Abstract
Metagenomic studies suggest that only a small fraction of viruses existing in nature have been identified and studied. Characterization of unknown viral genomes is hindered by many non-specific genomes populating any virus sample. Here, we report a new platform integrating drop-based microfluidics and computational analysis that enables purification of any single viral species from a complex, mixed virus sample and the retrieval of complete genome sequences. Using this platform, we retrieve the genome sequence of a 5243 bp dsDNA virus that was spiked into wastewater with > 96% sequence coverage and > 99.8% identity. This platform holds great potential for virus discovery as it allows enrichment and sequencing of previously undescribed viruses as well as known viruses.
Keywords: Microfluidics, Microemulsions, Viruses, Genome sequencing, High throughput screening
Viruses are the most abundant biological entities on Earth and significantly impact living organisms by causing diseases and shaping their immune systems. Despite their ubiquity and influence, less than 0.01% of viruses are sequenced[1]. Establishment of an extensive virus database is crucial to identify potential emerging infectious diseases[2] and to improve our understanding of virus diversity, ecology, adaption and evolution. The major roadblock to characterizing unknown viral genomes is the lack of technologies enabling efficient enrichment of various types of viruses. Enrichment of a target viral species is required for the most common virus samples such as environmental samples, which generally harbor diverse viral populations[3], or clinical samples where the amount of viral genomes is often lower than the amount of host genomes and the virions are localized to a small subset of cells in the tissue. An enrichment step is particularly crucial for viral genome sequencing because other abundant DNA in the sample such as genomic fragments of host DNA is often much larger than viral genomes and dominate the sequence space even with a small number of copies. Traditional enrichment methods for viruses include cell culture[4], immunoscreening[5] followed by sequence-independent PCR[6] and differential hybridization[7]. All of these methods are labor-intensive, inefficient and more importantly, only applicable to a limited subset of viruses. Recently, a flow cytometric method was developed to disperse single virions into microwells and obtain their individual genome sequences[8]. However, this method does not employ a selection strategy. A selection strategy allows efficient usage of sequencing power and enables rare virus sequencing with a reasonable sequencing cost and time.
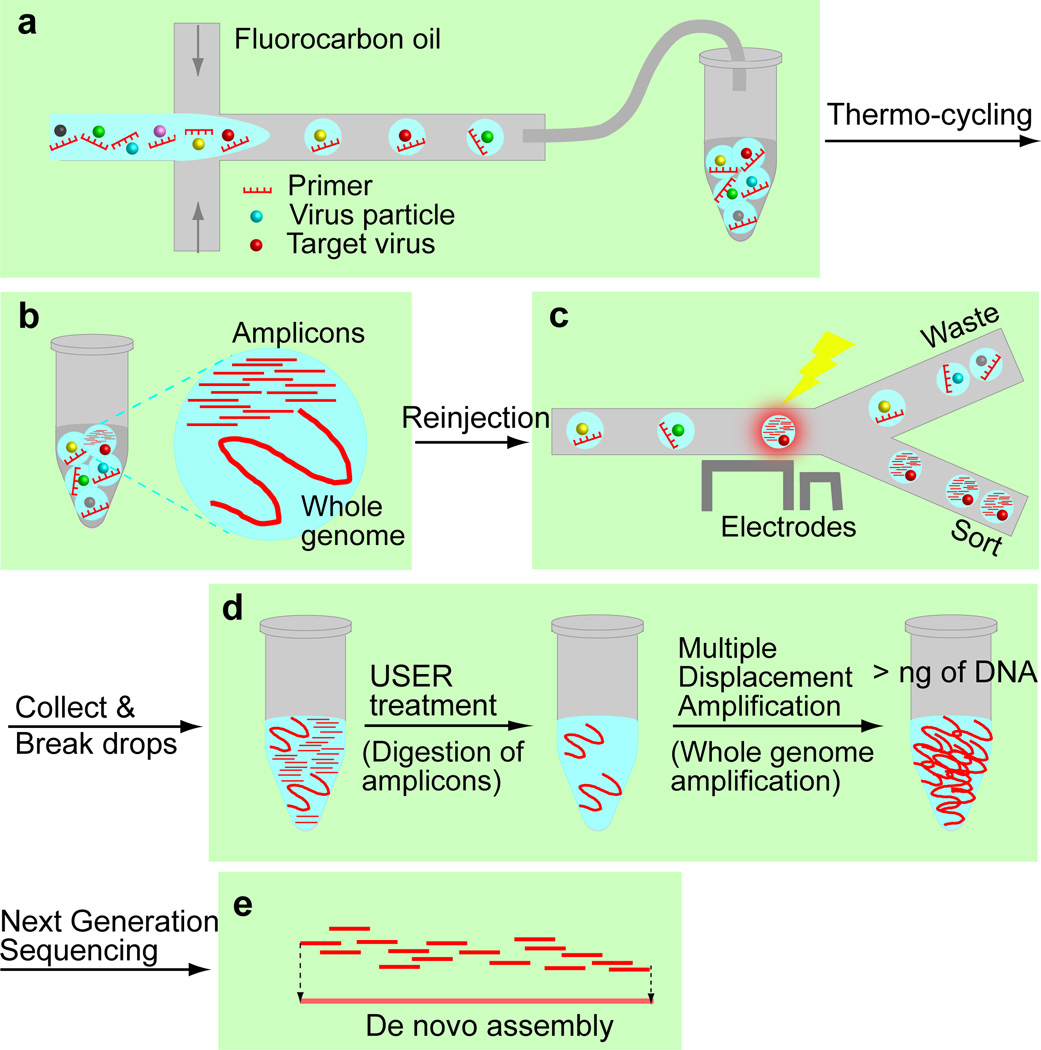
In this paper, we report the development of a platform to isolate and sequence any single viral species from a large genetic space of viral sequences. Our platform integrates drop-based microfluidics for high throughput single virus assays and sorting (Scheme 1a–c), molecular biology techniques for whole genome amplification of the selected viruses (Scheme 1d) and a computational pipeline for de novo assembly of viral genome sequence (Scheme 1e).
Scheme 1.
Identification of viral genomes from an environmental sample. a) Viruses are encapsulated into drops. b) After thermo-cycling, amplicons are generated in the drops containing the target virus. c) Drops are sorted based on their fluorescence intensities. d) The enriched virus solution is treated with USER to digest amplicons followed by whole genome amplification. e) The amplified products are sequenced and assembled using our computational pipeline.
Drop-based microfluidics offers unprecedented advantages, allowing high throughput screening of single cells or viruses with high sensitivity and minimal time[9]. Combining droplet digital PCR (ddPCR) with microfluidic sorting techniques, we selectively enrich viruses of interest from an environmental sample. Using a drop maker, we encapsulate an aqueous mixture containing a virus sample, PCR buffer, primers, dUTP/dATP/dCTP/dGTP and SYBR Green I. Encapsulation is performed so that the majority of drops contain no more than one virion. All virus types can be efficiently encapsulated into drops following the Poisson statistics as the size of viruses, 5 nm-3 µm, is considerably smaller than the size of drops, 25–100 µm, and virions are well-dispersed in wastewater samples (Figure S3). With primers specific to a target virus, amplicons are generated selectively in a drop containing a target virion after thermo-cycling. Primers can be designed for both known viruses and unknown viruses (Figure S4, see supporting information for details). In the PCR mixture, dTTP is replaced with dUTP so that amplicons generated during PCR contain dUTP in the place of dTTP. A dsDNA intercalating dye, SYBR Green I, is added to the mixture to stain amplicons and identify the drops containing a target virion.
After thermo-cycling the drops, we inject them into the microfluidic sorter and select the drops displaying an enhanced fluorescence signal[9c]. The selected virus solution is then treated with uracil-specific excision reagent (USER), which generates a nucleotide gap at the location of a uracil. During USER treatment, amplicons which contain dUTP are selectively digested into small pieces while the viral genomes which do not contain dUTP remain intact. Selective digestion of amplicons is important since they are over-represented in the enriched virus solution and would impede the characterization of the complete viral genome if left undigested.
Multiple displacement amplification (MDA) is performed on the USER treated virus solution to generate more than 1ng of clonal copies, an amount sufficient for DNA sequencing[10]. To suppress amplification of high molecular weight DNA contaminants in commercial MDA reagents[11], we treat the reagents with UV[12] and perform MDA reaction in a reduced reaction volume[13] of 4µL.
The amplified DNA products are sequenced using an Illumina Platform and the sequence reads are assembled to recover the genome sequence of the sorted viruses. A computational pipeline was developed for sequence data cleaning, de novo assembly and selection of the target virus contig. In the pipeline, low quality reads and human reads are removed and the remaining reads are assembled into contigs (CLC assembler, CLC bio). Meta-assembly is then performed using a homebuilt MATLAB code since CLC assembler occasionally yields contigs having 13–50 bp long sequence overlaps. The genome sequence of the enriched viruses is determined by selecting the contig having the highest relative abundance among the high quality contigs that do not originate from known organisms.
Previous metagenomic studies reveal that wastewater harbors diverse viral species including previously undescribed viruses with unknown viral sequences outnumbering known viral sequences by a factor of 10 to 1,000[3]. This indicates that raw sewage is an excellent source for previously undescribed viruses. To assess our platform for sequencing viral genomes from wastewater, we spike a well-characterized virus, SV40, into wastewater, select SV40 virions using the microfluidic platform, sequence the enriched sample and perform de novo assembly. We then verify whether the assembled genome sequence aligns with the SV40 genome and how much of the SV40 genome is covered by this sequence.
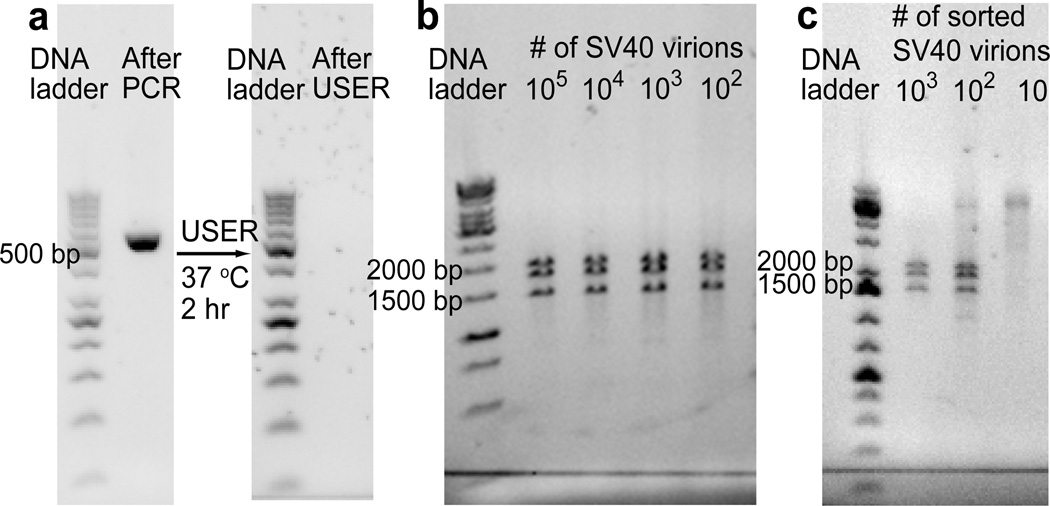
For enrichment of SV40, SV40/wastewater mixtures are encapsulated into 8 pL drops (Figure 1a). After thermo-cycling, a small fraction of the drops displays a high level of fluorescence under excitation at 470 nm indicating the presence of SV40 virions in those drops (Figure 1b). These drops are selected using the microfluidic sorter (Figure 1c). Amplicons in the collected solution are digested with USER (Figure 2a), followed by whole genome amplification using φ29 DNA polymerase. Restriction analysis allows us to quickly examine whether the amplified DNA contains the SV40 genome sequence without sequencing. A restriction enzyme, PvuII, cleaves SV40 at three sites to produce the DNA fragments of 1446, 1790 and 1997 bp (Figure 2b). The digestion test shows that at least 100 PCR positive drops are required to obtain a detectable amount of the SV40 genome sequence after MDA (Figure 2c). With fewer drops, the amplified DNA is not cleaved by PvuII and migrates as large DNA during gel electrophoresis.
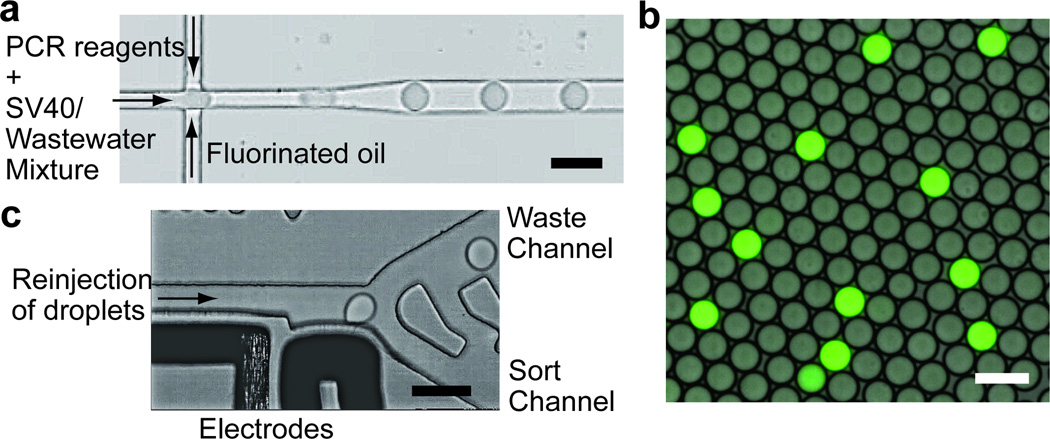
Figure 1.
a) A partial image of the drop-making device. b) A fluorescence image of drops after PCR. The fluorescence signal is from intercalation of SYBR green into amplicons. c) A partial image of the sorting device. Drops are sorted based on their fluorescence intensities. Scale bars = 50 µm.
Figure 2.
a) After PCR, 519 bp amplicons are produced in the drops containing SV40 virions. These amplicons incorporate dUTP and are digested after USER treatment. b, c) Restriction analysis is used to verify successful amplification of SV40 genome. b) After PvuII treatment, the genomic copies of SV40 are cleaved into three fragments of 1446, 1790 and 1997 bp. c) At least 100 PCR positive drops are required to obtain a detectable amount of the SV40 genome sequence by MDA.
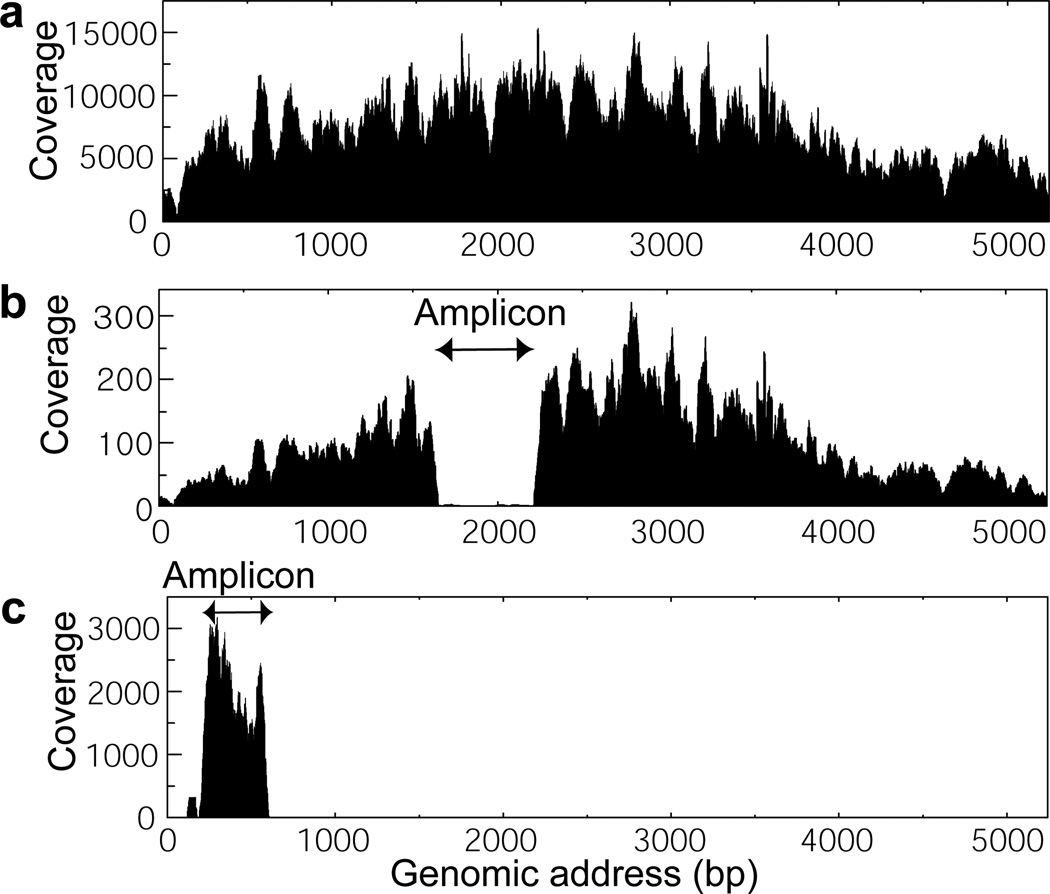
To further evaluate the effectiveness of our purification and amplification method, the amplified DNA is sequenced and analyzed. First, the percentage of SV40 mapped reads to total sequence reads is compared for 3% SV40/wastewater mixture and the selected SV40 samples (Sample 1, Table S1). Without sorting, the amplified sequences contain 0.004% of SV40 reads. The low percentage of SV40 reads is due to high molecular weight DNA in wastewater. During MDA, larger DNA is preferentially amplified over shorter DNA; thus, the sequence of high molecular weight contaminants is over-represented in the amplified product. This issue is particularly prominent and problematic in viral genome sequencing as viral genomes are generally much smaller than other abundant DNA in the sample. The percentage of SV40 reads increases to 94.0% and 98.6% for samples of 100 and 1000 selected drops, respectively. The sequencing results on the selected SV40 samples from other sets of sorting experiments also show high percentages of SV40 reads (Table S1). This confirms that our microfluidic platform effectively enriches target virions from a complex virus mixture. We also verify whether the SV40 reads cover the complete genome of SV40. The reference mapping results show that the complete genome sequence of SV40 is recovered after collecting as little as 100 positive drops (Figure 3a and S5). In two out of eleven samples, a portion of the SV40 genome corresponding to the amplicon sequence is absent (Figure 3b and S5). The absence may be due to the digested amplicon fragments, which can anneal to the SV40 genome and hinder DNA strand extension by φ29. The sequence mapping results highlight the importance of the amplicon digestion step for whole genome amplification. When the sorted virions are amplified without USER treatment, the amplified sequences only cover sequences from amplicons themselves (Figure 3c).
Figure 3.
a) A representative sequence coverage graph of the enriched SV40 samples. The sequence reads cover the entire SV40 genome. b) A sequence coverage graph of the sample lacking a large portion of the amplicon sequence (1761–2259). c) A sequence coverage graph of the sample where 259 bp (237–495) amplicons are not digested before MDA. Without USER treatment, only amplicon sequences are recovered after sequencing.
The sequence reads are assembled into contigs using our computational pipeline. The contigs are analyzed by a BLAST search to identify their origin and sequence homology to the reference genome. In all cases, the contig with the highest relative abundance aligns with the SV40 genome (Table S2). When the sequence reads cover the complete SV40 genome, the generated SV40 contigs cover > 97% of the genome sequence (Figure 3a and Table 1). The missing sequences are in the 72 bp repeat region of the SV40 genome. In our study, sequencing is performed with 50 bp single end read, which makes it challenging to distinguish repeating sequences larger than 50 bp. Therefore, the sequence coverage can be further improved by performing paired-end sequencing with longer reads. We obtain SV40 contigs that cover > 96% of the genome sequence even from the samples lacking a large portion of the amplicon sequence (Figure 3b and Table 1). To correct the absence of the amplicon sequence in these contigs, we characterized the amplicon sequence by performing Sanger sequencing on the sorted drops and manually inserted that sequence based on the sequence overlap. Our assembly technique is highly robust towards contamination. The samples amplified with the MDA reagents that are incompletely decontaminated have only 8.9% and 0.74% SV40 reads; however, the SV40 contigs from these samples cover > 95.8% of the genome with 100% sequence identity (Table 1). Sequence analysis on non-SV40 reads shows that over 99% of the contaminant sequences are from human, bacteria, and mouse, which are common contaminant sources in the lab environments (Figure S6). For de novo assembly of unknown viral genomes, raw sequencing data will be computationally cleaned by removing the common contaminant sequences. The high purity sequence reads will ensure accurate de novo assembly. Our assembly result indicates that a cheaper sequencing option can also be used for sequencing. De novo assembly was successful when the sequence coverage was higher than 50 (Table S1). Considering that the size of viral genomes is a few kb to hundreds of kb, Ion Torrent Personal Genome Machine, which reads 10 million bases per run, can be used for sequencing a small number of virus samples.
Table 1.
Summary of the SV40 contigs generated from the representative enriched virus samples
| Sample | # of sorted drops |
SV40 reads /Total reads (%) |
Contig length (bp) |
Sequence Coverage (%) |
Sequence Identity (bp) |
Missing Sequencec (locus) |
|---|---|---|---|---|---|---|
| 1-1a | 100 | 94.0 | 5066 | 96.6 | 5066 | 124–207 1648–1740 |
| 1-2 | 1000 | 98.6 | 5162 | 98.5 | 5162 | 125–205 |
| 2-1 | 1000 | 95.8 | 5134 | 97.9 | 5134 | 124–232 |
| 2-2 | 5000 | 98.8 | 5187 | 98.9 | 5186 | 151–207 |
| 3-1b | 100 | 8.9 | 5159 | 98.4 | 5158 | 124–207 |
| 3-2a,b | 1000 | 0.74 | 5016 | 95.7 | 5016 | 99–232 1648–1740 |
The sequence reads from these samples lack a large portion of the amplicon sequence (1761–2259 bp).
MDA reagents used for these samples are incompletely decontaminated.
Two 72 bp repeats reside in 107–250 bp.
In summary, using the platform, we retrieved > 97% of the target genome sequence after collecting as little as 100 virions. The total mass of DNA in 100 SV40 virions is 5×10−16 g. This value is orders of magnitude less than the amount of DNA in a single bacterium, which has the smallest genome among cells used for single cell sequencing.
Even though the platform is developed for DNA viruses, RNA viruses can also be enriched and sequenced. For RNA virus sequencing, the cDNA copies of genomes are encapsulated into drops instead of the genome itself. The genomic cDNA copies are synthesized by reverse-transcription with either oligo dT or random hexamers. We synthesized the genome cDNA copies of norovirus and show that various segments of the genome can be amplified from these cDNA copies (Table S4 and Figure S7). The genomic cDNA copies are then encapsulated into drops. After thermo-cycling of drops, a few drops display a high level of fluorescence under excitation at 470 nm (Figure S8). The PCR positive drops can be selected and processed following the same protocol as for the DNA viruses.
Lastly, we demonstrate the potential of our platform for unknown virus sequencing by designing primers specific to a previously undescribed virus related to human rhinovirus and performing ddPCR. From metagenomic analysis on a wastewater sample[3], we identified contigs showing 50–65% sequence homology to human rhinovirus. We designed primers for each contig (Table S5) and performed PCR on the reverse-transcribed wastewater sample. The size of amplicons generated from PCR agrees with the size predicted from the contig sequences (Figure S9). This result confirms the validity of our method for designing primers for unknown viruses. We then performed ddPCR with the designed primers and show that a small fraction of drops display high level of fluorescence signal under excitation at 470 nm (Figure S10). These PCR positive drops can be selected using our microfluidic sorter and the target viral genome can be sequenced following the same protocol as for the known viruses.
Our platform enables efficient isolation of single viral species from a mixture of other viruses and DNA contaminants. This eliminates the requirement for cell culture to prepare a homogeneous virus solution and allows genome sequencing of uncultivable viruses from an environmental sample. Therefore, our platform holds great potential for virus discovery and the establishment of an extensive genomic database. A comprehensive virus database will facilitate interpretation of metagenomic data by providing reference genomes, lead to a better understanding of virus diversity, ecology, adaptation and evolution, and enable the prediction of emerging infectious diseases caused by viruses. Our work represents an important step towards exploration of the viral universe. We expect that extensions of this work, enabling faster sorting to isolate rare viruses and sequencing of individual viruses, will further enhance the power of this new platform.
Supplementary Material
Acknowledgments
The research work of the authors is supported by the U.S. National Institute of Health grants, R21-AI101291 (D.A.W. and J. M.P.) and by Defense Advanced Research Projects Agency, HR0011-11-C-0093 (D.A.W. and J.M.P.).
Contributor Information
Dr. Hee-Sun Han, Department of Physics, School of Engineering and Applied Sciences, Harvard University, Cambridge, MA 02138, USA
Paul G. Cantalupo, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA 15260, USA
Dr. Assaf Rotem, Department of Physics, School of Engineering and Applied Sciences, Harvard University, Cambridge, MA 02138, USA
Dr. Shelley K. Cockrell, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA 15260, USA
Martial Carbonnaux, Department of Physics, School of Engineering and Applied Sciences, Harvard University, Cambridge, MA 02138, USA.
Prof. James M Pipas, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA 15260, USA
Prof. David A Weitz, Department of Physics, School of Engineering and Applied Sciences, Harvard University, Cambridge, MA 02138, USA.
References
- 1.Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, Zambrana-Torrelio CM, Solovyov A, Ojeda-Flores R, Arrigo NC, Islam A, Ali Khan S, Hosseini P, Bogich TL, Olival KJ, Sanchez-Leon MD, Karesh WB, Goldstein T, Luby SP, Morse SS, Mazet JAK, Daszak P, Lipkin WI. mBio. 2013;4:e00598–e00513. doi: 10.1128/mBio.00598-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantalupo PG, Calgua B, Zhao G, Hundesa A, Wier AD, Katz JP, Grabe M, Hendrix RW, Girones R, Wang D, Pipas JM. mBio. 2011;2:e00180–e00111. doi: 10.1128/mBio.00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leland DS, Ginocchio CC. Clin. Microbiol. Rev. 2007;20:49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choo Q, Kuo G, Weiner A, Overby L, Bradley D, Houghton M. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 6.a) Reyes GR, Kim JP. Mol. Cell. Probes. 1991;5:473–481. doi: 10.1016/s0890-8508(05)80020-9. [DOI] [PubMed] [Google Scholar]; b) Froussard P. Nucleic Acids Res. 1992;20:2900. doi: 10.1093/nar/20.11.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lisitsyn N, Lisitsyn N, Wigler M. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 8.Allen LZ, Ishoey T, Novotny MA, McLean JS, Lasken RS, Williamson SJ. PLoS One. 2011;6:e17722. doi: 10.1371/journal.pone.0017722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Proc. Natl. Acad. Sci. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret J-C, Marquez M, Klibanov AM, Griffiths AD, Weitz DA. Proc. Natl. Acad. Sci. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Mazutis L, Gilbert J, Ung WL, Weitz DA, Griffiths AD, Heyman JA. Nat. Protoc. 2013;8:870–891. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wang BL, Ghaderi A, Zhou H, Agresti J, Weitz DA, Fink GR, Stephanopoulos G. Nat. Biotechnol. 2014;32:473–478. doi: 10.1038/nbt.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW, Church GM. Nat. Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]; b) Marcy Y, Ishoey T, Lasken RS, Stockwell TB, Walenz BP, Halpern AL, Beeson KY, Goldberg SMD, Quake SR. PLoS Genet. 2007;3:e155. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Rodrigue S, Malmstrom RR, Berlin AM, Birren BW, Henn MR, Chisholm SW. PLoS One. 2009;4:e6864. doi: 10.1371/journal.pone.0006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blainey PC, Quake SR. Nucleic Acids Res. 2011;39:e19. doi: 10.1093/nar/gkq1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woyke T, Sczyrba A, Lee J, Rinke C, Tighe D, Clingenpeel S, Malmstrom R, Stepanauskas R, Cheng J-F. PLoS One. 2011;6:e26161. doi: 10.1371/journal.pone.0026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutchison CA, Smith HO, Pfannkoch C, Venter JC. Proc. Natl. Acad. Sci. 2005;102:17332–17336. doi: 10.1073/pnas.0508809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.