Abstract
Background
Establishment and maintenance of cell polarity is critical for normal embryonic development. Previously, it was thought that the echinoderm embryo remained relatively unpolarized until the first asymmetric division at the 16 cell stage. Here we analyzed roles of the cell polarity regulators, the PAR complex proteins, and how their disruption in early development affects later developmental milestones.
Results
We found that PAR6, aPKC and CDC42 localize to the apical cortex as early as the 2 cell stage and that this localization is retained through the gastrula stage. Interestingly, PAR1 also colocalizes with these apical markers through the gastrula stage. Additionally, PAR1 was found to be in complex with aPKC, but not PAR6. PAR6, aPKC, and CDC42 are anchored in the cortical actin cytoskeleton by assembled myosin. Furthermore, assembled myosin was found to be necessary to maintain proper PAR6 localization through subsequent cleavage divisions. Interference with myosin assembly prevented the embryos from reaching the blastula stage, while transient disruptions of either actin or microtubules did not have this effect.
Conclusions
These observations suggest that disruptions of the polarity in the early embryo can have a significant impact on the ability of the embryo to reach later critical stages in development.
Keywords: Deuterostome, polarity, PAR, myosin, actin, sea urchin
Introduction
Polarization of a developing embryo is critical to ensure proper segregation of developmental determinants throughout embryogenesis. Since their initial discovery in the C. elegans embryo, the PAR proteins have been found to regulate polarity in a wide variety of multicellular eukaryotes (Kemphues et al., 1988; Morton et al., 1992; Etemad-Moghadam et al., 1995; Guo and Kemphues 1995; Watts et al., 1996; Tabuse et al., 1998; Hung and Kemphues, 1999; Morton et al., 2002; Hao et al., 2006). They have now been extensively studied for their roles in the polarization of embryos, neurons, epithelial cells, stem cells and in some types of cancer (McCaffrey and Macara, 2011; Nance and Zallen, 2011; Ellenbroek et al., 2012; Lalli, 2012; Chen and Zhang, 2013).
The PAR proteins consist of core group of signaling proteins that cooperatively function to control polarity. PAR1 and PAR4 are serine-threonine kinases (Guo and Kemphues, 1995; Watts et al., 2000; McCaffrey and Macara, 2009). PAR2 is a nematode specific PAR protein; it is a RING finger domain protein that functions as an E3 ubiquitin ligase (Levitan et al., 1994; Boyd et al., 1996). PAR3 and PAR6 are both PDZ domain proteins that act as scaffolding proteins for the other PAR proteins as well as for other polarity regulators, such as aPKC (Etemad-Moghadam et al., 1995; Watts et al., 1996; Hung and Kemphues, 1999; McCaffrey and Macara, 2009). Lastly, PAR5 is a 14-3-3 protein that is recruited to phosphorylated serine and threonine residues (Morton et al., 2002). In addition, the kinase aPKC and the GTPase CDC42 have been found to play a significant role in the establishment and maintenance of polarity. PAR3, PAR6, and aPKC are known to interact in what is referred to as the PAR complex (Joberty et al., 2000; Lin et al., 2000; McCaffrey and Macara, 2009). CDC42 was later found to act upstream of the PAR complex through its interaction with PAR6 (Joberty et al., 2000; Lin et al., 2000; McCaffrey and Macara, 2009). aPKC is known to inhibit PAR6 activity, but through its association with CDC42, this repression is partially relieved (Goldstein and Macara, 2007). CDC42 has also been shown to be necessary for the proper localization of the PAR complex in the cell cortex and is well known regulator of the actin cytoskeleton (Goldstein and Macara, 2007; Iden and Collard, 2008).
Following fertilization PAR1 and PAR2 become enriched at the posterior cortex, while PAR3 and PAR6 are enriched in the anterior cortex in C. elegans embryos (Kemphues, 2000). Localization of the posterior proteins is necessary for the exclusion of the anterior proteins and vice versa (Kemphues, 2000). These mutual exclusion events ensure proper segregation of the PAR proteins and help to maintain the polarized domains found in the C. elegans embryo and are well conserved among other eukaryotes (Benton and Johnston, 2003; Suzuki et al., 2004). The acto-myosin cortex also plays a key role in maintaining the segregation of the anterior PAR proteins (Cowan and Hyman, 2004; Munro et al., 2004; Munro, 2006). Once the asymmetry of these proteins has been established, the PAR proteins help to coordinate the localization of the mitotic spindle and subsequent asymmetric division (Ahringer, 2003; Hao et al., 2010; Galli et al., 2011).
The PAR proteins have been found to play a conserved role in polarity during development across a multitude of species including Drosophila, Xenopus, mice, and humans (McCaffrey and Macara, 2009; McCaffrey and Macara, 2012). For example, the PAR proteins regulate neuroblast polarity as well as adheren junctions during gastrulation in Drosophila (Atwood et al., 2007; Harris and Peifer, 2007). In mouse embryos PAR3 and aPKC become polarized at the 8 cell stage of development and help to regulate cell divisions, while PAR6B plays a role in trophoectoderm formation and aPKC is involved in endoderm maturation in later development (Plusa et al., 2005; Vinot et al., 2005; Alarcon, 2010; Saiz et al., 2013). Additionally in Xenopus, PAR1 along with aPKC and 14-3-3 protein are critical for successful gastrulation (Kusakabe and Nishida, 2004; Hyodo-Miura et al., 2006). Other studies have also shown that aPKC is involved in polarized cell divisions during the blastula stage of development in Xenopus (Chalmers et al., 2003; Chalmers et al., 2005). Further work has shown that the apical localization of the PAR complex proteins are required for the asymmetric division at the 8 cell stage in ascidian embryos (Patalano et al., 2006). These studies in deuterostome embryos have mainly focused on later developmental milestones. However, unlike in protostome embryos, the role of the PAR proteins in the earliest cleavage stages and the resultant polarity has not been extensively studied in deuterostomes.
The sea urchin embryo has long been used as model system of deuterostome development (Ernst, 2011). These embryos were previously thought to remain relatively unpolarized until at least the 16 cell stage of development, the first time that they undergo asymmetric division. However, there is now mounting evidence to suggest that these embryos polarize earlier than originally thought. For example, lectin receptors have been shown to have an apical surface localization following the first cleavage (McCaig and Robinson, 1982). Schroeder also demonstrated that even at the 2 cell stage the apical surface is enriched in microvilli, whereas regions of cell-cell contact are relatively free of these actin-based protrusions even upon dissociation (Schroeder, 1988). Additionally, cadherin polarizes to new sites of cell-cell contact at the first division (Miller and McClay, 1997). More recently Burke and colleagues have shown that βC integrins are found only on the outer surface that is exposed to the extracellular matrix as early as the 2 cell stage of development and that these integrins associate with focal adhesion kinase, which is necessary for proper cortex formation (Burke et al., 2004; Chan et al., 2013). Lipid rafts are additionally polarized to the free cell surface following the first cleavage in sea urchin embryos (Ng et al., 2005; Alford et al., 2009). Developmental determinants such as dishevelled have also been found to have polarized localization in the vegetal cortex in the unfertilized egg and fertilized zygote (Weitzel et al., 2004; Leonard and Ettensohn, 2007; Peng and Wikramanayake, 2013).
Although the PAR proteins have been examined across a wide variety of species, it was not until recently that they were explored in sea urchins (Shiomi and Yamaguchi, 2008; Alford et al., 2009; Pruliere et al., 2011; Shiomi et al., 2012). PAR6, aPKC, and CDC42 all localize to the apical cortex as early as the 2 cell stage of development in these embryos and this apical polarity was found to be functional, as endocytosis was only observed on the apical surface even upon dissociation of the blastomeres (Alford, et al. 2009).
In this study we further examine the role of the PAR proteins during early development of the sea urchin embryo. Following first cleavage, the apical localization of the PAR complex proteins is retained through the gastrula stage. In addition, unlike in other systems, PAR1 colocalizes with these apical PAR proteins at the free cell surface (apical) through the gastrula stage. Furthermore, PAR1 is found in complex with aPKC but not with PAR6 during these developmental stages. We find that these proteins are anchored in the apical cortex specifically by assembled myosin, which likely organizes the actin cortex, and that perturbation in its localization during early cleavage stages severely impedes later development. These data demonstrate the importance of early polarity establishment in deuterostome embryos and the significant functional impact it has on later developmental events.
Results
The PAR complex proteins along with PAR1 localize to the apical cortex through the gastrula stage
Previous studies have shown that PAR6, aPKC, and CDC42 localize to the apical cortex as early as the 2 cell stage of development in the sea urchin embryo (Alford et al., 2009). These observations suggest that the early polarization of these proteins may be important for the proper development of the sea urchin embryo, similar to their role in other model systems (Nance and Zallen, 2011). In order to investigate further, we first examined the localization pattern of PAR6 in live embryos expressing PAR6-Venus encoded by microinjected PAR6-Ven mRNA. PAR6-Ven protein expression detected after the first few divisions and was found predominantly at the apical surface at the 16 cell stage and at the blastula and gastrula stages of development; there also was a downregulation of PAR6-Ven expression in the archenteron at the gastrula stage (Fig. 1A). We also detected PAR6-Ven in apical puncta and in confocal slices that were imaged at the apical surface encircling junctional complexes of blastula and gastrula epithelia (Fig. 1B, C). Immunofluoresence assays were then utilized in order to observe the localization pattern of PAR6 along with aPKC and CDC42 at early cleavage stages and in embryos at a higher resolution and for quantitative purposes.
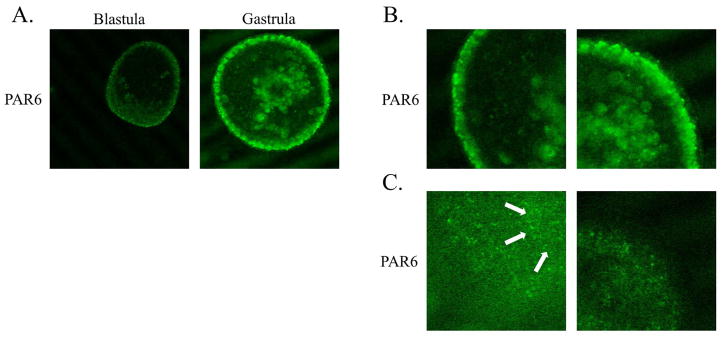
Figure 1.
PAR6 localizes to the apical cell surface in live blastula and gastrula stage embryos. P. lividus embryos were injected with PAR6-Ven mRNA prior to fertilization and PAR6-Ven protein expression and localization was analyzed at blastula and gastrula stages of development by confocal microscopy. PAR6-Ven was found at the apical cell surface at both stages of development (A). In addition, PAR6-Ven was diminished in amount in the archenteron of gastrula stage embryos (A). PAR6-Ven expression can be clearly seen in junctions in zoomed in images of gastrula stage embryos (B) and in z slices from the top of gastrula stage embryos (C). Additionally, arrows highlight PAR6-Ven localization to the apical junctional complex that encircles the epithelium at the apical surface (C).
The localization of endogenous PAR6 by immunofluoresence was identical to that found in living embryos expressing PAR6-Ven (Fig. 1, 3, 4). We found that PAR6, aPKC, and CDC42 remain localized to the apical cortex from the 2 cell stage through the 16 cell stage of development, the first asymmetric division in sea urchins (Fig. 3A, B). As aPKC was recently found to be involved in ciliogenesis in the sea urchin embryo (Pruliere et al., 2011), we also determined the localization of PAR6, aPKC, and CDC42 at the blastula stage of development. At the blastula stage, in addition to the presence of cilia, embryos have formed an epithelium and apical cell-cell junctions are present. We found that all these polarity proteins remain localized to the apical or free cell surface at the blastula stage (Fig. 3C, D). Additionally, these endogenous proteins can be observed in the cilia in the immunofluorescence experiments similar to the aPKC expression that was seen previously (Pruliere et al., 2011) (Fig. 3C, D). However, PAR6-Ven was not found in cilia (Fig. 1).
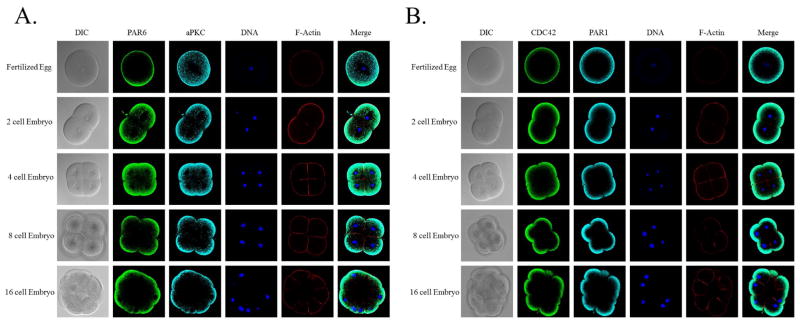
Figure 3.
PAR1 colocalizes with the PAR complex in the apical cortex through the blastula stage. L. pictus embryos were fixed and then stained for endogenous PAR6 and aPKC (A and C) or CDC42 and PAR1 (B and D). PAR6, aPKC, and CDC42 maintain the apical localization pattern initially seen at the 2 cell stage through the 16 cell stage (A and B). At these early cleavage divisions PAR1 also colocalizes with these known apical markers (B). This unique colocalization at the apical surface is further retained at the blastula stage after an epithelium has formed and junctions are present (C and D).
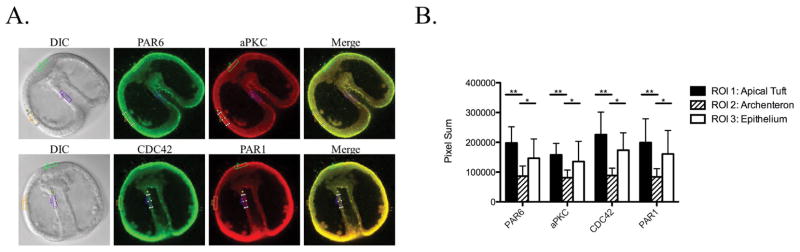
Figure 4.
The PAR complex along with PAR1 remain enriched in the apical epithelium of the ectoderm and are down regulated in the archenteron at the gastrula stage. L. pictus embryos were raised to the gastrula stage and then fixed and stained for endogenous PAR6, aPKC, CDC42, and PAR1 (A). These endogenous proteins were all found to have a significant decrease in expression in the archenteron (ROI 2) as compared to both the apical tuft (ROI 1) similar to what was seen for expression of PAR6-Ven in living embryos (Fig. 1) (**p≤ 0.001) and the surrounding epithelium (ROI 3) (*p≤ 0.05) (B). The fluorescence intensity (pixel sum in arbitrary units (AU)) of PAR6 was 197360.4±54422.1 in the apical tuft, 86450.5±34162.4 in the archenteron, and 146407.6±64839.5 in the epithelium (n=10 embryos, mean±SD). The fluorescence intensity of aPKC was 157470.9±38619.6 in the apical tuft, 81117.3±25727.6 in the archenteron, and 135381.9±67839.2 in the epithelium (n=10 embryos, mean±SD). The fluorescence intensity of CDC42 was 225433.3±75871.2 in the apical tuft, 88369.8±24944.3 in the archenteron, and 173483.6±58318.1 in the epithelium (n=10 embryos, mean±SD). The fluorescence intensity of PAR1 was 198961.5±79431.1 in the apical tuft, 83908.9±27518.6 in the archenteron, and 160788.5±78914.1 in the epithelium (n=10 embryos, mean±SD).
Having examined the localization of the PAR complex proteins and their upstream regulator, CDC42, we were interested in studying the localization pattern of a traditionally posterior polarity regulator, PAR1, in order to determine how it polarized in the sea urchin embryo (Fig. 3B). Using the predicted PAR1 sequence, we created a specific antibody against the S. purpuratus PAR1 protein (Cameron et al., 2009) that was found to identify the PAR1 protein at the correct molecular weight of 81.5 kD as well as a PAR1 dimer at 163 kD (Panneerselvam et al., 2006) (Fig. 2). Based on its annotation in the S, purpuratus genome, the predicted PAR1 protein contains the three conserved domains of PAR1 proteins: the kinase domain, the kinase associated domain, and the ubiquitin associated domain (Cameron et al., 2009). Immunofluoresence assays revealed that during early cleavage stages, PAR1 colocalizes with the PAR complex proteins in the apical cortex through the 8 cell stage (Fig. 3B). Furthermore, PAR1 retains this colocalization with the apical PAR complex at the 16 cell stage, after the first asymmetric division, and at the blastula stage, after junctions have formed within the epithelium (Fig. 3B, D). As actin staining can be clearly observed in both the apical and basolateral cortex in early cleavage stage embryos, this demonstrates that there is not a permeability issue during fixation; therefore, the PAR1 localization pattern that is observed in the apical cortex is not due to antibody exclusion from the basolateral cortex.
Figure 2.
Anti-PAR1 Antibody Design and Verification. A) The GLEAN prediction Sp-Mark3 amino acid sequence of Strongylocentrotus purpuratus. The antigen sequence is highlighted. This antigenic region was chosen because it was found specifically outside of the three conserved domains of the PAR1 protein: the kinase domain, the kinase associated domain, and the ubiquitin associated domain (Cameron et al., 2009). Commercial antibodies against these domains were found to be nonspecific. This region was additionally selected because it was conserved between both S. purpuratus and H. pulcherrimus (Cameron et al., 2009; Shiomi et al., 2008). B) Western blot of L. pictus extracts with the purified polyclonal rabbit anti-S. purpuratus PAR1 antibody.
In order to determine if PAR1 polarize to a distinctive region later in development in the sea urchin embryo, we also analyzed its localization pattern at the gastrula stage. mRNA transcripts of PAR1 are known to be expressed throughout embryogenesis, but appear to become more vegetal in localization following hatching from the fertilization envelope in the sea urchin species, Hemicentrotus pulcherrimus (Shiomi and Yamaguchi, 2008). During the gastrula stage the embryo is undergoing massive cellular rearrangements to form the three primary germ layers, the endoderm, mesoderm, and ectoderm. We find that PAR6, aPKC, CDC42, and PAR1 retain their localization to the apical surface at this stage (Fig. 4A). However, as was found with living embryos expressing PAR6-Ven, there is a significant downregulation of the expression of all of these proteins within the epithelium of the archenteron compared to both the apical tuft and the epithelium of the ectoderm (Fig. 1, 4B). There does appear to be a slight enrichment of these proteins in the apical tuft compared to the epithelium of the ectoderm, but this was not found to be statistically significant (Fig. 4B). The downregulation of these proteins in the archenteron may reflect that the PAR proteins have a stronger role in the maintenance of the epithelium in the ectoderm and apical tuft and a less significant role in the regulation of the movement of the archenteron into the blastocoel.
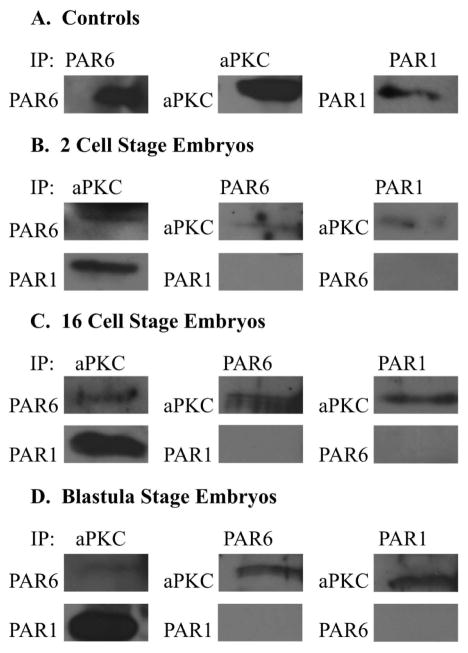
Because of this unique PAR1 localization, we performed co-immunoprecipitation assays in order to determine if PAR1 was interacting with the PAR complex proteins PAR6 and aPKC (Fig. 5). In other experimental models, PAR1 specifically localizes to the opposing regions of the PAR complex as aPKC and PAR1 typically actively exclude each other from the same domain (Benton and Johnston, 2003; Hurov and Piwnica-Worms, 2007). Previously, we have shown that PAR6 and aPKC in sea urchin embryos, as in other model systems, were associated with each other (Joberty et al., 2000; Lin et al., 2000; Alford et al., 2009; McCaffrey and Macara, 2009; Li et al., 2010). Co-immunoprecipitation assays at the 2 cell (first appearance of polarization to the apical cortex), 16 cell (first asymmetric division), and blastula (formation of an epithelium) stages were carried out in order to study the relationship of these proteins. We found that across all studied stages of development aPKC was in complex with both PAR6 and PAR1; however, PAR6 and PAR1 were not in complex with each other at any stage (Fig. 5).
Figure 5.
aPKC is in complex with PAR6 and PAR1 at the 2 cell, 16 cell and blastula stages of development. S. purpuratus embryos were grown to the stages indicated: 2 cell (B), 16 cell(C), and Blastula (D), then lysed and a co-immunoprecipitation assay was performed using the Dynabeads kit (Invitrogen). Western blots were run and samples were probed for PAR6, aPKC, and PAR1 as specified. aPKC was found to be in complex with both PAR6 and PAR1 at the 2 cell, 16 cell, and blastula stages of development. However, PAR6 and PAR1 did not associate with each other at any of the examined stages.
aPKC is not involved in micromere formation
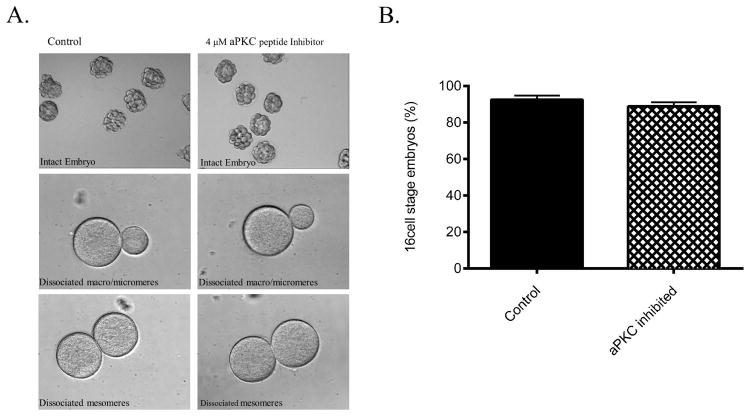
The PAR proteins were initially discovered based on their involvement in the regulation of the first asymmetric division in the C. elegans embryo and have since been found regulate the divisions of many other organisms at various stages of development (Kemphues et al., 1988; Etemad-Moghadam et al., 1995; Watts et al., 1996; Chalmers et al., 2003; Hao et al., 2010; Niessen et al., 2013). Previously we have shown that treatment with a specific peptide inhibitor of PKCζ resulted in the formation of multipolar spindles, short asters, and improper spindle rotation during early cleavages (Alford et al., 2009). Here we investigated whether or not aPKC is involved in the first asymmetric division in the sea urchin embryo, at the 16 cell stage of development. We find that in both intact embryos and in dissociated blastomeres treatment with the specific peptide inhibitor of PKCζ at the 8 cell stage did not affect the formation of mesomeres, macromeres, or micromeres at the 16 cell stage (Fig. 6). In order to ensure the efficacy of the peptide inhibitor, subsets of the same batches of embryos were tested to determine if earlier divisions at the 2 and 4 cell stages were still affected by the addition of the peptide inhibitor. As these divisions became asymmetric as we observed previously (Alford et al., 2009), this demonstrates that the peptide inhibitor is still active at the 2–4 cell stages and yet does not affect the first asymmetric division at the 16 cell stage of development. Furthermore, because the intact embryos divided normally in the presence of the peptide inhibitor of PKCζ and individual blastomeres of sea urchin embryos are known to follow their normal developmental pattern (Driesch, 1892), even upon separation, this confirms that aPKC activity is not required for the asymmetric division at the 16 cell stage.
Figure 6.
aPKC does not regulate the first asymmetric division in the sea urchin embryo. 4 μM PKCζ peptide inhibitor was added at the 8 cell stage of development in intact and dissociated embryos (A). Fertilization envelopes were retained in intact embryos as the PKCζ peptide inhibitor is able to penetrate the envelope (Alford et al., 2009), but had to be removed in dissociated embryos in order to manually dissociate the blastomeres. No effect was observed on the formation of micromeres, macromeres, or mesomeres compared to controls in trials of 100 embryos each (n=6). Normal 16 cell embryo formation is quantified (B) in controls (92.5±5.4) and treated groups (88.8±5.7) (mean ± SD). No significant difference was found between control and treated groups (p = 0.28).
Myosin assembly and cell-cell contact are required for the localization of the PAR complex to the apical cortex
In order to assess what anchors the PAR complex proteins in the apical cortex, we tested a series of small molecular inhibitors of the cytoskeleton. We first assessed inhibitors of the acto-myosin cortex as its involvement in the localization of the PAR complex has been well documented (Cowan and Hyman, 2004; Munro et al., 2004; Munro, 2006). Additionally, prior research has found that polarized plasma membrane rafts in sea urchin embryos, marked by the ganglioside GM1, are dependent on assembled myosin filaments (Ng et al., 2005; Alford et al., 2009; Gudejko et al., 2012). Embryos were treated with the various inhibitors at the 2 cell stage of development, after the PAR complex proteins have polarized to the apical cortex. We used specific small molecule inhibitors in order to dissect the different functions of myosin: blebbistatin as an inhibitor of myosin’s ATPase activity and ML-7 or ML-9 to inhibit myosin light chain kinase (MLCK). Phosphorylation of serine 19 on myosin light chain by MLCK results in both bipolar filament assembly and an enhancement of the actin activated ATPase activity (Bresnick, 1999). Thus, the effects of each activity of myosin, either myosin motor function or filament assembly, can be determined through the use of these specific inhibitors.
First, 2 cell stage embryos were treated with either ML-7 or ML-9 in order to inhibit bipolar filament assembly. ML-7 and ML-9 act as ATP (adenosine triphosphate) competitors in order to inhibit MLCK activity. These inhibitors have been used extensively in sea urchin embryos to study the activity of MLCK. One of the major phenotypes associated with MLCK inhibition in sea urchin embryos is the formation of bi-nucleate cells because while mitosis can proceed normally, the contractile ring is unable to form without myosin assembly and without a contractile ring cells do not undergo cytokinesis (Mabuchi and Takano-Ohmuro, 1990). Additional data in sea urchin embryos has shown that mono-phosphorylated regulatory light chain is required for cleavage furrow formation as well as for contraction of the furrow (Uehara et al., 2008). Further studies have found that myosin II is involved in coordination of the global activation of the cortex prior to the onset of cytokinesis and the maintenance of contractility in the furrow during division (Miyoshi et al., 2006). Additionally, MLCK activity has been shown to initiate myosin contractility in sea urchin embryos (Lucero et al., 2006). Here treatment with either ML-7 or ML-9, in addition to blocking contractile ring function, resulted in the cytoplasmic pooling of PAR6, aPKC, and CDC42 (Fig. 7A).
Figure 7.
Myosin assembly is required for maintaining the cortical localization of the PAR complex proteins. A). L. pictus embryos were fixed at the 2 cell stage after treatment with the various inhibitors for 15 minutes. Fixed embryos were stained for endogenous PAR6, aPKC, and CDC42. PAR6, aPKC, and CDC42 are apically localized in the cortex of the sea urchin embryo at the 2 cell stage of development. Inhibition of myosin light chain kinase (MLCK) by ML-7 resulted in cytoplasmic pooling of these cell polarity regulators. To a lesser extent this same effect was observed with ML-9, a less specific MLCK inhibitor. Treatment with a general Rho kinase inhibitor, H1152, an actin polymerization inhibitor, latrunculin B, an astral microtubule inhibitor, urethane, or a myosin ATPase inhibitor, blebbistatin did not affect the cortical localization of these proteins. B). L. pictus embryos were fixed and then stained for PAR6. Brightness and contrast were enhanced for visualization. PAR6 localizes to the cortex and apical cortex at the fertilized egg and 2 cell stages (i and iii). Treatment with ML-7 for 15 minutes at both the fertilized egg and 2 cell stages resulted in cytoplasmic pooling of PAR6 (ii and iv). Pulse treatment with ML-7 for 15 minutes at the fertilized egg and analysis of PAR6 localization at the next cleavage (2 cell) revealed PAR6 remained cytoplasmic (v).
We further dissected the role of myosin function by treating embryos with blebbistatin. Blebbistatin preferentially binds to the ATPase intermediate of myosin with ADP and phosphate bound at the active site. It then slows down the release of the phosphate and blocks the myosin heads in a low-actin affinity conformation (Kovacs et al., 2004). Unlike treatment with ML-7 or ML-9, which block the formation of the contractile ring, treatment with blebbistatin inhibits constriction of the contractile ring during cytokinesis (Miyoshi et al., 2006). Treatment with blebbistatin did not perturb the apical or cortical localization of the PAR complex (Fig. 7A). Because blebbistatin was not found to have an effect on PAR complex localization, these results indicate that it is bipolar filament assembly and not the ATPase activity of myosin that is necessary for the PAR complex localization.
To further confirm the specificity of the role of MLCK, we also treated the embryos with H1152, a Rho kinase (ROCK) inhibitor. ROCK, like MLCK, is also indirectly involved in the phosphorylation of myosin light chain; however, instead of directly phosphorylating myosin light chain like MLCK, ROCK phosphorylates myosin phosphatase. Phosphorylation of myosin phosphatase inhibits its activity and thus maintains myosin light chain phosphorylation. Treatment with H1152 inhibits the phosphorylation of myosin phosphatase and thus promotes the dephosphorylation of myosin light chain, which then prevents bipolar filament formation. Treatment with H1152 had no effect on the localization of these PAR complex proteins (Fig. 7A). These results reflect the specific requirement for MLCK activity in the localization of the PAR complex proteins and not just myosin light chain phosphorylation, as both ROCK and MLCK inhibition affect myosin light chain phosphorylation through different pathways.
In order to further analyze the specific role of myosin in this process, the other major cytoskeletal elements were examined. Myosin motors along with actin filaments are the dominant structural proteins in the cellular cortex. As the other dominant structural protein in the cortex, actin involvement was also analyzed by inhibition of actin polymerization with latrunculin B. Latrunculin B inhibits actin activity by causing the shortening and thickening of stress fibers (Schatten et al., 1986; Spector et al., 1983; Wakatsuki et al., 2001). We also inhibited astral microtubules with urethane to assess what role another major cytoskeletal component may play in anchoring the PAR complex specifically within the apical cortex (Strickland et al., 2005). Urethane shortens and destabilizes astral microtubules and promotes astral microtubule catastrophe (Strickland et al., 2005). Astral microtubules are also known to be involved in the maintenance of cortical PAR domains in C. elegans, although the role of microtubules in PAR polarization remains somewhat controversial (Ai et al., 2011; Nance and Zallen, 2011). Due to issues with lack of reversibility, cytochalasin D and nocodazole were not used as inhibitors of the actin and microtubule cytoskeleton, respectively. We found that neither latrunculin B or urethane treatment affected the localization pattern of the PAR6, aPKC, or CDC42 indicating a specific role for assembled myosin in PAR complex localization (Fig. 7A).
Once we determined that myosin bipolar filaments were involved in anchoring the PAR complex in the apical cortex, we next analyzed the stability of these disruptions in PAR complex localization by MLCK inhibition. We treated embryos at the fertilized egg stage of development with a short pulse of ML-7, washed out the ML-7 and then allowed the embryos to divide to the 2 cell stage before fixation and staining for PAR6, which we used as a marker of the PAR complex (Fig. 7B). We found that a short pulse of ML-7 at the fertilized egg stage resulted in the same cytoplasmic pooling of PAR6 as it does at the 2 cell stage (Fig. 7Bii, 7Biv). Additionally, we find that the pulse treatment with ML-7 at the fertilized egg stage causes PAR6 to remain cytoplasmic through the next cell division to the 2 cell stage (Fig. 7Bv). This suggests that the initial disruption is maintained because after only a short disruption in myosin assembly during the fertilized egg stage PAR6 did not return to the cortex at the 2 cell stage.
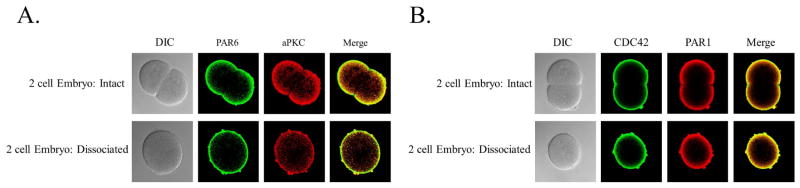
Previous studies had shown that PAR6 localization is also dependent on calcium mediated cell adhesion (Alford et al., 2009). Here, embryos were dissociated with gentle pipetting in CaFSW and PAR6 along with aPKC, CDC42, and PAR1 were examined in dissociated 2 cell stage embryos. Like first shown for PAR6, we found that all polarity proteins become evenly distributed throughout the cortex upon dissociation (Fig. 8).
Figure 8.
PAR6, aPKC, CDC42, and PAR1 are reliant upon calcium dependent cell adhesion for apical localization. L. pictus embryos were fixed and stained for endogenous PAR6 and aPKC (A) or CDC42 and PAR1 (B) in both intact and dissociated 2 cell embryos. All proteins were found to have a uniform distribution in the cortex upon dissociation.
Assembled myosin during early cleavage stages is necessary for blastula formation
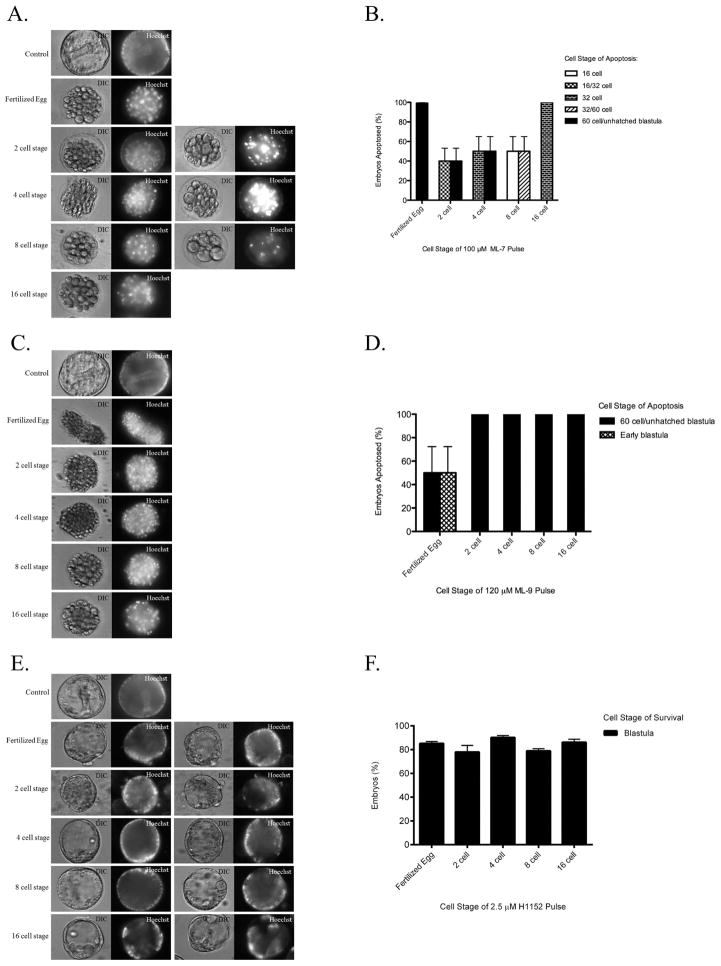
After determining that the PAR complex proteins were anchored in the cortex by assembled myosin and that these disruptions in localization could be maintained through the next division, we analyzed the effects of PAR disruption on later development. We pulse treated L. pictus embryos with the same cytoskeletal inhibitors (ML-7, ML-9, H1152, latrunculin B, urethane, and blebbistatin) for 15 minutes at the fertilized egg, 2 cell, 4 cell, 8 cell, and 16 cell stages of development and then monitored the progression of the embryos. Embryos were pulse treated with each of the cytoskeletal inhibitors as opposed to continuous treatments because each of these cytoskeletal inhibitors caused embryos to arrest either during mitosis or cytokinesis (data not shown). Therefore, experiments with continuous treatments would have assessed cell cycle defects, rather than developmental deficiencies. All inhibitors used in these experiments were found to be reversible in sea urchin embryos; they could be washed out and the embyos would continue through the cell cycle normally (data not shown). A 15 minute pulse of ML-7 or ML-9 was determined to be sufficient to disrupt the localization of PAR6, aPKC, and CDC42 (Fig. 7A). Embryos that were pulse treated with either ML-7 or ML-9 continued to cleave normally, but failed to reach the blastula stage when pulse treated at these early cleavage stages. Embryos underwent apoptosis prior to forming the polarized epithelium that is characteristic of the blastula stage (Fig. 9). This effect appears to be specific to assembled myosin, as cleavage stage embryos pulse treated with the other cytoskeletal inhibitors were able to reach the gastrula stage (Fig. 9, data not shown). Embryos pulse treated with either latrunculin B, urethane, or blebbistatin underwent normal gastrulation, similar to controls (data not shown). Embryos pulse treated with H1152 still reached the blastula stage, although there was slight variation in morphology as the epithelium appeared abnormal compared to the controls and these embryos did not undergo gastrulation (Fig. 9).
Figure 9.
Assembled myosin is required during early cleavage stages for blastula formation. 100 embryos were treated with 100 μM ML-7 (A) (n=12), or 120 μM ML-9 (C) (n=6), or 2.5μM H1152(E) (n=6), pulse for 15 minutes at various cell stages as indicated and allowed to develop until the control embryos had reached the gastrula stage (≥90% of controls reached gastrulation). Results are quantified (B,D, and F) (mean±SD). Embryos treated with the MLCK inhibitors, ML7 and ML-9, failed to reach the hatched blastula stage. Embryos treated with the Rho kinase inhibitor (H1152) reached the blastula with an abnormal morphology.
Actin localization is affected by myosin light chain kinase inhibition
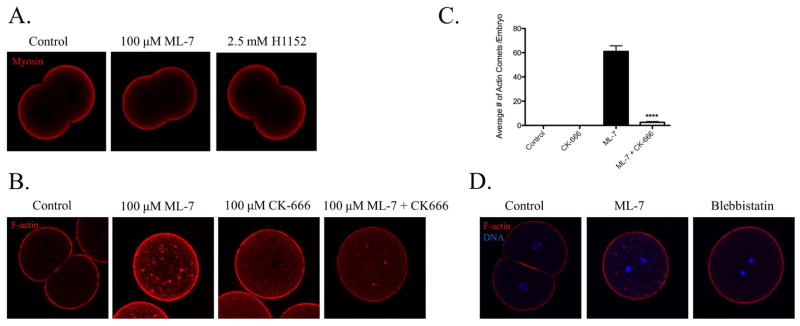
The role of acto-myosin cortex in the localization of the PAR complex has been well documented (Cowan and Hyman, 2004; Munro et al., 2004; Munro, 2006) and has been further characterized here as playing a role in the PAR complex localization in the sea urchin embryo. We additionally wanted to assess the mechanism by which MLCK activity was affecting the localization of the PAR complex proteins and subsequent development of the sea urchin embryo. We first examined the effect of MLCK inhibition on myosin localization in order to determine if the cortex itself was being disrupted and this was leading to the cytoplasmic localization of the PAR complex. Interestingly, in immunofluoresence assays of 2 cell stage embryos treated with either ML-7 or H1152, myosin localization remained unperturbed (Fig. 10A).
Figure 10.
Actin, but not myosin, localization is disrupted by myosin light chain kinase inhibition. A) L. pictus embryos were fixed and stained for endogenous myosin. Treatment with either ML-7 or H1152 did not affect the localization of myosin. B) L. pictus embryos were fixed and stained for endogenous actin. Brightness and contrast were enhanced for visualization. Treatment with ML-7 resulted in the appearance of actin comets (61±4.7 actin comets per cell) (mean ±SD). Inhibition of Arp2/3 by CK-666 in ML-7 treated zygotes decreased the number of comets observed (2.6±0.6 comets per cell) (mean±SD). C) Quantification of the number of comets observed per embryo. Inhibition of Arp2/3 in ML-7 treated zygotes leads to a significant decrease in the number of actin comets formed (****p<0.0001). D) Inhibition of MLCK, not myosin ATPase activity, causes the formation of actin comets. L. pictus embryos were treated 20 minutes post-fertilization with ML-7 or blebbistatin and were fixed when control embryos reached the 2 cell and stained for actin. Actin comets are seen in ML-7 treated, but not blebbistatin treated embryos.
Myosin motors along with actin filaments are the dominant structural proteins in the cellular cortex. Recent data demonstrates that assembled myosin forms clusters that act as network nucleators to organize and remodel the cortex (Ideses et al., 2013). Additional evidence suggests that inhibition of myosin can affect actin turnover because myosin motors are able to dissemble actin filaments (Murthy and Wadsworth, 2005; Haviv et al., 2008; Salbreux et al., 2012). The effects of ML-7 treatment on actin localization were thus studied in order to determine if assembled myosin regulated actin dynamics in cleavage stage sea urchin embryos and if this resulted in the cytoplasmic localization of the PAR complex. Treatment with ML-7 resulted in the formation of multiple F-actin puncta in the cytoplasm, which could be observed in both live embryos injected with Lifeact-GFP (Movie S1) and in fixed embryos stained with phalloidin (Fig. 10B). The F-actin puncta appear to be Arp2/3-actin comets, similar to those seen in mammalian cells infected with Listeria monocytogenes (Tilney and Portnoy, 1989). Z-stack timelapse images of Lifeact-GFP in live cell images show actin comets appearing to originate at the cell cortex (Movie S1). Comet formation seems to be specific to the activity of MLCK, as inhibition of myosin ATPase with blebbistatin did not show any signs of actin mislocalization (Fig. 10D). These data suggest MLCK inhibition leads to both an instability of the acto-myosin cortex and a resulting cytoplasmic localization of the PAR complex.
During infection by L. monocytogenes the proteins of the Arp2/3 complex are utilized for the polymerization of actin filaments leading to the formation of actin comets (Welch et al., 1997). In order to assess if the Arp2/3 complex is responsible for the actin comet formation we observed under MLCK inhibition, we utilized the Arp2/3 inhibitor CK-666. Treatment of L. pictus embryos at the fertilized egg stage with CK-666 resulted in the formation of binucleate cells with normal F-actin localization at cell cortex (Fig. 10B). Embryos treated with ML-7 had an average of 61±4.7 actin comets per cell; however, those treated with both ML-7 and CK-666 had a significant decrease in the average number of actin comets per cell (2.6±0.6 comets per cell, Fig. 10C). These data suggest that the actin comets that are formed upon inhibition of MLCK are dependent on the nucleation of actin filaments by Arp2/3.
Discussion
The development of an embryo is a tightly regulated process by which an entire organism is generated from a single fertilized egg. While the sea urchin embryo has long been used a model to study this process, little was known until recently about the development of early cell polarity during early cleavage stages. Here we find that the sea urchin embryo has a unique localization pattern of the PAR proteins, which despite its nontraditional localization pattern, is critical for proper development of the embryo. This suggests that like protostome embryos, deuterostomes polarize during early cleavages and that this polarity is required for normal development.
The PAR complex proteins, PAR6 and aPKC, as well as its upstream regulator, CDC42, colocalize to the apical cortex as early as the 2 cell stage of development in sea urchin embryos (Alford et al., 2009). We now find these proteins retain their apical localization in the cortex through the gastrula stage of development. While others have shown that aPKC is excluded from the vegetal pole at the 16 cell stage of development in the Hemicentrotus pulcherrimus species of sea urchin, we now find that the PAR complex retain its apical localization in both the micromere and macromeres of the vegetal pole (Pruliere et al., 2011). However, they are significantly reduced in amount in the archenteron of the gastrula. The colocalization of PAR6, aPKC, and CDC42 mirrors the results seen in other model organisms (Nance and Zallen, 2011; McCaffrey and Macara, 2012). However, the colocalization of PAR1 with these proteins in the apical cortex appears to be unique to the sea urchin embryo. Despite having an asymmetric division at the 16 cell stage, forming tight junctions in an epithelium at the blastula stage, and undergoing convergent extension movements during gastrulation PAR1 remains localized to the apical surface along with the other PAR proteins. Given the distinctive localization pattern of PAR1 in the sea urchin embryo, determining the localization pattern of other traditionally posterior polarity regulators will be imperative to explore. Lethal giant larvae (LGL) is known to regulate polarity in C. elegans, function redundantly with PAR2 in Drosophila, and is a predicted protein based on the sequenced sea urchin genome (Cameron et al., 2009; Beatty et al., 2010; Hoege et al., 2010; Prehoda and Bowerman, 2010; Beatty et al., 2013). In sea urchin embryos, it may be that other posterior polarity regulators, such as LGL, function to regulate polarity in areas of cell-cell contact.
Our results suggest that although the PAR proteins are polarized, the manner in which they polarize is distinct to the sea urchin embryo. Furthermore, the down regulation of the expression of these proteins in the archenteron may reflect a functional significance as they may be more actively involved in the formation and maintenance of the epithelium that is formed at the blastula stage. Recent data, however, has found the PAR6 regulates skeletogenesis and gut differentation in the Hemicentrotus pulcherrimus species of sea urchin (Shiomi et al., 2012). Inhibition of aPKC did not appear to effect archenteron ingression or spicule formation in the Paracentrotus lividus species of sea urchin (Pruliere et al., 2011). Future experiments could determine if there is a species specific or protein specific requirement for the PAR proteins during and post gastrulation.
Here we find that PAR1 is in complex with aPKC at the 2 cell, 16 cell, and blastula stages of development, which is surprising given the traditionally antagonistic behavior of these proteins (Hurov et al., 2004). Because PAR1 is a substrate of aPKC, it is perhaps less surprising that they are associated with one another as opposed to PAR6 and PAR1, which are not known to interact in other systems. In other model organisms aPKC phosphorylates PAR1 and PAR1 phosphorylates PAR3 as a means to exclude each other from their respective domains (Benton and Johnston, 2003; Hurov and Piwnica-Worms, 2007). These typically transient phosphorylation events may be an indicator of why PAR1 and aPKC are found in the same complex in the sea urchin embryo.
The cleavage to the 16 cell stage is an imperative step in the development of the sea urchin embryo. It is the time of the first asymmetric division and it lays down some important foundational work for later development events. At this stage β-catenin becomes nuclear only in the micromeres of the vegetal pole, which is critical later during gastrulation for endomesoderm specification (Wikramanayake et al., 1998; Weitzel et al., 2004; Wikramanayake et al., 2004). These micromeres have also recently been found to be germ line precursors (Juliano et al., 2006; Yajima and Wessel, 2012). aPKC has been found to regulate a number of other asymmetric divisions and we were interested in determining its involvement in the sea urchin embryo (Suzuki et al., 2002; Chalmers et al., 2003; Durgan et al., 2011; Niessen et al., 2013). While prior research has shown that inhibition of aPKC at early cleavages stages resulted in several spindle defects (Alford et al., 2009), we find here that it does not appear to regulate the first asymmetric division in sea urchin embryos. Given these data it may be that other polarity proteins besides aPKC are involved in this particular asymmetric division. Candidates include LGL, Scribble, Pins, Crumbs, and CDC42 (McCaffrey and Macara, 2012).
Previously, we have shown that MLCK activity is important in maintaining the stability of polarized plasma membrane domains in sea urchin embryos (Ng et al., 2005; Alford et al., 2009; Gudejko et al., 2012). Here we have demonstrated the importance of MLCK activity in maintaining the localization of the PAR proteins at the apical cortex, as treatment with ML-7 or ML-9 resulted in the cytoplasmic pooling of these proteins. This disruption in localization by inhibition of myosin assembly is sustained through the next division. Additionally, assembled myosin was found to be required during these early cleavage stages in the embryo in order to reach the blastula stage. Pulse treatments with ML-7 or ML-9 resulted in apoptosis of the embryos prior to becoming blastula. In contrast, treatment with other small molecule inhibitors of the actin or microtubule cytoskeleton did not perturb the localization of the PAR proteins nor did it alter the embryos’ ability to reach the gastrula stage of development. These data demonstrate the specificity of the MLCK activity and its necessity during early cleavage stages for normal development. These results also mirror our previous findings that demonstrated that aPKC activity was required in early cleavage stages for blastula formation (Alford et al., 2009). Collectively, these data suggest that both proper activity and cortical localization of the PAR complex in early development are necessary for blastula formation.
The reliance of calcium dependent adhesion to maintain the apical localization of PAR6, aPKC, CDC42 and PAR1 highlights the importance for these proteins to remain cortical, but not necessarily apical for proper development. Driesch showed in his classic experiments that dissociated early blastomeres of sea urchin embryos could develop into normal, albeit smaller, adults (Driesch, 1892). However, here we have shown that the cortical reorganization of the PAR proteins after myosin filament disruption results in apoptosis prior to blastula formation.
How the PAR proteins, critical regulators of cell polarity, interact and influence embryonic regulators of polarity, such as dishevelled, remains an important question to address. Dishevelled has been documented to interact with PAR proteins in Xenopus, C. elegans, and in cultured cells (Sun et al., 2001; Wharton, 2003; Dollar et al., 2005; Ossipova et al., 2005; Schlessinger et al., 2007; Terabayashi et al., 2008). In sea urchins, dishevelled has been extensively studied because of its vital importance for β catenin signaling and endomesoderm specification. In canonical signaling, dishevelled first becomes active at the 16 cell stage of development, which is when β catenin becomes nuclear in the micromeres of the vegetal pole (Kumburegama and Wikramanayake, 2007). The crosstalk between dishevelled and the PAR proteins may occur in early development in order to establish axis specification before individual blastomeres have committed to their respective cell fates. Others have examined the role of dishevelled in early cleavages stages in sea urchin embryos (Peng and Wikramanayake, 2013); however, how dishevelled and the PAR proteins collectively influence the polarity of the sea urchin embryo in early cleavage stages remains an area to explored.
Interestingly, we find that perturbations in MLCK activity during early cleavage stages do not disrupt myosin localization, but rather actin localization in the cortex of these embryos. These results support the idea that assembled myosin acts a molecular scaffold that can regulate cortical dynamics (Murthy and Wadsworth, 2005; Salbreux et al., 2012; Ideses et al., 2013). The formation of Arp 2/3 nucleated actin comets by treatment with ML-7 or ML-9 can be seen in both live and fixed embryos (Fig. 10, Movie S1). These changes in the structure of the cortex may be responsible for the cytoplasmic pooling of the PAR proteins that are observed with the same drug treatments and may indeed affect dishevelled vegetal cortical localization. These data connect the idea that myosin is a molecular scaffold that actively organizes the cortex and the participation of cortical actin in the PAR complex localization that is observed in other model systems.
These studies on the sea urchin embryo have explored how and when polarity is established and the impact of disturbing this polarity during stages of critical developmental decisions. From these data there is a clear role for the PAR proteins during the early development of the sea urchin embryo. The unconventional polarization of these proteins to the apical cortex during early cleavage stages is essential for blastula formation. These insights into the generation and maintenance of polarity are crucial to our understanding of how a cell or a developing embryo properly partitions components to ensure that the right function occurs in the correct location.
Experimental Procedures
Sea Urchin Embryo Culture
Lytechinus pictus and Stronglyocentrotus pupuratus (Marinus Scientific, Long Beach, CA) and Paracentrotus lividus (Villefranche-sur-Mer, France) gametes were obtained by intracoelomic injection of 0.5M KCl. Eggs were shed into artificial seawater (ASW) and sperm was dry collected. Eggs were swirled twice to expand the jelly coat. Sperm was diluted 1:1000 in ASW prior to use and added to a culture of eggs in ASW for fertilization. Fertilization was monitored by the formation of the fertilization envelope. In order to remove the fertilization envelope, L. pictus eggs were cultured in 4 mM para-aminobenzoic acid (PABA) and run through a 118 μm nytex. Fertilized embryos were then cultured at 15°C in either ASW, filtered seawater (FSW) or calcium-free seawater (CaFSW) as indicated. Embryos were treated with 100 μM ML-7 (Sigma-Aldrich), 120 μM ML-9 (Tocris Bioscience), 2.5 μM H1152 (Alexis-Biochemicals), 4 μM myristolated protein kinase c zeta peptide (PKCζ) inhibitor (Enzo Life Sciences), 25 nM latrunculin B (Sigma-Aldrich), 40 mM urethane (Sigma-Aldrich), 30 μM blebbistatin (Sigma-Aldrich) or 100 μM CK666 (Tocris Bioscience) at various cell stages as indicated. Embryos were stained with Hoechst (1:10,000; Life Technologies) as indicated and then were imaged using either a Nikon TE 200 inverted microscope or a Nikon TE2000 inverted microscope with a Yokogawa spinning disk head, both controlled by Metamorph software.
Live Cell Fluorescent Labeling
For F-actin imaging in live cells, the actin binding probe Lifeact was utilized (Riedl et al., 2008). Lifeact-mEGFP was isolated and amplified from the pEGFP-N1 mammalian expression vector and TOPO cloned into the bacterial expression vector pEXP5-CT (Life Technologies). Proper insertion of the fusion protein sequence was verified by restriction digestion and sequencing. Escherichia coli cells were then transformed with the pEXP5-CT-Lifeact-GFP plasmid and expression was induced with 1 mM IPTG for 4 hours at 37°C. His-tagged Lifeact-GFP was purified using affinity chromatography. Purified Lifeact-GFP was then diluted to 2 mg/ml in injection buffer (10 mM HEPES, 150 mM aspartic acid, pH 7.2). Lifeact-GFP was then microinjected into zygotes cultured in ASW using a Picospritzer III at 40 psi with 10 msec injections and imaged on a Leica DM I 6000 inverted microscope equipped with the Leica TCSSP5 confocal system.
Phallusia mammillata PAR6 (NCBI GenBank AY987398.1 (Patalano et al., 2006)) was subcloned into pSPE3 to create a C-terminal fusion construct with Venus. Phallusia mammillata PAR6 protein shares 69% sequence identity with the Stronglyocentrotus pupuratus PAR6 protein and both share the conserved PB1 and PDZ domains that are typical of PAR6 proteins. mRNA of PmPAR6::Venus was prepared using mMessage mMachine T3 transcription kit following the manusfacturers protocol. mRNA encoding PAR6::Ven (100–200 ng/μl) was microinjected at 1% volume into unfertilized Paracentrotus eggs and the fluorescence from embryos collected using an SP2 Leica confocal microscope.
PAR1 Antibody Generation
The GLEAN prediction Sp-Mark3 amino acid sequence of Strongylocentrotus purpuratus was utilized to generate a specific antibody against the PAR1 protein (Cameron et al., 2009). KFSRRSLVMAEPPSEYVKPR was used as the antigenic sequence for antibody production. Antibody generation in rabbits and purification was performed by Covance and then verified through both Western blot and immunofluoresence analysis (Fig. 2, 3).
Fixation and Immunofluorescence
Lytechinus pictus embryos were treated with inhibitors as indicated and then incubated in fixation buffer (3.2% formaldehyde, 0.125% glutaraldehyde, 0.2 M NaH2PO4H2O, 0.136 M NaCl) for 45 minutes. Following fixation, embryos were permeabilized in fixation buffer with 0.1% NP-40 for an additional 20 minutes and treated with 50 mM glycine for 15 minutes. Embryos were washed three times with phosphate buffered saline (PBS). Primary antibody, polyclonal goat anti-PARD6A (1:100; Santa Cruz, sc-14405), polyclonal goat anti-Cdc42 (1:50; Santa Cruz, sc-87), polyclonal rabbit anti-PKCζ (1:100; Santa Cruz, sc-216), polyclonal rabbit anti-S. purpuratus PAR1 (1:100) or polyclonal rabbit-anti-S. purpuratus egg myosin II (1:250) antibody were added in PBS+0.1% Trion (PBT) overnight at 4°C (Alford, et al. 2009; Gudejko, et al. 2012). Fixed embryos were then washed three times with PBT for 20 min. Secondary antibody, Alexa 488-conjugated donkey anti-goat (1:1000) (for anti-PAR6 and Cdc42 primary antibodies) or Alexa 647 or 555-conjugated donkey anti-rabbit (1:1000) (for anti-PKCζ, myosin II and PAR1 primary antibodies) were added in PBT and incubated for 3 hours at room temperature (Life Technologies). For some fixations, Alexa 546 conjugated phalloidin (1:500) and Hoechst (1:10,000) were added simultaneously with the secondary antibodies (Life Technologies). Fixed embryos were again washed three times with PBT for 20 min and mounted in mounting media (1:1 glycerol:PBS) and imaged on a Leica DM I 6000 inverted microscope equipped with the Leica TCSSP5 confocal system.
For immunofluoresence assays of gastrula stage embryos, a region of interest (ROI) of 6.488 × 26.199 μm2 was utilized to analyze the pixel intensity of three regions using the Leica LAS AF software: the apical tuft, the archenteron, and the epithelium. These data were analyzed in Microsoft Excel and GraphPad Prism. Statistical significance between each ROI was then determined using an unpaired t-test and a P-value of ≤ 0.05 was considered significant.
Similarly, for immunofluoresence assays examining actin comet formation, the number of comets were quantified using the Leica LAS AF software and then analyzed using Microsoft Excel and GraphPad Prism. Statistical significance was then determined using an unpaired t-test and a P-value of ≤ 0.05 was considered significant.
Co-immunoprecipitation
Co-immunoprecipitation assays were performed using the Dynabeads Co-immunoprecipitation Kit (Life Technologies). Polyclonal rabbit anti-S. purpuratus PAR1, polyclonal goat anti-PARD6A, and polyclonal rabbit anti-PKCζ antibodies were coupled to the Dynabeads per manufacture’s instructions. Each antibody was coupled to the Dynabeads at a final concentration of 10 μg/ml. S. purpuratus embryos were raised to developmental stages as indicated, pelleted, washed once with PBS, and then lysed in extraction buffer B (100 mM MgCl, 1X IP buffer, 150 mM NaCl, 1 mM DTT, and 500 μM PMSF) for 15 minutes on ice. The embryo lysis suspension was centrifuged at 2600 × g for 5 minutes at 4°C. 1.5 mg of antibody-coupled beads were washed with extraction buffer B, resuspended in the embryo lysate, and rotated for 30 minutes at 4°C. Beads were then washed 3 times with extraction buffer B and once with the last wash buffer for 5 minutes (1x LWB +0.02% Tween 20) at room temperature. Lastly, the beads were incubated with the elution buffer for 5 minutes at room temperature. The eluted supernatant was analyzed by Western blot.
Western Blot Analysis
L. pictus embryos were lysed in Laemmli’s SDS-Sample Buffer (250 mM Tris-HCl, pH 6.8, 8% SDS, 40% glycerol, 8% βME, and 0.02% Bromophenol) for polyclonal rabbit anti-PAR1 verification Western blots. These lysates or the co-immunoprecipitation samples were run on a 12% SDS-PAGE and then were transferred to an Immobilon-P membrane (Millipore) for Western blot analysis. Blots were blocked in 5% nonfat dry milk in Tris-buffered saline (TBS) with 0.1% Tween-20 (TBS-T) for 1 hour at room temperature. Blots were then incubated with polyclonal rabbit anti-S. purpuratus PAR1 (1:10,000), polyclonal goat anti-PARD6A (1:200), or polyclonal rabbit anti-PKCζ (1:200) in 5% milk in TBS-T overnight at 4°C. Blots were washed 3 times with TBS-T and then incubated in a 1:10,000 dilution of horseradish peroxidase(HRP) conjugated donkey-anti-rabbit or donkey-anti-goat (Amersham Bioscience) in 5% milk in TBS-T for 1 hour at room temperature. Secondary antibodies were preincubated with a cold acetone extraction of S. purpuratus eggs prior to use in order to minimize cross-reactivity. Blots were washed with TBS-T three times before the addition of the HRP substrate.
Supplementary Material
Inhibition of myosin light chain kinase by ML-7 induces actin comet formation. L. pictus embryos injected with Lifeact-GFP show strong localization of F-actin to the cell cortex. Upon treatment with ML-7 multiple actin comets can be seen within the cytoplasm.
Key points.
PAR6, aPKC, CDC42, and PAR1 remain apical through the gastrula stage.
aPKC is in complex with PAR6 and PAR1 and does not regulate micromere formation.
The PAR complex is anchored in the cortex by myosin assembly.
Disruption of early polarity impeded blastula formation.
Inhibition of myosin light chain kinase affects actin localization and is mediated by Arp 2/3 nucleation.
Acknowledgments
We would like to thank Josh Rosenberg and Bret Judson for microscopy support. We would also like to thank John Ahern, Nina Millman, and Megan Martik for help with experiments as well as Brad Shuster for insightful discussion. Additionally, we would like to thank Roland Wedlich-Soeldner for the generous gift of Lifeact-GFP. This work was supported in part by NIH grant GM093978 to DB.
References
- Ahringer J. Control of cell polarity and mitotic spindle positioning in animal cells. Curr Opin Cell Biol. 2003;15:73–81. doi: 10.1016/s0955-0674(02)00018-2. [DOI] [PubMed] [Google Scholar]
- Ai E, Poole DS, Skop AR. Long astral microtubules and RACK-1 stabilize polarity domains during maintenance phase in Caenorhabditis elegans embryos. PloS one. 2011;6:e19020. doi: 10.1371/journal.pone.0019020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon VB. Cell polarity regulator PARD6B is essential for trophectoderm formation in the preimplantation mouse embryo. Biol Reprod. 2010;83:347–358. doi: 10.1095/biolreprod.110.084400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford LM, Ng MM, Burgess DR. Cell polarity emerges at first cleavage in sea urchin embryos. Dev Biol. 2009;330:12–20. doi: 10.1016/j.ydbio.2009.02.039. [DOI] [PubMed] [Google Scholar]
- Atwood SX, Chabu C, Penkert RR, Doe CQ, Prehoda KE. Cdc42 acts downstream of bazooka to regulate neuroblast polarity through par-6 aPKC. J Cell Sci. 2007;120:3200. doi: 10.1242/jcs.014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty A, Morton D, Kemphues K. The C. elegans homolog of drosophila lethal giant larvae functions redundantly with PAR-2 to maintain polarity in the early embryo. Development. 2010;137:3995–4004. doi: 10.1242/dev.056028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty A, Morton DG, Kemphues K. PAR-2, LGL-1 and the CDC-42 GAP CHIN-1 act in distinct pathways to maintain polarity in the C. elegans embryo. Development. 2013;140:2005–2014. doi: 10.1242/dev.088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Johnston DS. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Boyd L, Guo S, Levitan D, Stinchcomb DT, Kemphues KJ. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development. 1996;122:3075–3084. doi: 10.1242/dev.122.10.3075. [DOI] [PubMed] [Google Scholar]
- Bresnick AR. Molecular mechanisms of nonmuscle myosin-II regulation. Curr Opin Cell Biol. 1999;11:26–33. doi: 10.1016/s0955-0674(99)80004-0. [DOI] [PubMed] [Google Scholar]
- Burke RD, Murray G, Rise M, Wang D. Integrins on eggs: The βC subunit is essential for formation of the cortical actin cytoskeleton in sea urchin eggs. Dev Biol. 2004;265:53–60. doi: 10.1016/j.ydbio.2003.09.025. [DOI] [PubMed] [Google Scholar]
- Cameron RA, Samanta M, Yuan A, He D, Davidson E. SpBase: The sea urchin genome database and web site. Nucleic Acids Res. 2009;37:D750–D754. doi: 10.1093/nar/gkn887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers AD, Pambos M, Mason J, Lang S, Wylie C, Papalopulu N. aPKC, Crumbs3 and Lgl2 control apicobasal polarity in early vertebrate development. Development. 2005;132:977–986. doi: 10.1242/dev.01645. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Strauss B, Papalopulu N. Oriented cell divisions asymmetrically segregate aPKC and generate cell fate diversity in the early Xenopus embryo. Development. 2003;130:2657. doi: 10.1242/dev.00490. [DOI] [PubMed] [Google Scholar]
- Chan D, Thomas C, Taylor V, Burke R. Integrins on eggs: Focal adhesion kinase is activated at fertilization, forms a complex with integrins, and is necessary for cortex formation and cell cycle initiation. Mol Biol Cell. 2013;24:3472–3481. doi: 10.1091/mbc.E13-03-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang M. The Par3/Par6/aPKC complex and epithelial cell polarity. Exp Cell Res. 2013;319:1357–1364. doi: 10.1016/j.yexcr.2013.03.021. [DOI] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Asymmetric cell division in C. elegans: Cortical polarity and spindle positioning. Annu Rev Cell Dev Biol. 2004;20:427–453. doi: 10.1146/annurev.cellbio.19.111301.113823. [DOI] [PubMed] [Google Scholar]
- Dollar GL, Weber U, Mlodzik M, Sokol SY. Regulation of lethal giant larvae by dishevelled. Nature. 2005;437:1376–1380. doi: 10.1038/nature04116. [DOI] [PubMed] [Google Scholar]
- Driesch H. Foundations of experimental embryology. Hafner; New York: 1892. The potency of the first two cleavage cells in echinoderm development. Experimental production of partial and double formations; pp. 38–50. [Google Scholar]
- Durgan J, Kaji N, Jin D, Hall A. Par6B and atypical PKC regulate mitotic spindle orientation during epithelial morphogenesis. J Biol Chem. 2011;286:12461. doi: 10.1074/jbc.M110.174235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenbroek SI, Iden S, Collard JG. Cell polarity proteins and cancer. Semin Cancer Biol. 2012;22:208–215. doi: 10.1016/j.semcancer.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Ernst SG. Offerings from an urchin. Dev Biol. 2011;358:285–294. doi: 10.1016/j.ydbio.2011.06.021. [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Guo S, Kemphues KJ. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- Galli M, Muñoz J, Portegijs V, Boxem M, Grill SW, Heck AJR, van den Heuvel S. aPKC phosphorylates NuMA-related LIN-5 to position the mitotic spindle during asymmetric division. Nat Cell Biol. 2011;13:1132–1138. doi: 10.1038/ncb2315. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: Fundamental players in animal cell polarization. Developmental Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudejko HF, Alford LM, Burgess DR. Polar expansion during cytokinesis. Cytoskeleton. 2012;69:1000–1009. doi: 10.1002/cm.21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. Par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Hao Y, Du Q, Chen X, Zheng Z, Balsbaugh JL, Maitra S, Shabanowitz J, Hunt DF, Macara IG. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical pins. Current Biology. 2010 doi: 10.1016/j.cub.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Boyd L, Seydoux G. Stabilization of cell polarity by the C. elegans RING protein PAR-2. Developmental cell. 2006;10:199–208. doi: 10.1016/j.devcel.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJC, Peifer M. aPKC controls microtubule organization to balance adherens junction symmetry and planar polarity during development. Developmental cell. 2007;12:727. doi: 10.1016/j.devcel.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haviv L, Gillo D, Backouche F, Bernheim-Groswasser A. A cytoskeletal demolition worker: Myosin II acts as an actin depolymerization agent. J Mol Biol. 2008;375:325–330. doi: 10.1016/j.jmb.2007.09.066. [DOI] [PubMed] [Google Scholar]
- Hoege C, Constantinescu AT, Schwager A, Goehring NW, Kumar P, Hyman AA. LGL can partition the cortex of one-cell Caenorhabditis elegans embryos into two domains. Current Biology. 2010;20:1296–1303. doi: 10.1016/j.cub.2010.05.061. [DOI] [PubMed] [Google Scholar]
- Hung TJ, Kemphues KJ. PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development. 1999;126:127–135. doi: 10.1242/dev.126.1.127. [DOI] [PubMed] [Google Scholar]
- Hurov J, Piwnica-Worms H. The par-1/MARK family of protein kinases: From polarity to metabolism. Cell cycle. 2007;6:1966–1969. doi: 10.4161/cc.6.16.4576. [DOI] [PubMed] [Google Scholar]
- Hurov JB, Watkins JL, Piwnica-Worms H. Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Current Biology. 2004;14:736–741. doi: 10.1016/j.cub.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Hyodo-Miura J, Yamamoto TS, Hyodo AC, Iemura SI, Kusakabe M, Nishida E, Natsume T, Ueno N. XGAP, an ArfGAP, is required for polarized localization of PAR proteins and cell polarity in Xenopus gastrulation. Developmental cell. 2006;11:69–79. doi: 10.1016/j.devcel.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Iden S, Collard JG. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat Rev Mol Cell Biol. 2008;9:846–859. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]
- Ideses Y, Sonn-Segev A, Roichman Y, Bernheim-Groswasser A. Myosin II does it all: Assembly, remodeling, and disassembly of actin networks are governed by myosin II activity. Soft Matter. 2013;9:7127–7137. [Google Scholar]
- Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- Juliano CE, Voronina E, Stack C, Aldrich M, Cameron AR, Wessel GM. Germ line determinants are not localized early in sea urchin development, but do accumulate in the small micromere lineage. Dev Biol. 2006;300:406–415. doi: 10.1016/j.ydbio.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Kemphues K. PARsing embryonic polarity. Cell. 2000;101:345–348. doi: 10.1016/s0092-8674(00)80844-2. [DOI] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng N. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Kovacs M, Toth J, Hetenyi C, Malnasi-Csizmadia A, Sellers JR. Mechanism of blebbistatin inhibition of myosin II. J Biol Chem. 2004;279:35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- Kumburegama S, Wikramanayake AH. Specification and patterning of the animal-vegetal axis in sea urchins by the canonical wnt signaling pathway. Signal Transduction. 2007;7:164–173. [Google Scholar]
- Kusakabe M, Nishida E. The polarity-inducing kinase par-1 controls Xenopus gastrulation in cooperation with 14-3-3 and aPKC. EMBO J. 2004;23:4190. doi: 10.1038/sj.emboj.7600381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli G. Crucial polarity regulators in axon specification. Essays Biochem. 2012;53:55–68. doi: 10.1042/bse0530055. [DOI] [PubMed] [Google Scholar]
- Lázaro-Diéguez F, Cohen D, Fernandez D, Hodgson L, van IJzendoorn SCD, Müsch A. Par1b links lumen polarity with LGN–NuMA positioning for distinct epithelial cell division phenotypes. The Journal of Cell Biology. 2013;203:251–264. doi: 10.1083/jcb.201303013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JD, Ettensohn CA. Analysis of dishevelled localization and function in the early sea urchin embryo. Dev Biol. 2007;306:50–65. doi: 10.1016/j.ydbio.2007.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan DJ, Boyd L, Mello CC, Kemphues KJ, Stinchcomb DT. Par-2, a gene required for blastomere asymmetry in Caenorhabditis elegans, encodes zinc-finger and ATP-binding motifs. Proc Natl Acad Sci U S A. 1994;91:6108–6112. doi: 10.1073/pnas.91.13.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kim H, Aceto DG, Hung J, Aono S, Kemphues KJ. Binding to PKC-3, but not to PAR-3 or to a conventional PDZ domain ligand, is required for PAR-6 function in C. elegans. Dev Biol. 2010;340:88–98. doi: 10.1016/j.ydbio.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T. A mammalian PAR-3–PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol. 2000;2:540–547. doi: 10.1038/35019582. [DOI] [PubMed] [Google Scholar]
- Lucero A, Stack C, Bresnick AR, Shuster CB. A global, myosin light chain kinase-dependent increase in myosin II contractility accompanies the metaphase-anaphase transition in sea urchin eggs. Mol Biol Cell. 2006;17:4093–4104. doi: 10.1091/mbc.E06-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi I, Takano-Ohmuro H. Effects of inhibitors of myosin light chain kinase and other protein kinases on the first cell division of sea urchin eggs. Dev Growth Differ. 1990;32:549–556. doi: 10.1111/j.1440-169X.1990.00549.x. [DOI] [PubMed] [Google Scholar]
- McCaffrey LM, Macara IG. Widely conserved signaling pathways in the establishment of cell polarity. Cold Spring Harb Perspect Biol. 2009;1:a001370. doi: 10.1101/cshperspect.a001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey LM, Macara IG. Epithelial organization, cell polarity and tumorigenesis. Trends Cell Biol. 2011;21:727–735. doi: 10.1016/j.tcb.2011.06.005. [DOI] [PubMed] [Google Scholar]
- McCaffrey LM, Macara IG. Signaling pathways in cell polarity. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaig C, Robinson K. The distribution of lectin receptors on the plasma membrane of the fertilized sea urchin egg during first and second cleavage. Dev Biol. 1982;92:197–202. doi: 10.1016/0012-1606(82)90163-4. [DOI] [PubMed] [Google Scholar]
- Miller JR, McClay DR. Characterization of the role of cadherin in regulating cell adhesion during sea urchin development. Dev Biol. 1997;192:323–339. doi: 10.1006/dbio.1997.8740. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Satoh SK, Yamada E, Hamaguchi Y. Temporal change in local forces and total force all over the surface of the sea urchin egg during cytokinesis. Cell Motil Cytoskeleton. 2006;63:208–221. doi: 10.1002/cm.20118. [DOI] [PubMed] [Google Scholar]
- Morton DG, Roos JM, Kemphues KJ. Par-4, a gene required for cytoplasmic localization and determination of specific cell types in Caenorhabditis elegans embryogenesis. Genetics. 1992;130:771–790. doi: 10.1093/genetics/130.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton DG, Shakes DC, Nugent S, Dichoso D, Wang W, Golden A, Kemphues KJ. The Caenorhabditis elegans par-5 gene encodes a 14-3-3 protein required for cellular asymmetry in the early embryo. Dev Biol. 2002;241:47–58. doi: 10.1006/dbio.2001.0489. [DOI] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Developmental cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Munro EM. PAR proteins and the cytoskeleton: A marriage of equals. Curr Opin Cell Biol. 2006;18:86–94. doi: 10.1016/j.ceb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Murthy K, Wadsworth P. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Current Biology. 2005;15:724–731. doi: 10.1016/j.cub.2005.02.055. [DOI] [PubMed] [Google Scholar]
- Nance J, Zallen JA. Elaborating polarity: PAR proteins and the cytoskeleton. Development. 2011;138:799. doi: 10.1242/dev.053538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MM, Chang F, Burgess DR. Movement of membrane domains and requirement of membrane signaling molecules for cytokinesis. Developmental cell. 2005;9:781–790. doi: 10.1016/j.devcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Niessen MT, Scott J, Zielinski JG, Vorhagen S, Sotiropoulou PA, Blanpain C, Leitges M, Niessen CM. aPKCλ controls epidermal homeostasis and stem cell fate through regulation of division orientation. J Cell Biol. 2013;202:887–900. doi: 10.1083/jcb.201307001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipova O, Dhawan S, Sokol S, Green J. Distinct PAR-1 proteins function in different branches of wnt signaling during vertebrate development. Developmental cell. 2005;8:829–841. doi: 10.1016/j.devcel.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Panneerselvam S, Marx A, Mandelkow EM, Mandelkow E. Structure of the catalytic and ubiquitin-associated domains of the protein kinase MARK/par-1. Structure. 2006;14:173–183. doi: 10.1016/j.str.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Patalano S, Pruliere G, Prodon F, Paix A, Dru P, Sardet C, Chenevert J. The aPKC-PAR-6-PAR-3 cell polarity complex localizes to the centrosome attracting body, a macroscopic cortical structure responsible for asymmetric divisions in the early ascidian embryo. J Cell Sci. 2006;119:1592–1603. doi: 10.1242/jcs.02873. [DOI] [PubMed] [Google Scholar]
- Peng CJ, Wikramanayake AH. Differential regulation of disheveled in a novel vegetal cortical domain in sea urchin eggs and embryos: Implications for the localized activation of canonical wnt signaling. PLOS ONE. 2013;8:e80693. doi: 10.1371/journal.pone.0080693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusa B, Frankenberg S, Chalmers A, Hadjantonakis AK, Moore CA, Papalopulu N, Papaioannou VE, Glover DM, Zernicka-Goetz M. Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J Cell Sci. 2005;118:505. doi: 10.1242/jcs.01666. [DOI] [PubMed] [Google Scholar]
- Prehoda KE, Bowerman B. Cell polarity: Keeping worms LeGaL. Curr Biol. 2010;20:R646–R648. doi: 10.1016/j.cub.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruliere G, Cosson J, Chevalier S, Sardet C, Chenevert J. Atypical protein kinase C controls sea urchin ciliogenesis. Mol Biol Cell. 2011 doi: 10.1091/mbc.E10-10-0844. mbc.E10-10-0844v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z. Lifeact: A versatile marker to visualize F-actin. Nature Methods. 2008;5:605–608. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiz N, Grabarek JB, Sabherwal N, Papalopulu N, Plusa B. Atypical protein kinase C couples cell sorting with primitive endoderm maturation in the mouse blastocyst. Development. 2013;140:4311–4322. doi: 10.1242/dev.093922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salbreux G, Charras G, Paluch E. Actin cortex mechanics and cellular morphogenesis. Trends Cell Biol. 2012;22:536–545. doi: 10.1016/j.tcb.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Schatten G, Schatten H, Spector I, Cline C, Paweletz N, Simerly C, Petzelt C. Latrunculin inhibits the microfilament-mediated processes during fertilization, cleavage and early development in sea urchins and mice. Exp Cell Res. 1986;166:191–208. doi: 10.1016/0014-4827(86)90519-7. [DOI] [PubMed] [Google Scholar]
- Schlessinger K, McManus EJ, Hall A. Cdc42 and noncanonical wnt signal transduction pathways cooperate to promote cell polarity. J Cell Biol. 2007;178:355. doi: 10.1083/jcb.200701083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TE. Contact-independent polarization of the cell surface and cortex of free sea urchin blastomeres. Dev Biol. 1988;125:255–264. doi: 10.1016/0012-1606(88)90209-6. [DOI] [PubMed] [Google Scholar]
- Shiomi K, Yamaguchi M. Expression patterns of three par-related genes in sea urchin embryos. Gene Expression Patterns. 2008;8:323–330. doi: 10.1016/j.gep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Shiomi K, Yamazaki A, Kagawa M, Kiyomoto M, Yamaguchi M. Par6 regulates skeletogenesis and gut differentiation in sea urchin larvae. Dev Genes Evol. 2012:1–10. doi: 10.1007/s00427-012-0409-5. [DOI] [PubMed] [Google Scholar]
- Spector I, Shochet NR, Kashman Y, Groweiss A. Latrunculins: Novel marine toxins that disrupt microfilament organization in cultured cells. Science. 1983;219:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- Strickland LI, Donnelly EJ, Burgess DR. Induction of cytokinesis is independent of precisely regulated microtubule dynamics. Mol Biol Cell. 2005:E05. doi: 10.1091/mbc.E05-04-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TQ, Lu B, Feng JJ, Reinhard C, Jan YN, Fantl WJ, Williams LT. PAR-1 is a dishevelled-associated kinase and a positive regulator of wnt signalling. Nat Cell Biol. 2001;3:628–636. doi: 10.1038/35083016. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Hirata M, Kamimura K, Maniwa R, Yamanaka T, Mizuno K, Kishikawa M, Hirose H, Amano Y, Izumi N. aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Current Biology. 2004;14:1425–1435. doi: 10.1016/j.cub.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Ishiyama C, Hashiba K, Shimizu M, Ebnet K, Ohno S. aPKC kinase activity is required for the asymmetric differentiation of the premature junctional complex during epithelial cell polarization. Journal of Cell Science. 2002;115:3565–3573. doi: 10.1242/jcs.00032. [DOI] [PubMed] [Google Scholar]
- Tabuse Y, Izumi Y, Piano F, Kemphues KJ, Miwa J, Ohno S. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development. 1998;125:3607–3614. doi: 10.1242/dev.125.18.3607. [DOI] [PubMed] [Google Scholar]
- Terabayashi T, Funato Y, Miki H. Dishevelled-induced phosphorylation regulates membrane localization of Par1b. Biochem Biophys Res Commun. 2008;375:660–665. doi: 10.1016/j.bbrc.2008.08.098. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, listeria monocytogenes. J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Hosoya H, Mabuchi I. In vivo phosphorylation of regulatory light chain of myosin II in sea urchin eggs and its role in controlling myosin localization and function during cytokinesis. Cell Motil Cytoskeleton. 2008;65:100–115. doi: 10.1002/cm.20246. [DOI] [PubMed] [Google Scholar]
- Vinot S, Le T, Ohno S, Pawson T, Maro B, Louvet-Vallée S. Asymmetric distribution of PAR proteins in the mouse embryo begins at the 8-cell stage during compaction. Dev Biol. 2005;282:307–319. doi: 10.1016/j.ydbio.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Wakatsuki T, Schwab B, Thompson NC, Elson EL. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J Cell Sci. 2001;114:1025–1036. doi: 10.1242/jcs.114.5.1025. [DOI] [PubMed] [Google Scholar]
- Watts JL, Etemad-Moghadam B, Guo S, Boyd L, Draper BW, Mello CC, Priess JR, Kemphues KJ. Par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development. 1996;122:3133–3140. doi: 10.1242/dev.122.10.3133. [DOI] [PubMed] [Google Scholar]
- Watts J, Morton D, Bestman J, Kemphues K. The C. elegans par-4 gene encodes a putative serine-threonine kinase required for establishing embryonic asymmetry. Development. 2000;127:1467. doi: 10.1242/dev.127.7.1467. [DOI] [PubMed] [Google Scholar]
- Weitzel HE, Illies MR, Byrum CA, Xu R, Wikramanayake AH, Ettensohn CA. Differential stability of β-catenin along the animal-vegetal axis of the sea urchin embryo mediated by dishevelled. Development. 2004;131:2947. doi: 10.1242/dev.01152. [DOI] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by arp 2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Wharton KA. Runnin’ with the dvl: Proteins that associate with Dsh/Dvl and their significance to wnt signal transduction. Dev Biol. 2003;253:1–17. doi: 10.1006/dbio.2002.0869. [DOI] [PubMed] [Google Scholar]
- Wikramanayake AH, Peterson R, Chen J, Huang L, Bince JM, McClay DR, Klein WH. Nuclear beta catenin dependent wnt8 signaling in the vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of the endoderm and mesoderm cell lineages. Genesis. 2004;39:194–205. doi: 10.1002/gene.20045. [DOI] [PubMed] [Google Scholar]
- Wikramanayake AH, Huang L, Klein WH. β-Catenin is essential for patterning the maternally specified animal-vegetal axis in the sea urchin embryo. Proc Natl Acad Sci U S A. 1998;95:9343. doi: 10.1073/pnas.95.16.9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. Autonomy in specification of primordial germ cells and their passive translocation in the sea urchin. Development. 2012;139:3786–3794. doi: 10.1242/dev.082230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y, Terabayashi T, Miki H. Par1b/MARK2 phosphorylates kinesin-like motor protein GAKIN/KIF13B to regulate axon formation. Mol Cell Biol. 2010;30:2206. doi: 10.1128/MCB.01181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Inhibition of myosin light chain kinase by ML-7 induces actin comet formation. L. pictus embryos injected with Lifeact-GFP show strong localization of F-actin to the cell cortex. Upon treatment with ML-7 multiple actin comets can be seen within the cytoplasm.