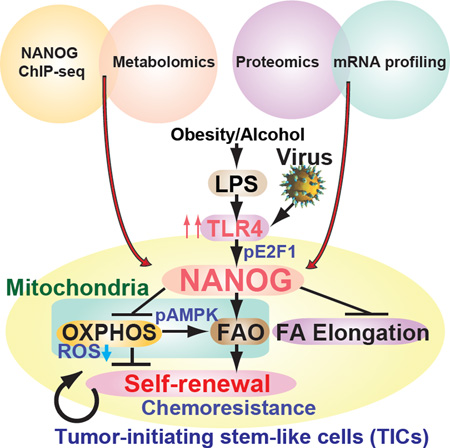
SUMMARY
Stem cell markers such as NANOG have been implicated in various cancers; however, the functional contribution of NANOG to cancer pathogenesis has remained unclear. Here, we show that Toll-like receptor 4 (TLR4) signaling phosphorylates E2F1 to transactivate NANOG. Down-regulation of Nanog reduces tumor progression. NANOG ChIP-seq identified genes associated with NANOG-dependent mitochondrial metabolic pathways to maintain tumor-initiating stem-like cells (TICs). The causal roles of NANOG in mitochondrial metabolic reprogramming occurred through the inhibition of oxidative phosphorylation (OXPHOS) with decreased production of mitochondrial ROS and activation of fatty acid oxidation (FAO), which was required for self-renewal and drug resistance. Restoration of OXPHOS activity and inhibition of FAO rendered TICs susceptible to a standard care chemotherapy drug, sorafenib. This study provides insights into the mechanisms of NANOG-mediated generation of TICs, tumorigenesis and chemo-resistance due to metabolic reprograming of mitochondrial functions.
Keywords: Tumor-initiating stem-like cells (TICs), NANOG, Metabolic reprogramming, OXPHOS, Fatty acid, self-renewal, HCC, Liver
Graphical Abstract
INTRODUCTION
Major risk factors for the third most deadliest cancer, hepatocellular carcinoma (HCC), are hepatitis C virus (HCV), alcoholism, and obesity (He et al., 2008; Okuda et al., 2002). Compelling evidence identifies a synergism between obesity/alcohol and HCV infection with the associated risk of developing HCC (Yuan et al., 2004). The risk of HCC increases from 8–12 to 48–54 by co-morbidities such as alcoholism or obesity (Yuan et al., 2004). Obesity and alcoholism increase gut permeability leading to endotoxemia, which in turn activates Toll-like receptor 4 (TLR4) in the liver with production of cytokines and an inflammatory response. This leads to subsequent development of obesity/alcohol-related liver disease (Hritz et al., 2008). Therefore, an in-depth understanding of the underlying molecular mechanisms regulating obesity/alcohol/HCV-induced hepatocarcinogenesis is essential for the development of improved therapeuties.
By using mice with liver-specific expression of the HCV NS5A protein, we demonstrated that mice fed alcohol for 12 months develop liver tumors in a TLR4-dependent manner (Chen et al., 2013). TLR4 is ectopically induced by the HCV viral protein NS5A in hepatocytes/hepatoblasts. Circulating endotoxin binds TLR4, activates hepatocytes/hepatoblasts and induces the stem cell marker NANOG. This process generates TLR4/NANOG-dependent, chemoresistant tumor-initiating stem-like cells (TICs; CD133+), which can induce HCC in mice (Chen et al., 2013).
TICs are rare, highly malignant cells that are present in diverse tumor types and play a central role in tumorigenesis, malignant progression, and resistance to chemotherapy (Machida et al., 2009; Rountree et al., 2008). Sorafenib, a multi-kinase inhibitor, is the most commonly used monotherapy agent for the treatment of HCC; however, resistance to sorafenib eventually occurs in patients (Villanueva et al., 2008). We recently reported that treatment with sorafenib made TICs more susceptible to tumor growth retardation, with a decrease in tumor size by ~55% when combined with knockdown of NANOG-inducible proto-oncogenes (including YAP1, which induces antioxidant gene programs) (Chen et al., 2013). However, the underlying mechanism of chemoresistance and self-renewal of TICs remains incompletly understood.
We hypothesized that NANOG promotes self-renewal ability, tumor-initiation property, and chemoresistance of TICs through metabolic reprogramming. Our studies showed that oxidative phosphorylation (OXPHOS) and fatty acid oxidation (FAO) were NANOG-mediated oncogenic pathways through metabolic reprogramming as demonstrated by NANOG ChIP-seq analysis and metabolomic profiling.
RESULTS
Microarray and proteomics analysis of three different liver disease models
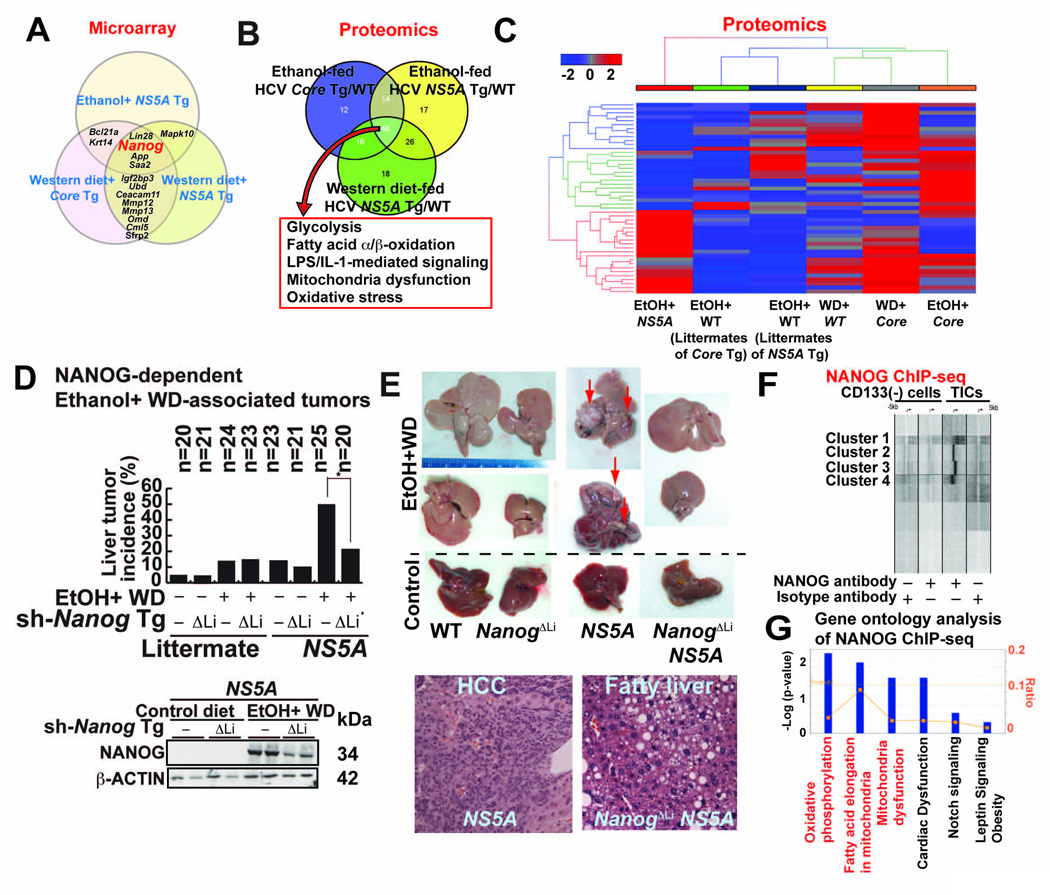
We profiled liver specimens from the alcohol- or obesity-HCV-induced tumor models using microarray and identified Nanog as the most consistently up-regulated gene (Fig. 1A). Using proteomics approach, we further showed that enzymes involved in glycolysis, fatty acid metabolism and mitochondrial respiration were similarly dysregulated in the three liver tumor models (Fig. 1B, 1C, S1A, S1B and Table S1B).
Fig. 1. NANOG plays a critical role in liver oncogenesis.
(A) Results of gene expression microarray comparing liver cancers arising from feeding of ethanol or Western diet (WD)-fed in HCV NS5A transgenic mice and WD+ HCV Core transgenic (Tg) mice. Venn diagram shows genes associated with each etiology and those shared among two or more liver cancer models.
(B) Summary of proteomic analysis of three mouse liver cancer models listed in A. All models showed similar metabolomic properties as shown in the Venn diagram.
(C) Heat map showing more extensive proteomic signatures in liver cancer models: alcohol+NS5A; alcohol alone, high cholesterol high- fat Western diet (WD); WD+HCV core gene, and alcohol + HCV core gene. Animals used were either wt, transgenic for either NS5A or core, as indicated.
(D) Western diet (WD) combined with alcohol increased tumor incidence in NS5A Tg mice compared to control. Upper panel-tumor incidence percentage. Lower panel-immunoblot of Nanog expression. Sh-Nanog Tg indicates animals receiving inducible transgene for NANOG silencing.
(E) Liver tumor formation in NANOG and NS5A Tg mice. Upper panel, liver tumors arising from NS5A and Nanog showing contributions of alcohol+Western diet. Knockdown of NANOG (ΔLi), as indicated, reduced tumor incidence in wt control and NS5A mice. Lower panel-liver histology showing pathology is increased following Nanog knockdown in NS5A Tg mice.
(F) NANOG ChIP analysis: comparison of promoter fragments from CD133− and CD133+ cell populations.
(G) Summary of gene ontology families identified by NANOG ChIP-seq analysis.
NANOG plays a critical role in liver oncogenesis
In addition to the effects of diet and alcohol on HCC in wt mice, nearly 50% of the HCV transgenic mice fed ethanol-containing Western diet (WD: high in cholesterol and saturated fat) developed liver tumors. This incidence was reduced by 80% in the liver-specific Nanog knockdown (ΔLi) cohort (Fig. 1D, 1E and Table S2 for histological scores), thus demonstrating the critical role of Nanog in tumor development via obesity/alcohol-HCV interactions.
To investigate the underlying mechanism of TIC-mediated tumorigenicity, we conducted a genome-wide transcriptional profiling of NANOG-promoter interactions in TICs with a ChIP-seq approach using a NANOG-specific antibody. We identified NANOG enrichment proximal to transcription start sites (TSS) in TICs compared to CD133(−) cells (Fig. 1F, Fig S1C and Table S3 for Nanog target genes). An Ingenuity Pathway analysis implicated the involvement of mitochondrial functions, including OXPHOS-related and FAO genes (Fig. 1G and Fig. S1D). Furthermore, a bioinformatics sequence analysis of NANOG-enriched promoter fragments showed the presence of other consensus binding sites similar to the STAT3-consensus-binding motif (Fig. S1E). NANOG has been shown to physically binds STAT3 (Torres and Watt, 2008). To demonstrate that NANOG induction resulted in increased occupancy of the STAT3 promoter; we used Nanog silenced cells which are in reduced STAT3-mediated transcriptional activity (Fig. S1F), indicating that Nanog interaction with STAT3 is functionally relevant.
The tumor incidence in several HCC mouse models is TLR4/NANOG-dependent
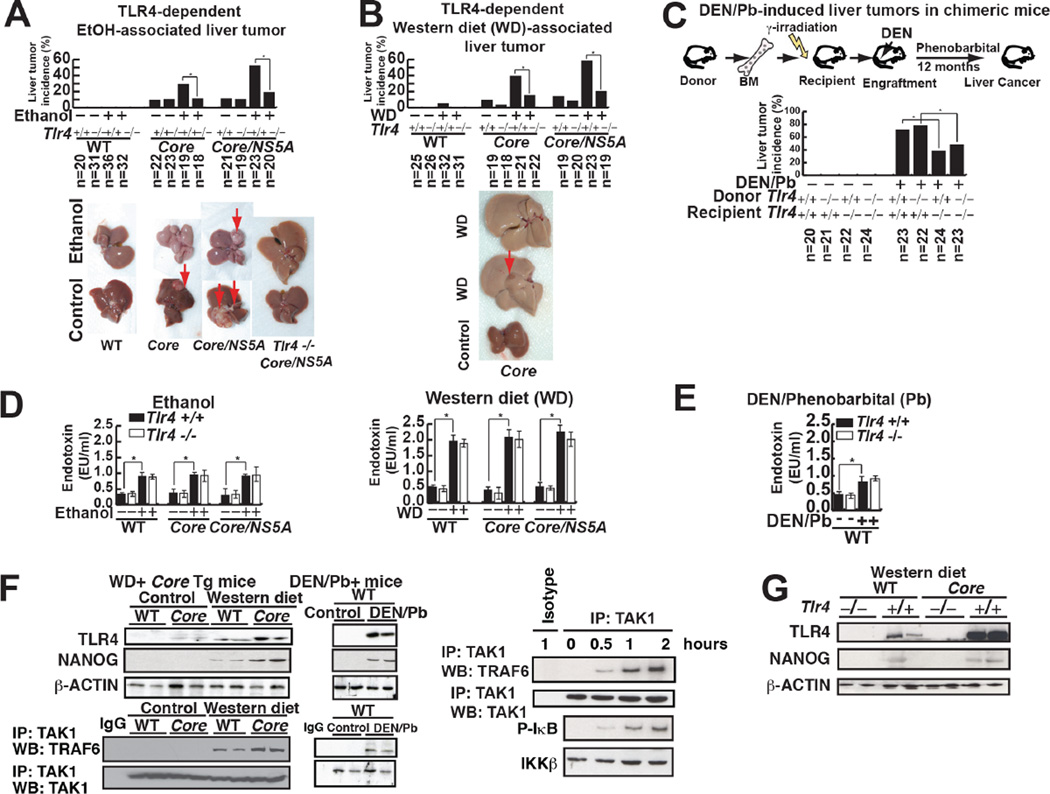
Long-term (12 months) feeding of alcohol diet or a Western diet induced liver tumors in overexpressing HCV non-structural protein NS5A (Majumder et al., 2002), HCV structural protein Core or Core/NS5A transgenic (Tg) mice. Liver tumor incidence was significantly reduced in mice with a Tlr4−/− background (Fig. 2A, B). Tumors from NS5A Tg mice fed WD expressed α-fetoprotein (AFP) (Fig. S2A) and histological analysis confirmed that morphological feature of tumors includes HCC and dysplastic nodules (Fig. S2B and Table S2 for histological scores). Plasma levels of lipopolysaccharide (LPS), a ligand of TLR4, were equally elevated in both Tlr4+/+ and Tlr4−/− mice fed a WD (Fig. 2D).
Fig. 2. The tumor incidence in several HCC mouse models is TLR4-dependent.
(A) Effect of alcohol feeding on tumor formation in Core and NS5A transgenic mice. Upper panel summary of liver tumor incidence among experimental animals: wt, HCV-Core (Core) and HCV-Core+NS5A (Core/NS5A) transgenic mice. Lower panel-the effect of ethanol feeding to wt and transgenic mice on liver tumor development.
(B) TLR4 is required for tumor development in Western diet fed (WD) HCV Tg mice. Upper panel-tumor incidence among wt and transgenic mice fed WD. Lower panel-the effect of WD on wt and Tg mice tumor development.
(C) TLR4-dependent TICs from DEN/Phenobarbital (Pb) and human HCC (non-viral etiology: no HCV) models. Chimeric mice were generated by transplantation of BM from Tlr4+/+ or Tlr4−/− mice into irradiated Tlr4+/+ or Tlr4−/− mice. DEN-Pb-induced tumor incidence is reduced by recipient TLR4 deficiency but not by donor Tlr4 deficiency. (D and E) Effect of alcohol and WD feeding on plasma endotoxin levels in transgenic mouse genotypes. Alcohol WD feeding equally elevated plasma endotoxin levels in all genotype mice. Serum endotoxin levels are elevated equally in both Tlr4+/+ and Tlr4−/− mice fed ethanol or WD (D) or diethylnitrosamine (DEN)/phenobarbital (Pb) treatment (E). Left panel-effect of alcohol feeding on single and double-transgenic animals. Right panel-effect of WD feeding on single and double transgenic animals. Plasma endotoxin levels were measured in wt, HCV-Core, HCV-NS5A and Tlr4−/− double Tg strains of the wt and single Tg animals, as indicated.
(F) TLR4 is induced in the liver tumor samples from the DEN/Phenobarbital HCC models. Activated TLR4 signaling is evident only in HCV Core fed Western diet (WD) as demonstrated by co-immunoprecipitation analysis showing TRAF6 interaction with TAK1. Induction of TLR4 and NANOG were detected in HCV Core Tg livers fed WD or DEN/Phenobarbital-treated mice.
(G) Tlr4 mutant mice fed Western diet for 12 months have less NANOG protein levels.
To determine whether resident liver cells (e.g., hepatocytes) or bone marrow (BM)-derived cells (Kupffer/lymphoid cells) are the primary site of TLR4-dependent oncogenic effects, cross-BM transplantation experiments were performed between Tlr4−/− and Tlr4+/+ mice prior to Diethylnitrosamine/Phenobarbital (DEN-Pb) treatment (Fig. 2C). Chimeric mice were generated by transplantation of BM from either Tlr4+/+ or Tlr4−/− mice into irradiated Tlr4+/+ or Tlr4−/− recipients, as previously described (Dapito et al., 2012). To test whether Tlr4−/− BM engrafted irradiated mice recipients lacked LPS-mediated IL-6 induction, we analyzed LPS-mediated IL-6 induction in splenocytes isolated from engrafted recipient mice (Fig. S2C and S2D). Transplantation of Tlr4−/− bone marrow cells into Tlr4+/+ mice had no influence on tumor incidence compared to transplantation of Tlr4+/+ BM cells into Tlr4+/+ mice. In contrast, transplantation of Tlr4+/+ BM cells into Tlr4−/− recipient mice resulted in significantly decreased tumor incidence, indicating that liver parenchymal cells, but not bone marrow-derived immune cells, were responsible for tumor development as previously described (Dapito et al., 2012) (Fig. 2C). Additionally the DEN-Pb-induced HCC tissues exhibited upregulated and activated TLR4 (Fig. 2F, right panel) with increased plasma endotoxin levels (Fig. 2E). These results demonstrate that elevated endotoxin and TLR4 levels in liver parenchymal cells promote HCC progression.
We previously showed that ectopic TLR4 expression in hepatocytes/hepatoblasts mediated by HCV NS5A activates the stem cell marker Nanog to promote HCC development (Chen et al., 2013). We found that TLR4-NANOG signaling is activated in other HCC models, such as HCV viral protein Core-WD and DEN-Pb-induced HCC models (Fig. 2F). We analyzed the expression levels of TLR4 and its downstream signaling proteins TAK1-TRAF6, revealing that TAK1-TRAF6-association is increased in WD-fed mice and carcinogen-injected mice (Fig. 2F). Ubiquitination links TAK1/TBP1/2 complex with TRAF6-TRIF complex. Accordingly, Core-Tlr4−/− mice fed a WD did not express NANOG (Fig. 2G). The absence of Tlr4 in WD fed mice, prevented tumorigenesis, thus supporting a causal link between Tlr4 and NANOG.
Lastly, we isolated TICs from HCC mouse models and analyzed whether TLR4 and NANOG influenced their tumor initiating property in immunocompromised mice. Silencing Tlr4 in TICs inhibited stemness gene expression as determined by qRT-PCR (Fig. S2G). Silencing Tlr4 or Nanog inhibited tumor growth in immune-compromised mice (Fig. S2H and S2I), indicating that TLR4-NANOG plays a key role for liver oncogenesis.
TLR4-TAK1/TBK1-mediated E2F1 phosphorylation transactivates NANOG through E2F1-binding sites
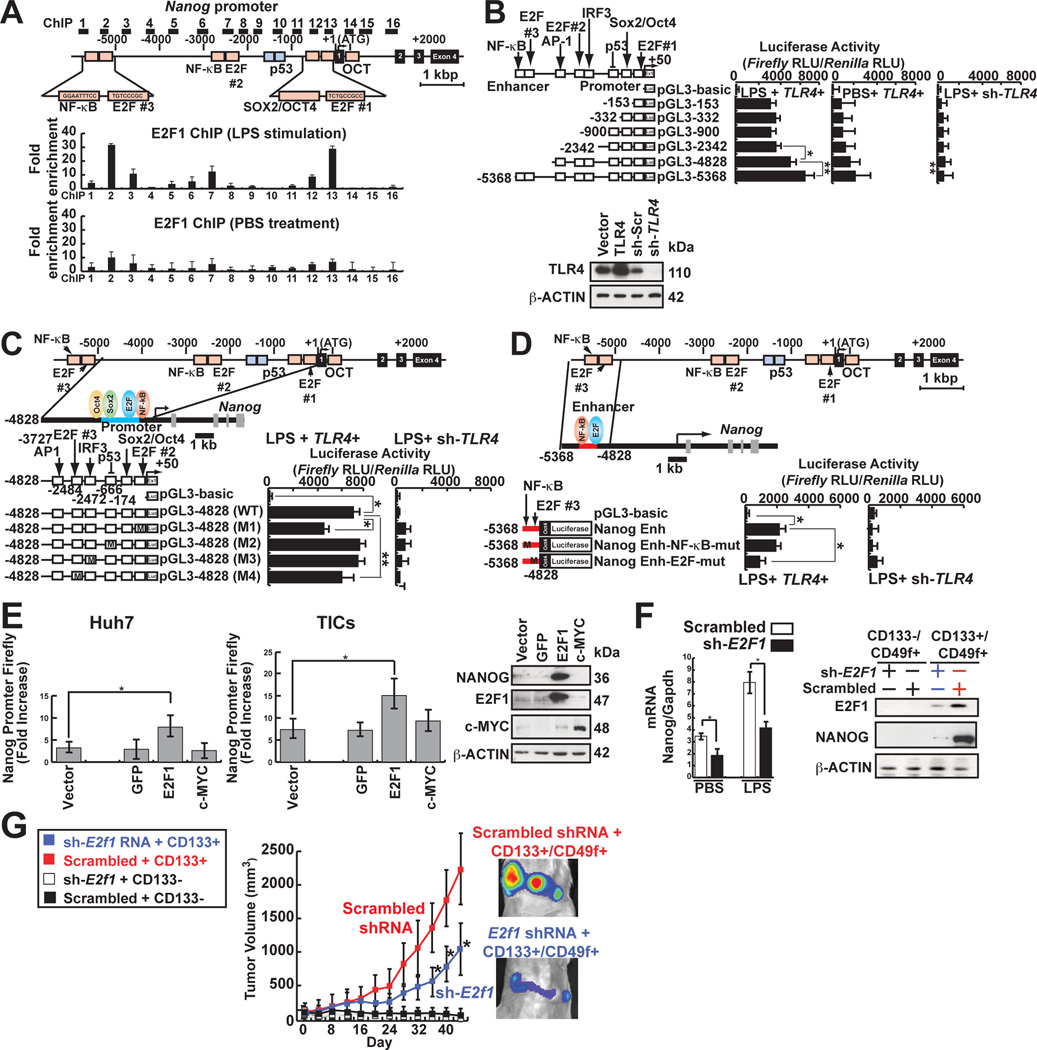
To understand the regulation of NANOG in TLR4 activation, endogenous NANOG promoter activity was monitored. HCV-infected Huh7 cells stimulated with LPS showed increased NANOG promoter activity (Fig. S3A and S3B). A genome-wide mapping of mouse ES cell transcription factors showed binding of E2F1, ESRRB, and TCFCP2I1 to the Nanog enhancer (Wang et al., 2007; Wu et al., 2006). To determine if E2F1 binds to its cis-elements, ChIP-qPCR analysis was performed. We observed E2F1 was enriched in the Nanog distal enhancer and the promoter proximal region in response to LPS (Fig. 3A). Deletion of the distal enhancer region between nucleotides −5368 to −4828 and the promoter proximal region from −153 to +50 containing E2F1 binding sites reduced Nanog promoter activity following LPS stimulation in Huh7 cells (Fig. 3B). Furthermore, base substitution mutations of the promoter proximal region (−153 to +50; Fig. 3C) and the distal enhancer (nt −5368 to −4828; Fig. 3D) reduced NANOG promoter activity.
Fig. 3. TLR4 signaling transactivates Nanog promoter through E2F1-binding sites.
(A) Nanog promoter ChIP assay using anti-E2F1 antibody following qPCR in TICs. E2F1 showed enrichment in Nanog promoter.
(B) Truncated promoter luciferase constructs were used to map the region responsive to TLR4 signaling. LPS-mediated Nanog promoter activity is compare with PBS (Vehicle)-treated cells to demonstrate Nanog promoter activity upon TLR4 stimulation (n=3). *: P < 0.05, **: P < 0.01.
(C and D) E2F1-binding sites were required for efficient Nanog transactivation (n=4). Four mutant-luciferase plasmids were constructed by in vitro mutagenesis “M” indicates the sites of mutation.
(D) E2F1 (blue oval) regulated Nanog enhancer for LPS-induced activation.
(E) Overexpression of E2F1 resulted in NANOG promoter activation. Gfp, E2F1 or c-MYC were overexpressed in Huh7 cells and examined for luciferase reporter activities in response to LPS stimulation.
(F) Silencing E2F1 reduced Nanog mRNA and protein levels in response to LPS.
(G) Silencing E2F1 reduced tumor growth in NOG mice.
E2F1 overexpression in TICs and Huh7 cells significantly increased Nanog promoter activity and protein level (Fig. 3E), indicating that E2F1 transcriptionally activates NANOG in TICs. Furthermore, shRNA-mediated knockdown of E2F1 reduced TLR4-induced NANOG transcriptional activation (Fig. 3F) and subsequent tumor development in immunocompromised NOG mice (Fig. 3G).
To determine if TLR4 signaling activates E2F1 via phosphorylation, we analyzed candidate adapter molecules/kinases in the TLR4 signaling cascade, namely TBK1, TAB1, IRF3, TRAF6 and TAK1. Using a lentiviral shRNA-mediated knockdown approach in TICs, we demonstrated that TLR4-activated TAK1 and TBK1 resulting in E2F1 phosphorylation at Ser337 and Ser332, respectively (Fig. S3C and S3D). Knockdown of either TAK1 or TBK1 in TICs did not significantly reduce TLR4-mediated NANOG induction (Fig. S3E). However, silencing of both TAK1 and TBK1 significantly reduced NANOG protein levels (Fig. S3F). Furthermore, transduction of the phospho-mimetic mutant of E2F1 (S332D/S337D) more efficiently transactivated NANOG in comparison to the wt E2F1 in sh-Tlr4-Huh7 cells (Fig. S3G). Overexpression of non-phosphorylatable mutant of E2F1 (S332A/S337A) inhibited induction of NANOG mRNA in Huh7 cells (Fig. S3G), indicating that phosphorylation of E2F1 is crucial for NANOG expression and full activation of Nanog-dependent promoters.
TICs were further analyzed following transduction of shRNAs targeting Tlr4 in combination with wild type E2F1 or E2F1 (S332A/S337A) mutants to test if constitutively active E2F1 transactivated NANOG (Fig. S3H). We observed that constitutively active E2F1 in both Tlr4+/+ TICs and in sh-Tlr4-TICs significantly induced NANOG expression while in shTlr4-TICs, E2F1 overexpression did not affect NANOG expression (Fig. S3H, left panel), consistent with a requirement of E2F1 post-translational modification downstream of TLR4 signaling to induce NANOG (Fig. S3I). Thus, these results demonstrated that TLR4-mediated activation of NANOG by LPS required phosphorylation of E2F1 at both S332 and S337 by TBK1 and TAK1, respectively (Fig. S3J).
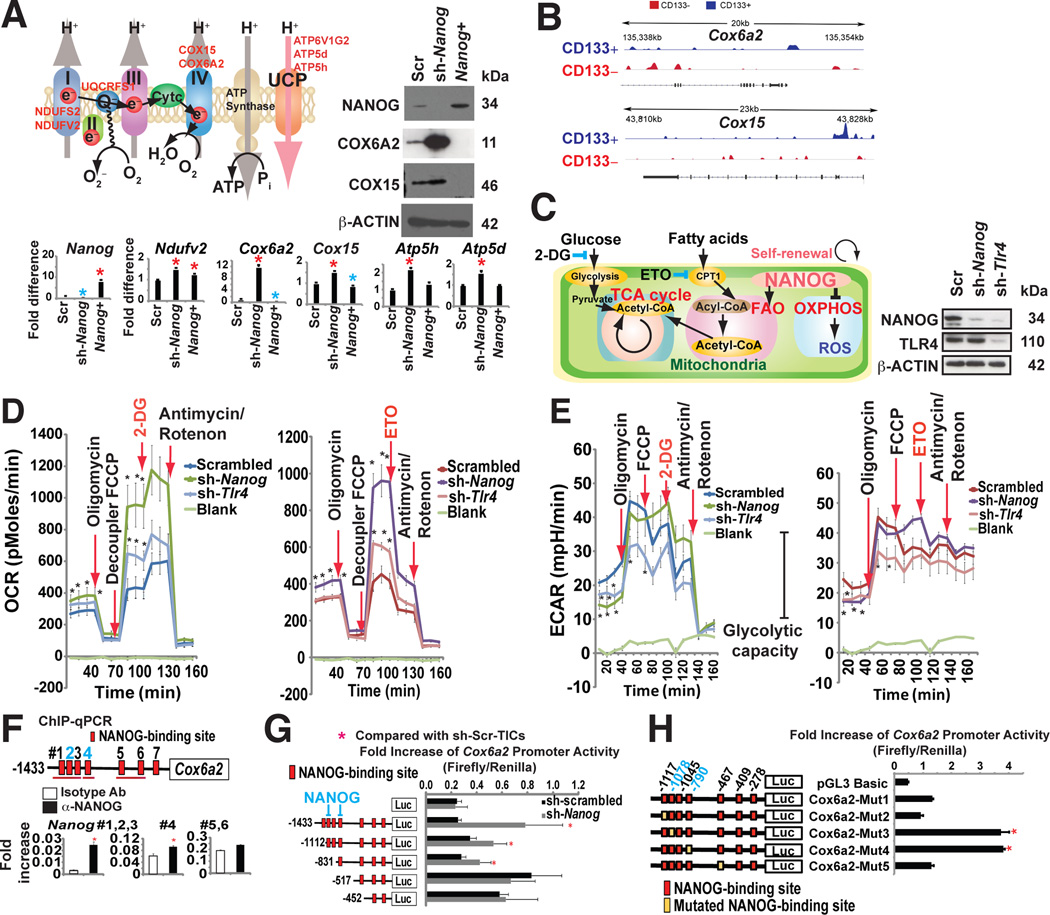
NANOG reduces mitochondrial OXPHOS
Although the NANOG regulon comprises a large number of genes, we examined the importance of metabolic genes, especially those participating in oxidative phosphorylation based on our gene ontology analysis of Nanog ChIP-seq results. Nanog overexpression decreased OXPHOS activity, whereas Nanog knockdown using shRNA significantly up-regulated the OXPHOS genes and corresponding respiratory activity in TICs (Fig. 4A).
Fig. 4. NANOG reduced mitochondrial OXPHOS preventing mitochondrial ROS production.
(A) Effect of NANOG on expression of selected OXPHOS enzymes. A representative model (inset) shows the putative relationship of Nanog silencing to corresponding increased OXPHOS gene expression in TICs.
(B) NANOG ChIP-seq analysis identified mitochondrial OXPHOS genes are major NANOG regulated genes.
(C) The Etomoxir (ETO: CPT1 inhibitor)-blocked component of oxygen consumption rate (OCR) and glycolysis inhibitor 2-deoxyglucose (2-DG)-blocked the glycolytic component of extracellular acidification rate. ETO inhibits CPT1 to block entry of fatty acid into mitochondria.
(D) Seahorse assays demonstrated that NANOG silencing promoted increased oxygen consumption rate (OCR). Effects of oligomycin, FCCP, 2-DG or ETO, and antimycin/rotenone on OCR were evaluated in the Nanog-silenced TICs (sh-Nanog) and scrambled shRNA-transduced TICs. Real-time measurement of OCR showed that ETO but not 2-DG abrogated FCCP-induced OCR. NANOG silencing switched between FAO and glucose utilization (an adult-like metabolic pattern).
(E) Silencing Nanog or Tlr4 reduced ECAR, demonstrating that inhibition of Nanog or Tlr4 reduced glycolytic activity. (Left and Right) ECAR of sh-Scr-TICs represented with a dark blue plot cells that were transduced with lenti-sh-Nanog is shown with a dark green line and lenti-sh-Tlr4 transduced cells are indicated by a light blue line (Left) ECAR represents the sum of FAO and glycolysis, respectively. ECAR measurement after ETO inhibition of β-oxidation showed a rapid decrease of glycolysis only in the sh-Nanog-TICs; yet, sh-Scr-TICs, ETO did not affect ECAR (glycolysis). (Right) ETO treatment does not affect ECAR in sh-Scr-TICs while ETO treatment inhibits ECAR in sh-Nanog-TICs, indicating that TICs are dependent on glycolysis, but inhibition of fatty acid import into mitochondria only inhibits glycolytic activity in sh-Nanog-TICs, but not sh-Scr-TICs.
(F) ChIP-qPCR of NANOG in Cox6a2 promoter of TICs.
(G) Truncation of Nanog promoter identified region responsive to NANOG-mediated inhibition (n=3, *: P < 0.05). Promoter activity increased by deletion of the promoter segment containing critical cis-element(s).
(H) Mutagenesis in NANOG binding sites (−1078 and −790) promotes Cox6a2 promoter activity (n=3, *: P < 0.05).
To test if NANOG regulates mitochondrial respiration, we examined the oxygen consumption rate (OCR) in TICs. The basal OCR rates increased in Nanog- or Tlr4-silenced TICs, compared to untransduced cells (Fig. 4D and Fig. S4A–C). Silencing Nanog reduced glycolytic activity as demonstrated by a decline of extracellular acidification rate (ECAR) (Fig. 4E). Addition of Etomoxir (ETO), an inhibitor of FA transporter (CPT1), antagonized the FCCP-induced OCR in Nanog silenced TICs (Fig. 4C–D), indicating NANOG inhibits mitochondrial respiration.
We the analyzed the expression of the cytochrome c oxidase subunit 6A (Cox6a2) gene since it was the most downregulated OXPHOS gene in our ChIP-seq (Fig. 4B) and qRT-PCR (Fig. 4A) analyses. Additionally, Nanog enrichment was observed in the Cox6a2 promoter of TICs via ChIP-qPCR analysis (Fig. 4F). Knockdown of Nanog in TICs increased transcription from the Cox6a2 promoter (nt −1433 to +17), which was suppressed in TICs. A deletion in the Cox6a2 promoter from nt −1433 to −518 significantly increased the Cox6a2 promoter activity in TICs (Fig. 4G), indicating that this region had negative-regulatory activity. Further bioinformatics analysis confirmed putative Nanog binding sites in the region spanning nt −1433 to −518. Mutations in the Nanog binding sites (−1078 and −790) restored Cox6a2 promoter activity in TICs (Fig. 4H). These results show that decreased mitochondrial activity in TICs resulted from Nanog-mediated repression of Cox6a2.
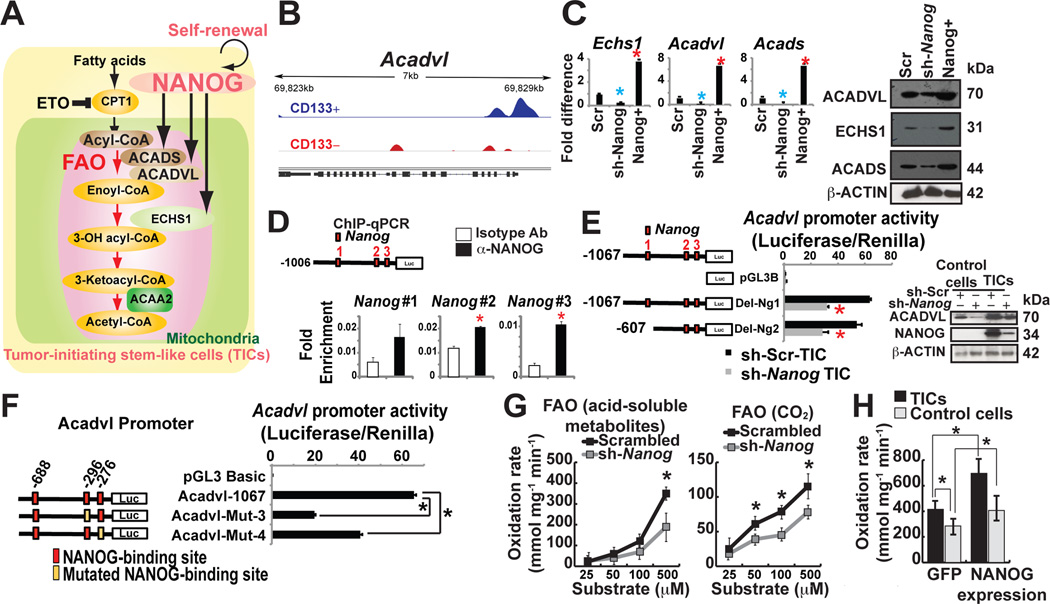
NANOG promotes mitochondrial FAO
Since silencing NANOG increased OXPHOS levels in TICs (Fig. 4D, left panel), we reasoned that NANOG should activate additional catabolic pathways, such as FAO, in order to meet the cellular energy demands (Fig. 5A). Based on our gene ontology analysis of the Nanog-ChIP-seq data, the lipid metabolism pathways appeared to be a critical property of TICs (Fig. 1G; Fig. 5B), suggesting that fatty acids were an alternative energy source (Fig. 5A). Targeted expression analysis by qRT-PCR and immunoblot analyses showed that genes associated with the FAO pathway (viz., Acadvl, Echs1, Acads) were significantly expressed in TICs; Nanog knockdown in TICs down-regulated both transcripts and protein levels of FAO associated genes (Fig. 5C).
Figure 5. NANOG promoted mitochondrial FAO.
(A) Hypothetical model of NANOG-mediated metabolic reprogramming.
(B) NANOG ChIP-seq analysis identified FAO genes are NANOG regulated genes (i.e., Acadvl).
(C) qRT-PCR and immunoblot analysis of NANOG-target FAO genes in sh-Nanog or Nanog-overexpressing TICs.
(D) ChIP-qPCR of NANOG in the Acadvl promoter in TICs.
(E, left) Acadvl promoter luciferase constructs used to map the region responsive to NANOG-mediated inhibition. (E, right) Protein levels of FAO enzymes (ACADVL) were measured by immunoblot in TICs expressing sh-NANOG.
(F) Mutations in NANOG binding sites in ACADVL promoter reduced ACADVL promoter activity.
(G) FAO was measured by incubation of extracts from sh-scrambled and sh-Nanog-TICs with [14C]palmitate; recovery of acid-soluble metabolites (G, left) and captured 14CO2 (G, right) (n = 5 per genotype, *P < 0.05).
(H) Overexpression of Nanog induced FAO oxidation rate. FAO was measured by incubation of [3H] palmitate in TICs transfected with NANOG-expression vectors and vector control one week after transfection of vectors expressing GFP or sh-Nanog (acid-soluble metabolites, n = 3–4 per category). *P < 0.05.
We focused our analysis of Acadvl in TICs since a significant enrichment of Nanog was observed in the Acadvl promoter by ChIP-seq (Fig. 5B), which was further validated by ChIP-qPCR analyses (Fig. 5D). In reporter assays, full-length Acadvl promoter (nt −1067 to +1), and a truncated promoter (nt −607 to +1) showed significant decreases in their activities when Nanog was silenced in TICs (Fig. 5E). Mutations in two of the three Nanog binding sites further reduced Acadvl promoter activity (Fig. 5F) indicating that Nanog binding sites proximal to the transcription start site were essential for Acadvl transactivation.
To determine if NANOG induced FAO in TICs, we used FAO flux analysis with 14C-radiolabeled-palmitic acid for production of acid-soluble 14C metabolites and 14CO2. NANOG+-TICs have significantly higher levels of FAO activity (oxidation rate) under physiological conditions (Fig. 5H). Silencing of Nanog in TICs resulted in significantly reduced FAO activity, indicating TICs dependence on NANOG for FAO activation (Fig. 5G).
To determine whether NANOG contributed to peroxisome FAO activity, we assayed the contribution of Nanog-target Acaa1a, a peroxisome FAO-related gene, by shRNA-transduced TICs. We observed Nanog silencing did not alter Acaa1a expression (Fig. S5A), thus substantiating our finding that, NANOG regulated the mitochondrial FAO genes of TICs for satisfying energy requirements.
We next determined if the observed effect of Nanog on Cox6a2, and activating effect on Acadvl were related to tumor incidence. Cox6a2, Cox15, Acadvl and Echs1 mRNAs were quantified by qRT-PCR in tumor-bearing mice and compared to non-tumor-bearing mice. This in vivo analysis further corroborated the ex vivo findings that mitochondrial FAO genes (Acadvl and Echs1) were turned on in the cancerous regions whilst not in non-cancerous liver tissues. In particular, tumors in endogenous cancer models exhibited lower levels of Cox6a2 and Cox15 (Fig. S5B–D). NANOG suppression in whole liver reduced hepatocarcinogenesis. Acadvl is overexpressed in a small fraction of tumors (8%) (Fig. S5B–D), demonstrating that not all tumor cells have elevated levels of ACADVL. These results strongly support our model that in TICs, NANOG regulates mitochondrial FAO gene expression to sustain energy production in TICs for tumor survival and growth.
PPARδ physically interacts with NANOG
TICs downregulated key transcription factors involved in cell differentiation. Based on our NANOG ChIP-seq analysis in TICs, the peroxisome proliferator-activated receptors (PPARs; nuclear receptor proteins which function as transcription factors in cell differentiation) were strongly associated with NANOG. To determine whether the expression of NANOG specifically regulated PPAR (α, δ, γ, γ2) and repressed their pro-differentiation activities, we performed Nanog loss or gain of function analysis.
Since activation of PPARδ is known to increase FAO in cells (Gutgesell et al., 2009) we next studied the induction of PPARδ upon overexpression of Nanog (Fig. S5E). Sequential-ChIP-qPCR analysis on the Acadvl locus in TICs demonstrated that both NANOG and PPARδ were co-enriched at the Acadvl locus (Fig. S5F) thereby suggesting a possible cooperative interaction to increase FAO in TICs. Co-immunoprecipitation of HA–NANOG (Fig. S5G) and PPARs demonstrated that NANOG bound specifically to PPARδ, but not to PPARγ, PPARγ2, or PPARa. Further truncation analysis of the Acadvl promoter demonstrated that the C-terminal end of PPARδ (aa 155–266), but not the N-terminal region (aa 1–155), interacted with NANOG (Fig. S5H). This indicated that the C-terminus of PPARδ (155–266) mediated interaction with NANOG for regulation of PPARδ-dependent transcription. Overexpression of either PPARδ or NANOG in TICs increased the rate of FAO (Fig. S5I). Conversely, Nanog silencing reduced the rate of FAO (Fig. S5I), whichwas consistent with the observation that NANOG interacted with PPARδ in the Acadvl locus to increase FAO.
We next performed metabolomics analysis to assesstricarboxylic acid cycle (TCA cycle) activity in TICs. NANOG-deficient TICs exhibited elevated external levels of glutamate, proline and α-ketoglutarate in growth media in comparison to those in scrambled shRNA- transduced TICs (Fig. S6A), whilst many other amino acids (e.g., threonine, asparagine and tryptophan) were reduced in media from NANOG-deficient TICs (Fig. S6A). This indicated that sh-Nanog TICs consumed more amino acids, reflecting a change in respiration activity. Several amino acids (glutamate, glutamate γ-methylester and proline) increased within TICs in comparison to those in normal hepatocytes while other amino acids (asparagine, glutamine and cysteine) were depleted (Fig. S6B and S6C). The oxidized form of antioxidant glutathione (GSH) was reduced in TICs in comparison to that of primary hepatocytes (Fig. S6C). Indeed, NANOG ChIP-seq analysis identified antioxidant genes involved in non-canonical glutamine metabolism (Got2 and Glutathione reductase, Gsr) (Fig. S6D). These observations suggested increased energy production occurred through the TCA cycle or alternatively decreased utilization of citrate for fatty acid synthesis was occurring in NANOG-deficient TICs (Fig. S6E and S6F).
To assess functionality of the mitochondrial TCA cycle in TICs, we tested the possibility that glutamine utilization (an anaplerosis fuel in cancer cells) in the TCA cycle occurred in TICs. The sh-Tlr4 or sh-Nanog silenced TICs showed reduced glutamine uptake as evident by decreased percentage of M3 and M4 glutamate (C2-C4) and M4 and M5 (C2-C5) fragments (Fig. S6G and S6H). However, sh-TLR4 or sh-Nanog silencing of these cells enhanced TCA cycle activity, as demonstrated by the increased generation of M0 and M1 glutamate in both fragments. These results showed that sh-Tlr4 or sh-Nanog silencing promoted glutamine oxidation in mitochondria. We also observed Nanog silencing induced increased glucose flux through the TCA cycle via pyruvate carboxylase pathway, but not PDH (Fig. S6I–M). Finally, glutamine withdrawal reduced cell growth rate in all TICs in the presence or absence of TLR4 or NANOG (Fig. S6Q). These results showed TICs have striking intrinsic differences in cellular metabolism, compared to normal cell types.
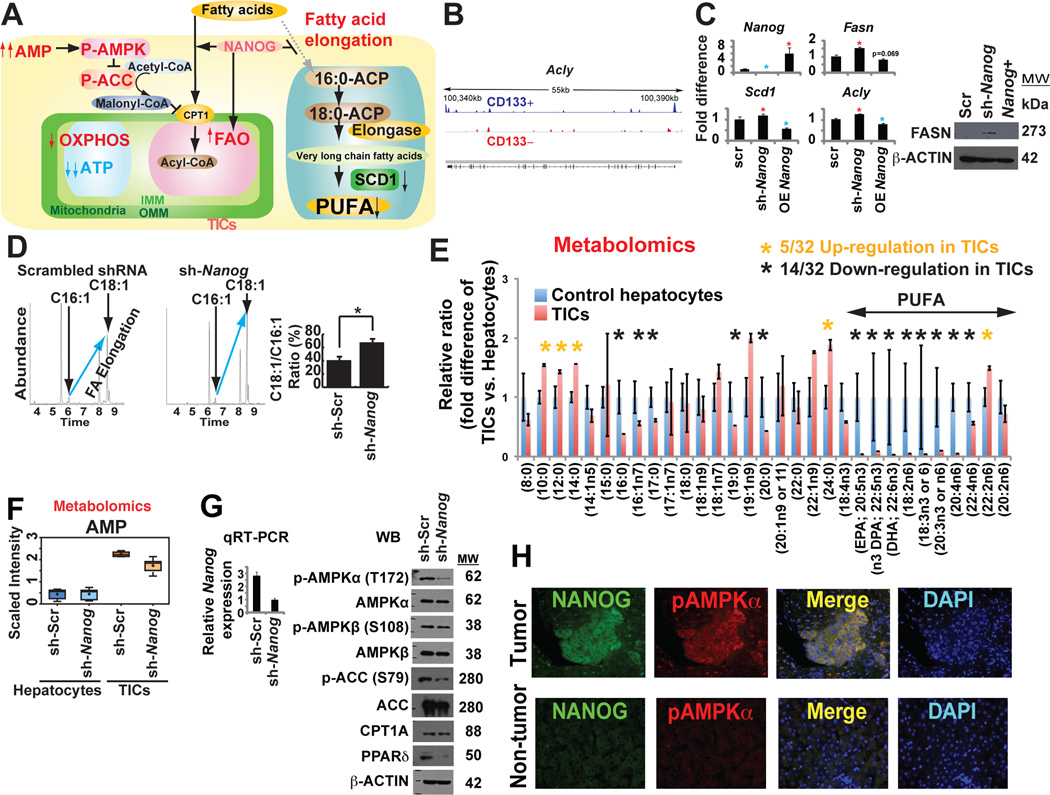
NANOG orchestrates mitochondrial metabolic reprogramming
We have observed that NANOG was important for OXPHOS inhibition and FAO activation. We studied the effect of NANOG on inhibition of fatty acid elongation enzymes by NANOG (Fig. 6A). ChIP-seq showed that NANOG was enriched in the Acly promoter (Fig. 6B). Moreover, fatty acid synthesis genes (Scd1, Fasn and Acly) were derepressed in sh-Nanog-TICs compared to sh-scrambled-TICs (Fig. 6C). Based on the fatty acid elongation profile determined by GC/MS, the rate of fatty acid elongation [ratio of oleate/palmitoleate (C18:1/C16:1 fatty acids)] increased in Nanog-silenced TICs (Fig. 6D), indicating that NANOG inhibited fatty acid elongation.
Fig. 6. NANOG inhibited mitochondrial fatty acid elongation and promotes AMP/ATP ratio increase with AMPKα phosphorylation.
(A) Schematic diagram of the proposed role of NANOG in mitochondrial metabolic reprogramming. AMPK activation in the increased ratio of AMP/ATP leads to phosphorylation of ACC to reduce malonyl CoA levels and thus increase mitochondrial fatty acid uptake (via de-repression of CPT1).
(B) NANOG ChIP-seq analysis identified that FAO elongation genes (i.e., Acly) were NANOG-regulated genes.
(C) qRT-PCR analysis of representative genes associated with fatty acid elongation and synthesis.
(D) Rate of fatty acid elongation was affected in Nanog silenced TICs using GC-MS with stable isotope 14C. The relative ratio of C18:1/C16:1 (oleate/palmitoleate) was determined from measured levels.
(E) Abnormal reduction of unsaturated long-chain or polyunsaturated fatty acids (PUFA) in TICs compared to those in hepatocytes. Metabolomics analyses were performed on mouse TICs and control hepatocytes transduced with sh-Nanog or scrambled shRNA control (n = 5 per group).
(F) Adenosine 5’-monophosphate (AMP) levels increased in TICs whereas Nanog silencing reduced AMP level as determined from metabolomics analysis.
(G) The sh-Nanog treatment of TICs affected phosphorylation of AMPKα and AMPKβ associated with ACC phosphorylation.
(H) Phospho-AMPK level was increased in human tumor tissues.
Further analysis of FAO in TICs was performed by examining fatty acid substrate utilization by metabolomic analysis. Mouse TICs were examined for their ability to metabolize free fatty acids of varying carbon length and C-C bond unsaturation. As summarized in figure 6E, TICs showed lower levels of long-chain FA (C16 to C22) utilization as compared to scrambled shRNA-transduced normal hepatocytes. These data indicated TICs differed from normal hepatocytes in their ability to metabolize long-chain fatty acids; in fact TICs may be more efficient at metabolizing longer chain fatty acids (>22). From these studies, we elucidated the causal roles of NANOG in mitochondrial metabolic reprogramming through activation of FAO, which was found to be required for self-renewal of TICs.
AMP/ADP is converted into ATP during OXPHOS. Metabolite analysis of TICs revealed that the AMP level was higher in scrambled shRNA-transduced TICs when compared to Nanog-silenced TICs (Fig. 6F), while the ATP level was lower in TICs, but higher in sh-Nanog-TICs (Fig. S6N). As Nanog-mediated inhibition of OXPHOS is expected to promote AMP/ADP accumulation and AMPK activation (Hawley et al., 1996), phosphorylation of AMPK was examined. The level of phospho-AMPKα was reduced in TICs following Nanog silencing (Fig. 6G), suggesting that NANOG inhibition of OXPHOS promoted the accumulation of AMP, leading to activation of AMPKα via phosphorylation (Hawley et al., 1996). IF microscopy showed phosphorylated AMPKα levels were increased in human TICs (Fig. 6H), consistent with metabolic AMP accumulation.
NANOG restoration in sh-Tlr4-TICs resulted in comparable levels of OCR compared to that of scrambled control TICs (Fig. S6O). ETO treatment reduced ATP production while glycolysis inhibitor (2-DG) did not significantly lower ATP production in TICs (Fig. S6P). Taken together, NANOG apparently plays an important role in the metabolic reprogramming of TICs through down-regulation of the OXPHOS pathway and up-regulation of the FAO pathway.
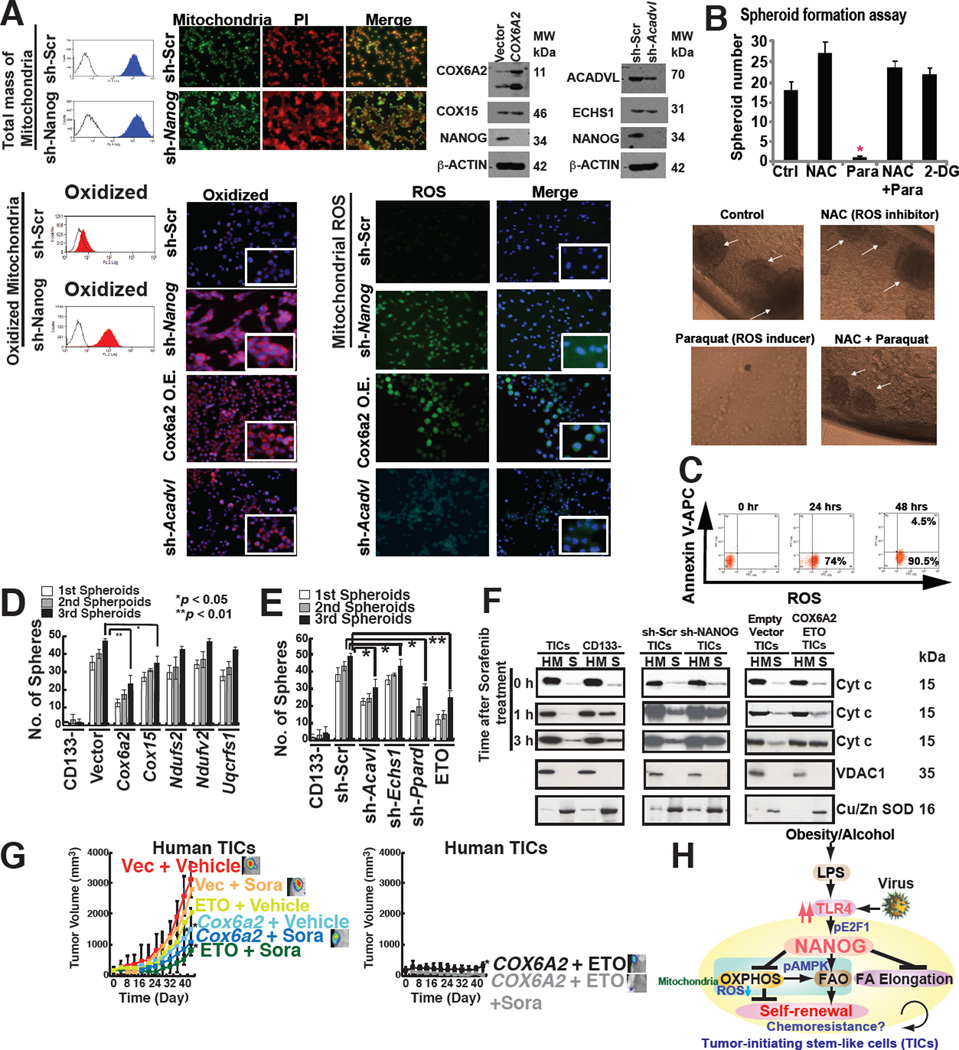
NANOG prevents mitochondrial ROS production and maintains self-renewal ability
We next evaluated the respiratory status of mitochondria with respect to reactive oxygen species (ROS), a major by-product of the mitochondrial respiratory chain, in TICs with or without Nanog silencing. We observed that more oxidatively active mitochondria were present in sh-Nanog TICs (Fig. 7A). Consequently ROS production was higher in Nanog-silenced TICs over scrambled shRNA-treated TICs (Fig. 7A, right), indicating that NANOG suppressed basal mitochondrial ROS production. Acadvl-silencing of TICs exhibited increased ROS production compared to control cells, indicating that downregulation of Acadvl promoted ROS accumulation (Fig. 7A, bottom). Glutamine withdrawal promoted ROS production in TICs (Fig. S7A and S7B), indicating that glutaminolysis-mediated antioxidant products reduced ROS levels and supported cell proliferation. Treatment of TICs with Paraquat– an inducer of ROS, increased ROS production and significantly reduced tumor spheroid formation of TICs (Fig. 7B and 7C). Similarly Nanog silencing reduced ROS-dependent spheroid formation (Fig. S7C). These results indicated that increased ROS production suppressed self-renewal ability.
Fig. 7. NANOG orchestrated TIC oncogenic and therapeutic resistance mechanisms via mitochondrial metabolic reprogramming.
(A) Mitochondrial ROS production increased in sh-Nanog TICs, but total mitochondrial levels were unchanged in TICs compared to sh-Nanog TICs.
(B) ROS inducer Paraquat (Para), but not ROS scavenger (NAC), inhibited spheroid formation, but minimal cell death induction was observed (C).
(D) Restoration of OXPHOS genes in TICs promoted self-renewal ability.
(E) Silencing OXPHOS genes and FAO genes inhibited spheroid formation.
(F) Mitochondrial cytochrome c release was increased by the combination of sorafenib and ETO treatment or overexpression of Cox6a2 in TICs. Cytochrome c release from mitochondria was analyzed by immunoblotting of the cytosol (soluble fraction) and mitochondria-rich (heavy membrane: HM) fractions of the cell lysates. TICs and CD133(−) cells transduced with sh-Nanog were lysed and fractionated into purified heavy membrane (HM) and cytosolic (S) fractions. The fractions were then probed for cytochrome c (Cyt c), VDAC1 and Cu/Zn SOD.
(G) Overexpression of Cox6a2 and ETO treatment abrogated drug-resistance and reduced tumor growth.
(H) A summary diagram depicting the proposed roles of TLR4/NANOG for metabolic reprogramming and genesis of TICs in liver oncogenesis due to alcohol and HCV. NANOG-induced chemotherapy-resistance occurred via mitochondrial metabolic reprogramming (suppression of mitochondrial OXPHOS and promotion of FAO).
To determine if NANOG-regulated OXPHOS gene(s) regulates mitochondrial metabolism, NANOG-regulated OXPHOS gene(s) were overexpressed in TICs. TICs transduced with Cox6a2 or Cox15 expression vectors showed two- and three-fold greater O2 consumption, respectively (Fig. S7E). In order to test if restoration of OXPHOS gene(s) inhibited self-renewal ability, spheroid formation assays were performed on serially passaged TICs following OXPHOS gene transduction (Fig. S7D). Indeed, overexpression of either COX6A2 or COX15 inhibited spheroid formation ability (Fig. 7D).
Along similar lines human TICs were transduced with shRNAs which targeted various FAO genes and serially-passaged for appearance of spheroid cell masses. We found that the spheroid numbers were reduced in the FAO gene-silenced group, indicating that the self-renewal ability of TICs was inhibited (Fig. 7E). Transduction of shRNAs-targeting Echs1 and Acadvl in TICs (Fig. S7F) inhibited the FAO activity of TICs (Fig. S7G). These results demonstrated the importance of NANOG in maintenance of TIC self-renewal ability by reducing mitochondrial respiration and ROS production.
NANOG orchestrates TIC oncogenicity and therapeutic resistance mechanisms via mitochondrial metabolic reprogramming
To address the effects of alteration of OXPHOS/FAO gene expression on the efficacy of the chemotherapeutic agent, sorafenib (Llovet and Bruix, 2008), we tested the roles of the NANOG-repressed OXPHOS gene (Cox6a2) or the FAO inhibitor, ETO, on sorafenib chemoresistance. We overexpressed OXPHOS genes or employed ETO as an inhibitor of FAO in TICs and assessed their effects on cellular sorafenib sensitivity in an orthotopic tumor transplantation model in alcohol-fed mice. These results indicated causal roles of NANOG repression on OXPHOS and elevated expression of FAO genes in chemoresistance in a human and mouse TICs-xenograft mouse model (Fig. 7G and S7H).
To test if restoration of OXPHOS and/or inhibition of FAO promote sorafenib-mediated apoptosis through the mitochondrial-pathway, cytochrome c release was examined in the mitochondria-enriched, heavy membrane fraction (HM) of total cell extract. We observed following sorafenib treatment, cytochrome c was translocated from mitochondria into the soluble fraction (cytoplasm) of hepatocytes within 1–3 hours post treatment while cytochrome c in TICs remained mostly in the heavy membrane (HM) fraction (mitochondria) (Fig. 7F). The silencing of Nanog or overexpression of Cox6a2 and/or addition of FAO inhibitor (ETO) enhanced cytochrome c release from mitochondria in response to sorafenib treatment (Fig. 7F). Purity of the mitochondria-enriched fractions (HM) and cytosolic fractions (S), fraction-specific proteins was confirmed by marker enzymes. Voltage-dependent anion channel 1 (VDAC1), a major outer mitochondrial protein was detected in HM fractions, whereas copper zinc superoxide dismutase (Cu/Zn SOD), a cytosolic protein, was found in cytosolic fractions (Fig. 7F), indicating that the cell fractionation process was effective and supported this model. These results demonstrated causal roles of NANOG-mediated repression of OXPHOS and induction of FAO genes in chemoresistance.
Non-TIC cancer cells (HepG2 and Hep3B) were transduced with sh-COX6A2 or ACADVL expression vector and protein levels were validated by immunoblots (Fig. S7I and S7J). Reduction of OXPHOS genes (i.e., Cox6a2) or overexpression of a FAO gene (i.e., Acadvl) in human, non-TIC HCC cell lines (HepG2 and Hep3B) promoted self-renewal ability (Fig. S7K) and NANOG expression (Fig. S7L) by spheroid formation assays and qRT-PCR analysis of NANOG. Thus, this relationship of OXPHOS gene expression to cell proliferation may be a more general property rather than restricted to TICs. Silencing and overexpression of Acadvl in non-TIC cancer cells conferred sorafenib resistance (Fig. S7M), indicating that alterations in the NANOG-OXPHOS pathway promotes a more stem-like property of cancer cells; thus rendering HCC more malignant and therapy-resistant.
DISCUSSION
Complementary NANOG ChIP-seq and metabolomics studies of TICs demonstrated that NANOG induced by TLR4 suppressed mitochondrial OXPHOS and activated FAO, thus inhibiting OCR and ROS production. This conferred a tumor chemoresistant state which could be abrogated by NANOG-targeted gene silencing (Fig. 7H). Our findings demonstrated a NANOG-dependent downstream effect on mitochondrial function in TICs that contributed to the mechanism of chemotherapy resistance. These metabolic reprogramming promoted self-renewal/oncogenesis, and explained how NANOG activation inhibited therapy-mediated apoptosis by quenching ROS production. Restoration of OXPHOS and activation of decreased FAO reduces tumorigenic capacity of TICs and increases susceptibility to chemotherapy.
As TICs rely on active FAO for their maintenance and function, FAO inhibitor suppresses self-renewal of leukemia-initiating cells (LICs) (Samudio et al., 2010). We experimentally reversed the effects of FAO gene silencing and restored the original TIC phenotype by overexpression of FAO genes (Fig. S7K, S7L). Thus the fate of stem cells is metabolically switched by FAO (Ito et al., 2012). Potential mechanisms by which elevation of FAO maintains self-renewal ability include: (i) shunting of long-chain FA away from lipid and cell membrane synthesis; (ii) downregulation of ROS through production of NADPH to avoid loss of TICs; and (iii) reduction of metabolic resistance to chemotherapy. By these criteria, NANOG function could be construed to serve as a gatekeeper for FAO activity.
In conclusion, we elucidated several mechanisms of actions of NANOG in the maintenance and chemotherapy resistance of TICs involving not only the direct activation of self-renewal via stemness genes, but also the subsequent metabolic reprogramming in these cells leading to amplification of TIC oncogenic activity and their overall survival. Our data showed that NANOG reprogramming of mitochondrial metabolism was indeed responsible for human TIC oncogenicity and chemoresistance. The metabolic bases of altered cell functions and cell fate in TICs define potentially new approaches for chemo-sensitization and elimination of TICs for more efficacious HCC therapies. These studies have led to a paradigm shift in our understanding the underlying basis of alcohol/HCV-associated cancer, thus facilitating future development of new personalized treatment strategies targeted towards NANOG+ TICs arising from obesity, alcohol, or HCV-related HCC.
Experimental Procedures
Detailed methods and analytical procedures are detailed in the Supplemental Experimental Procedures section.
Mice
HCV NS5A Tg mice (Majumder et al., 2002) were generated by Dr. Ratna B. Ray (St. Louis Univ.) on a C57BL/6 background. NS5A transgenic (Tg) and Tlr4 deficient mice (Jackson Lab) were intercrossed at least six times. HCV Core transgenic mice were generated in University of Southern California (USC) Transgenic Core facility (Machida et al., 2010). Mice were fed a Lieber-DeCarli diet containing 3.5% ethanol or isocaloric dextrin (Bioserv, Frenchtown, NJ) and/or high-cholesterol high-saturated fat diet, as indicated. To test the role of Nanog in hepatocytes/hepatoblasts in liver oncogenesis in alcohol-fed HCV mice, we overexpressed shRNA against Nanog in mice harboring CMV-loxP-Gfp-stop-loxP/U6-sh-Nanog and Albumin-Cre. In these mice the shRNA is conditionally expressed to knockdown Nanog (KD) in albumin-expressing cells (Yamaguchi et al., 2009).
Bone marrow transplantation
Bone marrow transplantation (BMT) was performed as previously described (Dapito et al., 2012; Seki et al., 2007) with modification from traditional protocols as previsouly described (Kisseleva et al., 2006; Tsung et al., 2005).
Treatment with alcohol Western diet or diethylnitrosamine/phenobarbital
High-cholesterol high-fat diet was used, containing very similar diet components (TD.03350: Harkan Teklad, Inc.) as previously described (Haluzik et al., 2004; Van Heek et al., 1997). Mice were fed alcohol Western diet (Dyets Inc. Cat#D710362) or dextrin control diet (Dyets Inc. Cat#D710027) for 12 months. This alcohol WD is modified from Lieber-DeCarli (L–D) alcohol diet and contains 3.5% ethanol, high-cholesterol and high-saturated fat (1%w/w chol, 21%Cal lard, 4%Cal corn oil, Dyets Inc.). For the chemical carcinogenesis mouse model, diethylnitrosamine (DEN) was intraperitoneally injected at four weeks of age and phenobarbital was fed in drinking water from eight weeks of age to euthanasia as previously established (Machida et al., 2010).
Endotoxin measurement
For endotoxin measurements, blood was collected from inferior vena cava with pyrogen-free heparin as previously described (Mathurin et al., 2000)
Isolation of human TICs
Isolation of CD133+/CD49f+ TICs from HCC tissues obtained from alcoholic patients with or without HCV infection, as previously described (Chen et al., 2013; Gripon et al., 2002).
Cell lines
TICs were grown in DMEM/F12 or Kubota medium for all experiments. HEK293T and Huh7 cells were cultured in DMEM (Cellgro) with 10% FBS and essential amino acid supplements.
Gene array and proteomic analysis of liver tumors
Systematic gene microarray and proteomic analyses were performed for dysplastic and normal tumors, to identify changes in known or unknown signaling pathways that are tightly associated with synergistic induction of liver tumor by Western diet (WD) or alcohol.
Gene ontology analysis
Genes which are differentially associated with NANOG in TICs or control cells were functionally analyzed in the context of gene ontology and molecular networks by using the Ingenuity pathway software.
Proteomics
Proteomic analysis was performed at the Proteomic Exploration Laboratory at California Institute of Technology, Pasadena, CA. In brief, the liver samples were lysed for protein extraction and extracted proteins were subjected to one-dimensional gel electrophoresis, and stained protein bands were used for in-gel trypsin digestion and MS sequencing.
ChIP-seq
Four pairs of TICs and Nanog-/CD133−/CD49f+ control cells (~1×105 per mouse) were isolated from four independent mouse liver tumors. ChIP was performed with NANOG antibody using CD 133(+) as well as CD133(−) cell lines following a standard protocol as suggested by the manufacturer (Millipore). To generate sequencing library constructs, ChIP DNA fragments (1–10 ng) were used for adapter ligation, gel purification and PCR, followed by ligation. ChIP-seq library constructions and high-throughput DNA sequencing was performed using Illumina HiSeq 2000 (Illumina, San Diego, CA, USA) using a 50bp SE reads at the USC Genomic Core.
XF24 extracellular flux analyzer for measurement of cellular OCR and ECAR
Measurement of extracellular flux of cellular bioenergetics is determined by a Seahorse XF96 Extracellular Flux Analyzer (Ferrick et al., 2008).
Stable-isotope carbon labeling is traced for flux analysis
Cells were cultured in DMEM/F12 medium (17.5 mM unlabeled glucose) supplemented with 7.5 mM [U13C6]-glucose (Cambridge Isotope Laboratories) for 48 hr and total ion chromatography of fatty acids was performed by stable isotope tracing using [U13C6]-glucose for 48 hr. Three independent replicates of 2 × 106 cells for each cell line were collected, and the cell pellets were suspended in 0.5 ml of water and lysed by sonication. Cell debris was separated by centrifugation and proteins precipitated by treating the clarified supernatant with 1 ml of cold acetone. The final supernatant was air-dried and the free glutamic acid was converted to its trifluoroacetamide butyl ester for GC-MS analysis (Lee, 1996). Rate of fatty acid synthesis is represented by Oleate C18:1/Palmitoleate C16:1 ratio, demonstrating that Nanog silencing reduces fatty acid chain elongation. In addition, CO2 production of TICs is very low, indicating that TLR4/NANOG induction in TICs inhibits oxygen consumption through inhibition of FA oxidation and TCA cycle entrance.
Fatty acid β-oxidation assay
Rates of fatty acid β-oxidation were determined, in which the rate of carbon dioxide production from the oxidation of [14C]palmitate was measured in Metabolomic Core facility of University of Southern California. Cells were cultured in the presence of [14C]palmitate–BSA complex and the released [14C]carbon dioxide trapped for 1 h at 37 °C onto filter paper soaked in 100 mM sodium hydroxide. The rate of β-oxidation was calculated as the amount of trapped [14C]carbon dioxide in relative units produced per mg protein per hour.
ATP production measurements
Relative ATP/cell assays were performed by Cell Titer-Glo (Promega) and CyQUANT(Invitrogen). Luminescence and fluorescence readings were consecutively measured after room-temperature incubation for 10 min.
Statistical Considerations
A two-tailed t-test was used for most comparisons, with p<0.05 considered statistically significant. Data are presented as mean±S.D.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Ratna Ray (Saint Louis Univ.) for providing HCV NS5A Tg mice, Dr. Hidekazu Tsukamoto for valuable suggestions, Ms. Akiko Ueno and Mr. Raul Lazaro, the animal core personnel for performing mouse experiments, Dr. Bangyan Stiles, Dr. Tin A. Than and Mr. Chad Nakagawa (USC) for critical reading, Raymond Wu (USC) for suggestions and reagents, Dr. Todd L. Sladek (Rosalind Franklin Univ., Illinois) for E2F1 mutant constructs, Dr. Carlo V. Catapano for PPARδ expression constructs. Nanog luciferase reporter constructs were obtained from Dr. Paul Robson and Dr. Ng Huck Hui at the Genome Institute of Singapore and Dr. Takashi Tada in Kyoto Univ. This project was supported by NIH grants 1R01AA018857-01, pilot project funding (5P30DK048522-13), P50AA11999 (Animal Core, Morphology Core, and Pilot Project Program) and R24AA012885 (Non-Parenchymal Liver Cell Core). This research is also supported by a Research Scholar Grant, RSG-12-177-01-MPC and pilot funding (IRG-58-007-48) from the American Cancer Society. Microscopy was performed by the Cell and Tissue Imaging Core of the USC Research Center for Liver Diseases (P30 DK048522). S.H. is supported by the Gordon and Betty Moore Foundation through grant GBMF775 and the Beckman Institute. Statistical analysis was performed by Dr. Susan Groshen and Ms. Lingyun Ji of the Norris Comprehensive Cancer Center Biostatistics Core supported by NIH/NCI P30 CA 014089. Animal imaging was performed by the USC Molecular Imaging Center supported by NIH/NVRR S10. Tissue pathological slide preparation was performed by Ms. Moli Chen of the Norris Comprehensive Cancer Center Translational Pathology Core. Liver tissues were obtained from The Liver Tissue Cell Distribution System (LTCDS) in Univ. Minnesota.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: K.M. and C.C. conceived of the study. C.C., K.M., D.U., J.X., L.S. obtained the data. V.P. analyzed ChIP-seq, microarray and proteomic data analysis. S.H. conducted and analyzed the proteomics experiments. S. T. contributed for drafting and discussion. C.C., K.M., L.M., and provided data management, provided statistical support. K.M. and C.C. conducted the data analysis and drafted the report. All authors interpreted the data and contributed to the final version of this report.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- Chen CL, Tsukamoto H, Liu JC, Kashiwabara C, Feldman D, Sher L, Dooley S, French SW, Mishra L, Petrovic L, et al. Reciprocal regulation by TLR4 and TGF-beta in tumor-initiating stem-like cells. The Journal of clinical investigation. 2013;123:2832–2849. doi: 10.1172/JCI65859. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov Today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. Infection of a human hepatoma cell line by hepatitis B virus. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutgesell A, Wen G, Konig B, Koch A, Spielmann J, Stangl GI, Eder K, Ringseis R. Mouse carnitine-acylcarnitine translocase (CACT) is transcriptionally regulated by PPARalpha and PPARdelta in liver cells. Biochim Biophys Acta. 2009;1790:1206–1216. doi: 10.1016/j.bbagen.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Haluzik M, Gavrilova O, LeRoith D. Peroxisome proliferator-activated receptor-alpha deficiency does not alter insulin sensitivity in mice maintained on regular or high-fat diet: hyperinsulinemic-euglycemic clamp studies. Endocrinology. 2004;145:1662–1667. doi: 10.1210/en.2003-1015. [DOI] [PubMed] [Google Scholar]
- Hawley SA, Davison M, Woods A, Davies SP, Beri RK, Carling D, Hardie DG. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J Biol Chem. 1996;271:27879–27887. doi: 10.1074/jbc.271.44.27879. [DOI] [PubMed] [Google Scholar]
- He N, Park K, Zhang Y, Huang J, Lu S, Wang L. Epigenetic inhibition of nuclear receptor small heterodimer partner is associated with and regulates hepatocellular carcinoma growth. Gastroenterology. 2008;134:793–802. doi: 10.1053/j.gastro.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Hritz I, Mandrekar P, Velayudham A, Catalano D, Dolganiuc A, Kodys K, Kurt-Jones E, Szabo G. The critical role of toll-like receptor (TLR) 4 in alcoholic liver disease is independent of the common TLR adapter MyD88. Hepatology. 2008;48:1224–1231. doi: 10.1002/hep.22470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee CH, et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18:1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva T, Uchinami H, Feirt N, Quintana-Bustamante O, Segovia JC, Schwabe RF, Brenner DA. Bone marrow-derived fibrocytes participate in pathogenesis of liver fibrosis. J Hepatol. 2006;45:429–438. doi: 10.1016/j.jhep.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Lee WN. Stable isotopes and mass isotopomer study of fatty acid and cholesterol synthesis. A review of the MIDA approach. Adv Exp Med Biol. 1996;399:95–114. doi: 10.1007/978-1-4613-1151-5_8. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312–1327. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida K, Tsukamoto H, Liu JC, Han YP, Govindarajan S, Lai MM, Akira S, Ou JH. c-Jun mediates hepatitis C virus hepatocarcinogenesis through signal transducer and activator of transcription 3 and nitric oxide-dependent impairment of oxidative DNA repair. Hepatology. 2010;52:480–492. doi: 10.1002/hep.23697. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Machida K, Tsukamoto H, Mkrtchyan H, Duan L, Dynnyk A, Liu HM, Asahina K, Govindarajan S, Ray R, Ou JH, et al. Toll-like receptor 4 mediates synergism between alcohol and HCV in hepatic oncogenesis involving stem cell marker Nanog. Proc Natl Acad Sci U S A. 2009;106:1548–1553. doi: 10.1073/pnas.0807390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder M, Ghosh AK, Steele R, Zhou XY, Phillips NJ, Ray R, Ray RB. Hepatitis C virus NS5A protein impairs TNF-mediated hepatic apoptosis, but not by an anti-FAS antibody, in transgenic mice. Virology. 2002;294:94–105. doi: 10.1006/viro.2001.1309. [DOI] [PubMed] [Google Scholar]
- Mathurin P, Deng QG, Keshavarzian A, Choudhary S, Holmes EW, Tsukamoto H. Exacerbation of alcoholic liver injury by enteral endotoxin in rats. Hepatology. 2000;32:1008–1017. doi: 10.1053/jhep.2000.19621. [DOI] [PubMed] [Google Scholar]
- Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, Weinman SA. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366–375. doi: 10.1053/gast.2002.30983. [DOI] [PubMed] [Google Scholar]
- Rountree CB, Senadheera S, Mato JM, Crooks GM, Lu SC. Expansion of liver cancer stem cells during aging in methionine adenosyltransferase 1A–deficient mice. Hepatology. 2008;47:1288–1297. doi: 10.1002/hep.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, Kaluarachchi K, Bornmann W, Duvvuri S, Taegtmeyer H, et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. The Journal of clinical investigation. 2010;120:142–156. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, Schwabe RF. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med. 2007;13:1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- Torres J, Watt FM. Nanog maintains pluripotency of mouse embryonic stem cells by inhibiting NFkappaB and cooperating with Stat3. Nat Cell Biol. 2008;10:194–201. doi: 10.1038/ncb1680. [DOI] [PubMed] [Google Scholar]
- Tsung A, Hoffman RA, Izuishi K, Critchlow ND, Nakao A, Chan MH, Lotze MT, Geller DA, Billiar TR. Hepatic ischemia/reperfusion injury involves functional TLR4 signaling in nonparenchymal cells. Journal of immunology. 2005;175:7661–7668. doi: 10.4049/jimmunol.175.11.7661. [DOI] [PubMed] [Google Scholar]
- Van Heek M, Compton DS, France CF, Tedesco RP, Fawzi AB, Graziano MP, Sybertz EJ, Strader CD, Davis HR., Jr Diet-induced obese mice develop peripheral, but not central, resistance to leptin. The Journal of clinical investigation. 1997;99:385–390. doi: 10.1172/JCI119171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, Tovar V, Roayaie S, Minguez B, Sole M, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972–1983. doi: 10.1053/j.gastro.2008.08.008. e1971–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZX, Kueh JL, Teh CH, Rossbach M, Lim L, Li P, Wong KY, Lufkin T, Robson P, Stanton LW. Zfp206 is a transcription factor that controls pluripotency of embryonic stem cells. Stem Cells. 2007;25:2173–2182. doi: 10.1634/stemcells.2007-0085. [DOI] [PubMed] [Google Scholar]
- Wu Q, Chen X, Zhang J, Loh YH, Low TY, Zhang W, Zhang W, Sze SK, Lim B, Ng HH. Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells. J Biol Chem. 2006;281:24090–24094. doi: 10.1074/jbc.C600122200. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kurimoto K, Yabuta Y, Sasaki H, Nakatsuji N, Saitou M, Tada T. Conditional knockdown of Nanog induces apoptotic cell death in mouse migrating primordial germ cells. Development. 2009;136:4011–4020. doi: 10.1242/dev.041160. [DOI] [PubMed] [Google Scholar]
- Yuan JM, Govindarajan S, Arakawa K, Yu MC. Synergism of alcohol, diabetes, and viral hepatitis on the risk of hepatocellular carcinoma in blacks and whites in the U.S. Cancer. 2004;101:1009–1017. doi: 10.1002/cncr.20427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.