Abstract
The murine double-stranded RNA-binding protein RAX and the human homolog PACT were originally characterized as activators of protein kinase R (PKR). Mice deficient in RAX show reproductive and developmental defects, including reduced body size, craniofacial defects and anterior pituitary hypoplasia. As these defects are not observed in PKR-deficient mice, the phenotype has been attributed to PKR-independent activities of RAX. Here we further investigated the involvement of PKR in the physiological function of RAX, by generating rax−/− mice deficient in PKR, or carrying a kinase-inactive mutant of PKR (K271R) or an unphosphorylatable mutant of the PKR substrate eIF2α (S51A). Ablating PKR expression rescued the developmental and reproductive deficiencies in rax−/− mice. Generating rax−/− mice with a kinase-inactive mutant of PKR resulted in similar rescue, confirming that the rax−/− defects are PKR dependent; specifically that the kinase activity of PKR was required for these defects. Moreover, generating rax−/− mice that were heterozygous for an unphosphorylatable mutant eIF2α provides partial rescue of the rax−/− defect, consistent with mutation of one copy of the Eif2s1 gene. These observations were further investigated in vitro by reducing RAX expression in anterior pituitary cells, resulting in increased PKR activity and induction of the PKR-regulated cyclin-dependent kinase inhibitor p21WAF1/CIP1. These results demonstrate that PKR kinase activity is required for onset of the rax−/− phenotype, implying an unexpected function for RAX as a negative regulator of PKR in the context of postnatal anterior pituitary tissue, and identify a critical role for the regulation of PKR activity for normal development.
Keywords: Prkra, Eif2ak2, Development, Hypoplasia, Cell cycle
Introduction
The Prkra gene encodes a double-stranded RNA-binding protein, identified and named independently as protein activator of the double-stranded RNA-dependent protein kinase (PKR) (PACT) in human [1], and PKR-associated protein X (RAX) in mouse [2]. Several key functions of PACT/RAX have been reported in addition to its role in PKR regulation, including a role in the production of small RNAs involved in RNA silencing [3–6], and in the innate immune response through modulating the activities of the retinoic acid inducible gene I [7]. Mutations in Prkra are also associated with Dyt16, a young-onset dystonia-parkinsonism disorder [8, 9], and more recently demonstrated with pure dystonia [10]. Early studies of this protein were focused on its ability to activate PKR in response to various stresses [2, 11–18]. Upon stress, PACT/RAX is phosphorylated, leading to interaction with and subsequent activation of PKR [19–23]. Activation of PKR promotes cell signal transduction [24, 25.] and results in inhibition of protein translation [26–28], which if sustained induces cell cycle arrest [29–31] and apoptosis [32, 33]. Although the function of PACT/RAX as an activator of PKR and retinoic acid inducible gene I, and its role in small RNA biogenesis have been previously described, its involvement in dystonia-parkinsonism and the overall physiological function of the protein are less clear.
Defects resulting from disruption of the rax gene in mice (Prkratm1Gsc, rax−/−) demonstrated the involvement of the protein in craniofacial and ear development, growth and fertility [34]. These phenotypes were duplicated in an ethylnitrosyl urea mutagenesis screen that generated a mutation (S130P) in the rax gene (Prkrarep), confirming the role of RAX in development [35]. Further investigation revealed that RAX-deficient mice developed hypoplastic anterior pituitaries resulting from reduced postnatal proliferation, and that all five hormone-producing lineages of the anterior pituitary are affected [36]. These anterior pituitary cells secrete hormones required for growth and sexual development; as such, their reduced proliferation and subsequent reduction in hormone levels has been assumed to be the cause of the developmental and reproductive anomalies observed in RAX-deficient mice [36].
In contrast to the rax−/− animal, two independent lines of mice with disruptions in the PKR gene (Eif2ak2tm1Cwe and Eif2ak2tm1Jcbe) have no overt developmental defects [37, 38], implicating RAX in physiological functions independent of PKR. Given the pituitary proliferation defect, the well-established cell-cycle and proliferation effects of PKR and the biochemical interaction between PACT/RAX and PKR, we sought to further investigate the role of PKR in the physiological function of RAX and its role in postnatal anterior pituitary proliferation by generating rax−/− animals that were also mutant for PKR and its substrate eIF2α. We also sought to develop a tractable cell culture model to test the role of RAX in anterior pituitary hypoplasia.
Results and Discussion
Eliminating PKR or PKR kinase activity rescues the developmental and reproductive deficiencies and anterior pituitary hypoplasia observed in rax−/− mice
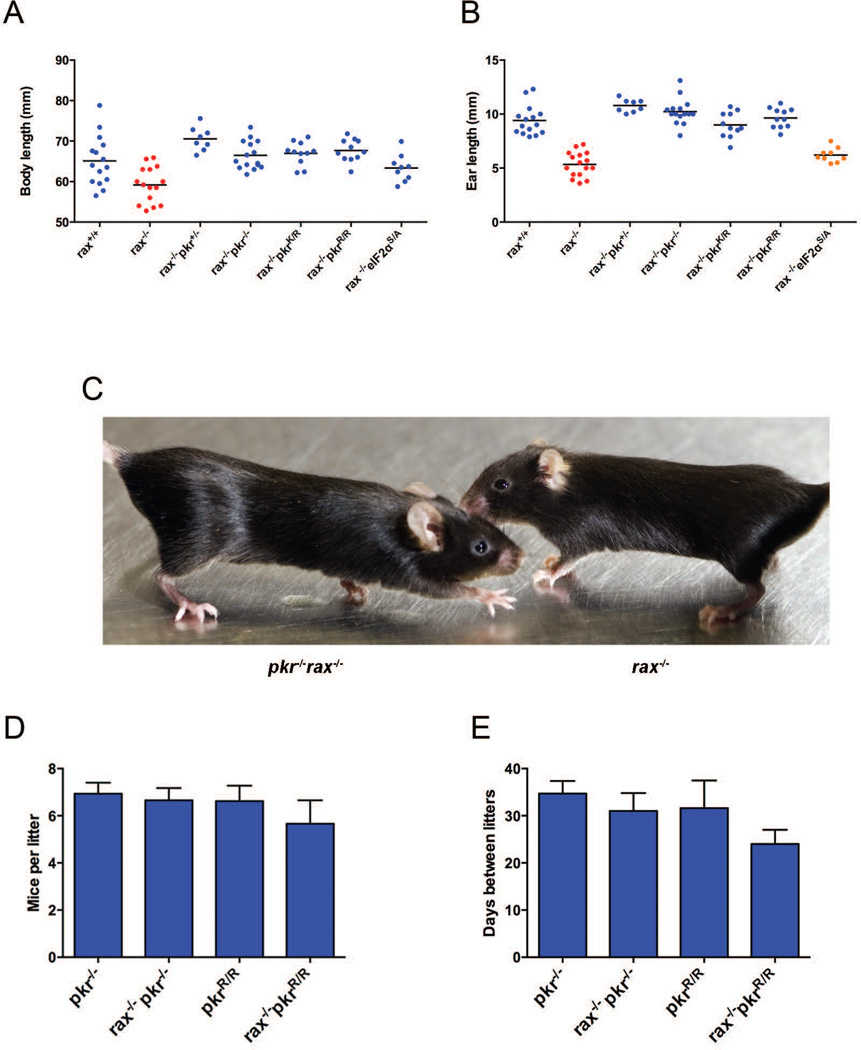
To investigate the role of PKR in the RAX-deficient mouse phenotype, rax−/−pkr−/− mice were generated. Moreover, to specifically assess the contribution of PKR kinase activity to the phenotype of rax−/− mice, PKR was replaced with the kinase-inactive K271R mutant (pkrR/R) [39–41] in rax−/− mice. Remarkably, both transgenic lines of mice (rax−/−pkr−/− and rax−/−pkrR/R) developed normally, showing complete rescue of body length (nose to tail base) (Figure 1A and C and Table 1) and ear size (helix to tragus) (Figure 1B and C and Table 1). This rescue was also observed in rax−/− mice that were heterozygous for pkr (rax−/−pkr+/−) and for the kinase-inactive mutant (rax−/−pkrK/R). Importantly, combined data from a wide age range of mice of both sexes shown in Figure 1 is convincing and rescue was as pronounced when analysis was restricted to specific ages and/or sexes of mice. The profound fertility defects seen in rax−/− mice were also wholly rescued, as litter sizes and intervals between consecutive litters for dams from rax−/−pkr−/− and rax−/−pkrR/R lines were equivalent to those seen for the pkr−/− and pkrR/R mice (Figure 1, D and E) and were statistically indistinguishable from those reported for the C57Bl/6 genetic background upon which the RAX and PKR mutations were introduced [42, 43]. Furthermore, rax−/−pkr+/−, rax−/−pkr−/−, rax−/−pkrK/R and rax−/−pkrR/R mice were born in the expected Mendelian frequencies from heterozygous intercrosses, and rax−/−pkr−/− and rax−/−pkrR/R intercrosses produced viable offspring that were used to continuously maintain these lines.
Figure 1.
Investigation of the rax−/− developmental phenotype in rax−/−pkr−/− and rax−/−pkrR/R mice. (A) Body length measured from nose to tail base or (B) ear size measured from helix to tragus in adult male and female mice. Data points show individual mice and lines indicate mean. p-values were determined by unpaired two-tailed t-test; see Table 1 for statistical analysis. In both (A) and (B), blue data points indicate statistically similar (p > 0.05) to wild type, red indicates statistically distinct from wild type (p < 0.05) and statistically similar (p > 0.05) to rax−/−, orange indicates statistically distinct (p < 0.05) from both wild type and rax−/− (intermediate phenotype). (C) rax−/− and pkr−/−rax−/− mice were photographed to illustrate phenotypic rescue. Fertility parameters, including (D) mice per litter and (E) intervals between consecutive litters for individual dams, were also determined. Data shown are mean ± SEM and were obtained from a minimum of three litters from at least four independent breeding pairs per genotype. No statistical difference (p > 0.05) was found between genotypes by one-way ANOVA or by comparing pairs of genotypes by unpaired two-tailed t-test.
Table 1.
Statistical analysis of the effect of mutating PKR or eIF2α for the developmental defects measured in the rax−/− mice.
| Body length | ||||||||
|---|---|---|---|---|---|---|---|---|
| p values | rax+/+ | rax+/− | rax−/− | rax−/−pkr+/− | rax−/−pkr−/− | rax−/−pkrK/R | rax−/−pkrR/R | rax−/−eIF2αS/A |
| rax+/+ | N/A | 0.8767 | 0.0054 | 0.029 | 0.4631 | 0.3637 | 0.2081 | 0.4392 |
| rax−/− | 0.0054 | 0.0005 | N/A | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | 0.0246 |
| Ear length | ||||||||
| p values | rax+/+ | rax+/− | rax−/− | rax−/−pkr+/− | rax−/−pkr−/− | rax−/−pkrK/R | rax−/−pkrR/R | rax−/−eIF2αS/A |
| rax+/+ | N/A | 0.2554 | <0.0001 | 0.013 | 0.0871 | 0.4241 | 0.6198 | <0.00001 |
| rax−/− | < 0.0001 | < 0.0001 | N/A | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | 0.0486 |
p values derived from an unpaired two-tailed t-test of body length and ear size between indicated groups. Shaded cells indicate p < 0.05.
We extended these transgenic mouse results by investigating the requirement for phosphorylation of the most well studied substrate of PKR, the α subunit of eukaryotic initiation factor 2 (eIF2α). To this end, we generated rax−/− mice that also carried an unphosphorylatable mutant of eIF2α, S51A. As this mutation causes early neonatal lethality, mutant eIF2α animals must be maintained as heterozygotes [44], therefore only a partial removal of eIF2α phosphorylation is attainable. RAX-deficient mice that are also heterozygous for the S51A mutation (rax−/−eif2αS/A) show statistically significant improvement of development as assessed by measures of body (Figure 1A and Table 1) and ear length (Figure 1B and Table 1). These results demonstrate that consistent with a requirement for PKR kinase activity, phosphorylation of eIF2α is required for the rax−/− developmental phenotypes. Interestingly, fertility was not restored in these mice, suggesting either eIF2α haplosufficiency, or that alternative PKR substrates are responsible for the reproductive defects in rax−/− mice.
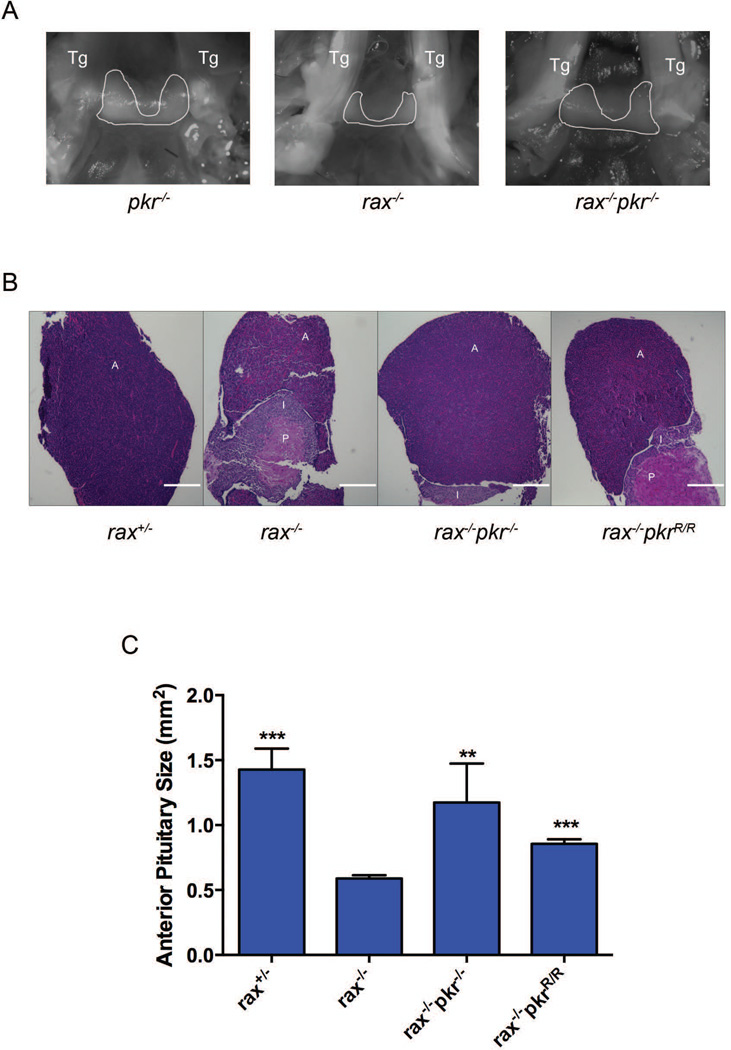
Many of the organism-level developmental defects observed in rax−/− mice have been attributed to defective anterior pituitary development in these animals [36]. The anterior pituitary undergoes rapid proliferation in neonatal mice, thereby expanding the cells that produce factors required for growth and reproduction (reviewed in [45]). To determine whether rescue of the developmental defects seen in rax−/−pkr−/− and rax−/−pkrR/R mice correlated with rescued pituitary development, the anterior pituitary in these mice was analyzed. Macroscopic analysis of the pituitary revealed a smaller anterior pituitary in rax−/− mice compared with pkr−/− or rax−/−pkr−/− controls (Figure 2A). This was recapitulated with subsequent histological analysis of pituitaries. Hematoxylin and eosin staining of pituitary sections from rax−/− mice showed a reduced anterior pituitary and rescue of this defect in rax−/−pkr−/− and rax−/−pkrR/R mice (Figure 2B, quantified in Figure 2C). Consistent with previous reports characterizing the RAX-deficient mouse phenotype, posterior pituitary size was not affected by removal of RAX or PKR, and removal of PKR in a wild-type background did not affect anterior pituitary phenotype [37, 38].
Figure 2.
Histological comparison of pituitary from wild-type, rax−/−, rax−/−pkr−/− and rax−/−pkrR/R mice. (A) Pituitaries were photographed in situ in mice during dissection (2× magnification). Trigeminal nerves that flank the pituitary are marked as Tg. The anterior pituitary is outlined in white. Hematoxylin and eosin stained longitudinal sections of pituitary from mice of the indicated genotypes were photographed at 5× magnification (B – representative samples shown) and the size of the anterior pituitary was calculated from micrographs (C). Scale bar indicates 0.1 mM. A, I and P indicate anterior, intermediate and posterior lobes of the pituitary, respectively. Data shown are mean ± SEM of multiple sections from at least two mice. p values were calculated relative to rax−/− using an unpaired two-tailed t-test. *** indicates p < 0.0001, ** indicates p < 0.01.
These compelling transgenic animal data do not correlate with the previously reported role for RAX as an inducer of PKR activity established in vitro, and the correlated assumption that the absence of a conspicuous developmental defect in pkr−/− mice precludes its involvement in the rax−/− developmental and reproductive defects. Rather, rescue of the defects of the RAX-ablated animal by removing PKR expression or kinase activity is consistent with RAX functioning as a PKR inhibitor. Bearing this in mind, it is interesting to compare the developmental phenotypes in rax−/− mice, and mice lacking the Protamine-1 (Prm-1) RNA-binding protein (PRBP, the mouse homolog of human TAR-RNA binding protein 2 (TARBP2)), which inhibits PKR activation and shares a double-stranded RNA-binding domain architecture with RAX. Notably, these mice (Tarbp2tm1Reb) do not demonstrate the craniofacial and ear malformations that are present in rax−/− mice, so the phenotypes are not equivalent. Pituitary hypoplasia was not investigated; however, prbp−/− mice also have reductions in body size and reproductive defects [46]. Interestingly, the reproductive defect in male prbp−/− mice was demonstrated to be due to a disruption of spermatid differentiation that was attributed to PRBP-dependent control of translation. In light of our data and the established role of PRBP as a PKR inhibitor, it would be interesting to test involvement of PKR in the prbp−/− phenotype and to investigate anterior pituitary development in these animals.
We note that another study has reported inhibition of PKR by PACT in the context of HIV infection, although this study consisted of an in vitro infection model in cells overexpressing PACT [47]. Here we describe for the first time evidence that endogenous RAX negatively regulates PKR in the physiologically relevant context of mouse development. This predicts inappropriate activation of PKR in the absence of RAX; however, our efforts to measure PKR activity in the pituitary by measuring autophosphorylation of PKR itself or phosphorylation of eIF2α were inconclusive. As such transient and likely developmentally dependent effects are difficult to capture in vivo, we sought to confirm this role of RAX in a tractable pituitary cell model.
RAX knockdown in anterior pituitary cells results in a static cell cycle distribution
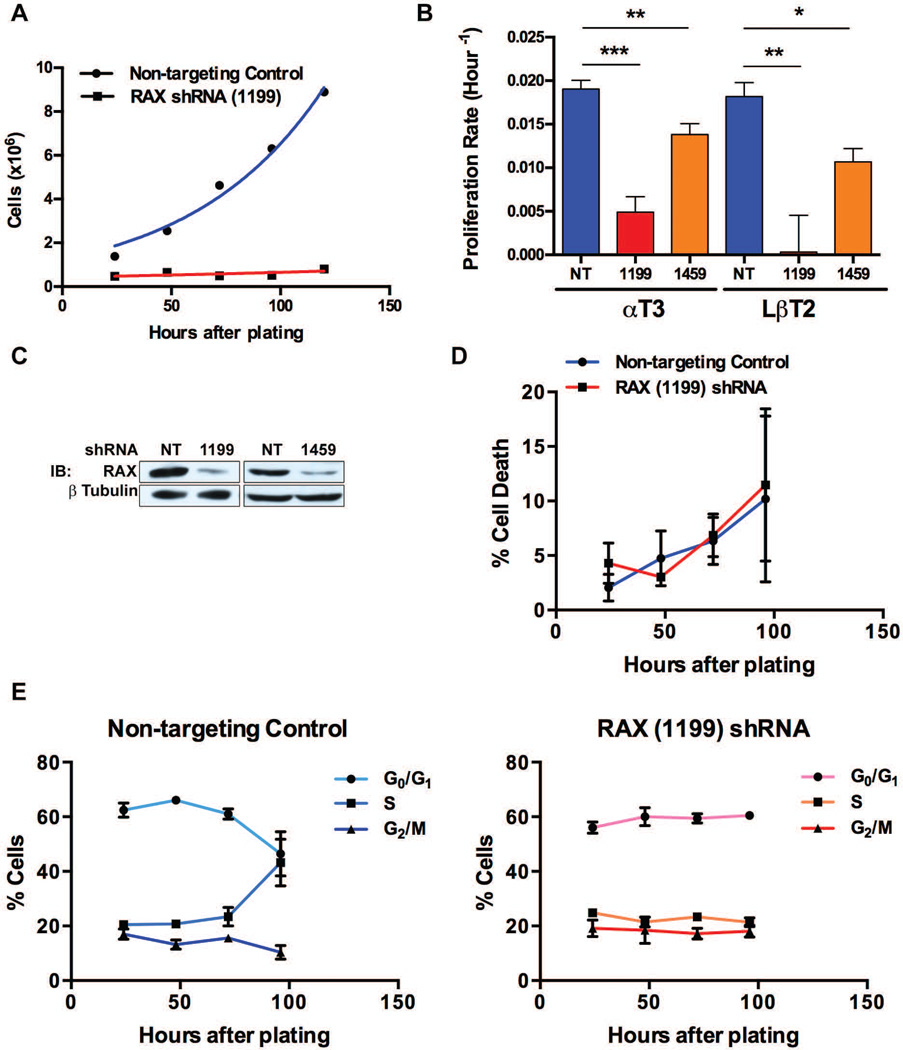
In order to directly investigate the underlying mechanism of our unexpected discovery in rax−/−pkr−/− mice, we sought to build on preliminary in vitro modeling of the RAX-deficient phenotype by Peters et al. [36], who investigated the proliferation defect both in rax−/− mice in vivo and by using small interfering RNA to knockdown RAX in vitro in the mouse gonadotroph cell line LβT2 [48] and the rat somatolactotroph cell line GH3 [49]. Impaired proliferation of each hormone-secreting cell-lineage has been seen in mouse anterior pituitary [36]; therefore we focused on a single lineage using the mouse gonadotroph cell lines αT3 [50] and LβT2 to investigate this observation in vitro. To investigate the function of RAX in anterior pituitary proliferation, we refined previous approaches by using lentivirus-delivered RAX short hairpin RNAs (shRNAs) to allow selection of transduced cells in the mouse pituitary cell lines. To establish this method, αT3 cells were infected with a lentivirus expressing RAX shRNA (1199), which targets the 3’ untranslated region of RAX, or a non-targeting control shRNA lentivirus. Following puromycin selection for shRNA expression, cells were plated in replicate wells then trypan blue stained and counted every 24 h to determine the number of viable cells. Introduction of RAX (1199) shRNA resulted in a reduction of αT3 cells compared to control cells over time (Figure 3A). This initial experiment (and all subsequent proliferation experiments) was repeated using the Celltiter96 cell proliferation assay (Promega) with a second independent shRNA, RAX shRNA (1459), that also targets the 3’ untranslated region of RAX, and was repeated in the second pituitary cell line LβT2. The proliferation rate, calculated by fitting growth kinetics data to an exponential growth equation, was significantly reduced following expression of either shRNA and in αT3 and LβT2 cells (Figure 3B). These experiments were repeated and data from at least three replicates are shown in Table 2. Reduced RAX levels were confirmed following selection by western blotting for RAX (Figure 3C). These data are in agreement with Peters et al. [36], demonstrating reduced proliferation following RAX knockdown in a cell culture model of the anterior pituitary hypoplasia observed in RAX-deficient mice. This validates the previously reported model and suggests that an intrinsic cellular function of RAX regulates proliferation, which is important considering the complex endocrine signaling involved in pituitary development.
Figure 3.
Cell cycle distribution of RAX knockdown αT3 cells. (A) αT3 cells expressing an shRNA construct targeting RAX (1199) or a non-targeting control (NT) were plated following 72 h of puromycin selection and counted at 24-h time points. (B) Proliferation of αT3 and LβT2 cells stably expressing one of two RAX shRNAs (1199 or 1459) or a non-targeting control was determined as in (A); data were then fit to the equation Y = Y0eKT, where T is hours after plating, Y is cell number (or absorbance) and Y0 is starting cell number (or absorbance), to determine the proliferation rate K. Results are shown as mean ± SEM, αT3 NT (n=7), 1199 (n=5), 1459 (n=5), LβT2 NT (n=4), 1199 (n=4), 1459 (n=3); * denotes p < 0.05, ** denotes p < 0.01, *** denotes p < 0.0001. See Table 2 for statistical analysis. (C) αT3 cells expressing one of two RAX shRNAs (1199 or 1459) or a non-targeting control were analyzed for RAX expression by western blot following 72 h of puromycin selection (T=0 h in time course shown in (A)). (D) Cells from the experiment shown in (E) were stained with trypan blue to determine viability. Results are shown as mean ± SEM (n=3). (E) αT3 cells expressing RAX (1199) shRNA or a non-targeting control were selected with puromycin and stained with PI at 24-h time points to measure DNA content and cell cycle distribution by flow cytometry. Percentage of cells in G0/G1, S and G2/M phases are shown as mean ± SEM (n=3).
Table 2.
Statistical analysis of proliferation rates in RAX knockdown cell lines.
| Cell line | shRNA | Proliferation
rate (h−1, mean ± SEM) |
p value (compared to NT) |
|---|---|---|---|
| Non-targeting control | 0.01903 ± 0.00102 | – | |
| αT3 | RAX (1199) | 0.00489 ± 0.00180 | < 0.0001 |
| RAX (1459) | 0.01381 ± 0.00126 | 0.0089 | |
| Non-targeting control | 0.01815 ± 0.00163 | – | |
| LβT2 | RAX (1199) | 0.00032 ± 0.004215 | 0.0076 |
| RAX (1459) | 0.01067 ± 0.00154 | 0.0233 |
Proliferation rate of αT3 and LβT2 cells stably expressing one of two RAX shRNAs (1199 or 1459) or non-targeting control (Figure 3B). Results are shown as mean ± SEM, αT3 NT (n=7), 1199 (n=5), 1459 (n=5), LβT2 NT (n=4), 1199 (n=4), 1459 (n=3). p values are derived from an unpaired two-tailed t-test of RAX shRNA infected cells compared to corresponding non-targeting controls.
To begin characterizing the proliferation defect seen in RAX knockdown pituitary cells, we first analyzed their cell cycle distribution. αT3 cells were infected with RAX shRNA (1199) lentivirus, puromycin selected and split into replicate wells. At 24-h intervals following puromycin selection, cells were collected to determine the DNA content by propidium iodide (PI) staining and were trypan blue stained to determine viability. Consistent with previous reports [36], there was no increase in cell death following RAX knockdown (Figure 3D). There was also no accumulation of cells in a specific phase of the cell cycle in RAX shRNA expressing cells. Cells expressing the non-targeting control, however, had a dynamic cell cycle distribution shifting towards S-phase over time following puromycin selection, while RAX knockdown cells showed a static distribution (Figure 3E).
RAX knockdown results in upregulation of the cyclin-dependent kinase inhibitor p21WAF1/CIP1
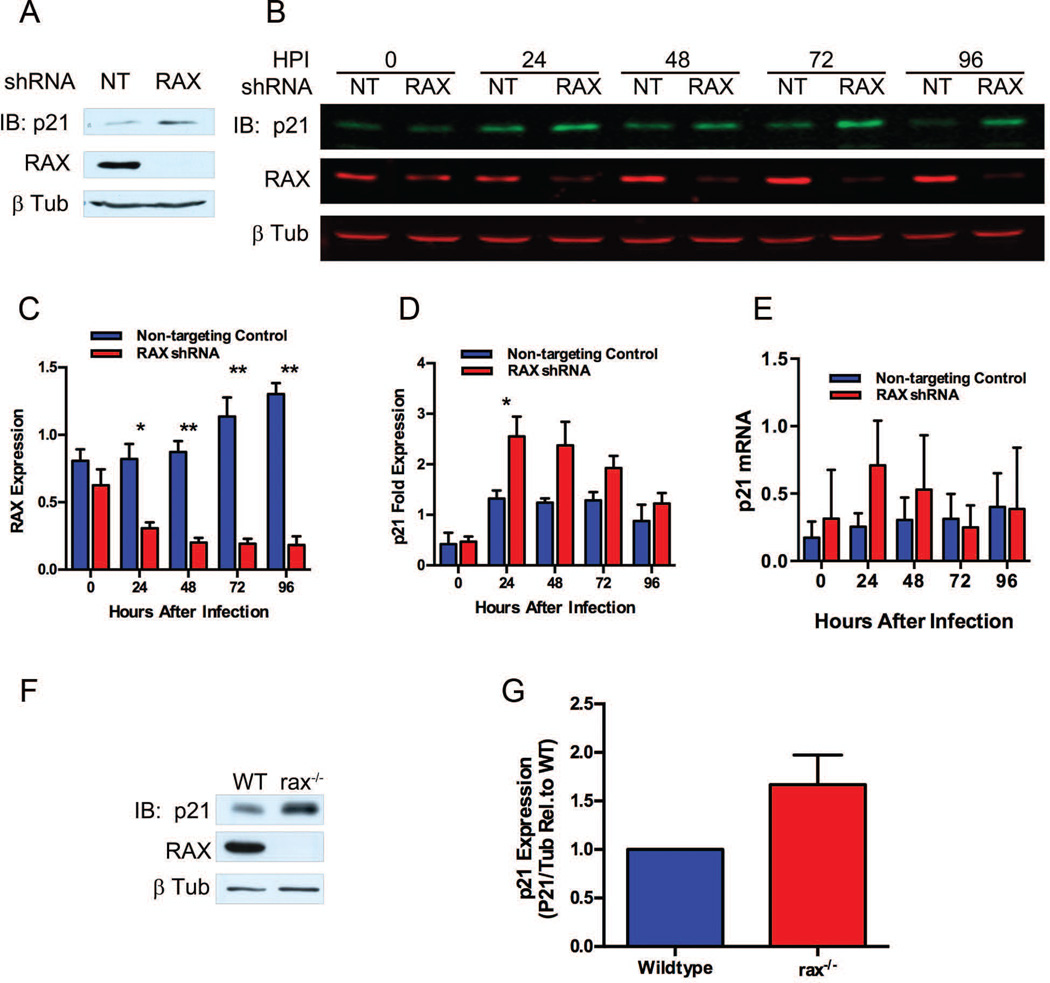
Activation of PKR has been shown to upregulate the cell-cycle inhibitor p21WAF1/CIP1 [31, 51], which can arrest the cell cycle in the G1 to S [52, 53], S to G2 [54] and G2 to M transitions [55]. A simultaneous block in transition between each of these phases is consistent with our cell cycle distribution data showing stasis without accumulation in any specific phase of the cell cycle in RAX knockdown cells (Figure 3E). Accordingly, we investigated p21WAF1/CIP1 expression levels in non-targeting control or RAX (1199) infected cells. Figure 4A demonstrates an increase in p21WAF1/CIP1 in RAX knockdown cells by western blot following 72 h of puromycin selection, consistent with the observed cell cycle defect. In order to rule out the possibility that the p21WAF1/CIP1 induction is a byproduct of puromycin selection, pituitary cells were infected with lentivirus at a high multiplicity of infection, eliminating the need for selection. Following infection at 3 transducing units (TU)/cell, protein lysates and RNA were harvested at 24-h intervals and subjected to quantitative western blot and to real-time PCR of RAX and p21WAF1/CIP1 protein and RNA, respectively. By 24 h post-infection, there is a clear reduction in RAX protein levels (Figure 4B and C) concurrent with an increase in p21WAF1/CIP1 protein (Figure 4B and D) of 2.555 fold (± 0.388 SEM, n=3) compared to control cells (p=0.0426). Measures of the levels of the p21WAF1/CIP1 transcript show there was no significant difference (p=0.0855, n=3) between cells with normal or reduced RAX expression (Figure 4E), indicating that p21 levels are regulated post-transcriptionally.
Figure 4.
p21WAF1/CIP1 expression following RAX knockdown. (A)Western blot of p21WAF1/CIP1 in shRNA-expressing αT3 cells following 72 h of puromycin. (B) Quantitative Odyssey® western blot of RAX and p21WAF1/CIP1 in αT3 cells infected with 3 TU/cell RAX (1199) shRNA or a non-targeting control (NT). (C) RAX expression level was quantitated from (B). (D) p21WAF1/CIP1 expression level was quantitated from (B). Data in (C) and (D) are shown as RAX or p21WAF1/CIP1 signal normalized to β tubulin relative to the average normalized signal from non-targeting control samples and shown as mean ± SEM (n=3); * denotes p < 0.05, ** denotes p < 0.01. (E) p21WAF1/CIP1 mRNA was quantified by real-time RT-PCR from cells used in (B). Data are shown as p21WAF1/CIP1 normalized to 18S rRNA. Results are shown as mean ± SEM (n=3). (F) Representative western blot of p21WAF1/CIP1 in WT and rax−/− pituitary from neonatal (14-day-old) mice. (G) Quantitation of western blots from WT and rax−/− pituitary from neonatal (14-day-old) mice, shown as mean ± SEM p21WAF1/CIP1 signal normalized to loading control, relative to wild type for each blot (n=6).
To test whether p21WAF1/CIP1 expression is increased in vivo, we assessed 14-day-old neonates, as this stage coincides with both increased RAX expression in wild-type mice and the onset of pituitary hypoplasia in rax−/− mice [36]. Pituitaries were harvested and p21WAF1/CIP1 expression analyzed by western blot. There was an increase in p21WAF1/CIP1 in rax−/− pituitary (Figure 4F); although not statistically significant (Figure 4G, p = 0.0794), this is consistent with observations from our in vitro experiments. This lack of statistical significance in the increase in p21WAF1/CIP1 expression in vivo may be a consequence of small age differences (less than 24 h) in the animals compared, particularly as RAX levels are rapidly changing in neonatal mice [36]. It is likely that a transient requirement for RAX in early postnatal pituitary proliferation defines a narrow temporal window in which to observe molecular characteristics of this phenotype directly, such as p21WAF1/CIP1 expression and PKR activation. In this case, such effects can be more reliably observed directly in vitro, without the variation between individual mice that may differ in age by up to 24 h.
As cell-cycle control is tightly regulated, it is unsurprising that p21WAF1/CIP1 is regulated at the transcriptional, post-transcriptional and post-translational levels. Several reported substrates of PKR (eIF2α, p53, the B56α regulatory subunit of protein phosphatase 2A, nuclear factor associated with RNA 1 and 2 (NFAR1/2), glycogen synthase kinase 3β (GSK-3β)) converge on regulation of p21WAF1/CIP1 at each of these levels. The lack of a detectable increase in p21WAF1/CIP RNA in RAX-deficient cells would appear to exclude p53 as the likely mechanism. At the post-transcriptional level, the p21WAF1/CIP1 mRNA is stabilized by NFAR [56, 57], which is regulated by PKR [58]. Expression of p21WAF1/CIP1 is controlled post-translationally through phosphorylation of certain residues, which inhibits the normally rapid protein degradation. Phosphorylation of these residues occurs downstream of c-Myc [59–61]. The activities of both B56α [62] and GSK-3β [63] decrease the stability of c-Myc, which would in turn increase the stability of p21WAF1/CIP, although GSK-3β can also phosphorylate p21WAF1/CIP directly to decrease protein stability [64]. The canonical activity of PKR, eIF2α phosphorylation, has also been shown to increase p21WAF1/CIP1 expression [65, 66]. This response also regulates AKT activity [67], which phosphorylates p21WAF1/CIP1, altering subcellular localization [68], which modulates its antiproliferative effects [66, 69].
While we see at least partial rescue of developmental defects in rax−/−eIF2αS/A mice, indicating that eIF2α phosphorylation is important in the RAX-deficient phenotype, we cannot discount the involvement of additional PKR substrates. As we were not able to reliably observe rescue of p21WAF1/CIP1 protein levels in rax−/−pkr−/− mice, it is possible that there are also PKR-independent effects, and that a “two-hit” model of both upregulation and activation of p21WAF1/CIP1 is required for the rax−/− phenotype. In this case, removal of PKR could remove the activation signal, while any PKR-independent upregulation of p21WAF1/CIP1 would go unnoticed in the overt phenotype. Such a scenario might account for the unchanged levels of p21WAF1/CIP1 protein in a mouse with phenotypic rescue. Alternatively, the RAX-dependent regulation of p21WAF1/CIP1 observed in cell lines may be immaterial in vivo. Additional analysis is required to establish the precise mechanism through which PKR activity induces the developmental defects in rax−/− mice.
RAX knockdown promotes PKR activation
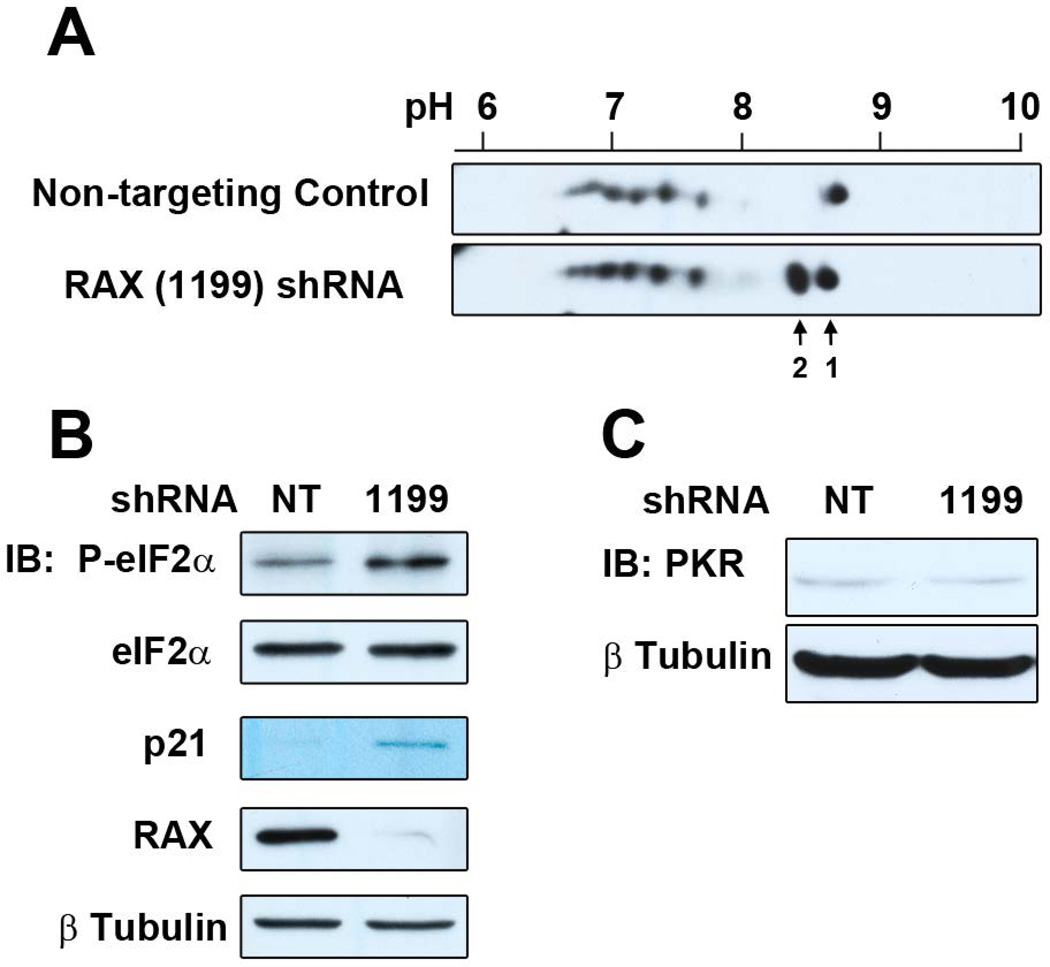
Activation of PKR proceeds by phosphorylation of multiple sites, for which only a few have site-specific phospho-antibodies. Therefore, to determine whether PKR is activated following RAX knockdown, we measured the isoelectric point (pI) of total PKR by two-dimensional western blot to assess the phosphorylation state of PKR. This method enables an unbiased assessment of post-translational modification of PKR and is potentially more sensitive than phosphorylated-residue specific PKR antibodies developed for investigating activation in the context of viral infection, during which PKR is both upregulated and strongly autophosphorylated. In addition, phosphorylation of eIF2α was probed by western blot using a phosphorylation-specific antibody. αT3 cells were infected with RAX (1199) shRNA or non-targeting control lentivirus (3 TU/cell) and protein samples were harvested 24 h post-infection for analysis of PKR and eIF2α phosphorylation. These data demonstrate a unique pI shift in the population of PKR molecules from cells with reduced RAX expression (Figure 5A, labeled spot 2, compared to unmodified PKR, labeled spot 1) concurrent with an increase in eIF2α phosphorylation (Figure 5B). The modified pI of PKR and associated increase in eIF2α phosphorylation observed in RAX knockdown αT3 cells substantiates the data from transgenic studies, which indicate that removal of RAX results in activation of PKR. Altered PKR levels would provide an alternative explanation for the increase in PKR activity without contradicting the well-established function of RAX as a PKR activator, particularly given the role of PACT/RAX in post-transcriptional gene regulation via micro RNAs. It is important to note, however, that we did not observe an increase in PKR protein levels in RAX knockdown cells (Figure 5C).
Figure 5.
PKR activation following RAX knockdown. (A) Protein lysates from αT3 cells infected with 3 TU/cell of either non-targeting control (NT) or RAX (1199) shRNA lentivirus were separated by isoelectric focusing using a pH 3–10 gradient prior to second-dimension SDS-PAGE and western blot for PKR (using Santa Cruz D-20 PKR antibody). Image has been cropped to the pH range between 6 and 9, and the expected molecular weight of PKR (~65 kDa). Arrow labeled 1 denotes the predicted pI of unmodified PKR (8.57); arrow labeled 2 denotes the shifted population. (B) Western blot for P-eIF2α of cells used for two-dimensional western blot in (A). (C) Western blot of total PKR from NT or RAX (1199) infected αT3 cells.
The observed shift in pI (~8.6 to ~8.3) in the absence of RAX corresponds to the predicted change of a single phosphorylation. However, this change has not been verified as phosphorylation and therefore could be due to a different post-translational modification. In either case, this indicates a low-level or alternate activation state of the kinase, rather than full activation arising from phosphorylation of several residues. Of particular interest are studies demonstrating tyrosine phosphorylation of PKR and phosphorylation by upstream kinases [70–72]. Keeping this in mind, the altered pI observed in RAX knockdown cells could indicate that RAX blocks activation of PKR by upstream regulatory pathways, rather than blocking autophosphorylation. In this way, RAX could function as an inhibitor of PKR activation by alternate mechanisms, while still functioning as an activator of conventional PKR autophosphorylation upon appropriate stress signals. Alternatively, RAX could promote dephosphorylation of specific PKR residues by phosphatases. This speculation could account for the opposing roles for RAX as an activator (previously established) or an inhibitor of PKR as demonstrated here. That PKR is involved in a variety of cell signaling responses implies that the kinase could be activated by alternative mechanisms. We present here the first description of the involvement of PKR in development. This aspect of PKR function has not previously been incorporated into our understanding of the physiological function of the kinase.
Anterior pituitary cells are amongst a select subset expressing particularly high levels of PACT/RAX [1], and are also the only cells in which we see a specific proliferation defect. There are many proteins involved in the cellular functions of PKR, and expression levels of these proteins may differ between different cell types. Therefore, the shift from the canonical PKR activation function of RAX to negative regulation of PKR activity may depend upon the stoichiometric ratios of RAX, PKR and other binding partners, such as TRBP/PRBP [4, 73, 74], DICER [3, 4], and other PKR-interacting proteins such as Hsp70, Hsp90 and p58IPK [75–77], as well as the relative expression levels of each of the downstream substrates of PKR. The phosphorylation status of PACT/RAX has also been demonstrated to alter its affinity for PKR and other PKR-interacting proteins. Therefore, the relative levels and activity of PACT/RAX kinase(s) and phosphatase(s) may influence this effect as well. It is likely that the relative expression level of each of the many proteins involved in this pathway fine-tunes the PKR response in a cell-type and context-dependent manner. While the precise mechanism of hypoplasia downstream of PKR requires further investigation, these results unequivocally demonstrate that aberrant PKR kinase activity results in the RAX-deficient phenotype, identifying a function for RAX as a negative regulator of PKR activity in the context of anterior pituitary development, thereby establishing the importance of controlling PKR activity and that regulation of PKR by RAX is critical for normal development.
Materials and Methods
Plasmids, cell lines, antibodies, mice and reagents
Mouse L929, αT3 and LβT2, and human HT1080 and HEK293T cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (Atlanta Biologicals, Atlanta, GA, USA) supplemented with glucose (4.5 g/l), penicillin (50 U/ml), streptomycin (50 μg/ml), L-glutamine (2 mM) and sodium pyruvate (1 mM) (henceforth referred to as complete DMEM – all additives were from Gibco, Gaithersburg, MA, USA). All cell culture incubations were at 37°C in 5% CO2 in a humidified incubator. Hexadimethrine bromide (polybrene) was obtained from Sigma-Aldrich (St. Louis, MO, USA) and prepared as an 8 mg/ml solution in water. pVSG-G was obtained from Clontech (Mountain View, CA, USA), pCMV-R8.74 was obtained from Addgene (Cambridge, MA, USA), pLKO.1-puro and pLKO.1 non-targeting control were obtained from Sigma-Aldrich. The following primary antibodies were used in 0.5% dry skim milk in TBS-Tween unless otherwise noted: full-length PACT/RAX [78], β-actin (Clone AC-15, Sigma-Aldrich, Catalog # A1978), P-eIF2α (Ser52) (Invitrogen, Carlsbad, CA, USA, Catalog # 44–728; membranes using P-eIF2α were blocked and probed in 5% BSA), eIF2α (Cell Signaling, Danvers, MA, USA, Catalog # 9722) and p21WAF1/CIP1 (F-5, Santa Cruz Biotechnology, Dallas, TX, USA, Catalog # sc-6246). Secondary antibodies were HRP-conjugated goat anti-rabbit (Catalog # 611–103–122) or HRP-conjugated goat anti-mouse (Catalog # 610–103–121) from Rockland Immunochemicals (Gilbertsville, PA, USA).
rax−/− (Prkratm1cs) [34], pkr−/− (Eif2ak2tm1Cwe) [37], pkrR/R (Eif2ak2tm1Ajs) [41] and eIF2αS/A (Eif2s1tm1Rjk) mice [79] are all previously described. Eif2s1tm1Rjk mice were a kind gift from Dr Randal Kaufman, Sanford-Burnham Medical Research Institute, La Jolla, California, USA. Mouse experiments were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of Cleveland Clinic (Approval Number ARC 08738).
Phenotypic analysis of rax−/−pkr−/− and rax−/−pkrR/R mice
To generate mice deficient in both RAX and PKR, rax+/− mice were crossed with pkr−/− mice to generate rax+/−pkr+/− mice, which were in turn intercrossed to generate rax−/−pkr−/− mice. To generate mice deficient in RAX and lacking PKR kinase activity (using the K271R mutant), rax+/− mice and pkrR/R mice were crossed to generate rax+/−pkrK/R mice, which were in turn intercrossed to generate rax−/−pkrR/R mice. To generate mice deficient in RAX with the eIF2αS/A mutation, rax+/− mice were crossed with eIF2αS/A mice. Resulting rax+/−eIF2αS/A mice were intercrossed, but viable rax−/−eIF2αA/A were not produced from these crosses.
To quantify developmental defects seen in rax−/− mice, ear size (from helix to tragus) and body length (from nose to tail base) were measured using Vernier calipers (Scienceware, Wayne, NJ, USA). Adult male and female mice aged between 5 and 34 weeks were used for measurements. Numbers of mice per litter and time between consecutive litters for dams were calculated from breeding records over a 2-year period.
For macroscopic analysis, adult age, sex-matched mice between 10 and 25 weeks old were dissected such that the pituitary was exposed, but remained connected to trigeminal nerves and the soft palate of the mouth. Pituitaries were photographed using an M60 Stereo microscope (Leica, Wetzlar, Germany) and camera system.
For histological analysis, whole pituitaries were harvested, still attached to the skull and trigeminal nerves (to retain morphology), from adult sex-matched mice aged between 10 and 25 weeks and fixed for 4 days in 10% neutral buffered formalin (Fisher Scientific, Hampton, NH, USA). Fixed pituitaries were removed from the residual skull, embedded in paraffin and sectioned longitudinally prior to hematoxylin and eosin staining. Sections were analyzed on a Leica Microsystems (Wetzlar, Germany) DMR upright microscope and photographed at 5× magnification using a Retiga EX cooled CCD camera (Spectracore, Ontario, NY, USA) and ImagePro Plus software (MediaCybernetics, Rockville, MD, USA). The size of anterior pituitary was quantified by analyzing micrographs using ImagePro Plus to select the relevant region of the micrograph and determine the number of pixels it contained. Pixel size was calibrated to physical size based on the objective lens used to take the micrograph, enabling calculation of the size of the anterior pituitary based on the number of pixels.
Cloning of RAX shRNA constructs
Annealed oligonucleotides were ligated into AgeI and EcoRI (New England Biolabs, Ipswich, MA, USA) digested pLKO.1-puro using Rapid DNA Ligation Kit (Roche, Basel, Switzerland). All clones were verified by sequencing. For pLKO.1-RAX shRNA (1199)-puro, expressing an shRNA targeting nucleotides 1199–1217 in the 3’UTR of the RAX mRNA (underlined in sequence), the DNA oligonucleotides RAX (1199) F (CCG GCC GTC AAC TTT CCA GAT TTC TCG AGA AAT CTG GAA AGT TGA CGG TTT TTG) and RAX (1199) R (AAT TCA AAA ACC GTC AAC TTT CCA GAT TTC TCG AGA AAT CTG GAA AGT TGA CGG) were annealed and ligated into pLKO.1-puro. For pLKO.1-RAX shRNA (1459)-puro, expressing an shRNA targeting nucleotides 1459–1477 in the 3’UTR of the RAX mRNA (underlined in sequence), the DNA oligonucleotides RAX (1459) F (CCG GTG TTG AGG CTT GTG ATG AAC TCG AGT TCA TCA CAA GCC TCA ACA TTT TTG) and RAX (1459) R (TTA ACA AAA ATG TTG AGG CTT GTG ATG AAC TCG AGT TCA TCA CAA GCC TCA ACA) were annealed and ligated into pLKO.1-puro.
Lentiviral packaging, concentration and determination of titer
Replication-deficient, VSV pseudotyped recombinant lentivirus was produced by cotransfection of pLKO.1-puro-derived plasmids with the packaging plasmid pCMV-dR8.74 and the pseudotyping plasmid pVSV-G using the calcium phosphate method into HEK293T cells. Lentivirus-containing supernatants were harvested three times every 12–16 h; the collections were pooled, filter-sterilized and titered as follows. HT1080 cells were infected with serial dilutions in complete DMEM of lentivirus supernatant with polybrene added to 8.0 μg/ml for 24 h. Medium was replaced with complete DMEM without polybrene for 48 h. Infected cells were selected for 7 days in complete DMEM containing 2.0 μg/ml puromycin, prior to fixation and staining with crystal violet. Puromycin-resistant colonies were counted to calculate titer. To concentrate lentivirus, supernatants prepared as described above were concentrated by ultracentrifugation at 47,000 × g for 2 h at 16°C in an SW28 rotor using a Beckman (Indianapolis, IN, USA) Optima L-100 XP ultracentrifuge. Supernatant was discarded following ultracentrifugation, and the lentivirus-containing pellet was resuspended in complete DMEM and titered as described.
Determination of cell proliferation
αT3 cells were infected with either pLKO.1 non-targeting control or pLKO.1 RAX shRNA (1199 or 1459) lentivirus. Lentiviral infection was carried out in complete DMEM containing 8.0 μg/ml polybrene for 24 h. Following shRNA lentivirus infection, inoculum was replaced with fresh complete DMEM without polybrene and cells were incubated for 48 h. Cells were then split into complete DMEM containing 2.0 μg/ml puromycin and cultured for 72 h. Preliminary experiments were performed by trypan blue staining and counting cells, while subsequent experiments were performed using the CellTiter 96 Non-Radioactive Cell Proliferation Assay (Promega, Madison, WI, USA, Cat# G4000). Cells were plated in replicate wells of 6-well dishes at 1×106 cells/well for trypan blue counting or in 96-well plates at 3.5×104 cells in 100 μl/well for Promega assay. For trypan blue counting, cells were trypsinized, pooled with media and washes to collect dead cells, centrifuged at 600 × g for 5 min at room temperature, resuspended in PBS containing 5 mM EDTA, stained with trypan blue and counted using a hemocytometer at 24-h intervals after plating. For Promega assays, 15 μl/well dye solution was added to each well, incubated for 1 h, then solubilized in stop/solubilization buffer for 48 h. Absorbance at 600 nm was measured in a Wallac Victor2 1420 multilabel reader (Perkin Elmer, Waltham, MA, USA). Data were analyzed for both methods by plotting number of cells or A600 for Promega assays over time, and fit to the exponential growth equation Y = Y0eKT (where Y = cell number or A600, Y0 = initial cell number or A600 and T = hours after plating) using the Graphpad Prism version 5.02 software package (GraphPad Software, San Diego CA) to calculate the proliferation rate (K).
Determination of cell cycle distribution
αT3 cells were infected with pLKO.1 non-targeting control or pLKO.1 RAX shRNA (1199) lentivirus and selected as previously described prior to splitting into replicate dishes. Every 24 h, one dish of non-targeting control cells and RAX shRNA (1199) cells was trypsinized, pooled with media and washes to collect dead cells, pelleted by centrifugation at 600 × g for 5 min at room temperature, trypan blue stained, and counted to determine cell number and percentage trypan blue positive cells. For each time point, 4×106 cells were pelleted, washed in cold PBS, fixed in cold 70% ethanol and stored at −20°C until all samples were ready for processing. Fixed cells were centrifuged at 1000 × g for 10 min at room temperature, washed twice in cold PBS and incubated for 10 min at room temperature in DNA extraction buffer (100 mM sodium phosphate dibasic, 20 mM sodium citrate, 0.5× PBS, pH 7.8). Cells were collected by centrifugation and resuspended in PI staining solution (50 μg/ml PI, 20 μg/ml RNase A in 1xPBS). Cells were incubated on ice in PI staining solution for 2–4 h, until samples were analyzed using a FACScan flow cytometer (Becton Dickenson Biosciences, San Jose, CA, USA) and analyzed using ModFit LT software (Verity Software, Topsham, ME, USA) to fit data to a model of DNA content.
Protein isolation and western blot
Protein was isolated from cultured cells by lysis in Triton X-100 lysis buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 5 mM 2-mercaptoethanol, 10% glycerol, supplemented with Complete protease inhibitor cocktail (Roche) and PhoSTOP phosphatase inhibitor cocktail (Roche)). Protein was isolated from mouse tissue by grinding whole tissue in Triton X-100 lysis buffer. Proteins were separated using SDS-PAGE and transferred to PVDF membrane (Immobilon, Millpore, Billerica, MA, USA) for western blotting. Secondary antibodies were detected using ECL (GE, Little Chalfont, Buckinghamshire, UK), ECL 2 (Pierce, Rockford, IL, USA) or ECL Prime (GE). Quantitation of ECL signal from p21WAF1/CIP1 western blots of primary tissue was performed using ImageQuant (Molecular Dynamics, Little Chalfont, Buckinghamshire, UK, version 5.2); signal was normalized to a loading control (β actin or β tubulin) and expressed as normalized signal relative to the normalized signal from the corresponding wild-type control sample.
Determination of p21WAF1/CIP1 expression kinetics by quantitative western blot and real-time RT-PCR
αT3 cells were infected with 3 TU/cell pLKO.1-puro non-targeting control or RAX (1199) shRNA lentivirus, as previously described in five replicate plates for each virus. Medium was replaced with complete DMEM without polybrene and cells from each set of replicates were harvested at 24-h intervals, washed in PBS and divided into two tubes for RNA and protein analysis. RNA was isolated from one tube using Roche Highpure Total RNA Isolation Columns; RNA (1.6 μg) was reverse transcribed in a 40 μl volume using the Superscript III reverse transcription kit with random hexamer primers (Invitrogen). Lack of an 18S rRNA PCR band was used to exclude genomic DNA contamination in all RNA samples prior to reverse transcription. To quantitate p21WAF1/CIP1 transcript levels, 4 ng cDNA per reaction was used as template in real-time PCR. Real-time PCR was conducted using the SYBR green PCR system (Applied Biosystems, Foster City, CA, USA) for two-step RT-PCR, except that the final reaction volume used was 12.5 μl. Accumulated PCR product was quantified by comparison to a standard curve and normalized to an 18S rRNA control reaction. Primer sequences for p21WAF1/CIP1 were as follows: 5’ - gaacatctcagggccgaaaacg, 3’ - ctaaggccgaagatggggaagag; and for 18S rRNA were 5’-attgacggaagggcaccaccag and 3’-caaatcgctccaccaactaagaacg. PCR cycling was as follows: 3 min 95°C, followed by 40 cycles of: 30 sec 95°C, 1 min 52°C, 30 sec 72°C. The second pellet from each time point was lysed in Triton X-100 lysis buffer as described above, 10 μg total protein was separated by SDS-PAGE and transferred to nitrocellulose membrane (Bio-Rad, Carlsbad, CA, USA, cat# 162–0146). Membranes were blocked in 5% milk in 1xTBS without Tween20, and probed with anti-p21WAF1/CIP1 (F5) at 1:100 or anti-PACT at 1:2000 and anti-β-tubulin at 1:20000 in 0.5% milk in TBS-T. Membranes were incubated with IRDye 680LT goat anti-rabbit secondary (Cat# 926–68021) and IRDye 800CW goat anti-mouse secondary (Cat# 926–32210) from Licor (Lincoln, NE, USA) at 1:20000 and 1:5000, respectively, in 0.5% milk in 1xTBS-T. Final washes were performed in 1xTBS without Tween20. Detection was performed using the Odyssey® system (Licor), and quantified using the Image Studio software package (Version 2.0, Licor).
Determination of PKR activation by two-dimensional PKR western blot and P-eIF2α western blot
αT3 cells were infected with 3 TU/cell with concentrated pLKO.1-puro non-targeting control or pLKO.1-RAX (1199)-puro lentivirus as for the p21WAF1/CIP1 expression assay as previously described. Following infection, cells were split into fresh complete media without polybrene for an additional 24 h. Cells were harvested and washed in PBS, split into two tubes and pelleted by centrifugation. One cell pellet from each sample was resuspended in 2D lysis buffer (4% CHAPS, 7 M urea, 2 M thiourea, 40 mM DTT, 0.002% Bromophenol Blue, 0.5% IPG Buffer pH 3–10 (GE Life Sciences, Pittsburgh, PA, USA)), passed through a 22G needle three times, and incubated on ice for 20 min. The second pellet from each sample was resuspended in Triton X-100 lysis buffer. For isoelectric focusing, 30 μg of each sample in 2D lysis buffer was applied to a 7 cm Immobiline IPG strip, pH 3–10 (GE Life Sciences). Strips were focused in ceramic strip holders using the following program at 15°C: 12 h rehydration at 30 V, 1 h at 300 V, linear gradient between 300 V and 1000 V over 30 min, linear gradient between 1000 V and 5000 V over 1 h and 20 min, 5000 V for 6500 V Hours. Strips were then held at 500 V overnight. Focused strips were equilibrated into Equilibration buffer (75 mM Tris-HCl pH 8.8, 6 M urea, 2% SDS, 30% glycerol, 0.002% Bromophenol Blue) + 1% DTT for 15 min, followed by equilibration into Equilibration buffer + 2.5% iodoacetamide for 15 min at room temperature with rocking. Strips were then subjected to second-dimension electrophoresis on an 8% SDS-PAGE gel. Gels were transferred to PVDF and probed with anti-PKR (D-20, Santa Cruz Biotechnology, 1:5000) in 0.5% milk in 1xTBS-T, followed by detection using goat anti-rabbit HRP secondary (1:20000) and ECL-prime (GE). Images have been cropped to display the relevant molecular weight and pI ranges. Triton X-100 lysates were used for conventional western blot with anti-P-eIF2α as described above.
Statistical analysis
All statistical analyses, including Student’s t-test and non-linear regression analysis, were performed using the Graphpad Prism version 5.02 software package (GraphPad Software, San Diego, CA, USA).
Acknowledgments
The authors wish to thank the Lerner Research Institute Biological Resources Unit for providing valuable assistance with animal husbandry, the Flow Cytometry core for assistance in analyzing shRNA-expressing cells for DNA content and cell cycle distribution, Jim Lang for assistance with photographing live mice and the Genomics core for assistance in sequencing plasmids. The authors would also like to thank Avanti Desai, Ying Zhang and Jaime Wetzel for technical assistance with animal husbandry, Peter Faber for advice regarding the kinase-dead PKR mice and Frances Cribbin for valuable editorial assistance. This work was supported by US NIH grant 5P01CA062220, Australian Research Council grant DP110102641 and the Victorian Government’s Operational Infrastructure Support Program.
Abbreviations
- DMEM
Dulbecco’s modified Eagle’s medium
- eIF2α
eukaryotic translation initiation factor 2 α subunit
- Eif2ak2
eIF2α kinase 2 (gene encoding PKR)
- GSK-3β
glycogen synthase kinase 3β
- NFAR
nuclear factor associated with RNA
- p21WAF1/CIP1
cyclin-dependent kinase inhibitor 1A
- PACT
protein activator of PKR
- pI
isoelectric point
- PI
propidium iodide
- PKR
interferon-inducible double-stranded RNA-dependent protein kinase (protein kinase R)
- PRBP
protamine-1 RNA binding protein
- Prkra
protein kinase R (PKR) activator (gene encoding PACT/RAX)
- RAX
PKR-associated protein X
- shRNA
short hairpin RNA
- TARBP2
human immunodeficiency virus-1 trans-activation response element RNA binding protein 2
- TU
transducing units
Footnotes
Author contributions
BKD and CLW contributed to the conception, design, execution, analysis and interpretation of the experiments. GCS contributed to the conception, design, analysis and interpretation of the experiments. PK, AJS and BRGW contributed to the generation of reagents, analysis and interpretation of the experiments. All authors contributed to the writing of the manuscript, and read and approved the final version.
References
- 1.Patel RC, Sen GC. PACT, a protein activator of the interferon-induced protein kinase PKR. Embo J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito T, Yang M, May WS. RAX, a cellular activator for double-stranded RNA-dependent protein kinase during stress signaling. J Biol Chem. 1999;274:15427–15432. doi: 10.1074/jbc.274.22.15427. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. Embo J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kok KH, Ng MH, Ching YP, Jin DY. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J Biol Chem. 2007;282:17649–17657. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 5.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kok KH, Lui PY, Ng MH, Siu KL, Au SW, Jin DY. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell host & microbe. 2011;9:299–309. doi: 10.1016/j.chom.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Camargos S, Scholz S, Simon-Sanchez J, Paisan-Ruiz C, Lewis P, Hernandez D, Ding J, Gibbs JR, Cookson MR, Bras J, Guerreiro R, Oliveira CR, Lees A, Hardy J, Cardoso F, Singleton AB. DYT16, a novel young-onset dystonia-parkinsonism disorder: identification of a segregating mutation in the stress-response protein PRKRA. Lancet Neurol. 2008;7:207–215. doi: 10.1016/S1474-4422(08)70022-X. [DOI] [PubMed] [Google Scholar]
- 9.Seibler P, Djarmati A, Langpap B, Hagenah J, Schmidt A, Bruggemann N, Siebner H, Jabusch HC, Altenmuller E, Munchau A, Lohmann K, Klein C. A heterozygous frameshift mutation in PRKRA (DYT16) associated with generalised dystonia in a German patient. Lancet Neurol. 2008;7:380–381. doi: 10.1016/S1474-4422(08)70075-9. [DOI] [PubMed] [Google Scholar]
- 10.de Carvalho Aguiar P, Borges V, Ferraz HB, Ozelius LJ. Novel compound heterozygous mutations in PRKRA cause pure dystonia. Mov Disord. 2015;30:877–878. doi: 10.1002/mds.26175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruvolo PP, Gao F, Blalock WL, Deng X, May WS. Ceramide regulates protein synthesis by a novel mechanism involving the cellular PKR activator RAX. J Biol Chem. 2001;276:11754–11758. doi: 10.1074/jbc.M011400200. [DOI] [PubMed] [Google Scholar]
- 12.Patel CV, Handy I, Goldsmith T, Patel RC. PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J Biol Chem. 2000;275:37993–37998. doi: 10.1074/jbc.M004762200. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert SJ, Duance VC, Mason DJ. Tumour necrosis factor alpha up-regulates protein kinase R (PKR)-activating protein (PACT) and increases phosphorylation of PKR and eukaryotic initiation factor 2-alpha in articular chondrocytes. Biochem Soc Trans. 2002;30:886–889. doi: 10.1042/bst0300886. [DOI] [PubMed] [Google Scholar]
- 14.Chen G, Ma C, Bower KA, Ke Z, Luo J. Interaction between RAX and PKR modulates the effect of ethanol on protein synthesis and survival of neurons. J Biol Chem. 2006;281:15909–15915. doi: 10.1074/jbc.M600612200. [DOI] [PubMed] [Google Scholar]
- 15.Peters GA, Hartmann R, Qin J, Sen GC. Modular structure of PACT: distinct domains for binding and activating PKR. Mol Cell Biol. 2001;21:1908–19020. doi: 10.1128/MCB.21.6.1908-1920.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett RL, Blalock WL, Abtahi DM, Pan Y, Moyer SA, May WS. RAX, the PKR activator, sensitizes cells to inflammatory cytokines, serum withdrawal, chemotherapy and viral infection. Blood. 2006;108:821–829. doi: 10.1182/blood-2005-11-006817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee ES, Yoon CH, Kim YS, Bae YS. The double-strand RNA-dependent protein kinase PKR plays a significant role in a sustained ER stress-induced apoptosis. FEBS Lett. 2007;581:4325–4332. doi: 10.1016/j.febslet.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Singh M, Fowlkes V, Handy I, Patel CV, Patel RC. Essential role of PACT-mediated PKR activation in tunicamycin-induced apoptosis. J Mol Biol. 2009;385:457–468. doi: 10.1016/j.jmb.2008.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peters GA, Li S, Sen GC. Phosphorylation of specific serine residues in the PKR activation domain of PACT is essential for its ability to mediate apoptosis. J Biol Chem. 2006;281:35129–35136. doi: 10.1074/jbc.M607714200. [DOI] [PubMed] [Google Scholar]
- 20.Bennett RL, Blalock WL, May WS. Serine 18 phosphorylation of RAX, the PKR activator, is required for PKR activation and consequent translation inhibition. J Biol Chem. 2004;279:42687–42693. doi: 10.1074/jbc.M403321200. [DOI] [PubMed] [Google Scholar]
- 21.Gupta V, Huang X, Patel RC. The carboxy-terminal, M3 motifs of PACT and TRBP have opposite effects on PKR activity. Virology. 2003;315:283–291. doi: 10.1016/s0042-6822(03)00589-0. [DOI] [PubMed] [Google Scholar]
- 22.Huang X, Hutchins B, Patel RC. The C-terminal, third conserved motif of the protein activator PACT plays an essential role in the activation of double-stranded-RNA-dependent protein kinase (PKR) Biochem J. 2002;366:175–186. doi: 10.1042/BJ20020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Peters GA, Ding K, Zhang X, Qin J, Sen GC. Molecular basis for PKR activation by PACT or dsRNA. Proc Natl Acad Sci U S A. 2006;103:10005–10010. doi: 10.1073/pnas.0602317103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar A, Haque J, Lacoste J, Hiscott J, Williams BR. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc Natl Acad Sci U S A. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar A, Yang YL, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams BR. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. Embo J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chong KL, Feng L, Schappert K, Meurs E, Donahue TF, Friesen JD, Hovanessian AG, Williams BRG. Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. EMBO J. 1992;11:1553–1562. doi: 10.1002/j.1460-2075.1992.tb05200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hershey JW. Protein phosphorylation controls translation rates. J Biol Chem. 1989;264:20823–20826. [PubMed] [Google Scholar]
- 28.Friedman RM, Metz DH, Esteban RM, Tovell DR, Ball LA, Kerr IM. Mechanism of interferon action: inhibition of viral messenger ribonucleic acid translation in L-cell extracts. J Virol. 1972;10:1184–1198. doi: 10.1128/jvi.10.6.1184-1198.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zamanian-Daryoush M, Der SD, Williams BRG. Cell cycle regulation of the double stranded RNA activated protein kinase, PKR. Oncogene. 1999;18:315–326. doi: 10.1038/sj.onc.1202293. [DOI] [PubMed] [Google Scholar]
- 30.Alisi A, Mele R, Spaziani A, Tavolaro S, Palescandolo E, Balsano C. Thr 446 phosphorylation of PKR by HCV core protein deregulates G2/M phase in HCC cells. J Cell Physiol. 2005;205:25–31. doi: 10.1002/jcp.20363. [DOI] [PubMed] [Google Scholar]
- 31.Bennett RL, Pan Y, Christian J, Hui T, May WS., Jr The RAX/PACT-PKR stress response pathway promotes p53 sumoylation and activation, leading to G(1) arrest. Cell cycle. 2012;11:407–417. doi: 10.4161/cc.11.2.18999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Der SD, Yang YL, Weissmann C, Williams BR. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc Natl Acad Sci U S A. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeung MC, Liu J, Lau AS. An essential role for the interferon-inducible, double-stranded RNA-activated protein kinase PKR in the tumor necrosis factor-induced apoptosis in U937 cells. Proc Natl Acad Sci U S A. 1996;93:12451–12455. doi: 10.1073/pnas.93.22.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rowe TM, Rizzi M, Hirose K, Peters GA, Sen GC. A role of the double-stranded RNA-binding protein PACT in mouse ear development and hearing. Proc Natl Acad Sci U S A. 2006;103:5823–5828. doi: 10.1073/pnas.0601287103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickerman BK, White CL, Chevalier C, Nalesso V, Charles C, Fouchecourt S, Guillou F, Viriot L, Sen GC, Herault Y. Missense mutation in the second RNA binding domain reveals a role for Prkra (PACT/RAX) during skull development. PLoS One. 2011;6:28537. doi: 10.1371/journal.pone.0028537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peters GA, Seachrist DD, Keri RA, Sen GC. The double-stranded RNA-binding protein, PACT, is required for postnatal anterior pituitary proliferation. Proc Natl Acad Sci U S A. 2009;106:10696–10701. doi: 10.1073/pnas.0900735106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang YL, Reis LF, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams BR, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. Embo J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abraham N, Stojdl DF, Duncan PI, Methot N, Ishii T, Dube M, Vanderhyden BC, Atkins HL, Gray DA, McBurney MW, Koromilas AE, Brown EG, Sonenberg N, Bell JC. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem. 1999;274:5953–5962. doi: 10.1074/jbc.274.9.5953. [DOI] [PubMed] [Google Scholar]
- 39.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 40.Katze MG, Wambach M, Wong ML, Garfinkel M, Meurs E, Chong K, Williams BR, Hovanessian AG, Barber GN. Functional expression and RNA binding analysis of the interferon-induced, double-stranded RNA-activated, 68,000-Mr protein kinase in a cell-free system. Molecular and cellular biology. 1991;11:5497–5505. doi: 10.1128/mcb.11.11.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irving AT, Wang D, Vasilevski O, Latchoumanin O, Kozer N, Clayton AH, Szczepny A, Morimoto H, Xu D, Williams BR, Sadler AJ. Regulation of actin dynamics by protein kinase R control of gelsolin enforces basal innate immune defense. Immunity. 2012;36:795–806. doi: 10.1016/j.immuni.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 42.The Jackson Laboratory. MPD:14938 Mouse Phenome Database web site. Bar Harbor, Maine, USA: The Jackson Laboratory; 2014. Reproductive performance survey, females of 33 inbred strains of mice. MPD:14938. Mouse Phenome Database. [Google Scholar]
- 43.The Jackson Laboratory. MPD:31403 Mouse Phenome Database web site. Bar Harbor, Maine, USA: The Jackson Laboratory; 2014. Breeding performance survey, females of 35 commonly used strains of JAX mice. MPD:31403. Mouse Phenome Database. [Google Scholar]
- 44.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 45.Ooi GT, Tawadros N, Escalona RM. Pituitary cell lines and their endocrine applications. Molecular and cellular endocrinology. 2004;228:1–21. doi: 10.1016/j.mce.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 46.Zhong J, Peters AH, Lee K, Braun RE. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat Genet. 1999;22:171–174. doi: 10.1038/9684. [DOI] [PubMed] [Google Scholar]
- 47.Clerzius G, Shaw E, Daher A, Burugu S, Gelinas JF, Ear T, Sinck L, Routy JP, Mouland AJ, Patel RC, Gatignol A. The PKR activator, PACT, becomes a PKR inhibitor during HIV-1 replication. Retrovirology. 2013;10:96. doi: 10.1186/1742-4690-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turgeon JL, Kimura Y, Waring DW, Mellon PL. Steroid and pulsatile gonadotropin-releasing hormone (GnRH) regulation of luteinizing hormone and GnRH receptor in a novel gonadotrope cell line. Mol Endocrinol. 1996;10:439–450. doi: 10.1210/mend.10.4.8721988. [DOI] [PubMed] [Google Scholar]
- 49.Tashjian AH, Jr, Yasumura Y, Levine L, Sato GH, Parker ML. Establishment of clonal strains of rat pituitary tumor cells that secrete growth hormone. Endocrinology. 1968;82:342–352. doi: 10.1210/endo-82-2-342. [DOI] [PubMed] [Google Scholar]
- 50.Windle JJ, Weiner RI, Mellon PL. Cell lines of the pituitary gonadotrope lineage derived by targeted oncogenesis in transgenic mice. Mol Endocrinol. 1990;4:597–603. doi: 10.1210/mend-4-4-597. [DOI] [PubMed] [Google Scholar]
- 51.Zamanian-Daryoush M, Der SD, Williams BR. Cell cycle regulation of the double stranded RNA activated protein kinase, PKR. Oncogene. 1999;18:315–326. doi: 10.1038/sj.onc.1202293. [DOI] [PubMed] [Google Scholar]
- 52.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 53.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 54.Flores-Rozas H, Kelman Z, Dean FB, Pan ZQ, Harper JW, Elledge SJ, O’Donnell M, Hurwitz J. Cdk-interacting protein 1 directly binds with proliferating cell nuclear antigen and inhibits DNA replication catalyzed by the DNA polymerase delta holoenzyme. Proc Natl Acad Sci U S A. 1994;91:8655–8659. doi: 10.1073/pnas.91.18.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guadagno TM, Newport JW. Cdk2 kinase is required for entry into mitosis as a positive regulator of Cdc2-cyclin B kinase activity. Cell. 1996;84:73–82. doi: 10.1016/s0092-8674(00)80994-0. [DOI] [PubMed] [Google Scholar]
- 56.Shi L, Zhao G, Qiu D, Godfrey WR, Vogel H, Rando TA, Hu H, Kao PN. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3’-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J Biol Chem. 2005;280:18981–18989. doi: 10.1074/jbc.M411034200. [DOI] [PubMed] [Google Scholar]
- 57.Schwaller J, Koeffler HP, Niklaus G, Loetscher P, Nagel S, Fey MF, Tobler A. Posttranscriptional stabilization underlies p53-independent induction of p21WAF1/CIP1/SDI1 in differentiating human leukemic cells. J Clin Invest. 1995;95:973–979. doi: 10.1172/JCI117806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harashima A, Guettouche T, Barber GN. Phosphorylation of the NFAR proteins by the dsRNA-dependent protein kinase PKR constitutes a novel mechanism of translational regulation and cellular defense. Genes Dev. 2010;24:2640–2653. doi: 10.1101/gad.1965010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perez-Roger I, Solomon DL, Sewing A, Land H. Myc activation of cyclin E/Cdk2 kinase involves induction of cyclin E gene transcription and inhibition of p27(Kip1) binding to newly formed complexes. Oncogene. 1997;14:2373–2381. doi: 10.1038/sj.onc.1201197. [DOI] [PubMed] [Google Scholar]
- 60.Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 61.Zhu H, Nie L, Maki CG. Cdk2-dependent Inhibition of p21 stability via a C-terminal cyclin-binding motif. J Biol Chem. 2005;280:29282–29288. doi: 10.1074/jbc.M407352200. [DOI] [PubMed] [Google Scholar]
- 62.Arnold HK, Sears RC. Protein phosphatase 2A regulatory subunit B56alpha associates with c-myc and negatively regulates c-myc accumulation. Mol Cell Biol. 2006;26:2832–2844. doi: 10.1128/MCB.26.7.2832-2844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee JY, Yu SJ, Park YG, Kim J, Sohn J. Glycogen synthase kinase 3beta phosphorylates p21WAF1/CIP1 for proteasomal degradation after UV irradiation. Mol Cell Biol. 2007;27:3187–3198. doi: 10.1128/MCB.01461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zu K, Bihani T, Lin A, Park YM, Mori K, Ip C. Enhanced selenium effect on growth arrest by BiP/GRP78 knockdown in p53-null human prostate cancer cells. Oncogene. 2006;25:546–554. doi: 10.1038/sj.onc.1209071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rajesh K, Papadakis AI, Kazimierczak U, Peidis P, Wang S, Ferbeyre G, Kaufman RJ, Koromilas AE. eIF2alpha phosphorylation bypasses premature senescence caused by oxidative stress and pro-oxidant antitumor therapies. Aging (Albany NY) 2013;5:884–901. doi: 10.18632/aging.100620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kazemi S, Mounir Z, Baltzis D, Raven JF, Wang S, Krishnamoorthy JL, Pluquet O, Pelletier J, Koromilas AE. A novel function of eIF2alpha kinases as inducers of the phosphoinositide-3 kinase signaling pathway. Mol Biol Cell. 2007;18:3635–3644. doi: 10.1091/mbc.E07-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rossig L, Jadidi AS, Urbich C, Badorff C, Zeiher AM, Dimmeler S. Akt-dependent phosphorylation of p21(Cip1) regulates PCNA binding and proliferation of endothelial cells. Mol Cell Biol. 2001;21:5644–5657. doi: 10.1128/MCB.21.16.5644-5657.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cmielova J, Rezacova M. p21Cip1/Waf1 protein and its function based on a subcellular localization [corrected] Journal of cellular biochemistry. 2011;112:3502–3506. doi: 10.1002/jcb.23296. [DOI] [PubMed] [Google Scholar]
- 70.Bleiblo F, Michael P, Brabant D, Ramana CV, Tai T, Saleh M, Parrillo JE, Kumar A, Kumar A. JAK kinases are required for the bacterial RNA and poly I:C induced tyrosine phosphorylation of PKR. International journal of clinical and experimental medicine. 2013;6:16–25. [PMC free article] [PubMed] [Google Scholar]
- 71.Su Q, Wang S, Baltzis D, Qu LK, Raven JF, Li S, Wong AH, Koromilas AE. Interferons induce tyrosine phosphorylation of the eIF2alpha kinase PKR through activation of Jak1 and Tyk2. EMBO Rep. 2007;8:265–270. doi: 10.1038/sj.embor.7400891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Su Q, Wang S, Baltzis D, Qu LK, Wong AH, Koromilas AE. Tyrosine phosphorylation acts as a molecular switch to full-scale activation of the eIF2alpha RNA-dependent protein kinase. Proc Natl Acad Sci U S A. 2006;103:63–68. doi: 10.1073/pnas.0508207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laraki G, Clerzius G, Daher A, Melendez-Pena C, Daniels S, Gatignol A. Interactions between the double-stranded RNA-binding proteins TRBP and PACT define the Medipal domain that mediates protein-protein interactions. RNA Biol. 2008;5:92–103. doi: 10.4161/rna.5.2.6069. [DOI] [PubMed] [Google Scholar]
- 74.Park H, Davies MV, Langland JO, Chang HW, Nam YS, Tartaglia J, Paoletti E, Jacobs BL, Kaufman RJ, Venkatesan S. TAR RNA-binding protein is an inhibitor of the interferon-induced protein kinase PKR. Proc Natl Acad Sci U S A. 1994;91:4713–4717. doi: 10.1073/pnas.91.11.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pang Q, Christianson TA, Keeble W, Koretsky T, Bagby GC. The anti-apoptotic function of Hsp70 in the interferon-inducible double-stranded RNA-dependent protein kinase-mediated death signaling pathway requires the Fanconi anemia protein, FANCC. J Biol Chem. 2002;277:49638–49643. doi: 10.1074/jbc.M209386200. [DOI] [PubMed] [Google Scholar]
- 76.Donze O, Abbas-Terki T, Picard D. The Hsp90 chaperone complex is both a facilitator and a repressor of the dsRNA-dependent kinase PKR. EMBO J. 2001;20:3771–3780. doi: 10.1093/emboj/20.14.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Polyak SJ, Tang N, Wambach M, Barber GN, Katze MG. The P58 cellular inhibitor complexes with the interferon-induced, double-stranded RNA-dependent protein kinase, PKR, to regulate its autophosphorylation and activity. J Biol Chem. 1996;271:1702–1707. doi: 10.1074/jbc.271.3.1702. [DOI] [PubMed] [Google Scholar]
- 78.Marques JT, White CL, Peters GA, Williams BR, Sen GC. The role of PACT in mediating gene induction, PKR activation, and apoptosis in response to diverse stimuli. J Interferon Cytokine Res. 2008;28:469–476. doi: 10.1089/jir.2007.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Scheuner D, Vander Mierde D, Song B, Flamez D, Creemers JW, Tsukamoto K, Ribick M, Schuit FC, Kaufman RJ. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11:757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]