Abstract
Scope
The objective was to investigate the metabolome changes in female rats gavaged with partially purified cranberry procyanidins (PPCP) using 1H NMR and UHPLC-Q-Orbitrap-HRMS metabolomics approaches, and to identify the contributing metabolites.
Methods and results
Twenty four female Sprague-Dawley rats were randomly separated into two groups and administered PPCP or partially purified apple procyanidins (PPAP) for 3 times using a 250 mg extracts/kg body weight dose. Plasma were collected six hours after the last gavage and analyzed using 1H NMR and UHPLC-Q-Orbitrap-HRMS. No metabolome difference was observed using 1H NMR metabolomics approach. However, LC-HRMS metabolomics data show that metabolome in plasma of female rats administered PPCP differed from those gavaged with PPAP. Eleven metabolites were tentatively identified from a total of 36 discriminant metabolic features based on accurate masses and/or product ion spectra. PPCP caused a greater increase of exogenous metabolites including p-hydroxybenzoic acid, phenol, phenol-sulfate, catechol sulphate, 3, 4-dihydroxyphenylvaleric acid, and 4′-O-methyl-(−)-epicatechin-3′-O-beta-glucuronide in rat plasma. Furthermore, the plasma level of O-methyl-(−)-epicatechin-O-glucuronide, 4-hydroxy-5-(hydroxyphenyl)-valeric acid-O-sulphate, 5-(hydroxyphenyl)-γ-valerolactone-O-sulphate, 4-hydroxydiphenylamine, and peonidin-3-O-hexose were higher in female rats administered with PPAP.
Conclusion
The metabolome changes caused by cranberry procyanidins were revealed using an UHPLC-Q-Orbitrap-HRMS global metabolomics approach. Exogenous and microbial metabolites were the major identified discriminate biomarkers.
Keywords: cranberries, 1H NMR, metabolomics, procyanidins, UHPLC-Q-Orbitrap-HRMS
1 Introduction
Procyanidins are oligomers and polymers of (−)-epicatechin or (+)-catechin [1]. The molecular weight of procyanidins is described by degree of polymerization (DP). Monomeric procyanidins are (−)-epicatechin or (+)-catechin. Procyanidins with DP 2, 3, or 4 are dimers, trimers, and tetramers, respectively. The most widely distributed procyanidins in foods are the B-type, which are linked through C4 → C8 and/or C4 → C6 interflavan bonds [2]. Examples of foods that contain exclusively B-type procyanidins are apples, pears, blueberries, and cocoa. A-type procyanidins are rare in foods and they have an additional ether interflavan bond between C2 →O →C7. Cranberries are among a few foods that contain A-type procyanidins. A previous study showed that procyanidins with at least one A-type bond accounted for more than 90% of trimers through undecamers in cranberry press cake [3]. Studies suggested that A-type procyanidins have greater and unique bioactivity compared with B-type [4]. For example, cranberries are known to prevent or mitigate urinary tract infection [5]. Such activity was attributed to A-type procyanidins but not B-type ones [4].
Metabolomics have been widely applied in clinical, pharmaceutical and toxicological studies for identification of biomarkers [6]. It assesses the metabolic changes in a global manner in order to monitor biological function alteration due to genetic modification, pathophysiological changes, or exogenous challenges [7]. Phytochemicals originating from foods are ingested, metabolized and absorbed in the gastrointestinal tract generating a characteristic metabolome profile, which may further alter endogenous metabolites. Metabolomics is an effective approach to distinguish the metabolome caused by different diets. It is expected to reveal the physiological effects of phytochemical intake and identify new candidate biomarkers. NMR and UHPLC-HRMS are the two most widely used metabolomics platforms [8]. Both techniques are able to detect hundreds or thousands of metabolites in biological samples. NMR spectroscopy has the advantage of being quantitative, highly reproducible, non-selective [8] and requiring minimal sample preparation [9], while UHPLC-HRMS is highly sensitive and able to identify the chemical structures of metabolites [8]. The high-dimensional data produced by a metabolomics study is often processed using multivariate statistical techniques such as PLS-DA and OPLS-DA to reduce the dimensionality of the data [10].
The mechanism by which cranberry procyanidins mitigate urinary tract infection remains elusive. A-type procyanidins from cranberry juice inhibited the adhesion of uropathogenic E. coli, whereas those from apple juice showed no activity. Anti-adhesion activity in human urine was detected following cranberry juice cocktail consumption, but not after consumption of apple juice [4]. We hypothesized that the metabolome changes caused by cranberry procyanidins in female rats may be different from those caused by apple procyanidins. The objective of this study is to identify molecular profile and putative biomarkers in plasma of female rats after intake of partially purified cranberry procyanidins (PPCP) using both 1H NMR and UHPLC-Q-Orbitrap-HRMS based global metabolomics approaches.
2 Materials and methods
2.1 Chemicals and materials
Freeze-dried cranberry powder was provided by Ocean Spray Cranberries, Inc. (Lakeville-Middleboro, MA, USA). Fresh granny smith apples were purchased from a local grocery store. LC-MS grade acetonitrile, methylene chloride, methanol, acetic acid, formic acid, and acetone were purchased from Fischer Scientific Co. (Pittsburgh, PA, USA). (−)-Epicatechin was provided by Sigma Chemical Co. (St. Louis, MO, USA). A mixture of partially pure procyanidin oligomers (monomers through nonamers) was provided by Mars Inc. (McLean, VA, USA). D2O (99.9% D) was provided by Cambridge Isotope Laboratories, Inc. (Tewksbury, MA, USA). Creatine-D3, L-leucine-D10, L-tryptophan-2, 3, 3-D3, caffeine-D3 were obtained from CDN Isotopes Inc. (Pointe-Claire, Quebec, Canada). Sephadex LH-20 resin was purchased from Sigma-Aldrich (St. Louis, MO, USA). Amberlite FPX 66 resin was a product of Rohm and Haas Co. (Philadelphia, PA, USA). Pooled quality control plasma samples used in NMR metabolomics were purchased from the American Red Cross and were collected over a period of about 2 weeks.
2.2 Extraction, purification and characterization of PPCP and PPAP
One hundred and twenty grams of freeze-dried cranberry powder was extracted with 1 L of methanol. The cranberry-methanol mixture was put into a beaker sealed with Parafilm M and sonicated for 30 min. After sonication the cranberry-methanol mixture was placed in darkness at room temperature for 48 h. Extracts obtained after vacuum filtration were combined and concentrated under a partial vacuum using a rotary evaporator at 45 °C. The concentrated extract was re-suspended in 20 mL of water and loaded onto a column packed with Amberlite FPX 66 resin. Column was eluted with 3 L of de-ionized water to remove free sugars and organic acids. Column was then eluted with 500 mL of methanol to recover cranberry phytochemicals absorbed on the resin. Methanol was evaporated using a SpeedVac Concentrator (Thermo scientific ISS110, Waltham, MA, USA) under a reduced pressure to yield dry cranberry sugar-free extract (5.40 g). The sugar-free extract (5.40 g) was suspended in 100 mL of 30% methanol and loaded onto a column (5.8 × 28 cm) packed with Sephadex LH-20, which was soaked in 30% methanol for over 4 hours before use. The column was eluted with 30% methanol to remove anthocyanins and phenolic acids, and then eluted with 70% acetone to yield partially purified cranberry procyanidins (3.95 g). To extract procyanidins from fresh apples, 5,000 g fresh granny smith apples were used. Fresh apples were stored at −20 °C and divided into two batches before the extraction. Each 2,500 g frozen apples were cut into small pieces and pulverized to get apple puree using a blender. Apple puree was mixed with 2 L of methanol and sonicated for 40 min in an ice bath. Then the apple puree-methanol suspension was placed in darkness at −10 °C for 48 h. Two batches of extracts obtained after vacuum filtration were combined and concentrated under a partial vacuum using a rotary evaporator at 45 °C. The concentrated extract was re-suspended in 50 mL of water and loaded onto a column packed with Amberlite FPX 66 resins. Column was eluted with 3 L of de-ionized water to remove free sugars and organic acids. Column was then eluted with 500 mL of methanol to recover apple procyanidins absorbed on the resin. Methanol was then evaporated using a SpeedVac Concentrator (Thermo scientific ISS110, Waltham, MA) under a reduced pressure to yield partially purified apple procyanidins (5.30 g). Procyanidins were analyzed using HPLC-FLD-MSn (details in supplementary data).
2.4 Animals and experiment design
Approval for animal study was sought through the Institutional Animal Care and Use Committee at the University of Florida (IACUC Study #201307837). Female Sprague Dawley (n=24, 220–280 g) were housed in the animal facility and acclimated for 5–7 days using a purified diet free of flavonoid compounds (D10012G, Research diet Inc., New Brunswick, NJ, USA). Two female rats were housed in a cage. After the acclimation period female rats were randomly divided into two groups with 12 female rats per group, and fasted for six hours before the metabolomics study. PPCP or PPAP were dispersed in water and administered by oral gavage at 0 and 12 hours using a dose of 250 mg extracts/kg body weight. Female rats had free access to food and water after dosing. At 24 hours, female rats were gavaged for a third time. Six hours after the 3rd gavage, female rats were anesthetized and blood samples were collected by cardiac puncture into vials containing sodium heparin using heparinized syringes. Blood collection time point was selected based on a previous study which showed that [14C] procyanidin B2 in rats reached a peak plasma concentration at Tmax 5–6 hours after oral administration [11]. Blood samples were centrifuged at 2,000 g for 10 min at 4 °C to obtain plasma. All plasma samples were aliquoted and kept in a −80 °C freezer until analyses.
2.5 1H NMR analyses
Plasma samples were thawed at 4°C in a cold room. Four hundred μL of saline solution (NaCl 0.9% in 10% D2O) was added to 200 μL of each plasma sample. The mixtures were vortexed for 1 minute and centrifuged at 16, 000 g for 15 min at 4°C and 550 μL of supernatant was transferred into 5 mm Bruker NMR tubes (Z105684 Bruker 96 well rack) using Gilson 215 Liquid Handler (Trilution software version 2.0). All 1H-NMR spectra were collected on a 600 MHz Avance II NMR spectrometer (Bruker Biospin, Rheinstetten, Germany) equipped with a 5 mm CryoProbe. A Bruker sampleJet operated by IconNMR in Topspin was used to record spectra automatically. 1D CPMG-presaturated spectra for plasma were recorded. Optimal probe tuning and matching, 90° pulse length, water offset, and receiver gain were adjusted on the representative sample. The probe was automatically locked to H2O+D2O (90%+10%) and shimmed for each sample. All NMR data were acquired at 300 K.
2.6 UHPLC-Q-Orbitrap-HRMS analyses
Frozen plasma samples (−80 °C) were thawed at room temperature. One plasma sample (50 μL) was mixed with 400 μL acetonitrile: acetone: methanol (8:1:1, v: v: v) to precipitate the proteins. Ten μL isotopically-labeled standard solution (40 μg/mL L-tryptophan-D3, 4 μg/mL L-leucine-D10, 4 μg/mL creatine-D3, 4 μg/mL caffeine-D3) was added to the above extraction mixture as internal standards. The sample was then vortexed and placed in a 4 °C refrigerator for 30 min to assist protein precipitation. Then the sample was centrifuged at 20,000 g for 10 min at <10 °C to pellet the protein. One hundred and twenty five μL of supernatant was transferred to a new 1 mL Eppendorf tube and dried under a gentle stream of Nitrogen (Organomation Associates, Inc., Berlin, MA, USA). Dried sample was reconstituted in 50 μL 0.1% formic acid in water and vortexted. The sample solution was placed in ice bath for 10–15 min and centrifuged at 20,000 g for 5 min at <10 °C to remove debris. The supernatant was transferred into a LC glass vial with fused glass insert for analyses. Three pooled quality control (QC) samples were prepared by mixing an equal volume of the supernatant from 24 rat plasma extracts. In addition, three neat QC samples were prepared by adding 20 μL of isotopically-labeled standard solution directly to three LC glass vials, respectively. To monitor the performance of data acquisition, run sequence was started with 3 blanks (0.1% formic acid in water), one neat QC, and one pooled QC followed by every 10 plasma samples to ensure instrument drift was minimal.
Chromatographic separation was performed on a Thermo Scientific-Dionex Ultimate 3000 UHPLC using an ACE Excel 2 C18-PFP column, 100 mm x 2.1 mm i.d., 2 μm (Advanced Chromatography Technologies, Aberdeen, UK). The mobile phase consisted of (A) water with 0.1% formic acid and (B) acetonitrile. The gradient was as follows: 0–3 min, 100% A isocratic; 3–13 min, 0–80% B linear; 13–16 min, 80% B isocratic; 16–16.5 min, 80–0% B linear; followed by 3 min of re-equilibration of the column before the next run. The flow rate was 350 μL/min and the injection volume was 4 μL. Before starting the sequence, UHPLC column was rinsed using acetonitrile and then equilibrated using water with 0.1% formic acid for 10 min. The UHPLC system was coupled to a Q Exactive™ Hybrid Quadrupole-Orbitrap High Resolution Mass Spectrometer (Thermo Fisher Scientific, San Jose, CA, USA). The MS acquisition was performed in negative ionization with a mass resolution of 70,000 at m/z 200 and separate injections were performed in a data-dependent (top 5) MS/MS mode with the full scan mass resolution reduced to 35,000 at m/z 200. The m/z range for all full scan analyses was 70–1000. Heated electrospray ionization (HESI) parameters were as follows: sheath gas flow 45 arb, auxiliary gas flow 10 arb, sweep gas flow 1 arb, spray voltage 3.5 kV, capillary temperature 320 °C, and probe temperature 350°C. In source CID (Collision-Induced Dissociation) was 2 eV. The mass spectrometer was calibrated using Pierce™ negative ion calibration solution (Thermo Fisher Scientific, San Jose CA, USA). To avoid possible bias, the sequence of injections for plasma samples was randomized.
2.7 Multivariate data processing and statistical analyses
All NMR spectra were phased and baseline corrected using NMRPipe [12], and then converted to FT (Fourier transformed) files. The FT files were imported into MATLAB (R2013B, the Mathworks, Inc., Natick, MA, USA). Spectra were referenced to the alanine peak at δ1.469 ppm and water resonance region (4.66–4.95 ppm) was excluded. Then the spectra were aligned and normalized in MATLAB. The resultant data set was imported into SIMCA (Version 13.0.3, Umetrics, Umea, Sweden) for multivariate statistical analyses. Data were mean-centered and Pareto scaled before PCA, PLS-DA and OPLS-DA analyses in SIMCA. LC-HRMS data were converted to mzXML using MSConvert from ProteoWizard [13] and then processed using MZmine 2.12 [14]. Peaks in each sample were extracted, deconvoluted, and deisotoped. Alignment (using join aligner) was conducted with a 10 ppm tolerance for m/z values and 0.2 min tolerance for retention time. Gap filling (using peak finder) was performed to fill in missing peaks. The resultant data set was imported into SIMCA (Version 13.0.3, Umetrics, Umea, Sweden) for multivariate statistical analyses. Data were mean-centered, Pareto scaled and log-transformed before PCA analysis. Data were mean-centered and log-transformed before PLS-DA and OPLS-DA analyses in SIMCA. Unsupervised PCA model was performed to initially examine intrinsic variation in the data set. Then supervised pattern recognition methods including PLS-DA and OPLS-DA [10] were used to extract maximum information on discriminant compounds from the data. Validation of the model was tested using 7-fold internal cross-validation and permutation tests for 200 times. To further evaluate the predictive ability of the PLS-DA and OPLS-DA models, an external validation procedure was performed [15]. The LC-HRMS metabolomics data set was split into a training set and a test set. Approximately 80% of the samples were randomly selected as the training set and the remaining 20% were treated as the test set. PLS-DA and OPLS-DA models were built based on the training set and then blindly predicted the classes of the samples in the test set. This procedure was repeated 30 times and a correct classification rate was calculated. For univariate analyses, mass spectral intensity data of selected metabolites which have been mean-centered and log-transformed were subjected to Welch’s t test. Benjamini–Hochberg procedure at α=0.01 [16] was conducted to control false discoveries. The univariate analyses were done using Microsoft Excel (Version 2010, Microsoft Corporation, Seattle, WA, USA).
3 Results and discussion
3.1 Procyanidin composition and content in PPCP and PPAP
Procyanidins were extracted and partially purified from cranberry powder or fresh apples. Figure S1 in supplementary data shows the HPLC fluorescence chromatograms of procyanidins in PPCP and PPAP. Oligomeric procyanidins with DP of 1–6 and high polymeric procyanidins in apples or cranberry powder were identified and quantified by HPLC-MSn. The content of total procyanidins in PPCP was 511 mg/g extracts, lower than that in PPAP (690mg/g extracts). Over 90% of procyanidin oligomers (dimer to tetramers) in PPCP were A-type (Table S1). Our results were consistent with a previous study which showed that procyanidins with at least one A-type bond accounted for more than 90% of trimers through undecamers in cranberry press cake [3]. Monomers through tetramers accounted for 45% of total procyanidin, with the rest being high polymers. A- and B-type pentamers and hexamers were identified but not quantified in PPCP due to peak overlapping (Figure S1). PPAP contained exclusively B-type procyanidins. Content of dimers through tetramers in PPAP were higher than those in PPCP (Table S1). Monomer through tetramers accounted for 65% of total procyanidins in PPAP, with about 13% being high polymers.
3.2 Quality control of multivariate analyses
In this study, the concept of biology QC which uses biological samples including plasma, urine or tissue as quality controls was adopted [17]. Biological QCs consisting of 4 replicates of pooled Red Cross plasma were analyzed together with rat plasma to validate NMR acquisition method. The PCA model was built to investigate the metabolome differences between QCs and rat plasma. The mechanism was based on the ability of the PCA model to cluster samples in an unsupervised approach. The PCA score plot (Figure S2) shows that the 4 replicates were segregated from experimental samples, indicating that the NMR data acquisition method was valid. Since variations between LC-HRMS injections and artifacts due to the order of acquisition and carry-over, sensitivity changes or ion suppression could occur during the experimental period [18]. Sample acquisition was randomized, and QC samples were used to monitor the instrument performance. Pooled QCs were further examined using multivariate statistic techniques. A PCA model was constructed to visualize any separation between three QCs. PCA score plot (Figure S4) demonstrates that the three QCs across the entire sequence were tightly clustered, suggesting a high quality of data acquisition.
3.3 NMR metabolomics analysis of rat plasma
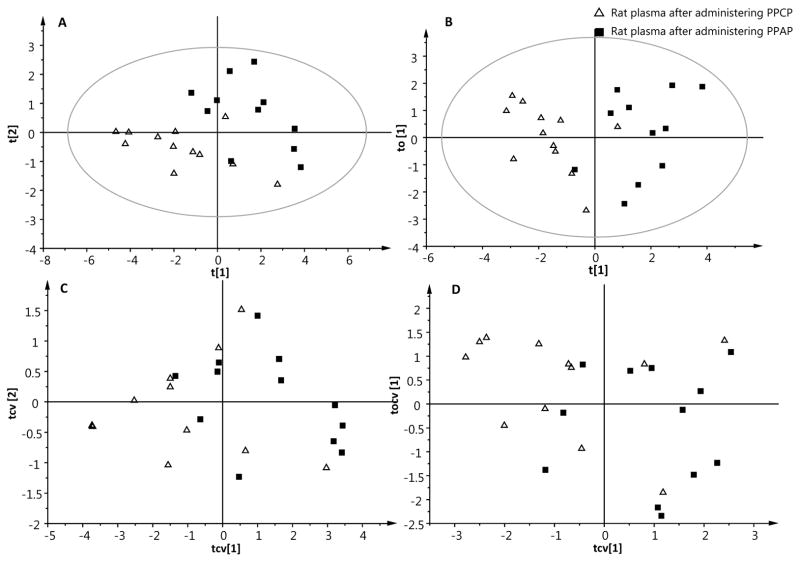
PCA model was built on NMR metabolomics data before supervised multivariate analyses. PCA score plot shows a separation between PPCP and PPAP, with one sample from PPAP group mixed with the group of PPCP (Figure S3). To further confirm and validate the metabolome differences between PPCP and PPAP, PLS-DA and OPLS-DA models were constructed. Two principal components were selected to build PLS-DA model; one principal component and one orthogonal component were used to construct OPLS-DA model. The R2X and R2Y of both models was 0.433 and 0.676, respectively (Table 1). R2 represents the goodness of fit, and the results indicated that about 43% of variance in X data matrix and 68% of variance in Y was explained by PLS-DA and OPLS-DA models. The high R2 values indicated the robustness of the supervised models [19]. Overfitting arises in PLS and OPLS models when the number of variables is much larger than that of observations, and could be a problem with any high-dimensional data. Accidental correlation between one or more variables becomes common for metabolomics data [20]. Internal cross-validation is thus the first step to test the predictability of the supervised models. If Q2 calculated from the cross-validation has a low value, then conclusion could be drawn that the supervised model does not have predictability. In the present study, 7-fold internal cross validation was performed on both PLS-DA and OPLS-DA models derived from NMR metabolomics data. Q2 obtained from cross-validation for PLS-DA and OPLS-DA was 0.254 and 0.291, respectively (Table 1). They were much lower than 0.5, a threshold value for a good multivariate model of metabolomics data [21]. Although a segregation between PPCP and PPAP was observed on the PLS-DA and OPLS-DA score plots (Figure 1A, 1B), the low Q2 values suggested that both models had poor predictability and the segregation was most likely due to overfitting. Misclassification that occurred during cross-validation (Figure 1C, 1D) also confirmed that NMR metabolomics data did not reveal a metabolome difference in rat plasma between PPCP and PPAP.
Table 1.
Summary of parameters for PCA, PLS-DA, and OPLS-DA models for rat plasma after administering PPCP or PPAP by oral gavage
| 1H NMR metabolomics | LC-HRMS metabolomics | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| PCA | PLS-DA | OPLS-DA | PCA | PLS-DA | OPLS-DA | |
| Na | 5 | 2 | 1Pc+1Od | 4 | 2 | 1Pc+1Od |
| R2X(cum)b | 0.783 | 0.433 | 0.433 | 0.516 | 0.428 | 0.428 |
| R2Y(cum)b | --- | 0.676 | 0.676 | --- | 0.995 | 0.995 |
| Q2(cum)b | 0.513 | 0.254 | 0.291 | 0.521 | 0.982 | 0.974 |
| *Correct Classification Rate | --- | --- | --- | --- | 100%±0 | 100%±0 |
N: number of components.
R2X (cum)and R2Y (cum) are the cumulative modeled variations in the X and Y matrix, respectively. Q2Y (cum) is the cumulative predicted variation in the Y matrix.
Predictive component.
Orthogonal component.
Figure 1.
The PLS-DA (A), OPLS-DA (B) score plots, PLS-DA (C) and OPLS-DA (D) cross-validated score plots derived from 1H NMR metabolomics. Triangle: rat plasma after administering PPCP. Filled box: rat plasma after administering PPAP. Each triangle or filled box represents an individual rat.
3.4 LC-HRMS metabolomics analysis of rat plasma
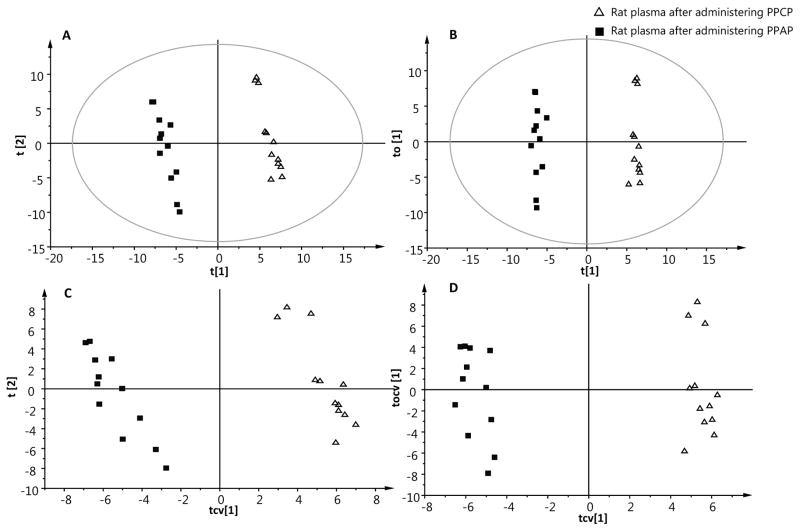
Similarly, LC-HRMS metabolomics data was analyzed using supervised models to reveal the metabolic changes of rat plasma after administering PPCP or PPAP. Figure 2A and 2B show a clear segregation between two groups on the score plot of both PLS-DA and OPLS-DA models. The advantage of OPLS-DA over PLS-DA is that the “structure noise” of data matrix which is unrelated to the variation of interest is filtered and described only by the orthogonal component. The variation of scientific interest is described in the predictive component. Therefore the interpretability of the resulting model is increased [10]. This was demonstrated in our previous study where OPLS-DA showed a better performance than PLS-DA model for human plasma and urine because it removed “structure noise” of data [22]. In the present study, PLS-DA models derived from LC-HRMS metabolomic data had high quality parameters which was not improved by OPLS-DA, suggesting low “structure noise” in the data set. PLS-DA had two principal components with an overall value of R2X and R2Y of 0.428 and 0.995, respectively (Table 1). Similarly, OPLS-DA generated one principal component and one orthogonal component. The R2X and R2Y of OPLS-DA model was 0.428 and 0.995 (Table 1). It shows that about 42% of variance in X data matrix and 99% of variance in Y data matrix was explained by both supervised models.
Figure 2.
The PLS-DA (A), OPLS-DA (B) score plots, PLS-DA (C) and OPLS-DA (D) cross-validated score plots derived from LC-HRMS metabolomics. Triangle: rat plasma after administering PPCP. Filled box: rat plasma after administering PPAP. Each triangle or filled box represents an individual rat.
To test for overfitting and the validity of PLS and OPLS models derived from LC-HRMS metabolomic data, three validation methods were used. Seven-fold internal cross validation was initially performed on both PLS-DA and OPLS-DA models. Predictability Q2 obtained from cross-validation was 0.982 and 0.974 for PLS-DA and OPLS-DA model, respectively. The high Q2 indicated both supervised models had excellent predictability. The cross-validated score plots (Figure 2C, 2D) show that no rat plasma from two groups was misclassified which was consistent with the internal validation result. In order to further confirm the predictability of PLS-DA and OPLS-DA models, permutation test was conducted. The class labels of PPCP and PPAP group were permuted and randomly assigned to different observations. Then a classification model was calculated with the permutated class labels. The procedure was repeated 200 times. R2 and Q2 within each model were calculated and a regression line was drawn. Ideally, all R2 and Q2 calculated from the permutation data should be lower than those from the actual data and the Q2-intercept value obtained from the regression line should be lower than 0.05 [23]. The rationale behind the permutation test is that the newly constructed classification models should not be able to predict the classes well with a wrong class label [24]. Figure S6 shows that the goodness of fit (R2) and predictive powder (Q2) of newly constructed models with permuted class labels were decreased compared to the actual model, indicating our supervised model is statistically valid and the achieved segregation between PPCP and PPAP was not due to overfitting. Cross-validation and permutation test provide a reasonable estimate of the predictability of a PLS or OPLS model [25]. However, external validation that uses an independent set of test data to evaluate predictability of a supervised model that is built on the training set is a more scrupulous and demanding method [25]. In the present study, the LC-HRMS data was split into a training set (approximate 80% of the samples) and a test set (remaining 20% of the samples) [15]. PLS-DA and OPLS-DA models were then constructed on the training data set and blindly classified the test set. This external validation procedure was repeated for 30 times and a correct classification rate was calculated by counting the correctly classified samples and divided by the total number of samples in the test set. Correct classification rate of 100% for both PLS-DA and OPLS-DA models (Table 1) indicated that the supervised models based on LC-HRMS metabolomics data had excellent predictability and were able to correctly predict the unknown samples. The validation tests suggested that the UHPLC-HRMS metabolomics approach was able to reveal the metabolome changes in female rats after administering PPCP compared with PPAP.
3.5 Discriminant metabolites identification
No modification of rat plasma metabolome was detected using 1H NMR-based metabolomics approach although it was proven to be an effective tool for metabolomics profiling in other studies [23, 26]. This was likely due to the inherent low sensitivity of NMR technique that failed to detect procyanidin metabolites in plasma. Untargeted UHPLC-HRMS metabolomics was a more sensitive method to reveal the metabolome differences and contributing markers.
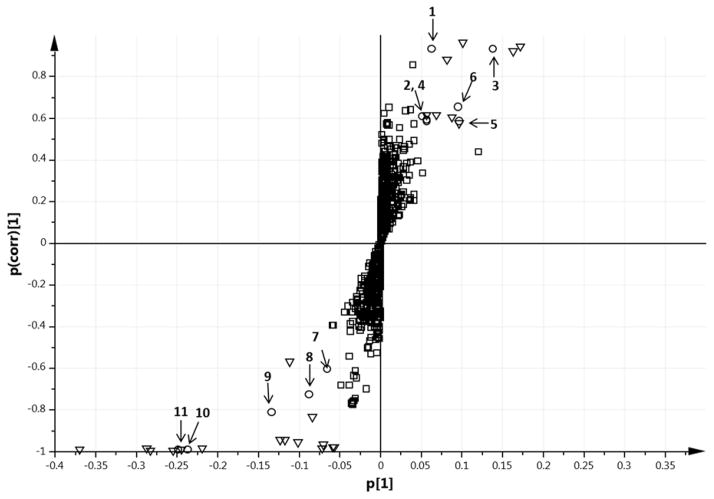
S-plot (Figure 3) is a statistical tool that visualize the variable influence in a projection-based model and discover the responsible metabolites. It is a scatter plot that combines the covariance (magnitude) and correlation loading (reliability) for the model variables [27]. It can be applied to projection-based models including OPLS, PLS or PCA. The x-axis in the S-plot describes the magnitude of each variable. The y-axis represents the reliability of each variable. The y-axis has a theoretical minimum of −1 and maximum of +1. Unless the variable variance is uniform, otherwise the scatter plot will look like an S-shape. At a p significance level of 0.05, a p(corr) of 0.5 was used as an arbitrary cutoff value to select the potential markers [19]. The markers with higher absolute p[1] and p(corr) values which are located on the upper right or lower left corner of the S-plot were the statistical relevant variables for explaining the separation between PPCP and PPAP. The variables in the middle of the S-plot did not show any relevance in the model. Variable importance for projection (VIP) is another statistical tool used to summarize the importance of X-variable both for X- and Y-models [25]. It is used to determine the relevance and importance of a variable in a projection-based model. The influence of each variable on the response is summed over all components and categorical responses, relative to the total sum of squares of the model. For a given model, there will be only one VIP-vector summarizing all components and Y-variables. This makes the VIP an appealing measure of the global effect of diet intervention. A threshold of VIP value ≥1 is usually considered appropriate for a metabolomics study [25]. In the present study, S-plot is used as the primary statistical tool for determining the significant metabolites. We compared the markers selected from S-plot to those selected using a VIP plot. All variables with VIP score >1 were plotted in Figure S7. Variables selected as significant ones from S-plot were colored in red. It was found that variables selected from S-plot had a higher VIP score (VIP>1.7). This result supported the reliability and effectiveness of S-plot, and indicated that S-plot is a more scrupulous method. These selected significant metabolites were then subjected to Welch’s t test, and the p-value obtained for each marker was smaller than 0.01. Benjamini–Hochberg procedure (α=0.01) was conducted to control false discoveries.
Figure 3.
S-plots associated with the OPLS-DA score plot of data derived from LC-HRMS of rat plasma after administering PPCP or PPAP. p[1] is the loading vector of covariance in the first principal component. p(corr)[1] is loading vector of correlation in the first principal component. Variables with |p| ≥0.05 and |p(corr)| ≥0.5 are considered statistically significant. Significant variables marked in circles were identified and numbered in Table 2. Unidentified significant variables marked in triangles were listed in Table S2. Non-significant variables were marked in squares.
A total of 1186 metabolic features were detected in rat plasma, among which 36 features were found to be discriminant metabolites on the basis of multivariate analysis (Figure 3). Eleven metabolites were identified based on their accurate masses and/or product ion spectra (Table 2). The other 25 unidentified metabolites were listed in Table S2. HMDB [28] and/or Phenol-Explorer [29] were searched to assist metabolite identification. One metabolite that was higher in rat plasma after PPCP was the ion at m/z 137.0246 [M-H]− producing a product ion at m/z 93.0339 [M-H-COO]− after MS/MS. It was tentatively identified as p-hydroxybenzoic acid as it matched in HMDB (Δ=0.0002 Da) and a previous publication [30]. The compound producing a [M-H]− ion at m/z 93.0337 [M-H]− was tentatively identified as phenol according to HMDB (Δ=0.0009 Da). The ion at m/z 172.9915 [M-H]− was tentatively identified as phenol sulfate which agreed with HMDB (Δ=0.0001 Da). The plasma level of catechol sulfate was elevated in female rats after administering PPCP. This metabolite was previously identified in human urine after drinking blackcurrant juice [31]. Identification of catechol sulfate was based on the accurate m/z 188.9863[M-H]−, product ion at m/z 109.0296 [M-H-sulphate]−, and HMDB match (Δ=0 Da). 3, 4-Dihydroxyphenylvaleric acid and 4′-O-methyl-(−)-epicatechin-3′-O-beta-glucuronide were also detected and tentatively identified in rat plasma after PPCP intake. It should be noted that the difference between detected mass and theoretical mass of 4′-O-methyl-(−)-epicatechin-3′-O-beta-glucuronide was 0.0707 Da, which was relatively higher than other mass error. However, 4′-O-methyl-(−)-epicatechin-3′-O-beta-glucuronide was tentatively assigned because it was the only database match that was biologically relevant and no standard was available for further identification. Furthermore, consumption of PPCP also decreased the plasma level of five metabolites. The metabolite producing a [M-H]− ion at m/z 479.1190 and a product ion at m/z 303.0885 [M-H-glucuronide]− was assigned as O-methyl-(−)-epicatechin-O-glucuronide by comparing with HMDB (Δ=0.0001 Da). A previous publication revealed that methylation of (−)-epicatechin occurred at 3′-position in rats [32]. Another study showed that glucuronidation of daidzein occurred at the 7 position after daidzein was incubated with Sprague-Dawley rat liver microsome [33]. The positions of glucuronidation and methylation in rats were markedly different from humans, mice, pigs, etc. In the present study, the substitution positions were not able to be further confirmed without NMR spectra of purified compounds. The metabolite producing a [M-H]− ion at m/z 289.0384 was putatively identified as 4-hydroxy-5-(hydroxyphenyl)-valeric acid-O-sulphate. The identification agreed with HMDB (Δ=0.0003 Da) and was described in a previous study [34]. 5-(hydroxyphenyl)-γ-valerolactone-O-sulphate was putatively identified based on its m/z at 271.0287 [M-H]− and HMDB match (Δ=0.0005 Da). The metabolite having a [M-H]− ion at m/z 184.0757 was putatively identified as 4-hydroxydiphenylamine according to the HMDB (Δ=0.0011 Da). 4-hydroxydiphenylamine is a metabolite of diphenylamine and found in stored apples [35]. The metabolite giving a [M-H]− ion at m/z 461.9787 and product ion at m/z 264.0330 [M-H-hexose-H2O]− was tentatively identified as peonidin-3-O-hexose. The exact type of hexose could not be determined due to lack of standard comparison. Previous studies showed that both peonidin-3-O-glactoside and peonidin-3-O-glucoside were found in rat plasma after they were administered with anthocyanin-rich extracts [36]. In the present study, the detection of peonidin-3-O-hexose in rat plasma after administering PPAP was likely due to the residual anthocyanins in PPAP.
Table 2.
Identification of discriminant metabolites for rat plasma after administering PPCP or PPAP by oral gavagea
| NO. | Detected Mass [M-H]− (MSMS) |
Retenti on Tim (min) |
p[1] (contribu tion) |
p(corr)[ 1] (confide nce) |
VIP | Metabolites Putative Identification |
Theoretical Mass [M-H]− |
Mass Difference (Da) |
Database ID |
PPCP VS. PPAP b |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 137.0246 (93.0339[M-H-COO]−) | 9.231 | 0.063 | 0.931 | 2.16 | p-hydroxybenzoic acid c | 137.0244 | 0.0002 | HMDB 00500 | ↑ |
| 2 | 172.9915 | 6.855 | 0.051 | 0.609 | 1.74 | phenyl sulfate | 172.9914 | 0.0001 | HMDB 60015 | ↑ |
| 3 | 188.9863 (109.0296 [M-H-sulphate]−) | 6.566 | 0.139 | 0.933 | 4.73 | catechol sulphate c | 188.9863 | 0 | HMDB 61713 | ↑ |
| 4 | 209.0904 | 9.525 | 0.057 | 0.587 | 1.99 | 3, 4-Dihydroxyphenylvaleric acid | 209.0819 | 0.0085 | HMDB 29233 | ↑ |
| 5 | 493.2059 | 7.572 | 0.098 | 0.584 | 3.22 | 4′-O-methyl-(−)-epicatechin-3′-O-beta-glucuronide | 493.1352 | 0.07075 | HMDB 29180 | ↑ |
| 6 | 93.0337 | 6.854 | 0.096 | 0.653 | 3.25 | phenol | 93.0346 | 0.0009 | HMDB 00228 | ↑ |
| 7 | 479.1194 (303.0885[M-H-glucuronide]−) | 7.463 | −0.065 | −0.608 | 2.24 | O-methyl-(−)-epicatechin-O-glucuronide c | 479.1195 | 0.0001 | HMDB 41659 | ↓ |
| 8 | 289.0384 | 7.095 | −0.088 | −0.725 | 3.06 | 4-hydroxy-5-(hydroxyphenyl)-valeric acid-O-sulphate c | 289.0387 | 0.0003 | HMDB 59976 | ↓ |
| 9 | 271.0287 | 7.793 | −0.134 | −0.812 | 4.68 | 5-(hydroxyphenyl)-gamma-valerolactone-O-sulphate | 271.0282 | 0.0005 | HMDB 599993 | ↓ |
| 10 | 184.0757 | 9.448 | −0.236 | −0.991 | 8.16 | 4-hydroxydiphenylamine | 184.0768 | 0.0011 | HMDB 32597 | ↓ |
| 11 | 461.9787 (264.0330[M-H-hexose-H2O]−) | 9.481 | −0.249 | −0.994 | 8.60 | peonidin-3-O-hexose | 462.1168 | 0.1380 | Phenol-Explorer | ↓ |
Benjamini–Hochberg procedure was conducted to control false discoveries at α=0.01. All metabolites had Welch t test p<0.01.
Arrows indicated a decrease or increase in metabolite level in female rats plasma after administering PPCP compared to PPAP
Procyanidins purified from cranberry powder were predominantly A-type, whereas PPAP contained exclusively B-type procyanidins. Procyanidins had a very low absorption rate in vivo and only a small portion of epicatechin and oligomeric procyanidins (DP<5) were able to be absorbed in the small intestine [2]. The majority of A- and B-type procyanidin oligomers and polymers were degraded by gut microbiota in the colon to produce microbial metabolites. More than half of discriminant metabolites detected in the present study corresponded to the phase II and microbial metabolites of procyanidins. A previous study showed that B-type procyanidin dimers were catabolized by microbial cleavage of C-ring and/or oxidation of A-ring, and further degraded into hydroxyphenyl-γ-valerolactone [37]. Phenylvalerolactones were then slowly dehydroxylated by bacteria to form phenylvaleric acids [38]. 5-(hydroxyphenyl)-γ-valerolactone-O-sulphate which was found to be decreased after rat receiving PPCP was formed after phase II metabolism of 5-(hydroxyphenyl)-γ–valerolactone. 4-hydroxy-5-(hydroxyphenyl)-valeric acid-O-sulphate was also decreased after ingesting PPCP. It was probably generated after dehydroxylation of dihydroxyphenyl-γ-valerolactone and further sulfation. 3, 4-Dihydroxyphenylvaleric acid was probably a dehydroxylation product from dihydroxyphenyl-γ-valerolactone. p-hydroxybenzoic acid was likely formed by progressive shortening the aliphatic chain by α-and β-oxidation of phenylvaleric acids [38]. Compared to extensive investigation on B-type dimers catabolism, limited data is available on the microbial catabolism of A-type procyanidins. An early study employed a pig cecum model and showed that, similar to B-type dimer catabolism, A-type procyanidins degradation was initiated by cleavage of C-ring followed by generation of various phenolic acids [39]. A-type procyanidins oligomers exhibited a more complicated pattern of hydroxylated catabolites probably due to their more rigid and complex interflavan ether bonds [39]. However, in the present study we failed to detect any metabolites that retain this unique ether linkage.
4 Concluding remarks
In conclusion, female Sprague-Dawley rat plasma metabolome differences between PPCP and PPAP were detected using an untargeted UHPLC-Q-Orbitrap-HRMS metabolomics approach but not a 1H NMR metabolomics approach. Our study is one of few publications that use two metabolomics tools. Compared to 1H NMR metabolomics, UHPLC-Q-Orbitrap-HRMS metabolomics is more effective to reveal the overall rat plasma metabolome modifications caused by PPCP or PPAP and identify the contributing makers. Discriminant metabolites including p-hydroxybenzoic acid, phenol, phenol-sulfate, catechol sulphate, 3, 4-dihydroxyphenylvaleric acid, 4′-O-methyl-(−)-epicatechin-3′-O-beta-glucuronide were significantly higher in rat plasma after PPCP intake. On the contrary, plasma level of several metabolites including O-methyl-(−)-epicatechin-O-glucuronide, 4-hydroxy-5-(hydroxyphenyl)-valeric acid-O-sulphate, 5-(hydroxyphenyl)-γ-valerolactone-O-sulphate, peonidin-3-O-hexose and 4-hydroxydiphenylamine were increased after female rats were gavaged with PPAP. The results obtained in this study highlight the importance of using an untargeted approach to reveal the metabolic impact of diet intervention. Development and complement of food metabolome database will be needed to obtain a full identification of biomarkers. Biochemical pathway alteration due to cranberry procyanidins intervention will also need to be elucidated in the future work.
Supplementary Material
Acknowledgments
NMR metabolomics data were collected at the Southeast Center for Integrated Metabolomics at the University of Florida (NIH/NIDDK 1U24DK097209-01A1) using the National High Magnetic Field Laboratory’s AMRIS Facility, which is supported by National Science Foundation Cooperative Agreement No. DMR-1157490 and the State of Florida. UHPLC-HRMS metabolomics data were collected at the Southeast Center for Integrated Metabolomics at the University of Florida. The authors thank Sandi Sternberg and Ramadan Ajredini for their help in sample preparation. Special thanks to Dr. Arthur S. Edison for facilitating the NMR experiment.
List of abbreviations
- CPMG
Carr-Purcell-Meiboom-Gill
- DP
degree of polymerization
- FT
Fourier transformed
- HESI
heated electrospray ionization
- HRMS
high resolution mass spectrometer
- OPLS-DA
orthogonal projection on latent structure-discriminant analysis
- PCA
principal component analysis
- PLS-DA
projection on latent structure-discriminant analysis
- PPCP
partially purified cranberry procyanidins
- PPAP
partially purified apple procyanidins
- QC
quality control
- VIP
variable importance to projection
Footnotes
The authors have declared no conflict of interest.
References
- 1.Gu L, Kelm MA, Hammerstone JF, Beecher G, et al. Screening of foods containing proanthocyanidins and their structural characterization using LC-MS/MS and thiolytic degradation. J Agric Food Chem. 2003;51:7513–7521. doi: 10.1021/jf034815d. [DOI] [PubMed] [Google Scholar]
- 2.Ou K, Gu L. Absorption and metabolism of proanthocyanidins. J Funct Foods. 2014;7:43–53. [Google Scholar]
- 3.Feliciano RP, Krueger CG, Shanmuganayagam D, Vestling MM, et al. Deconvolution of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry isotope patterns to determine ratios of A-type to B-type interflavan bonds in cranberry proanthocyanidins. Food Chem. 2012;135:1485–1493. doi: 10.1016/j.foodchem.2012.05.102. [DOI] [PubMed] [Google Scholar]
- 4.Howell AB, Reed JD, Krueger CG, Winterbottom R, et al. A-type cranberry proanthocyanidins and uropathogenic bacterial anti-adhesion activity. Phytochemistry. 2005;66:2281–2291. doi: 10.1016/j.phytochem.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 5.Vasileiou I, Katsargyris A, Theocharis S, Giaginis C. Current clinical status on the preventive effects of cranberry consumption against urinary tract infections. Nutr Res. 2013;33:595–607. doi: 10.1016/j.nutres.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 6.Lindon JC, Holmes E, Nicholson JK. Metabonomics techniques and applications to pharmaceutical research & development. Pharm Res. 2006;23:1075–1088. doi: 10.1007/s11095-006-0025-z. [DOI] [PubMed] [Google Scholar]
- 7.Griffin JL. The Cinderella story of metabolic profiling: does metabolomics get to go to the functional genomics ball? Philos Trans R Soc Lond, B, Biol Sci. 2006;361:147–161. doi: 10.1098/rstb.2005.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn WB, Broadhurst DI, Atherton HJ, Goodacre R, et al. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev. 2011;40:387–426. doi: 10.1039/b906712b. [DOI] [PubMed] [Google Scholar]
- 9.Beckonert O, Keun HC, Ebbels TM, Bundy J, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 10.Fonville JM, Richards SE, Barton RH, Boulange CL, et al. The evolution of partial least squares models and related chemometric approaches in metabonomics and metabolic phenotyping. J Chemometrics. 2010;24:636–649. [Google Scholar]
- 11.Stoupi S, Williamson G, Viton F, Barron D, et al. In vivo bioavailability, absorption, excretion, and pharmacokinetics of [14C] procyanidin B2 in male rats. Drug Metab Dispos. 2010;38:287–291. doi: 10.1124/dmd.109.030304. [DOI] [PubMed] [Google Scholar]
- 12.Delaglio F, Grzesiek S, Vuister GW, Zhu G, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 13.Chambers MC, Maclean B, Burke R, Amodei D, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pluskal T, Castillo S, Villar-Briones A, Orešič M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brindle JT, Antti H, Holmes E, Tranter G, et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nat Med. 2002;8:1439–1445. doi: 10.1038/nm1202-802. [DOI] [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995:289–300. [Google Scholar]
- 17.Gika HG, Theodoridis GA, Wingate JE, Wilson ID. Within-day reproducibility of an HPLC-MS-based method for metabonomic analysis: application to human urine. J Proteome Res. 2007;6:3291–3303. doi: 10.1021/pr070183p. [DOI] [PubMed] [Google Scholar]
- 18.Burton L, Ivosev G, Tate S, Impey G, et al. Instrumental and experimental effects in LC–MS-based metabolomics. J Chromatogr B. 2008;871:227–235. doi: 10.1016/j.jchromb.2008.04.044. [DOI] [PubMed] [Google Scholar]
- 19.Llorach R, Urpi-Sarda M, Jauregui O, Monagas M, et al. An LC-MS-based metabolomics approach for exploring urinary metabolome modifications after cocoa consumption. J Proteome Res. 2009;8:5060–5068. doi: 10.1021/pr900470a. [DOI] [PubMed] [Google Scholar]
- 20.Kemsley EK, Le Gall G, Dainty JR, Watson AD, et al. Multivariate techniques and their application in nutrition: a metabolomics case study. Br J Nutr. 2007;98:1–14. doi: 10.1017/S0007114507685365. [DOI] [PubMed] [Google Scholar]
- 21.Hawkins DM, Basak SC, Mills D. Assessing model fit by cross-validation. J Chem Inf Comput Sci. 2003;43:579–586. doi: 10.1021/ci025626i. [DOI] [PubMed] [Google Scholar]
- 22.Liu H, Tayyari F, Khoo C, Gu L. A 1 H NMR-based approach to investigate metabolomic differences in the plasma and urine of young women after cranberry juice or apple juice consumption. J Funct Foods. 2015;14:76–86. [Google Scholar]
- 23.Kang J, Choi MY, Kang S, Kwon HN, et al. Application of a 1H nuclear magnetic resonance (NMR) metabolomics approach combined with orthogonal projections to latent structure-discriminant analysis as an efficient tool for discriminating between Korean and Chinese herbal medicines. J Agric Food Chem. 2008;56:11589–11595. doi: 10.1021/jf802088a. [DOI] [PubMed] [Google Scholar]
- 24.Westerhuis JA, Hoefsloot HC, Smit S, Vis DJ, et al. Assessment of PLSDA cross validation. Metabolomics. 2008;4:81–89. [Google Scholar]
- 25.Eriksson L, Byrne T, Johansson E, Wikström C. Multi-and megavariate data analysis. MKS Umetrics AB; Umeå: 2006. [Google Scholar]
- 26.Graham SF, Holscher C, Green BD. Metabolic signatures of human Alzheimer’s disease (AD): 1H NMR analysis of the polar metabolome of post-mortem brain tissue. Metabolomics. 2014;10:744–753. [Google Scholar]
- 27.Wiklund S, Johansson E, Sjöström L, Mellerowicz EJ, et al. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem. 2008;80:115–122. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- 28.Wishart DS, Tzur D, Knox C, Eisner R, et al. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neveu V, Perez-Jimenez J, Vos F, Crespy V, et al. Phenol-Explorer: an online comprehensive database on polyphenol contents in foods. Database. 2010 doi: 10.1093/database/bap024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen PX, Bozzo GG, Freixas-Coutin JA, Marcone MF, et al. Free and conjugated phenolic compounds and their antioxidant activities in regular and non-darkening cranberry bean (Phaseolus vulgaris L.) seed coats. J Funct Foods. 2014 doi: 10.1016/j.jff.2014.10.032. [DOI] [Google Scholar]
- 31.Törrönen R, McDougall GJ, Dobson G, Stewart D, et al. Fortification of blackcurrant juice with crowberry: Impact on polyphenol composition, urinary phenolic metabolites, and postprandial glycemic response in healthy subjects. J Funct Foods. 2012;4:746–756. [Google Scholar]
- 32.Natsume M, Osakabe N, Oyama M, Sasaki M, et al. Structures of (−)-epicatechin glucuronide identified from plasma and urine after oral ingestion of (−)-epicatechin: differences between human and rat. Free Radical Biol Med. 2003;34:840–849. doi: 10.1016/s0891-5849(02)01434-x. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Song TT, Cunnick JE, Murphy PA, et al. Daidzein and genistein glucuronides in vitro are weakly estrogenic and activate human natural killer cells at nutritionally relevant concentrations. J Nutr. 1999;129:399–405. doi: 10.1093/jn/129.2.399. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Aloy M, Llorach R, Urpi-Sarda M, Jáuregui O, et al. A metabolomics-driven approach to predict cocoa product consumption by designing a multimetabolite biomarker model in free-living subjects from the PREDIMED study. Mol Nutr Food Res. 2014;59:212–220. doi: 10.1002/mnfr.201400434. [DOI] [PubMed] [Google Scholar]
- 35.Rudell DR, Mattheis JP, Fellman JK. Evaluation of diphenylamine derivatives in apple peel using gradient reversed-phase liquid chromatography with ultraviolet–visible absorption and atmospheric pressure chemical ionization mass selective detection. J Chromatogr A. 2005;1081:202–209. doi: 10.1016/j.chroma.2005.05.060. [DOI] [PubMed] [Google Scholar]
- 36.Ichiyanagi T, Shida Y, Rahman MM, Hatano Y, et al. Bioavailability and tissue distribution of anthocyanins in bilberry (Vaccinium myrtillus L.) extract in rats. J Agric Food Chem. 2006;54:6578–6587. doi: 10.1021/jf0602370. [DOI] [PubMed] [Google Scholar]
- 37.Stoupi S, Williamson G, Drynan JW, Barron D, et al. A comparison of the in vitro biotransformation of (−)-epicatechin and procyanidin B2 by human faecal microbiota. Mol Nutr Food Res. 2010;54:747–759. doi: 10.1002/mnfr.200900123. [DOI] [PubMed] [Google Scholar]
- 38.Sánchez-Patán F, Cueva C, Monagas M, Walton GE, et al. Gut microbial catabolism of grape seed flavan-3-ols by human faecal microbiota. Targetted analysis of precursor compounds, intermediate metabolites and end-products. Food Chem. 2012;131:337–347. [Google Scholar]
- 39.Engemann A, Hübner F, Rzeppa S, Humpf HU. Intestinal metabolism of two A-type procyanidins using the pig cecum model: detailed structure elucidation of unknown catabolites with Fourier transform mass spectrometry (FTMS) J Agric Food Chem. 2012;60:749–757. doi: 10.1021/jf203927g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.