Abstract
Background
Specification of cranial bone and dermal fibroblast progenitors in the supraorbital arch mesenchyme is Wnt/β-catenin signaling-dependent. The mechanism underlying how these cells interpret instructive signaling cues and differentiate into these two lineages is unclear. Twist1 is a target of the Wnt/β-catenin signaling pathway and is expressed in cranial bone and dermal lineages.
Results
Here, we show that onset of Twist1 expression in the mouse cranial mesenchyme is dependent on ectodermal Wnts and mesenchymal β-catenin activity. Conditional deletion of Twist1 in the supraorbital arch mesenchyme leads to cranial bone agenesis and hypoplastic dermis, as well as craniofacial malformation of eyes and palate. Twist1 is preferentially required for cranial bone lineage commitment by maintaining Wnt responsiveness. In the conditional absence of Twist1, the cranial dermis fails to condense and expand apically leading to extensive cranial dermal hypoplasia with few and undifferentiated hair follicles.
Conclusions
Thus, Twist1, a target of canonical Wnt/β-catenin signaling, also functions to maintain Wnt responsiveness and is a key effector for cranial bone fate selection and dermal condensation.
Keywords: skull bone, skin, craniofacial development
Introduction
Cranial neural crest cells (CNC) and cranial mesoderm cells migrate from the dorsal head to the supraorbital arch region and contribute to the cranial bone and dermal lineages (Jiang et al., 2002; Yoshida et al., 2008). Genetic lineage tracing experiments show that CNC have a common progenitor for cranial dermal and bone progenitors (Jiang et al., 2002; Tran et al., 2010; Yoshida et al., 2008). The cranial bone and dermal progenitors expand apically and become the skull bones and dermis of the cranial skin (Goodnough et al., 2012; Jiang et al., 2002; Tran et al., 2010; Yoshida et al., 2008). In mouse cranial bone development, skeletogenic mesenchyme progenitors express transcription factors Msx2 and Dlx5 from 11.5–13.5. Upon specification and commitment to the cranial bone lineage they begin to express Runx2 at E12.5 and Osterix (Osx) at E13.5 (Karsenty, 2008). Concomitantly, dermal progenitors under the surface ectoderm express pan-mesenchyme marker PDGFRalpa at E11.5 and Twist2, IGF, and Lef1 from E11.5–14.5 in the upper dermis (Atit et al., 2006; Driskell et al., 2013; Goodnough et al., 2014). However, it remains unclear what signals and transcription factors guide “osteo-dermo” progenitors to become distinct lineages.
Ectodermal Wnts and mesenchymal β-catenin are required for cranial bone and cranial dermal fate selection in the supraorbital arch (Day et al., 2005; Goodnough et al., 2014; 2012; Hill et al., 2005; Z. Jiang et al., 2013; Tran et al., 2010). Because of their very early expression in mouse cranial bone and dermal progenitors, Twist1 and Twist2 transcription factors are strong candidates for regulating lineage selection (Barnes and Firulli, 2009; M. S. Lee et al., 1999; Rice et al., 2000; Soo et al., 2002; Vincentz et al., 2013). Twist1 is also dependent on Wnt/β-catenin activity (Franco et al., 2011; Goodnough et al., 2012; Howe et al., 2003; Klapholz-Brown et al., 2007; Reinhold et al., 2006). The genetic basis of congenital dermal and bone defects is associated with a loss of Wnt signaling function or its genetic target Twist1/2 (Barrott et al., 2011; Ghouzzi et al., 1997; Grzeschik et al., 2007; Petti et al., 2011; Qin et al., 2012; Rose et al., 1997; Tukel et al., 2010; Wang et al., 2007).
Based on spatiotemporal genetic deletions of Twist1 in the developing mouse head, a variety of functions have been attributed to Twist1 during craniofacial development. Twist1 is required for directing the migration of CNC cells into the first branchial arch and differentiation of first arch derivatives (Bildsoe et al., 2009; Chen and Behringer, 1995; Soo et al., 2002). Conditional deletion of Twist1 in the Wnt1Cre expressing premigratory cranial neural crest leads to compromised cell survival and diminished craniofacial development (Bildsoe et al., 2009; Chen et al., 2007). Deletion of Twist1 in the premigratory cranial mesoderm leads to malformations of cranial-mesoderm derived skeleton and the mutant cells acquire epithelial-like morphology (Bildsoe et al., 2013). Both these conditional Twist1 mutants have secondary exencephaly that may affect the placement of cranial bone and dermal progenitors. Twist1 is also an inhibitor of cartilage formation in vitro (Reinhold et al., 2006). Deletion of Twist1 in specified embryonic osteoblast and dermal progenitors in the En1Cre lineage by E12.5 leads to diminished mineralized skull bones in the apex of the head and ectopic cartilage fate in the posterior interparietal bones (Goodnough et al., 2012). Similarly, deletion of β-catenin in cranial mesenchyme leads to cranial bone agenesis or conversion to cartilage fate. Despite these studies, it is unknown if Twist1 is a key effector of Wnt/β-catenin signaling in direct induction of cranial bone and dermal lineage fate selection.
To define the primary role of Twist1 in cranial bone and dermal development, we conditionally delete Twist1, preceding lineage selection of osteoblasts and dermal fibroblasts. Unlike previous studies, we used Twist2Cre to conditionally delete Twist1 at a later stage in development and circumvent defects in early differentiation of CNC and mesoderm progenitors. Here, we show Twist1 acts upstream and downstream of Wnt /β-catenin pathway in the cranial mesenchyme during lineage selection events. By deleting Twist1 in post-migratory supraorbital arch cranial mesenchyme, we identified its new role in cranial bone lineage specification and commitment in vivo and in vitro. Twist1 is required later in the dermal lineage for condensation and apical expansion of cranial dermis. Together, these results demonstrate that a circuit between Twist1 and Wnt signaling pathway is required for cranial bone lineage selection and may have redundant partners in the cranial dermal lineage.
RESULTS
Onset of Twist1 expression in cranial mesenchyme is dependent on ectodermal Wnts and mesenchymal β-catenin
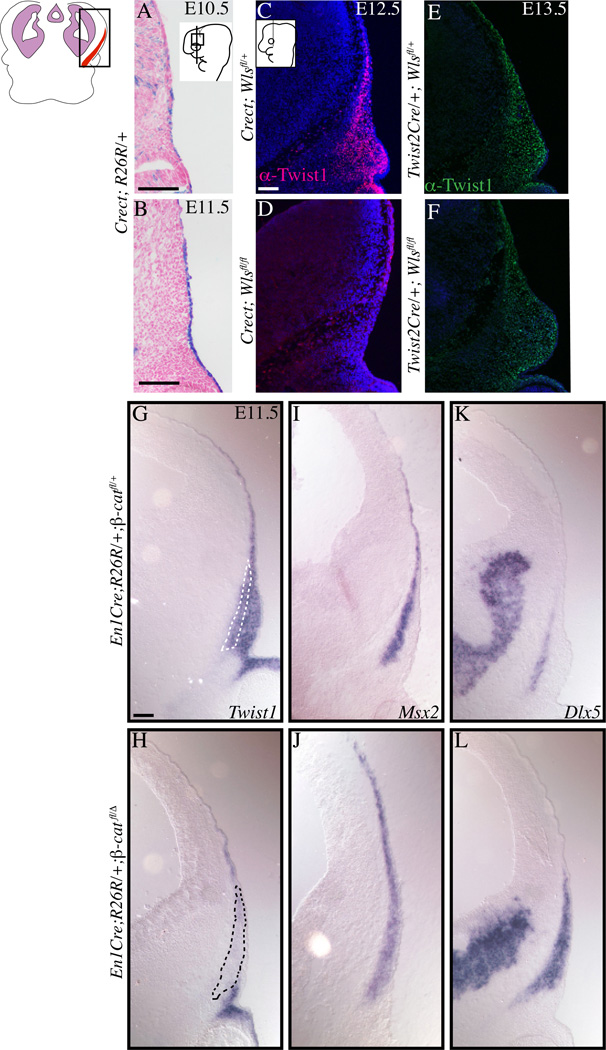
Given that Twist1 can be a target of the Wnt /β-catenin pathway and is one of the earliest transcription factor families expressed in cranial bone and dermal progenitors, we first examined the tissue source of Wnt ligands that are required for the onset of Twist1 expression. We previously demonstrated efficient deletion of Wntless (Wls) to eliminate Wnt ligand secretion in the early cranial ectoderm using Crect (Figure 1A,B) and in the supraorbital arch mesenchyme using Twist2Cre/Dermo1Cre (Figure 2B,D) between E10.5–12.5 (Carpenter et al., 2010; Goodnough et al., 2014; Reid et al., 2011; Tran et al., 2010; Yu, 2003). Twist1 protein was expressed in the cranial mesenchyme containing cranial bone and dermal progenitors at E12.5 in the Crect/+; Wlsfl/+ controls. However, Twist1 protein was absent in the cranial mesenchyme of Crect/+; Wlsfl/fl mutants (Figure 1C, D). Despite the efficient and similar timing of Wls deletion in the mesenchyme with Twist2Cre, Twist1 protein was still expressed in Twist2Cre/+; Wlsfl/fl mutants at E13.5 (Figure 1E, F). Twist1 protein expression was qualitatively diminished in the medial cranial mesenchyme of the Twist2Cre/+; Wlsfl/fl mutants, but quantification failed to achieve statistical significance (data not shown).
Figure 1. Ectoderm Wnts and mesenchymal β-catenin are required for Twist1 expression in cranial mesenchyme.
Coronal section of embryos stained with β-gal to show Crect lineage-marked cells in the supraorbital cranial ectoderm (A,B). Fluorescent immunohistochemistry of coronal sections showing expression of Twist1 protein in cranial bone and dermal progenitors in the conditional Wls mutants in the ectoderm (C,D) and mesenchyme (E,F). Twist1 protein expression is absent in the ectoderm Wls mutants, but not in the mesenchyme Wls mutants. In situ hybridization showing Twist1 mRNA expression requires mesenchymal β-catenin (G,H). Note that skeletogenic markers Msx2 and Dlx5 are not dependent on mesenchymal β-catenin (I–L). Control and mutants images were obtained at the same magnification. Scale bar=100um.
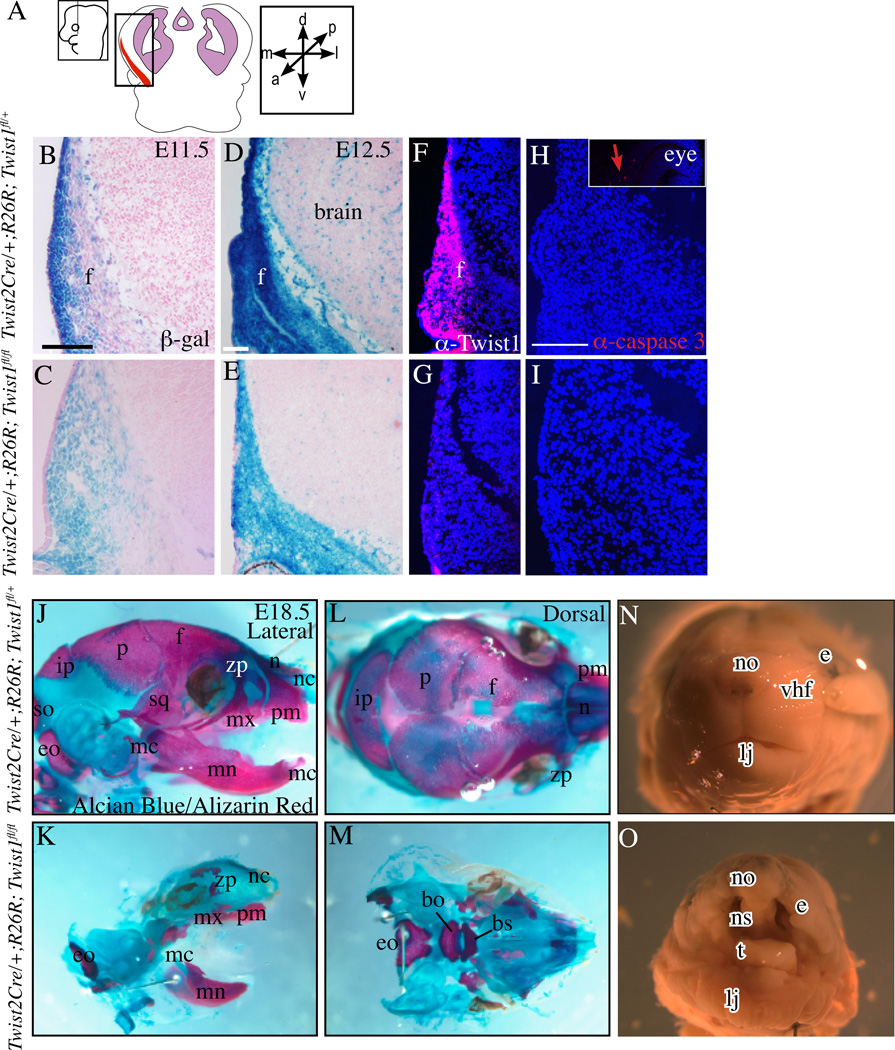
Figure 2. Absence of Twist1 in cranial mesenchyme leads to absence of skull bone.
Coronal section of embryos in the indicated plane and area of interest (A) stained with β-gal to show Twist2Cre lineage-marked cells in the cranial mesenchyme (B–E). Fluorescent immunohistochemistry showing expression of Twist1 and activated caspase3 protein in cranial bone and dermal progenitors in control embryos and conditional mutants at E12.5 (F–I). Positive control for caspase3 staining in the optic nerve (Inset in H). Lateral (J,K) and dorsal (L,M) views of whole-mount skeletal preps. Absence of mineralized frontal bone (f), parietal bone (p), interparietal bone (ip), squamous bone (sq), supraoccipital (so), nasal bones(n), zygomatic process(zp) in the mutants(J–M). Note the presence of smaller but mineralized exoccipital (eo), mandible (mn), maxilla (mx), and premaxilla (pm) bones in the mutants (J–M). The Meckel’s cartilage (mc) and nasal capsule (nc) are present (F–I). Morphology of E18.5 control and mutant embryos showing a shortened lower jaw (lj) with a protruding tongue (t) and the presence of a cleft palate below the nasal septum (ns) in the mutant (N,O). Other abbreviations: bo:basioccipital bone, bs:basisphenoid bone, e:eye, no:nose, vhf:vibrissae hair follicle. Scale bar =100um, control and mutant skull preps were imaged at the same magnification.
Twist1 protein expression lags behind the mRNA expression (Gitelman, 1997; Goodnough et al., 2012); therefore, we examined if cranial mesenchyme β-catenin activity was required for Twist1 mRNA expression. We conditionally deleted β-catenin in the supraorbital cranial mesenchyme by E11.5 using Engrailed1Cre(En1Cre) (Brault et al., 2001; Kimmel et al., 2000). Compared to controls, Twist1 mRNA was absent in En1Cre/+;RR; β-cateninfl/Δ mutant cells (Figure1G, H). The loss of Twist1 mRNA expression was consistent with our previous study on protein expression (Goodnough et al., 2012). However, the cranial mesenchyme in controls and β-catenin mutants expressed skeletogenic identity markers, Msx2 and Dlx5 (Figure 1I–L). Domains of Dlx5 and Msx2 expression were expanded, revealing acquisition of ectopic skeletogenic identity in the supraorbital arch mesenchyme in these mutants (Goodnough et al., 2012). These results together demonstrated that ectoderm Wnts and mesenchyme Wnt/β-catenin signaling are required for Twist1 mRNA expression in the cranial mesenchyme.
Twist1 is required for skull bone and cranial dermis formation
We tested the direct role of Twist1 as a key effector of lineage selection by conditionally deleting it in CNC-derived ectomesenchyme and head mesoderm (Figure 2A) preceding lineage selection. Twist2Cre recombined R26R-LacZ efficiently between E10.5–E11.5 in the cranial dermal mesenchyme and in the cranial osteoprogenitors between E11.5– E12.5 (Tran et al., 2010) (Figure 2B–D). Twist2Cre;R26R lineage-marked cells expressing β-galactosidase were present in the cranial mesenchyme of Twist1fl/+ controls and in conditional Twist1fl/fl mutants at E11.5 and E12.5 (Figure 2B–E). Twist1 deletion was complete in the cranial mesenchyme as confirmed by the absence of Twist1 protein expression in Twist2Cre; R26R; Twist1fl/fl mutants (Figure 2 F,G). We did not find notable differences in programmed cell death in the cranial mesenchyme at E12.5 (Figure 2H,I). We obtained mutant embryos between E16.5–18.5 in predicted Mendelian ratios indicating absence of early embryonic lethality of mutants. We examined the effect of Twist1 deletion on craniofacial development and head morphology. Whole-mount skeletal preps of E18.5 embryos stained with Alician blue and Alizarin Red showed preferential absence of mineralization in all bones of the skull vault and the nasal bones (Figure 2J–M). Twist2Cre lineage-descendants contribute to the maxilla(mx) and mandible(mn) bones in the jaw (Tran et al., 2010). Twist2 Cre; Twist1fl/fl mutants had smaller jaws, but ossified jaw bones. The mutants had a cleft palate as seen in whole mounts and in sections (Figures 2J,K, 3G,H).
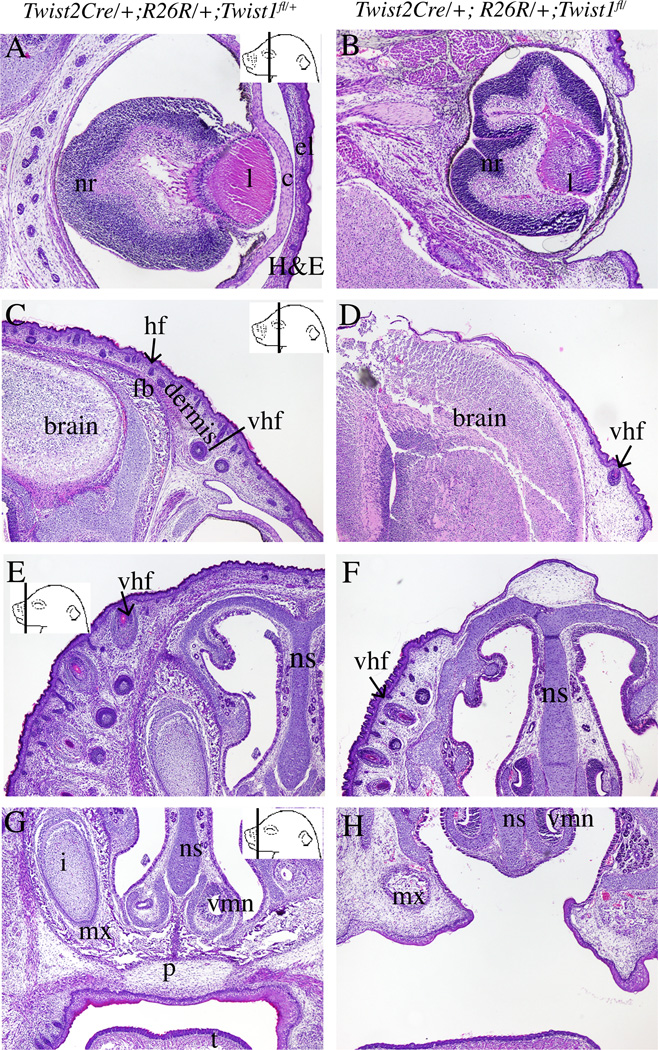
Figure 3. Craniofacial phenotypes in conditional Twist1 mutants.
Histology of coronal sections of E18.5 embryos stained with H&E (A–H). In the cranial region, the mutants lack a cornea(c) and eye lid (el) (A,B), and the frontal bone (C,D). The mutants have hypoplastic dermis (der) with fewer and less developed vibrissae and guard hair follicle primordia (hf) in the cranial and facial areas (C–F). The mutants have a cleft palate (p) compared to controls (G,H). Other abbreviations: i=incisor, l:lens, nr: neural layer of the retina, mx: maxilla, t: tongue, vmn:vomeronasal organ, vhf:vibrissae hair follicle. Control and mutant pairs of panels are displayed at the same magnification.
Histological analysis shows the loss of other craniofacial tissues. At E18.5, eye lid skin covers the eye in the controls and was absent in the mutants (Figure 3A,B). The cornea was also absent in the mutant (Figure 3A,B). H&E stained sections also confirmed the absence of the frontal bone (Figure 3C,D). In the cranial and facial regions, conditional Twist1 mutants had dermal hypoplasia with the absence of cranial dermis at the apex of the head (Figure 3C–F).
Overall, the conditional deletion of Twist1 in the Twist2Cre lineage affected the formation skull bones, nasal bone, craniofacial dermis, eye, and palate. The jaw bones were less affected.
Twist1 is required for condensation and expansion of cranial dermis
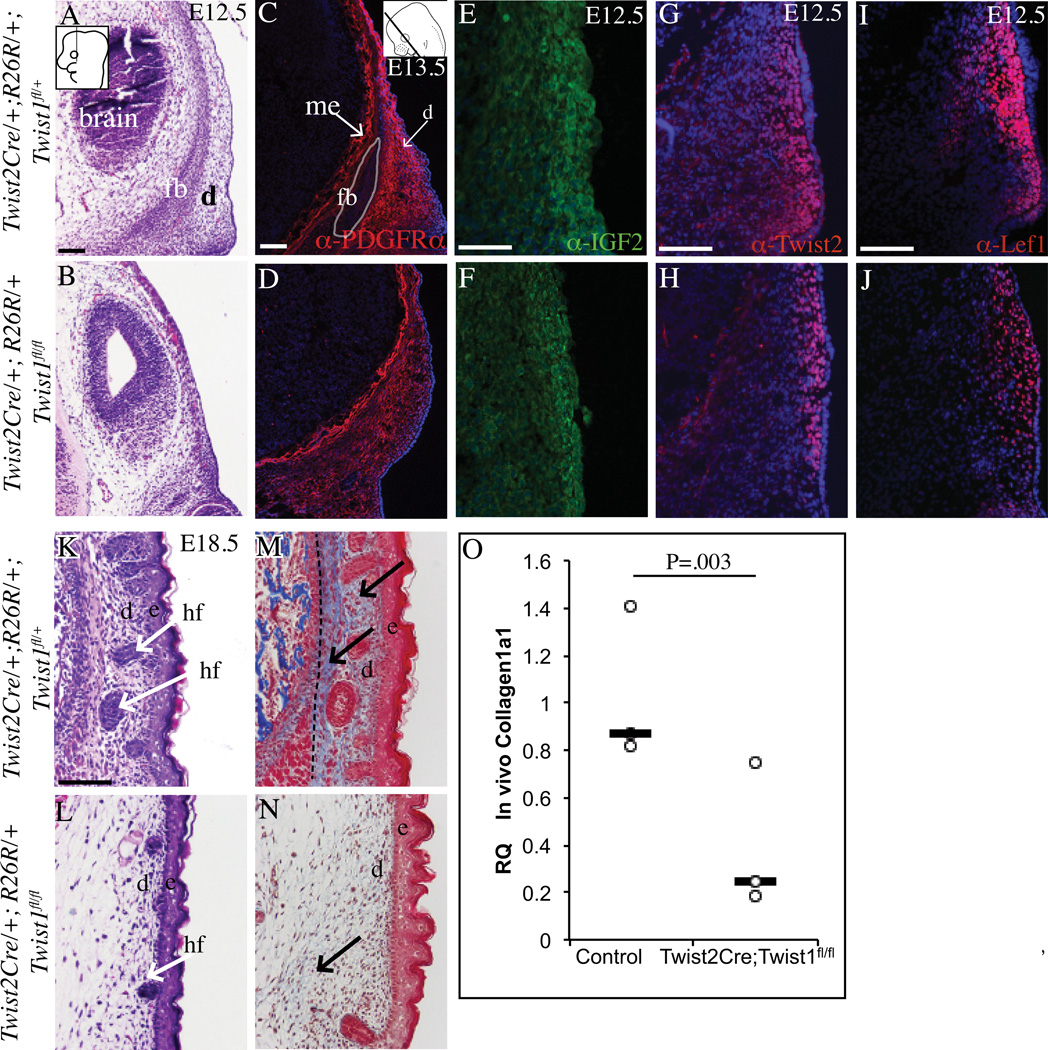
Twist1 conditional mutants had cranial and facial dermal hypoplasia and lacked apical expansion of the dermis, suggesting a primary defect in dermal development. Therefore, we examined the specification and differentiation of the dermal lineage. H&E staining confirmed the presence of cells in the dermal domain beneath the surface ectoderm at E12.5 in the supraorbital arch region (Figure 4A,B). In E13.5 controls, fibroblast precursor marker, platelet-derived growth factor receptor-α (PDGFRα) was expressed in the dermal domain and more medially in the meningeal progenitors, but diminished in the frontal bone primordia (Driskell et al., 2013) (Figure 4C). The frontal bone primordia does not develop properly and PDGFRα expression was seen broadly in the cranial mesenchyme of mutant embryos (Figure 4D). Insulin Growth factor 2(Igf2), Twist2/Dermo1, and Lef1 are some of the earliest markers of condensed upper dermal fibroblasts that support hair follicle formation (Atit et al., 2006; Goodnough et al., 2014). IGF2, Twist2, and Lef1 proteins were expressed in controls and mutants at E12.5 (Figure 4E, F), but mutants had smaller domains with loosely distributed cells (Figure 4G–J). Changes in dermal progenitor marker expression also occurred without changes in cell survival by E12.5 (Figure 2H, I) and cell proliferation. At E12.5, dermal progenitors also had comparable proliferation index to controls (34%±12.71% in controls versus 47%±10.00% SD in mutants, n=5). H&E staining of E18.5 skin revealed dense dermis with stage 2–4 hair follicles in controls (Figure 4K) (Paus et al., 1999). In the conditional Twist1 mutants, the dermis failed to condense and hair follicles were arrested in stage 1 (Figure 4L). H&E and Masson’s trichrome stain of E18.5 skin revealed thin collagen fibrils in the mutants compared to the dense fibrils in the controls (Figure 4K–N). Relative to control, Collagen1a1 mRNA quantity was also lower in conditional Twist1 mutant cranial mesenchyme and ectoderm by qRT-PCR (n=3, pooled from 2–4 embryos) (Figure 4O). The marker analysis suggests that the supraorbital region has fibroblast precursor identity and the dermal lineage was specified in the mutants. The upper dermal fibroblasts primarily failed to condense and differentiate sufficiently, thereby leading to craniofacial dermal hypoplasia and diminished hair follicle differentiation.
Figure 4. Absence of Twist1 in the cranial mesenchyme leads to cranial dermal hypoplasia.
Coronal sections of embryos stained with H&E (A,B, K, L) showing progressive dermal hypoplasia and stunted hair follicle primordia (hf) in the mutant compared to the controls.. Fluorescence immunohistochemistry with DAPI-stained nuclei (C–J) marking dermal progenitors and condensed dermis. Collagen deposition visualized by Masson’s Trichrome staining (blue) is markedly diminished in the upper and lower dermis of the mutants compared to the control (M, N). Relative quantity of Collagen1a1 mRNA in E12.5 control and conditional Twist1 mutant cranial mesenchyme and ectoderm (O) (n=3). Black hatched line demarcates the dermis from the muscle (M). Other abbreviations: fb:frontal bone primordia, d: dermal progenitors, hf: guard hair follicle. Scale bars =100um.
Twist1 is required for specification of cranial skull bone progenitors in the supraorbital mesenchyme
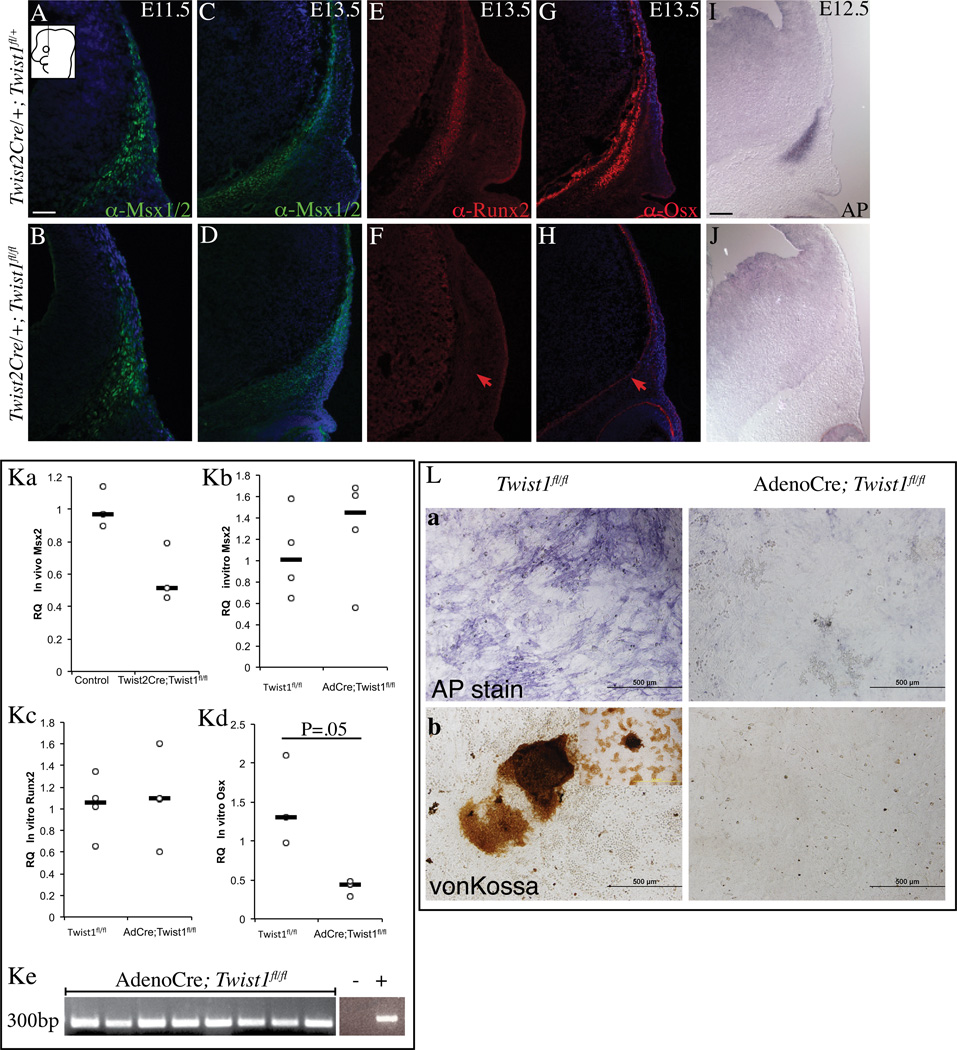
The complete agenesis of the skull vault suggests defects in the primordia of skull bones. Next, we examined if the patterning and specification events in the frontal bone primordia occurred without Twist1 in vivo. During lineage selection at E11.5, cells with skeletogenic identity expressed Msx2 protein, a target of the BMP pathway in craniofacial mesenchyme (Bonilla-Claudio et al., 2012; Vainio et al., 1993). Control and Twist1 mutants expressed Msx1/2 protein at E11.5 and E13.5 in the cranial mesenchyme (Figure 5A–D). Relative to control, quantity of Msx2 mRNA was modestly reduced in conditional Twist1 mutant cranial mesenchyme in vivo (n=3, pooled from 2–4 embryos) (Figure 5Ka). Early cranial bone lineage marker, Runx2 protein was expressed in the cranial mesenchyme of control E13.5 embryos and absent in the Twist1 mutant embryos (Figure 5E,F). Cranial bone progenitors in controls were committed and differentiated to the bone lineage by expressing Osx and Alkaline phosphatase (AP). In contrast, Twist1 mutant cranial mesenchyme lacked Osx protein and AP expression (Figure 5G–J). Importantly, notable differences in cell proliferation and cell death of the Twist1 mutant cranial mesenchyme did not precede changes in expression of marker genes (Figure 2 H,I). The proliferation index of Twist1 mutant cranial mesenchyme at E11.5 was comparable to controls (45%±13% in controls versus 50%±6.61% in mutants, n=4). Proliferation index of the cranial bone domain at E12.5 was 37% ± 8.17% SD in controls and were 32% ± 12.30% in Twist1 mutants (n=5). These results demonstrated that Twist1 mutant cranial mesenchyme preserved skeletogenic identity but specification of the cranial bone lineage failed to occur.
Figure 5. Deletion of Twist1 in cranial mesenchyme leads to loss of cranial bone lineage commitment in vivo.
Indirect immunofluorescence with DAPI-stained (blue) nuclei (A–H) and alkaline phosphatase (AP) staining (I, J) was performed on coronal mouse embryonic head sections at E13.5 or as indicated. Skeletogenic marker Msx1/2 (A–D) is present and markers of bone lineage commitment are absent in the mutant (E–J). Red arrows indicate changes in marker expression. Relative quantity of Msx2 in E12.5 control and conditional Twist1 mutant cranial mesenchyme and ectoderm (Ka). Relative quantity of osteoblast differentiation genes in primary calvarial osteoblast from control and Adeno-Cre infected Twist1fl/fl cultures at day 12 (Kb, Kc, Kd). Confirmation of Adeno-Cre mediate Twist1fl/fl allele recombination by PCR (Ke). Compared to control, Adeno-Cre infected Twist1fl/fl mutant cultures fail to differentiate and have few cells with alkaline phosphatase (AP) expression (La) with few Von-Kossa positive colonies (Lb) at day18 (L). Control and mutant pairs of panels are displayed at the same magnification. Scale bar =100microns (A–J) and 500microns (L).
We also tested whether Twist1 mediated calvarial osteoblast differentiation occurred in a cell-autonomous manner. Calvarial mesenchyme progenitor cells can faithfully recapitulate intramembranous ossification in culture under proosteogenic conditions (Hill et al., 2005; Marvaso and Bernard, 1977) (Figure 5L). We deleted Twist1 by using Adeno-Cre virus (AdCre) in primary calvarial osteoblast cultures from Twist1fl/fl embryos and deletion was confirmed by PCR (Figure 5Ke) (Chen et al., 2007; Hill et al., 2005). We found comparable expression of Msx2 and Runx2 in control and AdCre; Twist1fl/fl mutant cells (Figure 5Kb, 5Kc). Relative to control cultures, AdCre; Twist1fl/fl mutant cells had significantly lower expression of Osx (Figure 5Kd). After 18 days in osteogenic media, Twist1fl/fl control cultures expressed alkaline phosphatase and AdCre infected Twist1fl/fl cultures had substantially less differentiation (Figure 5La). Concomitantly, control Twist1fl/fl cultures formed large bone nodules that were dark brown by Von Kossa staining (75± 15 colonies, n=3) and AdCre infected Twist1fl/fl cultures had very few bone nodules (5 colonies ±6.5, n=3)(Figure 5Lb). Together these results demonstrated that Twist1 was cell-autonomously required for primary induction of cranial bone fate and differentiation in cells with skeletogenic identity.
Twist1 is required to sustain Wnt signaling responsiveness in the cranial bone progenitors
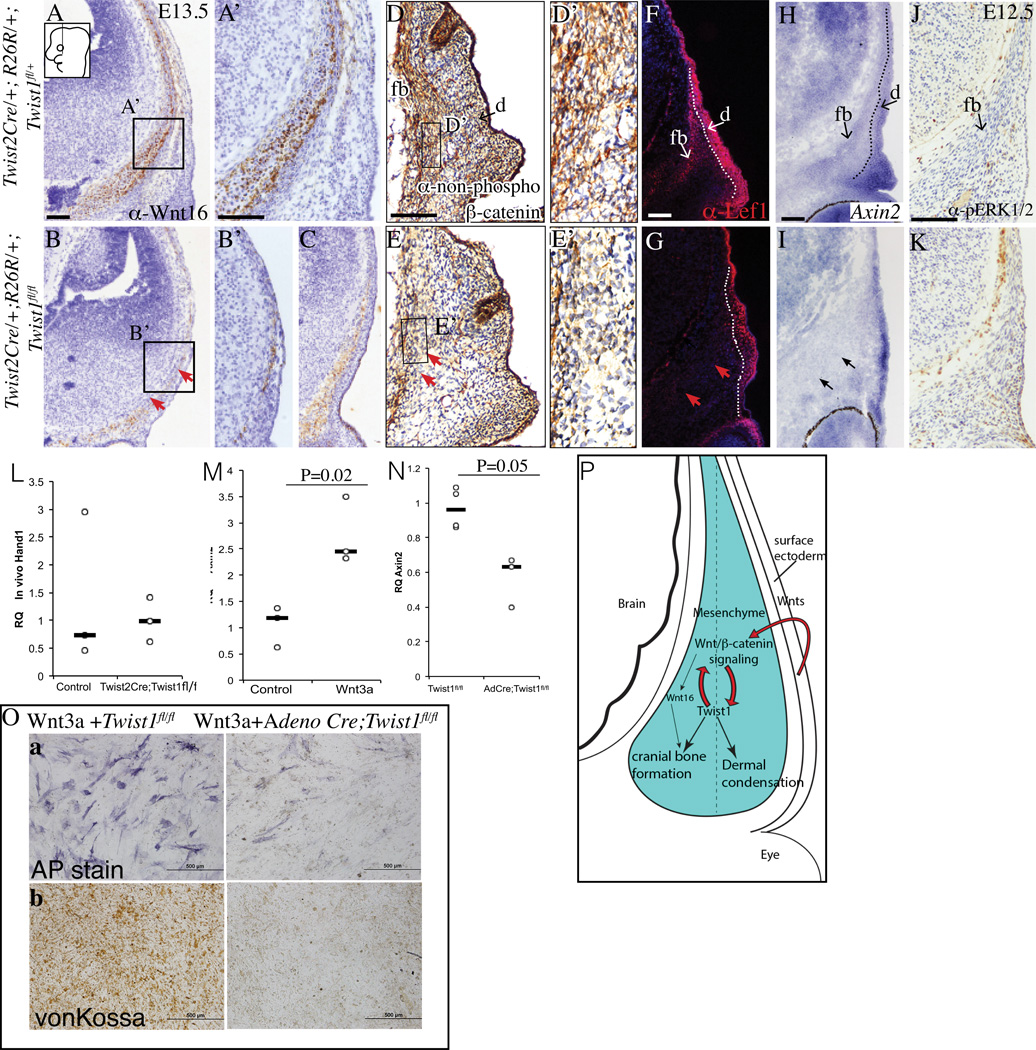
Wnt/β-catenin signaling pathway is known to be critical for cranial bone lineage commitment and dermal lineage selection in vivo (Day et al., 2005; Goodnough et al., 2012; Hill et al., 2005; Tran et al., 2010). Ectoderm Wnts and mesenchyme Wnt responsiveness are critical for lineage selection in the supraorbital region (Goodnough et al., 2014). Therefore, we tested if the conditional Twist1 mutants have altered expression of Wnt signaling pathway components and transduction in vivo. Wnt16, a canonical Wnt signaling pathway ligand, is expressed in cranial bone progenitors during cranial bone lineage commitment and is involved in intramembranous ossification of skull bones (Goodnough et al., 2014; Z. Jiang et al., 2013). Wnt16 protein was expressed in the osteoid of the developing frontal bone in the controls at E13.5 and was either diminished or retarded in Twist1 mutants (Figure 6A–C, A’, B’). Canonical Wnt signaling transduction was visualized by nuclear β-catenin and Lef1 protein and Axin2 mRNA expression in control cranial bone progenitors located medially at E13.5 (Figure 6D, D’, F, H). In contrast, nuclear β-catenin, Lef1, and Axin2 expression were markedly absent in the cranial bone progenitors of Twist1 mutants (Figure 6E, E’, G, I). Compared to the controls, the dermal domains of Lef1 protein and Axin2 mRNA were diminished in size in Twist1 mutants (Figure 6E, G). Twist1 can modulate FGF and BMP signaling in cranial suture formation (Connerney et al., 2008; Kim et al., 1998; Rice et al., 2000). We examined activation of FGF signaling in cranial bone progenitors by pERK1/2 immunohistochemistry (Figure 6J, K). We failed to find engagement of the FGF pathway in control cranial bone progenitors and in Twist1 mutants (Figure 6J,K) at E13.5. Similar to Msx2, expression of another BMP signaling target gene in craniofacial mesenchyme, Hand1 was also not significantly altered between control and Twist1 mutants in vivo (Figures 5Ka, 6L)(Bonilla-Claudio et al., 2012; Vainio et al., 1993).
Figure 6. Wnt responsiveness in skull bone progenitors requires expression of Twist1.
Immunostaining analysis of Wnt16 ligand expression (A–C, A’, B’), non-phospho-(active)-β-catenin (D,E, D’ E’), and Lef1 (F,G), in situ hybridization of Axin2 (H, I) on coronal sections showing diminished expression of Wnt16 and loss of canonical Wnt signal transduction in the frontal bone domain of Twist1 mutants (arrows in B,G,I) compared to controls. Immunostaining of pERK1/2 showing absence of FGF signaling in control and conditional mutants (J,K). Relative quantity of Hand1 in vivo in E12.5 cranial mesenchyme and ectoderm was comparable in control and mutants (n=3, L). Stimulation of Wnt signaling response by target gene Axin2 mRNA expression in wild-type primary calvarial osteoblasts after 24 hrs exposure to Wnt3a (M). Diminished Axin2 mRNA in AdCre; Twist1fl/fl primary calvarial osteoblasts culture compared to control. (N) (n=4 controls and 3 mutants). Alkaline Phosphatase (AP) and Von Kossa staining of primary calvarial mesenchyme cultures in osteogenic conditions with exogenous Wnt3a conditioned media showing arrest of differentiation in mutant cultures (O). Diminished expression of Wnt16 protein expression as seen in two different mutant embryos (B, C). Abbreviation: d:dermal progenitors, e: epidermis, fb:frontal bone primordia. Scale bar=100um.
Finally, we investigated effects of exogenous Wnt3a treatment in the calvarial osteoblast cultures. In wild-type calvarial osteoblasts in culture, exogenous Wnt3a treatment lead to induction of Wnt signaling as measured by Axin2 mRNA expression (Figure 6M). Compared to control, AdCre;Twist1fl/fl calvarial osteoblasts had diminished Wnt signaling responsiveness. (Figure 6N). In addition, exogenous Wnt3a failed to rescue the differentiation defect in AdCre;Twist1fl/fl calvarial osteoblast cultures (Figure 6O). Thus, Twist1 mediated Wnt-responsiveness was required cranial bone differentiation. Consistent with the diminished ossification after stabilization of β-catenin in vivo, prolonged Wnt activation in vitro also led to diminished mineralization in control calvarial osteoblast cultures (Figure 6Ob) (Day et al., 2005; Goodnough et al., 2012; Hill et al., 2005). In the control cultures without Wnt3a, osteoblast progenitors differentiated consistently and had numerous von Kossa positive large bone nodules (Figure 5Lb).
Taken together, our data support a new model in which Wnt signaling and responsiveness act upstream and downstream of Twist1 to regulate the progression of cranial bone differentiation and dermal condensation during embryonic development (Figure 6P).
Discussion
Wnt/β-catenin signaling acts as a molecular switch in the cranial mesenchyme while simultaneously effecting specification of cranial bone and dermal lineages and repressing cartilage cell fate (Day et al., 2005; Goodnough et al., 2012; Hill et al., 2005; Tran et al., 2010). Twist1 is a target of the Wnt signaling pathway during lineage selection in the cranial mesenchyme. Here, we show distinct tissue sources of Wnt ligands and Wnt signal transduction for the expression of Twist1. Unlike previous studies, we delete Twist1 in the post-migratory supraorbital arch mesenchyme and identify a preferential requirement of Twist1 in cranial bone lineage specification (Bildsoe et al., 2013; 2009; Chen et al., 2007). Twist1 is also required later in the craniofacial dermal lineage for condensation and apical expansion of the developing cranial dermis in vivo.
In this study, we identify a circuit where Wnt/β-catenin signaling is upstream and downstream of Twist1, and the circuit is required for cranial bone formation (Figure 6P). Recent ChIP-seq studies suggest Twist1 may regulate the Wnt signaling pathway components. Twist1 occupancy of DNA is associated with Wnt signaling pathway components in a tissue-specific manner (Lee et al., 2014; Vrljicak et al., 2012). Genes associated with the Wnt signaling pathway are also down-regulated in the Twist1 mutant cranial mesenchyme and in the Twist1 null artrioventricular canal (AVC) in the heart (Figure 6, Vrljicak et al., 2012). Consistently in our conditional Twist1 mutants, we find diminished Wnt responsiveness by non-phospho (active) β-catenin staining and expression of Wnt pathway components in the cranial mesenchyme such as Wnt16 ligand, Lef1, and Axin2 (Goodnough et al., 2014). In addition, our in vitro results show that Twist1-mediated Wnt responsiveness is required for calvarial osteoblast differentiation (Figure 6O). Therefore, these results suggest that Twist1 may regulate the Wnt signaling pathway at the level of Wnt ligands and/or the cellular Wnt responsiveness. Twist1 is also known to interact with FGF and BMP signaling in suture mesenchyme and endochondral bone formation, and haploinsufficiency of Twist1 leads to ectopic activation of these signaling pathways in suture mesenchyme and human adipose stem cells (Connerney et al., 2008; Kim et al., 1998; Morriss-Kay and Wilkie, 2005; Quarto et al., 2015; Rice et al., 2000). However, we do not observe changes in BMP signaling and FGF signaling in the Twist1 mutant frontal bone primordia between E12.5–E13.5 (Figures 4, 6). Defining the expression and role of additional Wnt ligands in tissue-restricted mutants will be the focus of future studies. Identifying the regulation of the relevant Wnt ligands by Twist1 will clarify how it regulates the Wnt signaling pathway during cranial bone differentiation.
Our functional analysis in vivo and in vitro demonstrates that Twist1 mutants have a cell-autonomous defect in committing to the skull bone lineage. Along with Twist1 regulating the Wnt pathway, our current study suggests that Twist1 may directly or indirectly transactivate key transcription factors in cranial bone formation. Twist1-ChIP-seq analysis of expressed transcripts in E10.5 mouse limb bud reveal DNA occupancy in the region associated with Runx2, suggesting transactivation by Twist1 (Lee et al., 2014). Our results are also consistent with studies on postnatal human calvarial osteoblasts in which deletion of the bHLH domain of the Twist1 gene lead to reduced Runx2 expression (Yousfi et al., 2002). Other studies in mouse development show a transient interaction between Twist1 protein and Runx2, leading to decreased binding at the osteocalcin promoter and diminished differentiation (Bialek et al., 2004). Future studies with Twist1 Chip-seq on embryonic cranial mesenchyme will reveal if Twist1 directly binds Runx2 and Osx loci. Collectively, these findings will demonstrate if Twist1 is the transcriptional regulator of cranial bone progenitor specification, thereby broadening our understanding of the factors that are important for tissue engineering skull bones.
Absence of Twist1 can lead to fate conversion and loss of mesenchyme identity (Bildsoe et al., 2013; Goodnough et al., 2012; Vincentz et al., 2013). Early deletion of Twist1 in mesoderm soon after gastrulation using the Mesp1Cre line show mutant cells clustered on either side of the open neural tube, and they acquired epithelial cell characteristics. In our study, the cranial mesenchyme in Twist1 mutants express markers of skeletogenic precursors and markers of dermal progenitors. The cranial mesenchyme derived from cranial neural crest and mesoderm fail to commit to cranial bone identity and remained Msx1/2+ and PDGRα+, and do not express markers of ectopic cell fates such as muscle or cartilage (data not shown). Compared to previous studies, timing of Twist1 deletion is later in our studies suggesting that maintenance of mesenchyme identity no longer requires Twist1. In contrast to the absence of cranial bone lineage commitment, Twist1 mutants express markers of cranial dermal fibroblasts lineage and canonical Wnt signaling in a smaller domain under the ectoderm. It is likely that redundant partners in the dermal domain are compensating for Twist1. For instance, the closely related protein Twist2/Dermo1 is expressed in cranial dermal progenitors, and may function redundantly with Twist1. Identifying the genetic program for dermal lineage identity is the subject of ongoing studies.
Twist1 mutant craniofacial dermis fails to condense and expand apically leading to dermal hypoplasia and stunted hair follicle development. Studies of Twist1 and its downstream target genes in different cell types have established roles in cell adhesion, migration, and ECM protein expression (Barnes and Firulli, 2009; Lee et al., 2014; Shelton and Yutzey, 2008; Vrljicak et al., 2012). Twist1 ChIP-seq on E10.5 mouse heart tissue reveal direct binding of Twist1 to Collagen1a1 and Collagen3a1 loci that are also expressed by fetal dermal fibroblasts (Collins et al., 2011; Kalluri and Zeisberg, 2006; Vrljicak et al., 2012). We find thinner collagen fibers and significantly less Collagen1a1 expression in the developing skin that may affect dermal condensation in the Twist1 mutant. The cellular mechanism and role of extracellular matrix (ECM) proteins underlying apical expansion of cranial dermis remain unexplored.
Defects in lineage selection and commitment may contribute to human congenital defects of the cranial dermis and skull bone as well as adjacent tissues. Fundamental knowledge generated from our study will be relevant to understanding human congenital defects in bone and dermis of known and unknown etiologies. In summary, our studies have provided key insights into how the Wnt signaling pathway and Twist1 transcription factor instruct the complex patterning of the cranial bones and dermis and bring us closer to a set of factors that together can regulate cell fate and condensation in a tissue engineering setting.
Experimental Procedures
Mice and Genotyping
Cre lines for the conditional functional studies include Engrailed1Cre (Kimmel et al., 2000), Twist2Cre/Dermo1Cre mice (Yu, 2003), and Crect(Reid et al., 2011) mice that were maintained on mixed genetic background. The timing of these Cre lines based on R26R LacZ recombination have also been previously described in supplementary figures (Goodnough et al., 2014; 2012; Tran et al., 2010). The β-catenin conditional loss-of-function allele β-cateninflox, the β-catenin null allele (β-cateninnull), and the Wntlessflox allele have been described previously (Brault et al., 2001; Carpenter et al., 2010; Haegel et al., 1995). Mice containing the conditional null Twist1 allele were obtained from MMRRC on C57BL/6 background (Chen et al., 2007) and crossed to R26R/R26R on CD1 background (Soriano, 1999). Twist2Cre/+;R26R/+; Twist1fl/+ males were mated with R26R/R26R;Twist1fl/fl females to generate conditional Twist1 mutant embryos of mixed genetic background. Mice and embryos were genotyped as described previously. Mice were time-mated, and the vaginal plug day was assigned as E0.5. At desired time points, embryos were harvested and processed for frozen or paraffin sections as previously described (Atit et al., 2006). For each experiment, a minimum of three mutants with litter-matched controls was studied. At least two to four litters were used for each functional analysis. Case Western Reserve Institutional Animal Care and Use Committee approved all animal procedures.
In situ hybridization, immunohistochemistry, and histology
Embryos were fixed in 4%PFA, cryopreserved or paraffin embedded, and sectioned at 8–12 um as previously described (Atit et al., 2006; Ohtola et al., 2008). In situ hybridization, β-galactosidase with eosin counter-staining, alkaline phosphatase staining, and immunohistochemistry were performed as described previously (Atit et al., 2006; Ohtola et al., 2008). In situ probes for Axin2(Addgene), Dlx5 (Liu et al., 1997) and Msx2 were generous gifts from Gail Martin, and Twist1 was shared by Richard R. Behringer. Primary antibodies for Msx1/2 (1:20, DSHB 4G1), Twist1 (1:500, Santa Cruz sc81417), Twist2 (1:200, Abcam66031), BrdU(1:25, Roche 1170376), non-phospho-β-Catenin (1:250, Cell Signaling D13A1), Myogenin (1:20, DSHB, 5FD), and Wnt16 (1:100, Santa Cruz, sc20268) proteins were used for immunofluorescence and brightfield immunohistochemistry. Primary antibodies against Runx2, Osx, activated Caspase3, TUNEL, IGF2, Sox9, and Lef1 were used as previously reported (Goodnough et al., 2014; 2012). Species-specific fluorescent and biotinylated secondary antibodies were used. Data from in situ hybridization were photographed using DIC/Nomarski Optics on the Zeiss Axioplan microscope with the SPOT camera system and software. Fluorescence and brightfield images were taken using an Olympus BX60 microscope and Olympus DP70 camera with CellSens Entry software. Images were processed and merged using Adobe Photoshop and InDesign.
Quantification of cell proliferation
Pregnant dams carrying E11.5 and E12.5 embryos were injected IP with 2.5mg of BrdU in PBS for a 90-minute pulse before dissection. Embryos were processed for paraffin sectioning and immunohistochemistry for BrdU as previously described (Ohtola et al., 2008). At E11.5, numbers of BrdU+ nuclei in the cranial mesenchyme between the ectoderm and brain were identified in images of 4–6 sections 50microns apart per sample (n=3). At E12.5, numbers of BrdU+ nuclei in a fixed area in the dermal and cranial bone domains were analyzed independently (n=4). Images were processed and analyzed in Adobe Photoshop CS6. Student’s t-test was used to determine statistical significance.
Alcian blue, Alizarin red and Mason’s trichrome staining
Staining of E18.5 wholemount skull preps was performed as previously reported (McLeod, 1980). Masson’s Trichrome staining on sections and Alizarin Red staining of cultures was done following standard protocols.
Primary cultures and adenovirus transfer
For primary osteoblast cultures, calvariae from E15.5 embryos were isolated from Twist1fl/fl mice and dedifferentiated in culture as previously described (Day et al., 2005; Hill et al., 2005). Briefly, calvariae were isolated and subjected to two sequential digests in 0.25% trypsin/EDTA for 5 min and 2% Liberase (Roche) in DMEM-F12 (Invitrogen) for 10 min at 37°C. Cells were grown in growth medium(BJGB media containing 10% FBS, 1x Antibiotics and Antimycotics) for two days. Osteoblast progenitors were infected with adenovirus Cre (AdCre) (250 MOI, University of Iowa, Virus Core) for 2 hours in 300ul of DMEM at 37°C. After two days, AdCre recombination was verified by PCR as previously described (Y.-T. Chen et al., 2007). Dedifferentiated calvarial mesenchyme progenitors were differentiated for 18 days in culture (growth medium supplemented with 5mM β-glycerophosphate and 100 ug/ml ascorbic acid) (Sigma-Aldrich). Cultures were stained for alkaline phosphatase and Von Kossa as previously described (Day et al., 2005; Hill et al., 2005).
RNA extraction, and qRT-PCR analysis
Total RNA was extracted from in vivo E12.5 control and Twist1 conditional mutant cranial mesenchyme and ectoderm (pooled 2–4 embryos, n=3) and E15.5 primary calavarial osteoblasts and processed for qRT-PCR analysis with 4ng of cDNA as previously described (Hamburg-Shields et al., 2015). Axin2 (MM00443610_m1), Collagen1a1(MM00801666_g1), Msx2 (MM00442992_m1) mRNA quantities were measured relative to Actb, using Taqman primers (Life Technologies). Runx2, Osterix, and Hand1 mRNA quantities were measured relative to GAPDH using SYBR mix (Applied Biosciences) as previously described (Bonilla-Claudio et al., 2012; Jiang et al., 2013). The sense and antisense primers used were as follows: mouse Runx2 sense 5‘CCGGGAATGATGAGAACTA 3’ and antisense 5’ ACCGTCCACTGTCACTTT 3’; mouse Osterix sense 5’ ACTCGTTGCCATCTAATCTTTCTG 3’ and antisense 5’ GGGAATCGAGGCGACAGC 3’; mouse Hand1 sense 5‘AGAGGAGACGCACAGAGAGCA 3’ and antisense 5’ TTGATCTTGGAGAGCTTGGTG 3’. Relative Msx2, Runx2, Osx mRNA quantity was measured on day 12 in E15.5 control and AdCre infected primary calvarial osteoblasts in culture. Relative Axin2 mRNA quantity was measured E15.5 control and AdCre infected primary calvarial osteoblast cultured in growth media supplemented with 15% Wnt3a conditioned media for 24hrs or four days (ATCC, CRL2647). Complete qRT-PCR data was depicted in univariate scatter plots as recently described (Weissgerber et al., 2015). Statistical significance was determined by Student t-test.
Acknowledgements
Sincere thanks to Nathaniel Mullin and Miarassa Steele for technical assistance, James Ferguson for the schematic, and to Veronique Lefebvre for Wnt3a conditioned media. Thanks to Drs. Ron Conlon, Diego Correa, Larry Needham for critical reading of the manuscript. Thanks to previous and current members of the Atit laboratory for excellent discussion and advice. Funding provided by NIH NIDCR R01 DE01870 (RA), NIDCR F31 DE020220 (LHG).
Footnotes
Competing Interests
The authors declare that they have no competing interests.
Authors’ contributions
LHG and RPA conceived experiments. LHG, GD, and RPA carried out the experiments. LHG, RPA, and GD analyzed the data. GD performed genotyping and immunohistochemistry. LHG, GD, and RPA generated figures. LHG, GD, and RA interpreted the data and wrote the manuscript. All authors had final approval of the submitted version.
References
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Developmental Biology. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- Barnes RM, Firulli AB. A twist of insight - the role of Twist-family bHLH factors in development. Int J Dev Biol. 2009;53:909–924. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrott JJ, Cash GM, Smith AP, Barrow JR, Murtaugh LC. Deletion of mouse Porcn blocks Wnt ligand secretion and reveals an ectodermal etiology of human focal dermal hypoplasia/Goltz syndrome. Proceedings of the National Academy of Sciences. 2011;108:12752–12757. doi: 10.1073/pnas.1006437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bildsoe H, Loebel DAF, Jones VJ, Chen Y-T, Behringer RR, Tam PPL. Requirement for Twist1 in frontonasal and skull vault development in the mouse embryo. Developmental Biology. 2009;331:176–188. doi: 10.1016/j.ydbio.2009.04.034. [DOI] [PubMed] [Google Scholar]
- Bildsoe H, Loebel DAF, Jones VJ, Hor ACC, Braithwaite AW, Chen Y-T, Behringer RR, Tam PPL. The mesenchymal architecture of the cranial mesoderm of mouse embryos is disrupted by the loss of Twist1 function. Developmental Biology. 2013;374:295–307. doi: 10.1016/j.ydbio.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla-Claudio M, Wang J, Bai Y, Klysik E, Selever J, Martin JF. Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development. 2012;139:709–719. doi: 10.1242/dev.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, Mcmahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Carpenter AC, Rao S, Wells JM, Campbell K, Lang RA. Generation of mice with a conditional null allele for Wntless. genesis. 2010;48:554–558. doi: 10.1002/dvg.20651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-T, Akinwunmi PO, Deng JM, Tam OH, Behringer RR. Generation of a Twist1 conditional null allele in the mouse. genesis. 2007;45:588–592. doi: 10.1002/dvg.20332. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes & Development. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- Collins CA, Kretzschmar K, Watt FM. Reprogramming adult dermis to a neonatal state through epidermal activation of β-catenin. Development. 2011;138:5189–5199. doi: 10.1242/dev.064592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerney J, Andreeva V, Leshem Y, Mercado MA, Dowell K, Yang X, Lindner V, Friesel RE, Spicer DB. Twist1 homodimers enhance FGF responsiveness of the cranial sutures and promote suture closure. Developmental Biology. 2008;318:323–334. doi: 10.1016/j.ydbio.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T, Guo X, Garrettbeal L, Yang Y. Wnt/β-Catenin Signaling in Mesenchymal Progenitors Controls Osteoblast and Chondrocyte Differentiation during Vertebrate Skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons Ben D, Charalambous M, Ferron SR, Herault Y, Pavlovic G, Ferguson-Smith AC, Watt FM. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504:277–281. doi: 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco HL, Casasnovas JJ, Leon RG, Friesel R, Ge Y, Desnick RJ, Cadilla CL. Nonsense mutations of the bHLH transcription factor TWIST2 found in Setleis Syndrome patients cause dysregulation of periostin. Int J Biochem Cell Biol. 2011;43:1523–1531. doi: 10.1016/j.biocel.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghouzzi ElV, Le Merrer M, Perrin-Schmitt F, Lajeunie E, Benit P, Renier D, Bourgeois P, Bolcato-Bellemin AL, Munnich A, Bonaventure J. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nat Genet. 1997;15:42–46. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- Gitelman I. Twist protein in mouse embryogenesis. Developmental Biology. 1997;189:205–214. doi: 10.1006/dbio.1997.8614. [DOI] [PubMed] [Google Scholar]
- Goodnough LH, Chang AT, Treloar C, Yang J, Scacheri PC, Atit RP. Twist1 mediates repression of chondrogenesis by beta-catenin to promote cranial bone progenitor specification. Development. 2012;139:4428–4438. doi: 10.1242/dev.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnough LH, DiNuoscio GJ, Ferguson JW, Williams T, Lang RA, Atit RP. Distinct Requirements for Cranial Ectoderm and Mesenchyme-Derived Wnts in Specification and Differentiation of Osteoblast and Dermal Progenitors. PLoS Genet. 2014;10:e1004152. doi: 10.1371/journal.pgen.1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik K-H, Bornholdt D, Oeffner F, König A, Del Carmen Boente M, Enders H, Fritz B, Hertl M, Grasshoff U, Höfling K, Oji V, Paradisi M, Schuchardt C, Szalai Z, Tadini G, Traupe H, Happle R. Deficiency of PORCN, a regulator of Wnt signaling, is associated with focal dermal hypoplasia. Nat Genet. 2007;39:833–835. doi: 10.1038/ng2052. [DOI] [PubMed] [Google Scholar]
- Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Lack of beta-catenin affects mouse development at gastrulation. Development. 1995;121:3529–3537. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- Hamburg-Shields E, DiNuoscio GJ, Mullin NK, Lafayatis R, Atit RP. Sustained β-catenin activity in dermal fibroblasts promotes fibrosis by up-regulating expression of extracellular matrix protein-coding genes. J. Pathol. 2015;235:686–697. doi: 10.1002/path.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8:727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Howe LR, Watanabe O, Leonard J, Brown AM. Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res. 2003;63:1906–1913. [PubMed] [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Developmental Biology. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- Jiang Z, den Hoff Von JW, Torensma R, Meng L, Bian Z. Wnt16 is involved in intramembranous ossification and suppresses osteoblast differentiation through the Wnt/β-catenin pathway. J. Cell. Physiol. 2013 doi: 10.1002/jcp.24460. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Rice DP, Kettunen PJ, Thesleff I. FGF-, BMP- and Shh-mediated signalling pathways in the regulation of cranial suture morphogenesis and calvarial bone development. Development. 1998;125:1241–1251. doi: 10.1242/dev.125.7.1241. [DOI] [PubMed] [Google Scholar]
- Kimmel RA, Turnbull DH, Blanquet V, Wurst W, Loomis CA, Joyner AL. Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes & Development. 2000;14:1377–1389. [PMC free article] [PubMed] [Google Scholar]
- Klapholz-Brown Z, Walmsley GG, Nusse YM, Nusse R, Brown PO. Transcriptional program induced by Wnt protein in human fibroblasts suggests mechanisms for cell cooperativity in defining tissue microenvironments. PLoS ONE. 2007;2:e945. doi: 10.1371/journal.pone.0000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MP, Ratner N, Yutzey KE. Genome-wide Twist1 occupancy in endocardialcushion cells, embryonic nerve sheath tumor cells. 2014;15:1–14. doi: 10.1186/1471-2164-15-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Lowe GN, Strong DD, Wergedal JE, Glackin CA. TWIST, a basic helix-loop-helix transcription factor, can regulate the human osteogenic lineage. J. Cell. Biochem. 1999;75:566–577. doi: 10.1002/(sici)1097-4644(19991215)75:4<566::aid-jcb3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Liu JK, Ghattas I, Liu S, Chen S, Rubenstein JL. Dlx genes encode DNA-binding proteins that are expressed in an overlapping and sequential pattern during basal ganglia differentiation. Dev Dyn. 1997;210:498–512. doi: 10.1002/(SICI)1097-0177(199712)210:4<498::AID-AJA12>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Marvaso V, Bernard GW. Initial intramembraneous osteogenesis in vitro. Am. J. Anat. 1977;149:453–468. doi: 10.1002/aja.1001490403. [DOI] [PubMed] [Google Scholar]
- McLeod MJ. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22:299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- Morriss-Kay GM, Wilkie AOM. Growth of the normal skull vault and its alteration in craniosynostosis: insights from human genetics and experimental studies. J. Anat. 2005;207:637–653. doi: 10.1111/j.1469-7580.2005.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtola J, Myers J, Akhtar-Zaidi B, Zuzindlak D, Sandesara P, Yeh K, Mackem S, Atit R. beta-Catenin has sequential roles in the survival and specification of ventral dermis. Development. 2008;135:2321–2329. doi: 10.1242/dev.021170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus R, Müller-Röver S, Van Der Veen C, Maurer M, Eichmüller S, Ling G, Hofmann U, Foitzik K, Mecklenburg L, Handjiski B. A comprehensive guide for the recognition and classification of distinct stages of hair follicle morphogenesis. J Invest Dermatol. 1999;113:523–532. doi: 10.1046/j.1523-1747.1999.00740.x. [DOI] [PubMed] [Google Scholar]
- Petti M, Samanich J, Pan Q, Huang CK, Reinmund J, Farooqi S, Morrow B, Babcock M. Molecular characterization of an interstitial deletion of 1p31.3 in a patient with obesity and psychiatric illness and a review of the literature. Am. J. Med. Genet. A. 2011;155A:825–832. doi: 10.1002/ajmg.a.33869. [DOI] [PubMed] [Google Scholar]
- Qin Q, Xu Y, He T, Qin C, Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res. 2012;22:90–106. doi: 10.1038/cr.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarto N, Senarath-Yapa K, Renda A, Longaker MT. TWIST1 silencing enhances in vitro and in vivo osteogenic differentiation of human adipose-derived stem cells by triggering activation of BMP-ERK/FGF signaling and TAZ upregulation. STEM CELLS. 2015;33:833–847. doi: 10.1002/stem.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BS, Yang H, Melvin VS, Taketo MM, Williams T. Ectodermal Wnt/beta-catenin signaling shapes the mouse face. Developmental Biology. 2011;349:261–269. doi: 10.1016/j.ydbio.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold MI, Kapadia RM, Liao Z, Naski MC. The Wnt-inducible transcription factor Twist1 inhibits chondrogenesis. J Biol Chem. 2006;281:1381–1388. doi: 10.1074/jbc.M504875200. [DOI] [PubMed] [Google Scholar]
- Rice DP, Aberg T, Chan Y, Tang Z, Kettunen PJ, Pakarinen L, Maxson RE, Thesleff I. Integration of FGF and TWIST in calvarial bone and suture development. Development. 2000;127:1845–1855. doi: 10.1242/dev.127.9.1845. [DOI] [PubMed] [Google Scholar]
- Rose CS, Patel P, Reardon W, Malcolm S, Winter RM. The TWIST gene, although not disrupted in Saethre-Chotzen patients with apparently balanced translocations of 7p21, is mutated in familial and sporadic cases. Human Molecular Genetics. 1997;6:1369–1373. doi: 10.1093/hmg/6.8.1369. [DOI] [PubMed] [Google Scholar]
- Shelton EL, Yutzey KE. Twist1 function in endocardial cushion cell proliferation, migration, and differentiation during heart valve development. Developmental Biology. 2008;317:282–295. doi: 10.1016/j.ydbio.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo K, O'Rourke MP, Khoo PL, Steiner KA, Wong N, Behringer RR, Tam PP. Twist function is required for the morphogenesis of the cephalic neural tube and the differentiation of the cranial neural crest cells in the mouse embryo. Developmental Biology. 2002;247:251–270. doi: 10.1006/dbio.2002.0699. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Tran TH, Jarrell A, Zentner GE, Welsh A, Brownell I, Scacheri PC, Atit R. Role of canonical Wnt signaling/β-catenin via Dermo1 in cranial dermal cell development. Development. 2010;137:3973–3984. doi: 10.1242/dev.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukel T, Sosic D, Al-Gazali LI, Erazo M, Casasnovas J, Franco HL, Richardson JA, Olson EN, Cadilla CL, Desnick RJ. Homozygous nonsense mutations in TWIST2 cause Setleis syndrome. Am. J. Hum. Genet. 2010;87:289–296. doi: 10.1016/j.ajhg.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio S, Karavanova I, Jowett A, Thesleff I. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell. 1993;75:45–58. [PubMed] [Google Scholar]
- Vincentz JW, Firulli BA, Lin A, Spicer DB, Howard MJ, Firulli AB. Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest. PLoS Genet. 2013;9:e1003405. doi: 10.1371/journal.pgen.1003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrljicak P, Cullum R, Xu E, Chang ACY, Wederell ED, Bilenky M, Jones SJM, Marra MA, Karsan A, Hoodless PA. Twist1 Transcriptional Targets in the Developing Atrio-Ventricular Canal of the Mouse. PLoS ONE. 2012;7:e40815. doi: 10.1371/journal.pone.0040815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Reid Sutton V, Omar Peraza-Llanes J, Yu Z, Rosetta R, Kou Y-C, Eble TN, Patel A, Thaller C, Fang P, Van Den Veyver IB. Mutations in X-linked PORCN, a putative regulator of Wnt signaling, cause focal dermal hypoplasia. Nat Genet. 2007;39:836–838. doi: 10.1038/ng2057. [DOI] [PubMed] [Google Scholar]
- Weissgerber TL, Milic NM, Winham SJ, Garovic VD. Beyond Bar and Line Graphs: Time for a New Data Presentation Paradigm. Plos Biol. 2015;13:e1002128. doi: 10.1371/journal.pbio.1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S. Cell lineage in mammalian craniofacial mesenchyme. Mechanisms of Development. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Yousfi M, Lasmoles F, Marie PJ. TWIST inactivation reduces CBFA1/RUNX2 expression and DNA binding to the osteocalcin promoter in osteoblasts. Biochem Biophys Res Commun. 2002;297:641–644. doi: 10.1016/s0006-291x(02)02260-x. [DOI] [PubMed] [Google Scholar]
- Yu K. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]