Abstract
It has been reported that mechanical vibrations of the magnetic resonance imaging scanner could produce spurious signal dropouts in diffusion‐weighted images resulting in artifactual anisotropy in certain regions of the brain with red appearance in the Directionally Encoded Color maps. We performed a review of the frequency of this artifact across pediatric studies, noting differences by scanner manufacturer, acquisition protocol, as well as weight and position of the subject. We also evaluated the ability of automated and quantitative methods to detect this artifact. We found that the artifact may be present in over 50% of data in certain protocols and is not limited to one scanner manufacturer. While a specific scanner had the highest incidence, low body weight and positioning were also associated with appearance of the artifact for both scanner types evaluated, making children potentially more susceptible than adults. Visual inspection remains the best method for artifact identification. Software for automated detection showed very low sensitivity (10%). The artifact may present inconsistently in longitudinal studies. We discuss a published case report that has been widely cited and used as evidence to set policy about diagnostic criteria for determining vegetative state. That report attributed longitudinal changes in anisotropy to white matter plasticity without considering the possibility that the changes were caused by this artifact. Our study underscores the need to check for the presence of this artifact in clinical studies, analyzes circumstances for when it may be more likely to occur, and suggests simple strategies to identify and potentially avoid its effects. Hum Brain Mapp 36:4745–4757, 2015. © 2015 Wiley Periodicals, Inc.
Keywords: diffusion tensor imaging, weight, position, quality, regrowth
INTRODUCTION
Diffusion Tensor Imaging (DTI) [Basser et al., 1994] is a quantitative Magnetic Resonance Imaging (MRI) technique used to study the structure and architecture of human brain tissue [Pierpaoli et al., 1996]. However, DTI data is known to suffer from numerous artifacts, which affect the quantitative accuracy and precision of tensor derived metrics [Jones et al., 2012; Pierpaoli, 2010]. Switching of the diffusion gradients produces vibrations in the gradient coils that propagate to other components of the MRI scanner, including the subject table. Although these vibrations may be exploited to measure tissue properties using magnetic resonance elastography [Gallichan et al., 2009], they are also known to produce an unwanted artifact in diffusion‐weighted images (DWI) [Gallichan et al., 2010; Hiltunen et al., 2006; Jones, 2010; Tournier et al., 2011]. The artifact manifests as increased diffusivity in the left‐right direction, mainly in parietal‐occipital regions [Gallichan et al., 2010; Liu and Liu, 2011] resulting in increased fractional anisotropy (FA) [Basser and Pierpaoli, 1996] and red appearance in Directionally Encoded Color (DEC) maps [Pajevic and Pierpaoli, 1999].
Although the artifact is documented and methods to avoid, detect, or correct the artifact have been proposed [Farzinfar et al., 2013; Gallichan et al., 2010; Koch and Finsterbusch, 2011; Mohammadi et al., 2012b; Scherrer and Warfield, 2012; Sharman et al., 2011], few investigators mention inspecting for it in their diffusion MRI studies [Jensen and Helpern, 2010; Mohammadi et al., 2012a]. Even when mentioned, it is often cursorily discussed as a possible confound for their findings [Aso et al., 2013; Budde et al., 2011; Mohammadi et al., 2012c; Mohammadi et al., 2013; Mueller et al., 2011; Rapacchi et al., 2011]. Also, all references to the artifact are related to a specific scanner (Siemens 3T Trio) although some studies do not report the scanner manufacturer, leaving open the possibility that it could be found on other scanners [Budde et al., 2011; Jones, 2010; Mohammadi et al., 2012c]. Systematic investigations have focused on quantifying the effects of the artifact on a single subject or gel phantom [Gallichan et al., 2010], or have measured the gradient‐induced vibration without assessing the presence of artifacts in the DWIs [Hiltunen et al., 2006], but prevalence of this artifact across a population of study participants has only been reported once for a Siemens scanner ranging from 1 to 54% across four sites [Farzinfar et al., 2013].
To achieve a more detailed understanding of the impact of this artifact in a clinical research setting, we conducted a retrospective review of our data and report the frequency of the artifact across datasets acquired on two scanners from different manufacturers (General Electric ≥ and Siemens). We identified the artifact by systematic visual inspection of the DWIs and the DEC maps and evaluated the only automated tool publicly available for the detection of this artifact (entropy tool within DTIPrep) [Farzinfar et al., 2013; Oguz et al., 2014], as well as another quantitative tool for general DWI artifact detection, RESTORE [Changet al., 2005]. In addition, we analyzed whether two factors—weight and positioning—contribute to the presence of the artifact. Our attention toward positioning was based on our observations during visual inspection that suggested that positioning might influence the presence of the artifact. Our investigation of weight was based on references that identified weight load as an influential factor. Specifically, Siemens addressed the vibration artifact by designing a new patient table that no longer has the table board in contact with the gradient coil [Liu and Liu, 2011]. In the patent for the new scanner bed, it was noted that the artifact may be reduced if the load on the table is greater than approximately 30 kg or if load distribution is adjusted. Thus, we hypothesized that greater weight and positioning away from center will be associated with decreased incidence of the artifact. On a prospective, single subject basis, we also implemented solutions previously suggested and investigated additional solutions for avoiding the artifact.
METHODS
Retrospective Analysis
We reviewed DWI data acquired on pediatric subjects from two scanner manufacturers (Siemens 3T Trio and General Electric ≥ Signa HDX). We examined whether weight and head position influenced the manifestation of the artifact. Two hundred thirty‐one children were included with three acquisition protocols resulting in a review of 484 datasets. The subject population included patients (autism or epilepsy) and typically developing controls. GE Data: One hundred eighty‐eight children ranging in age from 17 months to 10 years were scanned on a 1.5T GE Signa HDX. There were two acquisition protocols that were designed to reproduce the protocol used by the National Institutes of Health (NIH) MRI Study of Normal Brain Development (http://pediatricmri.nih.gov). Some children were scanned under both acquisition protocols and a subset had a repeat study for a total of 441 GE datasets. One hundred ninety‐one scans were acquired with the higher resolution protocol (High‐Res GE) of 60 directions at b = 1,100 s/mm2, 10 directions at b = 300 s/mm2, and 10 b = 0 s/mm2 for 80 brain volumes, 2.5‐mm isotropic voxels, 86 slices at 2.5‐mm thickness, and echo time/repetition time (TE/TR) 84.5/21,330 ms. Two hundred fifty scans were acquired with the lower resolution protocol (Low‐Res GE) six directions at b = 1,000 s/mm2 repeated four times, six directions at b = 500 s/mm2 repeated two times, and 6 b = 0 s/mm2 for 42 brain volumes, 3‐mm isotropic voxels, 86 slices at 3‐mm thickness, and TE/TR 71.9/12,408 ms. Seventy‐eight datasets were repeat studies (mean time between scans = 2.4 years; range = 15 days–3.63 years). This longitudinal data was examined to determine if the artifact persisted, disappeared, or appeared de novo at the second scan.
Siemens data
Forty‐three children were acquired on a 3T Siemens Trio ranging in age from 7 to 16 years. The Siemens DTI data were acquired with 30 directions at b = 1,000 s/mm2 and 5 b = 0 s/mm2, repeated twice for 70 brain volumes, 2.5‐mm isotropic voxels, 55 slices at 2.5‐mm thickness, Generalized Autocalibrating Partially Parallel Acquisition (GRAPPA) = 2, and TE/TR = 86/6,300 ms. Data were collected before and after a manufacturer upgrade, which did not include the updated scanner bed intended to eliminate the vibration artifact. The upgrade did include a gradient upgrade to the Siemens “TQ‐engine system,” which has maximum gradient amplitude of 45 mT/m for the longitudinal direction and 40 mT/m for the horizontal and vertical directions.
Detection of the artifact was done by three methods. One method was a systematic visual inspection of both DWI and DEC maps. Visual inspection was compared with a second automated detection method using the entropy tool within DTIPrep [Farzinfar et al., 2013; Oguz et al., 2014]. Lastly, we also evaluated the utility of a quantitative measure (chi‐squared) as a possible method of detecting outlier data using RESTORE robust tensor fitting [Chang et al., 2005; Chang et al., 2012].
For visual inspection, DEC maps were available following a correction pipeline using TORTOISE [Pierpaoli et al., 2010] to reduce effects of motion, eddy current distortions, and echo‐planar imaging (EPI) distortions. Corrections were performed in the native space of each subject. Following corrections, nonlinear tensor fitting was used to estimate the diffusion tensor and tensor derived metrics, which produced the DEC maps. Interrater reliability was measured using Cohen's kappa [Mackinnon, 2000] using the following criteria for determining whether the artifact was present:
Visual inspection of the DWIs for signal dropouts.
If a signal dropout is evident, confirm that the dropout occurs in the areas that are suggestive of the vibration artifact (parietal‐occipital) and consistently in a particular diffusion direction when looking across the entire acquisition for that subject. In contrast, bulk head motion has a different pattern of large or whole brain signal dropout occurring in any gradient direction, and within a volume, has a typical “venetian blind” or staircase pattern when viewed from an orthogonal plane.
Inspecting the DEC map for presence of a red hue in the parietal‐occipital region of the brain, as well as any other unusual patterns such as in inferior, ventral frontal regions as described by Gallichan and coworkers.
For the automated detection, we imported Low‐Res GE TORTOISE‐corrected data into DTIPrep. We only conducted the entropy analysis with the Low‐Res GE data because there was not enough artifact‐free data in the Siemens data to establish the acceptable range of entropy values. The mean and standard deviation of acceptable entropy values were established from a set of artifact‐free samples (n = 41). Those values were then used to calculate z‐scores to categorize the quality of the remaining Low‐Res GE DTI scans. We used categories as described by the entropy tool developers (z < 1.64 is Acceptable; z ≥ 1.64 is Suspicious; z ≥ 2.58 Unacceptable). To compare visual inspection with the entropy tool, we collapsed the suspicious/unacceptable categories to match the present/not present decision of a visual read. Sensitivity and specificity and the 95% confidence intervals (CI) of the entropy tool to detect the artifact as compared with visual inspection was calculated [Mackinnon, 2000].
Evaluation of chi‐squared was done by calculating mean chi‐squared values for two regions of interest (ROI) within the vibration artifact. One ROI was drawn on the slice of the DEC map that was within the core of the vibration artifact where the vibration artifact was most visually intense (i.e., strongest degree of anisotropy [redness]). The second ROI was drawn at the periphery of the artifact where the artifact was still visually evident but often was the last slice before it was no longer apparent. The mean chi‐squared values from each ROI were used to determine the percent of voxels in the entire brain for that subject that had a chi‐squared value greater than or equal to that value. With this data, we are able to get a sense of how specific the chi‐squared values might be in detecting abnormalities related to the vibration artifact (e.g., the percentage of voxels in the brain that were at or above the values within the vibration artifact).
In addition to detection, analysis focused on assessing the prevalence of the artifact within the clinical dataset and across time points for the subset of longitudinal data. We conducted a chi‐squared analysis to determine if prevalence of the artifact differed by protocol. A multivariate analysis of variance (MANOVA) was used to determine if weight and the six position parameters differed between the group with vibration and the group without vibration artifact across the different scanner protocols. Weight data in kilograms were collected at the time of scanning. Patient position data included translation and rotation. Translation was computed as the distance in millimeters (x, y, z) of the center of the central slice from the scanner isocenter, while rotation indicates the angle of the head from its native position to a standard AC_PC orientation (x rotation [pitch], y rotation [yaw], z rotation [roll]). Positioning information was obtained from the Digital Imaging and Communications in Medicine (DICOM) header and the parameters computed by the TORTOISE software [Pierpaoli et al., 2010] used for registering the subject data to standardized space. We also conducted a canonical linear discriminant analysis to determine if the presence of artifact could be predicted from a linear combination of these same variables.
Historical data analyzed in this study were acquired in studies approved by the Combined Neurosciences Institutional Review Board of the National Institutes of Health or the Institutional Review Board of the Children's National Medical Center. Parents provided informed consent and minors—who were able—provided assent.
Single Subject Evaluation of Strategies for Avoiding the Artifact
We prospectively tested factors that could affect the presence of the artifact to optimize our DTI acquisition on a Siemens 3T Trio scanner. We examined the effect of different degree of k‐space coverage in the acquisition, because Gallichan and coworkers suggested that acquiring full k‐space parallel‐accelerated data was the most feasible option for preventing the signal loss associated with the vibration artifact, while also managing increases in TE. Moreover, we also investigated head repositioning—at a more extreme angle than is typical—as a strategy to avoid the artifact.
This pilot testing was conducted on a single healthy subject (age 32 male) on a 3T Siemens Trio scanner. The following correction strategies were tested: (1) using full k‐space acquisition in place of the default three‐fourth k‐space acquisition, (2) increasing the TR, (3) using a dual spin‐echo [Reese et al., 2003] for acquisition, and (4) altering head position two ways. Six serial scans were acquired with a single‐shot EPI sequence using the following parameters: 41 diffusion directions, 31 b = 1,100 s/mm2, 5 b = 300 s/mm2, and 5 b = 0 s/mm2, and GRAPPA = 2, matrix = 96 × 96, Field of Vision (FOV) = 240 mm, slice thickness = 2.5 mm skip 0, 64 slices, TE/TR = 87/7,500 ms. The first scan was with no correction strategy acquired with three‐fourth k‐space. The second scan was modified for full k‐space acquisition. The third scan had full k‐space and TR = 11,000 ms. The fourth scan used a dual‐spin‐echo diffusion preparation period. The fifth scan altered head position by pitch ±30°. The sixth scan altered head position by roll ±45°.
RESULTS
Retrospective Analysis
Detection of the artifact
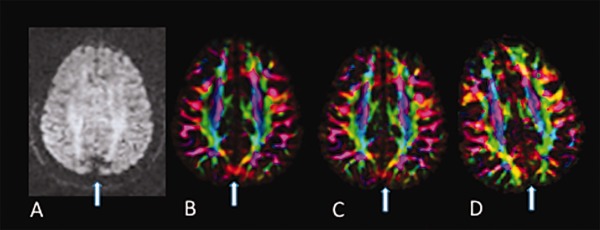
Interrater agreement of visual inspection was excellent (Cohen's Kappa = 0.81 (95% CI: 0.63–0.99)). Visual rating discrepancies were due to instances where the signal dropout was subtle. We found that the entropy tool had a high percentage of false negatives resulting in 10% sensitivity (95% CI: 0.04–0.20) and 95% specificity (95% CI: 0.90–0.98) using visual rating as a reference. The entropy tool often missed subtle cases of the artifact (Fig. 1). There were no instances of a false positive (i.e., identifying an artifact‐free dataset as abnormal), but in a few instances, the entropy tool identified abnormalities that were not specific to vibration artifact such as residual eddy distortion.
Figure 1.
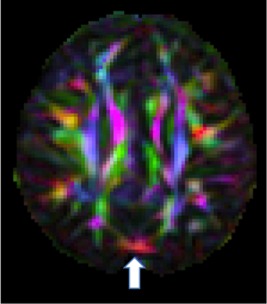
An example of a subtle artifact as seen in the DEC map of an axial slice in a GE dataset showing evidence of vibration artifact in the occipital area. The entropy tool of DTIPrep did not detect this artifact.
The range of chi‐squared values across core and periphery ROIs of the vibration artifact was large (Range: 0.74–60.86, Table 1). This was true for both GE and Siemens data. By definition, it is expected that regions with no artifact should have chi‐squared values near one. While on average, the chi‐squared values from artifact regions were higher than one, the standard deviation was large (Table 1). This was true for ROIs in the core of the artifact as well as at the periphery. As a result, many voxels in which the artifact was not present shared the same chi‐squared value as those with the artifact. At the extreme for one subject, 83% of brain voxels would have been identified as artifactual based on the chi‐squared mean value from the artifact ROI. Chi‐squared lacked specificity in detecting vibration abnormality in GE datasets as an average of 30.65% (SD 21.66%) of voxels shared the same or higher chi‐squared values as those mean values from the core ROI. Chi‐squared values were slightly more specific for Siemens data as an average of 7.39% (SD 10.93%) of voxels throughout the brain had the same or higher chi‐squared value as the mean chi‐squared value found in the core ROI. The lack of specificity is because there are instances when the vibration artifact is evident in the DEC map but the chi‐squared is normal (Fig. 2A,B). Moreover, even when an abnormality is present in both the DEC map (Fig. 3A) and chi‐squared map (Fig. 3B), after using RESTORE to remove the data contributing to the artifact, significant artifact persisted in the DEC map (Fig. 3C) despite having a normalized chi‐squared map (Fig. 3D).
Table 1.
Descriptives by scanner of chi‐square values within ROIs drawn within the vibration artifact
| Siemens | Lo‐Res GE | |
|---|---|---|
| Range of mean chi‐square values for ROIs with vibration artifact | 0.95–14.56 | 0.74–60.86 |
| Mean chi‐square in the core of the vibration artifact ROI (SD) | 6.08 (4.33) | 6.49 (8.28) |
| Mean chi‐square in the periphery of the vibration artifact ROI (SD) | 2.29 (0.797) | 5.49 (6.26) |
Figure 2.
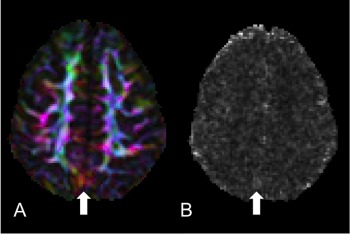
Lack of specificity of chi‐squared. (A) DEC map of an axial slice of a Siemens dataset where the vibration artifact is clearly visible yet subtle. (B) Chi‐squared map from the tensor fitting of the same slice showing no elevated chi‐squared values in the region of the artifact but some elevated values elsewhere from the artifact.
Figure 3.
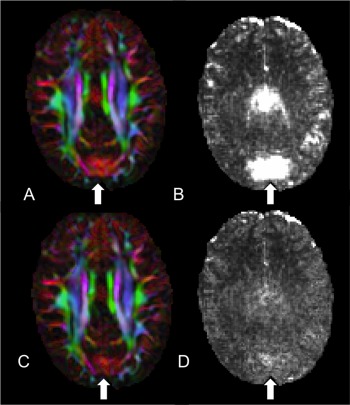
Assessing the use of RESTORE to identify and remediate the artifact. (A) DEC map where the vibration artifact is present. (B) Chi‐squared is elevated in the same region as (A). (C) DEC map still showing significant abnormalities yet the corresponding chi‐squared map (D) after tensor fitting with RESTORE showing no abnormal values.
Frequency of the artifact
By visual inspection, the artifact was identified in 18% of all pediatric datasets (n = 87 of 484). The artifact was more frequent in the Siemens data (48.8%) followed by the Low‐Res GE data (24.4%) then the High‐Res GE data (2.6%) (χ 2 = 65.33, P < 0.001, Table 2). A higher percentage of data acquired after the Siemens scanner gradient upgrade compared with preupgrade were affected by the artifact (68.9% postupgrade vs. 7% preupgrade, P < 0.001 Fisher's Exact Test (FET). For GE, 130 scans (52% of the data) were collected between August 2006 through August 2009 and all the artifacts occurred in that timeframe, prior to a software upgrade, while the other 120 scans collected between September 2009 through May 2011, after a software upgrade, had no artifact (Fig. 4).
Table 2.
Descriptives of rate of occurrence, weight, and positioning factors by scanning protocol
| Siemens | Lo‐Res GE | Hi‐Res GE | ||||
|---|---|---|---|---|---|---|
| 48.8% | 24.4% | 2.6% | ||||
| Incidence of artifact | No Artifact (n = 22) | Artifact (n = 21) | No artifact (n = 189) | Artifact (n = 61) | No artifact (n = 186) | Artifact (n = 5) |
| Parameters (Mean [SD]) | ||||||
| Weight (kg)a,b | 45.3 (16.1) | 34.1 (9.8) | 22.12 (8.14) | 18.05 (4.63) | n/a | n/a |
| x displacement (mm) | −2.20 (5.69) | −3.48 (9.40) | −3.23 (4.53) | −2.45 (4.15) | n/a | n/a |
| y displacement (mm)a | −37.58 (16.62) | −50.56 (13.48) | −33.51 (12.12) | −33.61 (8.26) | n/a | n/a |
| z displacement (mm)b | 13.62 (19.85) | 13.71 (11.04) | 17.20 (17.38) | 7.38 (13.40) | n/a | n/a |
| x rotation (degrees) | 4.57 (8.11) | 6.34 (5.51) | −1.39 (10.09) | −2.66 (9.67) | n/a | n/a |
| y rotation (degrees) | −0.07 (2.45) | −1.30 (2.82) | 0.25 (3.35) | −0.52 (4.49) | n/a | n/a |
| z rotation (degrees)b | −0.93 (2.35) | −0.94 (3.33) | −2.41 (5.07) | 0.04 (5.87) | n/a | n/a |
a P < 0.05 for Siemens.
b P < 0.01 for Lo‐Res GE.
Figure 4.
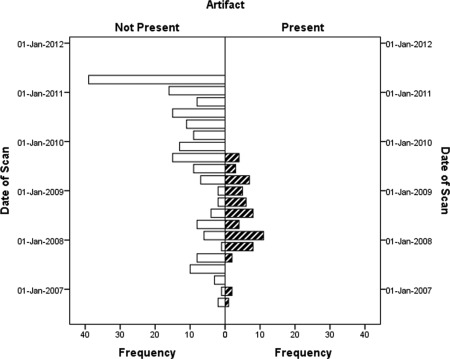
Presence of vibration artifact in Low‐Res GE data over time. No artifact was evident after September 2009 when a software upgrade occurred.
Seventy‐eight repeated scan datasets were reviewed for the longitudinal analysis. Forty‐three (55%) had no vibration artifact at either time point. Of the 35 that had the vibration artifact, one had it on both scans, 31 had the artifact at Scan 1 but not at the Scan 2 (Fig. 5), and three had the artifact only at the second time point.
Figure 5.
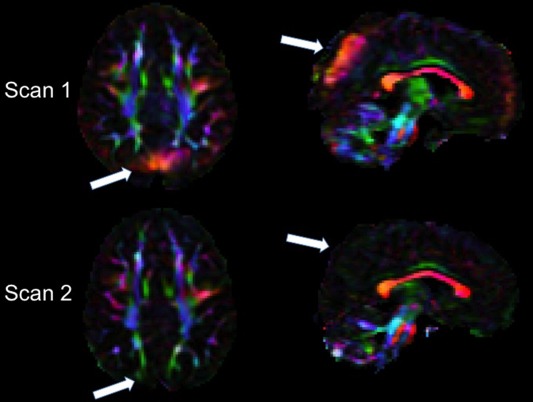
Example of inconsistent occurrence of the high anisotropy produced by the artifact in longitudinal scans of the same subject. Axial and sagittal views of the DEC map of Scan 1 (top) and Scan 2 acquired 34 months later (bottom) in the same child in the GE scanner. White arrows indicate brain regions affected by the vibration artifact in Scan 1 that show no evidence of the artifact in Scan 2.
Influence of weight and positioning
The High‐Res GE data were excluded from these analyses because they were nearly artifact‐free. Separate MANOVAs were performed on the Siemens and Low‐Res GE datasets. Descriptive data (Table 2) reveals that the range of positioning parameters was narrow with average displacement in any direction within 50 mm and average rotation within 8°. The data with vibration artifact differed from data with no artifact with respect to weight and positioning parameters for both the Siemens (F = 2.532, P = 0.035) and Low‐Res GE data (F = 5.911, P < .001). These parameters account for more of the variance between the artifact/no artifact groups in the Siemens protocol than in the GE protocol (Siemens = 0.364 vs. Low‐Res GE = 0.157). For both protocols, subjects with artifact weighed significantly less than subjects without artifact (Fig. 6). For Siemens only, subjects with vibration artifacts were positioned at a more negative y axis location with respect to the isocenter of the magnet. For the Low‐Res GE data, subjects with vibration artifacts were less displaced in the positive z direction and less rotated in the negative z rotation. We also tested whether these group differences remained using absolute values for the z rotation given that it should not matter if one is rotated left or right given the symmetry of the brain around that axis and because we noticed that the effect was largely due to a few outliers. Indeed, the z rotation finding was no longer significant when using absolute values. The displacement findings, however, held.
Figure 6.
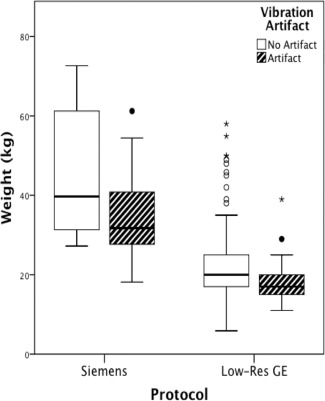
Boxplots of subjects’ weight (kilograms) grouped by scanning protocol and presence of the artifact. Dark lines within boxplot represent median values.
For the discriminant analysis, the value of the discriminant function was different for data with the artifact and data without the artifact for both Siemens (Wilks λ = 0.581, Chi‐squared = 18.198, df = 7, Canonical correlation = 0.647, P = 0.011) and Low‐Res GE data (Wilks λ = 0.878, Chi‐squared = 29.429, df = 7, Canonical correlation = 0.350, P < 0.001). However, consistent with MANOVA results, there were differences in findings by scanner. For Siemens, correlations between predictor variables and the discriminant function suggested that y displacement and weight, followed by z and y rotation were the best predictors of vibration artifact. The function extracted for Siemens data accounted for 72.2% of the variance for whether there was vibration artifact present or not, suggesting some added value for considering the linear combination of variables. The Siemens data function successfully predicted outcome for 76.9% of cases, with accurate predictions being made for 80.0% of data with no vibration artifact and 73.7% of data with vibration artifact. For Low‐Res GE data, weight and z displacement were the best predictors of vibration artifact, followed by y rotation, x displacement, and z rotation, respectively. The function extracted for Lo‐Res GE data only accounted for 13.9% of the variance for whether there was vibration artifact present or not. As a result, there was successful prediction of outcome for only 64.1% of cases, with accurate predictions being made for 59.9% of data with no vibration artifact and 76.3% of data with vibration artifact.
Single Subject Evaluation of Strategies for Avoiding the Artifact
For the single subject, the artifact was observed in the raw diffusion data (Fig. 7A) and DEC map (Fig. 7B). We found that the artifact was reduced when using full k‐space coverage, but not completely eliminated (Fig. 7C), and was eliminated by altering the head position with a +45° roll (Fig. 7D). Increasing the TR, setting the diffusion preparation to dual‐spin‐echo, and altering the pitch did not affect the artifact beyond the improvement made when using full k‐space acquisition.
Figure 7.
Effect of head rotation and k‐space coverage on the occurrence of the artifact in repeated scans of the same subject. (A) Raw diffusion data. Diffusion gradient direction was (x, y, z) = (0.9, −0.42, 0.11) (B) DEC map with original settings, (C) DEC map using full k‐space coverage, and (D) DEC map with altered head position.
DISCUSSION
We investigated the prevalence of a previously described vibration artifact in DWI data collected in pediatric clinical datasets on different scanners. We found that the prevalence of the artifact was present across multiple scanner manufacturers (not just Siemens) and affected by acquisition protocol. Even for the same scanner, software and hardware upgrades were strongly associated with changes in the incidence of the problem. Weight and positioning of the subject also were factors in its manifestation. While an automated software tool is available to detect the artifact (DTIPrep), on our data, it showed very low sensitivity and, therefore, in our view, visual inspection should still be the method of choice for detecting the artifact. Lastly, longitudinal data showed that the artifact is often inconsistently present in repeated scans of the same subject. We discuss the implications of our findings for interpreting DTI scans.
Having a better sense of the frequency of the artifact is important for determining the risk level of how this artifact may impact one's data and the potential for lost resources. Our results suggest that in the worst‐case scenario, a majority of one's data could be contaminated, but the risk is scanner and acquisition protocol dependent. Changing acquisition protocols to different resolutions or upgrading of sequences that presumably include gradient changes may impact the likelihood of having a vibration artifact. For example, the Siemens upgrade to the “TQ‐engine system” resulted in a significant increase of the problem. For the GE scanner, vibration artifacts were present only in the low‐resolution acquisitions and in a particular time window prior to a software upgrade. As Gallichan and coworkers suggested, k‐space coverage influences manifestation of the artifact. However, on the Siemens scanner during our pilot testing, increasing k‐space coverage did not completely eliminate the problem. On the GE scanner, there were changes to the amount of k‐space coverage between software versions before and after upgrade resulting in a decrease in artifact. A similar low resolution protocol on GE scanners was used by the NIH MRI Study of Normal Brain Development (http://pediatricmri.nih.gov). A systematic quality assessment performed in preparation of that database showed no instances of vibration artifact in data acquired on GE scanners with this protocol [Nayak et al., 2011]; however, data were collected with an earlier version of the software than that used in this study. Unfortunately, these software changes implemented by the manufacturers are often not known by the end user.
This study highlights that the artifact is not limited to a single scanner manufacturer. It would be informative for clinical research centers with other scanner manufacturers (e.g., Philips) to conduct a similar review to compare to our findings. Moreover, as new gradient systems and upgrades become available, we suggest that DTI data should be systematically inspected for this artifact. The artifact is relatively easy to identify because it is pervasive enough and consistent regarding location, with occipital‐parietal regions being mostly affected, although we and other investigators also found vibration artifacts in frontal areas. We found that viewing an entire volume across axial slices is optimal for checking for the artifact and were able to achieve very high interrater reliability with fairly simple criteria. There are several reasons for relying on visual inspection rather than an automated tool for identifying this artifact. As also noted by another group [Farzinfar et al., 2013], we highlight that vibration artifacts can result in regions of spurious anisotropy (with left‐right preferential orientation and red hue in the DEC maps) without an obvious abnormality in the residuals of the tensor fitting and the chi‐squared map. Essentially, the signal dropout in the x‐direction can increase with a function sufficiently compatible with the tensor model to prevent chi‐squared values from appearing abnormal (Fig. 2B). Even if an abnormality in the chi‐squared data is present, using RESTORE [Chang et al., 2005] removes the data most contributing to the abnormality and normalizes chi‐squared results (Fig. 3D). Unfortunately, in the case of vibration artifacts, significant artifacts may still be present in the DEC maps even if the chi‐squared map is normalized (Fig. 3C). Thus, chi‐squared values from within vibration artifact regions have high variability, which then diminishes the specificity of that metric as a helpful in detecting the vibration artifact.
The “coregressor” [Gallichan et al., 2010] approach is another method proposed to detect and potentially remediate the vibration artifact. We did not test this method because there is not software readily available to apply to large datasets. However, as a hypothesis for further investigation, we would expect this method to perform similarly to RESTORE because it attempts to detect abnormal signal attenuation that is not consistent with the tensor model, which is what RESTORE does, although the specific metric used is different.
While automated detection tools for this artifact would be useful to avoid the tedious visual inspection step, the only currently available tool, the entropy tool implemented in DTIPrep [Farzinfar et al., 2013], has limitations. First, it can only be used on a large dataset because it needs enough artifact‐free data to establish an appropriate range of entropy values. Thus, our Siemens dataset that had only 43 total datasets but half were impacted by the artifact could not undergo the training step. Second, while there was enough GE data to go through the entropy pipeline, the sensitivity of the tool was very low (10%). There are possible reasons for this unsatisfactory result. One reason is that the tool may require artifacts to be large in magnitude and spatial extension to be detected while the vibration artifact in our GE datasets was often subtle as compared with the Siemens datasets (Fig. 1 vs. 3A). In fact, a more recent paper from the developers of this tool reports the rates of detection of the entropy tool as 100% for severe artifact but as low as 67% for more subtle artifact [Oguz et al., 2014]. We consulted the developers to inquire about other possible explanation for our results and they suggested that a reason could be the mask used for the entropy computation. The developers used brain masking that removes all cerebrospinal fluid (CSF) regions as well as the usual skull stripping [Farzinfar et al., 2013], but this method of masking is not currently part of their DTIPrep automated pipeline [Styner, 2015].
We consider factors that contribute and strategies to address the artifact. Siemens addressed the problem by updating the scanner bed [Liu and Liu, 2011]; however, there is a report that the problem is improved but persists even following a scanner bed upgrade [Farzinfar et al., 2013], and many existing systems may still have the old bed configuration. We are aware that manufacturers have worked to improve equipment and software to reduce artifacts, which is evident from the timing of the GE data where the artifact was no longer evident in scans acquired at a later date. Increased weight was associated with avoiding the artifact for both GE and Siemens scanners. Weight was particularly influential for Siemens data, which is consistent with how the problem is described in Siemens’ own analysis of the problem in their patent. Our prospective pilot testing on a single subject indicated that altering the roll of head placement potentially affects the artifact. Specifically, the artifact was reduced with the head slightly rotated. Perhaps, repeating the scan with the head rotated could be used as a strategy for addressing the problem. Rotational parameters were not associated with the artifact in the archival review, perhaps because the range of rotation was quite narrow and only a few datasets exceeded 20° while the pilot testing was done at a more dramatic angle of about 45°. The direction of the displacement associated with frequency of the artifact differed by manufacturer/protocol. For Siemens, greater y displacement (i.e., having the subject's head further elevated and away from the bed) was associated with not having artifact, which may be due to having more padding under a child's head. Either the padding or being farther off the scanner bed may have disrupted the vibration's effect during the scan. For GE, greater z displacement (i.e., having the subject's head deeper in the scanner) was associated with not having the artifact. Nonetheless, our modeling with these factors did not explain all the variance—and certainly there are other factors that influence whether the vibration artifact is present, particularly for GE datasets. For Siemens, however, we accounted for a large portion of the variance and were able to predict 77% of cases accurately with information that is standardly known at the outset of a scanning session (weight and y position). Based on this data and our pilot testing, we suggest that altering the position of the head and weight load may reduce the effects of the vibrations within the brain and disrupt the directional bias of the artifact. In the Siemens scanner, repositioning (deeper in the MRI scanner, increased padding to elevate from the bed, and/or rotated from center) or adding of weight may be solutions in addition to changing parameters or doing a hardware replacement of the scanner bed.
Pediatric datasets may be prone to the artifact regardless of scanner type. One reason is because children weigh less. The Siemens patent demarcated a weight of 30 kg or less as the tipping point for when the load on the scanning bed is susceptible to vibration artifact. We found a weight of approximately 34 kg for our Siemens data as the mean weight for those cases with the artifact. In comparison, the artifact was less frequent in the GE data, which may be related to a different weight threshold (approximately 18 kg in our datasets) for when the scans are more susceptible to the artifact. However, there was overlap in the weights of those with and without the artifact, which, similar to our discriminant function, indicates that weight is not the only factor in determining whether the artifact is present. Interestingly, the one other study that examined this artifact also found a higher prevalence of vibration artifact in their youngest age group (6‐months old) even though they did not explicitly examine weight as a factor [Farzinfar et al., 2013]. Examination of all possible factors that contribute to the presence of the artifact are not within the scope of this study as this was largely a retrospective review of data. Future investigations would benefit from a more systematic approach that might include a more careful examination of the factors such as acquisitions at higher b‐values or higher field strength.
Implications for the Interpretation of Clinical Studies
The finding that weight affects the incidence of this vibration‐induced artifact suggests that care should be used in ruling out this confound when data from groups of subjects with different weight are compared. Moreover, our finding that this artifact manifested itself inconsistently in repeated scans of the same subject has important implications for the interpretation of the results of longitudinal diffusion MRI studies. Typically, changes in diffusion metrics, such as anisotropy, between time points are interpreted as indicative of changes in white matter structure and architecture. Given that this artifact could manifest itself inconsistently, it is important to confirm that regional changes in anisotropy are indeed caused by brain changes as opposed to the inconsistent presence of this—or any other—artifact. In this regard, we suspect that this artifact may have lead to misinterpretation of diffusion MRI findings in a previously published clinical study [Voss et al., 2006]. This case study claimed that DTI detected brain plasticity, in a 39‐year old man who spontaneously recovered from being in a minimally conscious state for 19 years. The patient underwent two scans conducted 18‐months apart. There was increased FA in the parietal‐occipital region with red appearance in the DEC map at Time 1 that was not present at Time 2. The differences in anisotropy between the two time points were interpreted by the authors as a “transitional stage of an ongoing process…of possible axonal regrowth.” The increased anisotropy with left‐right apparent fiber orientation found in this patient was very consistent to the manifestation of this artifact in our data (Fig. 5) and in the data reported in other publications [Gallichan et al., 2010; Jones, 2010; Tournier et al., 2011].
The notoriety of this case and the findings led to national and international media coverage [“Man Speaks after 19‐Year Silence,” 2003; BBC, 2003]. Beyond general public interest, the case report was also propagated within the scientific community as it has been cited 145 times using an electronic search performed in Web of Science (Science Citation Index All years 1900–2014; final search was completed June 10, 2014 using the title of the Voss article as the search criteria). While some citations were related to the length of survival for the patients, several citations of this study were for issues that had far‐reaching policy and economic impact including: a call to change medical dogma about the recovery possibilities of brain‐injured patients [Laureys et al., 2006; Schiff, 2009]; evidence for dedicating resources for systematic and longitudinal study of minimally conscious patients [Fins et al., 2007; Fins, 2011]; a “remarkable example of regenerative fibre growth” in support of pursuing drug development for traumatic brain injury [Mueller et al., 2009]; and a discussion point within an Institute of Medicine exploratory meeting to set policy [Fins, 2011; Fins et al., 2007]. Although some of the discussion within these citations called for caution and recognized that the neuroimaging evidence was preliminary as it was based on a single case study, none of the papers considered the possibility of misinterpretation of artifactual data.
The speed at which this article was propagated was assisted by the inherent interest of the case details; however, it also illustrates the danger of univocal clinical interpretation of DTI results. If we would have not considered the possibility of this artifact, we could have claimed “plasticity changes” in the brain of 34 of our subjects who demonstrated similar anisotropy changes in repeated scans. While increased FA can be caused by reorganization or regrowth of axons, alternative explanations must be considered as possible sources of DTI changes [Budde et al., 2011]. We cannot be certain that the findings of that paper were due to vibrational artifact; however, we feel that there is a high likelihood because of the location and plausibility of the findings. Interhemispheric parietal‐occipital white matter plasticity is unlikely as a physiological phenomenon but is exactly where the vibration artifact manifests. Moreover, given that the “axonal regrowth” was evident at the first time point and disappeared at the second time point, this is contrary to conventional understanding of neural reorganization principles where it is expected that regrowth should persist because it would be adaptive. Another possibility is that certain brains may have mechanical properties that make them more vulnerable to vibrational artifact. For example, the brain‐injured patient in the case report had a substantial degree of atrophy and, therefore, may have had a greater amount of cerebrospinal fluid compared to tissue. Similarly, young children may have larger water content in the brain parenchyma, which may lead to brains of younger subjects to have different mechanical properties from adults and be more susceptible to the occurrence of this DWI artifact for the same level of vibration. Conducting repeat scans with different positioning of the subject would have been a potential test to rule out an artifactual origin of the measured anisotropy. Repositioning the patient by adjusting the roll should not affect anisotropy caused by “axonal regrowth” and the pattern should be consistently present in subsequent scans.
CONCLUSION
This work reported a systematic analysis of the prevalence of a previously described vibration artifact in DWIs across a population. The most relevant findings include: (1) The artifact can be found in scanners other than Siemens Trio, in particular in pediatric subjects. (2) Children may be particularly susceptible because they weigh less than adults. (3) We investigate systematically how different factors (scanner, acquisition, weight, positioning) can affect the prevalence of this artifact. We identify head rotation as a new factor that may alter the mechanical coupling and ultimately the manifestation of this artifact. (4) We show that an automated method to identify the artifact was not as sensitive as visual inspection in GE data where the artifact is more subtle. (5) We confirm that postprocessing remedies that are highly effective at reducing other types of artifacts, such as the RESTORE approach, are largely ineffective in removing this artifact. (6) We also find that some of the solutions proposed to avoid the artifact, such as collecting full k‐space data, do not completely eliminate the artifact on our Siemens scanner, (7) Lastly, we address the potential for clinical misinterpretation by discussing a previously published case where changes consistent with this vibration artifacts have been interpreted as having unequivocally a biological origin. Our study underscores the need to check for the presence of this for the vibration artifact, we propose simple strategies to identify and potentially avoid its effects.
ACKNOWLEDGMENTS
The authors would like to acknowledge Jay Giedd, Audrey Thurm, and Susan Swedo for providing access to diffusion MRI data used in the retrospective analysis of vibration artifacts as well as participating parents and children. The diffusion data processing and analysis tools used for this work have been developed and implemented with contributions by employees of the Henry M. Jackson Foundation for the Advancement of Military Medicine Inc. The content of the information does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred. For purposes of this article, information includes news releases, articles, manuscripts, brochures, advertisements, still and motion pictures, speeches, trade association proceedings, etc. None of the authors have any conflicts of interest to report.
REFERENCES
- Aso T, Urayama S, Fukuyama H, Le Bihan D (2013): Comparison of Diffusion‐weighted fMRI and BOLD fMRI responses in a verbal working memory task. NeuroImage 67:25–32. (February): doi: 10.1016/j.neuroimage.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C (1996): Microstructural and physiological features of tissues elucidated by quantitative‐diffusion‐tensor MRI. J Magn Reson B 111:209–219. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D (1994): MR diffusion tensor spectroscopy and imaging. Biophys J 66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BBC (2003): US Man Wakes from 19‐Year Coma, July 9, sec. Americas. Available at: http://news.bbc.co.uk/2/hi/africa/3052433.stm.
- Budde MD, Janes L, Gold E, Turtzo LC, Frank JA (2011): The contribution of gliosis to diffusion tensor anisotropy and tractography following traumatic brain injury: Validation in the rat using fourier analysis of stained tissue sections. Brain 134:2248–2260. doi: 10.1093/brain/awr161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LC, Jones DK, Pierpaoli C (2005): RESTORE: Robust estimation of tensors by outlier rejection. Magn Reson Med 53:1088–1095. doi: 10.1002/mrm.20426. [DOI] [PubMed] [Google Scholar]
- Chang LC, Walker L, Pierpaoli C (2012): Informed RESTORE: A method for robust estimation of diffusion tensor from low redundancy datasets in the presence of physiological noise artifacts. Magn Reson Med 68:1654–1663. doi: 10.1002/mrm.24173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M Farzinfar, I Oguz, RG Smith, AR Verde, C Dietrich, A Gupta, ML Escolar, J Piven, S Pujol, C Vachet, S Gouttard, G Gerig, S Dager, RC McKinstry, S Paterson, AC Evans, MA Styner (2013): Diffusion imaging quality control via entropy of principal direction distribution. NeuroImage 82:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fins JJ (2011): Neuroethics, neuroimaging, and disorders of consciousness: Promise or peril? Trans Am Clin Climatol Assoc 122:336–346. [PMC free article] [PubMed] [Google Scholar]
- Fins JJ, Schiff ND, Foley KM (2007): Late recovery from the minimally conscious state–Ethical and policy implications. Neurology 68:304–307. doi: 10.1212/01.wnl.0000252376.43779.96. [DOI] [PubMed] [Google Scholar]
- Gallichan D, Robson MD, Bartsch A, Miller KL (2009): TREMR: Table‐resonance elastography with MR. Magn Reson Med 62:815–821. doi: 10.1002/mrm.22046. [DOI] [PubMed] [Google Scholar]
- Gallichan D, Scholz J, Bartsch A, Behrens TE, Robson MD, Miller KL (2010): Addressing a systematic vibration artifact in diffusion‐weighted MRI. Hum Brain Mapp 31:193–202. doi: 10.1002/hbm.20856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen J, Hari R, Jousmäki V, Müller K, Sepponen R, Joensuu R (2006): Quantification of mechanical vibration during diffusion tensor imaging at 3 T. NeuroImage 32:93–103. doi: 10.1016/j.neuroimage.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Jensen JH, Helpern JA (2010): MRI quantification of non‐gaussian water diffusion by kurtosis analysis. NMR Biomed 23:698–710. doi: 10.1002/nbm.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK (2010): Precision and accuracy in diffusion tensor magnetic resonance imaging. Top Magn Reson Imaging 21:87–99. [DOI] [PubMed] [Google Scholar]
- Jones DK, Knösche TR, Turner R (2012): White matter integrity, fiber count, and other fallacies: The do's and don'ts of diffusion MRI. NeuroImage 73:239–254. doi: 10.1016/j.neuroimage.2012.06.081. [DOI] [PubMed] [Google Scholar]
- Koch MA, Finsterbusch J (2011): Towards compartment size estimation in vivo based on double wave vector diffusion weighting. NMR Biomed 24:1422–1432. doi: 10.1002/nbm.1711. [DOI] [PubMed] [Google Scholar]
- Laureys S, Giacino JT, Schiff ND, Schabus M, Owen AM (2006): How should functional imaging of patients with disorders of consciousness contribute to their clinical rehabilitation needs? Curr Opin Neurol 19:520–527. doi: 10.1097/WCO.0b013e3280106ba9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KC, Liu XG (2011): Vibrationally decoupled patient table for use in magnetic resonance system, April. Available at: http://www.google.com/patents/US7924008.
- Mackinnon A (2000): A spreadsheet for the calculation of comprehensive statistics for the assessment of diagnostic tests and inter‐rater agreement. Comput Biol Med 30:127–134. [DOI] [PubMed] [Google Scholar]
- Man speaks after 19-year silence. (2003, July 7). Retrieved May 24, 2014, from http://www.cnn.com/2003/US/South/07/07/mute.no.more/
- Mohammadi S, Keller SS, Glauche V, Kugel H, Jansen A, Hutton C, Flöel A, Deppe M (2012a): The influence of spatial registration on detection of cerebral asymmetries using voxel‐based statistics of fractional anisotropy images and TBSS. PLoS One 7:e36851 doi: 10.1371/journal.pone.0036851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi S, Nagy Z, Hutton C, Josephs O, Weiskopf N (2012b): Correction of vibration artifacts in DTI using Phase‐encoding reversal (COVIPER). Magn Reson Med 68:882–889. doi: 10.1002/mrm.23308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi S, Nagy Z, Möller HE, Symms MR, Carmichael DW, Josephs O, Weiskopf N (2012c): The effect of local perturbation fields on human DTI: Characterisation, measurement and correction. Neuroimage 60:562–570. doi: 10.1016/j.neuroimage.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi S, Freund P, Feiweier T, Curt A, Weiskopf N (2013): The impact of post‐processing on spinal cord diffusion tensor imaging. NeuroImage 70:377–385. (April): doi: 10.1016/j.neuroimage.2012.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller BK, Mueller R, Schoemaker H (2009): Stimulating neuroregeneration as a therapeutic drug approach for traumatic brain injury. Br J Pharmacol 157:675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller K, Anwander A, Möller HE, Horstmann A, Lepsien J, Busse F, Mohammadi S, ML Schroeter, M Stumvoll, A Villringer, B Pleger (2011): Sex-Dependent Influences of Obesity on Cerebral White Matter Investigated by Diffusion-Tensor Imaging. PLoS ONE 6 Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3073967/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak A, Walker L, Pierpaoli C, Brain Development Cooperative Group (2011): Quality assessment in a DTI multicenter study. Proc Int Soc Mag Reson Med 19:2342. [Google Scholar]
- Oguz I, Farzinfar M, Matsui J, Budin F, Liu Z, Gerig G, Johnson HJ, Styner M (2014): DTIPrep: quality control of diffusion‐weighted images. Front Neuroinform 8:4. doi: 10.3389/fninf.2014.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajevic S, Pierpaoli C (1999): Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: Application to white matter fiber tract mapping in the human brain. Magn Reson Med 42:526–540. doi:. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C ( 2010): Artifacts in diffusion MRI. Diffusion MRI: Theory, Methods and Applications. New York: Oxford University Press; pp 303–318. [Google Scholar]
- Pierpaoli C, Jezzard P, Basser PJ, Barnett A, Di Chiro G (1996): Diffusion tensor MR imaging of the human brain. Radiology 201:637–648. doi: 10.1148/radiology.201.3.8939209. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C, Walker L, Irfanoglu MO, Barnett A, Basser P, Chang LC, Koay C, Pajevic S, Rohde G, Sarlls J (2010): TORTOISE: An integrated software package for processing of diffusion MRI data. Proc Int Soc Mag Reson Med 18:1597. [Google Scholar]
- Rapacchi S, Wen H, Viallon M, Grenier D, Kellman P, Croisille P, Pai VM (2011): Low B‐value diffusion‐weighted cardiac magnetic resonance imaging. Invest Radiol 46:751–758. doi: 10.1097/RLI.0b013e31822438e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese TG, O Heid, RM Weisskoff, VJ Wedeen (2003): Reduction of Eddy-Current-Induced Distortion in Diffusion MRI Using a Twice-Refocused Spin Echo. Magnetic Resonance in Medicine: Official Journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine 49:177–182. doi: 10.1002/mrm.10308. [DOI] [PubMed] [Google Scholar]
- Scherrer B, Warfield SK (2012): Retrospective local artefacts detection in diffusion‐weighted images using the random sample consensus (RANSAC) paradigm. In: 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI). Barcelona. pp 546–549. http://ieeexplore.ieee.org/xpls/abs_all.jsp?arnumber=6235606.
- Schiff ND (2009): Central thalamic deep‐brain stimulation in the severely injured brain. Ann N Y Acad Sci 1157:101–116. (doi:10.1111/j.1749‐6632.2008.04123.x. [DOI] [PubMed] [Google Scholar]
- Sharman MA, Cohen‐Adad J, Descoteaux M, Messé A, Benali H, Lehericy S (2011): Impact of outliers on diffusion tensor and Q‐ball imaging: Clinical implications and correction strategies. J Magn Reson Imaging 33:1491–1502. doi: 10.1002/jmri.22577. [DOI] [PubMed] [Google Scholar]
- Styner MA (2015): Entropy Results. January 2.
- Tournier J‐D, Mori S, Leemans A (2011): Diffusion tensor imaging and beyond. Magn Reson Med 65:1532–1556. doi: 10.1002/mrm.22924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HU Voss, AM Uluç, JP Dyke, R Watts, EJ Kobylarz, BD McCandliss, LA Heier, BJ Beattie, KA Hamacher, S Vallabhajosula, SJ Goldsmith, D Ballon, JT Giacino, ND Schiff (2006): Possible axonal regrowth in late recovery from the minimally conscious state. J Clin Invest 116:2005–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]


