Abstract
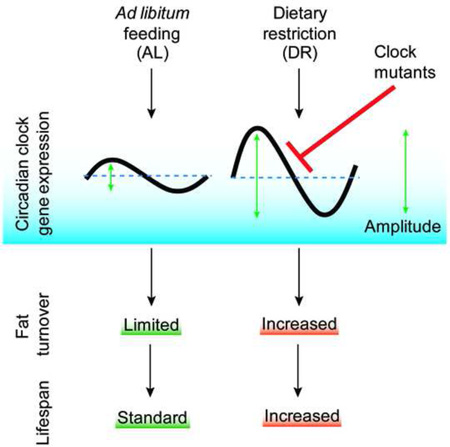
Endogenous circadian clocks orchestrate several metabolic and signaling pathways that are known to modulate lifespan, suggesting clocks as potential targets for manipulation of metabolism and lifespan. We report here that the core circadian clock genes, timeless (tim) and period (per), are required for the metabolic and lifespan responses to DR in Drosophila. Consistent with the involvement of a circadian mechanism, DR enhances the amplitude of cycling of most circadian clock genes, including tim, in peripheral tissues. Mass spectrometry-based lipidomic analysis suggests a role of tim in cycling of specific medium chain triglycerides under DR. Furthermore, overexpression of tim in peripheral tissues improves its oscillatory amplitude and extends lifespan under ad libitum conditions. Importantly, effects of tim on lifespan appear to be mediated through enhanced fat turnover. These findings identify a critical role for specific clock genes in modulating the effects of nutrient manipulation on fat metabolism and aging.
Graphical Abstract
Introduction
Circadian rhythms, such as daily cycles of sleep and activity and oscillations in metabolic, physiological, and endocrine functions, are vital to maintaining temporal homeostasis. These rhythms are controlled by endogenous clocks located in the brain and many peripheral tissues. Rhythmic activity of clock molecules drives cyclic expression of many other genes, resulting in rhythmic activities of metabolic and signaling pathways in mammals as well as in the fruit fly, Drosophila melanogaster (Buhr and Takahashi, 2013; Schibler and Sassone-Corsi, 2002; Xu et al., 2011). Disruption of circadian rhythms is associated with cancer and metabolic disorders that accelerate aging, including diabetes and obesity (Kondratov, 2007; Turek et al., 2005).
Metabolic homeostasis is intimately linked to longevity. Dietary restriction (DR), leads to metabolic reprogramming, which enhances fat turnover and mitochondrial function that is required for its protective effects on lifespan extension in the fly (Katewa et al., 2012). Similarly, in mice calorie restriction has been shown to increase fat turnover (Bruss et al., 2010). The improved cellular homeostasis upon DR delays the onset of a number of age-related diseases and aging in multiple species (Bishop and Guarente, 2007; Fontana et al., 2009; Kapahi et al., 2010; Mair and Dillin, 2008). Conservation of the protective effects of DR in Drosophila offers an opportunity to examine the fundamental molecular mechanisms of nutritional modulation of aging and age-related diseases (Katewa and Kapahi, 2011; Mair and Dillin, 2008; Tatar, 2007).
Given the well-established links between circadian rhythms and metabolism (DiAngelo et al., 2011; Green et al., 2008; Xu et al., 2011), we examined the role of circadian clocks in lifespan extension by DR in Drosophila. Circadian behavioral rhythms have been studied previously in the context of aging in both flies and mice, with studies showing that rhythms of rest:activity or sleep:wake break down with age (Koh et al., 2006; Turek et al., 2005). However, it is not known whether interventions that lead to life extension, such as DR, require functional circadian clocks, and if so by which mechanisms.
Here we show that circadian regulation is critical for DR dependent increase in lifespan. Flies mutant for circadian clock genes timeless or period showed an attenuated response to DR dependent changes in lifespan extension and fat metabolism. In support of the idea that clock genes contribute to effects of DR, we also show that DR improves the amplitude of circadian gene cycling. Previously, we showed that DR-dependent increase in fat metabolism is required for lifespan extension (Katewa et al., 2012). Now here we demonstrate that specific clock mutants can abrogate the changes in fat turnover upon DR, while overexpression of timeless enhances fat metabolism on ad libitum (AL) diet. Mass-spectrometry based lipidomic analysis of control and timeless mutant flies shows cycling of several triglycerides (TGs) under DR conditions. Specifically, we have identified a novel group of medium chain TGs that cycle under DR in a TIM dependent manner. Overexpression of tim improves fat turnover and extends lifespan on AL food, thereby mimicking the effects of DR.
Results
DR enhances the mRNA and protein oscillations of core clock genes
The oscillations of clock genes in peripheral tissues become weaker with age (Luo et al., 2012; Rakshit et al., 2012). If maintenance of these oscillations is important to delay aging, then DR may act by enhancing the amplitude of circadian oscillations. To assess how nutrients impact circadian clocks, we obtained daily mRNA expression profiles of clock genes timeless and period in control wild type Canton-S (CS) flies subjected to DR or AL feeding for 10 days. In Drosophila, restriction of yeast, particular amino-acids, or total calories increases the lifespan (Bruce et al., 2013; Mair et al., 2003; Min and Tatar, 2006). As reduction of yeast is sufficient to extend lifespan independent of the calorie content, DR is implemented mostly as restriction of dietary yeast (Chippindale et al., 1993; Kapahi et al., 2004; Katewa et al., 2012; Lee et al., 2008; Mair et al., 2005). In our study, the DR diet contains 0.5 % yeast extract and 5% sugar, while the AL diet contains 5% yeast extract and 5% sugar (additional details are provided in Supplemental Experimental Procedures).
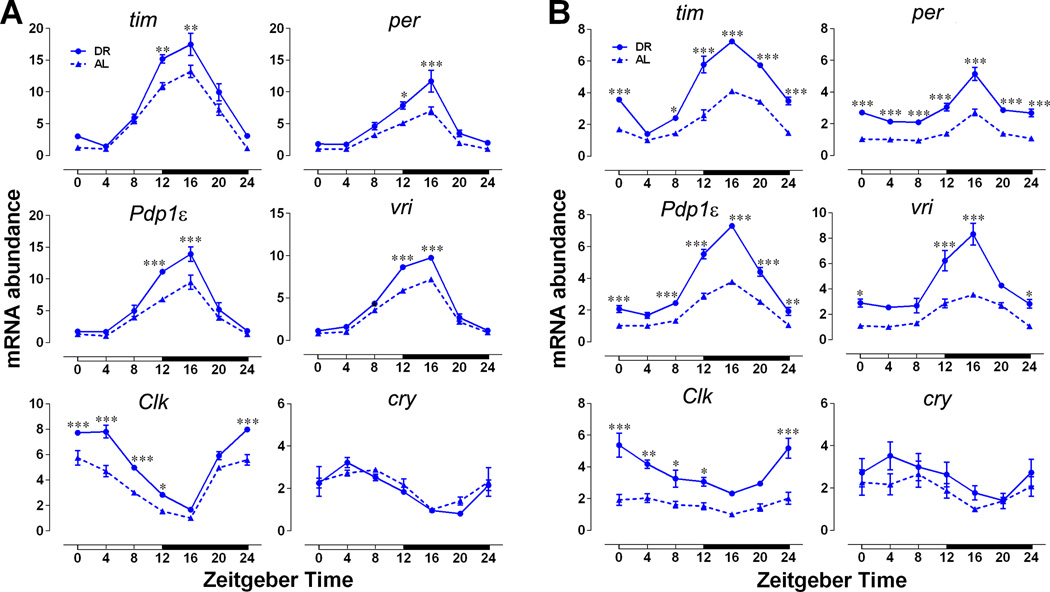
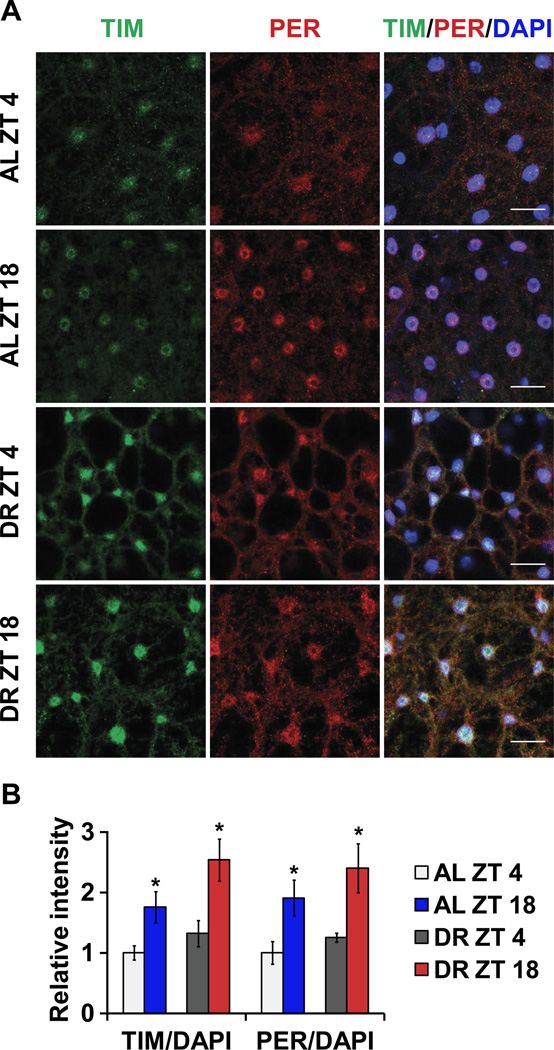
In flies, the timeless (tim) and period (per) genes are the major components of the cellular clock and are expressed rhythmically in several tissues as a result of negative feedback by their protein products (Zheng and Sehgal, 2008). The PER and TIM proteins negatively regulate transcription by inhibiting activity of the Clock (CLK) and cycle (CYC) transcriptional activators. We observed the expected daily oscillations of tim and per mRNA levels in animals on an AL diet with a trough at Zeitgeber Time 4 (ZT4) and peak at ZT16 in both heads (Figure 1A) and bodies of female flies (Figure 1B, for statistical analysis see Table S1). Flies subjected to DR had higher magnitude tim and per expression, in particular at the normal peak time of ZT 12–20, resulting in increased amplitude of cycling, compared to flies reared on an AL diet. This effect was particularly strong in body clocks (Figure 1B). We confirmed this further by measuring the cycling of tim and per in isolated fat bodies (Figure S1A). Male flies from either CS or w1118 also showed a similar increase in expression of tim and per mRNA upon DR (Figure S1B). However, we have used female flies for all other experiments as they typically show a stronger response to variation of yeast in the diet (Katewa et al., 2012; Vargas et al., 2010). We also observed that a minimum of 6 days of DR treatment is required to see a robust response in clock gene amplitude (data not shown). mRNA expression profiles of Par Domain Protein 1ε (Pdp1ε) and vrille (vri), were also significantly more robust in females on DR and the same was true for the amplitude of Clk oscillations (Figure 1A and B). DR did not have a significant effect on the daily oscillations of cryptochrome (cry), which encodes the Drosophila circadian photoreceptor and functions as a clock component in some peripheral tissues (Figure 1A and B) (Krishnan et al., 2001). Because the amplitude of clock gene oscillations declines with age, we also measured circadian gene expression in 33-day-old flies. While the overall amplitude was reduced with aging in both AL and DR conditions, DR flies sustained significantly higher oscillations of all clock genes (except cry) in both heads and abdomens (Figure S1C). To determine, if the increase in mRNA expression of clock genes is translated into more clock proteins, we measured the protein levels in adult fat bodies at ZT4 and ZT18. Control (w1118) female flies were fed AL or DR diet for 10 days, after which the adult fat bodies were removed, fixed and stained for TIM and PER proteins. As expected, levels of TIM and PER were significantly higher at ZT18 on both food types when compared to ZT4 (Figure 2A–B). Two-way-ANOVA indicated significant effect of diet (P= 0.0432) and time (P=0.0010) for TIM and significant effect of time (P=0.0019) for PER (Figure 2B). Combined the results (Figure 1 and Figure 2) suggest that DR not only increases the magnitude of mRNA expression of clock genes but it also increases the clock protein levels in peripheral tissues.
Figure 1. Increase in amplitude of circadian gene expression upon DR.
Daily relative mRNA concentration profiles of core clock genes in (A) heads and (B) bodies of control CS females fed on AL and DR diets for 10 days. Data are normalized to the trough (ZT4/16) values set at 1 for flies on AL diet for each age. White and black horizontal bars mark periods of light and dark respectively. Each data point represents mean ± SEM of three independent RNA samples. Statistical significance between AL and DR values was determined using two-way ANOVA with Bonferonni’s post hoc test, and is denoted by ***p< 0.001, **p<0.01, and *p<0.05 and is provided in Table S1. See also Figure S1.
Figure 2. Increase in magnitude of clock protein expression upon DR.
(A) TIM and PER expression in adult fat body at ZT 4 and 18 in control females fed AL and DR diets for 10 days. (left) TIM staining. (center) PER staining. (right) Merged images (TIM, PER and DAPI). Scale bars indicate 15 µm. (B) Quantification of both TIM and PER expressions in the nucleus. n = 90 from 6 fat bodies in each conditions. Error bars indicate SEM. (* p < 0.05 by t-test).
Circadian regulation is required for DR dependent increase in lifespan
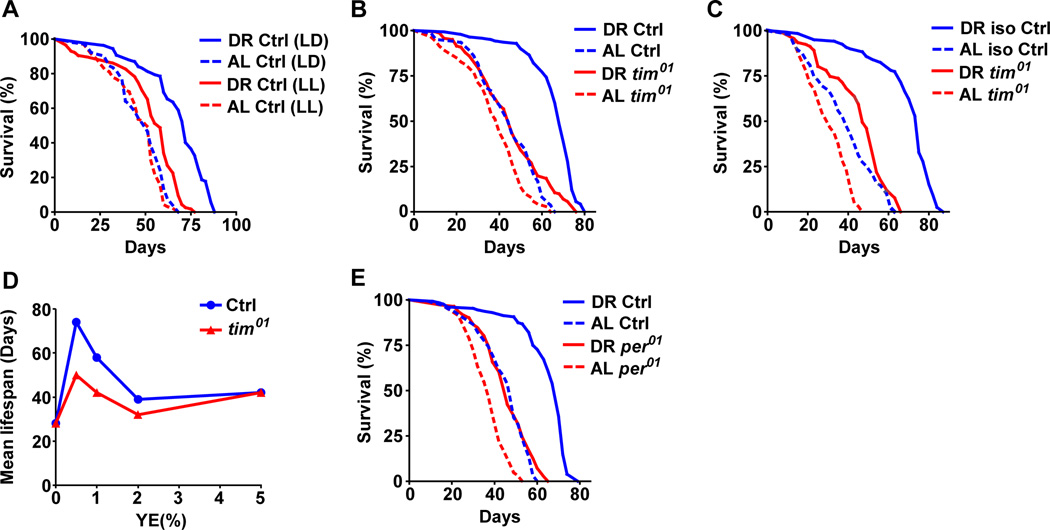
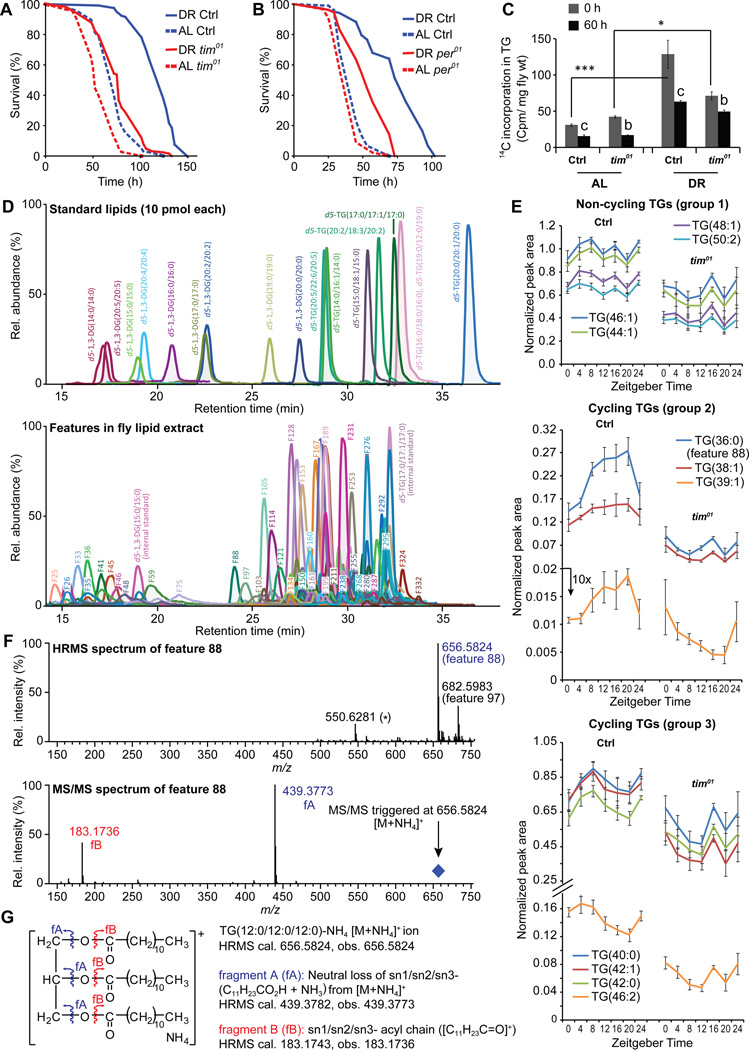
Next, we asked whether circadian clocks are required for DR-dependent responses by measuring the survival of female flies maintained under constant light conditions (LL), which disrupt the molecular clock and eliminate overt rhythms (Emery and Clayton, 2001). Wild-type CS female flies were maintained in incubators with 12h light: 12 h dark (LD) cycles or 24 h LL. Flies maintained in LD showed significantly greater extension in lifespan upon DR than did flies in LL (Figure 3A, diet*genotype interaction p value= 1.34E-07, for statistical analysis of all survival assays see Table S2 and for replications see Table S3). To verify that the reduction in DR-dependent lifespan extension was due to the loss of circadian regulation, we tested the effects of DR in flies lacking the timeless gene (tim01). tim01 mutants (outcrossed 8 times into the control CS background) maintained on DR showed a small increase in lifespan (10%) in contrast to a large extension in control flies (51%) (Figure 3B, interaction p value= 5.36E-06, Table S2 and S3). tim01 flies in another control (w1118) background also showed impaired lifespan extension upon DR compared to controls (Figure 3C, interaction p value= 6.68E-06,). For finer resolution of the changes in lifespan upon yeast variation, survival assays were also done at five different concentrations of yeast extract (YE) in the diet (Figure 3D, Table S2 and S3). Loss of TIM significantly decreased the increase in lifespan produced by lowering YE in the diet (Figure 3D). It had less effect at yeast concentrations that produced little to no change in lifespan. To determine whether reduced lifespan extension reflects a circadian function of tim, we also measured the effect of DR on lifespan in flies lacking the PERIOD (PER) protein, which is the partner of TIM in the molecular clock. Indeed the per01 mutant flies showed a similar reduction in DR dependent lifespan extension (Figure 3E, interaction p value= 1.85E-06, Tables S2 and S3). Surprisingly, despite showing a similar response in terms of mRNA expression (Figure S1B), male flies belonging to either tim01 or per01 groups were not significantly different from control animals on DR (Figure S2). Combined, the results indicate that DR improves temporal homeostasis in both sexes and that functional circadian clocks contribute to maximal DR-dependent lifespan extension in females.
Figure 3. Reduction of lifespan extension upon DR in flies with mutations in circadian clock genes.
DR mediated lifespan extension was reduced in (A) female flies maintained in 24 hr LL, (B) tim01 mutant (in CS background) under LD, and (C) tim01 mutant (in w1118 background) under LD. (D) tim01 flies showed reduced response to varying yeast extract concentration in the diet. (E) Lifespan extension upon DR was reduced in per01 mutants under LD. Statistical analysis of the survival curves, number of flies and data from additional independent repeats are provided in Tables S2–3. See also Figure S2.
Circadian clocks are required for DR dependent changes in fat metabolism
Enhanced triglyceride (TG) turnover is necessary for DR dependent lifespan extension in flies (Katewa et al., 2012). The improved fat metabolism in dietary-restricted flies also increases their survival in response to acute starvation (Katewa et al., 2012). To determine a role of circadian clocks in regulating fat metabolism, we first measured the starvation response of circadian mutant flies. Prior exposure to DR substantially increased the survival of control flies upon starvation, but had much less of an effect on tim01 (Figure 4A, interaction p value= 6.32E-05) and per01 mutants (Figure 4B, interaction p value= 1.34E-07). Thus per and tim mutants are impaired in the resistance to starvation that normally occurs following DR.
Figure 4. Mutations in circadian clock genes reduce triglyceride homeostasis upon DR.
Starvation resistance was reduced in tim01 (A) and per01 mutants (B). Statistical analysis of the survival curves, number of flies and data from additional independent repeats are provided in Tables S2–3. (C) tim01 mutant flies show reduced triglyceride turnover upon DR. Flies were fed AL and DR diet containing 14C labeled glucose for 24 hrs and the synthesis of TG was measured (shown as 0 hrs) by measuring the amount of 14C in the TG fraction of the isolated lipids from the flies. Breakdown was measured by a pulse-chase experiment where the 24 hrs fed flies were transferred to unlabelled food for 60 hrs and the remaining 14C label in TG fraction was measured (60 Hrs). Values are mean ± SEM of 5 independent preparations. Statistical significance was determined using Student’s t-test and is denoted between time points is denoted by a,b or c and by (*) between groups. c or *** indicates p< 0.001, b or ** p<0.01, and a or *p<0.05. (D–G) Mass spectrometry-based lipidomics to identify triglycerides (TGs) that cycle upon DR in a timeless-dependent fashion. (D) Extracted ion chromatograms (XICs) for (top) a mixture of synthetic d5-di and triglycerides (predominantly forming the corresponding [M+NH4]+ ions) used to establish and optimize conditions for HPLC-MS and –MS/MS analyses and (middle) ~253 features identified from HPLC-MS analysis of DR fly lipid extracts. For ease of visualization, each chromatogram is plotted to show a 3 min window around the peak of interest. Further, for fly lipid features (bottom), only exemplary peaks are labeled with the corresponding feature number (e.g. feature 88/F88) to avoid any overlap. (E) Levels of specific TGs (error bars, SEM) at different time points in control and tim01 flies maintained on DR food conditions, segregated in three groups based on their cycling nature: group 1: non-cycling, group 2: cycling, peak at night, and group 3: cycling, peak during day. (F) High resolution (HR) HPLC-MS/MS spectra for the most prominent cycling feature (F88). (G) Schematic annotations for the molecular ion ([M+NH4]+) and the observed fragment ions (Figure 4F), based on analysis of HRMS data of feature 88/TG(36:0). See also Figure S3 and S4.
We next asked whether the effect on starvation reflects altered synthesis or breakdown of TG. For this we measured fat turnover in tim01 mutant and control flies using a radiolabeled glucose tracer method (Katewa et al., 2012). Consistent with extended survival upon starvation, wild-type flies showed a 4-fold increase in the rate of TG synthesis after DR conditions, whereas the tim01 mutants showed only a 1.6-fold increase, indicating a reduced rate of TG synthesis in tim01 flies (Figure 4C). Furthermore, the control flies showed a 51% decrease of labeled TG after 60 hrs, whereas tim01 flies showed a 30% reduction suggesting decreased fat turnover in tim01 flies under DR (Figure 4C). These results support the idea that clock genes contribute to the enhanced synthesis and breakdown of TG’s upon DR.
To identify the TG that might cycle under DR and investigate their possible regulation by circadian clocks, we performed a detailed analysis of lipids on DR. A quantitative mass spectrometry approach was used to identify features (see Supplemental Experimental Procedures for details) in fly lipid extracts at different time points in control and tim01 flies maintained on DR food conditions. A preliminary analysis identified about 253 features (Figure 4D) from fly lipid extracts, and approximately 50 showed some cycling properties (had one clear peak and one trough in a 24 h time period) (Figure S3) in a tim-dependent fashion. Based on our previous results (Katewa et al., 2012) and the effect of tim01 on TG synthesis (Figure 4C), we focused on TGs and broadly classified them in three groups: Group 1 was non-cycling TGs, group 2 was cycling TGs with a peak in the night (at ZT16–20) and group 3 showed cycling with a peak at daytime (ZT4–8) (Figure 4E). Consistent with the involvement of the clock (Figure 4C), tim01 flies showed reduced levels of all TGs. Next, we used high resolution mass-spectrometry to identify specific TGs among the cycling features belonging to similar phased groups (Table S4) and observed that group 2 contained medium chain triglycerides (MCTs, containing medium chain fatty acids with 8 to 14 carbons). The most prominent tim-dependent cycling TG (feature 88, matched to TG(36:0) by high resolution MS analysis) was followed up with subsequent high resolution MS/MS analysis to elucidate it’s constituent fatty-acids (Figure 4F and 4G). Under our HPLC conditions, TGs most predominantly formed the corresponding [M+NH4]+ adduct ions (Figure 4D and Supplemental Experimental Procedures). When fragmented with a collision gas, triglyceride [M+NH4]+ adducts undergo a characteristic neutral loss of [RCO2H+NH3] (where RCO2H is a constituent fatty acid, e.g. R = C11H23 for the saturated C12-fatty acid, lauric acid), for each fatty acid component of the TG (King et al., 2015). Our MS/MS analyses revealed only a single [RCO2H+NH3] neutral loss fragment ion from feature 88, corresponding to [C11H23CO2H + NH3] (Figures 4F and 4G). To corroborate this, only a single fatty acyl chain fragment ion ([C11H23C=O]+), was further observed (Figures 4F and 4G). These data suggest that feature 88 is actually a symmetric TG, trilauryl glycerol (TLG), i.e. TG(12:0/12:0/12:0) (Figure 4G). Additionally, we found that the cycling property of TLG in flies is strictly tim-dependent upon DR food conditions (Figure 4E and S4).
Overexpression of timeless in peripheral tissues increases lifespan and fat metabolism on a rich nutrient diet
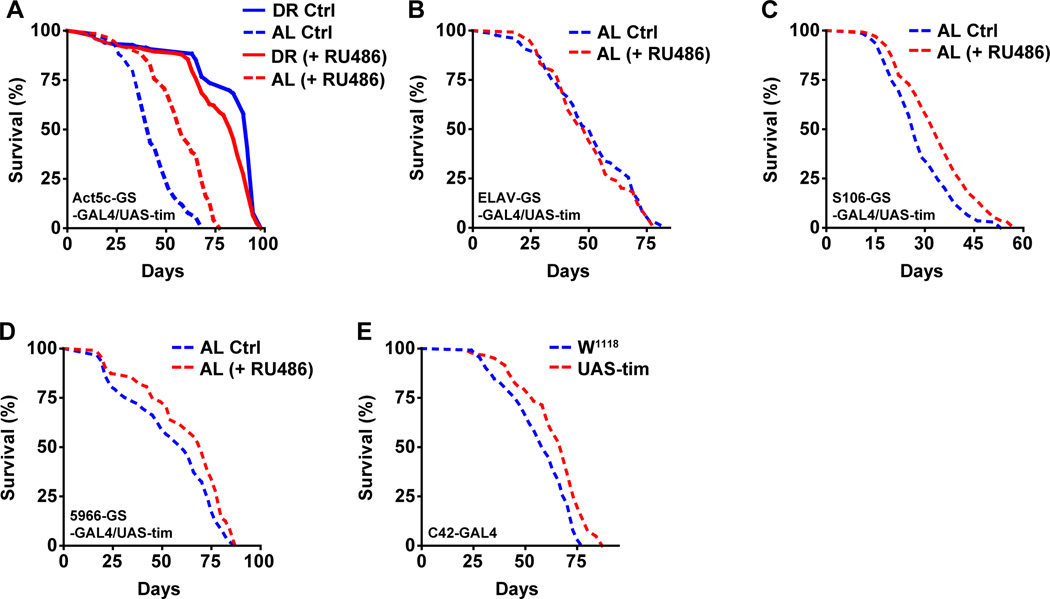
Aging causes a reduction in the amplitude of clock gene expression in peripheral tissues of both flies and mammals but it is not known whether increasing the amplitude might slow aging (Luo et al., 2012; Rakshit et al., 2012; Yamazaki et al., 2002). To determine whether increased expression or oscillations of tim could affect aging, we induced overexpression of tim using an Actin-5C-Gene switch-GAL4 driver (a drug inducible promoter that is expressed in whole flies), and measured survival. Expression of this driver is induced by adding RU486 to the medium, and so survival can be compared in induced and un-induced genetically identical strains. We observed a significant increase in survival in tim overexpressing flies on AL but not on DR diet. Flies on AL diet showed a 38% (16 day) increase in lifespan (Figure 5A, interaction p value= 6.44E-15), while flies on DR showed no significant difference in survival when tim was overexpressed in all tissues (Figure 5A, Table S2 and S3). We also tried overexpressing per (two different transgenes) in all tissues but did not see a comparable effect to tim overexpression (Figure S5A). This suggests either a tim specific effect or that it is limiting in some tissues. To identify the tissue responsible for lifespan extending effects of tim, we overexpressed tim in different tissues of the flies (by using tissue-specific promoter-driven expression) including neurons, fat bodies, gut and Malpighian tubules. Overexpression in neurons had no effect on lifespan (Figure 5B, interaction p value= 0.991 and S5B), whereas overexpression in all other tissues caused a small but significant increase in lifespan under AL but not under DR conditions (Figure S5C–E). Overexpression of tim in the fat body extended lifespan by 26% (Figure 5C, interaction p value= 4.76E-05), overexpression in gut extended lifespan by 14% (Figure 5D, interaction p value= 0.00152) and overexpression in tubules increased lifespan by 17% (Figure 5E, interaction p value= 0.00374). As the lifespan extension from individual tissues was lower than what we observed with whole body overexpression of tim (Figure 5A), we infer that multiple peripheral clocks contribute to the lifespan extension by tim overexpression.
Figure 5. Overexpression of tim increases survival in a diet dependent manner.
Kaplan Meier survival analysis of female flies upon tim overexpression under DR (solid line) and AL (dashed line) conditions, control flies (without RU486, blue) and overexpression flies (with RU486, red). (A) Overexpression of tim in whole body increases lifespan on an AL diet. (B) Overexpression of tim specifically in neurons has no effect on lifespan. (C–E) Overexpression of tim specifically in (C) fat body or (D) gut or (E) tubules increases lifespan on AL diet. Statistical analysis of the survival curves, complete genotype, number of flies and data from additional independent repeats are provided in Tables S2–3. See also Figure S5.
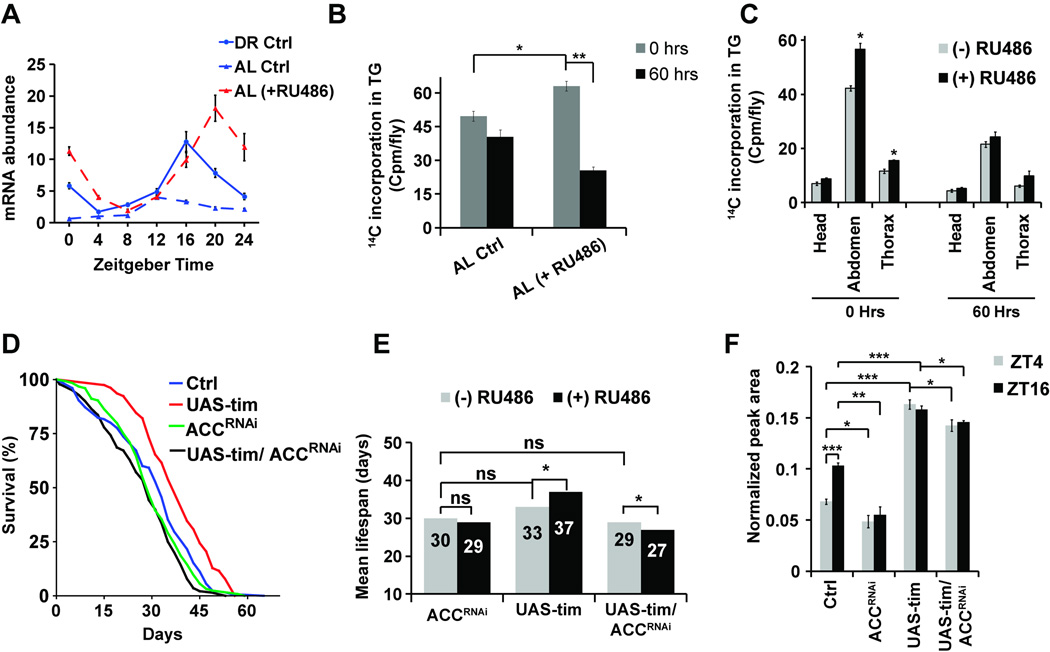
One caveat of the overexpression of any circadian gene is that the overexpressed mRNA may not follow the regular circadian expression pattern of the gene itself, so we also determined how the overexpression of tim affects the circadian nature of tim expression. Overexpression of tim increased the abundance of tim mRNA significantly at ZT16–20 in flies fed AL (Figure 6A) resulting in a 5-fold increase in the amplitude of the tim mRNA oscillation, similar to flies on DR, which showed a 3 fold increase in amplitude as peak and trough levels were elevated (Figure S6A). In addition, Clock was increased significantly (p>0.05) near the peak time at ZT4. However, this increase in rhythmicity was not observed for per suggesting a timeless specific effect of overexpression (Figure S6B). This could possibly explain why period overexpression did not increase lifespan on AL food, as per overexpression may not have maintained cyclic expression.
Figure 6. Overexpression of tim enhances fat metabolism, which mediates lifespan extension.
(A) Overexpression of tim in whole body increases relative abundance of tim mRNA and circadian amplitude of tim expression. The data are normalized to the trough (ZT4) levels seen in control flies on AL diet. Values are mean ± SEM of 3 independent preparations. (B) Overexpression of tim in whole body increases triglyceride turnover upon AL. Values are mean ± SEM of 4 independent preparations. Statistical significance was determined using Student’s t-test and is denoted by ***p< 0.001, **p<0.01, and *p<0.05. (C) Fat body specific overexpression of tim increases triglyceride synthesis specifically in the abdomen and thorax of the flies on AL diet. Values are mean ± SEM of 4–5 independent preparations. Statistical significance was determined using Student’s t-test and *denotes p<0.05. (D) Co-expression of tim overexpression and ACC RNAi in whole body abrogates the AL dependent increase in survival. Kaplan Meier survival analysis of female flies with tim overexpression under AL (with RU486, red) conditions, control flies (without RU486, blue), ACC RNAi (with RU486, green) and tim overexpression with ACC RNAi (with RU486, black). (E) The mean lifespan observed in survival curves shown in (6D). Statistical analysis of the survival curves, complete genotype and number of flies are provided in Table S2 and for an independent repeat see Figure S6. (F) Levels of trilauryl glycerol (TLG) are increased in flies overexpressing tim and are reduced upon ACC RNAi in whole body on AL diet. Values are mean ± SEM of 4 independent preparations. Statistical significance was determined using Student’s t-test and is denoted by *p<0.05, **p<0.01, ***p<0.001.
Next, we examined the effect of tim overexpression on fat metabolism. As DR increases fat metabolism (Katewa et al., 2012) and we observed that tim and per mutants showed diminished fat metabolism, we hypothesize that increasing clock oscillations results in more robust rhythmicity of fat metabolism. Overexpression of tim led to significantly higher de-novo TG synthesis from 14C labeled glucose and a higher rate of breakdown in flies on AL diet (Figure 6B), while it had little effect on flies maintained under DR (Figure S6C). Next, we examined whether tissue specific overexpression of tim modulates fat metabolism in the same or other tissues? tim overexpression in fat bodies increased fat synthesis and breakdown in fat bodies and to a smaller extent in the thorax but no effect was observed in heads (Figure 6C). The effect in thorax suggests a possibility of either an increased mobilization from fat bodies or a tissue autonomous effect of fat body clocks. Finally, to determine if fat metabolism is critical for the observed increase in lifespan under tim overexpression conditions, we measured survival in transgenic flies overexpressing tim while inhibiting Acetyl CoA Carboxylase (ACC). We have previously shown that upon inhibition of ACC there is a significant reduction of de novo synthesis of TG, leading to reduced fat turnover and a significant reduction in lifespan extension upon DR (Katewa et al., 2012). We observed that under AL conditions where tim overexpression increases lifespan, co-expression with ACC RNAi inhibits the observed extension in lifespan (Figure. 6D–E, S6D–E). We also evaluated if the levels of TLG are changing under this conditions. While ACC RNAi reduced the levels of TLG, the levels were significantly upregulated upon tim overexpression and co-expression with ACC RNAi resulted in a smaller but significant reduction (Figure 6F). Combined, the results indicate that increasing the rhythmic expression of tim mRNA is sufficient to increase fat turnover which mediates lifespan extension under rich nutrient conditions.
Discussion
Clocks are required for DR dependent lifespan extension
Within the last decade there have been important mechanistic insights into the protective effects of DR. Several genetic and metabolic pathways including the insulin/IGF-1 and target of rapamycin (TOR), AMPK and sirtuins have been shown to play a role in mediating DR responses (Bordone and Guarente, 2005; Kapahi et al., 2004; Mair and Dillin, 2008; Mair et al., 2011; Panowski et al., 2007). Our results show for the first time that circadian clock genes also play a role in modulating lifespan extension in response to DR. Flies that were maintained in constant light conditions, which disrupt the functioning of the molecular clock, showed a reduced response to DR in terms of lifespan extension. Significantly reduced response to DR was also found in flies mutant for either the tim or the per gene, further suggesting the importance of clocks for effects of DR on lifespan. As both per and tim mutant animals show some level of rhythmicity under normal 12hr LD (light-dark) cycle (Sehgal et al., 1994; Wheeler et al., 1993), the strong reduction in lifespan under constant light conditions and in the two circadian mutants argues that lifespan extension upon DR is not simply a consequence of altered circadian behaviors but more likely linked to clock controlled genes regulated by tim and per. It is important to note here that animals lacking clocks as well as those on constant light still display some lifespan extension, suggesting possible contributions of clock-independent pathways in the response to DR. This is not surprising as several pathways are likely to work in concert to mediate the maximal lifespan extension by DR in flies (Kapahi et al., 2010; Mair and Dillin, 2008). Though some circadian mutants are also slightly shorter lived on AL diet, we consistently observed that the reduction on DR food is higher in the circadian mutant animals. Additionally, it is reasonable to expect that certain manipulations such as circadian disruption that limits DR dependent lifespan may shorten lifespan under all diets as these cellular processes may be critical for ensuring a normal lifespan. Indeed, lifespan shortening effects are also observed for mutants of daf-16/dfoxO, which is a key effector of the lifespan extension produced by inhibiting the insulin signaling pathway in both C. elegans and Drosophila.
DR increases and maintains the magnitude of expression of clock genes and proteins
Our results demonstrate that DR increases the magnitude of circadian gene expression, which can attenuate the age-related loss of circadian oscillations that has been observed in several species (Asai et al., 2001; Rakshit et al., 2012; Yamazaki et al., 2002; Zhdanova et al., 2008). Aging is known to attenuate various circadian behaviors such as daily rhythms in hormone levels, body temperature, sleep/wake cycles etc. so conversely, improving oscillations might be expected to slow down the age-related decline in function (Koh et al., 2006; Touitou and Haus, 2000; Weinert and Waterhouse, 2007; Zhdanova et al., 2011). Longevity in golden hamsters was reduced with noninvasive disruption of circadian rhythmicity, but was increased in older animals when given suprachiasmatic implants that restored high amplitude rhythms in the animals (Hurd and Ralph, 1998). In Drosophila, overexpression of cryptochrome (cry) in all clock cells was shown to maintain strong rest/activity rhythms in older animals (Rakshit and Giebultowicz, 2013). However, overexpression of CRY in central clock neurons alone was not sufficient to restore rest/activity rhythms suggesting a possible role of peripheral clocks in delaying behavioral and physiological aging. Our results are in line with these observations as we show that DR improves the oscillations and levels of the clock gene expression and proteins in peripheral tissues. We also show that overexpression of tim in peripheral tissues increased fat turnover and increased lifespan even under AL food conditions. Although tim overexpression in most of the peripheral tissues tested increased lifespan, we observed that the lifespans of the control flies (without RU486) in this experiment were shorter than those of other controls, due to variability across GAL4 strains. Thus, at the present time we cannot distinguish between compensation by TIM for lifespan-shortening variations versus an ability to extend lifespan beyond normal. Nevertheless it is clear that TIM overexpression is beneficial for lifespan in, at least, some contexts.
The molecular mechanism of how DR enhances clock robustness will require future studies examining reciprocal links between nutrient sensing pathways and the clocks in various tissues. There are three possible explanation of how a reduction in dietary protein or yeast (in case of flies) leads to regulation of circadian clocks. The first is that yeast restriction could influence clocks through the inhibition of TOR/insulin signaling. It was previously shown that Drosophila foxo modulates stress sensitivity of circadian clocks (Zheng et al., 2007). Furthermore, they also showed a role of TOR/TSC and AKT in central clocks, likely through modulation of GSK3β, which phosphorylates central clock proteins (Zheng and Sehgal, 2010). Secondly, protein restriction modulates TOR/4EBP signaling (Kapahi et al., 2010; Katewa and Kapahi, 2010; Zid et al., 2009), which in turn could influence the translation of clock proteins. Finally, protein restriction influences the activity of nuclear de-acetylating proteins (such as HDACs, SIRT1 or SIRT6), which in turn could influence the transcriptional levels of clock dependent genes. Recently, Sassone-Corsi lab showed a role of SIRT1 and SIRT6 in regulating circadian gene expression in peripheral clocks (Masri et al., 2014).
Clocks regulate fat metabolism under DR
Circadian clocks are critical regulators of metabolism (Green et al., 2008; Kohsaka et al., 2007; Panda et al., 2002; Ramsey et al., 2007; Xu et al., 2011) and maintain temporal organization that optimizes various cellular functions (Kovac et al., 2009; Loboda et al., 2009; Ramsey et al., 2007). Genome-wide circadian expression profiling studies have uncovered potential connections between circadian clocks and many aspects of metabolism, including energy, carbohydrate, amino acid, lipid, and protein metabolism, as well as detoxification (Beaver et al., 2012; Panda and Hogenesch, 2004; Wijnen and Young, 2006). In flies, recent studies have highlighted the regulation of lipid and carbohydrate metabolism by circadian clocks (DiAngelo et al., 2011; Seay and Thummel, 2011; Xu et al., 2011). Although the levels of total TGs do not show circadian rhythmicity, flies mutant for circadian genes as such tim (tim01) and cry (cry01) show reduction in total levels of TGs (Seay and Thummel, 2011). Recently, we showed that upon DR, Drosophila melanogaster shift their metabolism towards increasing both fatty acid synthesis and breakdown and these changes are required for various responses to DR. Furthermore, flies that tend to increase fat turnover on AL food show increased lifespan (Katewa et al., 2012). However, it is not clear how flies undertake both increased breakdown and synthesis of TGs. Our results here support the idea that circadian clock genes play a critical role in this improved fat turnover. Both tim and per mutant flies showed reduced starvation resistance under DR conditions, and this was associated with reduced fat synthesis and reduced breakdown. Overexpression of tim on the other hand resulted in increased fat turnover under rich nutrient conditions in both whole flies or dissected tissues (Figure 6B–C). As the lifespan of tim01 female flies does not differ significantly from control flies on AL food, we reasoned that the most effective way of identifying the group of lipids related with lifespan effects in tim01 would be lipidomic analysis under DR food conditions. We found that one group of cycling lipids consists primarily of medium chain TGs (MCTs) (Figure 4E). One of the MCTs, trilauryl glycerol (TLG), showed an almost two fold increase between ZT4 and ZT16 under DR conditions and its levels were dependent on tim (Figure 4E, S4 and 6F). Reducing de novo TG synthesis by inhibition of ACC led to a reduction of the protective effects of tim overexpression, further supporting the critical role of fat turnover in lifespan extension under both DR (Katewa et al., 2012) as well as tim overexpression (Figure. 6E–F, S6D). The question of whether it is timeless alone or also other clock components that regulate DR response is still not clear. Although, the mRNA levels of other core clock genes are not influenced by timeless overexpression, our results showing that both tim and per mutants show reduction in DR dependent starvation resistance, and lifespan extension, indicates a role of circadian clocks in DR responses. However, while overexpression of timeless increases fat turnover and lifespan under AL conditions, period overexpression has no effect on lifespan. It is possible that per is not limiting for these effects, but tim could be the major driver here, with per mutants showing an effect only because of their effect on nuclear expression of TIM (Zheng and Sehgal, 2008).
MCTs are TGs containing medium chain fatty acids (MCFA, C8 to C12 length). MCTs were originally used for dietary treatment of malabsorption syndromes in humans because of their rapid absorption and breakdown compared to long chain TGs (LCT)(Scheig, 1968). In animals replacing dietary LCT by MCT causes a rise in energy expenditure, increases thermogenesis, depresses food intake and causes lowering of body fat (St-Onge and Jones, 2002). In humans also feeding MCT increases energy expenditure relative to LCT feeding (Mumme and Stonehouse, 2015). Based on these effects, MCT based interventions are being considered for dietary treatment of obesity and are proposed for weight-loss treatments (St-Onge and Jones, 2002). Despite being used in both animals and humans over several decades, ours is the first report of de novo synthesis of MCTs in response to DR in any animal. However, it’s still not clear why DR animals would enhance levels of MCTs? One hypothesis is that MCTs are faster to synthesize and easier to break down by cells for energy purposes. Another possibility is that as MCT feeding results in increased beta-oxidation of not only MCTs but also of LCTs (Ronis et al., 2013), this would lead to increase fatty acid turnover and will reduce the time available for fatty acids to undergo oxidation (lipid peroxidation etc). Finally, as their levels are regulated by clocks, changing concentrations of MCTs could act as signaling molecules and modulate activities of kinases or nuclear transcription factors. Support for this hypothesis comes from a recent study showing that MCT feeding ameliorates insulin resistance and inflammation in high fat diet-induced obese mice (Geng et al., 2015). In conclusion, our results strongly support a key role of clock regulated metabolic adaptation under DR and pave the way for future longevity studies involving manipulations of clocks via environmental or pharmacological interventions.
Experimental Procedures
Fly stocks, husbandry and survival assays: Fly stocks
The following fly strains were used: Act5C–GS-GAL4 (w; P{Act5C(−FRT)GAL4.Switch.PR}255B) (Ford et al., 2007), S1106 GAL4-(w1118; P{Switch1}106) (Roman et al., 2001), Elav-GS-GAL4 (yw;P{elav-Switch.O}GSG301) (Osterwalder et al., 2001), tim01 (Myers et al., 1995), per01 (Konopka and Benzer, 1971), UAS-tim, UAS-per24 (Yang and Sehgal, 2001), UAS-per10 (Stoleru et al., 2007), C42-GAL4 (w[*]; P{w[+mW.hs]=GawB}c42), and 5966-GS-GAL4 (Guo et al., 2014) and ACC- RNAi (w[1118];P{GD3482}v8105)(Katewa et al., 2012). All survival assays were carried out on AL or DR media as described previously (Katewa et al., 2012; Zid et al., 2009). Adult female flies were transferred within 2–3 days of eclosion to media differing only in the amount of yeast extract (YE) in the diet and were maintained at 25°C temperature, 60% humidity and 12 hr light and 12 hr dark conditions for their entire lifespan. About 25–30 mated females were maintained per vial and the flies were transferred every 2–3 days onto fresh media vials and deaths recorded. Additional details about various fly media recipes that were used in the study are provided in the Supplemental Experimental Procedures.
Circadian fly sample collections
Fly vials were removed every four hrs (starting at ZT0 (8.00 AM), ZT4, ZT8, ZT12, ZT16, ZT20 and ZT24 (next day 8.00 AM)) and were either frozen immediately (for mRNA measurements) or were kept on ice for fat body dissection. Flies selected for circadian analysis were transferred into individual racks during the light period so that removal of one vial during the night doesn't disturb other flies that would be frozen later in the night.
Fat turnover and starvation assay
After 10 days feeding on AL and DR media, 240 flies were transferred to AL or DR media with 2 µCi of 14C labeled glucose added on top of the media. After 24 hrs of feeding, half of the flies were snap-frozen in liquid Nitrogen (and is referred as 0 hrs sample). The other half was then transferred to fresh non-radioactive AL or DR media and were kept on this media for the next 60 hours and then immediately frozen (this samples were referred as 60 hrs sample). The frozen samples (20 flies/replicate) were homogenized in chloroform-methanol (2:1) and total lipid was extracted by the Folch method (Folch et al., 1957). Total lipid was resuspended in 500 µl of chloroform and was further fractionated into triglyceride fraction by using DSC-NH2 cartridges and different solvents as described previously (Katewa et al., 2012). The fractions were dried under nitrogen, re-suspended in the scintillation fluid, and counted in a scintillation counter. 0 hrs samples indicate the rate of incorporation of glucose in fatty acids and 60 hrs sample indicate the breakdown of the labeled fatty acids. For starvation assays female flies (day 10 on AL/DR media) were transferred to vials containing 1% agar. The flies were transferred to fresh vials every 24 hours and deaths were recorded every 6–12 hours.
Lipid extraction and High performance liquid chromatography (HPLC) - mass spectrometry (MS) sample preparation
Fly lipid extraction was performed based on a previously reported protocol (Hammad et al., 2011) with some modifications. Briefly, 5 flies for each time point for a genotype (control and tim01) were weighed, flash-frozen over liquid nitrogen, and subsequently homogenized ultrasonically using a Fisher Scientific’s 550 Sonic Dismembrator with 100 µL of 0.9% NaCl soution. Three 20 s pulses at amplitude setting 4 of the instrument (on ice) were sufficient to completely homogenize fly bodies. The homogenates were then transferred to clean glass vials and the original tubes washed twice with 50 µL of the 0.9% NaCl solution to ensure complete transfer. To this 1000 µL of 2:1 dichloromethane (DCM): methanol (containing 5 µM of each of the following internal standards: d5-1,3-DG(15:0/15:0) and d5-TG(17:0/17:1/17:0) was added and each sample vortexed for 5 times over a period of ~30 min (each 30 s long). Subsequently, the samples were centrifuged at 4,000 rpm for 10 min - at this point two clear layers separated and the fly debris was collected at the interphase of the layers. 500 µL of the lower organic layer was quantitatively collected using a clean glass syringe and 5 µL injected directly for qualitative high resolution HPLC-MS analysis, or diluted 1:5 with 1:1 DCM:methanol and then 5 µL injected for quantitative unit resolution HPLC-MS analysis without any further processing. Between each subsequent extract collection, the glass syringe was thoroughly washed with the 1:1 DCM:methanol solution. Additional details of HPLC-MS instrumentation and methods are provided in Supplemental Experimental Procedures.
HPLC-MS and –MS/MS analyses
A preliminary analysis revealed that the control/ZT 16 sample to contain high (or most prominently detectable, i.e. with the highest signal-to-noise ratio) intensities for most peaks and hence was thoroughly examined to tabulate a list of mass spectrometric features (~350), defined as a specific m/z at a particular retention time. The primary focus of our study was to identify cycling triglycerides (TGs); since a standard mix of synthetic d5-triglycerides (Figure 4D) showed predominantly [M+NH4]+ under our HPLC-MS conditions, care was taken to specifically tabulate the [M+NH4]+ adduct ions and eliminate features that indicated low (but still detectable) levels of the corresponding [M+H]+ or [M+Na]+ adducts. Further, features originating from solvent contaminants (and/or other exogenous sources) were carefully selected and removed prior to quantitative analysis. Subsequently, peak areas for ~253 features (Figure 4D) were computed from the unit resolution HPLC-MS data for fly lipid extracts from the QTRAP MS instrument across each of four replicates for 8 time points (ZT 0, 4, 8, 12, 16, 20, 24 h) of control and tim01 flies (total of 56 samples). Peak areas were subsequently normalized first by the fly weight for the sample (replicate), calculated prior to lipid extraction and then by the corresponding peak area of the DG-internal standard [M+NH4]+ adduct ion for each replicate run to account for variability during the biphasic lipid extraction process and/or sample to sample variability in MS response across ~60 hrs of continuous acquisition.
The cycling features were characterized further using the corresponding HRMS data acquired using the QTOF MS instrument (see Table S4) to annotate features to specific TGs and the most prominent cycling feature 88, TG(36:0) was subjected to high resolution MS/MS analysis (Figure 4F and 4G). The high resolution molecular ion and fragment ions were matched against LIPID MAPS’ (Sud et al., 2007) database using their online mass spectrometry tools (Fahy et al., 2007) for corresponding structural elucidation (Figure 4G).
Statistical Analysis
Circadian expression data were statistically analyzed with GraphPad Prism (v.5.0) and GraphPad Instat (v.3.0; San Diego, CA). qRT-PCR data for circadian gene expression were evaluated by two-way analysis of variance (ANOVA) with Bonferroni’s post hoc test. Other data were analyzed by Student’s t test. Each survival assay was repeated at least twice and the shown figures represent a typical curve. Survival curves were created using the product-limit method of Kaplan and Meier. The log-rank (Mantel-Cox) test was used to evaluate differences between survivals and determine P values. We used the Prism software package (GraphPad Software) to carry out statistical analysis and to determine lifespan values. Additionally, we have used Cox proportional hazards analysis implemented in the R package 'survival' to analyze the significance of the interaction between two variables in several of the survival outcomes. We report the probability that B1,2=0, from fitting the formula phenotype=B1*variable1+B2*variable2+B1,2*(variable1*variable2). The respective p values are included in the text.
Supplementary Material
Acknowledgments
We thank M. Kolipinski, J. Beck and M. Vargas for lifespan data collection and members of the Kapahi Lab for discussions and suggestions. We would also like to thank Prof. Daniel Promislow for help with survival analysis. This work was funded by grants from the American Federation for Aging Research (S.D.K), Larry L. Hillblom Foundation (S.D.K), NIH (R01AG038688, AG038012 & AG045835 (P.K.); R01 AG045830 (J.G.)), and from the Ellison Medical Foundation (A.S.). A.S. is an HHMI investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asai M, Yoshinobu Y, Kaneko S, Mori A, Nikaido T, Moriya T, Akiyama M, Shibata S. Circadian profile of Per gene mRNA expression in the suprachiasmatic nucleus, paraventricular nucleus, and pineal body of aged rats. J. Neuro. Res. 2001;66:1133–1139. doi: 10.1002/jnr.10010. [DOI] [PubMed] [Google Scholar]
- Beaver LM, Klichko VI, Chow ES, Kotwica-Rolinska J, Williamson M, Orr WC, Radyuk SN, Giebultowicz JM. Circadian regulation of glutathione levels and biosynthesis in Drosophila melanogaster. PLoS One. 2012;7:e50454. doi: 10.1371/journal.pone.0050454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat. Rev. Genet. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat. Rev. Mol. Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- Bruce KD, Hoxha S, Carvalho GB, Yamada R, Wang HD, Karayan P, He S, Brummel T, Kapahi P, Ja WW. High carbohydrate-low protein consumption maximizes Drosophila lifespan. Exp. Gerontol. 2013;48:1129–1135. doi: 10.1016/j.exger.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruss MD, Khambatta CF, Ruby MA, Aggarwal I, Hellerstein MK. Calorie restriction increases fatty acid synthesis and whole body fat oxidation rates. Am. J. Physiol. Endocrinol. Metab. 2010;298:E108–E116. doi: 10.1152/ajpendo.00524.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr ED, Takahashi JS. Molecular components of the Mammalian circadian clock. Handb. Exp. Pharmacol. 2013;217:3–27. doi: 10.1007/978-3-642-25950-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, van der Horst GT, Schellevis R, Nijman RM, Koerkamp MG, Holstege FC, Smidt MP, Hoekman MF. Insulin-FOXO3 signaling modulates circadian rhythms via regulation of clock transcription. Curr. Biol. 2014;24:1248–1255. doi: 10.1016/j.cub.2014.04.018. [DOI] [PubMed] [Google Scholar]
- Chippindale AK, Leroi AM, Kim SB, Rose MR. Phenotypic plasticity and selection in Drosophila life-history evolution. I. Nutrition and the cost of reproduction. J. Evol. Biology. 1993;6:171–193. [Google Scholar]
- DiAngelo JR, Erion R, Crocker A, Sehgal A. The central clock neurons regulate lipid storage in Drosophila. PLoS One. 2011;6:e19921. doi: 10.1371/journal.pone.0019921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery NJ, Clayton NS. Effects of experience and social context on prospective caching strategies by scrub jays. Nature. 2001;414:443–446. doi: 10.1038/35106560. [DOI] [PubMed] [Google Scholar]
- Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:W606–W612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Fontana L, Klein S, Holloszy JO. Effects of long-term calorie restriction and endurance exercise on glucose tolerance, insulin action, and adipokine production. Age (Dordr) 2010;32:97–108. doi: 10.1007/s11357-009-9118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford D, Hoe N, Landis GN, Tozer K, Luu A, Bhole D, Badrinath A, Tower J. Alteration of Drosophila life span using conditional, tissue-specific expression of transgenes triggered by doxycyline or RU486/Mifepristone. Exp. Gerontol. 2007;42:483–497. doi: 10.1016/j.exger.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng S, Zhu W, Xie C, Li X, Wu J, Liang Z, Xie W, Zhu J, Huang C, Zhu M, et al. Medium-chain triglyceride ameliorates insulin resistance and inflammation in high fat diet-induced obese mice. Eur. J. Nutr. Apr. 2015;25 doi: 10.1007/s00394-015-0907-0. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014;156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad LA, Cooper BS, Fisher NP, Montooth KL, Karty JA. Profiling and quantification of Drosophila melanogaster lipids using liquid chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 2011;25:2959–2968. doi: 10.1002/rcm.5187. [DOI] [PubMed] [Google Scholar]
- Hurd MW, Ralph MR. The significance of circadian organization for longevity in the golden hamster. J. Biol. Rhythms. 1998;13:430–436. doi: 10.1177/074873098129000255. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, Kockel L. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster . Proc. Natl. Acad. Sci. USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa SD, Demontis F, Kolipinski M, Hubbard A, Gill MS, Perrimon N, Melov S, Kapahi P. Intramyocellular Fatty-Acid Metabolism Plays a Critical Role in Mediating Responses to Dietary Restriction in Drosophila melanogaster . Cell Metab. 2012;16:97–103. doi: 10.1016/j.cmet.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa SD, Kapahi P. Dietary restriction aging 2009. Aging. Cell. 2010;9:105–112. doi: 10.1111/j.1474-9726.2010.00552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa SD, Kapahi P. Role of TOR signaling in aging and related biological processes in Drosophila melanogaster . Exp. Gerontol. 2011;46:382–390. doi: 10.1016/j.exger.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King BS, Lu L, Yu M, Jiang Y, Standard J, Su X, Zhao Z, Wang W. Lipidomic profiling of di- and tri-acylglycerol species in weight-controlled mice. PLoS One. 2015;10:e0116398. doi: 10.1371/journal.pone.0116398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Evans JM, Hendricks JC, Sehgal A. A Drosophila model for age-associated changes in sleep:wake cycles. Proc. Natl. Acad. Sci. USA. 2006;103:13843–13847. doi: 10.1073/pnas.0605903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Kondratov RV. A role of the circadian system and circadian proteins in aging. Ageing Res. Rev. 2007;6:12–27. doi: 10.1016/j.arr.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Kovac J, Husse J, Oster H. A time to fast, a time to feast: the crosstalk between metabolism and the circadian clock. Mol. Cells. 2009;28:75–80. doi: 10.1007/s10059-009-0113-0. [DOI] [PubMed] [Google Scholar]
- Krishnan B, Levine JD, Lynch MK, Dowse HB, Funes P, Hall JC, Hardin PE, Dryer SE. A new role for cryptochrome in a Drosophila circadian oscillator. Nature. 2001;411:313–317. doi: 10.1038/35077094. [DOI] [PubMed] [Google Scholar]
- Lee KP, Simpson SJ, Clissold FJ, Brooks R, Ballard JW, Taylor PW, Soran N, Raubenheimer D. Lifespan and reproduction in Drosophila: New insights from nutritional geometry. Proc. Natl. Acad. Sci. USA. 2008;105:2498–2503. doi: 10.1073/pnas.0710787105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda A, Kraft WK, Fine B, Joseph J, Nebozhyn M, Zhang C, He Y, Yang X, Wright C, Morris M, et al. Diurnal variation of the human adipose transcriptome and the link to metabolic disease. BMC Med. Genomics. 2009;2:7. doi: 10.1186/1755-8794-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Chen WF, Yue Z, Chen D, Sowcik M, Sehgal A, Zheng X. Old flies have a robust central oscillator but weaker behavioral rhythms that can be improved by genetic and environmental manipulations. Aging Cell. 2012;11:428–438. doi: 10.1111/j.1474-9726.2012.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- Mair W, Goymer P, Pletcher SD, Partridge L. Demography of dietary restriction and death in Drosophila. Science. 2003;301:1731–1733. doi: 10.1126/science.1086016. [DOI] [PubMed] [Google Scholar]
- Mair W, Morantte I, Rodrigues AP, Manning G, Montminy M, Shaw RJ, Dillin A. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470:404–408. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Piper MD, Partridge L. Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol. 2005;3:e223. doi: 10.1371/journal.pbio.0030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, Roqueta-Rivera M, Deng C, Osborne TF, Mostoslavsky R, et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell. 2014;158:659–672. doi: 10.1016/j.cell.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min KJ, Tatar M. Restriction of amino acids extends lifespan in Drosophila melanogaster . Mech. Ageing Dev. 2006;127:643–646. doi: 10.1016/j.mad.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Mumme K, Stonehouse W. Effects of medium-chain triglycerides on weight loss and body composition: a meta-analysis of randomized controlled trials. J. Acad. Nutr. Diet. 2015;115:249–263. doi: 10.1016/j.jand.2014.10.022. [DOI] [PubMed] [Google Scholar]
- Myers MP, Wager-Smith K, Wesley CS, Young MW, Sehgal A. Positional cloning and sequence analysis of the Drosophila clock gene, timeless. Science. 1995;270:805–808. doi: 10.1126/science.270.5237.805. [DOI] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. USA. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB. It's all in the timing: many clocks, many outputs. J. Biol. Rhythms. 2004;19:374–387. doi: 10.1177/0748730404269008. [DOI] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Rakshit K, Giebultowicz JM. Cryptochrome restores dampened circadian rhythms and promotes healthspan in aging Drosophila. Aging Cell. 2013;12:752–762. doi: 10.1111/acel.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakshit K, Krishnan N, Guzik EM, Pyza E, Giebultowicz JM. Effects of aging on the molecular circadian oscillations in Drosophila. Chronobiol. Int. 2012;29:5–14. doi: 10.3109/07420528.2011.635237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Marcheva B, Kohsaka A, Bass J. The clockwork of metabolism. Annu. Rev. Nutr. 2007;27:219–240. doi: 10.1146/annurev.nutr.27.061406.093546. [DOI] [PubMed] [Google Scholar]
- Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronis MJ, Baumgardner JN, Sharma N, Vantrease J, Ferguson M, Tong Y, Wu X, Cleves MA, Badger TM. Medium chain triglycerides dose-dependently prevent liver pathology in a rat model of non-alcoholic fatty liver disease. Exp. Biol. Med. (Maywood) 2013;238:151–162. doi: 10.1258/ebm.2012.012303. [DOI] [PubMed] [Google Scholar]
- Scheig R. Absoption of dietary fat: use of medium-chain triglycerides in malabsorption. Am. J. Clin. Nutr. 1968;21:300–304. doi: 10.1093/ajcn/21.4.300. [DOI] [PubMed] [Google Scholar]
- Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- Seay DJ, Thummel CS. The circadian clock, light, and cryptochrome regulate feeding and metabolism in Drosophila. J. Biol. Rhythms. 2011;26:497–506. doi: 10.1177/0748730411420080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Price JL, Man B, Young MW. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- Solon-Biet SM, McMahon AC, Ballard JW, Ruohonen K, Wu LE, Cogger VC, Warren A, Huang X, Pichaud N, Melvin RG, et al. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 2014;19:418–430. doi: 10.1016/j.cmet.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Onge MP, Jones PJ. Physiological effects of medium-chain triglycerides: potential agents in the prevention of obesity. J. Nutr. 2002;132:329–332. doi: 10.1093/jn/132.3.329. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Nawathean P, Fernandez MP, Menet JS, Ceriani MF, Rosbash M. The Drosophila circadian network is a seasonal timer. Cell. 2007;129:207–219. doi: 10.1016/j.cell.2007.02.038. [DOI] [PubMed] [Google Scholar]
- Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, Subramaniam S. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35:D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M. Diet Restriction in Drosophila melanogaster . In: Mobbs C, Yen K, Hof P, editors. Mechanisms of Caloric Restriction in Aging. Basel: Karger; 2007. pp. 115–136. [Google Scholar]
- Touitou Y, Haus E. Alterations with aging of the endocrine and neuroendocrine circadian system in humans. Chronobiol. Int. 2000;17:369–390. doi: 10.1081/cbi-100101052. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas MA, Luo N, Yamaguchi A, Kapahi P. A Role for S6 Kinase and Serotonin in postmating dietary switch and balance of nutrients in D. melanogaster . Curr. Biol. 2010;20:1006–1011. doi: 10.1016/j.cub.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert D, Waterhouse J. The circadian rhythm of core temperature: effects of physical activity and aging. Physiol. Behav. 2007;90:246–256. doi: 10.1016/j.physbeh.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Wheeler DA, Hamblen-Coyle MJ, Dushay MS, Hall JC. Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J. Biol. Rhythms. 1993;8:67–94. doi: 10.1177/074873049300800106. [DOI] [PubMed] [Google Scholar]
- Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- Xu K, DiAngelo JR, Hughes ME, Hogenesch JB, Sehgal A. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 2011;13:639–654. doi: 10.1016/j.cmet.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Effects of aging on central and peripheral mammalian clocks. Proc. Natl. Acad. Sci. USA. 2002;99:10801–10806. doi: 10.1073/pnas.152318499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- Zhdanova IV, Masuda K, Quasarano-Kourkoulis C, Rosene DL, Killiany RJ, Wang S. Aging of intrinsic circadian rhythms and sleep in a diurnal nonhuman primate, Macaca mulatta. J. Biol. Rhythms. 2011;26:149–159. doi: 10.1177/0748730410395849. [DOI] [PubMed] [Google Scholar]
- Zhdanova IV, Yu L, Lopez-Patino M, Shang E, Kishi S, Guelin E. Aging of the circadian system in zebrafish and the effects of melatonin on sleep and cognitive performance. Brain Res. Bull. 2008;75:433–441. doi: 10.1016/j.brainresbull.2007.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178:1147–1155. doi: 10.1534/genetics.107.088658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. AKT and TOR signaling set the pace of the circadian pacemaker. Curr. Biol. 2010;20:1203–1208. doi: 10.1016/j.cub.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Yang Z, Yue Z, Alvarez JD, Sehgal A. FOXO and insulin signaling regulate sensitivity of the circadian clock to oxidative stress. Proc. Natl. Acad. Sci. USA. 2007;104:15899–15904. doi: 10.1073/pnas.0701599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E–BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.