Abstract
The study of autophagy (‘self-eating’), a fundamental cell fate pathway involved in physiological and pathological subcellular processes, opens a new frontier in the continuous search for novel therapies for human asthma. Asthma is a complex syndrome with different disease phenotypes. Autophagy plays a central role in cell physiology, energy and metabolism, and cell survival. Autophagy’s hallmark is the formation of double-membrane autophagic autophagosomes, and this process is operational in airway epithelial and mesenchymal cells in asthma. Genetic associations between autophagy genes and asthma have been observed including single nucleotide polymorphisms in Atg5 which correlate with reduced lung function. Immune mechanisms important in asthma such as Th2 cells and eosinophils also manifest autophagy. Lastly, we address the role of autophagy in extracellular matrix deposition and fibrosis in asthmatic airways remodeling, a pathologic process still without effective therapy, and discuss potential pharmacologic inhibitors. We end by offering two opposing but plausible hypotheses as to how autophagy may be directly involved in airway fibrosis.
Keywords: asthma, autophagy, fibrosis, hypotheses, remodeling
Asthma is a chronic respiratory disease afflicting 200 million people worldwide and 27 million in the United States including children. In the United States alone, the annual cost of health care exceeds $56 billion, and it continues to grow. Asthma manifests varied symptoms including wheezing, breathlessness, and chest tightness, and interacts in a complicated manner with comorbid conditions such as gastroesophageal reflux disease, sinusitis, obstructive sleep apnea, and cardiac disease.
Asthma is a multifaceted illness, akin to a syndrome with related diseases under that umbrella, with biological and pathological pathways that are myriad. No single process in asthma explains the disease process in full. Familial asthma is a strong predictor of childhood asthma, signifying a possible genetic basis for the disease in some patients. Nevertheless, the genes which have been connected to asthma describe only a subgroup of the disease, and likely many undiscovered genetic variants exist in underappreciated pathways relevant to asthma.
Beyond single molecular pathways and genetic associations, increasing evidence from subcellular processes collectively known as ‘cell fate’ phenomena raises an important opportunity for discovery and translation to human disease. In this brief review, we focus on autophagy as one of these cell fate processes with the potential to advance novel therapies for human asthma. With respect to several lung diseases, in particular asthma, much work remains to be carried out in the quest to understand the role of autophagy in health and disease.
Autophagy: a major determinant of cell fate
Autophagy is an evolutionarily and highly conserved catabolic process whereby misfolded or unnecessary proteins and damaged organelles are directed to lysosomes for subsequent degradation. Its mechanism includes phagosome formation via specific steps known as initiation, elongation, and maturation (1). One of the most important hallmarks of autophagy is the formation of double-membrane autophagic vacuoles (AVs) or autophagosomes, which surround damaged cytosolic elements and organelles (e.g. endoplasmic reticulum, mitochondria, etc.) or protein, which then fuse with lysosomes to form autophagolysosomes.
The subcellular process of autophagy, meaning ‘self-eating’, has been the focus of intense research in the last few years. Autophagy is classified into three major types in eukaryotic cells: macroautophagy (in this article referred to as ‘autophagy’), microautophagy, and chaperone-mediated autophagy. The process of macro- and microautophagy both involve a dynamic mechanism of membrane rearrangement to engulf portions of the cytoplasm. Once engulfed, the membrane of the resulting autophagic body is lysed, and the resulting small molecules are transported back into the cytosol through membrane permeases for reuse (2, 3). Autophagy also plays critical roles in cellular growth and development, in the innate and adaptive immune system, and in programmed cell death (4–6).
Under oxidative conditions, autophagy effectively removes damaged endoplasmic reticulum (ER), peroxisomes, and mitochondria. Conversely, autophagy may also function as a cell-survival mechanism during periods of cell starvation, where cells self-digest to supply nutrients for the synthesis of essential proteins. The proteins PI3kinase, Akt1, and antiapoptotic BCL-2 family members suppress autophagy, while tumor suppressor proteins such as Beclin-1, Atg4c, BH-3 only proteins, and PTEN activate autophagy (7, 8).
While the morphological description of autophagy was first made in mammalian cells in the 1950s, recent extensive genetic work has identified 32 autophagy-related (Atg) genes to date (9–13). The Atg genes in yeast were discovered from genetic screening for mutants that affected protein turnover, peroxisome degradation, and delivery of a resident vacuolar hydrolase (the cytoplasm to vacuole targeting (Cvt) pathway). These are larger than Cvt vesicles to accommodate its specific cargo, the peroxisomes (14). The absence of autophagy can be deleterious whereas excessive levels of autophagy can be harmful, thus requiring tight regulation of the autophagic process as a whole (15, 16). Many orthologs of Atg gene products have been studied and have shown similar roles in higher eukaryotes including mammals (17, 18).
Multiple genes are involved in the regulation of autophagy in yeast and higher mammals including the aforementioned 32 autophagy-related genes (19). Among these, the microtubule- associated protein 1 light chain 3 (LC3; Atg8) is cleaved from a pro-form by Atg4 and then conjugated with phosphatidyl-ethanolamine by the sequential action of Atg7 and Atg3 (19). One of the essential steps in mammalian autophagy is LC3 lipidation [i.e. the conversion of LC3 from LC3-I (the free form) to LC3-II (the phosphatidyl-ethanolamine-conjugated form)], which is involved in autophagosome formation, a key event indicating autophagy flux in cells (1).
Atg12 is a small hydrophilic protein having structural homology to ubiquitin which covalently links to Atg5 (20), where the mode of conjugation of Atg12 and Atg5 is similar to that of ubiquitination. Atg7 activates Atg12 in an ATP-dependent manner (it functions as an ubiquitin-activating enzyme, E1), leading to the formation of a thioester bond between the C-terminal glycine in Atg12 and a cysteine residue in Atg7 (21). The C-terminal glycine in Atg12 is then transferred to the cysteine in Atg10 (it functions as a ubiquitin- conjugating enzyme, E2), forming a new thioester bond, and Atg7 is released (22). Finally, the C-terminal glycine in Atg12 forms an isopeptide bond with the ε-amino group of lysine 149 in Atg5, and Atg10 is in its free state again. Thus, the formation of the Atg12-Atg5 conjugate is indispensable to autophagosome formation (23). In mammalian cells, Atg5 and Atg12 are conjugated to each other in the same way as they are in yeast (24).
Atg8 (Aut7/Apg8) is the second ubiquitin-like protein essential for autophagy. Atg8 is a 117-amino acid protein and is present in the early isolation membranes, autophagosomes, and autophagic bodies (25), thus making it a good marker for studying membrane dynamics during autophagy. The protein Atg8 is mostly membrane-bound; approximately half of it is peripherally bound to the membrane and the other half behaves like an intrinsic membrane protein.
Atg8 can be activated by Atg7 (E1) in an ATP-dependent manner and transferred to a conjugating E2 enzyme, Atg3 (26). Atg7 activates two different ubiquitin-like proteins, Atg12 and Atg8, and assigns them to their proper E2 enzymes, Atg10 and Atg3, respectively. Final steps involve Atg8 interacting with phosphatidylethanolamine (PE), a highly abundant membrane phospholipid (26) undergoing a lipidation reaction which leads to a conformational change of Atg8 that is integral for the membrane dynamics of autophagy (27).
The cycle of conjugation and de-conjugation is important for the normal progression of autophagy. LC3 is the mammalian orthologue of Atg8, targets to the autophagosomal membranes in an Atg5-dependent manner, and remains there even after Atg12–Atg5 dissociates. Thus, LC3 is the only credible marker of the autophagosome in mammalian cells (28). LC3 is detected in 2 forms: LC3-I (18 kDa) and LC3-II (16 kDa) (29). Twenty-two amino acids in the C-terminus of the newly synthesized LC3 are cleaved immediately by the mammalian orthologue of the yeast cysteine proteinase Atg4, autophagin, to produce an active cytosolic form, LC3-I (30). Then with the catalysis of Atg7 and Atg3, LC3-I undergoes a series of ubiquitination-like reactions, and is modified to LC3-II. LC3-I is located in the cytoplasm, while LC3-II is a tightly membrane-bound protein and is attached to the preautophagosomal structure (PAS) and autophagosomes. The relative amount of membrane-bound LC3-II reflects the abundance of autophagosomes, so the induction and inhibition of autophagy can be monitored through measuring total and free LC3-II levels by means of an immunoassay (29).
In addition, the Atg12 and LC3 systems have a functional relationship. In Atg5−/− cells, LC3-II is not generated (31). As a result, LC3 cannot target the autophagosomal membranes. The recent generation of transgenic mice expressing green fluorescent protein (GFP) fused to LC3 provides a useful tool to investigate autophagy in various mammalian organs in vivo (32).
Atg1 is a serine/threonine protein kinase, which forms a protein complex with different regulatory proteins such as Atg13, Vac8, Atg17, and Cvt9. The composition of this complex is dynamic, and it may vary depending on nutrient conditions (33). Under nutrient-rich conditions, Atg13 is hyperphosphorylated so that its association with Atg1 is blocked. Conversely, under conditions of nutrient starvation or after treatment with rapamycin, Atg13 becomes partially dephosphorylated. This dephosphorylation leads to an Atg1–Atg13 interaction and subsequent generation of autophagosomes instead of Cvt vesicles and, thus, activates autophagy (34).
Autophagy and lung diseases
Autophagy has essential functions in numerous lung diseases including acute lung injury (ALI), pulmonary infectious diseases, pulmonary vascular disease, cystic fibrosis (CF), chronic obstructive pulmonary disease (COPD), and asthma.
Autophagy is probably involved in the pathogenesis of both ALI and sepsis (35, 36). The application of high oxygen therapy (hyperoxia) to preserve tissue oxygenation may be at least partially involved in the pathogenesis of ALI. This leads to enhanced production of reactive oxygen species (ROS) in critical care settings, which targets the pulmonary epithelium (37). Hyperoxia induces histological and biochemical hallmarks of autophagy in vivo, comprising autophagosome formation and LC3β-II accumulation (35). Pulmonary epithelial cell culture studies revealed that hyperoxia in the lung initiates reversible LC3β lipidation in this model (37). Therefore, LC3β may also be essential as a pro-survival arm in oxygen-induced cytotoxicity, and this highlights probable crosstalk between autophagy and apoptosis during oxygen toxicity (37).
Autophagy is likely one of the most important mechanisms involved in host defense against various micro-organisms including bacteria, viruses, and parasites (38–41). Recent evidence also suggests that autophagy is an important cellular mechanism in the promulgation of respiratory infections (42).
Autophagy has also been described in lung tissues derived from patients with various forms of pulmonary hypertension (PH), including pulmonary arterial hypertension (PAH) (43). It is not yet clear whether autophagy is playing a protective or harmful role in PH (44). Further studies are needed to clarify the functional role of autophagy in human adult PH.
Dysfunctional autophagosome clearance in CF may be responsible for the robust and sustained inflammatory responses observed in CF airways (45). It is also notable that inactivation of Beclin-1 is involved in the sequestration of PI3KC3 and accumulation of p62 in CF (46). Restoration of Beclin-1 or p62 reduction can salvage the trafficking of cystic fibrosis transmembrane conductance regulator (CFTR) mutant to the cell surface (46). Genetic targeting of p62 improves the therapeutic effect of CFTR channel activators (47). All of these data provide solid evidence regarding the role of autophagy in CF pathogenesis.
The cellular pathogenesis of COPD includes prolonged inflammatory responses in the lungs, exuberant mucus production, with subsequent loss of barrier integrity between cells, oxidative injury and faulty repair, and increased ER stress responses. Both apoptosis and autophagy of airway cells are also important processes in the pathogenesis of COPD (48). In addition, cellular senescence also plays a role in COPD, and ROS released during mitochondrial respiration plays an essential role in the progression of cellular senescence (49). Damaged mitochondria is a prerequisite for engulfment and degradation via mitochondria-selective autophagy also known as ‘mitophagy’ (50). Therefore, mitophagy could be a key mechanism in the development of COPD.
In asthma, autophagy may be playing a role in disease pathogenesis (51). Although little work has been carried out on the role of autophagy in human airway epithelium, we and others have begun to explore this further. The cytokine interleukin-13 (IL13) is a central player in epithelial Th2 responses in asthma including formation of mucin and secretion of the eotaxins, potent tissue eosinophil chemokines. Autophagy is essential for bronchial epithelial mucus secretion in a T helper type 2 (Th2) model of asthma (52). Furthermore, data from our laboratory suggest that autophagy rather than ER stress responses may contribute more to IL13-induced eotaxin-3 peptide secretion from human airway epithelial cells (53).
Autophagy genes in asthma – what do they mean?
Recently, genetic association of autophagy genes, including ULK1, SQSTM1, MAP1LC3B, beclin-1, and Atg5, was investigated in a family-based asthma study where correlations between autophagy genes and familial asthma were reported. Single nucleotide polymorphism (SNP) rs12212740G>A of the Atg5 gene was found to be strongly associated with asthma. Electron microscopy analysis of bronchial tissues (fibroblasts and epithelial cells) from a patient with moderate-to-severe asthma and a normal individual, looking for hallmarks of autophagy such as double-membrane autophagosomes, showed increased activation of autophagy in the asthmatic airways as compared to normal healthy control subjects (54). In another study, additional SNPs in Atg5 (rs12201458 and rs510432) were found to also be associated with childhood asthma (55).
The study by Poon et al. showed an association of the Atg5 SNP with asthma and reduced forced expiratory volume in 1 s (FEV1), a critical lung function parameter. The increased autophagosomes provided strong evidence that autophagic flux activity in asthmatic airway resident cells (such as the epithelium) is playing a key role in either the prevention or propagation of asthma – a mystery still not unraveled. The increase in overall autophagic flux could indicate either active autophagy, or instead, inhibition in the terminal stages of autophagy (phagosome–lysosome conjugation and later digestion of cellular material). However, it is important to note that these tissue markers of autophagy are mere correlations with lung function and asthma symptoms. Further research is required to address whether or not this important cellular process is causal in disease.
Our group has previously published an important role for autophagy and Atg5 in the fibrotic response in primary human airway smooth muscle (HASM) cells (56). We found that transforming growth factor-beta1 (TGFβ1)-induced autophagy is required for HASM cell production of collagen A1 and fibronectin, and that the silencing of key autophagy-inducing proteins Atg5 and Atg7 decreased this fibrotic response in HASM cells. The role of autophagy in this mechanism was also assessed using pharmacologic inhibition of autophagy flux using 3-methyladenine (3-MA) and bafilomycin-A1 (Baf-A1). Our findings were further confirmed by measuring LC3β lipidation, and using transmission electron microscopy to confirm the formation of double-membrane autophagosomes, and immunofluorescence staining to confirm autophagosome–lysosome fusion and the formation of LC3β punctae (56). This mechanism could have therapeutic application in asthma if targeting autophagy can mitigate TGFβ1-induced fibrotic responses in airways.
To shed light on this important observation, the first step is to investigate whether autophagy is playing a role in the pathogenesis of asthma (57). A thorough evaluation of the airway epithelium is vitally important, as is the submucosal mesenchyme including airway smooth muscle cells and fibroblasts [e.g. important in extracellular matrix (ECM) production]. Equally important is the assessment of autophagic responses in the immune response central to asthma pathogenesis.
Only with such a comprehensive approach to study both resident and inflammatory cells will we learn about the potentially divergent role(s) of autophagy in a disease as complex as human asthma. Because adverse structural changes named ‘airway remodeling’ remains an intractable problem in severe asthma, studying autophagy and its effect on cell fate and remodeling provides for an important new research opportunity that could positively impact human disease.
Autophagy and immune responses in asthma
One of the major hallmarks of asthma is chronic airway inflammation and lack of resolution of phlogistic signals. Many groups have investigated the role of adaptive immunity in airway inflammation, an antigen-dependent mechanism where T helper cells (CD4+) play a central role in disease pathogenesis. There are two major pathways in the T helper response including Th1 and Th2 which are characterized by their specific cytokine profiles. Investigations using the murine model and biological samples from asthmatic patients indicate that the Th2 immune response is essential in asthma. Asthma also involves an imbalance between Th1, Th2, and Th17 pathways where Th2 hyperactivation leads to persistent airway inflammation and the well-recognized asthma phenotype (57, 58).
Autophagy has recently been widely investigated in adaptive and innate immunity, and its function in these responses has been highlighted. Autophagy is responsible for transporting antigens on major histocompatibility complex (MHC) class II molecules to MHC class II-comprising sections (MIICs), which are catalyzed by lysosomes. Autophagy is an essential process for lymphocyte development and survival (59). Macroautophagy- deficient B and T cells show impaired differentiation and regulation. Further, conditional deletion of Atg5 in mice induces decreases in the number of T cells in the periphery and impairs proliferation upon antigenic stimulation. Autophagy activity is significantly higher in Th2-polarized T cells, and nutrient starvation increases autophagic activity in this model. Lastly, autophagy is also required for both central and peripheral tolerance to self-antigens where deletion of Atg5 in the mouse thymus gland enhances autoimmunity (57).
Patients with severe asthma manifest higher levels of autophagic activation in their sputum granulocytes, and peripheral blood eosinophils than patients with milder disease or healthy controls (51). In this study, the autophagy inhibitor 3-MA inhibited the expression of LC3-II and eosinophil cationic protein in HL-60 cells. Furthermore, dexamethasone did not alter autophagy levels in peripheral blood eosinophils from patients with asthma. The authors concluded that autophagy modulation could be a potentially novel target especially in steroid- and therapy-resistant severe asthma.
Autophagy and animal models of asthma
Ovalbumin (OVA) is widely used as an animal experimental model of asthma. Inhalation of OVA in mice increases levels of autophagic markers (60, 61), including beclin-1 in mouse airway tissues, which has been confirmed by Cy3-red staining (61). In OVA-challenged animals, both α-actin smooth muscle (α-SMA) and LC3A/B increased in the airway subepithelial space which highlights the role of autophagy, and possibly epithelial–mesenchymal transition (EMT) induction (61). According to these observations, the modulation of autophagy may have the potential to treat asthma. Regarding alternative animal models of allergic asthma, we found no significant publications using house dust mite antigen or other antigens in the study of autophagy.
Autophagy and extracellular matrix
Collagen deposition and the extracellular matrix (ECM) play an important role in tissue remodeling. Emerging data suggest that ECM can evoke autophagic regulatory responses in a variety of tissues via interaction with specific receptors (62). Several recent investigations show that autophagy may be very important in conventional and unconventional protein secretory pathways. Indeed, this line of evidence suggests that autophagy plays a role in unconventional mechanisms of protein secretion and trafficking via a mechanism named ‘autosecretion’ (63–65). This mechanism is considered one of the pathways allowing leaderless cytosolic proteins to exit the cell without overwhelming the ER-to-Golgi secretory conduit. Therefore, autophagy is possibly playing a role in the extracellular release of a subset of leaderless or addressless cytosolic proteins, which do not pass via the ER but can nonetheless be secreted by a pathway dependent on autophagy (64). As this mechanism regulates protein secretion, its impairment could also be relevant in the pathogenesis ECM-dependent diseases. In line with these findings, we recently showed that autophagy is necessary for ECM secretion in lung and heart fibroblasts (13, 56).
Remodeling and fibrosis in asthma
‘Airway remodeling’ is a term originally used to reflect the structural changes that normally take place during lung embryonic development; changes that are indispensable for healthy pulmonary development and function (66). In asthma, however, airway remodeling is typically used to describe pathological or adverse changes in the airway (67–69). These changes are due to complex and multifaceted cellular and molecular processes that alter the normal arrangement and structure of the airway wall ultimately leading to abnormal physiological function and symptoms of breathlessness and wheezing.
The distinguishing structural changes observed in the bronchial walls of asthma patients include epithelial goblet cell metaplasia and hyperplasia, mucus hypersecretion, increased ECM and subepithelial collagen deposition (i.e. reticular basement membrane (RBM) fibrosis), smooth muscle cell hyperplasia and hypertrophy, and vascular neoangiogenesis. Airway smooth muscle (ASM) is especially involved in asthma airway remodeling (69) which is observed in biopsies from asthmatic airways showing ASM hyperplasia and hypertrophy. Extracellular matrix deposition between muscle cells also contributes to increased ASM bundle mass and subepithelial fibrosis (69–71). Of these structural changes, smooth muscle hypertrophy and subepithelial fibrosis are the most difficult to treat, and often progress (11) despite current therapies, leading to fixed airflow obstruction and severe asthma.
It is commonly believed that the major effector cells involved in remodeling and fibrosis in asthma are the ASM, fibroblasts, and myofibroblasts, that is, mesenchymal cells (6, 68–70). The basement membrane which is produced by these cells and underlying mesenchymal layers can be congealed in asthma due to increased inflammatory cell infiltration, cytokine and growth factor release, ASM and fibroblast proliferation, and fibroblast differentiation (into myofibroblasts), as well as myofibroblast-mediated deposition of ECM proteins (68, 72).
Fibrosis is generally viewed as failed wound healing (usually in response to organ injury and inflammation) and results in deregulated and continuous ECM synthesis and accretion, concurrent with a possible decrease in the degradation of polymeric ECM components such as collagens (73). Continued fibrosis contributes to scar formation in the airways and lung, which eventually leads to a loss of lung function. Although a number of factors contribute to fibrogenesis (74), the major cytokines that promote and accelerate fibrosis are TGF-β1 and its downstream mediators, connective tissue growth factor (CTGF) and platelet-derived growth factor (PDGF), and endothelin-1 (ET-1) (72). Inflammatory cells, lung epithelial cells, and (myo)fibroblasts are mainly involved in TGF-β1 biosynthesis and secretion in the lung and upregulation of collagen production (75, 76).
Potential pharmacologic agents to modulate autophagy
The balance between inducing vs inhibiting autophagy will depend on a deeper understanding of the scope of autophagic activity over a time span not studied in most experiments. However, it is important to be aware of the agents currently available, and whether they can be applied to the clinical arena. Perhaps the most commonly used modulators of autophagy include rapamycin, Baf-A1, chloroquine, and 3-MA. However, a closer look at the broader literature reveals other potentially useful pharmacologic agents.
One of the earliest events in autophagy initiation is the formation of nucleation. The ULK1 and PIK3C3 complexes are involved in autophagy nucleation, followed by ULK1 movement and re-localization to target the Atg14-PIK3C3 complex. The PIK3C3 complex is responsible for the generation of phosphatidylinositol 3-phosphate (PtdIns3P) at the nucleation site with subsequent generation of a PtdIns3P-enriched environment. This supports the localization of the ULK1 complex and other Atg proteins at the growing membrane to trigger the assembly of PtdIns3P-binding effector proteins. These are all critical steps for the subsequent steps of autophagy (elongation and maturation) as described in previous sections.
In the past few years, significant progress has been made in developing PtdIns3K and PI3K inhibitors. One of the best classifications of PtdIns3K and PI3K is based on their target selectivity; they can be considered as pan-PI3K inhibitors, dual PI3K-mTOR inhibitors, and class or isoform-selective inhibitors (77–79). Wortmannin and LY294002 are pan-PI3K inhibitors; however, they are nonselective and therefore may not be ideal for clinical application (80, 81). Recently, pan- PI3K inhibitors with greater selectivity and less toxicity (e.g. BKM120, BAY80-6946) have been developed and may be better suited for use in clinical trials (80, 82).
In the past few years, new PIK3C3 inhibitors, including VPS34-IN1 (83), Compound 31 (a tetrahydropyrimidopyrimidinone derivative) (84), PIK-III, and SAR405 have been developed. Of these agents, Compound 31 and SAR405 exhibit the highest potency and selectivity toward their targets (84, 85).
As we have previously mentioned, mTOR inhibits Atg1/ULK1 function via phosphorylation of Atg1/ULK1 (86). On the other hand, ULK1 is also positively regulated by the cellular energy sensor AMP-activated protein kinase (AMPK) (87). Egan et al. defined ULK1’s consensus phosphorylation motif, demonstrated that ULK1 phosphorylates several autophagy components, and developed a ULK1 small molecule inhibitor (SBI-0206965). By modulating autophagy, this compound may be a potential therapy to inhibit fibrosis.
Although an assay exists for screening of Atg4 protease inhibitors (88), we are not aware of any experiments documenting specific inhibitors for autophagy related to fibrosis or asthma.
Seeing asthma through the lens of autophagy: a focus on airway fibrosis
Is autophagy required to initiate fibrosis in the airway and does it participate in airway remodeling in human asthma? Autophagy and fibrosis occur together in many diseases (89, 90). In hepatic cells, autophagy is necessary for the induction of fibrosis (90). It is possible that autophagy functions as a bulk degradation mechanism within the cell, thereby maintaining basal physiological conditions necessary for healthy cellular and organ function. Because fibrosis is a process that requires a high degree of ECM protein biosynthesis, cells that produce ECM need large energy reserves in order to provide sufficient ECM production (91). Autophagy can be a major source of energy and a pathway of cellular survival under conditions of illness or energy starvation. Therefore, autophagy can have several different and potentially opposite roles depending on the cell type involved, local tissue environment and cell signals, and energy requirements needed.
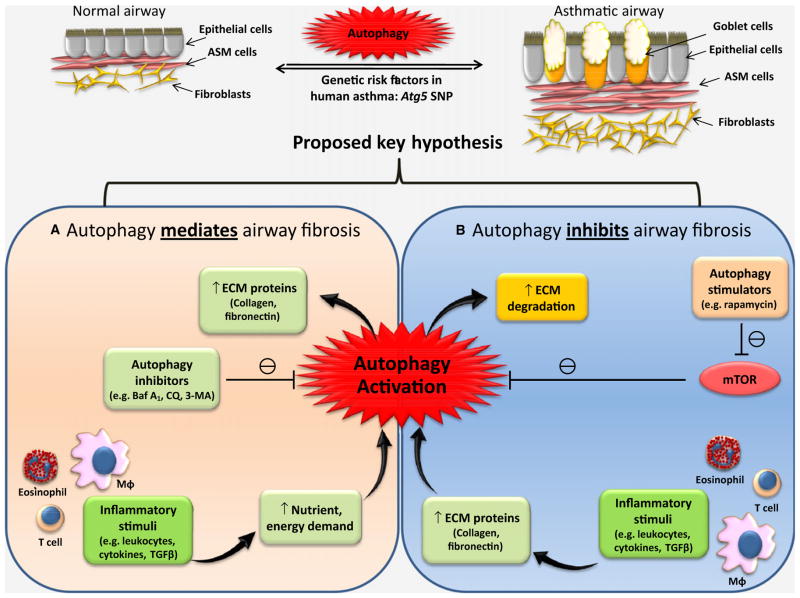
It is a plausible hypothesis that autophagy is a regulator of fibrosis and acts in parallel processes leading to cellular activation and enhanced ECM production (e.g. collagen, fibronectin) in airway mesenchymal cells, leading to airway thickening and rigidity (Fig. 1). This idea provides a novel framework for understanding fibrotic disease and the regulation of airway fibrosis. As a basic cell fate mechanism, autophagy’s role in fibrosis may occur in all tissues, and may not just be limited to the liver or airway. Because blocking autophagy in fibrogenic cells from different organs attenuates fibrogenesis, and because autophagy is an evolutionarily conserved and subcellular core pathway (76), it may contribute to the fibrotic responses observed in a wide range of tissues including the human airway in asthma (Fig. 1, panel A).
Figure 1.
Possible mechanisms of autophagy-dependent regulation of airway fibrosis: Autophagy may regulate airway fibrosis via two different mechanisms: Hypothesis (A) Autophagy is required for airway fibrosis by serving as a backup energy reserve providing the cellular resources needed during ECM protein biosynthesis. Hypothesis (B) Autophagy inhibits airway fibrosis via degradation of ECM proteins and their precursors. ASM = airway smooth muscle, ECM = extracellular matrix, Baf A1 = bafilomycin A1, CQ = chloroquine, 3- MA = 3-methyladenine, MΦ = macrophage, TGFβ = transforming growth factor-beta, mTOR = mammalian target of rapamycin.
Conversely, an alternative and equally plausible hypothesis is that autophagy may function to oppose or mitigate fibrosis if it is involved in the degradation of ECM proteins needed for fibrosis such as fibronectin and collagen (Fig. 1, panel B). Therefore, the occurrence of autophagy in the asthmatic lung may be a protective mechanism against the harmful effects of airway fibrosis, that is, as a compensatory mechanism against other aspects of remodeling. Thus, it is possible that the genetic correlation of Atg5 with familial asthma may be a developmental adaptation to reduce chronic airway remodeling. Therefore, correlations or associations of autophagy genes with lung function changes should be differentiated from causal mechanisms. Given our aforementioned preliminary findings (56), we predict the hypothesis proposed in panel (A) of the Figure is most likely to be true, at least in airway mesenchymal cells. However, further work is necessary to elucidate what such genetic associations mean with respect to clinically meaningful outcomes in asthma.
Despite our prediction, it is important to remain cautious regarding the full extent of the role of autophagy in the development of inflammation and fibrosis as it relates to asthma pathogenesis. Although no literature delineating a mechanism exists in support of a hypothesis relevant to asthma, it is worth reviewing what we know related to idiopathic pulmonary fibrosis (IPF), as the basic biology may be shared with airway fibrosis.
Although we are not aware of any literature relating ATG4B protease and asthma, some recent work in a model of pulmonary fibrosis may be relevant. Using an Atg4B-deficient mouse model, disruption of autophagy was shown to contribute to bleomycin-induced pulmonary fibrosis in vivo (92). In this study, the authors concluded that AtgB4 protease and autophagy are protective against bleomycin-induced inflammation and fibrosis. In another study evaluating airway epithelial and mesenchymal senescence, insufficient autophagy was thought to underlie the development of fibrosis (in a cell culture model of IPF and using lung specimens derived from IPF patients (93)). Whether or not these findings are relevant to human asthma remains an open question worthy of investigation.
The future
Future research should address the mechanism(s) leading to autophagy activation in asthma. Efforts to understand the role of autophagy will require cell-based in vitro experiments and in vivo animal models to address the many facets of asthma disease pathogenesis. Our proposed hypotheses address one key question, but clearly, many other hypotheses related to autophagy and inflammation, cytokine production, airway smooth muscle and fibroblast cell physiology, mucus production, immune cell mechanisms, lipid and metabolic responses, etc. in asthma, will require investigation.
If relevant, there is an opportunity to develop inhibitors or modulators of autophagy as a new therapeutic strategy for the treatment of asthma, where there are unmet and growing public health needs. Whether appropriate treatment regimens are administered via systemic or inhaled routes will depend on the extent of autophagy activity and the need to avoid unwanted drug side-effects (given autophagy’s central role in normal homeostatic cellular function).
Therefore, given that autophagy is truly a ‘double-edged’ sword, investigators should consider airway-targeted approaches such as the development of a novel inhaler, or nanoparticle-based cell-targeted methods to deliver drug payloads specifically to the epithelium or subepithelial airway fibroblasts. In the immediate future, we propose experiments using bafilomycin-A1 and chloroquine, well-known and established inhibitors of autophagy flux, the latter of which has been in clinical use for many years for different conditions. There is an opportunity to forge new roads in the journey to novel therapies for the treatment of human asthma, and we believe an exploration of autophagy is timely and warranted.
Acknowledgments
Funding
National Institutes of Health: 1K08HL114882-01A1 (AAZ). Manitoba Medical Service Foundation (SG).
Abbreviations
- 3-MA
3-methyladenine
- ALI
acute lung injury
- AMPK
AMP-activated protein kinase
- ASM
airway smooth muscle
- Atg
autophagy-related
- AVs
autophagic vacuoles
- Baf-A1
bafilomycin-A1
- CF
cystic fibrosis
- CFTR
cystic fibrosis transmembrane conductance regulator
- COPD
chronic obstructive pulmonary disease
- CQ
chloroquine
- CTGF
connective tissue growth factor
- Cvt
cytoplasm to vacuole targeting
- ECM
extracellular matrix
- EMT
epithelial–mesenchymal transition
- ER
endoplasmic reticulum
- ET-1
endothelin-1
- FEV1
forced expiratory volume in 1 s
- GFP
green fluorescent protein
- HASM
human airway smooth muscle
- IL13
interleukin-13
- IPF
idiopathic pulmonary fibrosis
- LC3
microtubule-associated protein 1 light chain 3
- MHC
major histocompatibility complex
- MIICs
MHC class II-comprising sections
- mTOR
mammalian target of rapamycin
- OVA
ovalbumin
- PAH
pulmonary arterial hypertension
- PAS
pre-autophagosomal structure
- PDGF
platelet-derived growth factor
- PE
phosphatidylethanolamine
- PH
pulmonary hypertension
- PtdIns3P
phosphatidylinositol 3-phosphate
- RBM
reticular basement membrane
- ROS
reactive oxygen species
- SNP
single nucleotide polymorphism
- TGFβ
transforming growth factor-beta
- Th2
T helper type 2
- α-SMA
α-actin smooth muscle
Footnotes
Author contributions
AAZ and SG conceived of this project, wrote, and edited the first and final drafts. AAZ re-wrote the draft with SG and conceived of the Fig. 1. BY edited the manuscript and helped refine Fig. 1. NJK provided expertise on the relevance to asthma, and helped edit the final versions of the manuscript. MP provided guidance related to cell fate pathways, in particular autophagy and as it relates to respiratory developmental biology. MP helped edit penultimate versions of the manuscript.
Conflicts of interest
None of the authors (AAZ, SG, BY, NJK, MP) report any conflict of interest with respect to the research reviewed or the ideas communicated in this manuscript.
References
- 1.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang F, Yang YP, Mao CJ, Cao BY, Cai ZL, Shi JJ, et al. Role of autophagy and proteasome degradation pathways in apoptosis of PC12 cells overexpressing human alphasynuclein. Neurosci Lett. 2009;454:203–208. doi: 10.1016/j.neulet.2009.03.027. [DOI] [PubMed] [Google Scholar]
- 3.Ghavami S, Shojaei S, Yeganeh B, Ande SR, Jangamreddy JR, Mehrpour M, et al. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Prog Neurobiol. 2014;112:24–49. doi: 10.1016/j.pneurobio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 5.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghavami S, Sharma P, Yeganeh B, Ojo OO, Jha A, Mutawe MM, et al. Airway mesenchymal cell death by mevalonate cascade inhibition: integration of autophagy, unfolded protein response and apoptosis focusing on Bcl2 family proteins. Biochim Biophys Acta. 2014;1843:1259–1271. doi: 10.1016/j.bbamcr.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Morselli E, Galluzzi L, Kepp O, Vicencio JM, Criollo A, Maiuri MC, et al. Anti- and pro-tumor functions of autophagy. Biochim Biophys Acta. 2009;1793:1524–1532. doi: 10.1016/j.bbamcr.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Ghavami S, Gupta S, Ambrose E, Hnatowich M, Freed DH, et al. Autophagy and heart disease: implications for cardiac ischemia- reperfusion damage. Curr Mol Med. 2014;14:616–629. doi: 10.2174/1566524014666140603101520. [DOI] [PubMed] [Google Scholar]
- 9.Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 10.Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- 11.Munakata M. Airway remodeling and airway smooth muscle in asthma. Allergol Int. 2006;55:235–243. doi: 10.2332/allergolint.55.235. [DOI] [PubMed] [Google Scholar]
- 12.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 13.Ghavami S, Cunnington RH, Gupta S, Yeganeh B, Filomeno KL, Freed DH, et al. Autophagy is a regulator of TGF-beta1-induced fibrogenesis in primary human atrial myofibroblasts. Cell Death Dis. 2015;6:e1696. doi: 10.1038/cddis.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci. 1999;112:4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- 15.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 16.Komatsu M, Ueno T, Waguri S, Uchiyama Y, Kominami E, Tanaka K. Constitutive autophagy: vital role in clearance of unfavorable proteins in neurons. Cell Death Differ. 2007;14:887–894. doi: 10.1038/sj.cdd.4402120. [DOI] [PubMed] [Google Scholar]
- 17.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 19.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 21.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 22.Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p- Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 25.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, et al. A ubiquitin- like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 27.Ichimura Y, Imamura Y, Emoto K, Umeda M, Noda T, Ohsumi Y. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J Biol Chem. 2004;279:40584–40592. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- 28.Yoshimori T. Autophagy: a regulated bulk degradation process inside cells. Biochem Biophys Res Commun. 2004;313:453–458. doi: 10.1016/j.bbrc.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 29.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 31.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stromhaug PE, Klionsky DJ. Approaching the molecular mechanism of autophagy. Traffic. 2001;2:524–531. doi: 10.1034/j.1600-0854.2001.20802.x. [DOI] [PubMed] [Google Scholar]
- 34.Scott SV, Nice DC, 3rd, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka A, Jin Y, Lee SJ, Zhang M, Kim HP, Stolz DB, et al. Hyperoxia-induced LC3B interacts with the Fas apoptotic pathway in epithelial cell death. Am J Respir Cell Mol Biol. 2012;46:507–514. doi: 10.1165/rcmb.2009-0415OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Y, Liu J, Wu YF, Lou J, Mao YY, Shen HH, et al. mTOR and autophagy in regulation of acute lung injury: a review and perspective. Microbes Infect. 2014;16:727–734. doi: 10.1016/j.micinf.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 37.Crapo JD. Morphologic changes in pulmonary oxygen toxicity. Annu Rev Physiol. 1986;48:721–731. doi: 10.1146/annurev.ph.48.030186.003445. [DOI] [PubMed] [Google Scholar]
- 38.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alavian SM, Ande SR, Coombs KM, Yeganeh B, Davoodpour P, Hashemi M, et al. Virus-triggered autophagy in viral hepatitis – possible novel strategies for drug development. J Viral Hepat. 2011;18:821–830. doi: 10.1111/j.1365-2893.2011.01530.x. [DOI] [PubMed] [Google Scholar]
- 40.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeganeh B, Rezaei Moghadam A, Tran AT, Rahim MN, Ande SR, Hashemi M, et al. Asthma and influenza virus infection: focusing on cell death and stress pathways in influenza virus replication. Iran J Allergy Asthma Immunol. 2013;12:1–17. [PubMed] [Google Scholar]
- 42.Yeganeh B, Ghavami S, Kroeker AL, Mahood TH, Stelmack GL, Klonisch T, et al. Suppression of influenza A virus replication in human lung epithelial cells by noncytotoxic concentrations bafilomycin A1. Am J Physiol Lung Cell Mol Physiol. 2015;308:L270–L286. doi: 10.1152/ajplung.00011.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SJ, Smith A, Guo L, Alastalo TP, Li M, Sawada H, et al. Autophagic protein LC3B confers resistance against hypoxia-induced pulmonary hypertension. Am J Respir Crit Care Med. 2011;183:649–658. doi: 10.1164/rccm.201005-0746OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lahm T, Petrache I. LC3 as a potential therapeutic target in hypoxia-induced pulmonary hypertension. Autophagy. 2012;8:1146–1147. doi: 10.4161/auto.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayer ML, Blohmke CJ, Falsafi R, Fjell CD, Madera L, Turvey SE, et al. Rescue of dysfunctional autophagy attenuates hyperinflammatory responses from cystic fibrosis cells. J Immunol. 2013;190:1227–1238. doi: 10.4049/jimmunol.1201404. [DOI] [PubMed] [Google Scholar]
- 46.Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina D, Settembre C, et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol. 2010;12:863–875. doi: 10.1038/ncb2090. [DOI] [PubMed] [Google Scholar]
- 47.De Stefano D, Villella VR, Esposito S, Tosco A, Sepe A, De Gregorio F, et al. Restoration of CFTR function in patients with cystic fibrosis carrying the F508del- CFTR mutation. Autophagy. 2014;10:2053–2074. doi: 10.4161/15548627.2014.973737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J, Yeganeh B, Ermini L, Post M. Sphingolipids as cell fate regulators in lung development and disease. Apoptosis. 2015;20:740–757. doi: 10.1007/s10495-015-1112-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marzetti E, Csiszar A, Dutta D, Balagopal G, Calvani R, Leeuwenburgh C. Role of mitochondrial dysfunction and altered autophagy in cardiovascular aging and disease: from mechanisms to therapeutics. Am J Physiol Heart Circ Physiol. 2013;305:H459–H476. doi: 10.1152/ajpheart.00936.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 51.Ban GY, Pham DL, Trinh HK, Lee SI, Suh DH, Yang EM, et al. Autophagy mechanisms in sputum and peripheral blood cells of patients with severe asthma: a new therapeutic target. Clin Exp Allergy. 2015 doi: 10.1111/cea.12585. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 52.Dickinson JD, Alevy Y, Malvin NP, Patel KK, Gunsten SP, Holtzman MJ, et al. IL13 activates autophagy to regulate secretion in airway epithelial cells. Autophagy. 2015 doi: 10.1080/15548627.2015.1056967. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeki AA, Ott S, Sandhu K, Ghavami S, Kenyon NJ. The complex roles of endoplasmic reticulum stress and autophagy in modulating Eotaxin-3 production and secretion from human airway epithelial cells. Am J Respir Crit Care Med. 2014;189:A5683. [Google Scholar]
- 54.Poon AH, Chouiali F, Tse SM, Litonjua AA, Hussain SN, Baglole CJ, et al. Genetic and histologic evidence for autophagy in asthma pathogenesis. J Allergy Clin Immunol. 2012;129:569–571. doi: 10.1016/j.jaci.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin LJ, Gupta J, Jyothula SS, Butsch Kovacic M, Biagini Myers JM, Patterson TL, et al. Functional variant in the autophagy- related 5 gene promotor is associated with childhood asthma. PLoS One. 2012;7:e33454. doi: 10.1371/journal.pone.0033454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghavami S, Yeganeh B, Serebrin A, Mutawe MM, Sharma P, McNeill KD, et al. Autophagy regulates TGF-beta1 induced fibrosis in human airway smooth muscle cells. Am J Respir Crit Care Med. 2011;183:A2110. [Google Scholar]
- 57.Jyothula SS, Eissa NT. Autophagy and role in asthma. Curr Opin Pulm Med. 2013;19:30–35. doi: 10.1097/MCP.0b013e32835b1150. [DOI] [PubMed] [Google Scholar]
- 58.Graham MT, Nadeau KC. Lessons learned from mice and man: mimicking human allergy through mouse models. Clin Immunol. 2014;155:1–16. doi: 10.1016/j.clim.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 59.Puleston DJ, Simon AK. Autophagy in the immune system. Immunology. 2014;141:1–8. doi: 10.1111/imm.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao Z, Pan P, Tan H, Tan Q, Wang Z, Su X, et al. Anti-nerve growth factor antibody reduces airway hyperresponsiveness in a mouse model of asthma by down-regulating the level of autophagy in lungs. Zhonghua Jie He He Hu Xi Za Zhi. 2014;37:507–511. [PubMed] [Google Scholar]
- 61.Cho IH, Choi YJ, Gong JH, Shin D, Kang MK, Kang YH. Astragalin inhibits autophagy- associated airway epithelial fibrosis. Respir Res. 2015;16:51. doi: 10.1186/s12931-015-0211-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neill T, Schaefer L, Iozzo RV. Instructive roles of extracellular matrix on autophagy. Am J Pathol. 2014 Aug;184:2146–2153. doi: 10.1016/j.ajpath.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol. 2010;188:527–536. doi: 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bruns C, McCaffery JM, Curwin AJ, Duran JM, Malhotra V. Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion. J Cell Biol. 2011;195:979–992. doi: 10.1083/jcb.201106098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deretic V, Jiang S, Dupont N. Autophagy intersections with conventional and unconventional secretion in tissue development, remodeling and inflammation. Trends Cell Biol. 2012;22:397–406. doi: 10.1016/j.tcb.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jeffery PK, Laitinen A, Venge P. Biopsy markers of airway inflammation and remodelling. Respir Med. 2000;94:S9–S15. doi: 10.1016/s0954-6111(00)90127-6. [DOI] [PubMed] [Google Scholar]
- 67.Sumi Y, Hamid Q. Airway remodeling in asthma. Allergol Int. 2007;56:341–348. doi: 10.2332/allergolint.R-07-153. [DOI] [PubMed] [Google Scholar]
- 68.Schaafsma D, McNeill KD, Mutawe MM, Ghavami S, Unruh H, Jacques E, et al. Simvastatin inhibits TGFbeta1-induced fibronectin in human airway fibroblasts. Respir Res. 2011;12:113. doi: 10.1186/1465-9921-12-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gerthoffer WT, Schaafsma D, Sharma P, Ghavami S, Halayko AJ. Motility, survival, and proliferation. Compr Physiol. 2012;2:255–281. doi: 10.1002/cphy.c110018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schaafsma D, Dueck G, Ghavami S, Kroeker A, Mutawe MM, Hauff K, et al. The mevalonate cascade as a target to suppress extracellular matrix synthesis by human airway smooth muscle. Am J Respir Cell Mol Biol. 2011;44:394–403. doi: 10.1165/rcmb.2010-0052OC. [DOI] [PubMed] [Google Scholar]
- 71.Stewart A. More muscle in asthma, but where did it come from? Am J Respir Crit Care Med. 2012;185:1035–1037. doi: 10.1164/rccm.201203-0457ED. [DOI] [PubMed] [Google Scholar]
- 72.Yeganeh B, Mukherjee S, Moir LM, Kumawat K, Kashani HH, Bagchi RA, et al. Novel non-canonical TGF-beta signaling networks: emerging roles in airway smooth muscle phenotype and function. Pulm Pharmacol Ther. 2013;26:50–63. doi: 10.1016/j.pupt.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 73.Gifford AH, Matsuoka M, Ghoda LY, Homer RJ, Enelow RI. Chronic inflammation and lung fibrosis: pleotropic syndromes but limited distinct phenotypes. Mucosal Immunol. 2012;5:480–484. doi: 10.1038/mi.2012.68. [DOI] [PubMed] [Google Scholar]
- 74.Sivakumar P, Ntolios P, Jenkins G, Laurent G. Into the matrix: targeting fibroblasts in pulmonary fibrosis. Curr Opin Pulm Med. 2012;18:462–469. doi: 10.1097/MCP.0b013e328356800f. [DOI] [PubMed] [Google Scholar]
- 75.Camoretti-Mercado B, Solway J. Transforming growth factor-beta1 and disorders of the lung. Cell Biochem Biophys. 2005;43:131–148. doi: 10.1385/CBB:43:1:131. [DOI] [PubMed] [Google Scholar]
- 76.Mehal WZ, Iredale J, Friedman SL. Scraping fibrosis: expressway to the core of fibrosis. Nat Med. 2011;17:552–553. doi: 10.1038/nm0511-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 78.Brana I, Siu LL. Clinical development of phosphatidylinositol 3-kinase inhibitors for cancer treatment. BMC Med. 2012;10:161. doi: 10.1186/1741-7015-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akinleye A, Avvaru P, Furqan M, Song Y, Liu D. Phosphatidylinositol 3-kinase (PI3K) inhibitors as cancer therapeutics. J Hematol Oncol. 2013;6:88. doi: 10.1186/1756-8722-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cleary JM, Shapiro GI. Development of phosphoinositide-3 kinase pathway inhibitors for advanced cancer. Curr Oncol Rep. 2010;12:87–94. doi: 10.1007/s11912-010-0091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Falasca M. Phosphoinositide 3-kinase pathway inhibitors: pharmacology, metabolism & drug development. Curr Med Chem. 2011;18:2673. doi: 10.2174/092986711796011210. [DOI] [PubMed] [Google Scholar]
- 82.McNamara CR, Degterev A. Small-molecule inhibitors of the PI3K signaling network. Future Med Chem. 2011;3:549–565. doi: 10.4155/fmc.11.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bago R, Malik N, Munson MJ, Prescott AR, Davies P, Sommer E, et al. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem J. 2014;463:413–427. doi: 10.1042/BJ20140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pasquier B, El-Ahmad Y, Filoche-Romme B, Dureuil C, Fassy F, Abecassis PY, et al. Discovery of (2S)-8-[(3R)-3-methylmorpholin- 4-yl]-1-(3-methyl-2-oxobutyl)-2-(trifluoromethyl)- 3,4-dihydro-2H-pyrimido[1,2-a] pyrimidin-6-one: a novel potent and selective inhibitor of Vps34 for the treatment of solid tumors. J Med Chem. 2015;58:376–400. doi: 10.1021/jm5013352. [DOI] [PubMed] [Google Scholar]
- 85.Ronan B, Flamand O, Vescovi L, Dureuil C, Durand L, Fassy F, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol. 2014;10:1013–1019. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- 86.Chan EY. mTORC1 phosphorylates the ULK1-mAtg13-FIP200 autophagy regulatory complex. Sci Signal. 2009;2:pe51. doi: 10.1126/scisignal.284pe51. [DOI] [PubMed] [Google Scholar]
- 87.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shu CW, Madiraju C, Zhai D, Welsh K, Diaz P, Sergienko E, et al. High-throughput fluorescence assay for small-molecule inhibitors of autophagins/Atg4. J Biomol Screen. 2011;16:174–182. doi: 10.1177/1087057110392996. [DOI] [PubMed] [Google Scholar]
- 89.Chu PM, Chen LH, Chen MT, Ma HI, Su TL, Hsieh PC, et al. Targeting autophagy enhances BO-1051-induced apoptosis in human malignant glioma cells. Cancer Chemother Pharmacol. 2012;69:621–633. doi: 10.1007/s00280-011-1747-0. [DOI] [PubMed] [Google Scholar]
- 90.Hernandez-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stoppoloni D, Politi L, Dalla Vedova P, Messano M, Koverech A, Scandurra R, et al. L-carnitine enhances extracellular matrix synthesis in human primary chondrocytes. Rheumatol Int. 2013;33:2399–2403. doi: 10.1007/s00296-012-2373-9. [DOI] [PubMed] [Google Scholar]
- 92.Cabrera S, Maciel M, Herrera I, Nava T, Vergara F, Gaxiola M, et al. Essential role for the ATG4B protease and autophagy in bleomycin-induced pulmonary fibrosis. Autophagy. 2015;11:670–684. doi: 10.1080/15548627.2015.1034409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Araya J, Kojima J, Takasaka N, Ito S, Fujii S, Hara H, et al. Insufficient autophagy in idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2013;304:L56–L69. doi: 10.1152/ajplung.00213.2012. [DOI] [PubMed] [Google Scholar]