Abstract
Epoxyketone proteasome inhibitors have attracted much interest due to their potential as anti-cancer drugs. While the biosynthetic gene clusters for several peptidyl epoxyketone natural products have recently been identified, the enzymatic logic involved in the formation of the terminal epoxyketone pharmacophore has been relatively unexplored. Here, we report the identification of the minimal set of enzymes from the eponemycin gene cluster necessary for the biosynthesis of novel metabolites containing a terminal epoxyketone pharmacophore in Escherichia coli, a versatile and fast-growing heterologous host. This set of enzymes includes a non-ribosomal peptide synthetase (NRPS), a polyketide synthase (PKS), and an acyl-CoA dehydrogenase (ACAD) homolog. In addition to the in vivo functional reconstitution of these enzymes in E. coli, in vitro studies of the eponemycin NRPS and 13C-labeled precursor feeding experiments were performed to advance the mechanistic understanding of terminal epoxyketone formation.
Keywords: terminal epoxyketone, biosynthesis, nonribosomal peptide/polyketide hybrid, acyl-CoA dehydrogenase, proteasome inhibitor
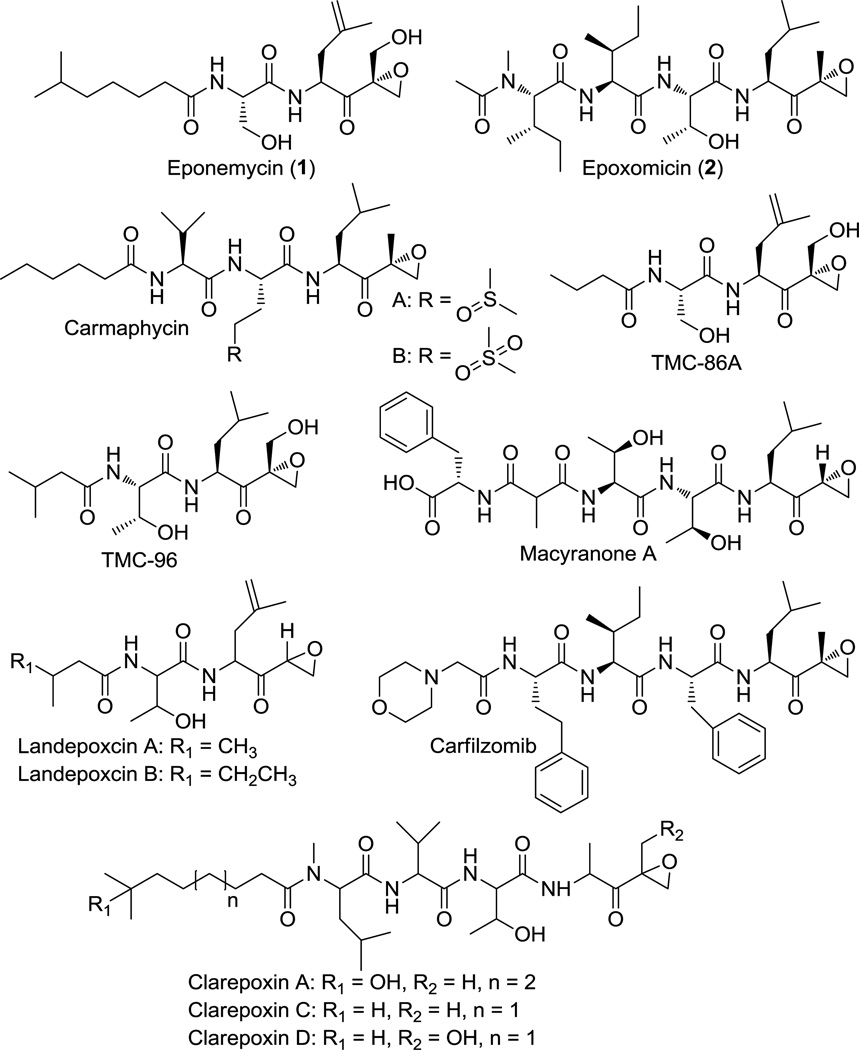
Peptidyl epoxyketones are a class of proteasome inhibitors consisting of a short peptide core and a terminal epoxyketone moiety. Over the past few decades, a variety of these natural products, including eponemycin (1), epoxomicin (2), and other related compounds, have been isolated from a wide range of bacterial sources (Scheme 1).[1–4] More recently, several additional epoxyketone proteasome inhibitors, specifically the clarepoxcins landepoxcins, and macyranone A, were discovered through metagenome mining[5] and NMR screening approaches.[6] Although these naturally-occurring peptidyl epoxyketones exhibit great structural diversity in the length and composition of their peptide core and can also differ in the substitution on the epoxide ring, all of them possess a common terminal epoxyketone pharmacophore, which is responsible for their high efficacy and specificity as proteasome inhibitors. In particular, the strongly electrophilic carbonyl and epoxide groups are highly susceptible to a double nucleophilic attack by the N-terminal threonine of the 20S proteasome, leading to the irreversible formation of a morpholino ring.[7–9] Consequently, these natural products have inspired the current development of anti-cancer, anti-inflammatory, and anti-parasitic drugs with a terminal epoxyketone warhead.[10–12] One such example is the synthetic epoxomicin analogue, carfilzomib (Scheme 1), which has been used as a treatment for relapsed and refractory multiple myeloma.[13–14]
Scheme 1.
Structures of peptidyl epoxyketone proteasome inhibitors.
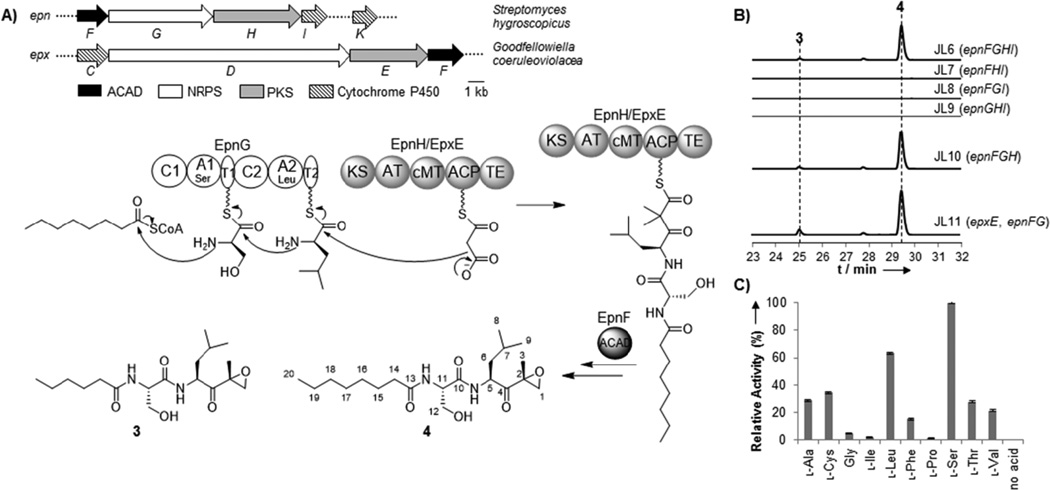
Given the potency and unique structure of these peptidyl epoxyketone natural products, there has been great interest in the biosynthetic origins of these compounds. In particular, the genetic basis for the biosynthesis of epoxomicin and eponemycin was recently identified from Goodfellowiella coeruleoviolacea and Streptomyces hygroscopicus, respectively (Figure 1A).[15] Moreover, gene clusters responsible for the biosynthesis of clarepoxins and landepoxins were recently identified in soil metagenomic libraries,[5] and the gene cluster responsible for the biosynthesis of macyranones was identified in the genome of Cystobacter fuscus MCy9118.[6] However, the specific enzymes and chemical logic governing the formation of the terminal epoxyketone pharmacophore have not yet been fully elucidated. Nonetheless, bioinformatics analysis has shown four genes to be conserved across the epoxomicin and eponemycin gene clusters, and these genes encode a non-ribosomal peptide synthetase (NRPS), i.e. EpnG/EpxD; a polyketide synthase (PKS), i.e. EpnH/EpxE; an acyl-CoA dehydrogenase (ACAD) homolog, i.e. EpnF/EpxF; and a cytochrome P450 monooxygenase, i.e. EpnI/EpxC. Based on this analysis, a biosynthetic pathway in which a hybrid NRPS-PKS assembly line generates a carboxylic acid that is subsequently modified by the ACAD and cytochrome P450 to form the terminal epoxyketone moiety was previously proposed.[15] Notably, however, the recently identified gene cluster for macyranone biosynthesis lacks a gene encoding a cytochrome P450, which led to an alternative acyl-carrier-bound proposed mechanism for epoxyketone formation involving the ACAD homolog, MynC, and possibly the type II thioesterase, MynH.[6]
Figure 1.
Biosynthesis of a peptidyl epoxyketone in E. coli. A) Schematic of the eponemycin (epn) and epoxomicin (epx) and proposed biosynthetic pathway in E. coli. B) Extracted ion chromatograms (3: m/z 357.2384 [M+H]+, 4: m/z 385.2697 [M+H]+) showing the presence and absence of 3 and 4 resulting from the heterologous expression of different combinations of EpnF,EpnG, EpnH/EpxE, and EpnI in E. coli. The calculated mass with a 10 ppm mass error tolerance was used. C) Characterization of the A-domain activities of EpnG. Error bars represent standard deviations from three replicates. A relative activity of 100% corresponds to 39k cpm.
In this work, we build upon the previous study of the biosynthesis of eponemycin and epoxomicin by turning to reconstitution in E. coli as a means to quickly and systematically identify the enzymes necessary for the generation of the terminal epoxyketone moiety. Due to its genetic tractability, fast growth rate, and well-characterized metabolism, E. coli is a particularly attractive host for studying the biosynthesis of natural products.[16–19] Furthermore, the reconstitution of NRPS-PKS activity in E. coli provides a superior platform for the overproduction and diversification of their products.[20–21] To determine the enzymes required for the biosynthesis of the terminal epoxyketone pharmacophore, we co-expressed the four genes from the eponemycin gene cluster (epnFGHI) proposed to be involved in E. coli on three plasmids. The plasmids were co-transformed into E. coli BAP1, which contains a chromosomal copy of the phosphopantetheinyl transferase encoding gene sfp,[22] to generate the strain, JL6. A negative control strain transformed with empty vectors was also constructed. Although the natural fatty acyl moiety in eponemycin is derived from 6-methyl heptanoic acid, which is uncommon in bacterial primary metabolism[23] and is presumably generated by dedicated enzymes encoded in the eponemycin biosynthetic gene cluster,[15] we reasoned that EpnG-activated serine could possibly be condensed with hexanoyl- or octanoyl-CoA produced endogenously by E. coli (Figure 1A). Consequently, cultures of JL6 were supplemented with 1 mM hexanoic acid or octanoic acid at the time of induction to promote the incorporation of these alternative fatty acyl groups into the assembly line. The culture extracts of JL6 and the control strain were analyzed by liquid chromatography-high resolution mass spectrometry (LC-HRMS)-based comparative metabolomics,[24] and two new compounds (3 and 4) with masses consistent with molecular formulae of C18H32N2O5 and C20H36N2O5, respectively, were detected in high abundance in the extracts of JL6 but were absent from the extracts of the control strain (Figures 1B and S1–2). Further HRMS/MS analysis confirmed 3 and 4 to be consistent with a C6 or C8 fatty acyl group condensed with serine, leucine, and malonate, but it did not give insight into the structure of the terminal moiety resulting from modifications on the malonate (Figures S1 and S2).
To reveal the exact molecular structure of new compounds, we then scaled up the production cultures and purified ~2 mg of the major product, 4, for NMR spectroscopic analysis. Compound 4 was purified by following the fractions using LC-MS, and the yield of 4 was ~0.2 mg/L. Analysis of the 1D (1H, 13C) and 2D (HSQC, COSY, HMBC) NMR spectra of purified 4 confirmed the presence of an acylated peptide composed of an eight-carbon fatty acyl chain, serine, and leucine connected to a methyl-substituted epoxyketone (Figures 1, S3–7, and Table S3). Notably, the proton and carbon shifts of the methyl-substituted epoxyketone and leucine residue are in strong agreement with those reported for a standard of 2, which has these same functionalities.[2] Moreover, the methyl-substituted quaternary epoxide ring was supported by HMBC correlations from the C3 methyl protons (δH 1.52) to both the quaternary carbon C2 (δC 59.5) and the oxygenated methylene carbon C1 (δC 52.7) and by HMBC correlations from the C1 protons (δH 2.91 and 3.28) to C2 and C3 (δC 16.9) (Figures 1, S7, and Table S3). HMBC correlations from the C1 and C3 protons to the ketone carbon C4 (δC 208.8) also confirmed the connectivity of the epoxide (Figures 1, S7, and Table S3). Thus, based on our HRMS and NMR analyses, we determined 4 to be a new terminal epoxyketone compound.
To probe the necessity of individual enzymes in the formation of 3 and 4, we systematically removed each of the four genes from JL6 and compared the product profiles of the resulting strains. Analysis of the culture extracts of these strains by LC-HRMS showed that the production of 3 and 4 was abolished in JL7, JL8, and JL9, which lack epnG, epnH, and epnF, respectively, but was maintained in JL10, which lacks epnI (Figure 1B). Moreover, LC-HRMS-based comparative metabolomics did not show any differences between the extracts of JL6 and JL10, and the titers of 3 and 4 were comparable in both strains, confirming that EpnI is not necessary for the formation of 4 in E. coli though its function remains unclear. These results demonstrate that the NRPS, PKS, and ACAD are sufficient for the biosynthesis of peptidyl epoxyketones in E. coli, and epoxide formation does not necessitate a cytochrome P450 monooxygenase. This result is also consistent with the recent study on macyranone biosynthesis in which the reported gene cluster did not contain any genes encoding for a cytochrome P450 nor could a close homolog of EpxC/EpnI be identified elsewhere on the C. fuscus MCy9118 genome.[6] In addition, the titers of 3 and 4 from JL11 in which epnH is replaced by epxE appeared to be slightly greater than those from JL10, demonstrating the interchangeability of EpnH and its homolog, EpxE (similarity/identity: 55%/44%), which is encoded in the epoxomicin biosynthetic gene cluster. Interestingly, no apparent small molecule intermediates could be detected by LC-HRMS-based comparative metabolomics analysis of the culture extracts of a strain expressing only the NRPS and PKS together, suggesting that the ACAD may act on the assembly line.
To complement the in vivo studies of EpnF, G, H, and I, we also attempted to perform in vitro characterization of these enzymes, beginning with EpnG, the dimodule NRPS that has a domain organization of C1-A1-T1-C2-A2-T2 (C, condensation; A, adenylation; T, thiolation) (Figures 1A and S8). The A domain substrate specificities of EpnG were tested using the well-established ATP-[32P]PPi exchange assay.[25] As expected, EpnG demonstrated a strong preference for l-serine and l-leucine though it was also able to activate l-cysteine, l-alanine, and l-threonine to some extent (Figure 1C). Unfortunately, the purification of active PKS’s, EpnH and EpxE, from E. coli or Streptomyces hosts was unsuccessful (Figure S8), which prevented further biochemical study of these enzymes.
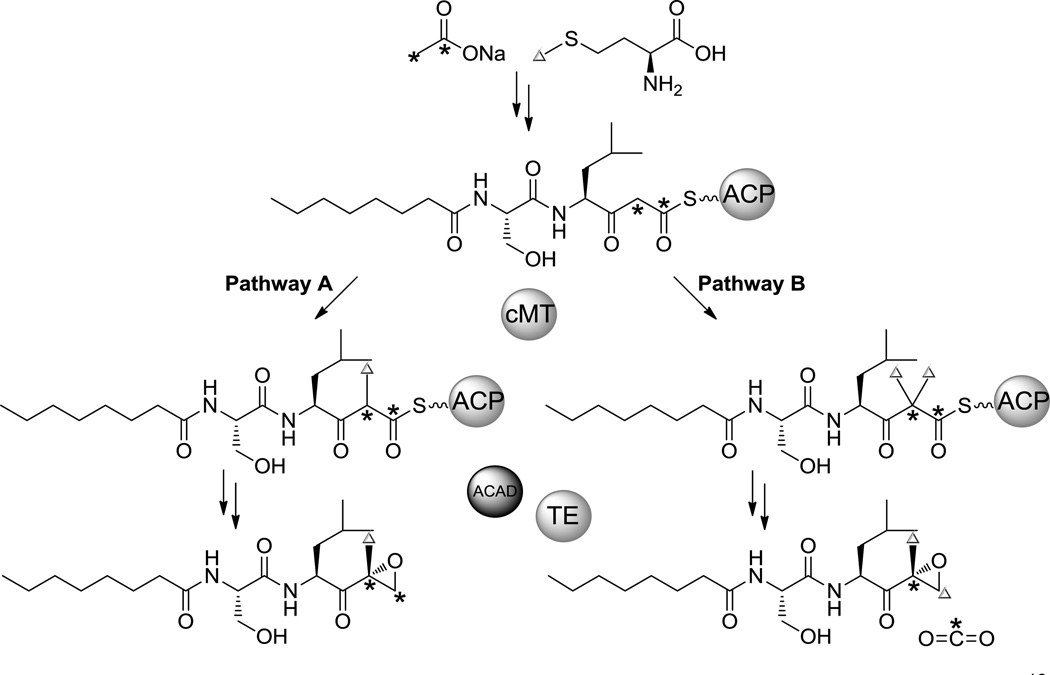
To better elucidate the biosynthetic mechanism for terminal epoxyketone formation, we took advantage of our E. coli heterologous expression system and used it to perform 13C-labeled precursor feeding studies. Two possible mechanisms (pathways A and B) for terminal epoxyketone formation have previously been put forth (Scheme 2).[15] Pathway A postulates the introduction of a single methyl group at C2 followed by the reduction of the carboxylic acid to an olefin that can then undergo epoxidation, while pathway B proposes dimethylation at C2 followed by decarboxylation of the carboxylic acid and subsequent epoxide installation.[15] To distinguish between these two possibilities, we fed [1,2-13C] sodium acetate to cultures of JL11, the highest producing strain, and compared the resulting 13C incorporation to that from unlabeled and [1-13C] sodium acetate feeding (Scheme 2 and Figures S9–10). Although the titers of labeled 3 and 4 were insufficient for NMR analysis, LC-HRMS analysis of 3 and 4 from the extracts of cultures fed with [1,2-13C] sodium acetate showed a significant increase in the ratio of the M+1 peak intensity to the M+ peak intensity compared to that of cultures fed with unlabeled and [1-13C] sodium acetate (Figures S9–10). Further HRMS/MS analyses and examination of the isotope distribution of fragments with and without the epoxyketone group indicated that this increase is the result of a single 13C atom from doubly labeled acetate being incorporated as the C2 carbon of 3 and 4 after decarboxylation (Scheme 2 and Figures S9–10). To evaluate the occurrence of dimethylation, we then separately fed [methyl-13C] l-methionine to cultures of JL11 as methionine is the precursor of the predicted methyl group donor, S-adenosylmethionine (SAM). This feeding experiment resulted in a significant increase in the ratio of the M+2 peak to the M+ peak for both 3 and 4, which is indicative of the incorporation of two 13C atoms, and additional HRMS/MS analyses showed that these 13C atoms were incorporated into the epoxyketone-containing fragment of 3 and 4 (Scheme 2 and S9–10). These feeding results thus support pathway B in which C2 is dimethylated by the cMT (C-methyltransferase) domain of the PKS and either following or in concert with the activity of the ACAD, decarboxylation of a terminal carboxylic acid takes place.
Scheme 2.
Two proposed pathways for terminal expoxyketone formation. Our 13C-labeled precursor feeding results support pathway B.
In summary, we have made headway in elucidating the biosynthesis of peptidyl epoxyketones, and in addition to generating new peptidyl epoxyketone products in E. coli, we systematically identified the minimal set of enzymes necessary for doing so. Specifically, through our reconstitution of the hybrid NRPS-PKS assembly line in E. coli, we found that only one additional modification enzyme, an ACAD homolog that presumably functions on the assembly line, is required for terminal epoxyketone formation. Furthermore, 13C-labeled precursor feeding studies suggest that the formation of the epoxyketone pharmacophore involves a dimethylation and a subsequent decarboxylation reaction. The work reported here demonstrates the first production of peptidyl epoxyketones by E. coli and paves the way for further biochemical characterization of the PKS and ACAD to understand their precise roles in forming the potent and unique terminal epoxyketone pharmacophore.
Experimental Section
The full experimental section is available in the Supporting Information file.
Supplementary Material
Acknowledgements
We thank for J. Pelton for assistance with NMR spectroscopic analysis. This research was financially supported by the Pew Scholars Program and the National Institutes of Health (DP2AT009148).
References
- 1.Sugawara K, Hatori M, Nishiyama Y, Tomita K, Kamei H, Konishi M, Oki T. J Antibiot. (Tokyo) 1990;43:8–18. doi: 10.7164/antibiotics.43.8. [DOI] [PubMed] [Google Scholar]
- 2.Hanada M, Sugawara K, Kaneta K, Toda S, Nishiyama Y, Tomita K, Yamamoto H, Konishi M, Oki T. J Antibiot. (Tokyo) 1992;45:1746–1752. doi: 10.7164/antibiotics.45.1746. [DOI] [PubMed] [Google Scholar]
- 3.Pereira AR, Kale AJ, Fenley AT, Byrum T, Debonsi HM, Gilson MK, Valeriote FA, Moore BS, Gerwick WH. Chembiochem. 2012;13:810–817. doi: 10.1002/cbic.201200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koguchi Y, Kohno J, Suzuki S, Nishio M, Takahashi K, Ohnuki T, Komatsubara S. J Antibiot. (Tokyo) 1999;52:1069–1076. doi: 10.7164/antibiotics.52.1069. [DOI] [PubMed] [Google Scholar]
- 5.Owen JG, Charlop-Powers Z, Smith AG, Ternei MA, Calle PY, Reddy BV, Montiel D, Brady SF. Proc. Natl. Acad. Sci. U. S. A. 2015;112:4221–4226. doi: 10.1073/pnas.1501124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keller L, Plaza A, Dubiella C, Groll M, Kaiser M, Muller R. J Am. Chem. Soc. 2015;137:8121–8130. doi: 10.1021/jacs.5b03833. [DOI] [PubMed] [Google Scholar]
- 7.Kim KB, Crews CM. Nat. Prod. Rep. 2013;30:600–604. doi: 10.1039/c3np20126k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groll M, Kim KB, Kairies N, Huber R, Crews CM. J Am. Chem. Soc. 2000;122:1237–1238. [Google Scholar]
- 9.Huber EM, Basler M, Schwab R, Heinemeyer W, Kirk CJ, Groettrup M, Groll M. Cell. 2012;148:727–738. doi: 10.1016/j.cell.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 10.Grawert MA, Groll M. Chem. Commun. (Camb.) 2012;48:1364–1378. doi: 10.1039/c1cc15273d. [DOI] [PubMed] [Google Scholar]
- 11.Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Ponder EL, Verdoes M, Asbjornsdottir KH, Deu E, Edgington LE, Lee JT, Kirk CJ, Demo SD, Williamson KC, Bogyo M. Chem. Biol. 2012;19:1535–1545. doi: 10.1016/j.chembiol.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn DJ, Chen Q, Voorhees PM, Strader JS, Shenk KD, Sun CM, Demo SD, Bennett MK, van Leeuwen FW, Chanan-Khan AA, Orlowski RZ. Blood. 2007;110:3281–3290. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCormack PL. Drugs. 2012;72:2023–2032. doi: 10.2165/11209010-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 15.Schorn M, Zettler J, Noel JP, Dorrestein PC, Moore BS, Kaysser L. ACS Chem. Biol. 2014;9:301–309. doi: 10.1021/cb400699p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaitzig J, Li J, Sussmuth RD, Neubauer P. ACS Synth. Biol. 2014;3:432–438. doi: 10.1021/sb400082j. [DOI] [PubMed] [Google Scholar]
- 17.Boghigian BA, Pfeifer BA. Biotechnol. Lett. 2008;30:1323–1330. doi: 10.1007/s10529-008-9689-2. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W, Li Y, Tang Y. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20683–20688. doi: 10.1073/pnas.0809084105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Zhu X, Seipke RF, Zhang W. ACS Synth. Biol. 2015;4:559–565. doi: 10.1021/sb5003136. [DOI] [PubMed] [Google Scholar]
- 20.Matthes D, Richter L, Muller J, Denisiuk A, Feifel SC, Xu Y, Espinosa-Artiles P, Sussmuth RD, Molnar I. Chem. Commun. (Camb.) 2012;48:5674–5676. doi: 10.1039/c2cc31669b. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Neubauer P. N Biotechnol. 2014;31:579–585. doi: 10.1016/j.nbt.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 22.Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 23.Janssen HJ, Steinbuchel A. Biotechnol. Biofuels. 2014;7:7. doi: 10.1186/1754-6834-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. Anal. Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryce GF, Brot N. Biochemistry. 1972;11:1708–1715. doi: 10.1021/bi00759a028. [DOI] [PubMed] [Google Scholar]
- 26.Ghisla S, Thorpe C. Eur. J. Biochem. 2004;271:494–508. doi: 10.1046/j.1432-1033.2003.03946.x. [DOI] [PubMed] [Google Scholar]
- 27.Vock P, Engst S, Eder M, Ghisla S. Biochemistry. 1998;37:1848–1860. doi: 10.1021/bi971827h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.