Abstract
The peptide T20, which corresponds to a sequence in the C-terminal segment of the HIV-1 trans-membrane glycoprotein gp41, is a strong entry inhibitor of HIV-1. It has been assumed that T20 inhibits HIV-1 infection by binding to the trimer formed by the N-terminal helical region (HR1) of gp41, preventing the formation of a six helix bundle by the N- and C-terminal helical regions of gp41. In addition to binding to gp41, T20 was found to bind to gp120 of X4 viruses and this binding was suggested to be responsible for an alternative mechanism for HIV-1 inhibition by this peptide. In the present study, T20 also was found to bind R5 gp120. Using NMR spectroscopy, the segments of T20 that interact with both gp120 and a gp120/CD4M33 complex were mapped. A peptide corresponding to the fourth constant region of gp120, sC4, was found to partially recapitulate gp120 binding to T20 and the segment of this peptide interacting with T20 was mapped. Our conclusion is that an amphiphilic helix on the T20 C-terminus, binds through mostly hydrophobic interactions, to a non-polar gp120 surface formed primarily by the C4 region. The ten to a thousand-fold difference between the EC50 of T20 against viral fusion and the affinity of T20 to gp120 implies that binding to gp120 is not a major factor in T20 inhibition of HIV-1 fusion. Nevertheless, this hydrophobic gp120 surface could be a target for anti-HIV therapeutics.
Keywords: T20, gp120, CCR5, HIV-1 inhibitors, NMR
Introduction
The fusion of HIV-1 with its target cell is mediated by the trans-membrane envelope protein gp41, three molecules of which together with the three molecules of the viral glycoprotein gp120 form the envelope spike of the virus. Initially gp120 binds to CD4, exposing sites on gp120 that subsequently bind to either the CCR5 or CXCR4 chemokine receptor, depending on the phenotype (R5 or X4) of the virus. R5 viruses are responsible for the vast majority of HIV-1 infections and are the predominant viruses in the asymptomatic phase of HIV-1 infection. Therefore, R5 viruses are a major target for anti-HIV-1 therapeutics.
Binding of gp120 to CCR5 or CXCR4 co-receptors causes a further conformational change that exposes gp41. An N-terminal segment of gp41, called the fusion peptide, inserts into the membrane of the target cell, creating a transient pre-hairpin intermediate conformation linking the viral and target cell membranes. Ultimately, the formation of a six-helix bundle hairpin structure by the gp41 trimer mediates the fusion event. Two helical segments, HR1 and HR2, located at the N- and C-terminal halves of gp41, respectively, form the six-helix bundle. Peptides corresponding to HR2, designated as C-peptides, bind to the trimer formed by the HR1 segments of gp41, preventing the formation of the six-helix bundle and thereby inhibiting HIV-1 fusion [1].
The peptide T20, which corresponds to a sequence in the C-terminal segment of gp41 (gp41638–673), is a strong entry inhibitor of HIV-1 with an EC50 of 0.5 ng/ml (0.1 nM range) [2, 3]. This 37-residue peptide is composed of a segment from HR2 and an adjacent conserved sequence from the membrane proximal external region, MPER, of gp41. T20 (Fuzeon, also called Enfuvirtide or DP-178 [4]) has been approved for treatment of HIV-1 infected patients and was the first in a class of anti-HIV-1 compounds that function as entry-inhibitors. It has been assumed that T20 shares the mechanism of fusion-inhibition of the gp41 C-peptides [1] described above. Indeed mutations in the 547GIV549 segment in HR1 confer resistance to T20, suggesting that the N-terminal half of T20 interacts with gp41 HR1 [5].
NMR studies of the conformation of T20 in aqueous solution revealed that the segment E657-F673 favors a helical conformation [6, 7]. Investigations of gp41 peptides [8] and the crystal structure of a gp41 molecule containing the membrane proximal region [9], indicate that the MPER region, which is part of T20, elongates and stabilizes the six-helix bundle, raising the possibility that the additional interactions from the MPER contribute to the biological activity of T20.
Several laboratories previously suggested additional mechanisms for HIV-1 neutralization by T20 [10–12] involving binding to gp120. The first indication of gp120 involvement in HIV-1 neutralization by T20 was the observation that the sensitivity to T20 is modulated by the V3 region and that X4 viruses are more effectively neutralized by T20 in comparison with R5 viruses [13]. These studies demonstrated that gp120 of X4 and dual-tropic X4/R5 viruses binds T20 with a dissociation constant in the µM range [11], which is one to three orders of magnitude weaker than the EC50 of T20 [2, 14]. gp120 of R5 viruses did not exhibit measurable binding to T20 [10, 11]. Antibodies that interact with the base of the V3 and with the CD4-induced epitope on gp120, CD4i, located in the fourth constant region C4, competed with T20 binding to gp120 [10]. Moreover, a chimeric strain in which R5 V3 was inserted into an X4 gp140 construct did not interact with T20, whereas the control X4 gp140 construct and a chimeric X4 gp120 construct with R5 V1/V2 inserts did [10]. These results and the fact that X4 and R5 viruses differ mostly in their V3 sequences led the authors to suggest, that V3 is a major determinant for T20 binding to gp120.
The dependence of the efficiency of T20 in HIV-1 neutralization on co-receptor specificity and on V3 prompted investigation of T20 interactions with the co-receptor binding site. Both the V3 and C4 regions form the binding site for CCR5 and CXCR4 co-receptors and the CD4-induced epitope, CD4i, raising the possibility that both regions may influence gp120 binding to T20. Haynes and co-workers showed that a C4-V3MN and C4-V389.6 peptides corresponding to the X4 MN and dual-tropic 89.6 viruses, respectively inhibited T20 binding to the dual-tropic R5/X4 virus 89.8 complexed with the A32 antibody, which mimics soluble CD4 (sCD4) binding to gp120 [11]. A C4 peptide alone, but not a V3 peptide, inhibited the binding of the gp12089.8-A32 complex to T20, although more weakly in comparison with C4-V3MN and C4-V389.6 peptides [11]. These findings demonstrate that C4 is the major determinant for T20 binding to X4 strains. However, V3 of X4 or dual-tropic gp120 can increase the binding affinity of T20 to gp120 and to C4 peptides [11].
Using overlapping 15-mers of HIV-1MN gp120-derived peptides, Jiang and co-workers mapped the C4 segment interacting with T20 to the sequence 417QCKIKQI423 in the β19 strand of gp120 [12]. C4 peptides that include 417QCKIKQI423 abrogate the cell-cell fusion inhibition mediated by T20 on cells infected with HIV-1IIIB with an EC50 ranging from 0.1–1.6 µM [12]. In addition to the C4 and V3 regions, Jiang and co-workers have shown that peptides from the trans-membrane domain of gp41 block the fusion-inhibition activity of T20 with an EC50 around 6 µM [12]. The anti-HIV-1 activity of the C34 peptide, which contains more acidic residues in comparison with T20, could not be blocked by any of the gp120 peptides that blocked the activity of T20 [12]. All of the above results lead to the conclusion that T20, in contrast to other C-peptides, may interact with multiple sites on gp120 and gp41 of X4 or dual tropic viruses but not on gp120 of R5 strains to contribute to the inhibition of viral fusion.
Based on these findings in the literature, in the present study we set out to characterize T20 binding to gp120 of R5 viruses, to map the T20 segments and the gp120 regions involved in the binding, and to assess the significance of the T20-gp120 interaction to the development of HIV-1 entry inhibitors. CCR5 utilizing viruses are almost exclusively involved in virus transmission, therefore, there is a great interest in identifying sites of vulnerability on HIV-1 R5 strains. Our results demonstrate that T20 binds to R5 gp120 containing or lacking the V3 region and in the presence or absence of a CD4-mimic peptide (CD4M33). Using NMR spectroscopy and carefully designed peptide surrogates, we determined the segment of T20 that interacts with both gp120 and a gp120/CD4M33 complex, and mapped the entire C4 segment of gp120 that interacts with T20. Our conclusion is that T20 binds, through mostly hydrophobic interactions, to a surface on gp120 formed primarily by the C4 region. The ten to a thousand-fold difference between the EC50 of T20 against viral fusion and the affinity of T20 to gp120 implies that binding to gp120 is not a major factor in T20 inhibition of HIV-1 fusion. Nevertheless, this hydrophobic gp120 surface on gp120 could be an additional target for the development of higher-affinity compounds that might serve as anti-HIV therapeutics.
Results
NN-T20-NITN binding to R5 gp120
NMR spectroscopy and surface plasmon resonance (SPR) were used to characterize T20 binding to gp120. To increase the solubility of T20 for NMR studies, two asparagine residues and the tetrapeptide segment NITN (corresponds to the native gp41 sequence) were added at the N- and C-termini of T20, respectively [6]. This modified T20 peptide was designated NN-T20-NITN. To examine the role of V3 in T20 binding and whether R5 gp120 binds T20, the interaction of NN-T20-NITN with gp120 of the HIV-1JR-FL R5 strain was tested using surface plasmon resonance (SPR). Two gp120 variants were used in the present study, mutgp120core and mutgp120core(+V3) [15]. As previously described, these gp120 variants contain four mutations that alleviate the aggregation tendency of the glycoprotein [15]. Both gp120 constructs were expressed in GnTI- HEK293S cells [16, 17] and, therefore, were homogenously glycosylated with Man5GlcNAc2 glycans at sites normally occupied by complex or hybrid glycans.
Immobilized biotinylated NN-T20-NITN was used for measuring gp120 binding. The SPR data (Table 1) indicated that the binding of NN-T20-NITN to gp120 of the R5 strain JR-FL with truncated V3, mutgp120core, was weak but significant i.e 57 ± 9 µM. Binding improved only slightly to gp120 containing V3, mutgp120core(+V3) [35 ± 4 µM] and also slightly upon the addition of the CD4 mimic peptide CD4M33 [18] to mutgp120core (KD=40 ± 9 µM). In contrast, complexation of CD4M33 to mutgp120core(+V3) improved NN-T20-NITN binding by 4.5-fold to a KD of 8 ± 2 µM, the strongest NN-T20-NITN binding observed in our studies. When NN-T20-NITN binding to mutgp120core(+V3) and mutgp120core in complex with CD4M33 was compared, we found that V3 improved NN-T20-NITN binding by 5-fold. However, it is possible that the stronger mutgp120core(+V3)/CD4M33 binding to NN-T20-NITN is in part the result of the previously observed aggregation of mutgp120core(+V3)/CD4M33 [15]. Therefore, this contribution of V3 to gp120 binding to NN-T20-NITN, in the presence of CD4M33, may or may not be significant.
Table 1.
Dissociation constants for gp120 complexes with NN-T20-NITN.
| Ligand | Binding affinity (KD) |
|---|---|
| mutgp120core | 57 ± 9 µM |
| mutgp120core/CD4M33 | 40 ± 9 µM |
| mutgp120core(+V3) | 35 ± 4 µM |
| mutgp120core(+V3)/CD4M33 | 8 ± 2 µM |
The NN-T20-NITN segments involved in interaction with R5 gp120
As mentioned above, the T20-NITN portion of the modified T20 peptide corresponds to residues 638–673 of gp41 of HIV-1. To map the segment within the NN-T20-NITN peptide that interacts with gp120 we used 1H-15N-HSQC measurements of uniformly 15N labeled (U-15N) NN-T20-NITN in the presence and absence of gp120. In general, the 1H-15N-HSQC spectrum of a 15N labeled peptide in complex with a much larger protein discriminates residues within the segment that interact with the protein from those peptide residues outside this region. In the peptide/protein complex, protons in residues of the peptide in the interacting segment are characterized by short T2 relaxation time similar to the protein protons and therefore, exhibit broad and weak 1H-15N-HSQC cross peaks. Peptide protons outside the interacting segment retain long T2 relaxation and exhibit sharp, thus strong, 1H-15N-HSQC cross peaks [19]. Discrimination on the basis of differences in T2-relaxation times and peak intensities has been termed “dynamic-filtering” [19, 20].
Earlier studies of NN-T20-NITN were carried out at pH 7.6 at which the peptide exhibited improved solubility in comparison with neutral pH [6]. The 1H-15N-HSQC spectrum of uniformly 15N-labeled NN-T20-NITN was very well resolved. Backbone assignments at 298 K, the temperature at which most measurements in the present study were carried out, were facilitated by a series of 1H-15N-HSQC spectra measured at different temperatures and referencing to assignments made previously at 277 K [6]. The combination of pH 7.6 and 298 K resulted in increased amide-proton exchange-rates and, therefore, not all the amide protons of NN-T20-NITN were observed. The majority of the cross peaks that could not be observed were at the N-terminus of the peptide, a segment which was found previously to be mostly unstructured, in contrast to the C-terminal segment that populated a helical conformation [6, 7].
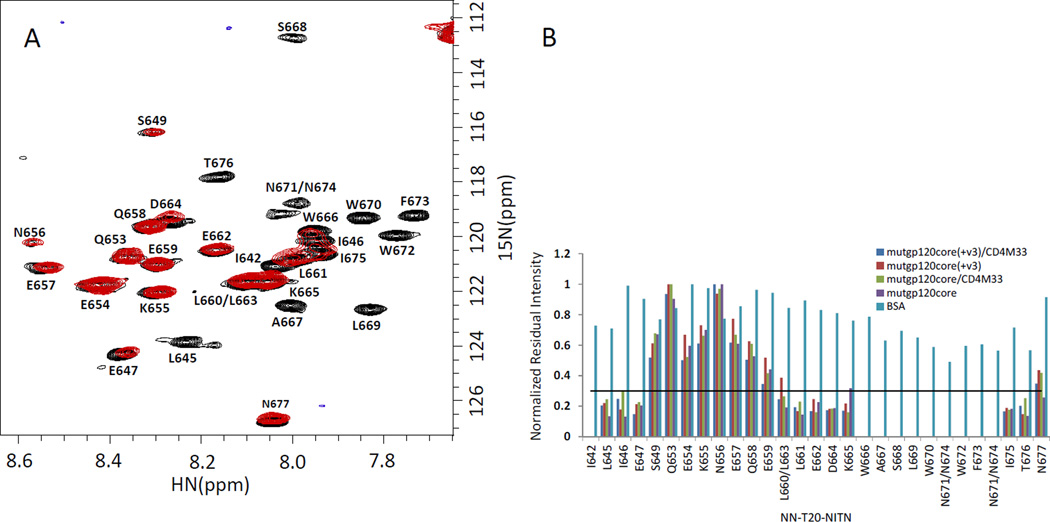
To optimize the gp120/NN-T20-NITN molar-ratio for the dynamic filtering measurements, the 1H-15N-HSQC spectra were recorded for solutions containing 0.5:1 and 1:1 molar ratios of mutgp120core and NN-T20-NITN. About ten cross peaks, for example W670, F673 and W672 in Fig. 1A and 1B, disappeared from the 1H-15N-HSQC spectrum of the 0.5:1 solution and a similar number exhibited considerably reduced cross-peak intensities. The other cross peaks retained strong intensities. The spectrum of the 1:1 molar ratio sample exhibited an overall reduction in signal-to-noise ratio (data not shown) probably due to aggregation. Therefore, we used 0.5:1 molar ratios of gp120 to NN-T20-NITN in the dynamic filtering measurements to discriminate between peptide residues in the segment that interacts with the protein and residues outside this segment. Some of the peptide residues that did not vanish completely but exhibited a significant reduction in cross peak intensities had minor changes in chemical shift in comparison to those in the free peptide. The disappearance of cross-peaks at a 0.5:1 molar ratio of mutgp120core to NN-T20-NITN suggests that the peptide is undergoing fast or medium exchange. Fast exchange would result in averaging of the chemical shifts and the T2 relaxation times of the free and bound peptide and a considerable loss in intensity for residues that interact tightly with the protein. Medium exchange can be an additional source of broadening for residues that exhibit change in chemical shift upon binding [21]. Peptide residues in regions that do not interact with the protein retain long T2 relaxation times and strong cross peak intensities and do not experience significant changes in chemical shift. For slow-exchange, no averaging of chemical shifts between bound and free peptide would occur and at 0.5:1 protein to peptide molar ratio the intensities of the amide cross peaks of peptide residues interacting with the protein would experience up to 50% reduction in cross peak intensities. This behavior was not observed for the NN-T20-NITN peptide bound to gp120.
Fig. 1.
Mapping the NN-T20-NITN segment interacting with gp120. (A) Overlay of the 1H-15N-HSQC spectrum of free 15N-labeled NN-T20-NITN ( black) and in the presence of 1:0.5 molar ratio of mutgp120core (red). (B) Normalized residual intensity for NN-T20-NITN amide-protons cross-peaks upon addition of a 50% molar ratio of gp120 variants or BSA. The intensity of the 1H-15N cross peak of a given residue when the peptide is in the presence of gp120 or BSA was divided by the intensity of the same residue in free NN-T20-NITN. The cross peak exhibiting the highest intensity ratio was used for normalization The solid black line represents a threshold of 0.3 for the normalized residual intensity below which the change in cross peak intensity is considered significant. All spectra were recorded for 40 µM concentration of NN-T20-NITN at 298 K in 20 mM D11-Tris-HCl pH 7.6, 50 mM NaCl and 95% H2O/5% D2O.
Similar NMR spectra were obtained when 40 µM U-15N-NN-T20-NITN was mixed with 20 µM of either: mutgp120core(+V3), mutgp120core(+V3)/CD4M33, mutgp120core or mutgp120core/CD4M33 (Fig. 1B). Based on the decrease in cross-peak intensities of U-15N- NN-T20-NITN upon gp120 binding, the NN-T20-NITN segment interacting with gp120 was mapped. Peak intensity in the presence of 50% molar ratio was compared to the free sample. The intensity of the 1H-15N cross peak of a given residue of the peptide in the presence of a gp120 variant or BSA (see below) was divided by the intensity of the same residue in the free NN-T20-NITN. Subsequently the cross peak exhibiting the highest intensity ratio was used for normalization of all other residues. Significant reduction in cross-peak intensity (>70%) is considered to be indicative of involvement in gp120 binding (Fig. 1B).
Approximately 20 cross peaks exhibited more than 70% decrease in intensity (Fig. 1B). These include residues L661-T676 and another much shorter segment located at the N-terminus of NN-T20-NITN. Very similar results were obtained for mutgp120core, mutgp120core(+V3)/CD4M33, mutgp120core and mutgp120core /CD4M33 (Fig. 1B), indicating that the V3 is not necessary for NN-T20-NITN binding. This observation is consistent with the SPR results.
In order to assess non-specific hydrophobic interactions, BSA was tested as an additional ligand (Fig. 1B). In general, the presence of BSA did not result in considerable loss of NN-T20-NITN cross-peak intensities. Although some peak-intensity loss was observed in the region spanning A667-N674, the effect was minor (usually 20–30% peak loss) compared with that caused by gp120 ligands, indicating that the interaction with gp120 is considerably stronger and perhaps more specific than that with BSA.
T20 residues that interact directly with gp120
The dynamic-filtering approach identifies the residues located in the peptide segment interacting with the protein but cannot discriminate between residues that are involved in direct interaction with the protein and residues in this segment that do not interact directly with the protein but are affected indirectly because of changes in the mobility of the interacting segment. The saturation-transfer-difference (STD) technique [22] enables identification of ligand-residues that interact directly with a macromolecule. A 1D STD spectrum measured for mutgp120core/CD4M33 complex in the presence of a 10-fold concentration of NN-T20-NITN (Fig. 2 red spectrum) revealed that certain aromatic residues as well as leucine and isoleucine residues of T20 exhibited the highest saturation transfer from the irradiated mutgp120core. Due to the overlap of the resonances of W666, W670, W672 and F673 we could not assign the STD signal observed to specific aromatic residues. However, since the four aromatic residues are located in the segment interacting with mutgp120core, as judged by the reduction in cross-peak intensities (Fig. 1), and since these are the only tryptophan and phenylalanine residues of NN-T20-NITN, it is likely that all or most of these aromatic residues interact directly with mutgp120core. Similarly, the overlap of the methyl resonances of isoleucine and leucine residues of T20 prevented their assignment to specific isoleucine or leucine residues interacting with NN-T20-NITN. The major segment interacting with mutgp120core contains L669, and the short N-terminal segment that interacts with mutgp120core contains I642, L645 and I646. Much weaker saturation transfer was observed for protons with resonances in the 2–3.5 ppm region (marked polar in Fig. 2; typical of side chains of polar residues such as Asp, Asn, Glu, Gln and Lys) suggesting that the side chains of these do not interact directly with mutgp120core. Thus, it seems that the residues interacting with mutgp120core form a hydrophobic surface consisting of all or most of the side chains of W666, A667, L669, W670, W672 and F673 at the C-terminal half of NN-T20-NITN.
Fig. 2.
1D–STD spectrum of NN-T20-NITN in the presence of mutgp120core/CD4M33. Reference and difference spectra are shown in blue and red, respectively. Peaks marked with arrows exhibit the most significant STD effect. The sample contained 14 µM mutgp120core/CD4M33 with a 10-fold excess of the NN-T20-NITN peptide. The experiment was carried out on AVIII800 at 298 K in 20 mM D11-Tris-HCl pH-7.6, 50 mM NaCl using on-resonance irradiation at −2 ppm.
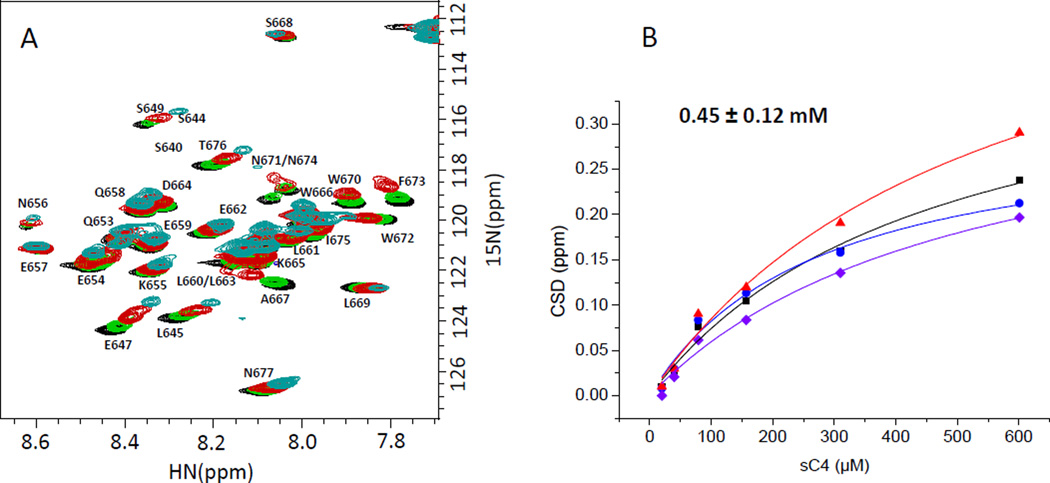
The NN-T20-NITN segment interacting with a C4 peptide
Peptides corresponding in sequence to the fourth constant region of gp120, C4, were previously found to abolish T20 binding to R5/X4 gp120 [11] and the HIV-1 inhibitory activity of T20 on X4 viruses [12]. We, therefore, wanted to map the T20 determinant interacting with C4 and to examine whether the same T20 determinant binds to both a C4 peptide and the gp120 protein. For this purpose, a U-15N labeled NN-T20-NITN sample was titrated at 298 K with a synthetic soluble C4 peptide, sC4, comprising the entire C4 segment (residues 417–444) of the R5 HIV-1 strain JR-FL with two additional residues at the N-terminus of the peptide. Cysteine 418 was replaced by serine in the sC4 peptide. Three serine residues were added at each terminus as a solubility tag (peptide sC4 in Table 2). A large number of NN-T20-NITN cross peaks exhibited a gradual change in their chemical shift upon titration with sC4 (Fig. 3A) and a KD of 450±120 µM was determined for sC4 binding to NN-T20-NITN at 298 K (Fig. 3B). A decrease in cross peak intensity was also observed for many cross peaks and was correlated with a large perturbation in chemical shift. The largest changes in chemical shift upon sC4 binding were observed in the C-terminal region of NN-T20-NITN, specifically for residues A667, N671, F673 and N674. E647 in the N-terminal region of NN-T20-NITN also exhibited a significant change in chemical shift. Considerable intensity loss upon sC4 binding was observed for residues K665, A667, W670 and W672. These observations suggested that the NN-T20-NITN segment interacting with sC4 comprises residues K665-F673 and is similar to the NN-T20-NITN segment interacting with gp120, indicating that the sC4 peptide partially recapitulates the binding of gp120 to NN-T20-NITN although with a one order larger KD.
Table 2.
Peptides used in the present study
| NN-T20-NITN | 636 NNYTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWFNITN677 |
| Bio-NN-T20-NITN | Biotin-GSG-636NNYTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWFNITN677 |
| sC4 | SSSPS-419RIKQIINMWQEVGKAMYAPPIRGQIR444-SSS |
| Rec-sC4 | PSSS-419RIKQIINMWQEVGKAMYAPPIRGQIR444-SSS |
Fig. 3.
Binding of sC4 peptide to NN-T20-NITN. (A) An overlay of the 1H-15N-HSQC spectra of 15N-labeled NN-T20-NITN titrated with SC4 (black: free 15N-labeled NN-T20-NITN; Green: after addition of sC4 in a 1:1 molar ratio; Red: 4:1 molar ratio; blue: 16:1 molar ratio of sC4 to 15N-labeled NN-T20-NITN). B. The plots of the changes in NN-T20-NITN chemical shifts for the four residues which showed the largest chemical shift deviations upon binding ( A667, N671 or N674, N674 or N671, F673). U-15N labeled NN-T20-NITN was at a concentration of 40 µM at 298 K, 20 mM D11-Tris-HCl pH 7.6, 50 mM NaCl.
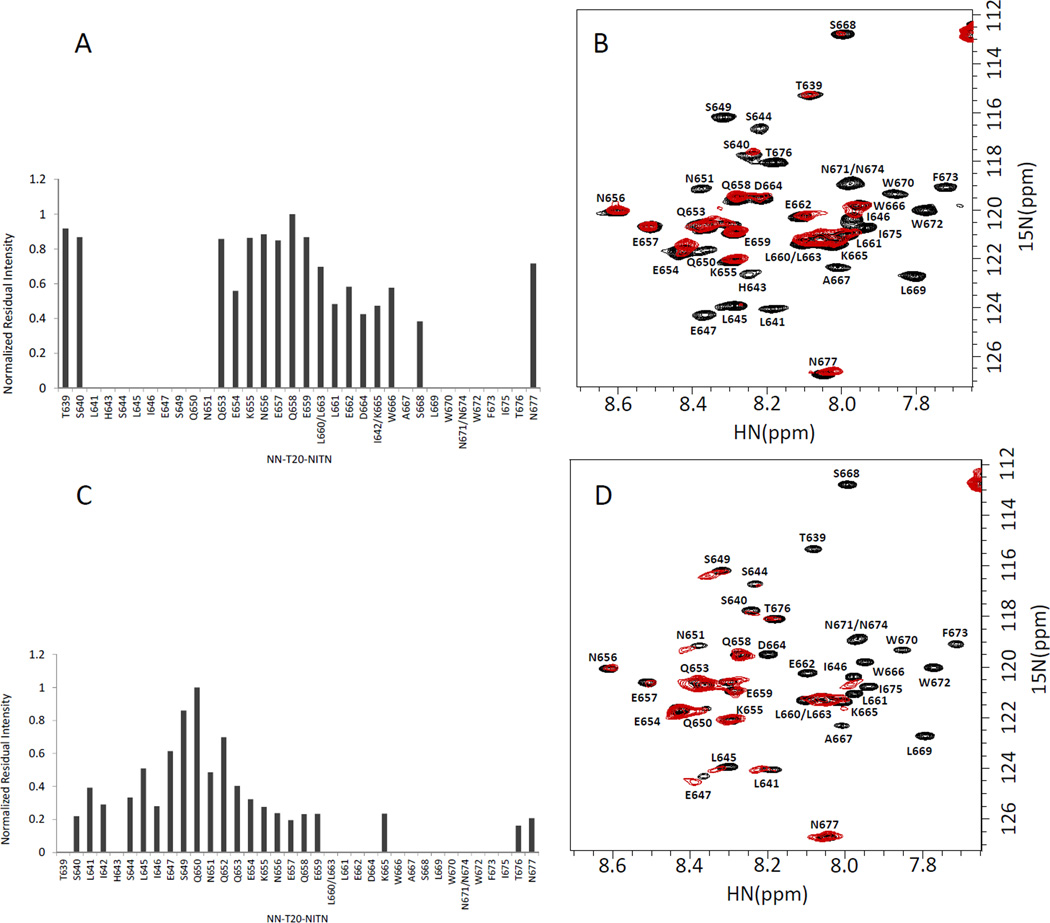
At 298 K the discrimination between NN-T20-NITN residues found in the segment interacting with sC4 and those outside this segment was not as pronounced as seen for NN-T20-NITN interacting with gp120 proteins. Moreover, as discussed below, the mapping of sC4 residues interacting with NN-T20-NITN could not be carried out under the conditions (pH 7.6; 298 K) used to study NN-T20-NITN/R5-gp120 interactions (Fig. 1) since many sC4 cross peaks were not observable under these measurement conditions. To map the interacting residues in NN-T20-NITN and sC4 under the same conditions, and to obtain better discrimination between interacting and non-interacting segments, the NMR measurements were repeated at 288 K, and pH 7. No NaCl was added to improve NN-T20-NITN solubility. According to the observed decrease in cross-peak intensities (Fig. 4), probably caused by broadening due to intermediate exchange [21], the segments of NN-T20-NITN interacting with sC4 were mapped to residues L641-N651 and A667-T676 of NN-T20-NITN. Under these measurement conditions a better definition of the N-terminal segment of NN-T20-NITN interacting with the sC4 peptide was obtained. The disappearance of NN-T20-NITN cross peaks measured for a 1:1 molar ratio of NN-T20-NITN and sC4 at 288 K suggests that the exchange is in the medium rate regime similar to that observed for the gp120/NN-T20-NITN complex, rather than the fast exchange observed for the two peptides complexed at 298 K.
Fig. 4.
Mapping the NN-T20-NITN segment interacting with a C4 peptide (sC4) and gp120. A. Normalized residual intensity for each residue in 15N-labeled NN-T20-NITN upon addition of a 1:1 molar ratio of sC4 at 42 µM. B. An overlay of the 1H-15N-HSQC spectrum of 15N-labeled NN-T20-NITN in the presence of 1:1 molar ratio of the C4 peptide sC4 (red) and the spectrum of the free NN-T20-NITN (black). C. Normalized residual intensity for each residue in 15N-labeled NN-T20-NITN upon addition of a 1:0.5 molar ratio of mutgp120core (+V3)/CD4M33 at 44 µM. D. An overlay of the 1H-15N-HSQC spectrum of 15N-labeled NN-T20-NITN in the presence of 1:0.5 molar ratio of mutgp120core (+V3)/CD4M33 (red) and the spectrum of free NN-T20-NITN (black). The normalized intensity was calculated by dividing the intensity of a specific 1H-15N cross peak of NN-T20-NITN in the presence of sC4 or mutgp120core (+V3)/CD4M33 by the intensity of the same residue in the spectrum of free NN-T20-NITN and then all of the relative intensities were normalized to the residue that exhibited the highest ratio. The measurements were done in 20 mM D11-Tris-HCL at pH 7, 288 K.
For direct comparison, the HSQC spectrum of U-15N labeled NN-T20-NITN in the presence of 50% molar concentration of gp120 was also measured at pH 7 and 288 K and a decrease in cross peak intensity upon gp120 binding was observed (Fig. 4). Similar to what was found with the sC4 peptide, the NN-T20-NITN segment A667-T676 at the C-terminal half of the peptide interacts with gp120. However, in the case of gp120 four additional residues, L660-D664, bind to the protein. On the other hand, the NN-T20-NITN N-terminal segment interacting with sC4 contains five additional residues (E647-N651) in comparison with the segment interacting with gp120 (S640-I646). Thus, the sC4 peptide partially recapitulates gp120 binding to NN-T20-NITN. Based on the level of intensity loss, at pH7 and 288 K the NN-T20-NITN N-terminal segment (S640-I646) seems to interact more weakly with gp120 compared with the NN-T20-NITN C-terminal segment (L660-I675).
Mapping the C4 region interacting with NN-T20-NITN
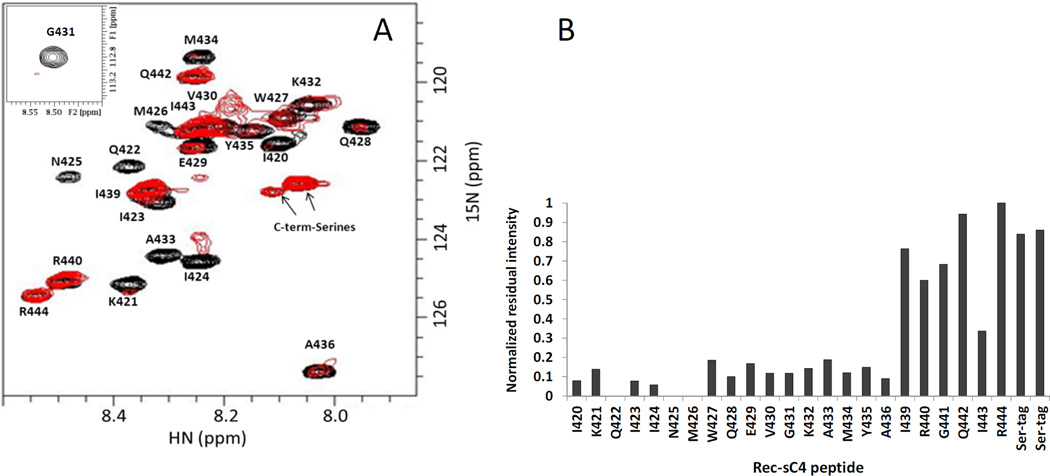
Given the findings with sC4, we choose to use it to define the C4 segment interacting with NN-T20-NITN. Recombinant soluble C4 peptide corresponding to the HIV-1JR-FL sequence was expressed in E. coli with serine residues added to the N and the C terminus as solubility tags (rec-sC4 in Table 2). Assignment of the peptide was obtained using 3D 15N-separated NOESY and TOCSY spectra of the peptide measured at pH 7 and 278 K with standard sequential assignment procedures. Titration of U-15N-sC4 with NN-T20-NITN was carried out at as above (288 K, pH 7). Under these conditions most of the sC4 amide cross peaks could be observed. Assignment of the HSQC spectra at 278 K could be easily transferred to 288 K by a gradual change in the measurement temperature of the HSQC spectrum. Addition of 15% molar excess of unlabeled NN-T20-NITN to the U-15N-rec-sC4 caused > 80% reduction in amide cross peak intensity of residues I420-A436 (Fig. 5A and 5B), suggesting that this segment forms the determinant that binds NN-T20-NITN.
Fig. 5.
Mapping the C4 segments interacting with NN-T20-NITN. (A) An overlay of the 1H-15N-HSQC spectrum of U-15N-rec-sC4 in the presence of 1:1.15 molar ratio of NN-T20-NITN (red) and the spectrum of free 15N-labeled rec-sC4 (black). (B) Normalized residual intensity for each residue in the rec-sC4 upon addition of a 15% molar excess of T20. The normalized intensity was calculated by dividing the intensity of a specific 1H-15N cross peak of rec-sC4 in the presence of NN-T20-NITN by the intensity of the same residue in the spectrum of free rec-sC4. The residue that exhibited the highest ratio was then used to normalize all relative intensities Rec-SC4 concentration was 45 µM at 288 K, 50 mM D11-Tris-HCl, pH 7.0.
Discussion
The interacting residues of T20 and the R5-gp120 C4 region
In the present study we have provided new insights into the interaction of T20 and R5-gp120 of HIV-1. We quantitated the binding of the NN-T20-NITN peptide to R5-gp120, demonstrating that this drug binds not only to X4 gp120, as was shown previously, but also to gp120 of R5 viruses that are the dominant cause of infection. We found that the binding of T20 to R5-gp120 is one order of magnitude weaker than reported for T20 binding to dual tropic X4/R5 gp120 [11]. Using high-resolution heteronuclear NMR we provide information on segments of T20 that interact with R5-gp120, and the R5-gp120 C4-residues that interact with NN-T20-NITN. The primary interaction between this entry inhibitor and gp120 involves a predominantly hydrophobic surface on gp120 (Fig. 6) and a hydrophobic surface of an amphiphilic helix at the C-terminal half of NN-T20-NITN (Fig. 7). Our study has shown that the core of gp120 is the dominant contributor to T20 binding and that the V3 region of the consensus JRFL R5 virus increases the binding of T20 to gp120 only by up to a factor 6. Since the core region of gp120 is mostly conserved between X4 and R5 viruses, our conclusions regarding T20 binding to the gp120 core of R5 viruses will likely hold for the gp120 core of X4 viruses.
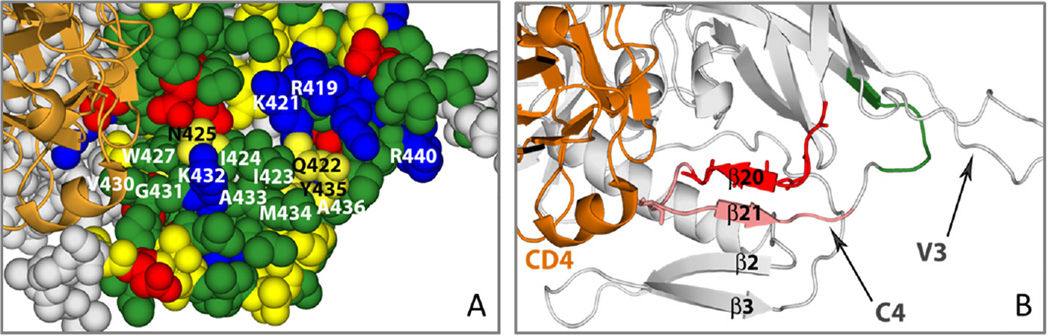
Fig. 6.
The gp120 C4 surface interacting with NN-T20-NITN. (A) Space filling representation of gp120. The residues in the C4 region and its vicinity are color coded according to charge and polarity. Negatively charged residues are colored red, polar yellow, hydrophobic green and positive blue. The C4 residues found by our NMR analysis to be involved in NN-T20-NITN binding are labeled. sCD4 is represented in orange ribbon to show where it binds gp120 and its position with respect to C4. (B) Ribbon presentation of the bridging sheet region of gp120, formed by β2, β3, β20 and β21 strands. The β20 and β21 form a surface that interacts with NN-T20-NITN. The segment that is colored red contains the residues that strongly interact with NN-T20-NITN and disappear in the NMR analysis upon the addition of NN-T20-NITN. The segment that is colored pink contains the residues that interact more moderately with NN-T20-NITN. The residues colored green (including V430) are the residues that were not significantly affected upon binding to NN-T20-NITN. The gp120 coordinates were taken from 2B4C.pdb.
Fig. 7.
Comparison of NN-T20-NITN and ECL2S binding surfaces to gp120. (A) Ribbon and surface representation of the T-20 binding surface taken from the gp41 trimetric structure (adapted from PDB 2×7R , Buzon et.al). (B) Ribbon and surface representation of the ECL2S binding surface (Abayev et al). Positively charged residues are colored blue, polar in yellow and hydrophobic in green. Residues participating in gp120 binding are labeled.
Our NMR measurements map the T20 region interacting with gp120 to the segment W666-N674 at the C-terminal half of T20. This segment forms the membrane-proximal external region of gp41 and is the target for HIV-1 broadly neutralizing antibodies such as 2F5 and 4E10 [23, 24]. The W666-N674 region of T20 forms an amphiphilic helix presenting a large hydrophobic surface on one side and a polar surface on the other side [6, 7, 9] (Fig. 7). On the basis of the STD experiments, we conclude that the hydrophobic surface created by the side chains of W666, L669, W670, W672 and F673 interacts directly with gp120.
The present study also identified a secondary NN-T20-NITN N-terminal segment that interacts with gp120. The binding of this N-terminal segment to gp120 is weaker than that of the amphiphilic C-terminal segment (W666-N674) as evidenced by higher residual mobility after gp120 binding (Fig. 1B and Fig. 4 C and D). This secondary site interacts with both mutgp120core and mutgp120core(+V3) (Fig. 1B) indicating that V3 is not involved in its binding.
A soluble peptide (sC4) comprising C4 residues 419–444 partially recapitulates the binding of gp120 lacking the V3 to NN-T20-NITN, but has a one-order of magnitude larger KD. There are similarities in the T20 segments that interact with gp120 and sC4 (Fig. 4). However, gp120 interacts with a shorter segment in the NN-T20-NITN N-terminal region and with a longer segment in the peptide’s C-terminal region. The C-terminal segment of NN-T20-NITN (W666-N674) is the major contributor to gp120 binding. These differences could arise from the increased flexibility and solvent exposure of the C4 peptide in comparison with those of this same region in the gp120 protein. Nevertheless, we felt it was reasonable to use sC4 to identify potential gp120 residues interacting with T20. HSQC experiments, using 15N-labeled sC4, mapped the C4 segment interacting with NN-T20-NITN to C4 residues I420-A436 (Fig. 5). We could not draw any conclusions regarding the participation of C418 and R419 in NN-T20-NITN binding since the first residue was replaced by serine and since the amide protons of both R419 and S418 could not be observed under the measurement conditions probably due to fast exchange.
Previously, Haynes and co-workers mapped the gp120 binding site for NN-T20-NITN to the C4 and V3 regions of X4 or dual tropic X4/R5 gp120, with C4 being the major contributor to T20 binding [11]. Using overlapping peptide sequences, Jiang and co-workers mapped the T20 binding determinant to the C4 segment 417QCKIKQI423 [12], which forms part of the co-receptor binding site. The 420IKQI423 sequence is present in our C4 peptide while the R5-C4 sequence contains arginine instead of a lysine at position 419 (see Table 3). C418 is conserved in both X4 and R5 viruses but, as mentioned above, was replaced by serine in the C4 peptide used in the present study. Our NMR results reveal that the interacting segment in C4 peptides is much more extensive than that found in the biochemical studies and mapped a continuous, mostly hydrophobic surface from residue 420 to 436 formed by the β20 and β21 strands of the C4 domain of gp120 as contributing to T20 binding (Fig. 6). Unlike T20, the C4 peptides do not have any tendency to form a helical conformation and do not adopt any well-defined secondary structure as judged from the deviations of the Hα chemical shifts from random coil values (data not shown). The βαβ supersecondary structures are a common motif found in proteins and it is possible that the amphiphilic helix formed by T20 could pack well against the β20-β21 hairpin and formed by the gp120 C4 region.
Table 3.
Sequence alignment of the C4 and V3 regions of gp120. Shown are the sequences of a representative R5 strain (JR-FL), X4 strain (MN) and dual-tropic (89.6, 89.6P). Shown also are the C4 peptides used in this study. Tag residues are italic and underlined.
The segment at the N-terminus of the C4 segment interacting with T20 is 90% identical in X4 and R5 viruses. This suggests that C4 of X4 and R5 viruses interact with T20 in the same manner. On the other hand, comparison of the V3 sequences of representative R5, X4 and dual-tropic X4/R5 viruses reiterates that the V3 sequences vary considerably between X4 and R5 viruses (Table 3), especially in their net charge, being +8 for the X4 MN strain and +3 for the R5 JR-FL strain. The large differences in the V3 sequences could modulate the affinity of gp120 for T20 and may explain the one order of magnitude difference in the binding of mutgp120core(+V3) of the R5 virus, JR-FL in complex with CD4M33, to T20 determined in the present study, compared to the binding to gp120 of the dual-tropic HIV-189.6 in complex with sCD4 [11]. Although this one order of magnitude difference in binding affinity to gp120 observed in the present study correlates well with the increased potency of T20 in neutralizing X4 in comparison with R5 viruses [13], other characteristics of R5 and X4 viruses may contribute to the observed differences in the T20 neutralizing potency.
Comparison to the gp120 surface interacting with antibodies targeting the CD4-induced epitope
The binding site for T20 on gp120, as deduced using sC4, is part of the CD4-induced surface, CD4i, which contains the bridging sheet formed by strands β2, β3, β20 and β21 [25] (Fig. 6). The later two strands, β20 and β21, are part of the C4 region of gp120. The CD4i determinant, which partially overlaps with the co-receptor binding site [26], is also the epitope for several antibodies such as 17b, CG10, X5, 48d and E51 [26–29]. Fab and Fv fragments of these antibodies neutralize diverse HIV-1 strains. However, partial occlusion of this site in the trimeric envelope spike, and steric hindrance preventing access of the large antibody molecules to this site after CD4 binding and the attachment of the virus to its target cell, limits the neutralizing activity of these antibodies. A common feature of CD4i–directed antibodies is that they interact with residues such as R419, 420I, K421 and Q422 in the β20 strand, which forms the N-terminal segment of C4. Some of these antibodies, such as CG10 and X5, interact also with the β20 strand, as does T20 [26]. The fact that the same site on gp120 is a target for T20 and for neutralizing antibodies directed against CD4i, suggests that if high affinity small molecules could be designed to target this region of gp120, they might be effective HIV-1 entry inhibitors.
Comparison with small molecule entry inhibitors
BMS-626529 and its predecessor BMS-373806 are HIV-1 entry inhibitors targeting gp120 [30, 31]. Site-directed mutagenesis and mechanistic studies suggest that these two compounds bind to the CD4 binding site, and its vicinity, and to the water channel formed after CD4 binding [30, 31]. V430, at the tip of the hairpin created by C4, as well as N425 and W427, are also part of the CD4 binding interface. K421, I423, I424 and N425 of gp120 C4 participate in forming the water channel. Thus, the C4 region of gp120 is a target for both T20 binding and also for HIV-1 entry inhibitors. The importance of C4 as a target for HIV-1 entry inhibitors is further supported by the discovery of the entry inhibitor 18A and the contribution of C4 residues I430 and M434 to its binding [32].
Comparison between T20 and HIV-1 neutralizing CCR5 ECL2 peptides
Peptides corresponding to the sequence of the putative second extra cellular loop, ECL2, of CCR5 were found to inhibit HIV-1 infection [33, 34]. Residues Q188-Q194 in an elongated ECL2 peptide (R168 to K197; ECL2S) were found to form an amphiphilic helix, which corresponds to the beginning of the fifth transmembrane helix in the crystal structure of CCR5 [35]. Residues Y187, F189, W190 and F193 of ECL2S form a hydrophobic surface of this amphiphilic helix that was found to interact directly with gp120 [35]. Although these interactions are not representative of the interactions of gp120 with ECL2 in the intact CCR5 receptor, they are crucial for the inhibition of HIV-1 by the ECL2 peptides.
The NN-T20-NITN peptide is mostly helical in the segment E657-L669 and partially helical in the segment W670-F673 [6]. The crystal structure of an HIV-1 gp41 construct, that included both the fusion peptide and membrane proximal external regions of the envelope protein, showed that the HR2 helix extends to residue 676 [9], which spans almost all of the NN-T20-NITN peptide. The T20 helical segment that interacts with gp120 consists of the 8-residue segment W666-F673 that is similar in length to the 7-residue ECL2 helical segment that interacts with gp120. Both helices are amphiphilic helices and present a large hydrophobic surface that would be capable of interacting with an opposing hydrophobic surfaces on gp120 (Fig. 7). These gp120 surfaces can serve as a target for HIV-1 entry inhibitors.
Conclusions
The KD of NN-T20-NITN binding to R5 mutgp120(+V3) is about two to three orders of magnitude higher than the IC50 of T20 against the virus [2] suggesting that binding alone to gp120 contributes only marginally to its inhibition of R5 viruses. The major mechanism of inhibition of R5 viruses by T20 is therefore, the inhibition of the formation of the six-helix bundle by the gp41 trans-membrane glycoprotein.
Our studies have revealed that T20 binds also to R5-gp120 and pinpointed specific areas on T20 that interact with specific areas on the C4 region of gp120. We found strong similarity between the T20 and the ECL2 surfaces interacting with mutgp120core. The inhibition of HIV-1 infection by these two peptides suggests that there is a hydrophobic site or sites of vulnerability in the envelope spike of HIV-1 that can be targeted by HIV-1 entry inhibitors. This gp120 region at least partially overlaps the co-receptor binding site.
Materials and Methods
Peptides’ Synthesis
CD4M33 was synthesized as described previously [17]. The peptide sC4 was synthesized and purified using similar procedures as described previously [36].
SPR measurements
Binding affinities (KD) were determined by SPR using a ProteOn XPR36 Protein Interaction Array System (Bio-Rad Haifa, Haifa, Israel). All experiments were carried out at 25 °C in SPR running buffer [20 mM Tris (pH 7.5), 50mM NaCl, 15mM MgCl2 0.05% NaN3 and 0.005% Tween]. Biotinylated-GSG-T20 was immobilized on a carboxymethylated dextran matrix chip precoated with streptavidin (ProteOn NLC sensor chip) at a flow-rate of 30 µl/sec to 212, 490 and 760 response units (RU). A blank sensor channel, with no peptide immobilized, served as a negative control and as a reference. Six threefold serial dilutions of gp120, mutgp120core and BSA (in the range 70–0.29 µM) or mutgp120core(+V3)/CD4M33 and mutgp120core /CD4M33 (in the range 33–0.135 µM) were prepared in running buffer and injected over the biotinylated NN-T20-NITN peptide immobilized surface at 60 µl/sec for 150 sec. Regeneration was performed between the different analytes by running 10 mM NaOH at 30 µl/sec for 50 sec contact time followed by an additional step with running buffer. Average measured KD values and standard deviations are shown for the binding measurements done at three levels of peptide immobilization. Data were analyzed with the ProteOn Manager software after subtraction of a blank channel using equilibrium analysis.
Expression and purification of gp120
Truncated 88–492gp120JR-FL lacking the V1 and V2 variable loops was expressed in HEK293 cells, two glycosylation sites, N301Q and T388A, were mutated resulting in loss of glycosylation at these sites. Additionally, four residues in the first α-helix (N99, E106, D107 and D113) were mutated to glutamine to create non-aggregating gp120, termed mutgp120core(+V3). A second construct, mutgp120core, in which residues R298–A330 of the V3 loop of mutgp120core(+V3) were replaced by a GAG sequence, termed mutgp120core, was also used. mutgp120core(+V3) and mutgp120core were expressed in HEK293 cells lacking N-acetyl glucosaminyl transferaseI enzymatic activity (GnTI− HEK293S cells) resulting in a protein homogenously glycosylated with Man5GlcNAc2 glycans at sites normally occupied by complex or hybrid glycans. In this study the carbohydrates were not removed by an enzymatic reaction. Expression and purification of the gp120 molecules were performed as described previously [15].
Expression and purification of C4 and NN-T20-NITN peptides
The NN-T20-NITN peptide was expressed as TrpΔLE fusion peptides containing an N-terminus poly-His Tag. The plasmid was transformed into E. coli strain BL-21(DE3) and grown at 37 °C in LB or minimal medium (M9) with 15N ammonium chloride for 15N labeled protein. The insoluble fraction was collected. Initially, the fusion peptide was purified on Ni-column by FPLC or gravity flow, followed by dialysis and lyophilization. The fusion protein was cleaved using a 400:1 molar ratio of CNBr to the fusion protein added at time 0 and again after 4 hours for a total of 8 hours cleavage in 70% TFA, 1 mg/ml fusion protein at room temperature. The cleavage fragments were recovered by rotary evaporation, and lyophilization, dissolved in 0.1% TFA 10% acetonitrile (10–20 ml) and loaded on a RP-HPLC C4 22 mm column on acetonitrile gradient 0.1% TFA, collected, lyophilized and analyzed by MassSpec (5179 Da for U-15N NN-T20-NITN, theoretical value 5181.5). Rec-sC4 was similarly expressed and purified, the fusion protein was cleaved at acetic conditions which cleaves between the Asp and Pro residues preceding the target peptide. 10% formic acid was used for 2.5 h at 80°C (3768 Da for U-15N- theoretical value 3766.3).
NMR Measurements
For mapping the NN-T20-NITN segment interacting with gp120, samples of 15N labeled NN-T20-NITN were dissolved in a 20 mM aqueous solution of D11-Tris-HCl buffer, pH 7.6, 5% D2O, 50 mM NaCl, 0.05% NaN3. mutgp120core(+V3), mutgp120core were dialyzed into NMR buffer and added in the presence or absence of CD4M33 to the 15N labeled NN-T20-NITN. All spectra were recorded for 40 µM concentration of NN-T20-NITN at 298 K in 20 mM D11-Tris-HCl pH 7.6, 50 mM NaCl 95% H2O/5% D2O, 0.05%NaN3. Samples were supplemented with 1 mM deuterated EDTA and protease inhibitor cocktail (Sigma-Aldrich P8340).
To map the NN-T20-NITN segments interacting with sC4, a concentrated solution of sC4 peptide was prepared in d6-DMSO and titrated to U-15N-NN-T20-NITN peptide at a concentration of 40 µM in 20 mM D11-Tris-HCl pH 7.6, 50 mM NaCl, 95% H2O/5% D2O, 0.05% NaN3 at 298 K. 1H-15N-HSQC spectra were also recorded for six points ranging from 20 µM-600 µM of sC4. Alternatively, U-15N labeled NN-T20-NITN was prepared at 37 µM concentration in 20 mM D11-Tris-HCl pH 7.0, 95% H2O/5% D2O, 0.05% NaN3 to which sC4 peptide from a concentrated solution in d6-DMSO was added to give a final 1:1 molar ratio. 1H-15N-HSQC was recorded at 288 K.
For Rec-sC4 resonance assignment, samples of 250 µM 15N labeled Rec-sC4 were dissolved in a 50 mM aqueous solution of D11-Tris-HCl buffer, pH 7.0, 5% D2O. 15N-separated NOESY and 15N-separated TOCSY at 278 K were used for sequential assignment using standard procedures. 1H-15N-HSQC spectra were recorded at 278 K, 283 K and 288 K.
To map the C4 segment interacting with NN-T20-NITN, a 45 µM solution of U-15N labeled Rec-sC4 peptide was prepared in 50 mM aqueous solution of D11-Tris-HCl buffer, pH 7.0, 5% D2O. 50 µM of unlabeled T20 from 100% d6-DMSO stock of 10 mM was added to the NN-T20-NITN solution resulting in a 15% molar excess.
All NMR spectra were measured on a Bruker AVIII800 spectrometer equipped with a 5 mm TCI cryoprobe. Data were processed and analyzed using the Topspin (Bruker-De), NMRDraw and NMRView programs.
NMR chemical shift perturbation analysis
Chemical shift perturbation (CSD) was calculated using the formula [Δ(δH)2 + Δ(δN/5)2]1/2, where Δ(δH) and Δ(δN) are the changes in 1H and 15N chemical shifts of NN-T20-NITN, respectively. CSD was plotted against sC4 peptide concentration and the dissociation constant of the complex was determined using a one-site binding model (Origin software).
In order to assess loss in 1H15N-HSQC cross peak intensity upon titration of ligand, the peak intensity of the peptide in the bound state was divided by the intensity of the corresponding peak in the free-state. The values were calibrated so that the peak resulting in the highest ratio was set to 1. The resulting “normalized residual intensity” was plotted for each visible residue.
1D saturation transfer difference (STD)
1D–STD was acquired on the AVIII-800 spectrometer using an irradiation power of 176 Hz for 2 sec at −2 ppm (or at 30 ppm for the control experiment). The sample contained 14 µM mutgp120core/CD4M33 with a 10-fold excess of the NN-T20-NITN peptide. The experiment was carried out at 298 K in 20mM D11-Tris-HCl pH-7.6, 50 mM NaCl supplemented with 1 mM benzamidine, 1 mM deuterated-EDTA and protease inhibitor cocktail (Sigma-Aldrich P8340).
Acknowledgement
We are most grateful to Meital Abayev for Rec-sC4 purification and Dr. Sabin Akabayov for figure preparation. ‡ This study was supported by Minerva and Israel Science Foundation grants, by NIH Grants GM53329 (J.A.) and GM22087 (F.N.) and by the Kimmelman Center. J.A. is the Dr. Joseph and Ruth Owades Professor of Chemistry. F.N is the Leonard and Esther Kurtz Term Professor at the College of Staten Island of the City University of New York. T.S. is the Incumbent of the Monroy-Marks Research Fellow Chair.
Abbreviations
- CCR5
human C-C chemokine receptor 5
- CD4
cluster of differentiation 4 CD4M33 a peptide mimic of CD4 used in this study
- gp120
the extra cellular subunit of the HIV-1 envelope protein
- HIV-1
human immuno-deficiency virus type 1
- HPLC
high pressure liquid chromatography
- HR1 and HR2
helical repeats 1 and 2, respectively, of gp41
- HSQC
hetero-nuclear single quantum coherence
- gp41
the trans-membrane HIV-1 envelope glycoprotein protein
- T-20
HIV fusion inhibitor corresponding to gp41 residues Y638–F673
- NN-T20NITN
A molecule corresponding to gp41 residues N636-N677 with the addition of two asparagine residues at the N-terminus
- mutgp120core
a gp120 core molecule that contains four mutations that alleviated the aggregation problem and which lacks V1, V2 and V3
- MPER
the membrane proximal external region of gp41
- NMR
nuclear magnetic resonance
- NOE
Nuclear Overhauser effect
- STD NMR
Saturation Transfer difference NMR
- sC4
a molecule corresponding to residues R419-R444 of gp120 flanked by SSSPS and SSS tags at the N-and C-terminus respectively
- RsC4
a molecule corresponding to residues R419-R444 of gp120 flanked by PSSS and SSS tags at the N-and C-terminus respectively
- SPR
surface plasmon resonance
- V1,V2 and V3
variable loops number 1, 2 and 3 of HIV-1 gp120
Footnotes
Author Contributions
AM and ZB helped to design the experiments, prepared reagents, carried out the experiments and analyzed the results. BA prepared essential reagents. TS helped setting up the experiments. AM, FN and JA discussed the results and wrote the manuscript. JA designed the experiments and helped in the analysis.
References
- 1.Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 2.Lawless MK, Barney S, Guthrie KI, Bucy TB, Petteway SR, Jr, Merutka G. HIV-1 membrane fusion mechanism: structural studies of the interactions between biologically-active peptides from gp41. Biochemistry. 1996;35:13697–13708. doi: 10.1021/bi9606962. [DOI] [PubMed] [Google Scholar]
- 3.Naider F, Anglister J. Peptides in the treatment of AIDS. Curr Opin Struct Biol. 2009;19:473–482. doi: 10.1016/j.sbi.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wild C, Greenwell T, Matthews T. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res Hum Retroviruses. 1993;9:1051–1053. doi: 10.1089/aid.1993.9.1051. [DOI] [PubMed] [Google Scholar]
- 5.Rimsky LT, Shugars DC, Matthews TJ. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J Virol. 1998;72:986–993. doi: 10.1128/jvi.72.2.986-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biron Z, Khare S, Quadt SR, Hayek Y, Naider F, Anglister J. The 2F5 epitope is helical in the HIV-1 entry inhibitor T-20. Biochemistry. 2005;44:13602–13611. doi: 10.1021/bi0509245. [DOI] [PubMed] [Google Scholar]
- 7.Biron Z, Khare S, Samson AO, Hayek Y, Naider F, Anglister J. A Monomeric 3(10)-Helix Is Formed in Water by a 13-Residue Peptide Representing the Neutralizing Determinant of HIV-1 on gp41(,) Biochemistry. 2002;41:12687–12696. doi: 10.1021/bi026261y. [DOI] [PubMed] [Google Scholar]
- 8.Noah E, Biron Z, Naider F, Arshava B, Anglister J. The membrane proximal external region of the HIV-1 envelope glycoprotein gp41 contributes to the stabilization of the six-helix bundle formed with a matching N’ peptide. Biochemistry. 2008;47:6782–6792. doi: 10.1021/bi7023139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buzon V, Natrajan G, Schibli D, Campelo F, Kozlov MM, Weissenhorn W. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010;6:e1000880. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan W, Craig S, Si Z, Farzan M, Sodroski J. CD4-induced T-20 binding to human immunodeficiency virus type 1 gp120 blocks interaction with the CXCR4 coreceptor. J Virol. 2004;78:5448–5457. doi: 10.1128/JVI.78.10.5448-5457.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alam SM, Paleos CA, Liao HX, Scearce R, Robinson J, Haynes BF. An inducible HIV type 1 gp41 HR-2 peptide-binding site on HIV type 1 envelope gp120. AIDS Res Hum Retroviruses. 2004;20:836–845. doi: 10.1089/0889222041725181. [DOI] [PubMed] [Google Scholar]
- 12.Liu S, Lu H, Niu J, Xu Y, Wu S, Jiang S. Different from the HIV fusion inhibitor C34, the anti-HIV drug Fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. The Journal of biological chemistry. 2005;280:11259–11273. doi: 10.1074/jbc.M411141200. [DOI] [PubMed] [Google Scholar]
- 13.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O’Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang D, Li W, Jiang S. Peptide fusion inhibitors targeting the HIV-1 gp41: a patent review (2009 – 2014) Expert opinion on therapeutic patents. 2014:1–15. doi: 10.1517/13543776.2014.987752. [DOI] [PubMed] [Google Scholar]
- 15.Moseri A, Schnur E, Noah E, Zherdev Y, Kessler N, Singhal Sinha E, Abayev M, Naider F, Scherf T, Anglister J. NMR observation of HIV-1 gp120 conformational flexibility resulting from V3 truncation. The FEBS journal. 2014 doi: 10.1111/febs.12839. [DOI] [PubMed] [Google Scholar]
- 16.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnur E, Noah E, Ayzenshtat I, Sargsyan H, Inui T, Ding FX, Arshava B, Sagi Y, Kessler N, Levy R, Scherf T, Naider F, Anglister J. The conformation and orientation of a 27-residue CCR5 peptide in a ternary complex with HIV-1 gp120 and a CD4-mimic peptide. J Mol Biol. 2011;410:778–797. doi: 10.1016/j.jmb.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin L, Stricher F, Misse D, Sironi F, Pugniere M, Barthe P, Prado-Gotor R, Freulon I, Magne X, Roumestand C, Menez A, Lusso P, Veas F, Vita C. Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nature biotechnology. 2003;21:71–76. doi: 10.1038/nbt768. [DOI] [PubMed] [Google Scholar]
- 19.Rosen O, Anglister J. Epitope mapping of antibody-antigen complexes by nuclear magnetic resonance spectroscopy. Methods Mol Biol. 2009;524:37–57. doi: 10.1007/978-1-59745-450-6_3. [DOI] [PubMed] [Google Scholar]
- 20.Weiss MA, Eliason JL, States DJ. Dynamic filtering by two-dimensional 1H NMR with application to phage lambda repressor. Proc Natl Acad Sci U S A. 1984;81:6019–6023. doi: 10.1073/pnas.81.19.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Mayer M, Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angewandte Chemie-International Edition. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 23.Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78:10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardoso RM, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, Burton DR, Wilson IA. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22:163–173. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 25.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody [see comments] Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding [see comments] Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 27.Xiang SH, Doka N, Choudhary RK, Sodroski J, Robinson JE. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res Hum Retroviruses. 2002;18:1207–1217. doi: 10.1089/08892220260387959. [DOI] [PubMed] [Google Scholar]
- 28.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiang SH, Wang L, Abreu M, Huang CC, Kwong PD, Rosenberg E, Robinson JE, Sodroski J. Epitope mapping and characterization of a novel CD4-induced human monoclonal antibody capable of neutralizing primary HIV-1 strains. Virology. 2003;315:124–134. doi: 10.1016/s0042-6822(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 30.Langley DR, Kimura SR, Sivaprakasam P, Zhou N, Dicker I, McAuliffe B, Wang T, Kadow JF, Meanwell NA, Krystal M. Homology models of the HIV-1 attachment inhibitor BMS-626529 bound to gp120 suggest a unique mechanism of action. Proteins. 2015;83:331–350. doi: 10.1002/prot.24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madani N, Perdigoto AL, Srinivasan K, Cox JM, Chruma JJ, LaLonde J, Head M, Smith AB, 3rd, Sodroski JG. Localized changes in the gp120 envelope glycoprotein confer resistance to human immunodeficiency virus entry inhibitors BMS-806 and #155. J Virol. 2004;78:3742–3752. doi: 10.1128/JVI.78.7.3742-3752.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herschhorn A, Gu C, Espy N, Richard J, Finzi A, Sodroski JG. A broad HIV-1 inhibitor blocks envelope glycoprotein transitions critical for entry. Nature chemical biology. 2014;10:845–852. doi: 10.1038/nchembio.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dogo-Isonagie C, Lam S, Gustchina E, Acharya P, Yang Y, Shahzad-ul-Hussan S, Clore GM, Kwong PD, Bewley CA. Peptides from second extracellular loop of C-C chemokine receptor type 5 (CCR5) inhibit diverse strains of HIV-1. The Journal of biological chemistry. 2012;287:15076–15086. doi: 10.1074/jbc.M111.332361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agrawal L, VanHorn-Ali Z, Berger EA, Alkhatib G. Specific inhibition of HIV-1 coreceptor activity by synthetic peptides corresponding to the predicted extracellular loops of CCR5. Blood. 2004;103:1211–1217. doi: 10.1182/blood-2003-08-2669. [DOI] [PubMed] [Google Scholar]
- 35.Abayev M, Moseri A, Tchaicheeyan O, Kessler N, Arshava B, Naider F, Scherf T, Anglister J. An extended CCR5 ECL2 peptide forms a helix that binds HIV-1 gp120 through non-specific hydrophobic interactions. The FEBS journal. 2015 doi: 10.1111/febs.13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moseri A, Tantry S, Sagi Y, Arshava B, Naider F, Anglister J. An optimally constrained V3 peptide is a better immunogen than its linear homolog or HIV-1 gp120. Virology. 2010;401:293–304. doi: 10.1016/j.virol.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]