Abstract
Inborn errors of metabolism (IEM) are not unlike common diseases. They often present as a spectrum of disease phenotypes that correlates poorly with the severity of the disease-causing mutations. This greatly impacts patient care and reveals fundamental gaps in our knowledge of disease modifying biology. Systems biology approaches that integrate multi-omics data into molecular networks have significantly improved our understanding of complex diseases. Similar approaches to study IEM are rare despite their complex nature. We highlight that existing common disease-derived datasets and networks can be repurposed to generate novel mechanistic insight in IEM and potentially identify candidate modifiers. While understanding disease pathophysiology will advance the IEM field, the ultimate goal should be to understand per individual how their phenotype emerges given their primary mutation on the background of their whole genome, not unlike personalized medicine. We foresee that panomics and network strategies combined with recent experimental innovations will facilitate this.
Keywords: omics, network biology, human genetic disease, metabolism
Introduction
The term ‘inborn errors of metabolism’ (IEM) was first coined in 1902 by Archibald Garrod, who is attributed to being the first to connect a human disorder with Mendel’s laws of inheritance (Garrod, 1902). It describes a class of inherited genetic diseases caused by mutations in genes coding for proteins that function in metabolism. The disease may be the result of the accumulation of toxic substrates or essential products being intolerably low. Although IEM occur in every biochemical pathway, historically they have been grouped in specific classes such as amino acidemias, organic acidurias and lysosomal storage disorders. An example of the latter is Gaucher disease (GD), which is caused by deficient activity of the lysosomal enzyme beta-glucocerebrosidase due to mutations in the encoding gene (GBA). As a consequence the substrate of GBA, glucosylceramide accumulates in the lysosome, especially of tissue macrophages of the liver, bone marrow and spleen thereby causing damage in hematological, skeletal and nervous systems (Baris et al., 2014). The incidence of IEM varies greatly and depends on the population. Some of the more frequent IEM are phenylketonuria (PKU) and medium-chain acyl-CoA dehydrogenase (MCAD) deficiency with respective incidences of 1 in 10,000 and 1 in 20,000 (Schulze et al., 2003; Wilcken et al., 2003). Most other IEM are much rarer with sometimes only a few or even one unique case diagnosed. Treatment has improved but often remains insufficient (Vernon, 2015).
IEM are not unlike complex disease
In a bird’s eye view IEM are Mendelian traits caused by single-gene mutations, which has led to the one gene-one disease paradigm. Garrod alluded to this by concluding that an individual with alkaptonuria, would either have the disease or not, and that there were essentially no shades of grey (Garrod, 1902). However, time has shown that IEM are also not unlike common diseases for the major reason they often present as a spectrum of disease phenotypes in which a clear correlation between the severity of mutation at the affected locus and the phenotype (genotype-phenotype correlation) is lacking (Dipple and McCabe, 2000a, b; Lanpher et al., 2006; Scriver and Waters, 1999).
The classic autosomal recessive disease PKU illustrates this oversimplification. Initially, mutations at the human phenylalanine hydroxylase locus (PAH) were deemed sufficient to explain the impaired function of the enzyme PAH, the associated metabolic phenotype, elevated plasma phenylalanine levels, and the resultant clinical phenotype, mental retardation (Scriver and Waters, 1999). However, PKU was subsequently found to arise from different genetic defects (e.g. tetrahydrobiopterin homeostasis), be influenced greatly by diet (e.g. protein intake) and importantly the PAH genotype and predicted effect on enzymatic function, often failed to consistently predict the extent of cognitive and metabolic phenotypes in the PKU patient. Importantly, PKU was not an exception to the rule as this oversimplification of one gene-one disease paradigm was also challenged in many other monogenic diseases. In summary, the prevailing view of the last two decades is that monogenic traits do conform to long-accepted ideas about the expression of major loci and their importance in determining parameters of phenotypes however, the associated features (such as cognitive behavior in PKU) are complex in nature and not unlike those in so-called complex traits (Scriver and Waters, 1999).
It has been over fifteen years since IEM have been viewed as complex traits (Dipple and McCabe, 2000a, b; Scriver and Waters, 1999). It is therefore surprising that despite avid application of unbiased systems biology and omics approaches to unravel complex diseases (Ritchie et al., 2015) there are few examples of their use in the IEM field. It appears as if complex methodologies are deemed not needed or not applicable. This is also despite the elegant words of Scriver and Waters, who pointed out that “genomes function in vivo, where much more than the major gene is expressed and where the whole organismal phenotype is more than the sum of the parts; it is an emergent property” (Scriver and Waters, 1999). This sentiment was also reinforced by Dipple et al, who stated that genotype to phenotype prediction would improve not just by understanding the individual modifying factors but also how they assembled into functional modules and how the system dynamics were affected (Dipple and McCabe, 2000b; Dipple et al., 2001). What better way to assess this emergent property than through multi-scale integrative network systems approaches (Figure 1A)?
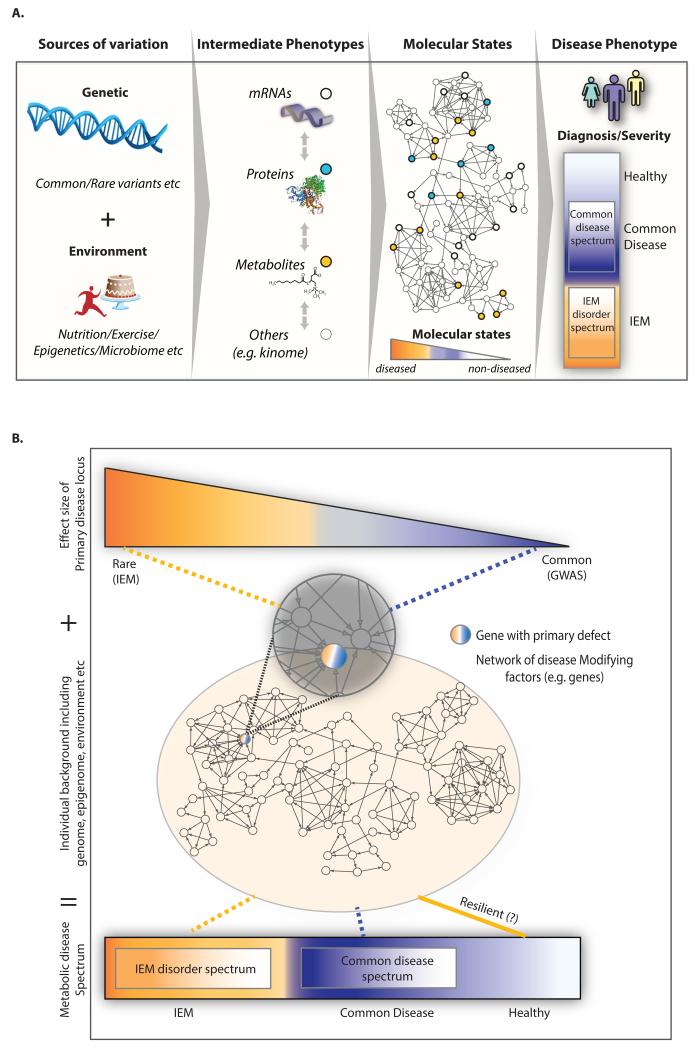
Figure 1. Inborn errors of metabolism (IEM) are increasingly viewed as complex diseases.
(A) IEM are not unlike common diseases as they often present as a spectrum of disease phenotypes that poorly correlate with the severity of the disease-causing mutations (genotype). The abandonment of the one gene-one disease idea implies that modifying factors such as environmental, epigenetic, and microbiome factors as well as additional genes contribute to the disease. It also means that IEM phenotypes are emergent properties of biological networks rather than the result of changes to single genes, metabolites or phenotypes alone. Thus we have to expand our understanding of the clinical expression of the IEM beyond a single gene level to that of a consequence of a set of molecular interactions (subnetwork). (B) IEM are being considered more and more alongside common disease as part of a spectrum of ‘errors’ in metabolism. In this spectrum, ‘classic’ IEM are on the on extreme and arise from a primary genetic variant influenced by modifier genes, while the common metabolic diseases are on the other extreme and are caused by multiple genetic variants with relatively small effect sizes. The variants in the primary disease locus in the context of the individual’s background such as genome, epigenome, and environmental exposures will ultimately determine the molecular state of the individual and an individual’s risk of disease and spectrum of phenotypic presentation. This view is highlighted by the Resilience project, whereby large scale genetic screening of general populations for a panel of rare disease causing mutations is hoping to uncover healthy individuals harboring rare genetic disease and the genetic modifiers that make them resilient to this disease.
The abandonment of the one gene-one disease idea has meant considering alternative explanations, such as the contribution of modifying factors (Dipple and McCabe, 2000a, b; Lanpher et al., 2006; Scriver and Waters, 1999). These modifying factors could include environmental, epigenetic, and microbiome factors as well as additional genes. In this review we focus on modifying genes and their associated biology (Figure 1A). The modifier gene concept was introduced already in 1941 by Haldane (Haldane, 1941) and several definitions have been given since then. We use the term to reflect a gene that can impact the phenotypic expression of the primary affected locus (Genin et al., 2008). Importantly the biological pathways affected by the modifying genes are not necessarily the same ones as that affected by the primary disease gene.
The lack of genotype to phenotype correlation greatly impacts the ability to predict a patient’s disease course. It also illustrates the existence of a fundamental gap in our knowledge of IEM disease pathophysiology, which impacts drug discovery. Thus the primary motivation for finding these modifiers and understanding their associated biology is the potential to improve clinical care and provide novel potential targets of therapeutic intervention beyond the primary disease-causing gene. Furthermore, genes that modify monogenic disease related phenotypes likely contribute to the development of common diseases in the general population and their identification will benefit understanding common disease pathophysiology (Blair et al., 2013; Lupski et al., 2011). These genes may have small effect size and go undetected in healthy individuals whereas their contribution to disease may be more easily unmasked on the background of a monogenic disorder (Cutting, 2010).
Current approaches and successes in finding modifier genes of IEM
The strategies used to date to identify genetic modifiers have included linkage and association studies. These studies have been approached either systematically where the whole genome is scanned or in a candidate gene way where focus is on known disease-associated biology (Genin et al., 2008). For example, candidate modifier genes in IEM could encode for other enzymes that function in the same biochemical pathway as the primary affected gene. In GD glucosylceramide synthesis enzymes are relevant candidate modifiers genes as they could theoretically modulate the substrate levels of the GBA enzyme thus potentially impacting on disease severity (Alfonso et al., 2013). Examples of diseases where modifiers have been successfully identified include the more common monogenetic diseases such as cystic fibrosis (Cutting, 2010; Gallati, 2014), sickle cell anemia (Lettre, 2012), thalassemias (Lettre, 2012) and most recently Huntington disease (Becanovic et al., 2015; Consortium, 2015b), which have incidences between 1:2,000 and 1:10,000. There are relatively fewer success stories in IEM, many of which have been by identified using candidate gene approaches such as in Smith-Lemli-Opitz syndrome (Lanthaler et al., 2013), inherited hemochromatosis (Ala and Schilsky, 2011) and GD (Alfonso et al., 2013; Lo et al., 2012; Mistry et al., 2002). Inherent challenges exist in successfully applying association or linkage approaches to genetic modifier discovery in IEM. First, many of the IEM are much rarer, yielding insufficient power to do unbiased, whole genome screens. In a recent unbiased GWAS study in GD which has an incidence of 1:50,000, not a single candidate gene met genome wide significance (Zhang et al., 2012). This limits the search to candidate gene approaches and genes of known disease relatedness. The generation of uniformly defined clinical phenotypes relevant for finding genetic modifiers is also problematic (Genin et al., 2008). Many patients are treated either through dietary interventions or enzyme replacement therapies thus impacting disease severity. Finally, population stratification greatly affects the ability to discover genetic modifiers of IEM (Genin et al., 2008) as it impacts chances of replication. Thus for reasons of a lack of power and statistical stringency, relieving candidate gene bias, expanding phenotype definitions, managing population stratification, and providing realistic frameworks to interpret IEM disease biology, we suggest that complementary approaches are needed to find genetic modifiers and modifying biology of IEM.
Studying IEM like complex disorders: A complementary approach to find genetic modifiers and modifying biology of IEM
The pursuit of genetic modifiers has established that IEM are more than just monogenic and not unlike common disease in complexity. This viewpoint has been further extended such that IEM phenotypes do not just form a spectrum within a specific disorder but that common diseases and IEM are actually part of a metabolic disease spectrum. In this view “genetic diseases represent a continuum with diminishing influence from a single primary gene influenced by modifier genes, to increasingly shared influence by multiple genes” (Dipple and McCabe, 2000a) (Figure 1B). In the allelic series hypothesis these variants can cause a disease phenotype at both ends of the spectrum (Blair et al., 2013; Lupski et al., 2011). A commonly used example of this phenomenon is familial hypercholesterolemia that occurs in two forms. Heterozygous familial hypercholesterolemia is relatively common and has an autosomal dominant inheritance pattern. It is caused by one mutated LDLR allele. Homozygous familial hypercholesterolemia is extremely rare and much more severe with cardiovascular disease in early childhood. It is caused by two mutated LDLR alleles and thus follows a recessive inheritance pattern (Brown and Goldstein, 1986). In support of this is the detection of significant comorbidities between common and Mendelian disorders determined through the mining medical records of over 110 million patients (Blair et al., 2013). These authors show that each common disease was comorbid with a diverse and unique combination of Mendelian diseases implicating this “Mendelian code” of loci in common disease pathogenesis (Blair et al., 2013). Indeed, they further show that common variants associated with common disease in GWAS are globally enriched for Mendelian loci, a finding previously reported by Lupski et al (Blair et al., 2013; Lupski et al., 2011).
Further rationale for this is driven by the fact that biochemical variation exists in each individual regardless of IEM diagnosis. For example in a recent GWAS performed on plasma metabolite levels in a healthy population, single nucleotide polymorphisms (SNPs) at the ACADM locus were found to associate with blood C8-carnitine levels (Shin et al., 2014), the same metabolite used for diagnosis of MCAD deficiency. The same GWAS study highlighted several other examples, such as an association between phenylalanine levels at a locus near PAH, which is mutated in PKU (Shin et al., 2014). Overall these observations highlight that a continuum of biochemical phenotypes is always present, where common variation at IEM loci may give rise to more subtle phenotypes, with variants in the middle of the spectrum falling just short of causing a recognizable IEM as found with rare/extreme variants (Figure 1B).
If IEM are like common disorders then we should also study IEM as common disorders, by taking advantage of the same approaches like multi-scale omics technologies and integrative network analysis or even the same datasets (Figure 2A). The vast array of omics technologies could greatly expand phenotypes of IEM beyond clinical portrayals to include different intermediate molecular phenotypes (Figure 2B). Network-based approaches that integrate these data could then provide the framework to connect the various phenotypes to their genetic modifiers. This is critical for understanding the clinical expression of the IEM beyond a single-gene level but rather as a consequence of a set of molecular interactions (subnetwork) (Dipple and McCabe, 2000a; Scriver and Waters, 1999). It is these subnetworks that are sensing DNA variations (rare and common) as well as environmental stimuli, and responding to these perturbations (either appropriately or not) through changes in individual biochemical (intermediary) phenotypes that ultimately influences the expressed clinical phenotype (Schadt, 2009). We propose that by identifying these subnetworks and their associated key molecular drivers, we can greatly impact our understanding of IEM disease biology in a similar way to that demonstrated for complex diseases (Schadt, 2009). A welcoming way to overcome the “rare disease rare data” hurdle in order to build these networks is actually repurposing the data collected from the general population and the molecular networks derived from them as the background to study molecular changes associated with IEM (Figure 2C). For the remainder of this review, we highlight how omics technologies and integrative network approaches can be advantageous to the IEM field.
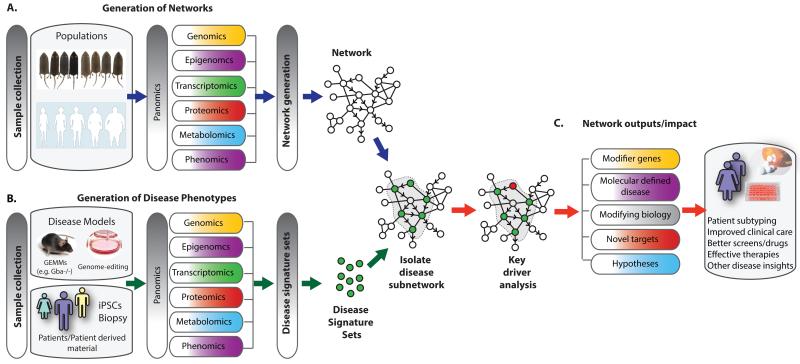
Figure 2. Studying IEM like complex disorders through adopting multi-scale omics technologies and network approaches.
We can study IEM as common disorders by taking advantage of approaches like multi-scale omics technologies and integrative network analysis and even by sharing datasets. (A) Omics data generated from samples collected in common populations of humans or model organisms can be integrated alongside public database information to generate predictive molecular networks. (B) We propose these networks can be repurposed and used as a reference or framework to associate the various IEM phenotypes, scored through multi-omics approaches on samples from IEM models and patients, to identify candidate genetic modifiers and modifying biology. For example, disease signature sets generated by various omics technologies on material derived from patients, patient-derived cells (iPSCs) or experimental model systems can be used to probe a reference network to reveal disease-associated subnetworks. (C) As these Bayesian networks have a causal predictive component they can be used to inform on key molecular drivers of the pathophysiology associated with the IEM. Genes within subnetworks can be nominated as key molecular drivers through statistical algorithms and functional and therapeutic insight can be derived through annotation of subnetwork gene members. Potential impacts of such network approaches to IEM include improving the presently poor correlation between disease severity and the primary mutated locus as well as overcoming the fundamental gap in our knowledge of disease modifying genes and biology.
Data-driven characterization of IEMs through multi-scale omics technologies
Metabolomics
Omics technologies provide a detailed and unbiased view on the changes within the biological layers between DNA and the ultimate presentation of the clinical phenotype. Thus application of these technologies to IEM experimental models, for example, would provide an efficient means to rapidly expand IEM biology beyond what is already known. Of the different omics technologies available, metabolomics would be most amenable to study IEM. Metabolomics is the comprehensive and systematic identification and quantification of metabolites in a biological sample. Such analysis would not only allow for measuring the primary accumulating and often diagnostic metabolites, it will also reveal all other ensuing changes in the metabolome such as those resulting from activation of alternative biochemical reactions. Those alternative biochemical reactions are important candidate modifiers as they can either assist in degrading an accumulating toxic metabolite or contribute to the production of unwanted toxic intermediates.
Biofluids such as plasma and urine are commonly used for these studies and are relatively easy to obtain in the context of a scientific experiment involving human subjects. In fact, measuring metabolites in body fluids is the domain of the clinical biochemist that performs diagnostic test for IEM. It is therefore surprising that untargeted metabolomics received little attention within the IEM field. Recently untargeted metabolomics was evaluated for its clinical utility in IEM diagnosis (Miller et al., 2015). In this retrospective study of samples from patients with a confirmed IEM, the diagnostic biomarker and many other disease-related metabolites were reliably detected in plasma (Miller et al., 2015). Such approaches when fully validated, will not only expedite diagnosis, but at the same time enable the identification of novel biomarkers and potential metabolites associated with phenotypic heterogeneity within a particular IEM (Miller et al., 2015) that signify modifying biology. Metabolomics can also play a crucial role in defining function of novel uncharacterized IEM genes. For example, exome sequencing identified SERAC1 mutations in MEGDEL syndrome, an IEM characterized by dystonia and deafness with Leigh-like syndrome, impaired oxidative phosphorylation and 3-methylglutaconic aciduria. Only a few clues on the function of SERAC1 were available. The presence of a conserved lipase sparked lipidomic analysis, which revealed that SERAC1 was crucial for phosphatidylglycerol remodeling, a phospholipid that is essential for both mitochondrial function and intracellular cholesterol trafficking (Wortmann et al., 2012).
Transcriptomics
Transcriptome analysis through microarray or RNA sequence analysis have evolved into powerful techniques to profile genome-wide changes in gene expression in a tissue of interest in either patient material where available or experimental model organisms such as knockout (KO) mice. In such assays tens of thousands of variables can be measured simultaneously, providing insights into a vast array of known and unknown biological processes. Computational approaches such as the generation of lists of differentially expressed genes when comparing diseased to non-disease samples, the construction of classifiers to predict membership into disease groups, the construction of gene networks to tease apart the relationships among the many variables, are then employed to translate the data into pathophysiological insights through identification of key pathways associated with the disease (Sieberts and Schadt, 2007; Wang et al., 2012). Given the potential mass of information generated from a single experiment, it is surprising that there are only few reports on the use of these methods to understand pathophysiology of IEM.
GD is one of the IEM that exemplifies the power of omics strategies to revealing novel insights (Mistry et al., 2010). Motivation for this stems from the challenge of the macrophage-centric view to explain unusually prevalent manifestations such as gammopathies, cancer risk pulmonary hypertension, cholesterol gallstones and Parkinson disease (PD) (Mistry et al., 2013). GD patients have an almost 37-fold greater risk of multiple myeloma as compared to the general population (Mistry et al., 2013) and approximately 5-10% of PD patients have GBA mutations making it one of the most important genetic predisposing risk factors identified to date (Beavan and Schapira, 2013). In an effort to determine the widespread effect of GBA deficiency on various immune cell populations, a mouse model with a conditional Gba deletion in cells of the hematopoietic and mesenchymal lineages was generated (Mistry et al., 2010). Importantly this mouse model recapitulated the human GD type 1 almost in its entirety, including differences in phenotypic severity as there were varying degrees of splenomegaly and hepatomegaly. Immunophenotyping and transcriptomic profiling revealed not only the dysfunction of macrophages but also aberrations in thymic T cell and dendritic cell development, suggesting that mechanisms other than macrophages may be worthwhile therapeutic targets (Mistry et al., 2010). The transcriptome dataset was further paired with the Connectivity Map (CMAP) in order to computationally perform drug-disease pairing, which is a strategy for repurposing existing therapies to new disease areas (Yuen et al., 2012). Not surprisingly, perhaps, CMAP ranked chemicals utilized in acute and chronic infections as the most relevant (Yuen et al., 2012).
Overall these studies demonstrate how omics approaches can be applied in terms of using detailed molecular and phenotypic characterization of experimental model systems to understand IEM, we likely just need to encourage more of them on other existing IEM models and where possible on IEM patient-derived material. A survey of the literature reveals that transcriptome datasets are available for only a handful of IEMs including methylmalonic acidemia and glycerol kinase deficiency (MacLennan et al., 2006; Manoli et al., 2013). Several datasets have been generated to study mitochondrial disorders, which represent a fairly common class of IEM characterized by respiratory chain defects (Skladal et al., 2003). A recent study of the transcriptome of muscle and fibroblasts of humans affected by a respiratory chain defect revealed common dysregulation of a nutrient-sensing signaling network (Zhang et al., 2013b). A meta-analysis of all mitochondrial transcriptome datasets identified several commonly dysregulated genes across diverse mitochondrial disease etiologies, models, and tissue types (Zhang and Falk, 2014).
Integration of omics data by network approaches to explain complex phenotypes
While potentially useful for prioritizing new candidate modifier genes of IEM, a list of biological entities altered in the IEM versus non-diseased state falls short of revealing its emergent properties. As the IEM phenotype is not just the sum of the parts, it is necessary to use additional methodologies to determine how the individual modifying factors assemble into functional modules and how the system dynamics are affected. Network models are one such framework amenable to exploring the context in which genes, gene products, metabolites and other components operate.
Different approaches to network modeling
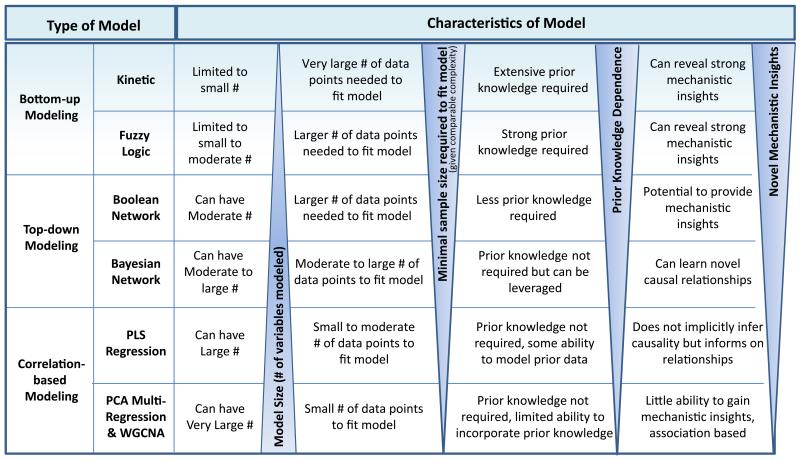
There are several possible representations of biological networks and a diverse array of mathematical models and datasets making those representations possible. For example, biological representations can be as simple as interaction networks where nodes are proteins and edges connecting the nodes represent physical interactions (Stelzl et al., 2005) to complex process descriptions where the networks are directed, sequential and mechanistic such as in a kinetic model, which describe the catalysis of substrates into products of a biochemical pathway (Le Novere, 2015). Although interaction type networks give a comprehensive view of a system such as genome transcriptional regulation in gene interaction networks (Neph et al., 2012), the process descriptions are generally on more focused biology, offering mechanistic insights and can be suitable for dynamic modeling (Le Novere, 2015). Thus the selection of modeling approaches to employ depends on a number of factors such as extent of prior knowledge required, dimensionality of the data to be modeled, the scale of data available to model, and the insight one is looking to derive from the data and the model (Figure 3).
Figure 3. A summary of different classes of mathematical modeling approaches that can be applied to biological data.
Networks represent a way to uncover relationships in data that may help elucidate causal relationships among molecular traits and biological processes and derive mechanistic insights into the causes of disease and other phenotypes of interest. They may also enable predictions of phenotypes.
At present, not all approaches are amenable to the IEM field, as some are highly dependent on large data-sets. However, for the reasons described above we can take advantage of existing datasets generated in complex diseases and use those network derived models in conjunction with IEM datasets as described below. Although an extensive review of the different types of network modeling approaches which could be applied to IEM data presently or as relevant datatypes become available is beyond the scope of this review, we nonetheless provide a brief synopsis of common methods followed by a case-study using one of these methods in order to ignite curiosity. Importantly, networks have already shown the potential to model disease relevant biology, whether from IEM or complex disease leading to novel, testable hypotheses. For example, a human disease network revealed that most disease genes are nonessential and do not have a tendency to encode hub proteins (Goh et al., 2007). Genome-scale networks were used to predict the phenotypic consequences of SNPs (Jamshidi and Palsson, 2006) and reveal known and novel biomarkers of IEM (Pagliarini and di Bernardo, 2013; Shlomi et al., 2009; Thiele et al., 2013).
Overview of biological network types
As summarized in Figure 3, network models can be defined as a spectrum that ranges from those models assuming the most complete knowledge of biological pathways (e.g. process descriptions) to those assuming no prior knowledge preferring instead to infer the network structures directly from the experimental data (e.g. interaction networks). The correlation based models are at the most extreme end of the distribution, requiring no prior knowledge. They are more exploratory in nature, seeking to understand the relationships that may exist between variables by elucidating the correlation structures in extensive datasets as a way to help understand key processes involved in complex phenotypes of interest. One draw-back is that they only reflect connections and influences on those connections; they do not explicitly infer causality and are not mechanistic. For example, correlation between genes using weighted coexpression network analysis (WGCNA) methods reveals whether genes are connected, such that if two genes are consistently up or down-regulated the chance that they share some regulatory feature or belong to the same biological process is higher (Le Novere, 2015; Zhang and Horvath, 2005).
Kinetic models are at the other extreme end of the distribution with respect to requiring extensive prior knowledge. They are typically represented as systems of ordinary differential equations and are thus fixed to the connectivity structure among the variables being modeled. A series of parameters are then fit from the data to define the model precisely. With these parameter estimates, the behavior of the system can be directly explored via simulations run on the model and thus provide for granular mechanistic insights such as predicting changes in protein or metabolite levels (Le Novere, 2015). The modeling of metabolic pathway flux and drug response are examples using this approach. Constraint-based modeling is a related approach as it also relies extensively on prior knowledge, however, it is much larger in biological scale and allows for genome-scale modeling of metabolism. In the case of metabolic networks no kinetic information is required, only network topology and uptake and secretion rates (Bordbar et al., 2014). Logic models represent another class of models that while requiring significant prior knowledge, includes an adaptive component that can be learned from the data thereby reducing the dependency on prior knowledge. They also maintain a simple and intuitive framework for understanding complex signaling networks yet still enable direct mechanistic insights to be derived from simulations on these models (Morris et al., 2010). Overall, kinetic, constraint-based and logic models are representative of bottom up modeling approaches which start with strong prior knowledge as to how pathways are put together with the flow of information through the system then being defined by the parameters on those pathways.
An intermediate type of network in terms of being a more flexible framework are Boolean networks that model biomolecules as binary variables that directly relate to state information such as activated or inhibited that is relevant to downstream biological processes (Albert and Thakar, 2014). The regulation of the different states represented are described in a parameter-free way, not by kinetic parameters thus providing for an approach that enables a more exploratory characterization of the dynamics of a complex system. Although the trade-off is providing less mechanistic insights compared to kinetic models, Boolean networks can represent many more variables.
Bayesian networks
An even more flexible framework for modeling complex biological processes is Bayesian networks. Bayesian networks are able to incorporate prior knowledge if so desired, yet are not dependent on it, and at the same time provide a way to learn regulatory relationships directly from the data. For example, genotype, gene expression, metabolomic data as well as literature-based knowledge can be integrated as priors into causal network models, which leads to an improvement in the accuracy of reconstructed networks (Zhu et al., 2012; Zhu et al., 2007; Zhu et al., 2008). Heuristic searching is used to construct networks comprised of many thousands of variables but equally large sets of data are required to effectively construct this type of model. Another draw-back is the ability to derive mechanistic insights since the causal relationships represented in these models are statistically inferred and not process defined. Another limitation of Bayesian networks relates to their ability to distinguish causal structures that have equivalent joint probability and conditional independence structures, known as the Markov equivalence. As statistically indistinguishable structures may reflect completely contradictory causal relationships, the severity of this problem is not minor. The Boolean and Bayesian network modeling approaches are examples of structure-based learning or top-down modeling approaches that seek to learn relationships directly from the data.
For the purposes of understanding complex systems where the relationships among the constituent components of the system are largely unknown, out of the broad spectrum of methods, Bayesian networks have emerged as a state-of-the-art approach (Chang et al., 2015; Zhu et al., 2012; Zhu et al., 2008). Bayesian networks strike a nice balance between being able to resolve mechanisms and structure and more broadly reflecting connections and their influences thus providing an efficient path for understanding information flow. The cohorts from which we and others have scored omics data and built networks are population based and have been derived in experimental model systems (e.g. F2 crosses of yeast or mice) or in humans and from many different tissue types. Importantly Bayesian networks have demonstrated to capture fundamental properties of complex systems in states that give rise to complex (diseased) phenotypes (Jansen et al., 2003; Lee et al., 2004; Schadt et al., 2008; Zhong et al., 2010; Zhu et al., 2004; Zhu et al., 2012; Zhu et al., 2007; Zhu et al., 2008). Furthermore several disease areas have successfully applied Bayesian network modeling approaches including chronic obstructive pulmonary disease, cancer, obesity, diabetes, inflammatory bowel disease, longevity, cardiovascular diseases and Alzheimer’s disease. From these a large number of novel targets have been identified and validated and the new insights considered to have greatly increased our understanding of these complex diseases (Argmann et al., 2009; Chen et al., 2008; Emilsson et al., 2008; Jostins et al., 2012; Lamb et al., 2011; Tran et al., 2011; Yang et al., 2009; Yoo et al., 2015; Zhang et al., 2013a).
A case study: Finding candidate genetic modifiers and modifying biology of GD using common population datasets and networks
The utility of Bayesian networks from common disease datasets to inform on IEM relies on demonstrating that IEM disease-oriented pathophysiology arises from molecular pathways that are not markedly atypical and actually reflect some extreme or alternate form of common physiology. We tested if this was the case for GD. For this we used a network generated from the F2 offspring of several inbred strains of mice including C57BL/6 and C3H mice (Figure 4A). These F2 populations are unlike KO mouse models, in that they have subtle genetic diversity at multiple loci and therefore mimic the natural range of DNA variation seen in humans (Argmann et al., 2005). The liver transcriptomes of these individual F2 mice were combined with their genetic information and organized into causal, predictive molecular interaction networks using the Bayes theorem (Chen et al., 2008; Schadt et al., 2005; Yang et al., 2006). We tested whether in this network there is a non-random, tight interconnection of genes affected in the Gba KO mouse model, which would indicate that the molecular pathways ascertained under complete ablation of Gba are related to common physiology. This is in contrast to finding that the interconnectivity is diffusely distributed across the whole network which might imply significant ‘new biology’ arises when Gba expression is depleted.
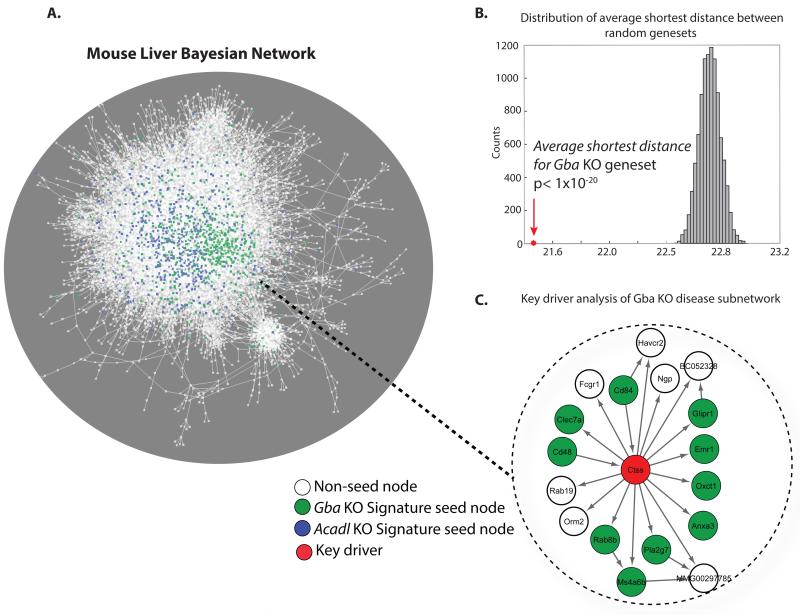
Figure 4. An example of a predictive molecular network, the mouse liver Bayesian network.
(A) A predictive molecular network generated from genomic and hepatic gene expression data scored in several hundred offspring from different F2 crosses of inbred strains of mice. The utility of networks from common disease datasets to inform on IEM relies on demonstrating that IEM disease-oriented pathophysiology arises from molecular pathways that are not markedly atypical and actually reflect some extreme or alternate form of common physiology. We tested if this was the case by probing the network with two different IEM model derived disease signature sets (seed set). One signature set was derived from the liver transcriptomic data generated in a Gba KO conditional mouse, an experimental model for Gaucher Disease (GD, green nodes) and the second was from the liver transcriptomic data generated in an Acadl KO mouse, a fatty acid oxidation deficient experimental model (FAO, blue nodes). (B) A histogram of the shortest path calculation for 104 randomly generated gene sets, of the same size as the disease signature sets, on the network in A. The arrow in the histogram represents the average shortest path of the GD signature gene set. The average shortest path for the FAO signature gene set was also significantly lower relative to random chance (data not shown). The low average shortest distance for the two disease signature sets relative to that of randomly derived gene sets indicates in network terms, a non-random, tight interconnection of genes in the network. In biological terms, this is suggestive that a significant part of the pathophysiology associated with IEM is indeed related to common physiology. (C) Key molecular drivers were nominated amongst the genes within the isolated GD subnetworks through statistical algorithms. The isolated Gba KO subnetwork highlights one nominated key driver, Cathepsin S (Ctss) in red.
Of the 1161 genes that were found differentially expressed in the livers of Gba KO mice (Mistry et al., 2010), 584 were represented in our mouse liver Bayesian network. For these genes we calculated the pair-wise shortest path within the network and compared that to the average shortest path of 104 randomly selected sets of 584 genes. On average, the shortest path for the genes in the Gba KO-associated gene set were much lower than expected by random chance(P < 0.001) (Figure 4B). In network terms, the low average shortest distance for the GD disease signature set, relative to random chance, indicates a non-random, tight interconnection of genes in the network. In biological terms, this means that a significant part of the GD pathophysiology is indeed related to common physiology.
In order to demonstrate that our observations are not restricted to GD, we applied the pair-wise shortest path algorithm to a differentially expressed gene set obtained from livers of mice with a defect in mitochondrial fatty acid oxidation (FAO, long-chain acyl-CoA dehydrogenase KO mice)(Houten et al., 2013; Kurtz et al., 1998). We also observed a low average shortest distance for this FAO gene set relative to that of randomly derived gene sets indicating strong connectivity of this gene set in the same liver mouse Bayesian network (P < 0.001, Figure 4B). Combined these data show that we can employ data-driven approaches and re-purpose existing causal predictive networks that have shown benefit to the common metabolic disease field for use in the IEM field. Networks based on mouse models serve an advantage in that material of all sorts can be profiled in controlled ways that make generation of the networks, as well as signature sets from models that represent the IEM, feasible. We do however suggest in the next section that molecular characterization of IEM patient material should be explored in a systematic way and could be used similarly.
Given the above, we now demonstrate that Bayesian networks can be used to inform on key molecular drivers of the pathophysiology associated with the IEM, in our case GD. Within the identified GD subnetwork (Figure 4C), some of the predicted key drivers include lysosomal-associated proteins such as Cathepsin S (Ctss) (Hsing and Rudensky, 2005), a Rab GTPase, Rab7b (Yao et al., 2009) and the PD associated gene Atp13a2 (Dehay et al., 2012). Importantly these genes and their subnetwork could be directly used to formulate novel hypotheses in terms of GD pathophysiology and set the stage for understanding modifying disease biology in a data-driven approach.
Gaining insight into disease through studying rare disease-associated subnetworks
Omics and network approaches in IEM also have clear translational value. By intersecting IEM-associated subnetworks with databases of other disease and drug-related signatures, we can better characterize and subtype diseases and gain insights into the full gamut of metabolic disease. In our example, querying the GD subnetwork against other disease databases revealed a significant enrichment in the genes of the macrophage enriched molecular network (MEMN) module (Chen et al., 2008; Emilsson et al., 2008)(>4-fold, p<0.05). The MEMN is associated with macrophage-related function and is significantly enriched for genes testing statistically causal for various complex metabolic disease traits including adiposity, lipid levels and insulin sensitivity (Chen et al., 2008). The MEMN has been subsequently shown as relevant for a great diversity of diseases including inflammatory bowel disease, Alzheimer’s disease, asthma, chronic obstructive pulmonary disease, and heart disease (Emilsson et al., 2008; Jostins et al., 2012; Wang et al., 2012; Zhang et al., 2013a). Canonical pathway enrichment analysis showed that the MEMN is indeed enriched in lysosomal genes and human phenotypes related to hepato- and spleno-megaly. The MEMN showed further relevance to GD as it is enriched for a liver transcriptome signature derived from mice treated with a compound that reduces the synthesis of glucosylceramide, the precursor of the more complex glycosphingolipids and the metabolite that accumulates in GD. Interestingly, this compound essentially normalizes gene expression of a genetically obese (ob/ob) mouse to one of a lean (C57BL/6) mouse (Bijl et al., 2009).
Overall, we isolated a subnetwork that could recreate the known associations of GD with macrophage and lysosome function and glycosphingolipid biology. Importantly, this subnetwork could shed light on the potential mechanisms for the clinical diversity seen in GD patients and highlights novel candidate modifier genes, given the associations of these genes with other complex diseases. These findings also serve to highlight the potential of an IEM to shed light on common disease biology. By associating a GD-derived molecular signature to a module of genes linked to common diseases (MEMN), we could reinforce the importance of macrophage function as well as assign lysosomal dysfunction as pathophysiological processes. Indeed glycosphingolipid synthesis inhibitors are anti-diabetic (Aerts et al., 2007) and lysosomal dysfunction has been associated with obesity (Gabriel et al., 2014; Xu et al., 2013).
Moving beyond genetics of gene expression based datasets and networks
Genetics of gene expression is widely used in the common disease world. However, models that incorporate information from other scales of biology such as that captured by metabolomics, proteomics, phenomics and kinomics are not yet common place in any disease area. This is despite the fact that these intermediate phenotype levels are considered to be equally important with respect to informing on the how the genetic variants ultimately impact on the clinical phenotype (Ritchie et al., 2015). Thus while our strategy in Figure 2 begins with a differentially expressed gene set derived in a GD mouse model, projected onto a molecular-based Bayesian network, an alternative strategy could be to use differentially expressed metabolites that associate with GD severity in conjunction with other biological networks. The delay in incorporating other omics data layers into integrative approaches has in part been caused by technological challenges, but several resource initiatives are now demonstrating feasibility. For example in the BXD mouse genetic reference population, dozens of strains have been co-characterized at the genomic, transcriptomic, proteomic, metabolomic and phenomic level (Andreux et al., 2012; Civelek and Lusis, 2014; Wu et al., 2014), with data being made publically available through GeneNetwork (Rosen et al., 2007). In humans large scale metabolomics datasets are starting to accumulate (Shin et al., 2014) and tissues are being collected as part of consortiums for the potential of panomics to be added (Consortium, 2015a).
The next generation outlook for IEM: Understanding the phenotype of the individual patient
While understanding disease pathophysiology in general will advance the IEM field, the ultimate goal should be to understand per individual how their phenotype emerges given their primary mutation on the background of their whole genome, not unlike personalized medicine. This essentially means working on a case by case manner and solving for an n of 1 case each time. In our next generation outlook for IEM, we argue that studying individual cases will become feasible by using multi-scale omics approaches and network models. This is because deep molecular profiling using an array of all available omics technologies combined with recent experimental innovations will yield results that depend less on de novo discovery of associations by providing a more comprehensive and holistic context in which to figure out a given case.
Whole exome sequencing is extremely powerful at detecting a Mendelian disease gene in a single patient (Bamshad et al., 2011) and has become more common place in the diagnostic arena and being applied especially in cases of unknown etiology. This is useful in IEM where many genes can cause similar disease phenotypes such as mitochondrial disorders (Carroll et al., 2014; Lieber et al., 2013), but also aids in finding the molecular cause when a phenotype is atypical for a known disease gene (Ratbi et al., 2015), or when a disease gene is associated with an unexpected metabolic pathway (Houten et al., 2014). Many of the new IEM genes however, encode sparsely or uncharacterized proteins, which offer limited disease insight and no treatment options. At the same time, newborn screening has identified many cases in IEM where the clinical significance of the defect is uncertain (Andresen et al., 2001; Gallant et al., 2012). Thus while not yet greatly contributing to our molecular understanding of IEM, newborn screening and genomics is currently impacting the IEM field with respect to increasing the number of IEM and patients to study.
Novel developments will help to understand the individual IEM patient
Although it may appear as if omics approaches are currently creating more questions than it answers there is hope. Enthusiasm for omics approaches and network frameworks in IEM medicine is derived from evidence in the cancer arena, which has demonstrated that present day technology is sufficiently advanced to tackle individual cases in order to understand the molecular drivers of those cases. In this sense, there are several cases now in which molecular profiling of a given individual cancer patient highlighted driver genes that indicated treatments that in turn were given and helped or even cured the patient (Schadt et al., 2014). Of course, cancers are not IEM, however, from a genetic standpoint; there is some similarity to IEM given the occurrence of variants with big effect sizes that make for profound perturbations leading to significant physiological changes. Similar to the initiatives of the cancer field then, we will need to start investing in the generation of panomic and next generation type of phenotyping data of existing patients in order to better understand disease (Weaver et al., 2014).
While in the case of cancers, tumor samples are readily available for multi-omics profiling, technology has evolved so that profiling of relevant disease tissues, outside of tumor biology is also feasible. With induced pluripotent stem cell (iPSC) technology we now have the ability to take patient fibroblasts, reprogram them into iPSC cells and then re-direct the cell lineage towards relevant disease cell types from which detailed molecular profiling can be performed (Inoue et al., 2014). For example, in a recent study iPSC-derived cardiomyocytes were used to study the pathophysiology underlying the cardiomyopathy observed in Barth syndrome, a mitochondrial disorder caused by a defect in cardiolipin synthesis due to mutations in TAZ. The iPSC-derived cardiomyocytes were used for a series of state-of-the-art assays including a heart-on-chip model that assesses contraction force (Wang et al., 2014). With single cell technologies we also now have the ability to do transcriptomics measures when biopsy material is available in limiting quantities (Macaulay and Voet, 2014). From a translational sense, these resources as well as advanced genome editing techniques such as CRISPR/Cas9 and modified RNA strategies facilitate downstream validation assays of hypotheses and potential therapeutic interventions (Hsu et al., 2014). Alternative data types such as metabolomics are also now feasible on large scale and on many tissue types. Thus in the elucidation of the molecular and biochemical basis of new IEM, multi-scale approaches such as genomics and metabolomics that are layered into informative networks will play synergistic roles (Figure 2).
Another way by which omics will be impacting on IEM is through the recently launched Resilience Project (http://resilienceproject.me/), whereby large scale genetic screening of general populations for a panel of rare disease causing mutations is hoping to uncover healthy individuals harboring rare genetic disease and the genetic modifiers that make them resilient to this disease (Friend and Schadt, 2014). The screening panel includes at least 195 genes known to cause a variety of IEM, such as PKU and MCAD deficiency. The premise of this approach is the ability to identify those mutations, which potentially have bigger effect sizes in the modifying genetics given a more extreme phenotype (i.e. absence of clinical symptoms) (Figure 1B). This is one potential way to solve the issue of lack of power using GWAS approaches in IEM patients as highlighted at the beginning of this perspective. Overall, these systems approaches are expected to provide insight in pathophysiology by identifying potential modifiers and their associated biology as well as offer new treatment options (Friend and Schadt, 2014). The initiative of resilience is not unlike the solving unique cases. Thus there is seemingly a paradigm shift happening towards individualized medicine that will undoubtedly benefit many disease areas (IEM to common disease to cancers) in terms of being able to understand the molecular drivers of those cases like never before.
Finally as in the case of cancer, our ability to make sense of the various scored data types will require a more network oriented view whereby a single patient is studied in the context of many. For this we will need to tackle organizing the digital universe of information from the IEM by building models as a way to capture knowledge and understanding from the vast seas of data as is done in cancer through the use of the cancer genome atlas datasets (TCGA Research Network: http://cancergenome.nih.gov/). We will for example need to construct predictive disease network models that can (i) render subtypes of IEM, leading towards biomarkers that can accurately stratify patient populations, and (ii) provide insights into disease networks that help resolve causal relationships among genes and phenotypes, leading to potential therapeutic targets. There are many network formats possible as described above and their use will depend on their performance and may ultimately require hybrid type models. With all these networks in hand, the final step is to use them to inform individual IEM cases in terms of prediction of disease severity, pathophysiological mechanisms and potential therapies.
Potential limitations and future directions
While we predict that pathophysiology associated with IEM is not necessarily a new form of biology and therefore minable in the general population, we do not know whether this holds for all IEM, even more so for different aspects of their pathophysiology. Another potential limitation is that the range of gene expression for an enzyme does not directly reflect metabolic flux through that enzyme. This is because enzymes do not work in isolation, but are kinetically linked to other enzymes via their substrates and product (Kacser and Burns, 1981). In a network framework dominated by gene expression datasets this could be problematic. Therefore network refinement by incorporating knowledge and other datatypes will be needed for the purpose of investigating IEM when using general population datasets.
To interrogate the reference networks we will rely on signatures derived from the characterization of the IEM preferably at multiple scales of biology. The rationale for this is that intermediate phenotypes are more proximal to the expression of the disease and help in making mechanistic links. At present however, we do not know which layers of information will reveal the most impactful biomarkers or modifiers so we need to aim to be as broad as possible. Although the IEM field has realized that collecting natural history is crucial to gather more phenotypic data on these diseases, they are often not collected in conjunction with other multiscale layers, even as basic as keeping a repository of DNA from such a cohort, which would be essential for the validation of the predicted modifiers. This means populating biobanks with various samples from IEM patients is imperative followed by expanding clinical characterizations to include other omics layers on these samples. Finally, we would like to modernize what Garrod conveyed to his medical students. He often assured them that they do not all need to become biochemists but rather they needed to know something of the biochemical approach to disease, we would suggest one also now needs to know something of the multi-scale biology approach to disease.
Acknowledgements
We thank J.M. Aerts and A.J. Verhoeven for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aerts JM, Ottenhoff R, Powlson AS, Grefhorst A, van Eijk M, Dubbelhuis PF, Aten J, Kuipers F, Serlie MJ, Wennekes T, et al. Pharmacological inhibition of glucosylceramide synthase enhances insulin sensitivity. Diabetes. 2007;56:1341–1349. doi: 10.2337/db06-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ala A, Schilsky M. Genetic modifiers of liver injury in hereditary liver disease. Semin Liver Dis. 2011;31:208–214. doi: 10.1055/s-0031-1276648. [DOI] [PubMed] [Google Scholar]
- Albert R, Thakar J. Boolean modeling: a logic-based dynamic approach for understanding signaling and regulatory networks and for making useful predictions. Wiley interdisciplinary reviews Systems biology and medicine. 2014;6:353–369. doi: 10.1002/wsbm.1273. [DOI] [PubMed] [Google Scholar]
- Alfonso P, Navascues J, Navarro S, Medina P, Bolado-Carrancio A, Andreu V, Irun P, Rodriguez-Rey JC, Pocovi M, Espana F, et al. Characterization of variants in the glucosylceramide synthase gene and their association with type 1 Gaucher disease severity. Human mutation. 2013;34:1396–1403. doi: 10.1002/humu.22381. [DOI] [PubMed] [Google Scholar]
- Andresen BS, Dobrowolski SF, O’Reilly L, Muenzer J, McCandless SE, Frazier DM, Udvari S, Bross P, Knudsen I, Banas R, et al. Medium-chain acyl-CoA dehydrogenase (MCAD) mutations identified by MS/MS-based prospective screening of newborns differ from those observed in patients with clinical symptoms: identification and characterization of a new, prevalent mutation that results in mild MCAD deficiency. Am J Hum Genet. 2001;68:1408–1418. doi: 10.1086/320602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreux PA, Williams EG, Koutnikova H, Houtkooper RH, Champy MF, Henry H, Schoonjans K, Williams RW, Auwerx J. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. 2012;150:1287–1299. doi: 10.1016/j.cell.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argmann C, Dobrin R, Heikkinen S, Auburtin A, Pouilly L, Cock TA, Koutnikova H, Zhu J, Schadt EE, Auwerx J. Ppargamma2 is a key driver of longevity in the mouse. PLoS Genet. 2009;5:e1000752. doi: 10.1371/journal.pgen.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argmann CA, Chambon P, Auwerx J. Mouse phenogenomics: the fast track to “systems metabolism”. Cell Metab. 2005;2:349–360. doi: 10.1016/j.cmet.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nature reviews Genetics. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- Baris HN, Cohen IJ, Mistry PK. Gaucher disease: the metabolic defect, pathophysiology, phenotypes and natural history. Pediatric endocrinology reviews: PER. 2014;12(Suppl 1):72–81. [PMC free article] [PubMed] [Google Scholar]
- Beavan MS, Schapira AH. Glucocerebrosidase mutations and the pathogenesis of Parkinson disease. Annals of medicine. 2013;45:511–521. doi: 10.3109/07853890.2013.849003. [DOI] [PubMed] [Google Scholar]
- Becanovic K, Norremolle A, Neal SJ, Kay C, Collins JA, Arenillas D, Lilja T, Gaudenzi G, Manoharan S, Doty CN, et al. A SNP in the HTT promoter alters NF-kappaB binding and is a bidirectional genetic modifier of Huntington disease. Nat Neurosci. 2015;18:807–816. doi: 10.1038/nn.4014. [DOI] [PubMed] [Google Scholar]
- Bijl N, Sokolovic M, Vrins C, Langeveld M, Moerland PD, Ottenhoff R, van Roomen CP, Claessen N, Boot RG, Aten J, et al. Modulation of glycosphingolipid metabolism significantly improves hepatic insulin sensitivity and reverses hepatic steatosis in mice. Hepatology. 2009;50:1431–1441. doi: 10.1002/hep.23175. [DOI] [PubMed] [Google Scholar]
- Blair DR, Lyttle CS, Mortensen JM, Bearden CF, Jensen AB, Khiabanian H, Melamed R, Rabadan R, Bernstam EV, Brunak S, et al. A nondegenerate code of deleterious variants in Mendelian loci contributes to complex disease risk. Cell. 2013;155:70–80. doi: 10.1016/j.cell.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordbar A, Monk JM, King ZA, Palsson BO. Constraint-based models predict metabolic and associated cellular functions. Nature reviews Genetics. 2014;15:107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- Carroll CJ, Brilhante V, Suomalainen A. Next-generation sequencing for mitochondrial disorders. British journal of pharmacology. 2014;171:1837–1853. doi: 10.1111/bph.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R, Karr JR, Schadt EE. Causal inference in biology networks with integrated belief propagation. Pac Symp Biocomput. 2015:359–370. [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhu J, Lum PY, Yang X, Pinto S, Macneil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nature reviews Genetics. 2014;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, G. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015a;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, G.M.o.H.s.D.G.-H. Identification of Genetic Factors that Modify Clinical Onset of Huntington’s Disease. Cell. 2015b;162:516–526. doi: 10.1016/j.cell.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting GR. Modifier genes in Mendelian disorders: the example of cystic fibrosis. Ann N Y Acad Sci. 2010;1214:57–69. doi: 10.1111/j.1749-6632.2010.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay B, Ramirez A, Martinez-Vicente M, Perier C, Canron MH, Doudnikoff E, Vital A, Vila M, Klein C, Bezard E. Loss of P-type ATPase ATP13A2/PARK9 function induces general lysosomal deficiency and leads to Parkinson disease neurodegeneration. Proc Natl Acad Sci U S A. 2012;109:9611–9616. doi: 10.1073/pnas.1112368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipple KM, McCabe ER. Modifier genes convert “simple” Mendelian disorders to complex traits. Mol Genet Metab. 2000a;71:43–50. doi: 10.1006/mgme.2000.3052. [DOI] [PubMed] [Google Scholar]
- Dipple KM, McCabe ER. Phenotypes of patients with “simple” Mendelian disorders are complex traits: thresholds, modifiers, and systems dynamics. Am J Hum Genet. 2000b;66:1729–1735. doi: 10.1086/302938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipple KM, Phelan JK, McCabe ER. Consequences of complexity within biological networks: robustness and health, or vulnerability and disease. Mol Genet Metab. 2001;74:45–50. doi: 10.1006/mgme.2001.3227. [DOI] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- Friend SH, Schadt EE. Translational genomics. Clues from the resilient. Science. 2014;344:970–972. doi: 10.1126/science.1255648. [DOI] [PubMed] [Google Scholar]
- Gabriel TL, Tol MJ, Ottenhof R, van Roomen C, Aten J, Claessen N, Hooibrink B, de Weijer B, Serlie MJ, Argmann C, et al. Lysosomal Stress in Obese Adipose Tissue Macrophages Contributes to MITF-Dependent Gpnmb Induction. Diabetes. 2014;63:3310–3323. doi: 10.2337/db13-1720. [DOI] [PubMed] [Google Scholar]
- Gallant NM, Leydiker K, Tang H, Feuchtbaum L, Lorey F, Puckett R, Deignan JL, Neidich J, Dorrani N, Chang E, et al. Biochemical, molecular, and clinical characteristics of children with short chain acyl-CoA dehydrogenase deficiency detected by newborn screening in California. Mol Genet Metab. 2012;106:55–61. doi: 10.1016/j.ymgme.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Gallati S. Disease-modifying genes and monogenic disorders: experience in cystic fibrosis. Appl Clin Genet. 2014;7:133–146. doi: 10.2147/TACG.S18675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrod AE. The incidence of alkaptonuria: a study in chemical individuality. Lancet. 1902;2:1616–1620. [PMC free article] [PubMed] [Google Scholar]
- Genin E, Feingold J, Clerget-Darpoux F. Identifying modifier genes of monogenic disease: strategies and difficulties. Human genetics. 2008;124:357–368. doi: 10.1007/s00439-008-0560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. The relative importance of principal and modifying genes in determining some human diseases. J Genet. 1941;41:147–157. [Google Scholar]
- Houten SM, Denis S, te Brinke H, Jongejan A, van Kampen AH, Bradley EJ, Baas F, Hennekam RC, Millington DS, Young SP, et al. Mitochondrial NADP(H) deficiency due to a mutation in NADK2 causes dienoyl-CoA reductase deficiency with hyperlysinemia. Hum Mol Genet. 2014;23:5009–5016. doi: 10.1093/hmg/ddu218. [DOI] [PubMed] [Google Scholar]
- Houten SM, Herrema H, te Brinke H, Denis S, Ruiter JP, van Dijk TH, Argmann CA, Ottenhoff R, Muller M, Groen AK, et al. Impaired amino acid metabolism contributes to fasting-induced hypoglycemia in fatty acid oxidation defects. Hum Mol Genet. 2013;22:5249–5261. doi: 10.1093/hmg/ddt382. [DOI] [PubMed] [Google Scholar]
- Hsing LC, Rudensky AY. The lysosomal cysteine proteases in MHC class II antigen presentation. Immunological reviews. 2005;207:229–241. doi: 10.1111/j.0105-2896.2005.00310.x. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Nagata N, Kurokawa H, Yamanaka S. iPS cells: a game changer for future medicine. EMBO J. 2014;33:409–417. doi: 10.1002/embj.201387098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshidi N, Palsson BO. Systems biology of SNPs. Molecular systems biology. 2006;2:38. doi: 10.1038/msb4100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, Emili A, Snyder M, Greenblatt JF, Gerstein M. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacser H, Burns JA. The molecular basis of dominance. Genetics. 1981;97:639–666. doi: 10.1093/genetics/97.3-4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz DM, Rinaldo P, Rhead WJ, Tian L, Millington DS, Vockley J, Hamm DA, Brix AE, Lindsey JR, Pinkert CA, et al. Targeted disruption of mouse long-chain acyl-CoA dehydrogenase gene reveals crucial roles for fatty acid oxidation. Proc Natl Acad Sci USA. 1998;95:15592–15597. doi: 10.1073/pnas.95.26.15592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JR, Zhang C, Xie T, Wang K, Zhang B, Hao K, Chudin E, Fraser HB, Millstein J, Ferguson M, et al. Predictive genes in adjacent normal tissue are preferentially altered by sCNV during tumorigenesis in liver cancer and may rate limiting. PloS one. 2011;6:e20090. doi: 10.1371/journal.pone.0020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanpher B, Brunetti-Pierri N, Lee B. Inborn errors of metabolism: the flux from Mendelian to complex diseases. Nature reviews Genetics. 2006;7:449–460. doi: 10.1038/nrg1880. [DOI] [PubMed] [Google Scholar]
- Lanthaler B, Steichen-Gersdorf E, Kollerits B, Zschocke J, Witsch-Baumgartner M. Maternal ABCA1 genotype is associated with severity of Smith-Lemli-Opitz syndrome and with viability of patients homozygous for null mutations. European journal of human genetics: EJHG. 2013;21:286–293. doi: 10.1038/ejhg.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novere N. Quantitative and logic modelling of molecular and gene networks. Nature reviews Genetics. 2015;16:146–158. doi: 10.1038/nrg3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Date SV, Adai AT, Marcotte EM. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- Lettre G. The search for genetic modifiers of disease severity in the beta-hemoglobinopathies. Cold Spring Harbor perspectives in medicine. 2012;2 doi: 10.1101/cshperspect.a015032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber DS, Calvo SE, Shanahan K, Slate NG, Liu S, Hershman SG, Gold NB, Chapman BA, Thorburn DR, Berry GT, et al. Targeted exome sequencing of suspected mitochondrial disorders. Neurology. 2013;80:1762–1770. doi: 10.1212/WNL.0b013e3182918c40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SM, Choi M, Liu J, Jain D, Boot RG, Kallemeijn WW, Aerts JM, Pashankar F, Kupfer GM, Mane S, et al. Phenotype diversity in type 1 Gaucher disease: discovering the genetic basis of Gaucher disease/hematologic malignancy phenotype by individual genome analysis. Blood. 2012;119:4731–4740. doi: 10.1182/blood-2011-10-386862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay IC, Voet T. Single cell genomics: advances and future perspectives. PLoS Genet. 2014;10:e1004126. doi: 10.1371/journal.pgen.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan NK, Rahib L, Shin C, Fang Z, Horvath S, Dean J, Liao JC, McCabe ER, Dipple KM. Targeted disruption of glycerol kinase gene in mice: expression analysis in liver shows alterations in network partners related to glycerol kinase activity. Hum Mol Genet. 2006;15:405–415. doi: 10.1093/hmg/ddi457. [DOI] [PubMed] [Google Scholar]
- Manoli I, Sysol JR, Li L, Houillier P, Garone C, Wang C, Zerfas PM, Cusmano-Ozog K, Young S, Trivedi NS, et al. Targeting proximal tubule mitochondrial dysfunction attenuates the renal disease of methylmalonic acidemia. Proc Natl Acad Sci U S A. 2013;110:13552–13557. doi: 10.1073/pnas.1302764110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Kennedy AD, Eckhart AD, Burrage LC, Wulff JE, Miller LA, Milburn MV, Ryals JA, Beaudet AL, Sun Q, et al. Untargeted metabolomic analysis for the clinical screening of inborn errors of metabolism. Journal of inherited metabolic disease. 2015 doi: 10.1007/s10545-015-9843-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry PK, Liu J, Yang M, Nottoli T, McGrath J, Jain D, Zhang K, Keutzer J, Chuang WL, Mehal WZ, et al. Glucocerebrosidase gene-deficient mouse recapitulates Gaucher disease displaying cellular and molecular dysregulation beyond the macrophage. Proc Natl Acad Sci U S A. 2010;107:19473–19478. doi: 10.1073/pnas.1003308107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry PK, Sirrs S, Chan A, Pritzker MR, Duffy TP, Grace ME, Meeker DP, Goldman ME. Pulmonary hypertension in type 1 Gaucher’s disease: genetic and epigenetic determinants of phenotype and response to therapy. Mol Genet Metab. 2002;77:91–98. doi: 10.1016/s1096-7192(02)00122-1. [DOI] [PubMed] [Google Scholar]
- Mistry PK, Taddei T, vom Dahl S, Rosenbloom BE. Gaucher disease and malignancy: a model for cancer pathogenesis in an inborn error of metabolism. Critical reviews in oncogenesis. 2013;18:235–246. doi: 10.1615/critrevoncog.2013006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MK, Saez-Rodriguez J, Sorger PK, Lauffenburger DA. Logic-based models for the analysis of cell signaling networks. Biochemistry. 2010;49:3216–3224. doi: 10.1021/bi902202q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Stergachis AB, Reynolds A, Sandstrom R, Borenstein E, Stamatoyannopoulos JA. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150:1274–1286. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini R, di Bernardo D. A genome-scale modeling approach to study inborn errors of liver metabolism: toward an in silico patient. J Comput Biol. 2013;20:383–397. doi: 10.1089/cmb.2012.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratbi I, Falkenberg KD, Sommen M, Al-Sheqaih N, Guaoua S, Vandeweyer G, Urquhart JE, Chandler KE, Williams SG, Roberts NA, et al. Heimler Syndrome Is Caused by Hypomorphic Mutations in the Peroxisome-Biogenesis Genes PEX1 and PEX6. Am J Hum Genet. 2015 doi: 10.1016/j.ajhg.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Methods of integrating data to uncover genotype-phenotype interactions. Nature reviews Genetics. 2015;16:85–97. doi: 10.1038/nrg3868. [DOI] [PubMed] [Google Scholar]
- Rosen GD, Chesler EJ, Manly KF, Williams RW. An informatics approach to systems neurogenetics. Methods in molecular biology. 2007;401:287–303. doi: 10.1007/978-1-59745-520-6_16. [DOI] [PubMed] [Google Scholar]
- Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461:218–223. doi: 10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Buchanan S, Brennand KJ, Merchant KM. Evolving toward a human-cell based and multiscale approach to drug discovery for CNS disorders. Frontiers in pharmacology. 2014;5:252. doi: 10.3389/fphar.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, GuhaThakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. Mapping the genetic architecture of gene expression in human liver. PLoS biology. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A, Lindner M, Kohlmuller D, Olgemoller K, Mayatepek E, Hoffmann GF. Expanded newborn screening for inborn errors of metabolism by electrospray ionization-tandem mass spectrometry: results, outcome, and implications. Pediatrics. 2003;111:1399–1406. doi: 10.1542/peds.111.6.1399. [DOI] [PubMed] [Google Scholar]
- Scriver CR, Waters PJ. Monogenic traits are not simple: lessons from phenylketonuria. Trends in genetics: TIG. 1999;15:267–272. doi: 10.1016/s0168-9525(99)01761-8. [DOI] [PubMed] [Google Scholar]
- Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T, Cabili MN, Ruppin E. Predicting metabolic biomarkers of human inborn errors of metabolism. Molecular systems biology. 2009;5:263. doi: 10.1038/msb.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberts SK, Schadt EE. Moving toward a system genetics view of disease. Mamm Genome. 2007;18:389–401. doi: 10.1007/s00335-007-9040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skladal D, Halliday J, Thorburn DR. Minimum birth prevalence of mitochondrial respiratory chain disorders in children. Brain. 2003;126:1905–1912. doi: 10.1093/brain/awg170. [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Thiele I, Swainston N, Fleming RM, Hoppe A, Sahoo S, Aurich MK, Haraldsdottir H, Mo ML, Rolfsson O, Stobbe MD, et al. A community-driven global reconstruction of human metabolism. Nature biotechnology. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LM, Zhang B, Zhang Z, Zhang C, Xie T, Lamb JR, Dai H, Schadt EE, Zhu J. Inferring causal genomic alterations in breast cancer using gene expression data. BMC systems biology. 2011;5:121. doi: 10.1186/1752-0509-5-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon HJ. Inborn Errors of Metabolism: Advances in Diagnosis and Therapy. JAMA Pediatr. 2015;169:778–782. doi: 10.1001/jamapediatrics.2015.0754. [DOI] [PubMed] [Google Scholar]
- Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IM, Zhang B, Yang X, Zhu J, Stepaniants S, Zhang C, Meng Q, Peters M, He Y, Ni C, et al. Systems analysis of eleven rodent disease models reveals an inflammatome signature and key drivers. Molecular systems biology. 2012;8:594. doi: 10.1038/msb.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver JM, Ross-Innes CS, Fitzgerald RC. The ‘-omics’ revolution and oesophageal adenocarcinoma. Nature reviews Gastroenterology & hepatology. 2014;11:19–27. doi: 10.1038/nrgastro.2013.150. [DOI] [PubMed] [Google Scholar]
- Wilcken B, Wiley V, Hammond J, Carpenter K. Screening newborns for inborn errors of metabolism by tandem mass spectrometry. N Engl J Med. 2003;348:2304–2312. doi: 10.1056/NEJMoa025225. [DOI] [PubMed] [Google Scholar]
- Wortmann SB, Vaz FM, Gardeitchik T, Vissers LE, Renkema GH, Schuurs-Hoeijmakers JH, Kulik W, Lammens M, Christin C, Kluijtmans LA, et al. Mutations in the phospholipid remodeling gene SERAC1 impair mitochondrial function and intracellular cholesterol trafficking and cause dystonia and deafness. Nat Genet. 2012;44:797–802. doi: 10.1038/ng.2325. [DOI] [PubMed] [Google Scholar]
- Wu Y, Williams EG, Dubuis S, Mottis A, Jovaisaite V, Houten SM, Argmann CA, Faridi P, Wolski W, Kutalik Z, et al. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158:1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, Ferrante AW., Jr. Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816–830. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, Torosyan G, Majid S, Falkard B, Kleinhanz RR, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41:415–423. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Liu X, Li D, Chen T, Cai Z, Cao X. Late endosome/lysosome-localized Rab7b suppresses TLR9-initiated proinflammatory cytokine and type I IFN production in macrophages. J Immunol. 2009;183:1751–1758. doi: 10.4049/jimmunol.0900249. [DOI] [PubMed] [Google Scholar]
- Yoo S, Takikawa S, Geraghty P, Argmann C, Campbell J, Lin L, Huang T, Tu Z, Feronjy R, Spira A, et al. Integrative Analysis of DNA Methylation and Gene Expression Data Identifies EPAS1 as a Key Regulator of COPD. PLoS Genet. 2015;11:e1004898. doi: 10.1371/journal.pgen.1004898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen T, Iqbal J, Zhu LL, Sun L, Lin A, Zhao H, Liu J, Mistry PK, Zaidi M. Disease-drug pairs revealed by computational genomic connectivity mapping on GBA1 deficient, Gaucher disease mice. Biochem Biophys Res Commun. 2012;422:573–577. doi: 10.1016/j.bbrc.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013a;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Statistical applications in genetics and molecular biology. 2005;4 doi: 10.2202/1544-6115.1128. Article 17. [DOI] [PubMed] [Google Scholar]
- Zhang CK, Stein PB, Liu J, Wang Z, Yang R, Cho JH, Gregersen PK, Aerts JM, Zhao H, Pastores GM, et al. Genome-wide association study of N370S homozygous Gaucher disease reveals the candidacy of CLN8 gene as a genetic modifier contributing to extreme phenotypic variation. American journal of hematology. 2012;87:377–383. doi: 10.1002/ajh.23118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Falk MJ. Integrated transcriptome analysis across mitochondrial disease etiologies and tissues improves understanding of common cellular adaptations to respiratory chain dysfunction. The international journal of biochemistry & cell biology. 2014;50:106–111. doi: 10.1016/j.biocel.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tsukikawa M, Peng M, Polyak E, Nakamaru-Ogiso E, Ostrovsky J, McCormack S, Place E, Clarke C, Reiner G, et al. Primary respiratory chain disease causes tissue-specific dysregulation of the global transcriptome and nutrient-sensing signaling network. PloS one. 2013b;8:e69282. doi: 10.1371/journal.pone.0069282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am J Hum Genet. 2010;86:581–591. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Lum PY, Lamb J, GuhaThakurta D, Edwards SW, Thieringer R, Berger JP, Wu MS, Thompson J, Sachs AB, et al. An integrative genomics approach to the reconstruction of gene networks in segregating populations. Cytogenet Genome Res. 2004;105:363–374. doi: 10.1159/000078209. [DOI] [PubMed] [Google Scholar]
- Zhu J, Sova P, Xu Q, Dombek KM, Xu EY, Vu H, Tu Z, Brem RB, Bumgarner RE, Schadt EE. Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate cell regulation. PLoS biology. 2012;10:e1001301. doi: 10.1371/journal.pbio.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wiener MC, Zhang C, Fridman A, Minch E, Lum PY, Sachs JR, Schadt EE. Increasing the power to detect causal associations by combining genotypic and expression data in segregating populations. PLoS computational biology. 2007;3:e69. doi: 10.1371/journal.pcbi.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhang B, Smith EN, Drees B, Brem RB, Kruglyak L, Bumgarner RE, Schadt EE. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet. 2008;40:854–861. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]