Abstract
Endogenous hydrogen polysulfides (H2Sn, n > 1) have been recognized as important regulators in sulfur-related redox biology. H2Sn can activate tumor suppressors, ion channels, and transcription factors with higher potency than H2S. While H2Sn are drawing increasing attention, their exact mechanisms of actions are still poorly understood. A major hurdle in this field is the lack of reliable and convenient methods for H2Sn detection. In this work we report a H2Sn-mediated benzodithiolone formation under mild conditions. This reaction takes advantage of the unique dual-reactivity of H2Sn as both a nucleophile and an electrophile. It opens a door for better understanding H2Sn's chemical biology. Based on this reaction, three fluorescent probes (PSP-1~3) were synthesized and evaluated. Among the probes prepared, PSP-3 showed a desirable off-on fluorescence response to H2Sn and high specificity. It was successfully applied in visualizing intracellular H2Sn.
Keywords: hydrogen polysulfides, imaging agents, fluorescence, cyclization, fluorescent probes
Reactive sulfur species (RSS) are a group of sulfur-containing molecules that play regulatory roles in biological systems. Important RSS include biothiols and S-modified protein cysteine adducts. RSS also include hydrogen sulfide (H2S) and sulfane sulfurs (such as cysteine persulfides (R-S-SH) and polysulfides (R-S-Sn-S-R', n > 0)). Understanding RSS's mechanisms of actions has become a very active research area in modern chemical biology, particularly from the methodological point of view.[1] Among various RSS, hydrogen polysulfides (H2Sn, n > 1) have attracted particular attention, mainly due to their involvement in H2S-related redox biology.[2] H2Sn can be generated from endogenous H2S upon reacting with reactive oxygen species (ROS).[3] H2S can also react with other sulfane sulfurs such as S8 to form H2Sn.[2d,3c,4] Moreover, H2Sn may have their own biosynthetic pathways. For example, while cystathionine γ-lyase (CSE)-mediated cystine metabolism can produce cysteine persulfide,[5] it may also catalyze direct generation of H2Sn from cystine. A series of recent works suggested that H2Sn might be a new group of signaling molecules.[2,3c,4] It was found that H2Sn can activate tumor suppressors, ion channels, and transcription factors with higher potency than H2S.[4] Some physiological activities that were originally assigned to H2S may actually be mediated by H2Sn. One such example is S-sulfhydration.[1a,5,6] This post-translational modification was previously thought to be the result of H2S. However, recent results have demonstrated that H2Sn are more effective than H2S in S-sulfhydration.[2,3b–c]
Although H2Sn have now been recognized as potent physiological mediators, a number of issues remain to be clarified, such as the production and degradation pathways of H2Sn, their regulatory mechanisms, and potential physiological stimuli that induce those regulatory mechanisms. To address these issues, it is critical to develop effective methods for H2Sn detection. The traditional method is to measure UV absorption peaks at 290–300 nm and 370 nm, which has low sensitivity and is not applicable for biological detections. Fluorescence assays could be useful because of their high sensitivity and spatiotemporal resolution capability. While fluorescent probes for H2S have been extensively studied,[7] the probes for H2Sn are still underdeveloped due to limited known chemistry of H2Sn.
Our laboratory has initiated a program to study new chemistry/reactions of H2Sn and develop reaction-based fluorescent probes for H2Sn. In 2014, we reported the first H2Sn-specific probes (DSP, Scheme 1) which employed a 2-fluoro-5-nitro-benzoic ester template to trap H2Sn and promote an intramolecular cyclization to release a fluorophore.[8] DSP probes, such as DSP-3, showed satisfactory sensitivity and selectivity for H2Sn. Several other groups have adopted the same template to develop H2Sn probes with interesting properties.[9] However, a drawback of DSP is that 2-fluoro-5-nitro-benzoic esters can also react with biothiols. Although such reactions do not turn on fluorescence, the consumption of the probes is a problem. In theory, the reaction product, i.e. thioether 1, can further react with H2Sn to switch on fluorescence. We found that the reaction was somewhat slow and non-productive when H2Sn concentration was low (at μM levels). Therefore, high loading of DSP probes may be needed for biological detections. Very recently our lab discovered another H2Sn-specific aziridine opening reaction and developed a probe (AP) based on this reaction.[10] AP showed excellent selectivity for H2Sn, it did not react with biothiols or H2S in physiological concentrations. However, due to limited options for aziridine-based fluorophores, the sensitivity of AP for H2Sn was not optimal and attempts to use AP for imaging H2Sn in cells were not successful. Obviously further improvement of the probes is needed. Herein we report the design, synthesis, and evaluation of a series of new fluorescent probes (PSP1-3) that showed high sensitivity and selectivity for H2Sn and can be used for endogenous H2Sn imaging.
Scheme 1.
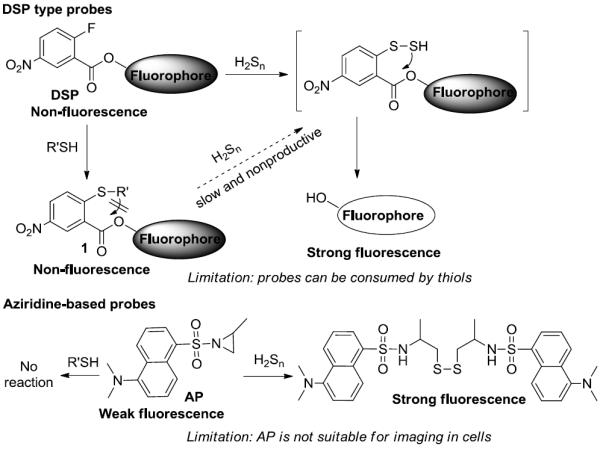
Previous designs of H2Sn-specific fluorescent probes.
The key of developing optimum probes is to identify a highly specific H2Sn recognition unit. Ideally, such a recognition unit should only react with H2Sn and not react with other sulfur species, especially biothiols. From a chemistry perspective, H2Sn are quite different from thiols or H2S. The estimated pKa values of H2Sn are in the range of 3 to 5.[11] For comparison, the pKa values of H2S and biothiols are in the range of 7 to 9.2. Under physiological pH, H2Sn are expected to be weak acids, and stronger and more reactive nucleophiles than biothiols and H2S due to alpha-effects. In addition, H2Sn belong to the sulfane sulfur family. A character of sulfane sulfurs is that they can function as electrophiles and react with certain nucleophiles.[12] Overall, H2Sn have a unique dual-reactivity and can act as both nucleophiles and electrophiles. Taking advantage of this property, we envisioned template 2 might be specific for H2Sn (Scheme 2). In 2, a thioester was employed to trap the nucleophilicity of H2Sn. One might worry that the thioester group could also react with biothiols as the reactions between thioesters and thiols are known, for example in the well-known Native Chemical Ligation.[13] However, those reactions are mostly for synthetic purposes with high concentrations of reactants and special solvent conditions. The relatively low concentrations of biothiols in biological systems and the mild, neutral, and aqueous environments may make the reactions slow and non-productive. In addition, we expected the manipulation of R groups (steric and electronic effects) in 2 could significantly differentiate its reaction rates toward thiols and H2Sn. If H2Sn could selectively react with an appropriate thioester group, the product, i.e. 3, should further react with H2Sn (now serving as an electrophile) to form 4. The following spontaneous cyclization should release the fluorophore. Overall, this process would be specific for H2Sn.
Scheme 2.
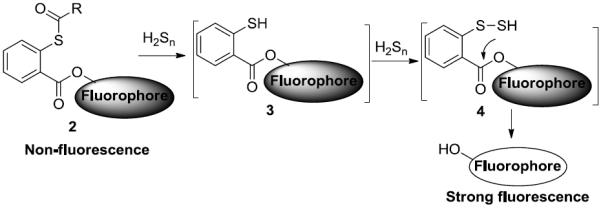
The design of dual-reactivity based probes.
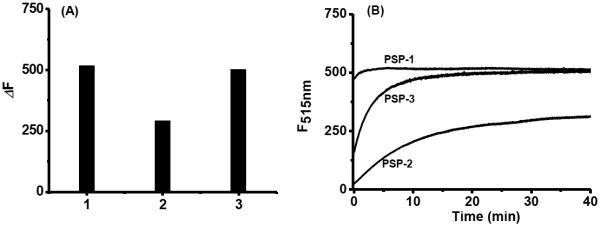
Based on this idea, we synthesized three probes (PSP-1, PSP-2, and PSP-3, Scheme 3). Three different R groups (Me, tBu, and Ph) were used in order to explore the effects of acyl groups on the reactivity of thioesters toward biothiols. We first tested their fluorescence properties and responses to H2Sn in PBS buffer. Freshly prepared Na2S2 solutions were used as the equivalents of H2Sn. All three probes showed almost no fluorescence emission (ΦPSP-1 = ΦPSP-2 = 0.02; ΦPSP-3 = 0.01) due to the protection of the two hydroxyl groups of fluorescein. Upon the treatment of Na2S2 (5 eq) for 30 min, the probes gave significant fluorescence enhancements (Figure 1A). PSP-1 and PSP-3 exhibited stronger fluorescence increases than PSP-2. Presumably the bulky t-butyl group of PSP-2 decreased the reactivity toward to H2Sn. We also tested time-dependent fluorescence changes of the probes in the presence of Na2S2 (Figure 1B). The maximum emission intensities of PSP-1 and PSP-3 were reached within 2 min and 10 min respectively, indicating fluorescence turn-on was fast. The fluorescence turn-on of PSP-2 was slower (~20 min), again suggesting the steric effect played a role in its reactivity. For the purpose of reproducibility, a reaction time of 30 min was employed in all of the following experiments.
Scheme 3.
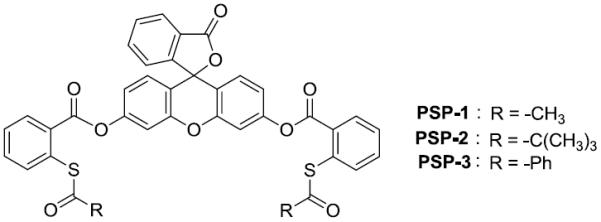
The structures of PSP probes.
Figure 1.
(A) Fluorescence intensity increases (ΔF) of PSP (10 μM) with Na2S2 (50 μM): (1) PSP-1; (2) PSP-2; (3) PSP-3. (B) Time-dependent fluorescence intensity changes of PSP (10 μM) in the presence of Na2S2 (50 μM).
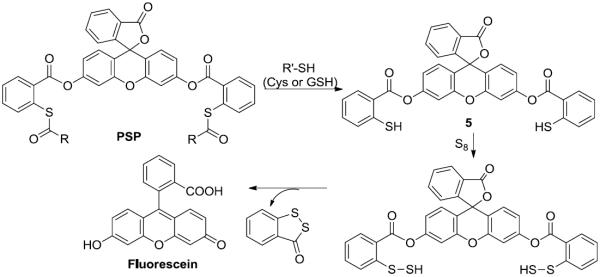
Next we studied the stability of the probes in the presence of biothiols. As shown in Scheme 4, if the probes undergo thioester exchange with biothiols, the resultant product 5 should be sensitive to sulfane sulfurs like elemental sulfur (S8) and produce strong fluorescence. Therefore, measuring fluorescence of the probes in a mixture of thiols and S8 should be suitable for evaluating the stability of the probes to thiols.
Scheme 4.
Possible reactions of PSP probes with biothiols and S8.
We then tested fluorescence responses of the probes (10 μM) to Cys (1 mM) and GSH (5 mM) in the absence or presence of S8 (50 μM). As shown in Figure 2, Cys, GSH, or S8 alone did not induce any responses. The mixtures of biothiols (GSH or Cys) and S8 produced strong fluorescence for PSP-1. The same mixtures caused almost no fluorescence increase for PSP-2 and PSP-3. As a positive control, the thioester exchange product 5 gave strong fluorescence for S8 under the same conditions. These results confirmed our hypothesis that the acyl group of thioester could affect its reactivity towards thiols. PSP-1 appeared to be labile to biothiols while PSP-2 and PSP-3 were stable. Therefore, the use of PSP-2 and PSP-3 should not be affected by cellular biothiols.
Figure 2.
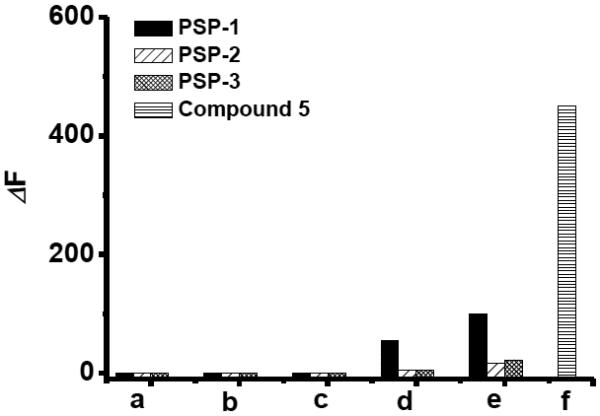
Fluorescence increase value (ΔF) of each probe (10 μM) in the presence of biothiols and S8: (a) Probe + 5 mM GSH; (b) Probe + 1 mM Cys; (c) Probe + 50 μM S8; (d) Probe + 5 mM GSH + 50 μM S8; (e) Probe + 1 mM Cys + 50 μM S8; (f) 10 μM Compound 5 + 50 μM S8.
Having demonstrated the excellent reactivity and stability of PSP-3, this probe was selected for further studies. We next validated its selectivity for H2Sn over other common RSS including Hcy, GSSG, H2S, SO32−, S2O32−, SO42−, and CH3SSSCH3. As shown in Figure 3A, no fluorescence increase was observed for these RSS (columns 2–8). Only Na2S2 and Na2S4 triggered strong fluorescence enhancements (columns 9 and 10). Additionally, It is known that H2Sn could be generated from the reactions between H2S and ROS. Therefore, we wondered if the probe could sense in situ H2Sn formation from H2S and ROS. As shown in Figure 3B, the responses of PSP-3 to a series of ROS including hydrogen peroxide (H2O2), hypochlorite (ClO−), superoxide (O2−), hydroxyl radical (•OH), and singlet oxygen (1O2) were first tested. These ROS did not induce any fluorescence enhancements (columns 2–6). When H2S was added to these ROS, a strong fluorescence signal was observed from ClO− (column 9), but not from other ROS. These results agreed with previous discoveries that H2Sn can be efficiently derived from H2S and ClO−, but other ROS are not effective for H2Sn generation under these concentrations.[3,8–10] The responses of PSP-3 in the presence of other representative amino acids were also evaluated and no responses were observed (Figure S1).
Figure 3.
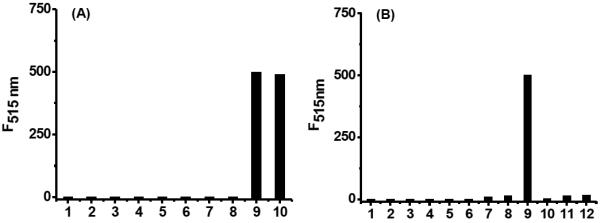
(A) Fluorescence intensity of 10 μM PSP-3 in the presence of various RSS: (1) probe alone; (2) 100 μM Hcy; (3) 100 μM GSSG; (4) 200 μM Na2S; (5) 100 μM Na2S2O3; (6) 100 μM Na2SO3; (7) 100 μM Na2SO4; (8) 100 μM CH3SSSCH3; (9) 50 μM Na2S2; (10) 50 μM Na2S4. (B) Fluorescence intensity of PSP-3 (10 μM) in the presence of ROS (with or without H2S). (1) probe alone; (2) 200 μM H2O2; (3) 50 μM ClO−; (4) 50 μM O2−; (5) 50 μM •OH; (6) 50 μM 1O2; (7) 200 μM H2O2 + 50 μM Na2S; (8) 200 μM H2O2 + 100 μM Na2S; (9) 50 μM ClO− + 100 μM Na2S; (10) 50 μM O2− + 100 μM Na2S; (11) 50 μM •OH + 100 μM Na2S; (12) 50 μM 1O2 + 100 μM Na2S.
To further evaluate the sensitivity of PSP-3 for H2Sn, a series of varied concentrations of Na2S2 (0 ~ 50 μM) were tested. The fluorescence intensities were linearly related to the concentrations of Na2S2 in the range of 0 ~ 20 μM (Figure S2). The detection limit was calculated to be 3 nM. This is the most sensitive probe for H2Sn reported so far. To clarify fluorescence turn-on mechanism, we studied the reactions between the probes and Na2S2. All of the reactions gave fluorescein and benzodithiolone as the isolated products with good yields (80% ~ 90%) (Figure S3). As such, we believe the fluorescence turn-on is initiated by H2Sn-mediated thioester cleavage. The resulting thiolate further reacts with H2Sn and promotes cyclization to form benzodithiolone and release the fluorophore.
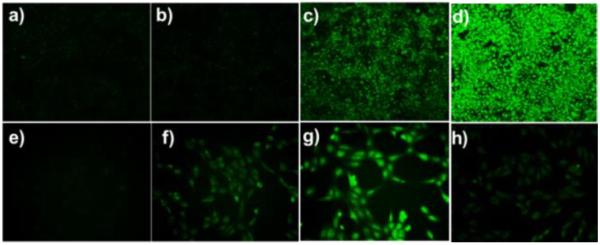
Next we wondered if PSP-3 could be used for imaging H2Sn in cells. We first validated the capability of PSP-3 in visualizing exogenous H2Sn. As shown in Figure 4, HeLa, RAW264.7, and Vero cells were respectively incubated with PSP-3 (5 μM) for 25 min. Extracellular probe was washed off, and no significant fluorescence was observed in cells. However, when cells were treated with Na2S2 or Na2S4 (30 μM) for 20 min, strong fluorescence was observed. These results demonstrated that PSP-3 has good cell permeability and could be used to monitor H2Sn in cells. In addition, cell viability assay demonstrated that PSP-3 has no cytotoxicity to cells (Figure S4).
Figure 4.
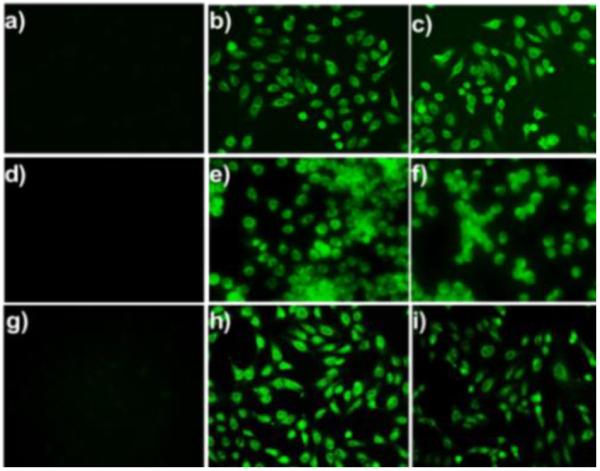
Fluorescence images of exogenous H2Sn in HeLa (a–c), RAW264.7 (d–f), and Vero (g–i). Cells were incubated with PSP-3 (5 μM) for 25 min, then washed and subjected to different treatments. (a, d, and g) controls (no Na2S2 or Na2S4); (b, e, and h) treated with 30 μM Na2S2; (c, f, and i) treated with 30 μM Na2S4.
H2Sn are short-lived species and can readily decompose in buffers. In this regard, in-situ generation of H2Sn from H2S and ClO− provides a more reliable and sustainable system for H2Sn production. Herein PSP-3 was used to monitor in-situ generation of H2Sn in cells. As shown in Figure S5, neither ClO− nor H2S gave noticeable fluorescence responses while the mixture of H2S/ClO− led to a strong fluorescence enhancement, which was even brighter than those obtained with exogenous H2Sn. These results confirmed that in-situ generation of H2Sn by H2S and ClO- is a more effective system than simply using Na2S2 (or Na2S4) to maintain H2Sn levels in cells.
Having demonstrated the capability of PSP-3 in detecting exogenous H2Sn in cells, we then sought to use PSP-3 to monitor endogenous H2Sn formation. Our recent studies found that CSE-overexpression causes significant elevation of persulfide and polysulfide levels in cells.[5] Therefore, CSE over-expressed cells provide a suitable system for endogenous H2Sn production. As shown in Figure 5a–d, normal and CSE-overexpressed[5] COS-7 cells were separately treated with PSP-3 for 30 min. As expected, CSE-overexpressed cells induced much-enhanced fluorescent signals as compared to normal cells. In fact, appreciable amount of H2Sn was directly generated from a recombinant CSE in a cell-free reaction mixture containing cystine as a substrate (Figure S6). These results demonstrate that PSP-3 is suitable for detecting endogenous H2Sn. Additionally, Kimura et al recently described a new H2S biosynthetic pathway from D-Cys in mammalian cells, which occurs predominantly in the kidney and the cerebellum.[14] This process seems to be regulated by 3-mercaptopyruvate sulfurtransferase (3MST) and D-amino acid oxidase (DAO). It was suggested that H2S produced from D-Cys might be stored as bound sulfane sulfur and the levels of sulfane sulfur can be significantly enhanced by D-Cys. In theory, H2Sn could be produced from the reactions between H2S and sulfane sulfurs. Therefore D-Cys stimulation in Vero cells may be a useful system of endogenous H2Sn generation and this system was used to further validate the probe's sensitivity for endogenous H2Sn. As shown in Figure 6e–h, the cells were incubated with PSP-3 (5 μM) for 25 min at 37 °C. The control (Figure 5e) showed almost no fluorescence. When cells were treated with 30 μM D-Cys, a modest but obvious fluorescence was observed (Figure 5f). When the concentration of D-Cys was increased to 1 mM, we observed strong fluorescence (Figure 5g). Moreover, when the cells were first treated with sodium benzoate, a DAO inhibitor, the stimulation by D-Cys (1 mM) led to significantly weakened fluorescence. Taken together, these results indicate that D-Cys and DAO may be responsible for cellular H2Sn production.
Figure 5.
Fluorescence images of endogenous H2Sn in COS-7 (a–d) and Vero (e–h). (a) Cells not treated with PSP-3; (b) Normal cells treated with PSP-3; (c) CSE-overexpressed cells treated with PSP-3; (d) Cells treated with 20 μM Na2S2. (e–g) Cells were incubated with PSP-3 (5 μM) for 25 min, washed, and subjected to different treatments. (e) cells were incubated with FBS-free media for 30 min; (f) cells were incubated with 30 μM D-Cys for 30 min; (g) cells were incubated with 1 mM D-Cys for 30 min; (h) cells were pre-treated with sodium benzoate (250 μM) for 30 min, then washed and incubated with PSP-3 (5 μM) for 25 min, and then washed and incubated with 1 mM D-Cys for 30 min.
In summary, we report here the development of three new fluorescent probes (PSP-1, PSP-2, PSP-3) based on the unique dual-reactivity of H2Sn. Among the probes prepared, PSP-3 showed desirable off–on fluorescence response to H2Sn and high specificity. PSP-3 was also successfully applied in visualizing both exogenous and endogenous H2Sn in cells. This novel probe overcomes the major limitations of previously reported H2Sn probes, and it is expected to serve as a useful tool in understanding the physiological functions of H2Sn.
Supplementary Material
Acknowledgements
This work was supported by an ACS Teva Scholar Grant and the NIH (R01HL116571). This work was also supported in part by Grants-in-Aid for Scientific Research and Grants-in-Aid for Scientific Research on Innovative Areas from MEXT of Japan.
References
- [1].a) Paulsen CE, Carroll KS. Chem. Rev. 2013;113:4633. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Seo YH, Carroll KS. Angew. Chem. Int. Ed. 2011;50:1342. doi: 10.1002/anie.201007175. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2011;123:1378. [Google Scholar]; c) Poole TH, Reisz JA, Zhao W, Poole LB, Furdui CM, King SB. J. Am. Chem. Soc. 2014;136:6167. doi: 10.1021/ja500364r. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Miljkovic JL, Kenkel I, Ivanovic-Burmazovic I, Filipovic MR. Angew. Chem. Int. Ed. 2013;52:12061. doi: 10.1002/anie.201305669. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013;125:12283. [Google Scholar]; e) Yang X, Guo Y, Strongin M. Angew. Chem. Int. Ed. 2011;50:10690. doi: 10.1002/anie.201103759. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2011;123:10878. [Google Scholar]
- [2].a) Greiner R, Pálinkás Z, Bäsell K, Becher D, Antelmann H, Nagy P, Dick TP. Antioxid. Redox Signal. 2013;19:1749. doi: 10.1089/ars.2012.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Toohey JI, Cooper AJL. Molecules. 2014;19:12789. doi: 10.3390/molecules190812789. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ono K, Akaike T, Sawa T, Kumagai Y, Wink D, Tantillo DJ, Hobbs AJ, Nagy P, Xian M, Lin J, Fukuto JM. Free Radical Biol. Med. 2014;77:82. doi: 10.1016/j.freeradbiomed.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kimura H. Antioxid. Redox Signal. 2015;22:362. doi: 10.1089/ars.2014.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Mishanina TV, Libiad M, Banerjee R. Nat. Chem. Biol. 2015;11:457. doi: 10.1038/nchembio.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Kabil O, Motl N, Banerjee R. Biochim. Biophys. Acta. 2014;1844:1355. doi: 10.1016/j.bbapap.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Nagy P, Winterbourn CC. Chem. Res. Toxicol. 2010;23:1541. doi: 10.1021/tx100266a. [DOI] [PubMed] [Google Scholar]; b) Nagy P, Pálinkás Z, Nagy A, Budai B, Tóth I, Vasas A. Biochim. Biophys. Acta. 2014;1840:876. doi: 10.1016/j.bbagen.2013.05.037. [DOI] [PubMed] [Google Scholar]; c) Kimura Y, Mikami Y, Osumi K, Tsugane M, Oka J, Kimura H. FASEB J. 2013;27:2451. doi: 10.1096/fj.12-226415. [DOI] [PubMed] [Google Scholar]
- [4].Kimura H. Proc. Jpn. Acad., Ser. B. 2015;91:131. doi: 10.2183/pjab.91.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ida T, Sawa T, Ihara H, Tsuchiya Y, Watanabe Y, Kumagai Y, Suematsu M, Motohashi H, Fujii S, Matsunaga T, Yamamoto M, Ono K, Devarie-Baez NO, Xian M, Fukuto JM, Akaike T. Proc. Natl. Acad. Sci. USA. 2014;111:7606. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].a) Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH. Sci. Signal. 2009;2:ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sen N, Paul BD, Gadalla MM, Mustafa AK, Sen T, Xu R, Kim S, Snyder SH. Mol. Cell. 2012;45:13. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhang D, Macinkovic I, Devarie-Baez NO, Pan J, Park C, Carroll KS, Filipovic MR, Xian M. Angew. Chem. Int. Ed. 2014;53:575. doi: 10.1002/anie.201305876. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2014;126:586. [Google Scholar]
- [7].a) Li X, Gao X, Shi W, Ma H. Chem. Rev. 2014;114:590. doi: 10.1021/cr300508p. [DOI] [PubMed] [Google Scholar]; b) Lin VS, Chen W, Xian M, Chang CJ. Chem. Soc. Rev. 2015;44:4596. doi: 10.1039/c4cs00298a. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhou X, Lee S, Xu Z, Yoon J. Chem. Rev. 2015;115:7944. doi: 10.1021/cr500567r. [DOI] [PubMed] [Google Scholar]
- [8].Liu C, Chen W, Shi W, Peng B, Zhao Y, Ma H, Xian M. J. Am. Chem. Soc. 2014;136:7257. doi: 10.1021/ja502968x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Zeng L, Chen S, Xia T, Hu W, Li C, Liu Z. Anal. Chem. 2015;87:3004. doi: 10.1021/acs.analchem.5b00172. [DOI] [PubMed] [Google Scholar]; b) Gao M, Yu F, Chen H, Chen L. Anal. Chem. 2015;87:3631. doi: 10.1021/ac5044237. [DOI] [PubMed] [Google Scholar]
- [10].Chen W, Rosser EW, Zhang D, Shi W, Li Y, Dong W, Ma H, Hu D, Xian M. Org. Lett. 2015;17:2776. doi: 10.1021/acs.orglett.5b01194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Hoffmann MR. Environ. Sci. Technol. 1977;11:61. [Google Scholar]; b) Schwarzenbach G, Fischer A. Helv. Chim. Acta. 1960;43:1365. [Google Scholar]
- [12].a) Wood JL. Methods Enzymol. 1987;143:25. doi: 10.1016/0076-6879(87)43009-7. [DOI] [PubMed] [Google Scholar]; b) Chen W, Liu C, Peng B, Zhao Y, Pacheco A, Xian M. Chem. Sci. 2013;4:2892. doi: 10.1039/C3SC50754H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Macmillan D. Angew. Chem. Int. Ed. 2006;45:7668. doi: 10.1002/anie.200602945. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2006;118:7830. [Google Scholar]
- [14].Shibuya N, Koike S, Tanaka M, Ishigami-Yuasa M, Kimura Y, Ogasawara Y, Fukui K, Nagahara N, Kimura H. Nat. Commun. 2013;4:1366. doi: 10.1038/ncomms2371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.