Abstract
Introduction
Histone deacetylase (HDAC) proteins, which counter the activity of histone acetyltransferases (HATs), are necessary for normal muscle atrophy in response to several pathophysiological conditions. Despite this, it remains unknown whether a common or unique transcriptional profile of HDAC and HAT genes exist during the progression of muscle atrophy.
Methods
Muscles were harvested from cast immobilized, denervated, or nutrient deprived animals for qRT-PCR analysis of HDAC and HAT gene expression.
Results
The mRNA levels of Hdac2, Hdac4, Hdac6, Sirt1, p300, Cbp and Pcaf increased, and Hdac7 decreased, in skeletal muscle in each experimental model of muscle atrophy. Hdac1 and Hdac3 were increased only in cast immobilized and denervated muscles.
Conclusion
While specific HDACs and HATs are increased in multiple models of muscle atrophy, increased expression of class I HDACs was unique to muscle disuse, reinforcing that specific HDAC inhibitors may be more effective than pan-HDAC inhibitors at countering muscle atrophy.
Keywords: muscle wasting, histone deacetylase, histone acetyltransferase, muscle disuse, denervation
Introduction
Histone deacetylase (HDAC) proteins remove acetyl groups from target proteins, and recently a critical role of specific HDAC proteins has emerged in the regulation of skeletal muscle atrophy.1–5 Indeed, of the 11 HDAC and 7 Sirtuin proteins, data supports the direct involvement of HDACs 1, 4, 5, 6 and SIRT1 in the regulation of muscle atrophy during various catabolic conditions.1–3,6 Despite these findings, the results following treatment with HDAC inhibitors to counter muscle atrophy have not yielded similar results.7,8 These contrasting findings may be because the HDAC inhibitor used in these studies, trichostatin A (TSA), is a pan HDAC inhibitor and too non-specific. In this regard, increasing evidence supports the notion each HDAC has unique protein targets with non-redundant functions9. Thus, in future studies it will be important to move towards the use of more specific HDAC inhibitors. However, before this can be considered, a comprehensive analysis of how individual HDACs, and their counterpart histone acetyltransferase (HATs), change during various atrophy conditions is necessary. Therefore, the purpose of the current study was to compare the expression changes of various HDACs and HATs in skeletal muscle during multiple atrophying conditions to determine if a common or unique transcriptional profile exists.
Materials and Methods
C57BL/6 mice (nutrient deprivation and cast immobilization) weighing ~20g, or Sprague Dawley rats (denervation) weighing ~175g were purchased from Charles River Laboratories (Wilmington, Massachusetts) and used for all animal experiments which were approved by the University of Florida Institutional Animal Care and Use Committee.
Mice were cast immobilized as described previously,10 and muscles were removed 4 or 10 days later. Food was withheld from nutrient deprived mice for 3 days and then muscles were harvested. Denervated rats underwent bilateral sciatic nerve transection, as described by others,11 and muscles were harvested after 3 and 7 days. Muscles (n=6 per group) were removed and processed for RNA isolation using a TRIzol-based method as previously described.10 cDNA was generated from 1 μg of RNA and used as a template for quantitative RT-PCR using various primers for p300, Cbp, Pcaf, Gcn5, Moz, Hdacs1-11, and Sirt1.
Results
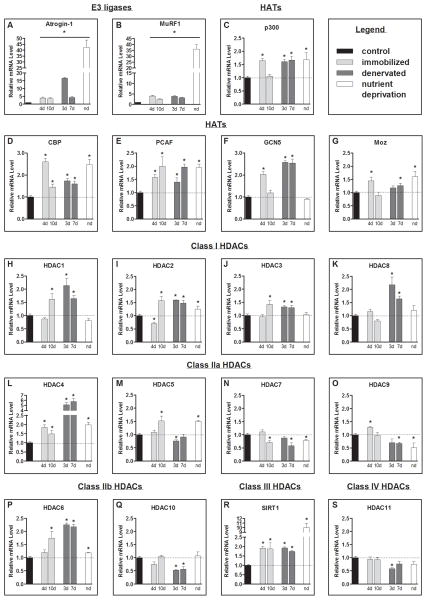
Since the atrophy and ubiquitin proteasome pathway biomarkers, atrogin-1 and MuRF1 showed a typical increase in gene transcription (Figure 1A–B), we next measured the mRNA level of the HATs, p300, Cbp, Pcaf, Gcn5 and Moz, and Hdacs1-11 and Sirt1 (Figure 1C–S). p300, Cbp and Pcaf were significantly increased in all models of muscle atrophy, at all time-points measured except for p300 following 10-days of immobilization (Figures 1C–E). Gcn5 was significantly increased following 4-days of immobilization, and 3 and 7-days of denervation but not during nutrient deprivation (Figure 1F). Moz was significantly increased following 4-days of immobilization, 7-days of denervation and nutrient deprivation (Figure 1G). Hdac2, Hdac4, Hdac6 and Sirt1 significantly increased in all conditions, and Hdac1 and Hdac3 increased in response to cast immobilization and denervation, but not nutrient deprivation (Figure 1H–J, 1L, 1P and 1R). Hdac7 was significantly decreased in all conditions and Hdac9 was significantly decreased in response to nutrient deprivation and 7-days of denervation (Figures 1N and 1O).
Figure 1.
Atrophy gene expression profile. Relative mRNA levels of; the E3 ligases (A) atrogin-1 (Mm_026346.3/Rn_133521.1) and (B) MuRF1 (Mm_001039048.2/Rn_080903.1), the HATs: (C) p300 (E1A binding protein p300; Mm_177821.6/Rn_576312.4), (D) Cbp (CREB (cAMP-responsive element binding protein)-binding protein; Mm_001025432.1/Rn_133381.3), (E) Pcaf (P300/CBP-associated factor; Mm_001177798.1/Rn_001107442.1), (F) Gcn5 (general control of amino-acid synthesis; Mm_029090.3/Rn_001107050.1), and (G) Moz (Monocytic leukemia zinc-finger protein; Mm_001081149.1/Rn_001100570.1); the class I Hdacs: (H–K) Hdac1 (Mm_008228.2/Rn_001025409.1), Hdac2 (Mm_008229.2/Rn_053447.1), Hdac3 (Mm_010411.2/Rn_053448.1), and Hdac8 (Mm_027382.3/Rn_001126373.2), class IIa Hdacs: (L–O) Hdac4 (Mm_207225.1/Rn_343629.4), Hdac5 (Mm_001077696/Rn_053450.1), Hdac7 (Mm_001204275.1/Rn_345868.4), Hdac9 (Mm_024124.3/Rn_001200045.1), class IIb Hdacs: (P& Q) Hdac6 (Mm_001130416.1/Rn_228753.5) and Hdac10 (Mm_199198.2/Rn_001035000.1), the class III Hdac: (R) Sirt1 (Mm_001159589.1/Rn_001080493.1) and the class IV Hdac: (S) Hdac11 (Mm_144919.2/Rn_001106610.2) from the soleus muscle of control and 4 day (4d) and 10 day (10d) immobilized mice, tibialis anterior (TA) muscle of control and 3 day (3d) and 7 day (7d) denervated rats, and nutrient deprived (nd) mice. All values were normalized to 18s. Bars represent means ± SE for 6 muscles/group. *P<0.05 vs control.
Discussion
In the current study we demonstrate that p300, Cbp, Pcaf, Hdac2, Hdac4, Hdac6 and Sirt1 mRNA levels all increase, and Hdac7 mRNA decreases, in skeletal muscle in response to three different models of muscle atrophy. The common increase in the mRNA levels of Hdac4 and Hdac6 is interesting given their direct implication in the regulation of skeletal muscle mass. Indeed, overexpression of HDAC4 is sufficient to cause muscle fiber atrophy6 and skeletal muscle specific knockout/knockdown of HDAC4 attenuates denervation-induced muscle atrophy.1,6 However, knockdown of HDAC4 does not attenuate fasting-induced muscle atrophy6 demonstrating in the absence of HDAC4 other mechanisms still drive muscle wasting during this condition. In addition, recent data also shows HDAC6 is causative in muscle atrophy associated with both denervation and chronic angiotensin II signaling.4,12.
The current work also demonstrates Hdac1, Hdac2 and Hdac3 are commonly increased in response to cast immobilization and denervation, suggesting increased expression of these class I HDACs is associated with conditions of muscle disuse. In support of this, recent work from our lab showed inhibition of class I HDACs, via treatment of mice with MS-275 (Entinostat), inhibits cast immobilized-induced muscle atrophy and weakness.3
The most well characterized role of HDACs/HATs is through the regulation of gene transcription via deacetylation-acetylation of histone proteins.13 In this regard deacetylation of histones causes a decrease in gene transcription whereas acetylation of histones increases gene transcription.14–16 Thus, HDACs could cause muscle atrophy, in part, through decreased transcription of genes required for the maintenance of muscle mass. Alternatively HDACs/HATs could regulate muscle mass via the regulation of atrophy-related transcription factors by either acting as co-factors within multimolecular transcriptional complexes17 and/or by directly acetylating/deacetylating such transcription factors. In this latter regard, the nuclear-cytosolic localization and/or DNA-binding activities of the transcription factors Forkhead boxO, nuclear factor-kappaB and CCAAT/enhancer-binding protein beta are all regulated via acetylation-deacetylation3,5,12,18–20 and each is required for muscle atrophy during various conditions.21–24 Another potential mechanism is via the regulation of protein stability. Indeed deacetylation-acetylation can either promote or block the protein degradation25 or change protein-protein interactions since acetylation of lysine residues can create docking sites for other proteins.26
Conclusion
The current study is the first to comprehensively examine the gene expression changes of 12 HDACs and 5 HATs in skeletal muscle during three independent models of muscle wasting. Our findings have established that Hdac2, Hdac4, Hdac6 and Sirt1 are commonly increased in all 3 models of muscle atrophy, while Class I HDACs are commonly increased in models of disuse atrophy. We are well aware that changes in protein expression, localization and, most importantly, substrate interactions, dictate the downstream consequences of HDACs, not changes in mRNA. However, the biological consequences of the identified common mRNA transcriptional profile may be inferred from the published work demonstrating that Hdac1, Hdac2, Hdac4 and Hdac6 are required for normal muscle atrophy in response to various conditions. Therefore selective chemical probes for these specific HDACs may provide the greatest chance of success in inhibiting muscle atrophy.
Acknowledgments
We thank Sarah Judge, Ph.D. for critical reading and editing of the manuscript. This work was supported by U.S. National Institute of Arthritis and Musculoskeletal and Skin Diseases [grant number R01AR060209 to A.R. Judge]. A.W. Beharry is supported by a T32 from the National Institute of Child Health and Human Development Grant T32-HD-043730.
Abbreviations
- CBP
CREB (cAMP-responsive element binding protein)-binding protein
- GCN5
General control of amino-acid synthesis
- HAT
Histone Acetyltransferase
- HDAC
Histone Deacetylase
- p300
E1A binding protein p300
- PCAF
P300/CBP-associated factor
- MOZ
Monocytic leukemia zinc-finger protein
- TA
Tibialis anterior
- TSA
Trichostatin A
Footnotes
Conflict of Interests
No conflict of interests
References
- 1.Moresi V, Williams AH, Meadows E, Flynn JM, Potthoff MJ, McAnally J, Shelton JM, Backs J, Klein WH, Richardson JA, Bassel-Duby R, Olson EN. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143(1):35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee D, Goldberg AL. SIRT1 by blocking the activities of FoxO1 and 3 inhibits muscle atrophy and promotes muscle growth. J Biol Chem. 2013 doi: 10.1074/jbc.M113.489716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beharry AW, Sandesara PB, Roberts BM, Ferreira LF, Senf SM, Judge AR. HDAC1 activates FoxO and is both sufficient and required for skeletal muscle atrophy. J Cell Sci. 2014;127(Pt 7):1441–1453. doi: 10.1242/jcs.136390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demos-Davies KM, Ferguson BS, Cavasin MA, Mahaffey JH, Williams SM, Spiltoir JI, Schuetze KB, Horn TR, Chen B, Ferrara C, Scellini B, Piroddi N, Tesi C, Poggesi C, Jeong MY, McKinsey TA. HDAC6 contributes to pathological responses of heart and skeletal muscle to chronic angiotensin-II signaling. American journal of physiology Heart and circulatory physiology. 2014;307(2):H252–258. doi: 10.1152/ajpheart.00149.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Senf SM, Sandesara PB, Reed SA, Judge AR. p300 Acetyltransferase activity differentially regulates the localization and activity of the FOXO homologues in skeletal muscle. Am J Physiol Cell Physiol. 2011;300(6):C1490–1501. doi: 10.1152/ajpcell.00255.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bongers KS, Fox DK, Ebert SM, Kunkel SD, Dyle MC, Bullard SA, Dierdorff JM, Adams CM. Skeletal Muscle Denervation Causes Skeletal Muscle Atrophy Through a Pathway that Involves Both Gadd45a and HDAC4. Am J Physiol Endocrinol Metab. 2013 doi: 10.1152/ajpendo.00380.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonetto A, Penna F, Minero VG, Reffo P, Bonelli G, Baccino FM, Costelli P. Deacetylase inhibitors modulate the myostatin/follistatin axis without improving cachexia in tumor-bearing mice. Curr Cancer Drug Targets. 2009;9(5):608–616. doi: 10.2174/156800909789057015. [DOI] [PubMed] [Google Scholar]
- 8.Benny Klimek ME, Aydogdu T, Link MJ, Pons M, Koniaris LG, Zimmers TA. Acute inhibition of myostatin-family proteins preserves skeletal muscle in mouse models of cancer cachexia. Biochem Biophys Res Commun. 2010;391(3):1548–1554. doi: 10.1016/j.bbrc.2009.12.123. [DOI] [PubMed] [Google Scholar]
- 9.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5(10):981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 10.Senf SM, Dodd SL, McClung JM, Judge AR. Hsp70 overexpression inhibits NF-kappaB and Foxo3a transcriptional activities and prevents skeletal muscle atrophy. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2008;22(11):3836–3845. doi: 10.1096/fj.08-110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2007;21(1):140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- 12.Ratti F, Ramond F, Moncollin V, Simonet T, Milan G, Mejat A, Thomas JL, Streichenberger N, Gilquin B, Matthias P, Khochbin S, Sandri M, Schaeffer L. Histone Deacetylase 6 Is a FoxO Transcription Factor-dependent Effector in Skeletal Muscle Atrophy. J Biol Chem. 2015;290(7):4215–4224. doi: 10.1074/jbc.M114.600916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408(6811):495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 15.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 16.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72(1):73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 17.Legube G, Trouche D. Regulating histone acetyltransferases and deacetylases. EMBO reports. 2003;4(10):944–947. doi: 10.1038/sj.embor.embor941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen LF, Greene WC. Regulation of distinct biological activities of the NF-kappaB transcription factor complex by acetylation. J Mol Med (Berl) 2003;81(9):549–557. doi: 10.1007/s00109-003-0469-0. [DOI] [PubMed] [Google Scholar]
- 19.Cesena TI, Cardinaux JR, Kwok R, Schwartz J. CCAAT/enhancer-binding protein (C/EBP) beta is acetylated at multiple lysines: acetylation of C/EBPbeta at lysine 39 modulates its ability to activate transcription. J Biol Chem. 2007;282(2):956–967. doi: 10.1074/jbc.M511451200. [DOI] [PubMed] [Google Scholar]
- 20.Bertaggia E, Coletto L, Sandri M. Posttranslational modifications control FoxO3 activity during denervation. Am J Physiol Cell Physiol. 2012;302(3):C587–596. doi: 10.1152/ajpcell.00142.2011. [DOI] [PubMed] [Google Scholar]
- 21.Senf SM, Dodd SL, Judge AR. FOXO signaling is required for disuse muscle atrophy and is directly regulated by Hsp70. Am J Physiol Cell Physiol. 2010;298(1):C38–45. doi: 10.1152/ajpcell.00315.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai D, Frantz JD, Tawa NE, Jr, Melendez PA, Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ, Shoelson SE. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119(2):285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 23.Reed SA, Sandesara PB, Senf SM, Judge AR. Inhibition of FoxO transcriptional activity prevents muscle fiber atrophy during cachexia and induces hypertrophy. FASEB J. 2012;26(3):987–1000. doi: 10.1096/fj.11-189977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang G, Jin B, Li YP. C/EBPbeta mediates tumour-induced ubiquitin ligase atrogin1/MAFbx upregulation and muscle wasting. EMBO J. 2011;30(20):4323–4335. doi: 10.1038/emboj.2011.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadoul K, Boyault C, Pabion M, Khochbin S. Regulation of protein turnover by acetyltransferases and deacetylases. Biochimie. 2008;90(2):306–312. doi: 10.1016/j.biochi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nature reviews Drug discovery. 2014;13(5):337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]