Abstract
Protein arginine phosphorylation is a post-translational modification (PTM) that is important for bacterial growth and virulence. Despite its biological relevance, the intrinsic acid lability of phosphoarginine (pArg) have impaired studies of this novel PTM. Herein, we report for the first time the development of phosphonate-amidines and sulfonate-amidines as isosteres of pArg and then use these mimics as haptens to develop the first high affinity sequence independent anti-pArg specific antibody. Employing this anti-pArg antibody, we further showed that arginine phosphorylation is induced in Bacillus subtilis during oxidative stress. Overall, we expect this antibody to see widespread use in analyzing the biological significance of arginine phosphorylation. Additionally, the chemistry reported here will facilitate the generation of pArg mimetics as highly potent inhibitors of the enzymes that catalyze arginine phosphorylation/dephosphorylation.
Keywords: phosphoarginine, haptens, phosphonate-amidine, antibodies, N-phosphorylation
Protein phosphorylation regulates numerous biological processes and intracellular signaling pathways in both eukaryotic and prokaryotic organisms.[1] In eukaryotes, the phosphorylation of serine, threonine and tyrosine residues, i.e. O-phosphorylation, represents a paradigm for the control of cell signaling cascades.[2] Apart from these phosphate monoesters, it is well established that histidine, arginine, and lysine are N-phosphorylated.[3] In contrast to O-phosphorylation, the N—P phosphoramidate bond is acid labile and thus is readily hydrolyzed under acidic conditions.[4] This acid lability is due to the protonation of the bridging nitrogen, which induces considerable lengthening and thereby weakening of the N—P bond.[3c, 5] Despite the challenging nature of the N—P bond, chemical biologists have recently begun to focus on the development of suitable tools for analyzing protein N-phosphorylation.[3d, 6] For example, Muir and colleagues recently reported the development of the first pan-phospho-histidine (pHis) specific antibody using a stable triazole-based pHis analog as the hapten.[6c]
Protein arginine phosphorylation was originally found to occur on histone proteins.[7] More recently, the first protein arginine kinase was identified in bacteria,[8] sparking the subsequent identification of more than 100 phosphoarginine (pArg) modification sites in the Gram-positive model organism Bacillus subtilis using an arginine phosphatase mutant strain.[9] Given that this modification is even more abundant than O-phosphorylation in B. subtilis, arginine phosphorylation likely represents a ubiquitous regulatory mechanism that plays a key role in cell physiology and survival under specific conditions.[9b] Moreover, the protein arginine kinase McsB was shown to be important for the virulence of Gram-positive pathogens such as Staphylococcus aureus,[10] thus highlighting the antibacterial potential of targeting this protein modification.
Current methods to detect cellular protein arginine phosphorylation rely on cumbersome mass spectrometric analyses that are carried out under acidic conditions leading to a time dependent loss of signal.[9, 11] Additionally, these techniques are technically challenging and are not readily available to all researchers in the field. Furthermore, they do not allow for the rapid and facile detection of the changes in arginine phosphorylation that occur in response to cell signalling events. Given the limitations of current methods, we sought to develop a facile method to visualize and enrich for arginine phosphorylated proteins. Towards that goal, we report the design and synthesis of non-cleavable phosphonate- and sulfonate-amidines as isosteres of pArg and demonstrate their use as haptens to generate the first high affinity sequence independent anti-pArg specific antibody.
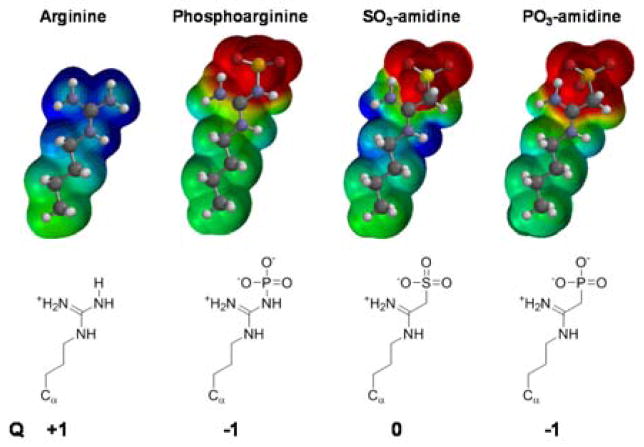
Previous attempts to generate anti-pArg antibodies using classical immunizations of mice and rabbits failed; most likely because the acid labile N—P bond is cleaved during endosomal/lysosomal processing of the immunogen.[12] An in vitro phage display approach was used to generate an antibody targeting a phosphoarginine containing peptide.[12] While this antibody is useful for in vitro studies with recombinant proteins,[12] its low affinity for pArg (see below) has precluded its use in cellular studies. To circumvent these drawbacks and obtain high affinity phosphoarginine-specific antibodies, we set out to synthesize a non-cleavable, acid stable pArg mimetic, hypothesizing that such a hapten would allow for the generation of a sequence independent high affinity anti-pArg antibody. With this goal in mind, we further hypothesized that amidines derivatized with either a sulfonate or a phosphonate might act as stable analogues of pArg (Figure 1) - the acid labile N—P bond is replaced with a stable, non-hydrolyzable C—P bond. Under physiological conditions, phosphoarginine exists in a zwitterionic form, containing a net charge of −1, with the positive charge distributed over the three nitrogens in the planar guanidinium moiety, whereas the two negative charges are localized to the oxygen anions of the phosphoryl group (Figure 1). Comparison of the calculated electrostatic charge potentials highlights the electronic and geometric similarities between phosphoarginine and its anticipated analogues.
Figure 1.
Structural representation of the electrostatic map of the side chain of arginine, phosphoarginine and its stable analogs SO3-amidine as well as PO3-amidine. The formal net charge of the individual compounds is denoted by Q.
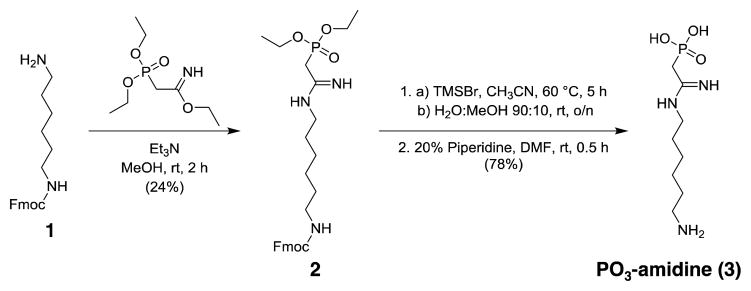
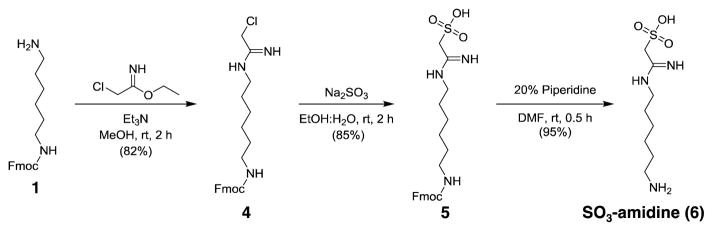
Phosphonate-amidine-hexylamine (PO3-amidine, 3), and sulfonate-amidine-hexylamine (SO3-amidine, 6), were prepared as described in Schemes 1 and 2, respectively. In the case of PO3-amidine, Fmoc protected 1,6-diaminohexane (1) was reacted with freshly prepared ethyl 2-(diethoxyphosphoryl)acetimidate to generate compound 2 that was deprotected over two steps, using TMSBr to deprotect the ethyl esters and piperidine to remove the Fmoc group, yielding the final product PO3-amidine (3) (Scheme 1 and Figure S1–S6). The synthesis of the SO3-amidine derivative involves the reaction of Fmoc-1,6-diaminohexane (1) with chloroacetimidate to obtain the Cl-acetamidine derivative 4. The sulfonate group was installed by the displacement of the chloride with sodium sulfite to synthesize 5. After Fmoc deprotection, the resulting SO3-amidine (6) was obtained in good yield. The detailed synthetic procedures and compound characterizations are provided in the supporting material (Figure S7–S12). Since the phosphoryl group in phosphoarginine exists as the doubly negative charged form (pKa2 = 4.5) under conditions of physiological pH,[3c] we evaluated the protonation state of the phosphonate group in PO3-amidine. Typically, the second ionization in alkylphosphonates occurs at pH ≥ 8.[13] However, based on 31P NMR pH titration experiments (Figure S13), the pKa2 of PO3-amidine was determined to be 6.0, indicating that the second oxygen in the phosphono group ionizes under physiological pH conditions. Interestingly, the pKa2 of PO3-amidine is similar to aminomethylphosphonate (pKa2 = 5.9) and thus highlights the strong pKa lowering effect of the closely connected amidine moiety.
Scheme 1.
Synthesis of PO3-amidine (3) (2-((6-aminohexyl)amino)-2-iminoethyl)phosphonic acid.
Scheme 2.
Synthesis of SO3-amidine (6) 2-((6-aminohexyl)amino)-2-iminoethane-1-sulfonic acid.
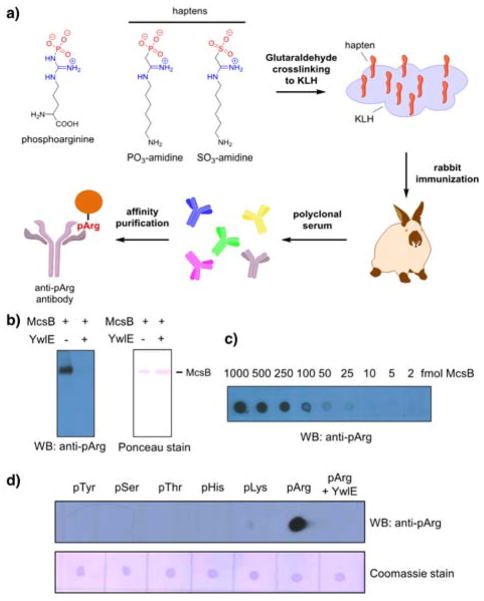
The haptens thus generated were conjugated to KLH carrier protein via their free amines, which are present distal from the SO3-amidine and PO3-amidine groups, and then individually injected into rabbits to raise polyclonal antibodies targeting these moieties (Figure 2a). After immunization, the resulting sera were tested for phosphoarginine recognition in Western Blot analysis (Figure 2b and Figure S14a). The serum, obtained from rabbits immunized with PO3-amidine, showed strong recognition of phosphoarginine (Figure 2b, Figure S15), thereby validating this functional group as a potent mimetic of phosphoarginine. By contrast, immunization of rabbits with SO3-amidine-KLH conjugates only elicited SO3-amidine specific antibodies that did not show any significant cross reactivity to phosphoarginine (Figure S14b).
Figure 2.
Generation of phosphoarginine specific antibodies using a non-cleavable hapten approach. a) Strategy to develop anti-pArg antibodies. b) Western Blot of autophosphorylated McsB treated plus/minus YwlE using anti-pArg antibody. The ponceau stain was included as the loading control. c) The sensitivity of the anti-pArg antibody was determined by dot blot analysis using serial dilutions of autophosphorylated McsB. d) The specificity of the anti-pArg antibody was monitored by dot blot analysis towards pTyr, pSer, pThr, pHis, pLys and pArg modified BSA. The coomassie stain was included as the loading control.
To obtain highly purified anti-pArg antibody, the crude rabbit anti-serum from PO3-amidine immunized rabbits was affinity purified over an agarose column containing immobilized arginine phosphorylated lysozyme (Figure S16 and S17). The resulting antibody directly recognizes pArg with high affinity and the limit of detection is ~10 fmol of autophosphorylated McsB (Figure 2c). Moreover, direct comparison with the previously described phage display generated antibody revealed a more than 10-fold higher sensitivity for the anti-pArg antibody developed using the hapten approach (Figure S18). To evaluate the selectivity of this anti-pArg antibody, we performed dot blot assays using distinct phosphoproteins comprising pTyr, pThr, pSer, pHis, pLys and pArg (Figure 2d). Notably, the antibody only recognized arginine phosphorylated protein and not the other phosphoproteins. Further highlighting the specificity of this antibody, pretreatment of the arginine phosphorylated protein with the protein arginine phosphatase YwlE completely abolished the signal. Western blot analyses further showed that this anti-pArg antibody selectively recognizes arginine phosphorylated McsB and lysozyme, but not their dephosphorylated counterparts, thereby confirming the general utility and the context independent nature of this pan-phosphoarginine targeted antibody (Figure S19).
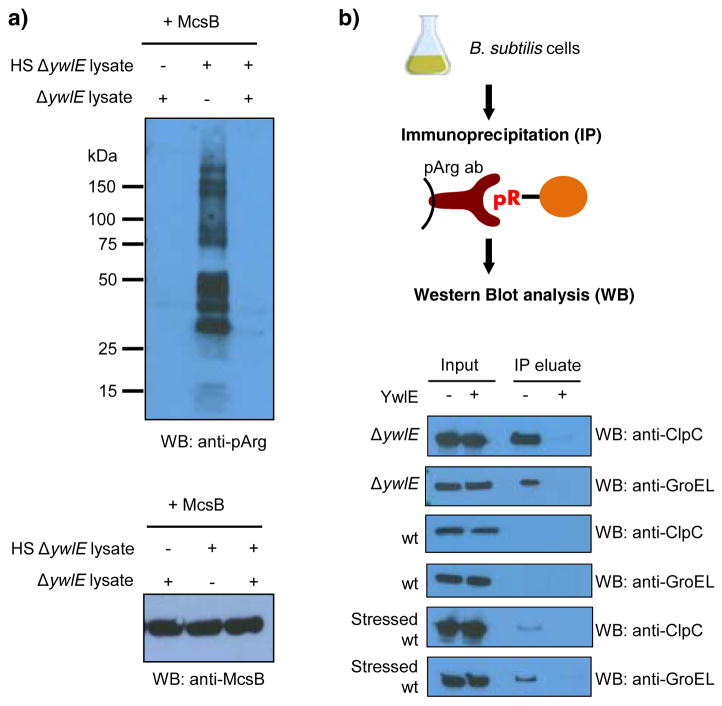
With this data in hand, we next tested the recognition of arginine phosphorylated proteins in lysates incubated with the protein arginine kinase McsB. Interestingly, McsB was barely active in non-stressed cell lysates, but shows strong phosphorylation activity in heat shocked lysates, as evidenced by the emergence of numerous arginine phosphorylated protein bands (Figure 3a). Surprisingly, McsB activity is also severely diminished in a sample containing a mixture of non-stressed and stressed lysates (Figure 3a, lane 3), indicating the presence for a potential inhibitor of McsB in non-stressed cell lysates.
Figure 3.
Detection of arginine phosphorylated proteins using the anti-pArg antibody in cellular extracts. a) Western Blot analysis of arginine phosphorylation in non-stressed and heat shocked (HS) cell lysates derived from B. subtilis ΔywlE strain incubated with recombinant McsB. For the loading control, equal McsB loading was verified by western blot analysis using anti-McsB antibody. b) Strategy to detect endogenous phosphoarginine in phosphoarginine containing proteins using immunoprecipitation. The results below show the presence or absence of immunoprecipitated, arginine phosphorylated ClpC and GroEL obtained from distinct cellular samples. Abbreviations: ΔywlE, B. subtilis ΔywlE strain; wt, B. subtilis wild type strain.
To further demonstrate the utility of our anti-pArg antibody, we next tested whether it could be used to enrich for proteins that are endogenously arginine phosphorylated. For these experiments, we grew a B. subtilis ΔywlE strain that is known to harbor arginine phosphorylated proteins,[9] and used the anti-pArg antibody to immunoprecipitate pArg containing proteins. As a proof of concept, we screened for the chaperone proteins ClpC and GroEL, which are known endogenous substrates of McsB mediated arginine phosphorylation.[9] As can be seen in Figure 3b, the anti-pArg antibody efficiently immunoprecipitates ClpC and GroEL. Importantly, both signals were lost upon pre-incubation of the lysates with recombinant arginine phosphatase YwlE. These results further confirm and highlight the specificity of this anti-pArg antibody assay (Figure 3b). Moreover, we were curious to determine whether we could detect arginine phosphorylated ClpC or GroEL in wild type cells. However, using the same assay setup we were unable to observe any signal for arginine phosphorylation of these chaperones. Since we had noticed that recombinant McsB activity is compromised in non-stressed lysates, we next tested whether the phosphorylation of these proteins was induced upon the addition of the cellular stressor diamide, which was previously shown to increase the level of arginine phosphorylation in a B. subtilis ΔywlE strain.[9b] For these studies, wild type B. subtilis cells were treated with diamide for 30 minutes, and then cells were lysed and the phosphorylation status of ClpC and GroEL monitored by immunoprecipitation and Western blotting. Notably, both proteins were detected using cell lysates derived from stressed wild type cells, while there was no signal in non-stressed cells (Figure 3b). We also performed the same experiment using the previously described phage display generated anti-pArg antibody. Notably, this low affinity antibody was not able to enrich these arginine phosphorylated proteins (Figure S20) highlighting the requirement for a high affinity antibody. Overall, these data indicate that arginine phosphorylation is a highly regulated PTM that is induced upon cellular stress in B. subtilis wild type cells.
In summary, we identified PO3-amidine as a suitable isostere of a phosphoarginine and used it to raise the first high affinity pan-phosphoarginine specific antibodies. Interestingly, the closely related SO3-amidine failed to elicit antibodies towards phosphoarginine. Although the overall geometry between SO3-amidine and PO3-amidine is very similar, the sulfonate group only contains a single formal negative charge whereas a phosphonate harbors two. It is interesting to note that subtle differences such as replacing a single sulfur atom by phosphorus in the hapten can have a great impact on antibody recognition and thus further highlights the selectivity of the generated antibody. Furthermore, we were able to employ this anti-pArg antibody to visualize arginine phosphorylated proteins in cell lysates and to detect endogenously arginine phosphorylated proteins. The biological applications of this anti-pArg antibody highlight the utility of this novel tool for cellular studies to analyze the physiological impact and regulation of protein arginine phosphorylation in diverse organisms. Moreover, we are currently analyzing the scope of SO3- and PO3-amidines as potential inhibitors of YwlE and McsB.
Supplementary Material
Acknowledgments
We thank Dr Ben Shen (Scripps Florida) for excellent support and providing access to high resolution mass spectrometry; and Dr. Vinayak Gupta (Scripps Florida) for technical assistance. This work was supported in part by NIH grant GM079357 (to P.R.T.), and by the Austrian Science Fund (FWF): J 3548-B21 (to J.F.).
Footnotes
Supporting information for this article is given via a link at the end of the document.
Contributor Information
Dr. Jakob Fuhrmann, Email: jfuhrman@scripps.edu.
Prof. Dr. Paul R. Thompson, Email: paul.thompson@umassmed.edu.
References
- 1.a) Hunter T. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]; b) Kobir A, Shi L, Boskovic A, Grangeasse C, Franjevic D, Mijakovic I. Biochim Biophys Acta Gen Subj. 2011;1810:989–994. doi: 10.1016/j.bbagen.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 3.a) Matthews HR. Pharmacol Ther. 1995;67:323–350. doi: 10.1016/0163-7258(95)00020-8. [DOI] [PubMed] [Google Scholar]; b) Attwood PV, Piggott MJ, Zu XL, Besant PG. Amino Acids. 2007;32:145–156. doi: 10.1007/s00726-006-0443-6. [DOI] [PubMed] [Google Scholar]; c) Besant PG, Attwood PV, Piggott MJ. Curr Protein Pept Sci. 2009;10:536–550. doi: 10.2174/138920309789630598. [DOI] [PubMed] [Google Scholar]; d) Kee JM, Villani B, Carpenter LR, Muir TW. J Am Chem Soc. 2010;132:14327–14329. doi: 10.1021/ja104393t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sickmann A, Meyer HE. Proteomics. 2001;1:200–206. doi: 10.1002/1615-9861(200102)1:2<200::AID-PROT200>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 5.a) Ruben EA, Chapman MS, Evanseck JD. J Am Chem Soc. 2005;127:17789–17798. doi: 10.1021/ja054708v. [DOI] [PubMed] [Google Scholar]; b) Denehy E, White JM, Williams SJ. Inorg Chem. 2007;46:8871–8886. doi: 10.1021/ic700687t. [DOI] [PubMed] [Google Scholar]
- 6.a) Hofmann FT, Lindemann C, Salia H, Adamitzki P, Karanicolas J, Seebeck FP. Chem Commun. 2011;47:10335–10337. doi: 10.1039/c1cc13341a. [DOI] [PubMed] [Google Scholar]; b) McAllister TE, Webb ME. Org Biomol Chem. 2012;10:4043–4049. doi: 10.1039/c2ob25517k. [DOI] [PubMed] [Google Scholar]; c) Kee JM, Oslund RC, Perlman DH, Muir TW. Nat Chem Biol. 2013;9:416–U428. doi: 10.1038/nchembio.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Bertran-Vicente J, Serwa RA, Schumann M, Schmieder P, Krause E, Hackenberger CPR. J Am Chem Soc. 2014;136:13622–13628. doi: 10.1021/ja507886s. [DOI] [PubMed] [Google Scholar]; e) Fuhs SR, Meisenhelder J, Aslanian A, Ma L, Zagorska A, Stankova M, Binnie A, Al-Obeidi F, Mauger J, Lemke G, Yates JR, 3rd, Hunter T. Cell. 2015;162:198–210. doi: 10.1016/j.cell.2015.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Fuhrmann J, Subramanian V, Thompson PR. ACS Chem Biol. 2013;8:2024–2032. doi: 10.1021/cb4001469. [DOI] [PubMed] [Google Scholar]
- 7.a) Wakim BT, Aswad GD. J Biol Chem. 1994;269:2722–2727. [PubMed] [Google Scholar]; b) Wakim BT, Grutkoski PS, Vaughan AT, Engelmann GL. J Biol Chem. 1995;270:23155–23158. doi: 10.1074/jbc.270.39.23155. [DOI] [PubMed] [Google Scholar]; c) Fuhrmann J, Clancy KW, Thompson PR. Chem Rev. 2015;115:5413–5461. doi: 10.1021/acs.chemrev.5b00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuhrmann J, Schmidt A, Spiess S, Lehner A, Turgay K, Mechtler K, Charpentier E, Clausen T. Science. 2009;324:1323–1327. doi: 10.1126/science.1170088. [DOI] [PubMed] [Google Scholar]
- 9.a) Elsholz AKW, Turgay K, Michalik S, Hessling B, Gronau K, Oertel D, Mader U, Bernhardt J, Becher D, Hecker M, Gerth U. Proc Natl Acad Sci U S A. 2012;109:7451–7456. doi: 10.1073/pnas.1117483109. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Schmidt A, Trentini DB, Spiess S, Fuhrmann J, Ammerer G, Mechtler K, Clausen T. Mol Cell Proteomics. 2014;13:537–550. doi: 10.1074/mcp.M113.032292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wozniak DJ, Tiwari KB, Soufan R, Jayaswal RK. Microbiology. 2012;158:2568–2576. doi: 10.1099/mic.0.060749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trentini DB, Fuhrmann J, Mechtler K, Clausen T. Mol Cell Proteomics. 2014;13:1953–1964. doi: 10.1074/mcp.O113.035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuhrmann J, Mierzwa B, Trentini DB, Spiess S, Lehner A, Charpentier E, Clausen T. Cell Rep. 2013;3:1832–1839. doi: 10.1016/j.celrep.2013.05.023. [DOI] [PubMed] [Google Scholar]
- 13.Freedman LD, Doak GO. Chemical reviews. 1957;57:479–523. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.