Abstract
Heroin addiction is heritable, but few specific genetic variants have been reproducibly associated with this disease. The zinc finger protein 804A (ZNF804A) gene is a biologically plausible susceptibility gene for heroin addiction, given its function as a transcription factor in human brain. Novel associations of two common ZNF804A single nucleotide polymorphisms (SNPs), rs7597593 and rs1344706, with heroin addiction have been reported in Han Chinese. Both SNPs have also been implicated for regulating ZNF804A expression in human brain, including the addiction-relevant dorsolateral prefrontal cortex. In this independent replication study, we tested the rs7597593 and rs1344706 SNP genotypes and their corresponding haplotypes for association with heroin addiction using cases drawn from the Urban Health Study and population controls: total N=10,757 (7,095 European Americans and 3,662 African Americans). We independently replicated both ZNF804A SNP associations in European Americans: the rs7597593-T (P=0.016) and rs1344706-A (P=0.029) alleles both being associated with increased risk of heroin addiction, consistent with the prior report. Neither SNP was associated in African Americans alone, but meta-analysis across both ancestry groups resulted in significant associations for rs1344706-A (P=0.016, odds ratio [95% confidence interval] = 1.13 [1.02–1.25]) and its haplotype with rs7597593-T (P=0.0067, odds ratio [95% confidence interval] = 1.16 [1.04–1.29]). By demonstrating consistent associations across independent studies and diverse ancestry groups, our study provides evidence that these two ZNF804A SNPs and their risk haplotype are among the few replicable genetic associations with heroin addiction.
Keywords: ancestry, genetic association study, haplotype, heroin, opioid, replication, rs1344706, rs7597593, Urban Health Study, ZNF804A
Introduction
Addiction to heroin and other opioid drugs is a growing public health concern (Substance Abuse and Mental Health Service Administration, 2013) with profound economic consequences (Mark et al., 2001). Heroin addiction is heritable (Mistry et al., 2014), and although a few single nucleotide polymorphisms (SNPs) in the opioid receptor genes OPRM1 (Hancock et al., in press) and OPRD1 (Nelson et al., 2014) have been reproducibly associated, the specific genetic variants contributing to heroin addiction remain largely unknown.
Novel associations between SNPs in the zinc finger protein 804A (ZNF804A) gene and heroin addiction (Sun et al., in press) have suggested that this gene may exert pleiotropic effects influencing multiple psychiatric-related phenotypes. ZNF804A was the first genome-wide significant finding reported for schizophrenia (O’Donovan et al., 2008), and since then, ZNF804A SNP associations have been widely implicated for schizophrenia and other psychiatric disorders in populations of European and Asian ancestry (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013, International Schizophrenia Consortium et al., 2009, Riley et al., 2010, Schwab et al., 2013, Steinberg et al., 2011, Williams et al., 2011, Xiao et al., 2011). ZNF804A is abundantly expressed in human brain (Tao et al., 2014) and is predicted to encode a transcription factor that directly interacts with genes related to dopaminergic transmission (Girgenti et al., 2012), a neural mechanism known to drive both schizophrenia (Nieratschker et al., 2010) and addiction (Hyman et al., 2006). In extending ZNF804A SNP associations to heroin addiction, Sun et al. (in press) tested six ZNF804A SNPs in Han Chinese (N=3,922) and found that two of the most robust SNPs for psychosis were associated with heroin addiction, rs7597593 and rs1344706 (lowest uncorrected P=0.023 for single SNP tests and 0.0035 for their haplotype test). Both of these SNPs have also been implicated for regulating ZNF804A expression in postmortem human brain (Guella et al., 2014, Riley et al., 2010, Zhang et al., 2011), with rs1344706 being implicated specifically in the dorsolateral prefrontal cortex—a highly relevant brain region for addiction (Goldstein & Volkow, 2011). Our study focused on these two regulatory SNPs and tested their associations for independent replication with heroin addiction in 7,095 European Americans and 3,662 African Americans.
Materials and Methods
Study participants
Heroin addiction cases were drawn from the Urban Health Study (UHS) of street-recruited people who reported past 30-day injection of an illicit drug (verified by signs of venipuncture) from the San Francisco Bay area between 1986 and 2005 (Kral et al., 2001, Kral et al., 2003). The current study focused on European Americans and African Americans who met the Office of National Drug Control Policy definition of heroin abuse, having injected 10+ times in the past 30 days (Morral et al., 2000, Rhodes et al., 2000). This level of heroin abuse is highly correlated with clinical dependence levels on the Severity of Dependence Scale (Gossop et al., 1992, Strang et al., 1999) and with DSM-IV heroin abuse/dependence (American Psychiatric Association, 1994) based on our analyses of the National Survey on Drug Use and Health data (Hancock et al., in press). These UHS participants, henceforth referred to as heroin addiction cases, reported abusing heroin an average of 80.9 times in the past 30 days and thus were very likely dependent on heroin.
The UHS heroin addiction cases were compared to population controls assembled together from six study cohorts in the database of Genotypes and Phenotypes (dbGaP), as previously described (Hancock et al., in press). Exclusions were made for DSM-IV opioid dependence, where the data were available in the two study cohorts ascertained for addiction. Controls from the other four cohorts were ascertained for phenotypes unrelated to addiction, where we expected minimal phenotype misclassification for heroin addiction.
All study protocols received Institutional Review Board approval at their respective sites, and all study participants provided informed consent.
Genotyping and Quality Control
As detailed elsewhere (Hancock et al., in press, Johnson et al., 2015), stored serum samples from the UHS were restored using the Illumina Formalin-Fixed Paraffin-Embedded kit to maximize the quality of genomic DNA and then genotyped on the Illumina Omni1-Quad BeadChip, Controls were genotyped on one of three Illumina platforms (Omni1-Quad, 1M-Duo, or Omni2.5). Both ZNF804A SNPs, rs7597593 located in intron 1 and rs1344706 located in intron 2, were assayed across these Illumina platforms and thus genotyped in all cases and controls. Both ZNF804A SNPs passed all quality control thresholds, thus having call rate ≥90%, minor allele frequency ≥1%, and Hardy Weinberg equilibrium P≥1×10−4.
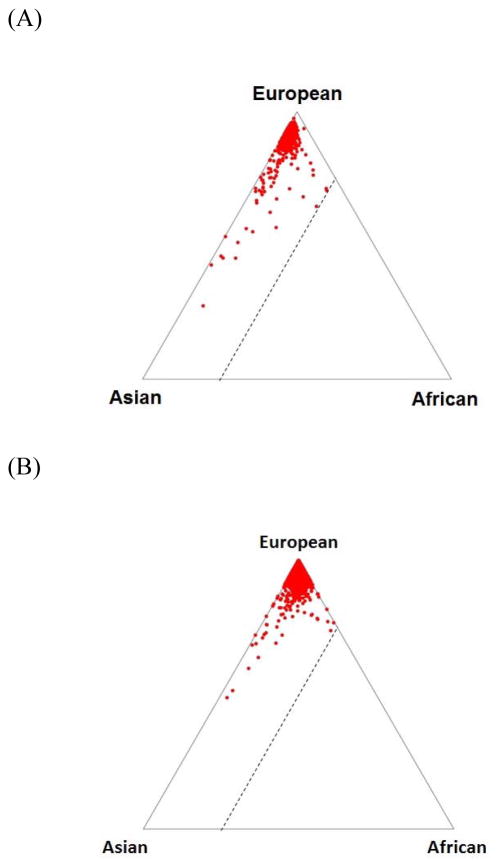
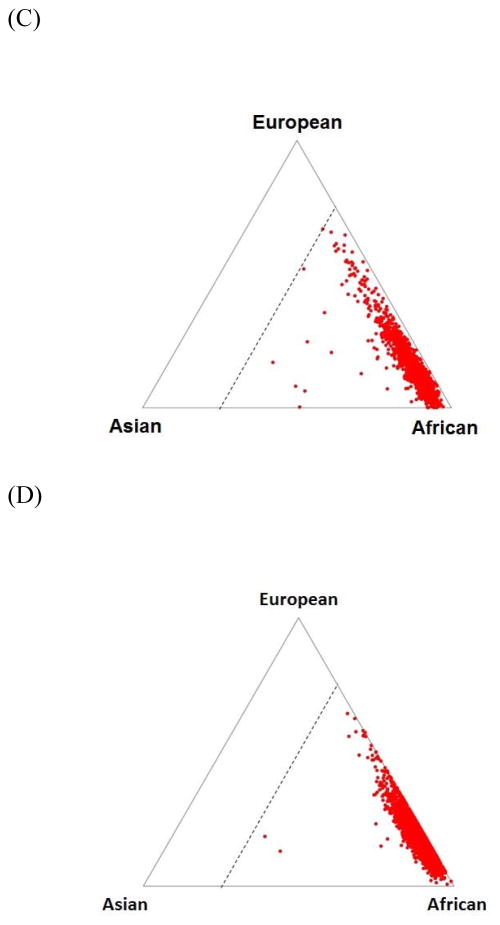
The genotyped participants’ ancestral proportions were determined using the STRUCTURE program (Pritchard et al., 2000) with reference to HapMap populations. Following exclusions for European Americans having >25% African ancestry and African Americans having <25% African ancestry, the heroin addiction case and control datasets closely resembled the expected ancestral proportions (Figure 1). Participants were also excluded for call rate <90%, sample duplication, gender discordance, excessive homozygosity, and first-degree relatedness. Our final analysis dataset included 7,095 European Americans (711 cases and 6,384 controls) and 3,662 African Americans (1,293 cases and 2,369 controls), as shown in Table 1.
Figure 1.
Ancestral proportions estimated using the STRUCTURE program with reference to HapMap populations and 10,000 randomly selected HapMap phase III SNPs. Ancestral proportions, as used for participant-level quality control, are shown for (A) European American heroin addiction cases, (B) European American controls, (C) African American heroin addiction cases, and (D) African American controls. Triangle vertices represent West Africans (denoted YRI in HapMap), European Americans (CEU), and East Asians (CHB), and triangle edges indicate the ancestral proportions with the dotted line representing 25% African ancestry. (A)
Table 1.
Characteristics of the heroin addiction cases and controls, by ancestry.
| Ancestry group | Heroin addiction | No. genotyped participants passing quality control | No. (%)female | No. (%) cases reporting past 30-day abuse of other drug(s) in addition to heroin | |||
|---|---|---|---|---|---|---|---|
| Any other drug | Specific drugs* | ||||||
| Cocaine | Marijuana | Amphetamines and other stimulants | |||||
| European Americans | Cases | 711 | 135 (19.0) | 264 (37.1) | 221 (31.1) | 4 (0.6) | 64 (9.0) |
| Controls | 6,384 | 3,505 (54.9) | NA | NA | NA | NA | |
| African Americans | Cases | 1,293 | 372 (28.8) | 611 (47.3) | 590 (45.6) | 24 (1.9) | 36 (2.8) |
| Controls | 2,369 | 913 (38.5) | NA | NA | NA | NA | |
NA, not applicable.
Abuse (10 or more times in the past month) of different drug categories was not mutually exclusive.
Statistical analyses
Additive SNP genotypes were tested for association with heroin addiction, separately by ancestry, using logistic regression models in SAS® software (SAS Institute, Cary, North Carolina) adjusted for sex and principal component eigenvectors. In each ancestry group, 10 eigenvectors were generated using EIGENSTRAT (Price et al., 2006); among these, the three eigenvectors which together explained >90% of the variance in heroin addiction case/control status were included as covariates in our regression models to circumvent any potential bias due to population stratification.
To infer the haplotypes occurring between the two ZNF804A SNPs, we phased the study genotypes in each ancestry group using the ShapeIT (version 2) program (Howie et al., 2012) with all 1000 Genomes reference haplotype panels, 500 conditioning states, recommended effective population sizes of 15,000 for African Americans and 11,418 for European Americans, and default settings for all other program options specified. Logistic regression models were used to test the two-SNP haplotypes (coded as dummy variables) for association with heroin addiction, adjusted for sex and eigenvectors. Ancestry-specific SNP and haplotype association results were combined using fixed-effects, inverse variance-weighted meta-analysis, the most commonly used approach for large-scale genetic association meta-analysis (Panagiotou et al., 2013). Fixed-effects methods, which optimize power for discovery analyses (Pereira et al., 2009), assume that heterogeneity across study-specific results are due to random error rather than true population differences; the inverse variance weighting involves taking the inverse of each study-specific standard error estimate and accounting for the direction of association (i.e., sign of the β) to generate combined regression coefficients.
Results
The ZNF804A SNP association results are shown in Table 2. Rs7597593-T was the minor allele in European Americans (frequency=38.9%, similar to the 39.5% observed in Han Chinese [Sun et al., in press]) but the major allele in African Americans (frequency=63.8%). Rs1344706-A was the major allele in both the European Americans (frequency=61.1%) and African Americans (frequency=91.5%), compared to its 48.4% frequency in the Han Chinese (Sun et al., in press). Both SNPs were associated at P<0.05 in European Americans with heroin addiction cases being 1.15 and 1.14 times as likely to carry the rs7597593-T and rs1344706-A alleles, respectively, compared with controls. Neither allele was significantly associated in the African Americans. However, consistent directions of association were observed across both ancestry groups for rs1344706-A, and it was the only SNP that was significantly associated in the multiancestry meta-analysis (P=0.016).
Table 2.
ZNF804A SNP associations with heroin addiction.
| SNP | Coded allele | Position on chromosome 2 (NCBI build 37) | European Americans (N=711 cases and 6,384 controls) | African Americans (N=1,293 cases and 2,369 controls) | Meta-analysis | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Coded allele frequency (%) | P | OR (95% CI) | Coded allele frequency (%) | P | OR (95% CI) | P | OR (95% CI) | |||
| rs7597593 | T* | 185,533,580 | 38.9 | 0.016 | 1.15 (1.03–1.29) | 63.8 | 0.67 | 0.98 (0.89–1.08) | 0.088 | 1.05 (0.97–1.13) |
| rs1344706 | A | 185,778,428 | 61.1 | 0.029 | 1.14 (1.01–1.29) | 91.5 | 0.27 | 1.10 (0.93–1.32) | 0.016 | 1.13 (1.02–1.25) |
CI, confidence interval; OR, odds ratio
T is the complementary allele to the A allele reported by Sun et al. (in press).
Of the 10,757 participants in our dataset, 45.8% were female (Table 1). We tested both SNPs for interaction with sex, given prior suggestive evidence of such interaction involving rs7597593 and schizophrenia (Zhang et al., 2011). We found no evidence for SNP-by-sex interaction in either the ancestry-specific analyses or meta-analysis (results not shown).
High D′ values between rs7597593 and rs1344706 indicated strong linkage disequilibrium (D′=1 in the African [denoted AFR] and D′=0.96 in the European [denoted EUR] panels from 1000 Genomes). Weaker r2 values between the two SNPs (r2=0.12 in AFR and r2=0.34 in EUR) were constrained by their differing allele frequencies.
The two-SNP haplotype associations with heroin addiction are presented in Table 3. Haplotypes carrying the non-risk rs1344706-C allele were collapsed, due to the low frequency of rs1344706-C/rs7597593-T (<1%) in both ancestry groups, and used as the reference. The haplotype carrying both risk alleles (rs1344706-A/rs7597593-T) was significantly associated with increased risk of heroin addiction in the European Americans-specific analysis (P=0.0064) and the multiancestry meta-analysis (P=0.0066), while the haplotype carrying rs1344706-A without rs7597593-T was not associated.
Table 3.
ZNF804A two-SNP haplotype associations with heroin addiction.
| rs7597593- rs1344706 haplotype | European Americans (N=711 cases and 6,384 controls) | African Americans (N=1,293 cases and 2,369 controls) | Meta-analysis | |||||
|---|---|---|---|---|---|---|---|---|
| Frequency (%) | P | OR (95% CI) | Frequency (%) | P | OR (95% CI) | P | OR (95% CI) | |
| T-A | 38.3 | 0.0064 | 1.20 (1.05–1.37) | 63.5 | 0.39 | 1.08 (0.90–1.30) | 0.0066 | 1.16 (1.04–1.29) |
| C-A | 22.9 | 0.55 | 1.05 (0.90–1.22) | 28.1 | 0.14 | 1.16 (0.95–1.40) | 0.16 | 1.09 (0.97–1.23) |
| C-C or T-C | 38.8 | Reference | 8.5 | Reference | Reference | |||
CI, confidence interval; OR, odds ratio
Table 1 shows that a large portion of our heroin addiction cases also reported abuse (using 10 or more times in the past month) of other drugs, predominantly cocaine. We repeated the haplotype association analyses for heroin addiction after excluding the cases with comorbid cocaine abuse, leaving 490 EA cases and 703 AA cases for comparison to the population control sets. With this reduced sample, the rs1344706-A/rs7597593-T haplotype remained associated in the European Americans (P=0.014, odds ratio [95% confidence interval] = 1.21 [1.04–1.41]); its multiancestry meta-analysis had P=0.065 and a similar magnitude of association (odds ratio [95% confidence interval] = 1.12 [0.9–1.27]) in comparison to the full sample (Table 3).
Discussion
Our study independently replicated the associations of the ZNF804A SNPs rs7597593 and rs1344706 and their haplotype with heroin addiction. Such replicable genetic associations with heroin addiction are limited. This replication may also be viewed as a generalization, as our results showed that the associations initially reported in Han Chinese extend to other ancestries, particularly European ancestry. Sun et al. (in press) first identified these heroin addiction associations in a Han Chinese dataset (N=1,035 DSM-IV defined cases and 2,887 community-based controls). Consistent with the prior report, both the rs7597593-T and rs1344706-A alleles and their corresponding haplotype were associated with increased risk of heroin addiction in our European American dataset. No statistically significant SNP or haplotype associations were observed in our African American dataset, but rs1344706-A and its risk haplotype with rs7597593-T presented consistent directions across the ancestry groups and remained associated at P<0.05 in our multiancestry meta-analyses. Similar magnitudes of rs1344706-A association have now been observed across diverse ancestries: odds ratios of 1.16 in Han Chinese (Sun et al., in press) and 1.13 in our meta-analysis of European Americans and African Americans.
The mechanism underlying ZNF804A function in human brain and heroin addiction risk is not known. However, rs1344706 has strong potential for influencing the ZNF804A gene, and its further characterization may help to elucidate the disease-relevant mechanism. The rs1344706-A risk allele maintains the predicted binding sites for two transcription factors expressed in the brain (Myt1L and POU3F1/Oct-6), and it has been associated with increased ZNF804A mRNA expression in postmortem dorsolateral prefrontal cortex, a highly relevant brain region for addiction (Goldstein & Volkow, 2011), from psychiatrically normal controls (Riley et al., 2010). An allelic specific expression study further suggested that rs1344706 directly affects ZNF804A expression in this brain region (Guella et al., 2014). ZNF804A functions as a transcription factor that regulates genes in the dopaminergic pathway (Girgenti et al., 2012), so increased ZNF804A mRNA expression would be expected to alter downstream effects on dopamine release and synthesis and contribute to heroin addiction risk. Additionally, ZNF804A is most highly expressed in human brain during the fetal period, and rs1344706 has been shown to affect expression of a novel splice variant of ZNF804A that is specific to fetal brain (Tao et al., 2014). This splicing mechanism could underlie the rs1344706 association with heroin addiction.
Rs1344706 may also influence structure of addiction-relevant brain regions. In reporting the rs1344706 association with heroin addiction, Sun et al. (in press) showed that its risk allele and haplotype were associated with greater gray matter volume across several human brain regions; they detected an interaction with heroin addiction specifically in the left sensorimotor cortex, a region that is relevant for developing and preserving addiction, whereby the risk haplotype was associated with greater gray matter volume in heroin abusers but lower grey matter volume in controls. This association involving rs1344706 and grey matter by disease state is consistent with prior findings in healthy individuals (Voineskos et al., 2011) and schizophrenia cases and controls (Nenadic et al., 2015). These studies further linked rs1344706-A to reduced cognitive function (Sun et al., in press, Voineskos et al., 2011).
Rs7597593 has also been implicated for influencing ZNF804A mRNA expression, but the association was found only in females (Zhang et al., 2011). We did not find any evidence for sex-by-genotype interactions in our study. Overall, we replicated the previously observed rs7597593 association with heroin addiction in European Americans, but not in African Americans. Rs7597593 may tag a different causal variant with varying linkage disequilibrium across the ancestral groups, which may be identified by fine mapping this region using a large sample of African Americans.
Our results support ZNF804A as one of the few susceptibility genes for heroin addiction with replicable associations and provide further evidence that this gene has pleiotropic effects on multiple psychiatric diseases. We found that the two-SNP haplotype association with heroin addiction remained after excluding cases with comorbid cocaine abuse, albeit to a lesser extent with the reduced sample size. Given that heroin and cocaine abuse were highly comorbid in the UHS, follow-up in other cohorts will be needed to assess the pleiotropic or independent associations of ZNF804A SNPs with addiction to heroin and other drugs. Taken together with prior work showing that these disease-associated SNPs influence ZNF804A gene regulation in human brain, the ZNF804A gene region merits future study to establish the underlying mechanism that confers risk on heroin addiction.
Acknowledgments
This study was supported by the National Institute of Drug Abuse grant numbers R33 DA027486 and R01 DA026141. Heroin addiction cases were drawn from the Urban Health Study and genotyped at the Center for Inherited Disease Research at Johns Hopkins University. Their data are deposited at dbGaP (phs000454.v1.p1).
Controls for comparison to the Urban Health Study heroin addiction cases were drawn from the following six cohorts in dbGaP. (1) Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the National Institutes of Health (NIH) Genes, Environment and Health Initiative (GEI) (U01 HG004422). SAGE is one of the genome-wide association studies (GWAS) funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the NCBI. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research (CIDR), was provided by the NIH GEI (U01 HG004438), the National Institute on Alcohol Abuse and Alcoholism, National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). The datasets used for the analyses described in this manuscript were obtained via dbGaP accession number phs000092.v1.p1.
(2) The authors acknowledge the contribution of data from “Genetic Architecture of Smoking and Smoking Cessation” accessed through dbGaP accession number phs000404.v1.p1. Funding support for genotyping, which was performed at CIDR, was provided by X01 HG005274. CIDR is fully funded through a federal contract from the NIH to The Johns Hopkins University, contract number HHSN268200782096C. Assistance with genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Funding support for collection of datasets and samples was provided by COGEND (P01 CA089392) and the University of Wisconsin Transdisciplinary Tobacco Use Research Center (P50 DA019706 and P50 CA084724).
(3) Funding support for the GWAS of Ischemic Stroke study was provided through the NIH GEI (U01 HG004436). The GWAS of Ischemic Stroke study is one of the GWAS funded as part of GENEVA under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the NCBI. Funding support for genotyping, which was performed at The Johns Hopkins University CIDR, was provided by the NIH GEI (U01 HG004438) and NIH contract number HHSN268200782096C. Field work for this project was supported by a Cooperative Agreement with the Division of Adult and Community Health, Centers for Disease Control and Prevention; the National Institute of Neurological Disorders and Stroke (NINDS) and the NIH Office of Research on Women’s Health (ORWH) (R01 NS045012); Office of Research and Development, Medical Research Service, Department of Veterans Affairs; and the University of Maryland General Clinical Research Center (M01 RR165001), National Center for Research Resources, NIH. This study used samples from the NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds). The datasets used for the analyses described in this manuscript were obtained via dbGaP accession number phs000292.v1.p1.
(4) Funding support for the GENEVA Prostate Cancer study was provided through the National Cancer Institute (R37 CA054281, R01 CA063464, P01 CA033619, U01 CA0136792, U01 CA098758, and RC2 CA148085) and the National Human Genome Research Institute (U01 HG004726). Assistance with phenotype harmonization, SNP selection, data cleaning, meta-analyses, data management and dissemination, and general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004789). The datasets used for the analyses described in this manuscript were obtained from dbGaP at phs000306.v2.p1.
(5) This work utilized in part data from the NINDS dbGaP database from the CIDR NeuroGenetics Research Consortium (NGRC) Parkinson’s disease study (accession number phs000196.v2.p1).
(6) For the “High Density SNP Association Analysis of Melanoma: Case-Control and Outcomes Investigation,” research support to collect data and develop an application to support this project was provided by P50 CA093459, P50 CA097007, R01 ES011740, and R01 CA133996 (dbGaP accession number phs000187.v1.p1).
LBJ and the spouse of NLS are listed as inventors on Issued U.S. Patent 8,080,371, “Markers for Addiction,” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. Otherwise, the authors have no conflicts of interest.
References
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-IV. 4. American Psychiatric Association; Washington DC: 1994. [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgenti MJ, LoTurco JJ, Maher BJ. ZNF804a regulates expression of the schizophrenia-associated genes PRSS16, COMT, PDE4B, and DRD2. PLoS One. 2012;7:e32404. doi: 10.1371/journal.pone.0032404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein RZ, Volkow ND. Dysfunction of the prefrontal cortex in addiction: neuroimaging findings and clinical implications. Nat Rev Neurosci. 2011;12:652–669. doi: 10.1038/nrn3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossop M, Griffiths P, Powis B, Strang J. Severity of dependence and route of administration of heroin, cocaine and amphetamines. Br J Addict. 1992;87:1527–1536. doi: 10.1111/j.1360-0443.1992.tb02660.x. [DOI] [PubMed] [Google Scholar]
- Guella I, Sequeira A, Rollins B, Morgan L, Myers RM, Watson SJ, Akil H, Bunney WE, Delisi LE, Byerley W, Vawter MP. Evidence of allelic imbalance in the schizophrenia susceptibility gene ZNF804A in human dorsolateral prefrontal cortex. Schizophr Res. 2014;152:111–116. doi: 10.1016/j.schres.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock DB, Levy JL, Gaddis NC, Glasheen C, Saccone NL, Page GP, Hulse GK, Wildenauer D, Kelty EA, Schwab SG, Degenhardt L, Martin NG, Montgomery GW, Attia J, Holliday EG, McEvoy M, Scott RJ, Bierut LJ, Nelson EC, Kral AH, Johnson EO. Cis-Expression Quantitative Trait Loci Mapping Reveals Replicable Associations with Heroin Addiction in OPRM1. Biol Psychiatry. doi: 10.1016/j.biopsych.2015.01.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EO, Hancock DB, Gaddis NC, Levy JL, Page G, Novak SP, Glasheen C, Saccone NL, Rice JP, Moreau MP, Doheny KF, Romm JM, Brooks AI, Aouizerat BE, Bierut LJ, Kral AH. Novel Genetic Locus Implicated for HIV-1 Acquisition with Putative Regulatory Links to HIV Replication and Infectivity: A Genome-Wide Association Study. PLoS One. 2015;10:e0118149. doi: 10.1371/journal.pone.0118149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kral AH, Bluthenthal RN, Lorvick J, Gee L, Bacchetti P, Edlin BR. Sexual transmission of HIV-1 among injection drug users in San Francisco, USA: risk-factor analysis. Lancet. 2001;357:1397–1401. doi: 10.1016/S0140-6736(00)04562-1. [DOI] [PubMed] [Google Scholar]
- Kral AH, Lorvick J, Gee L, Bacchetti P, Rawal B, Busch M, Edlin BR. Trends in human immunodeficiency virus seroincidence among street-recruited injection drug users in San Francisco, 1987–1998. Am J Epidemiol. 2003;157:915–922. doi: 10.1093/aje/kwg070. [DOI] [PubMed] [Google Scholar]
- Mark TL, Woody GE, Juday T, Kleber HD. The economic costs of heroin addiction in the United States. Drug Alcohol Depend. 2001;61:195–206. doi: 10.1016/s0376-8716(00)00162-9. [DOI] [PubMed] [Google Scholar]
- Mistry CJ, Bawor M, Desai D, Marsh DC, Samaan Z. Genetics of Opioid Dependence: A Review of the Genetic Contribution to Opioid Dependence. Current Psychiatry Rev. 2014;10:156–167. doi: 10.2174/1573400510666140320000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morral AR, McCaffrey D, Iguchi MY. Hardcore drug users claim to be occasional users: drug use frequency underreporting. Drug Alcohol Depend. 2000;57:193–202. doi: 10.1016/s0376-8716(99)00048-4. [DOI] [PubMed] [Google Scholar]
- Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand FL, Henders AK, Wallace L, Todorov AA, Schrage AJ, Madden PA, Degenhardt L, Martin NG, Montgomery GW. Association of OPRD1 polymorphisms with heroin dependence in a large case-control series. Addict Biol. 2014;19:111–121. doi: 10.1111/j.1369-1600.2012.00445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenadic I, Maitra R, Basmanav FB, Schultz CC, Lorenz C, Schachtzabel C, Smesny S, Nothen MM, Cichon S, Reichenbach JR, Sauer H, Schlosser RG, Gaser C. ZNF804A genetic variation (rs1344706) affects brain grey but not white matter in schizophrenia and healthy subjects. Psychol Med. 2015;45:143–152. doi: 10.1017/S0033291714001159. [DOI] [PubMed] [Google Scholar]
- Nieratschker V, Nothen MM, Rietschel M. New Genetic Findings in Schizophrenia: Is there Still Room for the Dopamine Hypothesis of Schizophrenia? Front Behav Neurosci. 2010;4:23. doi: 10.3389/fnbeh.2010.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Williams NM, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Gejman PV, Cichon S, Nothen MM, Gill M, Corvin A, Rujescu D, Kirov G, Owen MJ, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Cloninger CR Molecular Genetics of Schizophrenia C. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Panagiotou OA, Willer CJ, Hirschhorn JN, Ioannidis JP. The power of meta-analysis in genome-wide association studies. Annu Rev Genomics Hum Genet. 2013;14:441–465. doi: 10.1146/annurev-genom-091212-153520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira TV, Patsopoulos NA, Salanti G, Ioannidis JP. Discovery properties of genome-wide association signals from cumulatively combined data sets. Am J Epidemiol. 2009;170:1197–1206. doi: 10.1093/aje/kwp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes W, Layne M, Johnston P, Hozik L. Office of National Drug Control Policy, editor. What America’s Users Spend on Illegal Drugs 1988–1998. Washington, D.C: 2000. [Google Scholar]
- Riley B, Thiselton D, Maher BS, Bigdeli T, Wormley B, McMichael GO, Fanous AH, Vladimirov V, O’Neill FA, Walsh D, Kendler KS. Replication of association between schizophrenia and ZNF804A in the Irish Case-Control Study of Schizophrenia sample. Mol Psychiatry. 2010;15:29–37. doi: 10.1038/mp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab SG, Kusumawardhani AA, Dai N, Qin W, Wildenauer MD, Agiananda F, Amir N, Antoni R, Arsianti T, Asmarahadi A, Diatri H, Djatmiko P, Irmansyah I, Khalimah S, Kusumadewi I, Kusumaningrum P, Lukman PR, Mustar L, Nasrun MW, Naswati S, Prasetiyawan P, Semen GM, Siste K, Tobing H, Widiasih N, Wiguna T, Wulandari WD, Benyamin B, Wildenauer DB Indonesian Schizophrenia Genetics C. Association of rs1344706 in the ZNF804A gene with schizophrenia in a case/control sample from Indonesia. Schizophr Res. 2013;147:46–52. doi: 10.1016/j.schres.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Steinberg S, Mors O, Borglum AD, Gustafsson O, Werge T, Mortensen PB, Andreassen OA, Sigurdsson E, Thorgeirsson TE, Bottcher Y, Olason P, Ophoff RA, Cichon S, Gudjonsdottir IH, Pietilainen OP, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Athanasiu L, Suvisaari J, Lonnqvist J, Paunio T, Hartmann A, Jurgens G, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Breuer R, Moller HJ, Giegling I, Glenthoj B, Rasmussen HB, Mattheisen M, Bitter I, Rethelyi JM, Sigmundsson T, Fossdal R, Thorsteinsdottir U, Ruggeri M, Tosato S, Strengman E, Genetic R, Kiemeney LA, Melle I, Djurovic S, Abramova L, Kaleda V, Walshe M, Bramon E, Vassos E, Li T, Fraser G, Walker N, Toulopoulou T, Yoon J, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Jonsson EG, Terenius L, Agartz I, Petursson H, Nothen MM, Rietschel M, Peltonen L, Rujescu D, Collier DA, Stefansson H, St Clair D, Stefansson K Outcome in P. Expanding the range of ZNF804A variants conferring risk of psychosis. Mol Psychiatry. 2011;16:59–66. doi: 10.1038/mp.2009.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strang J, Griffiths P, Powis B, Gossop M. Heroin chasers and heroin injectors: differences observed in a community sample in London, UK. Am J Addict. 1999;8:148–160. doi: 10.1080/105504999305956. [DOI] [PubMed] [Google Scholar]
- Substance Abuse and Mental Health Service Administration; U. S. Department of Health and Health Services, editor. Results from the 2012 National Survey on Drug Use and Health: Summary of National Findings. Rockville, MD: Substance Abuse and Mental Health Services Administration; 2013. NSDUH Series H-46, HHS Publication No. (SMA) 13-4795) [Google Scholar]
- Sun Y, Zhao LY, Wang GB, Yue WH, He Y, Shu N, Lin QX, Wang F, Li JL, Chen N, Wang HM, Kosten TR, Feng JJ, Wang J, Tang Y, Liu SX, Deng GF, Diao GH, Tan YL, Han HB, Lin L, Shi J. ZNF804A variants confer risk for heroin addiction and affect decision making and gray matter volume in heroin abusers. Addict Biol. doi: 10.1111/adb.12233. in press. [DOI] [PubMed] [Google Scholar]
- Tao R, Cousijn H, Jaffe AE, Burnet PW, Edwards F, Eastwood SL, Shin JH, Lane TA, Walker MA, Maher BJ, Weinberger DR, Harrison PJ, Hyde TM, Kleinman JE. Expression of ZNF804A in human brain and alterations in schizophrenia, bipolar disorder, and major depressive disorder: a novel transcript fetally regulated by the psychosis risk variant rs1344706. JAMA Psychiatry. 2014;71:1112–1120. doi: 10.1001/jamapsychiatry.2014.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineskos AN, Lerch JP, Felsky D, Tiwari A, Rajji TK, Miranda D, Lobaugh NJ, Pollock BG, Mulsant BH, Kennedy JL. The ZNF804A gene: characterization of a novel neural risk mechanism for the major psychoses. Neuropsychopharmacology. 2011;36:1871–1878. doi: 10.1038/npp.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HJ, Norton N, Dwyer S, Moskvina V, Nikolov I, Carroll L, Georgieva L, Williams NM, Morris DW, Quinn EM, Giegling I, Ikeda M, Wood J, Lencz T, Hultman C, Lichtenstein P, Thiselton D, Maher BS, Malhotra AK, Riley B, Kendler KS, Gill M, Sullivan P, Sklar P, Purcell S, Nimgaonkar VL, Kirov G, Holmans P, Corvin A, Rujescu D, Craddock N, Owen MJ, O’Donovan MC Molecular Genetics of Schizophrenia Collaboration (MGC), International Schizophrenia Consortium (ISC), SGENE-plus GROUP. Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol Psychiatry. 2011;16:429–441. doi: 10.1038/mp.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Li W, Zhang H, Lv L, Song X, Yang Y, Li W, Yang G, Jiang C, Zhao J, Lu T, Zhang D, Yue W. To the editor: association of ZNF804A polymorphisms with schizophrenia and antipsychotic drug efficacy in a Chinese Han population. Psychiatry Res. 2011;190:379–381. doi: 10.1016/j.psychres.2011.05.031. [DOI] [PubMed] [Google Scholar]
- Zhang F, Chen Q, Ye T, Lipska BK, Straub RE, Vakkalanka R, Rujescu D, St Clair D, Hyde TM, Bigelow L, Kleinman JE, Weinberger DR. Evidence of sex-modulated association of ZNF804A with schizophrenia. Biol Psychiatry. 2011;69:914–917. doi: 10.1016/j.biopsych.2011.01.003. [DOI] [PubMed] [Google Scholar]