Abstract
Chronic rhinosinusitis (CRS) has been known as a disease with strong infectious and inflammatory components for decades. The recent advancement in methods identifying microbes has helped implicate the airway microbiome in inflammatory respiratory diseases such as asthma and COPD. Such studies support a role of resident microbes in both health and disease of host tissue, especially in the case of inflammatory mucosal diseases. Identifying interactive events between microbes and elements of the immune system can help us to uncover the pathogenic mechanisms underlying chronic rhinosinusitis. Here we provide a review of the findings on the complex upper respiratory microbiome in CRS in comparison to healthy controls. Furthermore, we have reviewed the defects and alterations of the host immune system that interact with microbes and could be associated with dysbiosis in CRS.
Introduction
Chronic rhinosinusitis (CRS) is a common inflammatory disease of the upper airways that afflicts about one tenth of US adults[1, 2]. The chronic inflammation in the sinus and nasal mucosa in CRS results in a constellation of symptoms that can have a significant negative effect on an individual’s quality of life [3].
It has been known for a long time that the membranes of the nasal mucosa are home to a large number of microorganisms such as fungi, bacteria and viruses that are found in subjects both with and without sinus disease. CRS is regarded as a disease of inflammation rather than infection. However, numerous studies have demonstrated roles for commensal resident microbes and pathogens, or their associated products, in the initiation and/or progression of mucosal inflammation[4, 5].
Microbes in the human body, like microbes in any other living organism, tend to adapt to their environment and to changes in that environment. Commensal microorganisms and their metabolites maintain the stability of their habitat and the health of their host in what is often viewed as a symbiotic system [6],[7]. For example symbiotic microbiota interact with gut immune cells to regulate intestinal homeostasis [8]. It is noteworthy that the effects of the microbiota are not limited to its immediate location but rather can impact the entire system of the host. For example, a healthy microbiome in the gastrointestinal system is necessary for development and training of a normal immune system[9, 10]. A disrupted microbiome in the gut, especially early in life, has been linked to the development of obesity and multiple inflammatory conditions in the rest of the body [11]. Of note in the context of this review, the gut microbiome has also been linked to multiple allergic diseases including asthma and eczema [12–14].
Multiple environmental factors – including birth mode, feeding patterns, diet, hygiene, and other life style measures – can alter the microbiome and affect its influence on the development of inflammatory conditions[15]. It is noteworthy that development of a healthy microbiome in the body is dependent on the individual’s exposure to a surrounding environment rich in microbes. Moreover, diversity of microbial exposure is inversely correlated with development of allergic and inflammatory conditions such as asthma and allergic rhinitis[16–18]. Various studies have reported findings that microbes are protective against allergic diseases, which was in part the basis for the “hygiene hypothesis”[19]. The protective effect of living on a farm against the development of allergy and asthma has been known for decades [20]. This protective role has been suggested to occur at least partially via exposure to a more diverse group of microorganisms in the farm environment[16, 21, 22]. In mouse models, administration of certain bacteria, including ones that were isolated from farm environments, produced a significant reduction in the allergic inflammatory reaction in lungs, with a decrease in the number of eosinophils and mucus-producing goblet cells in airway, and reduction in allergy-induced airway hyperreactivity[23–25]. Also, exposure to dust from animal-enriched environments has been shown to change the gut microbiome in mice and to consequently protect them from allergen-mediated airway disease. Fujimura et al confirmed that the bacteria from dust from houses associated with a dog are responsible for this effect[26].
A disturbance in the composition of the local microbiome has been shown in asthma and COPD[27, 28]. Multiple studies have shown major differences in the microbiome of the lung captured by bronchoscopy or induced sputum in asthmatic versus non-asthmatic individuals[28–30]. Furthermore, the airway microbiome was shown to affect the response to corticosteroid therapy in asthma[31].
It is possible that the local microbiome could be an epiphenomenon.
In this scenario, the inflammation, or components of host defense, elicited as a result of the disease condition create an environment in which certain microbes can or cannot grow and reproduce. Consequently, the microbiome can sustain and promote a pre-existing inflammation that has been induced primarily by the underlying disease [32]. Whether microbiome changes are a cause or an effect of inflammatory disease remains to be answered for multiple chronic inflammatory conditions associated with disrupted epithelia. For example, superinfection with S. aureus is one of the most common complications seen in atopic dermatitis. However, it is not clear whether skin inflammation triggers the changes in composition of colonizing microorganisms, allowing overgrowth of Staphylococcus, whether Staphylococcus overgrowth is an initiating event that affects the abundance of other species and triggers the inflammation, or both.
In the case of CRS, pathogen colonization and microbiota imbalance might be initial causes of the chronic immune response and inflammation. On the other hand, a dysfunctional immune barrier, inflamed mucosal epithelium and obstructed sinuses resulting from allergic or non-allergic inflammation may promote a condition suitable for secondary bacterial overgrowth and dysbiosis, setting the stage for chronic rhinosinusitis.
Studying the microbiome might be a useful approach for finding answers to some of the unsolved questions about CRS pathogenesis and for improving our knowledge about the inflammatory processes in this disease. Multiple studies have investigated the association of CRS with local microbial structures isolated from the sinuses or nasal cavities. Also, recent studies have used genomic analyses to evaluate nasal and sinus bacterial and fungal microbiomes in CRS, and have found associations between certain microorganisms and CRS. However these associations do not establish causality between the presence of these microbes in the airways and the development of disease. The present review aims to provide a summary of these findings by discussing the results of studies on the microbiome in CRS. We also briefly review the literature on the microbiome in asthma, another respiratory disease that is often comorbid with CRS.
The upper respiratory tract microbiome in chronic sinusitis
The association between CRS and commensal or pathogenic microbes cultured from the nasal cavity and paranasal sinuses has been under investigation for many decades. The sinuses were found to be colonized by microbial flora in both normal and disease conditions in the 1990s[33]. Multiple microorganisms isolated from sinus and nose in CRS patients, such as coagulase-negative staphylococci [34] have been identified and have been proposed as pathogens that mediate infectious processes in CRS and that result in chronic inflammation in nasal and sinus mucosa. This view has led to sinusitis (including both acute and chronic) to be the number one indication for the use of antibiotics. Furthermore, some studies have evaluated possible mechanisms by which selected bacteria could influence or initiate an immunologic cascade resulting in the chronic mucosal inflammation seen in CRS. The association between Staphylococcus aureus (S. aureus) superantigen specific IgE and skewing towards a Th2 milieu and eosinophilia in CRS is a notable example[35].
During the past two decades, it has been shown that conventional culture-based methods are not sufficient to elucidate the complex ecology of the bacterial populations of the human body. This was based, to a large extent, on studies in the mid-1980s when a group of investigators amplified the highly conserved 16S ribosomal ribonucleic acid (rRNA) bacterial gene from samples of organisms isolated from deep sea regions. They found that less than 1% of the organisms from those isolations could be cultured[36]. This method was later applied to microbiota studies in humans and other mammals, uncovering a vast and impressive ecosystem of microorganisms in virtually all epithelia.
Here we will focus on the recent studies in which genetic techniques have been used to characterize the local microbiome of the nasopharynx or sinuses with the goal of discovering an association with CRS. It is notable that such studies are fraught with challenges like recent use of antibiotics and steroids, exacerbations and sampling techniques. Another significant challenge results from a recent appreciation of the variability in constitutive epithelial host defense molecule secretion based on the region of the nose and sinuses[37]. Consistently, Yan et al have shown a significant variation in the microbial composition of anterior nares compared to middle meatus and sphenoethmoidal recess in healthy individuals[38]. Interestingly, they found no difference in the bacterial composition of the two latter sites. Relatively little is stated in published studies about the region from which microbiome samples were collected, so it is difficult to evaluate to what extent this concern applies to the available literature. Such challenges to the interpretation of findings must be kept in mind while considering the complex relationship between disease and the microbiome in CRS.
Summary of data from culture studies
A diverse group of bacteria have been identified in chronic rhinosinusitis based on culture methods. We have briefly reviewed the results of some of these studies below.
In 1998, Beil et al. published the results of bacterial cultures from endoscopically collected samples at the time of surgery in 174 CRS patients. The most commonly cultured bacteria were Coagulase-negative staphylococci (36%), followed by S. aureus (25%), Streptococcus viridans (8.3%), Corynebacterium (4.6%), and anaerobes (6.4%)[34]. Jiang et al. obtained culture samples from the ethmoid sinus by inserting a cotton-tip stick through a cannula placed in the ethmoid cavity after removing the ethmoid bulla. In this study, 39 CRS patients from Taiwan were included. They reported a positive culture rate of 60.9%. The most frequently isolated bacteria in this study were Streptococcus viridans, Klebsiella pneumoniae, Proteus mirabilis, and Haemophilus parainfluenzae [39].
In a retrospective study of nasal and sinus culture data obtained from 83 patients with CRS, positive cultures were reported in 71% of cases. The most common bacteria were reported as coagulase-negative staphylococci (31% of isolates), Haemophilus influenzae (H. Influenza) (25%), Streptococcus pneumoniae (S. pneumoniae) (12%), Moraxella catarrhalis (M. catarrahalis) (10%), Pseudomonas aeruginosa (P. aeruginosa) (7%), alpha-hemolytic streptococci (5%), and S. aureus (3%)[40].
Brook et al reported the results of cultures from sinus aspirates obtained during surgery in 6 patients with CRS. Polymicrobial flora was found in all specimens. The predominant organisms in this study were Prevotella spp.(13 isolates), followed by Fusobacterium nucleatum (11), Peptostreptococcus spp. (6), Staphylococcus spp (5), H. Influenza (2) and S. pneumoniae (2)[41].
A more recent study on a cohort of CRSwNP patients from Korea showed 58.0% positive aerobic and 8.6% positive anaerobic cultures in samples from the middle meatus, and 48% and 18.5% positive aerobic and anaerobic cultures in samples of maxillary sinus. The most commonly isolated species were S. aureus, H. influenzae, and S. pneumoniae and the predominant anaerobic organisms were Prevotella and Peptostreptococcus[42].
Despite the discrepancies among these results, which could be due to differential culture(including use of anaerobic cultures) and sampling methods, most of these culture-based analyses have reported evidence for staphylococci and streptococci spp. along with H. Influenza as predominantly detected bacteria in CRS. In the studies that have also evaluated anaerobes in sinus, Prevotella and Peptostreptococcus were commonly found.
It is noteworthy that multiple studies have shown a lack of correlation between nasal cultures with sinus cultures, which further implicates differences in sampling region as a major cause of discrepancy seen in microbial studies in CRS, regardless of analytical method[43]. Furthermore, as mentioned previously, a significant proportion of organisms from different isolations can not be cultured[36]. This makes culture-based studies less informative compared to PCR-based analyses.
Bacterial microbiome
The initial studies that used PCR-based methods to analyze bacteria-specific 16s ribosomal deoxyribonucleic acid (DNA) fragments and coding rRNA in CRS were limited by their use of a small number of primer sets focused on the capture of known bacterial flora of the upper respiratory tract. Moreover, these methods were time consuming and labor intensive [44],[45]. However, these studies expanded our knowledge beyond studies that were based on culture. Paju et al used 3 primer sets and found positive bacterial DNA results in 5 out of 11 CRS patients [45]. Keech et al, analyzed 64 samples from 32 CRS cases and focused on primer sets capturing DNA from Staphylococcus aureus (S. aureus), Staphylococcus epidermidis (S. epidermidis), Streptococcus pneumoniae (S. pneumoniae), Pseudomonas aeruginosa (P. aeruginosa), Moraxella catarrhalis, Haemophilus influenzae (H. influenzae), and Streptococcus pyogenes. They found that 62% of CRS patients were positive for at least one of these bacteria, while parallel culture method had found positive growth in 50% of cases[44]. In addition, except for Pseudomonas aeruginosa, all the tested bacteria were found in at least 3 cases[44]. Power et al used PCR-denaturing gradient gel electrophoresis (PCR-DGGE) and species-specific primers analyzing middle meatus aspirates from 6 patients with chronic rhinosinusitis. This study detected S.aureus and S. pneumoniae in 4 out 6 patients, the parallel cultures showed positive culture for at least one bacterium in all samples and was consistent with the PCR-DGGE results. The comparison between PCR-DGGE and cultures showed that PCR based method was able to detect a greater diversity of bacteria [46].
Since 2010, more advanced microbiome analytical methods, ones that are able to detect multiple bacteria simultaneously, have been used to study CRS [47][48]. Results of these studies are summarized below and in Table I.
Table 1.
Results of studies on Nasal microbiome in CRS
| Bacterial Microbiome Studies | ||||||||
|---|---|---|---|---|---|---|---|---|
| Name and year of publication |
Study participants |
Sample type |
Analysis method |
Microbiome analysis Results | ||||
| Microbial Diversity |
Microbial Abundance |
Differences at Phylum level |
Differences at order, family or genus level |
Differences at Species level |
||||
| Stephenson et al, 2010 (48) | 18 CRS 9 controls |
Anterior ethmoid mucosal biopsy | Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) | - | - | - | Propionibacterium found in 83% of CRS vs. 67% of controls(no statistics available) Diaphorobacter spp. And Peptoniphilus only reported in CRS(78% and 72% respectively) | Staphylococcus aureus found in 50% of CRS vs 100% of controls (no statistics available) |
| Stressmann et al, 2011(49) | 70 samples from 43 CRS No control |
Mucosal biopsy | 16S-rDNA sequencing and terminal restriction fragment length polymorphism (T-RFLP) | No control | - | - | Most abundant: Pseudomonas, Citrobacter, Haemophilus, Propionibacterium, Staphylococcus and Streptococcus | - |
| Abreu et al, 2012(53) | 7 CRS 7control |
Endoscopically guided maxillary sinus brushing | 16S rRNA PhyloChip Phylogenetic microarray approach | Decreased in CRS:
|
Not different | - | Decreased:order Lactobacillales | Increased:Corynebacterium tuberculostearicum Decreased: Lactobacillus Sakai, Carnobacterium alterfunditum, Enterococcus mundtii, and Pediococcus pentosaceus |
| Feazel et al, 2012(51) | 15 CRS (2 wNP and 13sNP) 5 controls |
Middle meatus swabs | 16-rDNA pyrosequencing and comparison with Silva version 104 | Diversity was not different, A trend towards lower evenness in CRS | Not different | - | - | Increased: a trend towards increase for Staphylococcus aureus |
| Aurora et al, 2013(52) | 30 CRS 12control |
Lavage of the middle meatus | 16S rRNA sequenced and submitted to NCBI; analyzed using scripts in the QIIME (Quantitative Insights into Microbial Ecology) | Increased diversity in CRS: 2333 in controls vs 3780 in CRS | Increased in CRS | No difference at phylum level | - | Increased Corynebacterium accolens, Curtobacterium species S22, Pseudomonas DT3-61, Staphylococcus aureus Pseudomonas aeruginosa Decreased: Alicycliphilus and Cloacibacterium |
| Boase et al, 2013(50) | 38 CRS and 6 controls |
Ethmoid sinus mucosal tissue | Ibis T5000 analysis PCR coupled with electrospray ionization mass spectrometry | Trends towards increased diversity; mean isolates per patient in controls was 2, in CRSsNP was 2.5 and in CRSwNP was 3.2 | Increased in CRS | - | - | Increased: Staphylococcus aureus in abundance and frequency Less frequently: Propionibacterium acnes |
| Choi et al, 2014(54) | 8 CRS (5wNP and 3sNP) 3 controls |
Nasal lavage (NAL) fluid | 16S-rDNA high-throughput pyrosequencing followed by identification using ExTaxon database. | Decreased diversity in CRS | Increased abundance in CRS | Increased: Proteobacteria Decreased: Bacteroidetes decreased, from 25.42% to 7.37% |
Increased: Staphylococcus, Corynebacterium and Propionibacterium Decreased: Prevotella, Streptococcus and Veillonella *Pseudomonas increased in CRSwNP compared with CRSsNP |
Increased: Staphylococcus epidermidis, Pseudomonas monteilii Decreased: Prevotella melaninogenica Staphylococcus aureus increased in CRSwNP compared to CRSsNP |
| Ramakrishnan et al, 2015(58) | 56 CRS and 26 controls |
Swabs from Ethmoid region obtained during surgery | 16-rDNA pyrosequencing and comparison with Silva version 111. | Not reported between CRS and Controls, Diversity was associated with optimal surgical outcome in CRS | No difference | No difference | Decreased: Propionibacterium and Peptoniphilus at genum level *Multiple Genera different when comparing CRS with and without asthma(detailed in the text) |
Not reported between CRS and controls |
| Fungal Microbiome Studies | ||||||||
| Study participants | Sample type | Analysis method | Microbial Diversity | Microbial Abundance | Differences at Phylum level | Differences at order, family or genus level | Differences at Species level | |
| Aurora et al. 2013(52) | 30 CRS 12 controls |
Lavage of the middle meatus | 18S rRNA deep sequencing | Increased diversity in CRS | - | Increased: Ascomycota Decreased: Basidiomycota |
- | Increased: Cryptococcus neofromans, Rhodosporidium diabovatum, Davadiella Tassaiana Decreased: Malassezia-uncultured stramenopile |
| Boase et al 2013(50) | 38 CRS and 6 controls | Ethmoid sinus mucosal tissue | Ibis T5000 analysis PCR coupled with electrospray ionization mass spectrometry | Fungi were only detected in 3 CRSwNP cases and in none of the control or CRSsNP | - | - | - | Only 3 positive samples; 2 CRSwNP cases had Aspergillus Fumigatus and 1 CRSwNP case had Penicilium Chrysogenum |
| Cleland et al. 2014(88) | 23 CRS 11 controls |
Middle meatus or anterior ethmoid swabs | 18S rDNA tag-encoded FLX amplicon pyrosequencing | Not different | - | - | Increased: Scutellospora 33 genera had differential abundance in CRS vs. Controls |
- |
In a study by Stephenson et al, mucosal biopsies were assessed using bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) of species-specific 16S rRNA. This method was able to identify up to 20 microorganisms in some individuals and all 18 patients included in the study showed evidence of multiple microbial colonization. Anaerobic bacteria, including Diaphorobacter and Peptoniphilus, were identified as predominant species, found in 78% and 72% of CRS cases, respectively, but not identified in control cases. Interestingly, S. aureus was found in half of the CRS cases, while it was detected in all controls[48].
Stressmann et al used 16S rRNA sequencing and terminal restriction fragment length polymorphism (T-RFLP) to analyze bacterial profiles in 70 tissue and mucus samples obtained from 43 CRS cases. This study had no control subjects. Species belonging to 34 different bacterial genera were identified in the CRS series. The most commonly found bacteria belonged to the following genera: Pseudomonas, Citrobacter, Haemophilus, Propionibacterium, Staphylococcus, and Streptococcus, with Pseudomonas aeruginosa being the most frequent species[49].
Boase et al used sinus mucosal tissue harvested from the ethmoid sinuses to investigate the microbiome at multiple levels by bacterial and fungal cultures, Ibis T5000 analysis (PCR coupled with electrospray ionization mass spectrometry) as well as biofilm analysis[50]. Thirty-eight CRS and 6 control subjects were studied. Using the Ibis method they were able to identify 33 different bacterial species in CRS patients, with a mean of 3.0 (2.0–4.0) species per patient, and 5 different organisms in controls, with a mean of 2.0 (1.0–3.0) per subject. CRS patients had significantly increased bacterial abundance compared to control patients. S. aureus was the most prevalent organism in CRS patients, followed by S. epidermidis and Propionibacterium acnes (P. acnes). S. aureus had a ten-fold increased abundance in CRS patients; S. epidermidis and P. acnes had similar abundances in the microbiome of CRS and control subjects. In control patients, P. acnes was the most commonly detected organism (5/6, 85%), followed by S. epidermidis, which was present in 4/6 (67%). Anaerobic species were detected in 47% of CRS cases, and 83% of controls. Furthermore, this study showed poor agreement between the Ibis biosensor and culture data for anaerobes. They concluded that conventional culture methods are selective for more abundant, rapidly growing aerobic organisms like S. aureus and S. epidermidis[50].
In 2012, Feazel et al studied 21 middle meatus swabs from a series of 15 CRS patients and 5 controls. A total of 57,407 pyrosequencing reads were generated in all samples (interquartile range, 682–3296 per specimen). The most commonly detected sequences in all subjects were from coagulase-negative staphylococci (21/21 specimens) followed by Corynebacterium species (Corynebacterium spp.) (18/21), Propionibacterium acnes (16/21), and S. aureus (14/21). In the vast majority of specimens (95.2%) more than half of the sequences belonged to known upper respiratory bacterial genera[51]. All of these subjects had positive bacterial cultures and the more prevalent sequence types (>15%) corresponded to the cultured organisms in 65% of cases. In comparison to controls, CRS patients had significantly fewer bacterial types and less even distribution of genus-level sequence types. Evenness is calculated based on relative distribution of bacterial types in the samples. S aureus was selectively enriched in CRS in this study. Furthermore, in this study chronic rhinosinusitis with asthma was associated with lower microbial diversity and higher abundance of S. aureus [51].
In another study, lavage samples from the middle meatus of 30 patients with CRS and 12 control individuals were tested for cytokine levels, immune cells, and bacterial and fungal microbiomes. In contrast to the previous study [51], the diversity of bacterial types was higher in CRS patients compared to controls. The most abundant species (in both CRS and in healthy controls) was an uncultured Cyanobacterium, followed by a Curtobacterium species, which was present in 29 patients, and present at lower abundance in the controls. Corynebacterium accolens was the most abundant species of bacteria (it was significantly increased in CRS relative to controls). In contrast, Alicycliphilus and Cloacibacterium were decreased in CRS patients compared to controls[52].
Abreu et al used a 16S rRNA PhyloChip to test for 8500 bacterial taxa in endoscopically guided brush samples of mucosal surfaces of the maxillary sinus. The bacterial profile was amplified sufficiently in 7 cases in each group (CRS and control) that it could be analyzed by microarray. CRS patients had significantly lower bacterial evenness, richness (number of bacterial types detected), and Shannon’s diversity (a metric calculated using richness and evenness indices). They detected pathogenic members of Pseudomonadaceae, Lachnospiraceae, Ralstoniaceae, Mycobacteriaceae, and Helico-bacteriaceae in CRS as well as healthy subjects. Lactobacillales such as Lactobacillus sakei (L. Sakei), as well as Carnobacterium alterfunditum, Enterococcus mundtii, and Pediococcus pentosaceus were reduced in abundance in CRS and there was only a single taxon, Corynebacterium tuberculostearicum (C. tuberculostearicum), that was increased in abundance in CRS. Interestingly, Lactobacillaceae, Enterococcaceae, Aerococcaceae, and Streptococcaceae, were associated with lower symptom score (Sino-nasal outcome test, SNOT-22), while Corynebacteria tuberculostearicum was associated with higher SNOT-22 score [53].
In a more recent study from Korea, Choi et al used nasal lavage fluid samples from 8 CRS patients and 3 controls. Bacterial 16S-rDNA amplicons were analyzed by high-throughput pyrosequencing on a Roche 454 GS-FLX platform. They also isolated and studied the contents of bacteria-derived extracellular vesicles (EV) [54]. These vesicles contain bacterial biological components like bioactive lipids, proteins, and nucleic acids, which could be delivered to other cells[55]. EV from Staphylococcus aureus have been shown to have a role in pathogenesis of atopic dermatitis[56]. Choi et al showed that CRS patients had significantly greater bacterial abundance and lower diversity, which held true when analyzing the bacterial or EV portion of samples. Bacterial and EV compositions strongly correlated. At the Phylum level, CRS patients had higher Proteobacteria and decreased Bacteroidetes. An analysis of the species showed increased S. epidermidis and decreased Prevotella melaninogenica in patients with CRS compared to control cases. Regarding the EV composition, EV from Pseudomonas monteilii, Corynebacterium spp., and Propionibacterium spp. were increased, while EV from Prevotella spp., Streptococcus spp., and Veillonella spp., were decreased in patients with CRS [54]. Comparison of CRSwNP and CRSsNP cases showed that CRSwNP cases had increased Pseudomonas spp. at the genus level and S. aureus at the species level compared with CRSsNP, both in bacteria and EVs [54].
Bucholtz et al studied 31 polyps, 6 turbinates, and 4 sinus biopsy samples using 16S rDNA. The bacterial DNA was rarely found inside any of these tissues. Streptococcus spp. DNA was seen inside the tissue in one polyp sample and Pseudomonas aeruginosa DNA was isolated from one maxillary sinus biopsy[57]. These negative results could be due to the sampling and analysis methods used in this study. A more recent study has found evidence of intracellular invasion by S. aureus in 39% of cases [58].
Recently a larger study on Microbiome of CRS was published; this study took a step forward and evaluated the microbiome not only in association with CRS, but also the phenotypic features and surgical outcome of patients[59]. Nasal swabs from the ethmoid region were examined in 56 CRS patients and 26 controls. The bacterial microbiome of CRS and control patients was similar in terms of biodiversity and at the phylum level, however there were a couple of differences at the genus level; Propionibacterium and Porphyromonas were both decreased in CRS compared with control cases. They found no difference between CRSsNP and CRSwNP cases in terms of bacterial community alterations.
The subgroup analyses showed that the sinus microbiome of CRS patients with asthma had significant differences compared with non-asthmatic CRS cases at both the Phylum and Genus levels. The first figure in that paper shows that the distribution of bacteria at the Phylum level in asthmatic CRS cases was more similar to controls than non-asthmatic patients with CRS. Furthermore at the genus level, Prevotella, which had a relative abundance of only 1.5% in controls and 0.6% in asthmatic CRS cases, had a significantly higher relative abundance of 7.7% in non-asthmatic CRS. A similar pattern was observed for Staphylococcus, which had a trend towards a lower abundance in CRS compared to controls, but a trend toward higher abundance in non-asthmatics within the CRS group. We find these findings rather thought-provoking as to why asthma should push back the microbiome of CRS patients closer to the healthy comparison group. However there were multiple genera that were different between asthmatic CRS and non-asthmatic CRS which were not reported to be distinguishing factors between CRS and controls, making direct comparisons difficult. Interestingly higher diversity and higher abundance of Actinobacteria at the phylum level and Corynebacterium at the genus level were associated with better outcomes after surgery[59].
All of the above studies showed some differences in the microbial community composition of the upper airways between CRS patients and non-CRS controls. However, there are major variabilities and discordances among the studies’ results (Table 1). Overall, most studies have shown increased bacterial abundance in CRS [50, 52, 54]. The bacterial microbiome in the respiratory tract tends to have a high diversity, while the absolute quantity of each species may be low. Therefore, defining the species with higher relative abundance in the bacterial community could be as important as the overall bacterial abundance.
The diversity of the microbiome has been proposed to be essential for sustaining a healthy microenvironment. Chronic inflammation in other diseases like inflammatory bowel disease has been associated with reduced diversity in bacterial composition[60, 61]. However, the results on bacterial diversity in CRS are varied and likely to be complicated by repeated antibiotic use in CRS patients. Two of the studies showed decreased[53, 54] diversity, while others showed either a significant increase in diversity [52], a trend toward increased diversity[50], or no difference at all[51],[59]. In the most recent study by Ramakrishnan et al., diversity was not different between CRS and controls, although in the CRS group it was predictive of better outcome[59]. The discrepancy among studies in terms of diversity might have been related to the use of antibiotics prior to and around the time of sampling for microbial analyses or based on regional microbial differences in the nasal cavity referred to above. Middle meatus and sphenoethmoidal recess regions have been shown to have bacterial composition and diversity similar to eachother but significantly higher compared to anterior nares[38]. In this regard, using lavage for microbiome analysis instead of direct sampling from a single region of the nasal cavity would complicate the analyses.
Role of S. Aureus in CRS
A meta-analysis of literature from 1990 to 2006 on culture-based bacterial analyses in rhinosinusitis revealed S. Aureus as one of the three most common pathogens cultured in acute rhinosinusitis[62]. Similarly S. aureus is a commonly found organism in chronic rhinosinusitis, observed consistently in multiple culture-based and PCR-based microbial studies[50, 51, 63, 64]. S. aureus frequently colonizes the nose of healthy subjects as well[38, 48, 65]. However, the abundance of this bacterium is increased in CRS[50]. Therefore, it seems the contribution of S. aureus to pathogenesis of CRS goes beyond just colonization of the nasal mucosal membranes. In one study, S. Aureus was found more frequently in nasal mucosa of CRSwNP compared to CRSsNP [54], the increased S. aureus colonization was observed in the nasal polyps in previous culture based reports[35]. In contrast, Ramakrishnan et al. showed a higher abundance but similar frequency of S. Aureus in CRSwNP compared to CRSsNP[66]. Sachse et al. used peptide nucleic acid-fluorescence in situ hybridization for detection of S. aureus and showed evidence for invasion of S. aureus into the epithelial layer in CRSwNP but not CRSsNP patients[67]. In context of loss of epithelial integrity seen in chronic rhinosinusitis[68, 69], the authors hypothesized that S. aureus can invade the epithelial layer and move into the submucosal space and potentially play a pathogenic role in formation of nasal polyps in CRSwNP patients. The authors also showed that S. aureus Newman could survive intra-cellularly for 48 hours in nasal polyp epithelial cells and induce the production of IL-6 from these cells in an in vitro cell culture model[67]. Tan NC. et al. showed evidence for intracellular S. aureus in 20 of 51 (39%) of patients. Intracellular S. aureus was associated with higher risk of late clinical and microbiological relapse[58]. How S. aureus can survive in nasal polyp cells in vivo and contribute to the pathogenesis of CRSwNP requires further investigation.
It has been proposed that S. aureus-derived enterotoxins can possibly act as super-antigens and induce a polyclonal stimulation resulting in eosinophilic inflammation and IgE formation. In a study by Bachert et al, almost half of the nasal tissue homogenates derived from NPs had detectable concentrations of specific IgE to S. aureus-derived enterotoxins, including A and/or B(SEA and SEB). The presence of sIgE to SEA and SEB were reportedly associated with total IgE concentration in tissue and local polyclonal IgE against inhalant allergens. The S. aureus-derived enterotoxins-specific IgE antibodies were also linked to higher eosinophilic inflammation, asthma[35],[70] and aspirin sensitivity [70]. Clark et al found an association between positive S. aureus cultures and higher total serum IgE and severity of disease in CRSwNP based on Lund-Mackay tomography score[64]. Another possible contributing factor to invasion of S. aureus could be pre-infection with herpes simplex virus type 1 (HSV1). Wang et al showed evidence suggestive of a significant damage to the nasal epithelium by HSV1 infection that could consequently facilitate invasion of S. aureus into the nasal mucosa [71].
All of this evidence supports a contribution of S. aureus in pathogenesis or worsening of CRS, especially in CRSwNP, which suggests that this bacterium could be a potential therapeutic target in CRSwNP. However it seems that a defect in mucosal barrier and the innate immune system in CRS may predispose individuals to S. aureus invasion and potentially increased enterotoxin production, further promoting inflammation, thus closing a positive feedback loop. The fact that treatment of CRS with prolonged antibiotics that cover S. aureus is only a partial remedy for CRS, and does not improve multiple symptoms like congestion or loss of smell[72], shows that merely eliminating S. aureus temporarily is not sufficient to break the cycle.
Biofilm formation in rhinosinusitis
A biofilm is a complex polymicrobial community of fungi or bacteria surrounded by an exopolysaccharide matrix produced by these microorganisms. Formation of biofilms is a defense mechanism for microbes that can protect themselves from antibiotics and from the host immune system[73]. Biofilm formation in sinonasal mucosa has been studied in CRS in multiple studies[74, 75]. It is associated with recurrence of disease, poor response to treatment and unfavorable outcome after surgery[75],[76, 77]. Biofilm formation has also been linked to environmental factors such as smoking[78]. Furthermore, diverse bacterial biofilms have been attributed to the pathogenesis and phenotypes of CRS [79]. Study of biofilms requires specialized techniques and most of the bacterial species that form them do not grow well in culture[80].
Boase et al compared a molecular PCR-based method, culture and FISH (fluorescent in situ hybridization) for detection of bacteria in sinonasal mucosal tissue. They used S. aureus as a comparitor and showed that use of a PCR-based method (as opposed to conventional cultures) could sensitively detect S. aureus, which was also present in biofilms in these cases[50]. It is not clear how the presence of biofilms will affect the microbiota and microbiome analyses of the nasal cavity and sinuses. More studies are needed to determine whether different sampling and digestion methods and advanced microbiome analysis are able to capture the microbial DNA from biofilms for detection and inclusion of these microorganisms in the analysis.
In 2008, use of baby shampoo nasal irrigation was proposed as a potential treatment for CRS. The proposed mechanism was based on its ability as a surfactant to inhibit biofilm formation. One percent Johnson’s Baby Shampoo was found to inhibit in vitro formation of P. aeruginosa biofilms; however, it had no ability to eradicate preformed Pseudomonas biofilms[81]. Furthermore, a study in healthy individuals showed that this treatment could increase the mucociliary clearance time (MCT)[82]. The first study in 2008 showed a significant improvement in symptoms of thickened nasal discharge and postnasal drainage in patients who remained symptomatic despite medical and surgical therapy for CRS and drew attention to this therapy as a mucolytic agent [81]. However another study in 2013 found no significant differences in overall subjective symptoms between patients who used diluted shampoo as surfactant versus the group who had used hypertonic saline irrigation solution in the early postoperative period[83]. Furthermore a higher number of patients reported significant side effects with surfactant[83].
The fungal microbiome (mycobiome) in sinusitis
Study of nonbacterial microbiota in the nose has been an area of interest in recent years. The association of fungi with CRS has been investigated in multiple studies, with controversial results. In the late 1990s, fungal cultures identified colonization in more than 90% of CRS patients and drew significant attention to this field[84]. In this way, several fungal species were identified in CRS[84]. But further studies showed evidence of fungal colonization in the nose in more than 90% of healthy controls from the same population [85]. Interestingly, studies in other populations showed a significantly lower level of fungal colonization in CRS. Similar fungal culture techniques yielded positive results in 13% of a Turkish[86] and 36% of a Malaysian [87] CRS series. Fungal organisms have also been found in biofilm formations in CRS [75]. Studies using antifungal treatments in CRS have primarily failed to support a clinical use of this approach[88, 89].
More recent studies that used organism-specific primers for PCR analyses of fungi were able to give a more complete picture of the fungal microbiome (mycobiome) in the nose and paranasal sinuses, but the results of these studies have engendered controversies as well. Cleland et al. analyzed nasal swabs of 23 CRS patients and 11 controls. They used 18S ribosomal DNA (rDNA) fungal tag-encoded FLX amplicon pyrosequencing [90]. They showed that CRS cases had greater fungal richness than controls (mean sample richness of 12.14 vs. 8.18 ). This richness decreased significantly following sinus surgery. In that study, Malassezia was the most common and abundant species and was found in all CRS cases at surgery [90]. Aurora et al. [52] analyzed the mycobiome by deep sequencing of 18S rDNA. They found qualitatively similar mycobiomes in CRS and controls, but greater diversity and abundance of fungi in the CRS group. Cryptococcus neoformans was the most abundant fungus in both CRS and control cases, although it was more prevalent in the CRS group (90% vs 61%). In their study, Malassezia was the 3rd most common species and was seen more commonly in controls (4.7% vs. 1.4%). Boase et al. [50] focused on pathogenic taxa using 16 primer pairs for PCR, which could survey nearly all pathogenic fungal species, but could not find evidence of pathogenic fungal DNA in most of the cases studied. Three CRSwNP cases were positive (two for Aspergillus fumigatus and one for Bipolaris papendorfii), while all CRSsNP cases and healthy controls were negative.
Viral microbiome and CRS
A majority of CRS exacerbations occur in seasons with a high prevalence of viral infections [91]. Using multiplex PCR for respiratory viruses, Cho et al [92] found viral nucleic acid sequences in 64% of sinus scrapings and 50% of nasal lavage samples in the CRS group, which was significantly higher than controls (30% and 14% in scraping and lavage respectively). In their study, rhinovirus (RV) was the most frequently detected virus. In a study from China, test of the scraped epithelial cells from the middle meatus revealed an evidence for respiratory viruses in 68.66%, 73.77% and 75.47% of CRSwNP, CRSsNP and control cases, which was not statistically different[93]. In 2006 Jang et al [94] showed that 21% of turbinate epithelial cell samples from CRS patients were positive for rhinovirus detected by reverse-transcription PCR. In that study, RV was not detected in the lavage of any of these patients. In contrast, Wood et al [95]did not find any evidence for respiratory viruses in 13 CRS patients tested by PCR for common respiratory viruses including, but not limited to, rhinovirus, parainfluenza viruses 1, 2, and 3, and respiratory syncytial virus (RSV).
It is noteworthy that studies of experimentally-induced viral infections in CRS are limited. In an in vitro study, NPs and turbinate epithelial cells from CRSwNP patients and healthy controls did not show any differential response in induction of IL-6 and IL-8 upon infection with RV [96]. However, another study showed that stimulation of cultured epithelial cells from CRSsNP cases with toll-like receptor (TLR) 3 agonist in the presence of cigarette smoke extract causes an exaggerated production of chemokine (C-C motif) ligand 5 (CCL5) and human beta-defensin-2 (hBD-2). These researchers proposed that conjoint cigarette smoking and viral infection could contribute to CRS exacerbation and eosinophilic inflammation[97].
The role of rhinovirus infection in asthma exacerbation has been well established and the similarities between the two diseases suggest a role for viral infection in triggering exacerbations of CRS, a hypothesis that calls for further investigation. However, current literature in this field is not conclusive.
Analyzing the sino-nasal microbiome: the disparity and the controversies
The disparities in organism profiling results found in the above studies might be reflective of differences in methods of collection or analysis, which can influence the results of a microbiome study. Controversies were also seen in previous decades in the literature of studies using culture-based methods, mainly due to insensitivity and variability of these methods in capturing microorganisms. This insensitivity is attributed to non-optimal culture conditions, oxygen exposure, or disruption of symbiotic interactions while culturing organisms.
As briefly discussed above, multiple factors could influence the microbiome studies that include, but are not limited to, regional variations of the microbiome (nasal cavity vs. sinuses) [98],[37], antimicrobial use around the time of sampling [53, 99], sampling methods (lavage, tissue biopsy, swab etc.), disease subgroups [54], age [65], severity and phenotypes of included cases or genetic background of patients. The area in the nasal cavity where the swab or biopsy is taken is of utmost importance. As the goal of most research studies of the microbiome is to elucidate the pathogenesis of disease, it is important to sample the microbes from a region that is most involved or representative of the disease. For example lavage would provide a diluted sample containing the microbes from almost all areas in the sino-nasal cavity, and potentially pharyngeal area, and could even be contaminated by skin microbes. Similarly, samples from the anterior nares and inferior turbinate are likely not representative of the sinus microbiome. The differences in sampling techniques could significantly influence the findings including the detection of anaerobes, which could have different abundance in various locations of sinonasal cavity.
Although surgically collected sinus tissue is the best representation, collecting a large series of cases along with healthy controls is more difficult and would limit that data primarily to surgical patients with more severe disease. Potentially, careful swab sampling of the middle meatus with the guidance of endoscopy could be an acceptable alternative.
Furthermore, there are critical factors – nucleic acid extraction, amplicon sequencing, and data analysis – that can influence the accuracy of metagenomic studies based on 16S ribosomal RNA gene (rDNA) sequencing [100]. In the 1970s it was discovered that the 16S ribosomal RNA (16S rRNA), which is a component of the 30S small subunit of prokaryotic ribosomes, could be used for phylogenetic classification. The 16S rRNA is present in all bacteria and contains highly conserved regions flanked by variable regions, which permit phylogenetic classification of the different microbial species[101]. With methods focused on analyzing 16S rDNA (the rRNA gene) for bacteria and 18S rDNA for fungi, the bacterial and fungal DNA base sequences can be analyzed without including human genomic DNA sequences. Although the results of older studies might vary depending on the primer sets, most above mentioned studies in the past 3–4 years have implemented more advanced methods that have the capacity to capture most bacterial or fungal DNA in the sample. However, major differences, including sampling techniques and data analyses mentioned above, exist that could contribute to the variability observed in these studies. The most advanced methods for studying the microbiome based on next generations sequencing are currently available; these methods do not require a cloning step before sequencing analysis and are the basis for high throughput parallel sequencing. However no CRS microbiome studies have yet used these advanced methods. The statistical method used for analysis of data is another important variable in accuracy of data. The data generated from microbiome experiments are multi-dimensional resulting from the large volume of variables. The statistical models are used to explain a clinical or biological endpoint in association with this high-dimensional data. Therefore, the data analysis method could be one of the most important factors playing a role in the discrepancies mentioned above.
Another reason for differences in published findings could be regional variation of the microbiota in the sinonasal cavity. The airway epithelium secretes multiple antimicrobial peptides (AMP); these AMPs are secreted both constitutively and in response to bacteria-derived and other pathogen-derived components; in return, host AMP can affect the microbiota on the adjacent mucosal surface. Different regions of the nasal cavity – inferior turbinate (IT) and uncinate tissue (UT) – have significantly different profiles of innate host defense molecules[37]. IT is the proximal point of contact of inhaled air containing microbial and pathogen components in close proximity to the nares; in contrast, the uncinate process is exposed to drainage of sinuses and is representative of sinus epithelium. The increased expression of gland-derived antimicrobial proteins – e.g. short palate lung and nasal epithelium clone -1 (SPLUNC1) and lactoferrin in uncinate tissue – could be due to differences in the microbiota in the proximal (IT) and distal (UT) areas of the sinonasal mucosa, or alternatively may present a highly regionalized mucosal surface that is inhospitable to many microorganisms.
The respiratory microbiome in asthma
Chronic rhinosinusitis and asthma are both chronic diseases characterized by respiratory mucosal inflammation, often rich in eosinophils. Approximately half of CRS patients also suffer from asthma, asthmatics are much more likely to have CRS than the general population, and the two diseases share an association with allergic diseases. There are similarities in the deviations of the microbiome from control subjects in these diseases as well.
Although acute viral and bacterial infections of the respiratory tract are long-known triggers of asthma exacerbations, the existence of a lower airway commensal microbiome was not discovered until recently [28]. Since then, multiple studies have evaluated the lung microbial residents in asthmatics in search of a causal relationship between these microorganisms and asthma[102]. However, the results of culture-based analyses were inconclusive and controversial, most probably due to the reasons described above in CRS studies, such as sampling technique and insensitivity of the methods. More recently, some studies have applied PCR-based methods aimed at detecting species-specific nucleic acids, and have supported an association between asthma and the increased presence of certain bacteria. For example, the levels of atypical bacteria like Chlamydophila pneumoniae and Mycoplasma pneumoniae in induced sputum or bronchoalveolar lavage are significantly higher in asthmatic patients compared to controls[103, 104]. Moreover the lung microbiota composition and diversity, specifically the abundance of certain phylotypes such as Comamonadaceae, Sphingomonadaceae and Oxalobacteraceae, have been correlated with the degree of bronchial hyperresponsiveness. [29].
Colonization with Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis or a combination of these organisms in neonatal samples from lung is associated with increased risk of asthma later in life[105]. This provides evidence of a persistent effect of microbial colonization of airways on initiation or progression of inflammatory respiratory disease. Along the same lines, multiple longitudinal studies have shown an association between viral respiratory illness with wheezing due to RSV and RV and subsequent development of asthma later in life[106–109].
Comparing CRS and asthma microbiomes
Similar to the finding of Choi et al. for upper airways of CRS cases[54], an increased abundance of bacteria of the phylum Proteobacteria [28, 30] and a decreased abundance of bacteria of the phylum Bacteroidetes has been reported for asthmatic lungs [28]. The number of bacteria of the phylum Bacteroidetes is also decreased in the upper airways of asthmatics compared to healthy controls[110]. A genus of bacteria found more abundantly in asthma was Haemophilus, which belongs to the phylum Proteobacteria. Haemophilus was more frequently detected in the lower airways of adult asthmatics [28] and in the oropharynx of toddlers with wheezing[111]. In CRS studies, Haemophilus was found in 13% [50] and 17% [48]of CRS cases but none of control cases[50]. H. Influenza was also one of the most frequently cultured bacteria in earlier culture-based studies of CRS[40],[41].
There are similar changes in the fungal microbiome in the two diseases. Mycobiome analysis of induced sputum in asthmatics has shown an association with the abundance of Malassezia pachydermatis[112]. As mentioned previously, Malassezia genus was found in the upper airways of CRS patients. However, the results are controversial: in one study Malassezia was detected in all CRS cases at time of surgery [90], while in another report it was found at a higher frequency in healthy upper airways [52].
In asthma, different bacteria have been associated with specific phenotypes of disease; pathogens like Moraxella catarrhalis and members of Haemophilus or Streptococcus genera have been associated with steroid resistant neutrophilic asthma[113]. The only species that was found to be associated with nasal polyposis, an important hallmark of disease phenotype in CRS, is S. aureus, which has been found more frequently in CRSwNP compared to CRSsNP [54]. S. aureus has also been associated with asthma in a series of CRS patients[51]. These findings from microbiome analyses are in line with previously found associations of S. aureus and specific IgE to its enterotoxins in nasal polyposis and asthma[35]. Ramakrishnan et al. showed that the relative abundance of genera Provotella, Fusobacterium and Campylobacter in upper airways was decreased in CRS with asthma compared with CRS without asthma, while Acinetobacter and Ralstonia were relatively increased in nasal microbiome of asthmatics CRS cases[59].
Genetic factors influencing microbiomes
The effect of host genetic factors on Microbiota is another area of unmet research need. Diversity in host genetic factors can influence recognition and binding of microbes to the body surfaces, which can provide differential growth conditions[114]. Furthermore the intensity of immune reactions to microorganisms could be different based on genetic factors[115]. There might be individualized predispositions to development of inflammation in response to microbes or to microbial components based on an individual’s immune response, which may result in the initiation or progression of inflammation and development of diseases like CRS or asthma. Hsu et al comprehensively reviewed the data on genetic polymorphisms and variants associated with CRS [116]. There are a few genetic variants in elements of the innate immune system associated with CRS – including the Nitric Oxide Synthase (NOS) pathway [117] and TLR-2 [110] – that could directly or indirectly influence the microbiome.
Host factors and the microbiome in CRS
The interplay between the innate and adaptive immune systems, the microbial communities and inflammation in sinonasal mucosa is rather complex and not completely understood yet. Th2 inflammation in eosinophilic CRS has been associated with down regulation of elements of innate immunity. Reductions in antimicrobial peptides like human beta-defensin 2(hBD-2) and surfactant protein-A (SP-A) and decreased expression of TLR9 have been described in CRS. These alterations can influence the defense against microbial pathogens and affect microbial colonization[118]. On the other hand, colonization with fungi and specific bacteria like S. aureus has been linked to maladaptive Th2 responses[35]. Microbes and their products are ligands for the innate immune receptors that are also present on inflammatory granulocytes like basophils and mast cells that are found in CRSwNP[119][120]. Activation of these cells through these receptors could result in recruitment of other inflammatory cells involved in Th2 response and progression of the inflammation in CRS.
In the following paragraphs, we have briefly summarized the findings regarding changes in the mucosal immune system in CRS that can influence the microbial communities in the sinonasal cavity.
Multiple elements of the innate immune system, including antimicrobial peptides and proteins, and pattern recognition receptors, have been studied in CRS[121],[122]. Sinus mucosal cells including submucosal mixed seromucinous glands, and epithelial cells produce a host of proteins and peptides with antimicrobial functions[123]. These molecules include enzymes (e.g. lysozyme and complement components), permeabilizing peptides (e.g. defensins, cathelicidins, bacterial permeability-increasing protein [BPI], and palate, lung, and nasal epithelium clone [PLUNC]), collectins (e.g. mannose-binding lectin[MBL], surfactant protein-A[SP-A] and surfactant protein-D[SP-D]), and binding and neutralizing proteins (e.g. lactoferrin and serum amyloid A [SAA])[124].
In normal conditions, one would anticipate an increase in the inducible peptides in response to increased microbial colonization or infection. Any reduction in the level of these molecules can potentially influence microbial colonization. Human sinonasal epithelial cells isolated from CRS patients were shown to have decreased expression of human beta-defensin 2(hBD-2), which belongs to the defensin family[118]. Defensins are one of the main antimicrobial peptide families involved in the microbiome of the respiratory epithelium[125]. And the reduced expression of hBD-2 could potentially affect the abnormal microbial colonization and immune responses in CRSwNP.
Furthermore, multiple members of the PLUNC family, including SPLUNC1 and long palate lung and nasal epithelium clone-2 (LPLUNC2), were reduced, along with reduced lysozyme and lactoferrin, in nasal polyps relative to uncinate tissue of healthy controls[126], which is reflective of the decreased number of submucosal glands in NP [126]. These findings suggest that decreased numbers of glands in nasal polyp leads to a localized defect in the production and release of glandular innate defense molecules, which can subsequently affect the microbial colonization of nose and sinuses in these individuals.
S100 proteins are involved in epithelial defense and repair, and have antimicrobial functions[127]. A few studies have shown a significant reduction in S100 proteins including S100A7 and S100A8/A9 in CRSsNP and CRSwNP [69, 128], which can lead to a diminished innate immune response.
Ficolins and collectins like MBL, SP-A and SP-D are soluble pattern recognition receptors. In vitro culture of human sinonasal epithelial cells with IL-4 or IL-13 reduced the expression of SP-A[118], which could compromise innate immunity in CRSwNP in which these cytokines are expressed. However, IHC staining of SP-A and SP-D in a clinical study showed similar expression of these two molecules in CRSwNP and controls[129]. In contrast, SP-A was reported to be increased in the sinus tissue of CRSsNP patients[130].
Pattern recognition receptors, including NOD-like receptors, Toll-like receptors and bitter taste receptors are important players in host and microbial interactions in nasal and sinus mucosa. Toll-like receptors (TLR 1 through TLR10) are expressed in sinonasal epithelium [131, 132]. Signaling through TLRs results in production of cytokines and chemokines along with antimicrobial peptides. Lane et al. showed that TLR2 mRNA expression was increased in CRS in general[121]. However, expression of TLR2 and TLR9 were decreased in patients with early recurrence of polyps after surgery [121]. Furthermore, TLR9 was decreased in CRSwNP[133]. The TLR2 receptor recognizes a wide spectrum of microbial organisms including gram-negative and gram-positive bacteria, and fungi, which are found in abundance in CRS, and TLR-9 is important in recognition of DNA from bacteria and viruses. Reduced expression of these receptors may link early recurrence of disease with possible deficiency in host defense responses.
Bitter taste receptors signal by increasing intracellular calcium flux and increasing nitric oxide (NO) production and ciliary beat frequency. A common polymorphism in a bitter taste receptor (TAS2R38) is associated with gram-negative infection and biofilm formation and has been correlated with positive cultures for gram-negative bacteria like P. aeruginosa in CRS [134]. This polymorphism results in reduced signaling through the receptor, thus decreasing NO production and ciliary beat frequency, which are important in host immune defense against gram-negative organisms, including P. aeruginosa [134], which is commonly found in CRS [49, 52, 57, 76].
IL-22 plays an important role in mucosal integrity and in immunity against bacteria in the intestine and lung [135, 136]. This cytokine elicits multiple innate immune responses through epithelial cells, maintains mucosal homeostasis [137], and limits the allergic response and production of Th2 cytokines [138]. The main source of Il-22 in airways has been shown to be type 3 innate immune cells (ILC3) [138]. It was recently shown that production of IL-22 in large intestine by ILC3 is induced by commensal gut microbiota, specifically Clostridia [139]. A polymorphism in the IL-22 receptor (IL-22R) has been associated with severe CRS [140]. Furthermore, in patients with recalcitrant CRSwNP, the expression of IL-22R in sinus mucosal epithelial cells is significantly reduced [141]. However Delwitt et al. reported an increased level of this receptor in CRSsNP[142]. These data collectively point to mutual interactions of IL-22 and microbiota, and the reduced level of the receptor for IL-22 in CRSwNP could be an important factor in the pathogenesis of this disease.
Therapeutic use of Microbiota in CRS
Introducing microbial species in the form of probiotics, or inducing a transition in benefit of certain commensal bacteria by fecal transplant have been used in the treatment of multiple gastrointestinal diseases that are associated with altered gut Microbiota such as clostridium difficile infection and inflammatory bowel disease [143, 144]. It is not clear whether altering the microbiome of the nasal cavity towards a more sustained and symbiotic combination in humans is feasible or has any therapeutic potential. One study tried to investigate a role for transitioning of the nasal microbiome for prevention or treatment in a mouse sinusitis model. Intranasal inoculations of S. epidermis plus S. aureus showed a significant decrease in the number of periodic acid-Schiff (PAS)-positive goblet cells in comparison with mice inoculated with S. aureus in this mouse model of sinusitis[145]. In a similar experimental model, Abreu et al. co-administered L. sakei with C. tuberculostearicum in mouse sinuses. They found significantly lower goblet cell hyperplasia and mucin hypersecretion in the sinus of these mice compared with those inoculated only with C. tuberculostearicum. This was suggestive that L. sakei protects the sinus epithelium, possibly through competitive inhibition of C. tuberculostearicum [53]. Whether these results could have any therapeutic implication in human disease remains to be investigated.
Summary and conclusions
Although the number of publications on microbiome in CRS has been increasing in the past few years, differences in experimental protocols, sampling and data analysis complicate comparisons. Except few species like S. Aureus that have been studied at length, it is rather difficult to extrapolate a consistent conclusion on the microbial changes in CRS from the existing data. However, available studies show that the microbiome of the nasal cavity differs significantly between CRS patients and controls and most studies have shown increased bacterial abundance in CRS. The results on bacterial diversity in CRS are varied, which is possibly due to antibiotic use prior to and around the time of sampling for microbial analyses or based on the regional microbial differences in the nasal cavity. Furthermore, there is a paucity of data on the viral microbiome in CRS, which can potentially affect the host inflammatory response as well as bacterial and fungal microorganisms.
Multiple inflammatory pathways and host defense elements are altered in CRS that could contribute to the observed microbiome differences. The observed decrease in number of glands and their associated antimicrobial peptides like SPLUNC1, LPLUNC2, lysozymes and lactoferrin in nasal polyps could potentiate the colonization and invasion of microbes to the adjacent mucosal membranes. Furthermore the nasal epithelium in CRS patients produces reduced amounts of other important AMPs like hBD-2, S100A7 and S100A8/9. Some of the pattern recognition receptors in charge of recognizing pathogen-associated molecules like TLR2 have also been found at reduced levels in refractory CRS. These defects, along with the loss of barrier function in the epithelium, further predispose the nasal and sinus mucosa to invasion by microbes like S. Aureus that would otherwise be non-pathogen residents.
However, it still remains to be determined if the alterations seen in nasal microbiome are cause or effect of this chronic inflammatory disease. Multiple microbial products are major triggers of inflammation; LPS and bacterial surface molecules trigger pattern-recognition receptors that are commonly found on innate immune system inflammatory cells like macrophages, epithelial cells and dendritic cells (DC). DCs have been described to be elevated in CRS tissue [146][147]. Furthermore, cells like basophils and mast cells that are found at increased numbers in nasal polyps and uncinate tissue of CRSwNP patients[119][120] possess pattern recognition receptors including TLR2[148][149] and formyl-methionyl-leucyl-phenylalanine(fMLP) receptors [150] that activate these cells upon microbial exposure and can potentially be a bridge between innate and adaptive immune response in CRS. These mechanisms can all link the mucosal inflammation that is the hallmark of CRS pathogenesis to the microbiome and point towards the importance of studying the nasal microbiome in association with inflammatory elements.
Acknowledgments
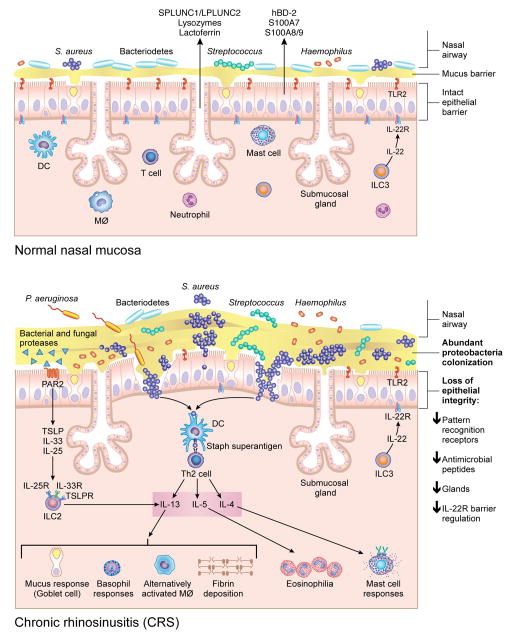
We would like to thank Mrs. Jacqueline Schaffer, MAMS for the professional illustration of figure1.
Figure 1. A Model for changes in the nasal microbiome and mucosal immune response in CRSwNP.
Although multiple microorganisms colonize healthy nasal mucosa membranes, as shown in the top figure, there are changes that occur in the microbiome in patients with CRSwNP, including increased S. aureus abundance, decreased Bacteriodetes and decreased diversity, as shown in the bottom figure. These changes, along with loss of epithelial integrity, decreased pattern recognition molecules, decreased mucosal glands and decreased antimicrobial peptide production in the nasal polyp and sinus tissue, can potentially provide an environment that promotes invasion of microorganisms across the mucosal barrier. Enterotoxins produced by S. aureus can act as superantigens and promote Th-2 inflammation, resulting in production of cytokines such as IL-13, IL-4 and IL-5 that further recruit and activate inflammatory cells such as eosinophils, mast cells, basophils and alternatively activated macrophages. Bacterial and fungal proteases can induce production of thymic stromal lymphopoietin (TSLP) through Protease-Activated Receptor-2(PAR-2)[151], which will subsequently result in activation of innate lymphoid type 2 cells (ILC-2) that further produce IL-5 and IL-13.
RPS is supported in part by the Ernest S. Bazley Foundation, U19 AI106683, R37 HL068546 and R01 HL078860 from the National Institute of Health. AK is supported by R01 AT007143-05, R01 AA023417-02 and R01 AA020216-05.
References
- 1.Anand VK. Epidemiology and economic impact of rhinosinusitis. The Annals of otology, rhinology & laryngology Supplement. 2004;193:3–5. doi: 10.1177/00034894041130s502. [DOI] [PubMed] [Google Scholar]
- 2.Pleis JR, Ward BW, Lucas JW. Summary health statistics for U.S. adults: National Health Interview Survey, 2009. Vital and health statistics Series 10, Data from the National Health Survey. 2010:1–207. [PubMed] [Google Scholar]
- 3.Gliklich RE, Metson R. The health impact of chronic sinusitis in patients seeking otolaryngologic care. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery. 1995;113:104–109. doi: 10.1016/S0194-59989570152-4. [DOI] [PubMed] [Google Scholar]
- 4.Bachert C, Gevaert P, van Cauwenberge P. Staphylococcus aureus enterotoxins: a key in airway disease? Allergy. 2002;57:480–487. doi: 10.1034/j.1398-9995.2002.02156.x. [DOI] [PubMed] [Google Scholar]
- 5.Shin SH, Ponikau JU, Sherris DA, Congdon D, Frigas E, Homburger HA, Swanson MC, Gleich GJ, Kita H. Chronic rhinosinusitis: an enhanced immune response to ubiquitous airborne fungi. The Journal of allergy and clinical immunology. 2004;114:1369–1375. doi: 10.1016/j.jaci.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS biology. 2006;4:e337. doi: 10.1371/journal.pbio.0040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 8.Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Berard M, Kleinschek M, Cua D, Di Santo JP, Eberl G. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nature immunology. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 9.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, Kambayashi T, Larosa DF, Renner ED, Orange JS, Bushman FD, Artis D. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nature medicine. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny KM, Finlay BB. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO reports. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penders J, Gerhold K, Stobberingh EE, Thijs C, Zimmermann K, Lau S, Hamelmann E. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. The Journal of allergy and clinical immunology. 2013;132:601–607. e608. doi: 10.1016/j.jaci.2013.05.043. [DOI] [PubMed] [Google Scholar]
- 13.van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, Reijmerink NE, Dompeling E, van den Brandt PA, Ferreira I, Mommers M, Thijs C. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. The Journal of allergy and clinical immunology. 2011;128:948–955. e941–943. doi: 10.1016/j.jaci.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 14.Penders J, Stobberingh EE, van den Brandt PA, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy. 2007;62:1223–1236. doi: 10.1111/j.1398-9995.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- 15.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nature reviews Immunology. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrlander C, Heederik D, Piarroux R, von Mutius E, Group GTS. Exposure to environmental microorganisms and childhood asthma. The New England journal of medicine. 2011;364:701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 17.Pakarinen J, Hyvarinen A, Salkinoja-Salonen M, Laitinen S, Nevalainen A, Makela MJ, Haahtela T, von Hertzen L. Predominance of Gram-positive bacteria in house dust in the low-allergy risk Russian Karelia. Environmental microbiology. 2008;10:3317–3325. doi: 10.1111/j.1462-2920.2008.01723.x. [DOI] [PubMed] [Google Scholar]
- 18.Lynch SV, Wood RA, Boushey H, Bacharier LB, Bloomberg GR, Kattan M, O’Connor GT, Sandel MT, Calatroni A, Matsui E, Johnson CC, Lynn H, Visness CM, Jaffee KF, Gergen PJ, Gold DR, Wright RJ, Fujimura K, Rauch M, Busse WW, Gern JE. Effects of early-life exposure to allergens and bacteria on recurrent wheeze and atopy in urban children. The Journal of allergy and clinical immunology. 2014;134:593–601. e512. doi: 10.1016/j.jaci.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Mutius E, Radon K. Living on a farm: impact on asthma induction and clinical course. Immunology and allergy clinics of North America. 2008;28:631–647. ix–x. doi: 10.1016/j.iac.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 21.van Strien RT, Engel R, Holst O, Bufe A, Eder W, Waser M, Braun-Fahrlander C, Riedler J, Nowak D, von Mutius E, Team AS. Microbial exposure of rural school children, as assessed by levels of N-acetyl-muramic acid in mattress dust, and its association with respiratory health. The Journal of allergy and clinical immunology. 2004;113:860–867. doi: 10.1016/j.jaci.2004.01.783. [DOI] [PubMed] [Google Scholar]
- 22.Sordillo JE, Hoffman EB, Celedon JC, Litonjua AA, Milton DK, Gold DR. Multiple microbial exposures in the home may protect against asthma or allergy in childhood. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2010;40:902–910. doi: 10.1111/j.1365-2222.2010.03509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Debarry J, Garn H, Hanuszkiewicz A, Dickgreber N, Blumer N, von Mutius E, Bufe A, Gatermann S, Renz H, Holst O, Heine H. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. The Journal of allergy and clinical immunology. 2007;119:1514–1521. doi: 10.1016/j.jaci.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 24.Hagner S, Harb H, Zhao M, Stein K, Holst O, Ege MJ, Mayer M, Matthes J, Bauer J, von Mutius E, Renz H, Heine H, Pfefferle PI, Garn H. Farm-derived Gram-positive bacterium Staphylococcus sciuri W620 prevents asthma phenotype in HDM- and OVA-exposed mice. Allergy. 2013;68:322–329. doi: 10.1111/all.12094. [DOI] [PubMed] [Google Scholar]
- 25.Vogel K, Blumer N, Korthals M, Mittelstadt J, Garn H, Ege M, von Mutius E, Gatermann S, Bufe A, Goldmann T, Schwaiger K, Renz H, Brandau S, Bauer J, Heine H, Holst O. Animal shed Bacillus licheniformis spores possess allergy-protective as well as inflammatory properties. The Journal of allergy and clinical immunology. 2008;122:307–312. 312 e301–308. doi: 10.1016/j.jaci.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 26.Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC, Boushey HA, Zoratti E, Ownby D, Lukacs NW, Lynch SV. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:805–810. doi: 10.1073/pnas.1310750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han MK, Huang YJ, Lipuma JJ, Boushey HA, Boucher RC, Cookson WO, Curtis JL, Erb-Downward J, Lynch SV, Sethi S, Toews GB, Young VB, Wolfgang MC, Huffnagle GB, Martinez FJ. Significance of the microbiome in obstructive lung disease. Thorax. 2012;67:456–463. doi: 10.1136/thoraxjnl-2011-201183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, Moffatt MF, Cookson WO. Disordered microbial communities in asthmatic airways. PloS one. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, Woyke T, Allgaier M, Bristow J, Wiener-Kronish JP, Sutherland ER, King TS, Icitovic N, Martin RJ, Calhoun WJ, Castro M, Denlinger LC, Dimango E, Kraft M, Peters SP, Wasserman SI, Wechsler ME, Boushey HA, Lynch SV National Heart L, Blood Institute’s Asthma Clinical Research N. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. The Journal of allergy and clinical immunology. 2011;127:372–381. e371–373. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. The Journal of allergy and clinical immunology. 2013;131:346–352. e341–343. doi: 10.1016/j.jaci.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goleva E, Jackson LP, Harris JK, Robertson CE, Sutherland ER, Hall CF, Good JT, Jr, Gelfand EW, Martin RJ, Leung DY. The effects of airway microbiome on corticosteroid responsiveness in asthma. American journal of respiratory and critical care medicine. 2013;188:1193–1201. doi: 10.1164/rccm.201304-0775OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Legatzki A, Rosler B, von Mutius E. Microbiome diversity and asthma and allergy risk. Current allergy and asthma reports. 2014;14:466. doi: 10.1007/s11882-014-0466-0. [DOI] [PubMed] [Google Scholar]
- 33.Jiang RS, Liang KL, Jang JW, Hsu CY. Bacteriology of endoscopically normal maxillary sinuses. The Journal of laryngology and otology. 1999;113:825–828. doi: 10.1017/s0022215100145311. [DOI] [PubMed] [Google Scholar]
- 34.Biel MA, Brown CA, Levinson RM, Garvis GE, Paisner HM, Sigel ME, Tedford TM. Evaluation of the microbiology of chronic maxillary sinusitis. The Annals of otology, rhinology, and laryngology. 1998;107:942–945. doi: 10.1177/000348949810701107. [DOI] [PubMed] [Google Scholar]
- 35.Van Zele T, Gevaert P, Watelet JB, Claeys G, Holtappels G, Claeys C, van Cauwenberge P, Bachert C. Staphylococcus aureus colonization and IgE antibody formation to enterotoxins is increased in nasal polyposis. The Journal of allergy and clinical immunology. 2004;114:981–983. doi: 10.1016/j.jaci.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 36.Vossbrinck CR, Maddox JV, Friedman S, Debrunner-Vossbrinck BA, Woese CR. Ribosomal RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature. 1987;326:411–414. doi: 10.1038/326411a0. [DOI] [PubMed] [Google Scholar]
- 37.Seshadri S, Rosati M, Lin DC, Carter RG, Norton JE, Choi AW, Suh L, Kato A, Chandra RK, Harris KE, Chu HW, Peters AT, Tan BK, Conley DB, Grammer LC, Kern RC, Schleimer RP. Regional differences in the expression of innate host defense molecules in sinonasal mucosa. The Journal of allergy and clinical immunology. 2013;132:1227–1230. e1225. doi: 10.1016/j.jaci.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, Relman DA. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell host & microbe. 2013;14:631–640. doi: 10.1016/j.chom.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang RS, Hsu CY, Leu JF. Bacteriology of ethmoid sinus in chronic sinusitis. American journal of rhinology. 1997;11:133–137. doi: 10.2500/105065897782537179. [DOI] [PubMed] [Google Scholar]
- 40.Chan J, Hadley J. The microbiology of chronic rhinosinusitis: results of a community surveillance study. Ear, nose, & throat journal. 2001;80:143–145. [PubMed] [Google Scholar]
- 41.Brook I, Frazier EH, Foote PA. Microbiology of chronic maxillary sinusitis: comparison between specimens obtained by sinus endoscopy and by surgical drainage. Journal of medical microbiology. 1997;46:430–432. doi: 10.1099/00222615-46-5-430. [DOI] [PubMed] [Google Scholar]
- 42.Kim HJ, Lee K, Yoo JB, Song JW, Yoon JH. Bacteriological findings and antimicrobial susceptibility in chronic sinusitis with nasal polyp. Acta oto-laryngologica. 2006;126:489–497. doi: 10.1080/00016480500437385. [DOI] [PubMed] [Google Scholar]
- 43.Benninger MS, Appelbaum PC, Denneny JC, Osguthorpe DJ, Stankiewicz JA. Maxillary sinus puncture and culture in the diagnosis of acute rhinosinusitis: the case for pursuing alternative culture methods. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery. 2002;127:7–12. doi: 10.1067/mhn.2002.124847. [DOI] [PubMed] [Google Scholar]
- 44.Keech DR, Ramadan H, Mathers P. Analysis of aerobic bacterial strains found in chronic rhinosinusitis using the polymerase chain reaction. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery. 2000;123:363–367. doi: 10.1067/mhn.2000.108204. [DOI] [PubMed] [Google Scholar]
- 45.Paju S, Bernstein JM, Haase EM, Scannapieco FA. Molecular analysis of bacterial flora associated with chronically inflamed maxillary sinuses. Journal of medical microbiology. 2003;52:591–597. doi: 10.1099/jmm.0.05062-0. [DOI] [PubMed] [Google Scholar]
- 46.Power DA, Burton JP, Chilcott CN, Tagg JR, Dawes PJ. Non-culture-based analysis of bacterial populations from patients with chronic rhinosinusitis. Journal of clinical microbiology. 2005;43:5822–5824. doi: 10.1128/JCM.43.11.5822-5824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewenza S, Charron-Mazenod L, Cho JJ, Mechor B. Identification of bacterial contaminants in sinus irrigation bottles from chronic rhinosinusitis patients. Journal of otolaryngology - head & neck surgery = Le Journal d’oto-rhino-laryngologie et de chirurgie cervico-faciale. 2010;39:458–463. [PubMed] [Google Scholar]
- 48.Stephenson MF, Mfuna L, Dowd SE, Wolcott RD, Barbeau J, Poisson M, James G, Desrosiers M. Molecular characterization of the polymicrobial flora in chronic rhinosinusitis. Journal of otolaryngology - head & neck surgery = Le Journal d’oto-rhino-laryngologie et de chirurgie cervico-faciale. 2010;39:182–187. [PubMed] [Google Scholar]
- 49.Stressmann FA, Rogers GB, Chan SW, Howarth PH, Harries PG, Bruce KD, Salib RJ. Characterization of bacterial community diversity in chronic rhinosinusitis infections using novel culture-independent techniques. American journal of rhinology & allergy. 2011;25:e133–140. doi: 10.2500/ajra.2011.25.3628. [DOI] [PubMed] [Google Scholar]
- 50.Boase S, Foreman A, Cleland E, Tan L, Melton-Kreft R, Pant H, Hu FZ, Ehrlich GD, Wormald PJ. The microbiome of chronic rhinosinusitis: culture, molecular diagnostics and biofilm detection. BMC infectious diseases. 2013;13:210. doi: 10.1186/1471-2334-13-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feazel LM, Robertson CE, Ramakrishnan VR, Frank DN. Microbiome complexity and Staphylococcus aureus in chronic rhinosinusitis. The Laryngoscope. 2012;122:467–472. doi: 10.1002/lary.22398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aurora R, Chatterjee D, Hentzleman J, Prasad G, Sindwani R, Sanford T. Contrasting the microbiomes from healthy volunteers and patients with chronic rhinosinusitis. JAMA Otolaryngol Head Neck Surg. 2013;139:1328–1338. doi: 10.1001/jamaoto.2013.5465. [DOI] [PubMed] [Google Scholar]
- 53.Abreu NA, Nagalingam NA, Song Y, Roediger FC, Pletcher SD, Goldberg AN, Lynch SV. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med. 2012;4:151ra124. doi: 10.1126/scitranslmed.3003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi EB, Hong SW, Kim DK, Jeon SG, Kim KR, Cho SH, Gho YS, Jee YK, Kim YK. Decreased diversity of nasal microbiota and their secreted extracellular vesicles in patients with chronic rhinosinusitis based on a metagenomic analysis. Allergy. 2014;69:517–526. doi: 10.1111/all.12374. [DOI] [PubMed] [Google Scholar]
- 55.Tetta C, Ghigo E, Silengo L, Deregibus MC, Camussi G. Extracellular vesicles as an emerging mechanism of cell-to-cell communication. Endocrine. 2013;44:11–19. doi: 10.1007/s12020-012-9839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hong SW, Kim MR, Lee EY, Kim JH, Kim YS, Jeon SG, Yang JM, Lee BJ, Pyun BY, Gho YS, Kim YK. Extracellular vesicles derived from Staphylococcus aureus induce atopic dermatitis-like skin inflammation. Allergy. 66:351–359. doi: 10.1111/j.1398-9995.2010.02483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bucholtz GA, Salzman SA, Bersalona FB, Boyle TR, Ejercito VS, Penno L, Peterson DW, Stone GE, Urquhart A, Shukla SK, Burmester JK. PCR analysis of nasal polyps, chronic sinusitis, and hypertrophied turbinates for DNA encoding bacterial 16S rRNA. American journal of rhinology. 2002;16:169–173. [PubMed] [Google Scholar]
- 58.Tan NC, Foreman A, Jardeleza C, Douglas R, Vreugde S, Wormald PJ. Intracellular Staphylococcus aureus: the Trojan horse of recalcitrant chronic rhinosinusitis? International forum of allergy & rhinology. 3:261–266. doi: 10.1002/alr.21154. [DOI] [PubMed] [Google Scholar]
- 59.Ramakrishnan VR, Hauser LJ, Feazel LM, Ir D, Robertson CE, Frank DN. Sinus microbiota varies among chronic rhinosinusitis phenotypes and predicts surgical outcome. The Journal of allergy and clinical immunology. doi: 10.1016/j.jaci.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 60.Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, Timmis KN, Schreiber S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Payne SC, Benninger MS. Staphylococcus aureus is a major pathogen in acute bacterial rhinosinusitis: a meta-analysis. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2007;45:e121–127. doi: 10.1086/522763. [DOI] [PubMed] [Google Scholar]
- 63.Sachse F, Becker K, Rudack C. Incidence of staphylococcal colonization and of the 753Q Toll-like receptor 2 variant in nasal polyposis. American journal of rhinology & allergy. 2010;24:e10–13. doi: 10.2500/ajra.2010.24.3416. [DOI] [PubMed] [Google Scholar]
- 64.Clark DW, Wenaas A, Citardi MJ, Luong A, Fakhri S. Chronic rhinosinusitis with nasal polyps: elevated serum immunoglobulin E is associated with Staphylococcus aureus on culture. International forum of allergy & rhinology. 2011;1:445–450. doi: 10.1002/alr.20079. [DOI] [PubMed] [Google Scholar]
- 65.Ramakrishnan VR, Feazel LM, Gitomer SA, Ir D, Robertson CE, Frank DN. The microbiome of the middle meatus in healthy adults. PloS one. 2013;8:e85507. doi: 10.1371/journal.pone.0085507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramakrishnan VR, Feazel LM, Abrass LJ, Frank DN. Prevalence and abundance of Staphylococcus aureus in the middle meatus of patients with chronic rhinosinusitis, nasal polyps, and asthma. International forum of allergy & rhinology. 3:267–271. doi: 10.1002/alr.21101. [DOI] [PubMed] [Google Scholar]
- 67.Sachse F, Becker K, von Eiff C, Metze D, Rudack C. Staphylococcus aureus invades the epithelium in nasal polyposis and induces IL-6 in nasal epithelial cells in vitro. Allergy. 65:1430–1437. doi: 10.1111/j.1398-9995.2010.02381.x. [DOI] [PubMed] [Google Scholar]
- 68.Soyka MB, Wawrzyniak P, Eiwegger T, Holzmann D, Treis A, Wanke K, Kast JI, Akdis CA. Defective epithelial barrier in chronic rhinosinusitis: the regulation of tight junctions by IFN-gamma and IL-4. The Journal of allergy and clinical immunology. 2012;130:1087–1096. e1010. doi: 10.1016/j.jaci.2012.05.052. [DOI] [PubMed] [Google Scholar]
- 69.Richer SL, Truong-Tran AQ, Conley DB, Carter R, Vermylen D, Grammer LC, Peters AT, Chandra RK, Harris KE, Kern RC, Schleimer RP. Epithelial genes in chronic rhinosinusitis with and without nasal polyps. American journal of rhinology. 2008;22:228–234. doi: 10.2500/ajr.2008.22.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bachert C, Gevaert P, Holtappels G, Johansson SG, van Cauwenberge P. Total and specific IgE in nasal polyps is related to local eosinophilic inflammation. The Journal of allergy and clinical immunology. 2001;107:607–614. doi: 10.1067/mai.2001.112374. [DOI] [PubMed] [Google Scholar]
- 71.Wang X, Zhang N, Glorieux S, Holtappels G, Vaneechoutte M, Krysko O, Zhang L, Han D, Nauwynck HJ, Bachert C. Herpes simplex virus type 1 infection facilitates invasion of Staphylococcus aureus into the nasal mucosa and nasal polyp tissue. PloS one. 2012;7:e39875. doi: 10.1371/journal.pone.0039875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Van Zele T, Gevaert P, Holtappels G, Beule A, Wormald PJ, Mayr S, Hens G, Hellings P, Ebbens FA, Fokkens W, Van Cauwenberge P, Bachert C. Oral steroids and doxycycline: two different approaches to treat nasal polyps. The Journal of allergy and clinical immunology. 125:1069–1076. e1064. doi: 10.1016/j.jaci.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 73.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clinical microbiology reviews. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hekiert AM, Kofonow JM, Doghramji L, Kennedy DW, Chiu AG, Palmer JN, Leid JG, Cohen NA. Biofilms correlate with TH1 inflammation in the sinonasal tissue of patients with chronic rhinosinusitis. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery. 2009;141:448–453. doi: 10.1016/j.otohns.2009.06.090. [DOI] [PubMed] [Google Scholar]
- 75.Foreman A, Psaltis AJ, Tan LW, Wormald PJ. Characterization of bacterial and fungal biofilms in chronic rhinosinusitis. American journal of rhinology & allergy. 2009;23:556–561. doi: 10.2500/ajra.2009.23.3413. [DOI] [PubMed] [Google Scholar]
- 76.Bendouah Z, Barbeau J, Hamad WA, Desrosiers M. Biofilm formation by Staphylococcus aureus and Pseudomonas aeruginosa is associated with an unfavorable evolution after surgery for chronic sinusitis and nasal polyposis. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery. 2006;134:991–996. doi: 10.1016/j.otohns.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 77.Psaltis AJ, Weitzel EK, Ha KR, Wormald PJ. The effect of bacterial biofilms on post-sinus surgical outcomes. American journal of rhinology. 2008;22:1–6. doi: 10.2500/ajr.2008.22.3119. [DOI] [PubMed] [Google Scholar]
- 78.Goldstein-Daruech N, Cope EK, Zhao KQ, Vukovic K, Kofonow JM, Doghramji L, Gonzalez B, Chiu AG, Kennedy DW, Palmer JN, Leid JG, Kreindler JL, Cohen NA. Tobacco smoke mediated induction of sinonasal microbial biofilms. PloS one. 2011;6:e15700. doi: 10.1371/journal.pone.0015700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Foreman A, Wormald PJ. Different biofilms, different disease? A clinical outcomes study. The Laryngoscope. 2010;120:1701–1706. doi: 10.1002/lary.21024. [DOI] [PubMed] [Google Scholar]
- 80.Foreman A, Singhal D, Psaltis AJ, Wormald PJ. Targeted imaging modality selection for bacterial biofilms in chronic rhinosinusitis. The Laryngoscope. 2010;120:427–431. doi: 10.1002/lary.20705. [DOI] [PubMed] [Google Scholar]
- 81.Chiu AG, Palmer JN, Woodworth BA, Doghramji L, Cohen MB, Prince A, Cohen NA. Baby shampoo nasal irrigations for the symptomatic post-functional endoscopic sinus surgery patient. American journal of rhinology. 2008;22:34–37. doi: 10.2500/ajr.2008.22.3122. [DOI] [PubMed] [Google Scholar]
- 82.Isaacs S, Fakhri S, Luong A, Whited C, Citardi MJ. The effect of dilute baby shampoo on nasal mucociliary clearance in healthy subjects. American journal of rhinology & allergy. 2011;25:e27–29. doi: 10.2500/ajra.2011.25.3583. [DOI] [PubMed] [Google Scholar]
- 83.Farag AA, Deal AM, McKinney KA, Thorp BD, Senior BA, Ebert CS, Jr, Zanation AM. Single-blind randomized controlled trial of surfactant vs hypertonic saline irrigation following endoscopic endonasal surgery. International forum of allergy & rhinology. 2013;3:276–280. doi: 10.1002/alr.21116. [DOI] [PubMed] [Google Scholar]
- 84.Ponikau JU, Sherris DA, Kern EB, Homburger HA, Frigas E, Gaffey TA, Roberts GD. The diagnosis and incidence of allergic fungal sinusitis. Mayo Clinic proceedings. 1999;74:877–884. doi: 10.4065/74.9.877. [DOI] [PubMed] [Google Scholar]
- 85.Braun H, Buzina W, Freudenschuss K, Beham A, Stammberger H. ‘Eosinophilic fungal rhinosinusitis’: a common disorder in Europe? The Laryngoscope. 2003;113:264–269. doi: 10.1097/00005537-200302000-00013. [DOI] [PubMed] [Google Scholar]
- 86.Hidir Y, Tosun F, Saracli MA, Gunal A, Gulec M, Yetiser S. Rate of allergic fungal etiology of chronic rhinosinusitis in Turkish population. European archives of oto-rhino-laryngology : official journal of the European Federation of Oto-Rhino-Laryngological Societies. 2008;265:415–419. doi: 10.1007/s00405-007-0475-x. [DOI] [PubMed] [Google Scholar]
- 87.Goh BS, Gendeh BS, Rose IM, Pit S, Samad SA. Prevalence of allergic fungal sinusitis in refractory chronic rhinosinusitis in adult Malaysians. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery. 2005;133:27–31. doi: 10.1016/j.otohns.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 88.Sacks PL, Harvey RJ, Rimmer J, Gallagher RM, Sacks R. Topical and systemic antifungal therapy for the symptomatic treatment of chronic rhinosinusitis. The Cochrane database of systematic reviews. 2011:CD008263. doi: 10.1002/14651858.CD008263.pub2. [DOI] [PubMed] [Google Scholar]
- 89.Ebbens FA, Scadding GK, Badia L, Hellings PW, Jorissen M, Mullol J, Cardesin A, Bachert C, van Zele TP, Dijkgraaf MG, Lund V, Fokkens WJ. Amphotericin B nasal lavages: not a solution for patients with chronic rhinosinusitis. The Journal of allergy and clinical immunology. 2006;118:1149–1156. doi: 10.1016/j.jaci.2006.07.058. [DOI] [PubMed] [Google Scholar]
- 90.Cleland EJ, Bassioni A, Boase S, Dowd S, Vreugde S, Wormald PJ. The fungal microbiome in chronic rhinosinusitis: richness, diversity, postoperative changes and patient outcomes. International forum of allergy & rhinology. 2014;4:259–265. doi: 10.1002/alr.21297. [DOI] [PubMed] [Google Scholar]
- 91.Rank MA, Wollan P, Kita H, Yawn BP. Acute exacerbations of chronic rhinosinusitis occur in a distinct seasonal pattern. The Journal of allergy and clinical immunology. 2010;126:168–169. doi: 10.1016/j.jaci.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cho GS, Moon BJ, Lee BJ, Gong CH, Kim NH, Kim YS, Kim HS, Jang YJ. High rates of detection of respiratory viruses in the nasal washes and mucosae of patients with chronic rhinosinusitis. J Clin Microbiol. 51:979–984. doi: 10.1128/JCM.02806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liao B, Hu CY, Liu T, Liu Z. Respiratory viral infection in the chronic persistent phase of chronic rhinosinusitis. The Laryngoscope. 124:832–837. doi: 10.1002/lary.24348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jang YJ, Kwon HJ, Park HW, Lee BJ. Detection of rhinovirus in turbinate epithelial cells of chronic sinusitis. American journal of rhinology. 2006;20:634–636. doi: 10.2500/ajr.2006.20.2899. [DOI] [PubMed] [Google Scholar]
- 95.Wood AJ, Antoszewska H, Fraser J, Douglas RG. Is chronic rhinosinusitis caused by persistent respiratory virus infection? International forum of allergy & rhinology. 1:95–100. doi: 10.1002/alr.20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang JH, Kwon HJ, Chung YS, Lee BJ, Jang YJ. Infection rate and virus-induced cytokine secretion in experimental rhinovirus infection in mucosal organ culture: comparison between specimens from patients with chronic rhinosinusitis with nasal polyps and those from normal subjects. Archives of otolaryngology--head & neck surgery. 2008;134:424–427. doi: 10.1001/archotol.134.4.424. [DOI] [PubMed] [Google Scholar]
- 97.Yamin M, Holbrook EH, Gray ST, Harold R, Busaba N, Sridhar A, Powell KJ, Hamilos DL. Cigarette smoke combined with Toll-like receptor 3 signaling triggers exaggerated epithelial regulated upon activation, normal T-cell expressed and secreted/CCL5 expression in chronic rhinosinusitis. The Journal of allergy and clinical immunology. 2008;122:1145–1153. e1143. doi: 10.1016/j.jaci.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 98.Jiang RS, Lin JF, Hsu CY. Correlation between bacteriology of the middle meatus and ethmoid sinus in chronic sinusitis. The Journal of laryngology and otology. 2002;116:443–446. doi: 10.1258/0022215021911248. [DOI] [PubMed] [Google Scholar]
- 99.Liu CM, Soldanova K, Nordstrom L, Dwan MG, Moss OL, Contente-Cuomo TL, Keim P, Price LB, Lane AP. Medical therapy reduces microbiota diversity and evenness in surgically recalcitrant chronic rhinosinusitis. International forum of allergy & rhinology. 2013;3:775–781. doi: 10.1002/alr.21195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R. Experimental and analytical tools for studying the human microbiome. Nature reviews Genetics. 2012;13:47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proceedings of the National Academy of Sciences of the United States of America. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang YJ, Boushey HA. The microbiome in asthma. The Journal of allergy and clinical immunology. 2015;135:25–30. doi: 10.1016/j.jaci.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Specjalski K, Jassem E. Chlamydophila pneumoniae, Mycoplasma pneumoniae infections, and asthma control. Allergy and asthma proceedings: the official journal of regional and state allergy societies. 2011;32:9–17. doi: 10.2500/aap.2011.32.3431. [DOI] [PubMed] [Google Scholar]
- 104.Martin RJ, Kraft M, Chu HW, Berns EA, Cassell GH. A link between chronic asthma and chronic infection. The Journal of allergy and clinical immunology. 2001;107:595–601. doi: 10.1067/mai.2001.113563. [DOI] [PubMed] [Google Scholar]
- 105.Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bonnelykke K, Brasholt M, Heltberg A, Vissing NH, Thorsen SV, Stage M, Pipper CB. Childhood asthma after bacterial colonization of the airway in neonates. The New England journal of medicine. 2007;357:1487–1495. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- 106.Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE, Printz MC, Lee WM, Shult PA, Reisdorf E, Carlson-Dakes KT, Salazar LP, DaSilva DF, Tisler CJ, Gern JE, Lemanske RF., Jr Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. American journal of respiratory and critical care medicine. 2008;178:667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sigurs N, Aljassim F, Kjellman B, Robinson PD, Sigurbergsson F, Bjarnason R, Gustafsson PM. Asthma and allergy patterns over 18 years after severe RSV bronchiolitis in the first year of life. Thorax. 2010;65:1045–1052. doi: 10.1136/thx.2009.121582. [DOI] [PubMed] [Google Scholar]
- 108.Kusel MM, de Klerk NH, Kebadze T, Vohma V, Holt PG, Johnston SL, Sly PD. Early-life respiratory viral infections, atopic sensitization, and risk of subsequent development of persistent asthma. The Journal of allergy and clinical immunology. 2007;119:1105–1110. doi: 10.1016/j.jaci.2006.12.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kotaniemi-Syrjanen A, Vainionpaa R, Reijonen TM, Waris M, Korhonen K, Korppi M. Rhinovirus-induced wheezing in infancy--the first sign of childhood asthma? The Journal of allergy and clinical immunology. 2003;111:66–71. doi: 10.1067/mai.2003.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Park H, Shin JW, Park SG, Kim W. Microbial Communities in the Upper Respiratory Tract of Patients with Asthma and Chronic Obstructive Pulmonary Disease. PloS one. 2014;9:e109710. doi: 10.1371/journal.pone.0109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cardenas PA, Cooper PJ, Cox MJ, Chico M, Arias C, Moffatt MF, Cookson WO. Upper airways microbiota in antibiotic-naive wheezing and healthy infants from the tropics of rural Ecuador. PloS one. 2012;7:e46803. doi: 10.1371/journal.pone.0046803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.van Woerden HC, Gregory C, Brown R, Marchesi JR, Hoogendoorn B, Matthews IP. Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: a community based case control study. BMC infectious diseases. 2013;13:69. doi: 10.1186/1471-2334-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Green BJ, Wiriyachaiporn S, Grainge C, Rogers GB, Kehagia V, Lau L, Carroll MP, Bruce KD, Howarth PH. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PloS one. 2014;9:e100645. doi: 10.1371/journal.pone.0100645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kellam P, Weiss RA. Infectogenomics: insights from the host genome into infectious diseases. Cell. 2006;124:695–697. doi: 10.1016/j.cell.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nibali L, Henderson B, Sadiq ST, Donos N. Genetic dysbiosis: the role of microbial insults in chronic inflammatory diseases. Journal of oral microbiology. 2014;6 doi: 10.3402/jom.v6.22962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hsu J, Avila PC, Kern RC, Hayes MG, Schleimer RP, Pinto JM. Genetics of chronic rhinosinusitis: state of the field and directions forward. The Journal of allergy and clinical immunology. 2013;131:977–993. 993 e971–975. doi: 10.1016/j.jaci.2013.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang Y, Endam LM, Filali-Mouhim A, Bosse Y, Castano R, Desrosiers M. Polymorphisms in the nitric oxide synthase 1 gene are associated with severe chronic rhinosinusitis. American journal of rhinology & allergy. 2011;25:e49–54. doi: 10.2500/ajra.2011.25.3588. [DOI] [PubMed] [Google Scholar]
- 118.Ramanathan M, Jr, Lee WK, Spannhake EW, Lane AP. Th2 cytokines associated with chronic rhinosinusitis with polyps down-regulate the antimicrobial immune function of human sinonasal epithelial cells. American journal of rhinology. 2008;22:115–121. doi: 10.2500/ajr.2008.22.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mahdavinia M, Carter RG, Ocampo CJ, Stevens W, Kato A, Tan BK, Kern RC, Conley DB, Chandra R, Hulse KE, Suh LA, Norton JE, Peters AT, Grammer LC, 3rd, Schwartz LB, Schleimer RP. Basophils are elevated in nasal polyps of patients with chronic rhinosinusitis without aspirin sensitivity. The Journal of allergy and clinical immunology. 2014;133:1759–1763. doi: 10.1016/j.jaci.2013.12.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Takabayashi T, Kato A, Peters AT, Suh LA, Carter R, Norton J, Grammer LC, Tan BK, Chandra RK, Conley DB, Kern RC, Fujieda S, Schleimer RP. Glandular mast cells with distinct phenotype are highly elevated in chronic rhinosinusitis with nasal polyps. The Journal of allergy and clinical immunology. 130:410–420. e415. doi: 10.1016/j.jaci.2012.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lane AP, Truong-Tran QA, Schleimer RP. Altered expression of genes associated with innate immunity and inflammation in recalcitrant rhinosinusitis with polyps. American journal of rhinology. 2006;20:138–144. [PMC free article] [PubMed] [Google Scholar]
- 122.Tieu DD, Kern RC, Schleimer RP. Alterations in epithelial barrier function and host defense responses in chronic rhinosinusitis. The Journal of allergy and clinical immunology. 2009;124:37–42. doi: 10.1016/j.jaci.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wu X, Peters-Hall JR, Ghimbovschi S, Mimms R, Rose MC, Pena MT. Glandular gene expression of sinus mucosa in chronic rhinosinusitis with and without cystic fibrosis. American journal of respiratory cell and molecular biology. 2011;45:525–533. doi: 10.1165/rcmb.2010-0133OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schleimer RP, Kato A, Kern R, Kuperman D, Avila PC. Epithelium: at the interface of innate and adaptive immune responses. The Journal of allergy and clinical immunology. 2007;120:1279–1284. doi: 10.1016/j.jaci.2007.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hiemstra PS. The role of epithelial beta-defensins and cathelicidins in host defense of the lung. Experimental lung research. 2007;33:537–542. doi: 10.1080/01902140701756687. [DOI] [PubMed] [Google Scholar]
- 126.Seshadri S, Lin DC, Rosati M, Carter RG, Norton JE, Suh L, Kato A, Chandra RK, Harris KE, Chu HW, Peters AT, Tan BK, Conley DB, Grammer LC, Kern RC, Schleimer RP. Reduced expression of antimicrobial PLUNC proteins in nasal polyp tissues of patients with chronic rhinosinusitis. Allergy. 2012;67:920–928. doi: 10.1111/j.1398-9995.2012.02848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Glaser R, Harder J, Lange H, Bartels J, Christophers E, Schroder JM. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nature immunology. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 128.Tieu DD, Peters AT, Carter RG, Suh L, Conley DB, Chandra R, Norton J, Grammer LC, Harris KE, Kato A, Kern RC, Schleimer RP. Evidence for diminished levels of epithelial psoriasin and calprotectin in chronic rhinosinusitis. The Journal of allergy and clinical immunology. 2010;125:667–675. doi: 10.1016/j.jaci.2009.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Woodworth BA, Lathers D, Neal JG, Skinner M, Richardson M, Young MR, Schlosser RJ. Immunolocalization of surfactant protein A and D in sinonasal mucosa. American journal of rhinology. 2006;20:461–465. doi: 10.2500/ajr.2006.20.2892. [DOI] [PubMed] [Google Scholar]
- 130.Lee HM, Kang HJ, Woo JS, Chae SW, Lee SH, Hwang SJ. Upregulation of surfactant protein A in chronic rhinosinusitis. The Laryngoscope. 2006;116:328–330. doi: 10.1097/01.mlg.0000194223.22763.5f. [DOI] [PubMed] [Google Scholar]
- 131.Vandermeer J, Sha Q, Lane AP, Schleimer RP. Innate immunity of the sinonasal cavity: expression of messenger RNA for complement cascade components and toll-like receptors. Archives of otolaryngology--head & neck surgery. 2004;130:1374–1380. doi: 10.1001/archotol.130.12.1374. [DOI] [PubMed] [Google Scholar]
- 132.Lane AP, Truong-Tran QA, Myers A, Bickel C, Schleimer RP. Serum amyloid A, properdin, complement 3, and toll-like receptors are expressed locally in human sinonasal tissue. American journal of rhinology. 2006;20:117–123. [PMC free article] [PubMed] [Google Scholar]
- 133.Ramanathan M, Jr, Lee WK, Dubin MG, Lin S, Spannhake EW, Lane AP. Sinonasal epithelial cell expression of toll-like receptor 9 is decreased in chronic rhinosinusitis with polyps. American journal of rhinology. 2007;21:110–116. doi: 10.2500/ajr.2007.21.2997. [DOI] [PubMed] [Google Scholar]
- 134.Lee RJ, Xiong G, Kofonow JM, Chen B, Lysenko A, Jiang P, Abraham V, Doghramji L, Adappa ND, Palmer JN, Kennedy DW, Beauchamp GK, Doulias PT, Ischiropoulos H, Kreindler JL, Reed DR, Cohen NA. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. The Journal of clinical investigation. 2012;122:4145–4159. doi: 10.1172/JCI64240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K, Husain S, Kreindler JL, Dubin PJ, Pilewski JM, Myerburg MM, Mason CA, Iwakura Y, Kolls JK. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nature medicine. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nature medicine. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 137.Eidenschenk C, Rutz S, Liesenfeld O, Ouyang W. Role of IL-22 in microbial host defense. Current topics in microbiology and immunology. 2014;380:213–236. doi: 10.1007/978-3-662-43492-5_10. [DOI] [PubMed] [Google Scholar]
- 138.Taube C, Tertilt C, Gyulveszi G, Dehzad N, Kreymborg K, Schneeweiss K, Michel E, Reuter S, Renauld JC, Arnold-Schild D, Schild H, Buhl R, Becher B. IL-22 is produced by innate lymphoid cells and limits inflammation in allergic airway disease. PloS one. 2011;6:e21799. doi: 10.1371/journal.pone.0021799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, Tjota MY, Seo GY, Cao S, Theriault BR, Antonopoulos DA, Zhou L, Chang EB, Fu YX, Nagler CR. Commensal bacteria protect against food allergen sensitization. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Endam LM, Bosse Y, Filali-Mouhim A, Cormier C, Boisvert P, Boulet LP, Hudson TJ, Desrosiers M. Polymorphisms in the interleukin-22 receptor alpha-1 gene are associated with severe chronic rhinosinusitis. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery. 2009;140:741–747. doi: 10.1016/j.otohns.2008.12.058. [DOI] [PubMed] [Google Scholar]
- 141.Ramanathan M, Jr, Spannhake EW, Lane AP. Chronic rhinosinusitis with nasal polyps is associated with decreased expression of mucosal interleukin 22 receptor. The Laryngoscope. 2007;117:1839–1843. doi: 10.1097/MLG.0b013e31811edd4f. [DOI] [PubMed] [Google Scholar]
- 142.Detwiller KY, Smith TL, Alt JA, Trune DR, Mace JC, Sautter NB. Differential expression of innate immunity genes in chronic rhinosinusitis. American journal of rhinology & allergy. 2014;28:374–377. doi: 10.2500/ajra.2014.28.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mattila E, Uusitalo-Seppala R, Wuorela M, Lehtola L, Nurmi H, Ristikankare M, Moilanen V, Salminen K, Seppala M, Mattila PS, Anttila VJ, Arkkila P. Fecal transplantation, through colonoscopy, is effective therapy for recurrent Clostridium difficile infection. Gastroenterology. 2012;142:490–496. doi: 10.1053/j.gastro.2011.11.037. [DOI] [PubMed] [Google Scholar]
- 144.Wang ZK, Yang YS, Chen Y, Yuan J, Sun G, Peng LH. Intestinal microbiota pathogenesis and fecal microbiota transplantation for inflammatory bowel disease. World journal of gastroenterology : WJG. 2014;20:14805–14820. doi: 10.3748/wjg.v20.i40.14805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cleland EJ, Drilling A, Bassiouni A, James C, Vreugde S, Wormald PJ. Probiotic manipulation of the chronic rhinosinusitis microbiome. International forum of allergy & rhinology. 2014;4:309–314. doi: 10.1002/alr.21279. [DOI] [PubMed] [Google Scholar]
- 146.Shi LL, Song J, Xiong P, Cao PP, Liao B, Ma J, Zhang YN, Zeng M, Liu Y, Wang H, Cui YH, Huang SK, Liu Z. Disease-specific T-helper cell polarizing function of lesional dendritic cells in different types of chronic rhinosinusitis with nasal polyps. American journal of respiratory and critical care medicine. 2014;190:628–638. doi: 10.1164/rccm.201402-0234OC. [DOI] [PubMed] [Google Scholar]
- 147.Poposki JA, Peterson S, Welch K, Schleimer RP, Hulse KE, Peters AT, Norton J, Suh LA, Carter R, Harris KE, Grammer LC, Tan BK, Chandra RK, Conley DB, Kern RC, Kato A. Elevated presence of myeloid dendritic cells in nasal polyps of patients with chronic rhinosinusitis. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2015;45:384–393. doi: 10.1111/cea.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schroeder JT. Basophils: emerging roles in the pathogenesis of allergic disease. Immunological reviews. 2011;242:144–160. doi: 10.1111/j.1600-065X.2011.01023.x. [DOI] [PubMed] [Google Scholar]
- 149.Shelburne CP, Abraham SN. The mast cell in innate and adaptive immunity. Adv Exp Med Biol. 716:162–185. doi: 10.1007/978-1-4419-9533-9_10. [DOI] [PubMed] [Google Scholar]
- 150.Siraganian RP, Hook WA. Mechanism of histamine release by formyl methionine-containing peptides. Journal of immunology. 1977;119:2078–2083. [PubMed] [Google Scholar]
- 151.Kouzaki H, O’Grady SM, Lawrence CB, Kita H. Proteases induce production of thymic stromal lymphopoietin by airway epithelial cells through protease-activated receptor-2. J Immunol. 2009;183:1427–1434. doi: 10.4049/jimmunol.0900904. [DOI] [PMC free article] [PubMed] [Google Scholar]