regions using Fast Fourier Transform (FFT, spec.pgram in R) with frequencies between 1/500 and 1/100 bases. See Supplemental Experimental Procedures for details. Intensity values for the 120–280 bp frequency range were determined from smooth FFT periodograms. S-shaped Pearson correlation between GE values and FFT intensities was observed around the major inter-nucleosome distance peak, along with a pronounced negative correlation in the 193–199 bp frequency range. The mean intensity in this frequency range was correlated with the average intensity with log2-transformed GE values for downstream analysis.
Supplementary Material
Acknowledgements
We thank D. May, J. Vierstra, M. Maurano, and members of the Shendure lab for helpful discussions. This work was funded in part by an NIH Director's Pioneer Award (1DP1HG007811 to J.S.). J.S. is an investigator of the Howard Hughes Medical Institute. A patent application has been filed for aspects of the methods disclosed here (M.W.S., M.K., and J.S.: “Methods of determining tissues and/or cell types giving rise to cell-free DNA, and methods of identifying a disease or disorder using same”; PCT/US2015/042310).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization: M.W.S., M.K., and J.S.; Methodology: M.W.S. and M.K.; Software: M.K.; Investigation: M.W.S. and R.M.D.; Formal Analysis: M.W.S., M.K., and A.J.H.; Visualization: M.W.S., M.K., and A.J.H.; Writing – Original Draft: M.W.S. and J.S.; Writing – Review & Editing: M.W.S., M.K., A.J.H., and J.S.; Data Curation: M.K.; Funding Acquisition: J.S.; Supervision: J.S.
Accession Numbers
The NCBI GEO accession for the aligned reads of the 60 samples as well as the peak calls presented in this paper is GSE71378.
REFERENCES
- Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AKC, Chiu RWK, Lo YMD. Cell-free nucleic acids in plasma, serum and urine: a new tool in molecular diagnosis. Ann. Clin. Biochem. 2003;40:122–130. doi: 10.1258/000456303763046030. [DOI] [PubMed] [Google Scholar]
- Chang CP-Y, Chia R-H, Wu T-L, Tsao K-C, Sun C-F, Wu JT. Elevated cell-free serum DNA detected in patients with myocardial infarction. Clinica Chimica Acta. 2003;327:95–101. doi: 10.1016/s0009-8981(02)00337-6. [DOI] [PubMed] [Google Scholar]
- Chiu RWK, Chan KCA, Gao Y, Lau VYM, Zheng W, Leung TY, Foo CHF, Xie B, Tsui NBY, Lun FMF, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc. Natl. Acad. Sci. USA. 2008;105:20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen P-Y, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J. Clin. Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. USA. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer—A survey. Biochimica Et Biophysica Acta (BBA) - Reviews on Cancer. 2007;1775:181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney DJ, McVicker G, Pai AA, Fondufe-Mittendorf YN, Lewellen N, Michelini K, Widom J, Gilad Y, Pritchard JK. Controls of nucleosome positioning in the human genome. PLoS Genet. 2012;8:e1003036. doi: 10.1371/journal.pgen.1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeazzi M, Morozzi G, Piccini M, Chen J, Bellisai F, Fineschi S, Marcolongo R. Dosage and characterization of circulating DNA: present usage and possible applications in systemic autoimmune disorders. Autoimmun. Rev. 2003;2:50–55. doi: 10.1016/s1568-9972(02)00101-5. [DOI] [PubMed] [Google Scholar]
- Gansauge M-T, Meyer M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc. 2013;8:737–748. doi: 10.1038/nprot.2013.038. [DOI] [PubMed] [Google Scholar]
- García-Olmo DC, Picazo MG, Toboso I, Asensio AI, García-Olmo D. Quantitation of cell-free DNA and RNA in plasma during tumor progression in rats. Mol. Cancer. 2013;12:8. doi: 10.1186/1476-4598-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco FA, Hainsworth JD. Introduction: unknown primary cancer. Seminars in Oncology. 2009;36:6–7. doi: 10.1053/j.seminoncol.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Harshman SW, Young NL, Parthun MR, Freitas MA. H1 histones: current perspectives and challenges. Nucleic Acids Res. 2013;41:9593–9609. doi: 10.1093/nar/gkt700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdenrieder S, Stieber P, Chan LYS, Geiger S, Kremer A, Nagel D, Lo YMD. Cell-free DNA in serum and plasma: comparison of ELISA and quantitative PCR. Clinical Chemistry. 2005;51:1544–1546. doi: 10.1373/clinchem.2005.049320. [DOI] [PubMed] [Google Scholar]
- Koh W, Pan W, Gawad C, Fan HC, Kerchner GA, Wyss-Coray T, Blumenfeld YJ, El-Sayed YY, Quake SR. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc. Natl. Acad. Sci. USA. 2014;111:7361–7366. doi: 10.1073/pnas.1405528111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, O'Shaughnessy J, Kinzler KW, Parmigiani G, Vogelstein B, et al. Detection of Chromosomal Alterations in the Circulation of Cancer Patients with Whole-Genome Sequencing. Sci Transl Med. 2012;4:162ra154–162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YMD, Chan KCA, Sun H, Chen EZ, Jiang P, Lun FMF, Zheng YW, Leung TY, Lau TK, Cantor CR, et al. Maternal Plasma DNA Sequencing Reveals the Genome-Wide Genetic and Mutational Profile of the Fetus. Sci Transl Med. 2010;2:61ra91–61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am. J. Hum. Genet. 1999;64:218–224. doi: 10.1086/302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui YYN, Chik K-W, Chiu RWK, Ho C-Y, Lam CWK, Lo YMD. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clinical Chemistry. 2002;48:421–427. [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouliere F, Messaoudi ElS, Pang D, Dritschilo A, Thierry AR. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol. Oncol. 2014;8:927–941. doi: 10.1016/j.molonc.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C-T, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen JS, Valen E, Velazquez AMV, Parker BJ, Rasmussen M, Lindgreen S, Lilje B, Tobin DJ, Kelly TK, Vang S, et al. Genome-wide nucleosome map and cytosine methylation levels of an ancient human genome. Genome Res. 2014;24:454–466. doi: 10.1101/gr.163592.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainer TH, Wong LKS, Lam W, Yuen E, Lam NYL, Metreweli C, Lo YMD. Prognostic use of circulating plasma nucleic acid concentrations in patients with acute stroke. Clinical Chemistry. 2003;49:562–569. doi: 10.1373/49.4.562. [DOI] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambuy Y, De Angelis I, Ranaldi G, Scarino ML, Stammati A, Zucco F. The Caco-2 cell line as a model of the intestinal barrier: influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell Biol. Toxicol. 2005;21:1–26. doi: 10.1007/s10565-005-0085-6. [DOI] [PubMed] [Google Scholar]
- Savitzky A, Golay M. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964;36:1627–1639. [Google Scholar]
- Schep AN, Buenrostro JD, Denny SK, Schwartz K, Sherlock G, Greenleaf WJ. Structured nucleosome footprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 2015;25 doi: 10.1101/gr.192294.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh T-Y, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder TM, Khush KK, Valantine HA, Quake SR. Universal noninvasive detection of solid organ transplant rejection. Proc. Natl. Acad. Sci. USA. 2011;108:6229–6234. doi: 10.1073/pnas.1013924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teif VB, Vainshtein Y, Caudron-Herger M, Mallm J-P, Marth C, Höfer T, Rippe K. Genome-wide nucleosome positioning during embryonic stem cell development. Nat. Struct. Mol. Biol. 2012;19:1185–1192. doi: 10.1038/nsmb.2419. [DOI] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2012;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, Wang H, John S, Sandstrom R, Stamatoyannopoulos JA. Coupling transcription factor occupancy to nucleosome architecture with DNase-FLASH. Nat. Meth. 2013;11:66–72. doi: 10.1038/nmeth.2713. [DOI] [PubMed] [Google Scholar]
- Wimberger P, Roth C, Pantel K, Kasimir-Bauer S, Kimmig R, Schwarzenbach H. Impact of platinum-based chemotherapy on circulating nucleic acid levels, protease activities in blood and disseminated tumor cells in bone marrow of ovarian cancer patients. Int. J. Cancer. 2010;128:2572–2580. doi: 10.1002/ijc.25602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.